Abstract
With one exception (Glikoreijevic et al., Mol. Biochem. Parasitol. 2008; 159: 7 – 23) all previous quantification of chloroquine (CQ) potency vs. P. falciparum has been by growth inhibition assays, meaning potency is defined as cytostatic potential and quantified by IC50 values. In this study we investigate the cytocidal potency of CQ and other common quinoline antimalarial drugs (quantified as LD50). Similar to results from assays for cytostatic potency, we are able to readily distinguish drug resistant from drug sensitive P. falciparum parasites as well as different degrees of resistance. However, we find that fold-resistance to CQ and other quinoline drugs quantified via LD50 ratios differs quite dramatically from fold resistance calculated via IC50 ratios. Also, importantly, we find that verapamil chemoreversal of CQ resistance differs when quantified via cytocidal vs. cytostatic assays, as do patterns of “multidrug” resistance in well-known laboratory strains of P. falciparum. The results have important implications for development of new antimalarial drugs and for fully defining the genetic loci that confer clinically relevant antimalarial drug resistance phenomena.
Keywords: malaria, drug resistance, verapamil, LD50, SyberGreen, multidrug resistance
Introduction
Virtually all quantification of antimalarial drug potency vs. Plasmodium falciparum has been via long term continuous growth inhibition assays in the constant presence of drug (e.g. [1–8]). These assays report relative parasite growth in live merozoite blood culture via the use of various reporters, typically either 3H-hypoxanthine incorporation, staining of newly synthesized nucleic acid with SYBR Green I or other intercalating dyes, or enzymatic assays that quantify metabolic activity of intraerythrocytic malarial parasites. Formally, these are cytostasis assays that quantify the cytostatic potential of the antimalarial drug as an IC50 value, which is the concentration of drug that inhibits growth by fifty percent relative to growth in the absence of drug.
Many important concepts originate from these data. For example, distinction between chloroquine (CQ) sensitive (CQS) vs. chloroquine resistant (CQR) malaria is usually quantified using these assays. Fold-resistance to CQ for CQR strains or isolates is calculated as a ratio of IC50 and is generally found to be ~ 5 – 10-fold [1 – 8]. Using cytostasis assays as a phenotypic measure, genetic loci that confer or further modulate CQR have been identified [3, 4, 6] and a few proteins encoded by key genes within these loci (e.g. pfcrt and pfmdr1) have been investigated [4, 9, 10, 11]. The biochemistry of these proteins forms the basis of most of what is currently known regarding the molecular mechanism of resistance to quinoline antimalarial drugs [12].
However, such a characterization of CQR is incomplete. A malarial patient being examined in the clinic is typically infected with ~ 1011 – 1012 parasites. When the infection is caused by CQS parasites and CQ is administered, the parasitemia is reduced by > 102 fold in less than a day indicating that successful antimalarial drug therapy does not merely prevent parasite growth but also kills malarial parasites. If a patient infected with a CQR malarial parasite is given a similar dose of CQ, the patient will not show this fast, pronounced drop in parasitemia. This implies that CQR parasites are resistant to the cytocidal effects of CQ. We suggest that quantification of parasite susceptibility to antimalarial drug cytocidal effects is an essential, but usually neglected, aspect of antimalarial drug design and drug resistance research.
One reason for this neglect is that there have been only a few attempts to develop assays that conveniently measure cytocidal potency of antimalarial drugs vs. malarial parasites [13,14]. Only one to our knowledge has attempted to quantify cytocidal effects via calculating explicit LD50 values [14]. However, this previous study relied on visual counting of parasites using a light microscope and is thus quite tedious, not amenable to high-throughput format, and has several sources of error as summarized in Discussion. Here, we use a simple, inexpensive, fluorescence-based assay to calculate LD50 for diffusible antimalarial drugs. We define several striking, previously overlooked characteristics of CQR malarial parasites. These characteristics suggest that further analysis of genetic loci associated with antimalarial drug resistance may be warranted.
Materials and Methods
Materials
P. falciparum strains were obtained from the MR4 (Manassas, VA), deposited by T. E. Wellems (HB3 & Dd2) and D. Walliker (7G8). Type O+ pooled human whole blood and off-the-clot, heat inactivated human serum were provided by BioChemed Services (Winchester, VA). Custom 5% O2/5% CO2/90% N2 gas blend was from Robert’s Oxygen (Rockville, MD). SYBR Green I was from Invitrogen (Carlsbad, CA) and CQ diphosphate was from MP Biomedicals (Solon, OH). All other chemicals were reagent grade or better and were purchased from Sigma-Aldrich (St. Louis, MO).
Cell Culture
P. falciparum strains were cultured in human erythrocytes at 2% hematocrit, following Trager and Jensen’s method [15] with a few modifications. The parasites were maintained at 37°C in complete media composed of RPMI 1640 supplemented with 10% O+ human serum, 25 mM HEPES pH 7.4, 24 mM NaHCO3, 11 mM glucose, 0.75 mM hypoxanthine and 20 µg/L gentamicin sulfate under 5% O2/ 5% CO2/ 90% N2 atmosphere. When needed, cultures were synchronized three times via the sorbitol method [16] and allowed to pass through one RBC cycle prior to use.
LD50 Assay
Drug stocks were initially prepared by dissolving the drugs in either deionized water (chloroquine diphosphate, verapamil hydrochloride) or 50% ethanol (amodiaquine dihydrochloride dihydrate, mefloquine hydrochloride, primaquine diphosphate, quinidine hydrochloride monohydrate, quinine free base). Serial drug dilutions were made using complete media and 100 µl aliquots were transferred to 96-well clear-bottom black plates. After adding 100 µl of P. falciparum culture (2% parasitemia, 2% final hematocrit), assays were initiated by incubating the plates at 37°C under 5% O2/ 5% CO2/ 90% N2 for 6 hrs. Following the drug incubation, plates were centrifuged with an Eppendorf 5804 centrifuge fitted with an A-2-DPW rotor (Hauppauge, New York) at 700g for 3 minutes. Drug-containing medium was removed and cell pellets were washed three times with 200 µl of medium each time, using the same centrifuge settings, and then resuspended in the same volume of media. Earlier work [14], in which visual inspection of parasites was used to estimate LD50 for parasites incubated with drug, showed that three washings completely removed intracellular CQ.
Washed plates were then incubated at 37 °C for ≥ 48 hr (see Results). For companion cytostasis assays, parallel plates were set-up whereby parasites were continuously exposed to lower concentrations of the same drugs. After staining surviving parasites with SYBR Green I for 1 hour [1], fluorescence (excitation and emission of 485 nm and 538 nm, respectively) was measured with a Gemini EM Max microplate reader from Molecular Devices (Sunnyvale, CA) fitted with a 530 nm long – pass filter. As described in earlier work [1], linear standard curves that convert measured fluorescence to known parasitemia were prepared on the day each plate was analyzed. Background controls included fluorescence from non-infected RBC. Data points (parasitemia vs. control parasites not exposed to drug) were curve fitted with a sigmoidal function using SigmaPlot 9.0 (San Jose, CA) to determine LD50 and IC50 values. In general, LD50 values reported are the average of at least three separate assays (three separate treatments vs. different bolus doses of drug), with each assay done in triplicate (nine determinations total).
Results
In a high-throughput antimicrobial growth inhibition assay, dilute aliquots of microbe are placed in appropriate media harboring serial dilutions of a candidate antimicrobial drug and the cells are allowed to proliferate until the aliquot growing in the absence of drug (zero drug control) reaches plateau. The relative growth for the other aliquots is then quantified in some fashion and plotted vs. concentration of drug in the medium. The plot is fit to a sigmoid, and IC50 (mid point of the sigmoidal curve fit) is calculated [1]. For laboratory P. falciparum strains HB3 (CQS, S. America) and Dd2 (CQR, S. E. Asia), IC50 values for CQ are in our hands found to be 10 – 20 nM and 125 – 175 nM, respectively, via versions of a SYBR Green assay that detects relative growth via fluorescence-based quantification of newly synthesized parasite DNA [1]. Absolute CQ IC50 values for various P. falciparum strains vary across the literature, but most data show that Dd2 CQR parasites are ~10-fold resistant to CQ regardless the reference CQS strain and the absolute IC50 values obtained in a given growth inhibition format. In general, if CQ sensitivity as quantified by CQ IC50 is “x” for a reference CQS parasite, then it is “ 5x – 15x” for all known CQR parasite strains and isolates, assuming the same method for each IC50 calculation.
Cytocidal potency of antimalarial drugs can in theory be quantified in multiple ways, but what must be common to all approaches is that the drug is not delivered continuously but delivered in bolus fashion, meaning as a relatively short pulse of drug. Those cells not killed by the drug pulse can then be quantified in one of several ways (see Discussion). In such an assay, cell survival (not relative cell growth in the continuous presence of drug) is then plotted vs. concentration of bolus drug dose. Cytocidal assays often yield results that are time-dependent since the cytotoxic potency of many drugs depends on the duration of the bolus dose.
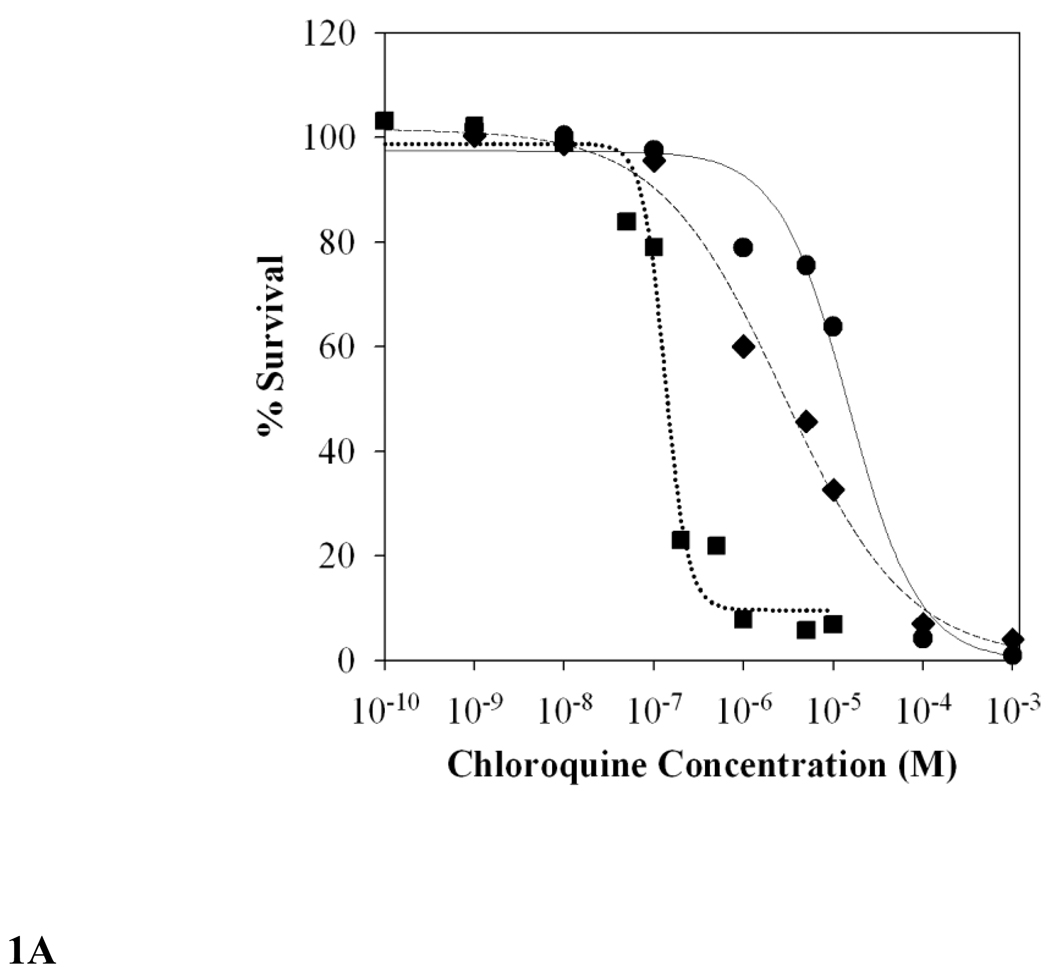
Figure 1 presents typical results from our cytocidal assay wherein survival from bolus doses of CQ is quantified vs. dose (Fig. 1A) and time (Fig. 1B). Survival is measured as SYBR Green I fluorescence 48 hrs post drug exposure (see Methods), but other approaches are in theory possible as described in the Discussion section. As described in Methods, LD50 are calculated from sigmoidal fits to raw data as shown in Fig. 1A.
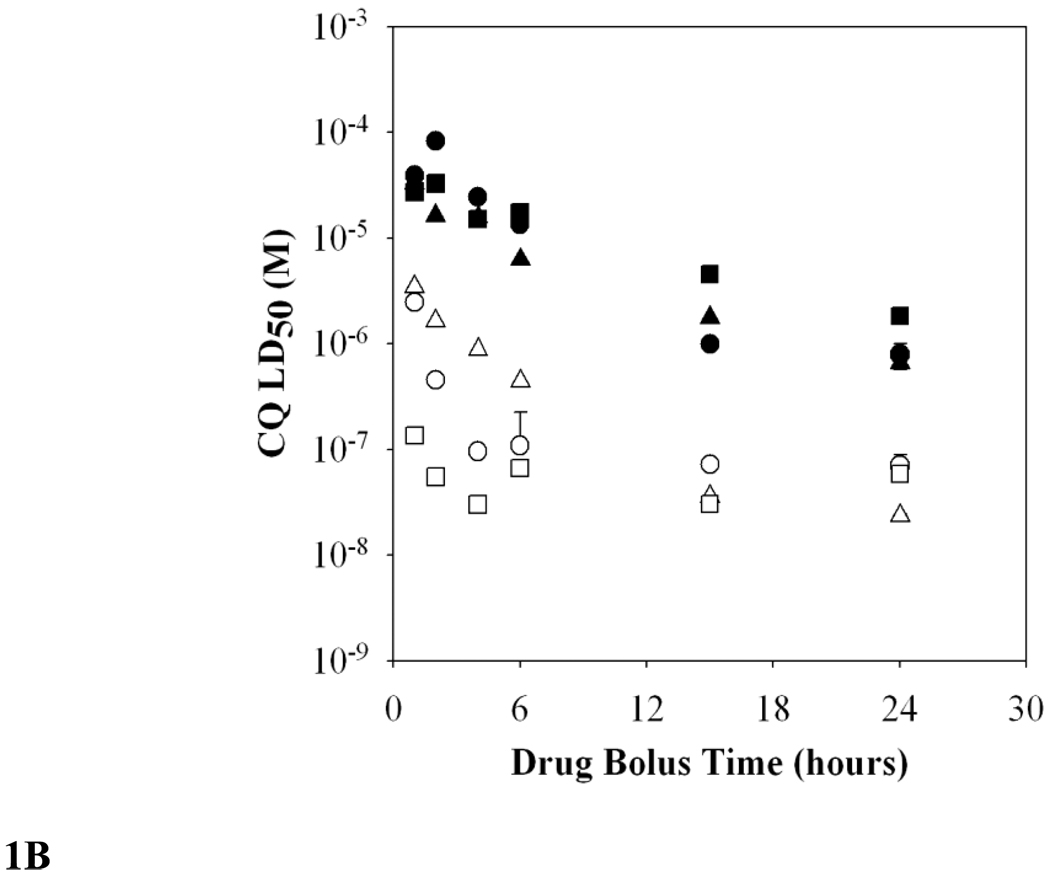
FIG. 1.
A. Representative survival curves for asynchronous HB3 (squares, dotted line), Dd2 (circles, straight line), and 7G8 (diamonds, dashed line) iRBC parasites incubated vs variable bolus CQ (x axis) for 6 hours. Fluorescence data of CQ-treated parasites were converted to % parasitemia using standard curves (see Methods) and then expressed as percent survival relative to parasites not exposed to drug. Data points shown were averaged from three independent experiments, each done in triplicate (9 determinations in total), and then fit to a sigmoidal function [y = y0 + a/(1 + (x/x0)b)] using SigmaPlot 9.0. CQ LD50 values were calculated to be 126 nM (HB3), 15,669 nM (Dd2), and 3,991 nM (7G8) (see Table 1).
B. Measured CQ LD50 decreases with increasing drug exposure time. While Dd2 (filled symbols) did not show large LD50 differences between the different stages, at short bolus times HB3 trophozoites (open squares) are more sensitive to CQ than are rings (open triangles) or asynchronous culture (open circles).
Fig. 1B plots LD50 values (y-axis) vs duration of bolus CQ dose in hours (x-axis). As expected, this plot shows that the duration of drug exposure affects the calculated CQ LD50 for both CQS strain HB3 (open symbols) and CQR strain Dd2 (closed symbols) parasites. Very short exposure times result in higher LD50, up to several millimolar for HB3 and ~100 mM for Dd2 at 1–2 hr exposure. But as the drug exposure approaches 6 hrs, calculated LD50 values begin to plateau, such that 15 hr and 24 hr exposure times yield more similar LD50 relative to 6 hr LD50. Using synchronized cultures, we note different parasite stage dependencies for time dependent LD50, with Dd2 stages being less variable in their sensitivity to CQ toxicity vs. strain HB3 stages (Fig. 1B, squares vs circles vs triangles). For example, at 2–4 hr exposure times, CQS HB3 trophozoites are more sensitive to CQ toxicity relative to ring stages (25-fold more sensitive) whereas CQR strain Dd2 trophozoites and rings show similar LD50.
To standardize this assay for high-throughput conditions, we chose a 6 hr bolus dose period and used asynchronous cultures. This duration is convenient and is near the point where calculated CQ LD50 begin to plateau, such that increasing time further only modestly changes computed LD50. Table 1 presents summary 6 hr bolus dose LD50 data for HB3 and Dd2 strains as well as strain 7G8 (CQR, S. America). Strain 7G8 is particularly informative to analyze alongside Dd2 since its CQR phenotype is known to be distinct relative to Dd2 [7]. For example, 7G8 parasites are known to have lower CQ IC50 relative to Dd2 (the strain is less CQR) and also to be less susceptible to chemoreversal by verapamil (VPL). Data (Table 1) shows that LD50 for 7G8 is indeed less than Dd2 LD50 as might be expected from IC50 data. However, the fold difference in LD50 is much larger than that typically observed for the corresponding IC50 ratios (Table 2). That is, the relative degree of CQR, which can be quantified as a “resistance factor” or “Rf” (right hand side, Tables 1 & 2), depends upon whether CQ susceptibility for the strains is expressed via LD50 or IC50 ratios (Dd2/HB3 and 7G8/HB3, right hand side, table 1 and 2 respectively).
TABLE 1.
LD50 for asynchronous HB3, Dd2, and 7G8 vs several antimalarial drugsa
| Drug | Experimental LD50, nM (standard error mean) |
Experimental Rf | |||
|---|---|---|---|---|---|
| HB3 | Dd2 | 7G8 | Dd2/HB3 | 7G8/HB3 | |
| Chloroquine | 126.3** ## (30) | 15669.0+ (2411) | 3990.7 (681) | 124.1 | 31.6 |
| Chloroquine +5 µM VPL | 124.5** ## (36) | 13458.5+ (2080) | 2694.6 (703) | 108.1 | 21.6 |
| Amodiaquine | 37.0 (7) | 51.4 (4) | 65.1 (9) | 1.4 | 1.8 |
| Mefloquine | 476.9 (109) | 416.6 (100) | 225.3 (27) | 0.9 | 0.5 |
| Quinidine | 423.7** # (76) | 27238.2++ (3389) | 1133.6 (142) | 64.3 | 2.7 |
| Quinine | 7121.4* # (1161) | 27308.1 (3798) | 26272.8 (6438) | 3.8 | 3.7 |
Each assay was independently performed 3–6 times, with each experiment done in triplicate (9–18 total) as described in methods. Independent t-tests were employed to identify statistically significance across a given row.
For HB3 vs. Dd2: * indicates p < 0.01, ** (p < 0.001).
For HB3 vs. 7G8, # (p < 0.01), ## (p < 0.001);
and for Dd2 vs. 7G8, + (p < 0.01), ++ (p < 0.001).
All other comparisons across a given row yielded 0.01 < p <0.5, except MQ LD50 for HB3 vs. Dd2 and QN LD50 for Dd2 vs. 7G8, where p > 0.5.
TABLE 2.
Literaturea and experimental resistance factors (Rf) from IC50 values for asynchronous HB3, Dd2, and 7G8 parasites
| Drug | Literature IC50, nM | Literature Rf | Experimental Rf | ||||
|---|---|---|---|---|---|---|---|
| HB3 | Dd2 | 7G8 | Dd2/ HB3 |
7G8/ HB3 |
Dd2/ HB3 |
7G8/ HB3 |
|
| Chloroquine | 22.7 | 225.9 | 167.5 | 10.0 | 7.4 | 9.3 | 5.9 |
| Chloroquine + VPL | 25.0 | 75.1 | 137.1 | 3.0 | 5.5 | 2.1 | 2.8 |
| Amodiaquine | 10.0 | 27.7 | 34.2 | 2.8 | 3.4 | 2.3 | 5.1 |
| Mefloquine | 21.0 | 30.4 | 14.2 | 1.4 | 0.7 | 1.1 | 0.2 |
| Quinidine | 17.8 | 119.1 | 44.6 | 6.7 | 2.5 | 1.3 | 0.5 |
| Quinine | 129.3 | 277.7 | 126.5 | 2.1 | 1.0 | 1.3 | 0.7 |
Numbers presented are averages of values reported in other papers: (i) Sidhu,A.B., D. Verdier-Pinard, and D. A. Fidock. 2002. Science 298:210–213. (ii) Lakshmanan, V., P. G. Bray, D. Verdier-Pinard, D. J. Johnson, P. Horrocks, R. A. Muhle, G. E. Alakpa, R. H. Hughes, S. A. Ward, D. J. Krogstad, A. B. Sidhu, and D. A. Fidock. 2005. EMBO J 24:2294–2305. (ii) Fidock, D. A., T. Nomura, A. K. Talley, R. A. Cooper, S. M. Dzekunov, M. T. Ferdig, L. M. B. Ursos, A. B. S. Sidhu, B. Naude, K. W. Deitsch, X. – Z. Su, J. C. Wootton, P. D. Roepe, and T. E. Wellems. 2000. Molecular Cell 6:861–871. (iv) Mehlotra, R. K., H. Fujioka, H. Fujioka, P. D. Roepe, O. Janneh, L. M. Ursos, V. Jacobs-Lorena, D. T. McNamara, M. J. Bockarie, J. W. Kazura, D. E. Kyle, D. A. Fidock, and P. A. Zimmerman. 2001. Proc Natl Acad Sci USA 98:12689–12694. (v) Mu, J., M. T. Ferdig, X. Feng, D. A. Joy, J. Duan, T. Furuya, G. Subramanian, L. Aravind, R. A. Cooper, J. C. Wootton, M. Xiong, and X. Z. Su. 2003. Mol Microbiol 49:977–989. (vi) Sanchez, C. P., J. E. McLean, W. Stein, and M. Lanzer. 2004. Biochemistry 43:16365–16373. (vii) Wellems, T. E., L. J. Panton, I. Y. Gluzman,V. E. do Rosario, R. W. Gwadz, A. Walker-Jonah, and D. J. Krogstad. 1990. Nature 345:253–255.
CQS HB3 parasites are, as expected, not resistant to plasma levels of CQ because they show LD50 that are ≤ 1/5th plasma [CQ]. In contrast, as expected, CQR parasites show LD50 above what would be found to be plasma [CQ] for many patients, consistent with inefficient clearance of CQR parasites within hours of CQ therapy. In the case of CQR strain Dd2, resistance to CQ toxicity is quite profound, with LD50 near 15 µM. CQR 7G8 parasites are noticeably less resistant (~4-fold less resistant) to CQ toxicity vs. CQR Dd2 parasites (Table 1, CQ Rf of 32 vs. 124, respectively).
Table 2 shows a summary of previously determined IC50 for CQ as well as other quinoline antimalarial drugs vs. these strains. Relative CQR calculated for 7G8 and Dd2 via LD50 (Rf = 34, 124, respectively) contrasts quite strikingly with the < 2-fold decreased resistance for 7G8 vs. Dd2 when resistance is quantified by IC50 ratio (Table 2, CQ Rf of 6 vs. 9, respectively). That is, the distinction between degree of CQR for strains 7G8 and Dd2 is much more dramatic when quantified via LD50.
In order to test whether relative resistances for other drugs also differed when quantified by LD50 vs IC50 data, cytocidal assays were performed as for CQ. Rf values computed by LD50 ratios for these drugs reveal additional very interesting new information. For example, when quantified by IC50 data, in our hands the rank order degree of drug resistances for CQR Dd2 parasites are CQR > AQR > QDR ~ QNR > MQR (column 7, table 2; note that some other studies show higher QDR for this strain, see “literature Rf” column 5 table 2), but this changes to CQR > QDR >> QNR > AQR > MQR when resistance is quantified by LD50 data (column 5, table 1). Similarly, although the precise Rf values differ, CQR 7G8 parasites also show a profile that is CQR > AQR > QNR ~ QDR > MQR via IC50 data, that changes to CQR >> QNR ~ QDR > AQR > MQR when quantified by LD50 (we again note some small differences between our IC50 Rf vs. those that can be calculated from the literature, see caption to table 2). As shown elsewhere [7], the different drug selection histories for these two CQR strains clearly promotes different multidrug resistance phenotypes (meaning, different rank order resistance to different drugs). However, what has been missed until now is that relative sensitivity to cytocidal vs. cytostatic effects of these drugs can differ within a given strain and also differ when comparing multiple CQR strains (e.g. Dd2 vs. 7G8).
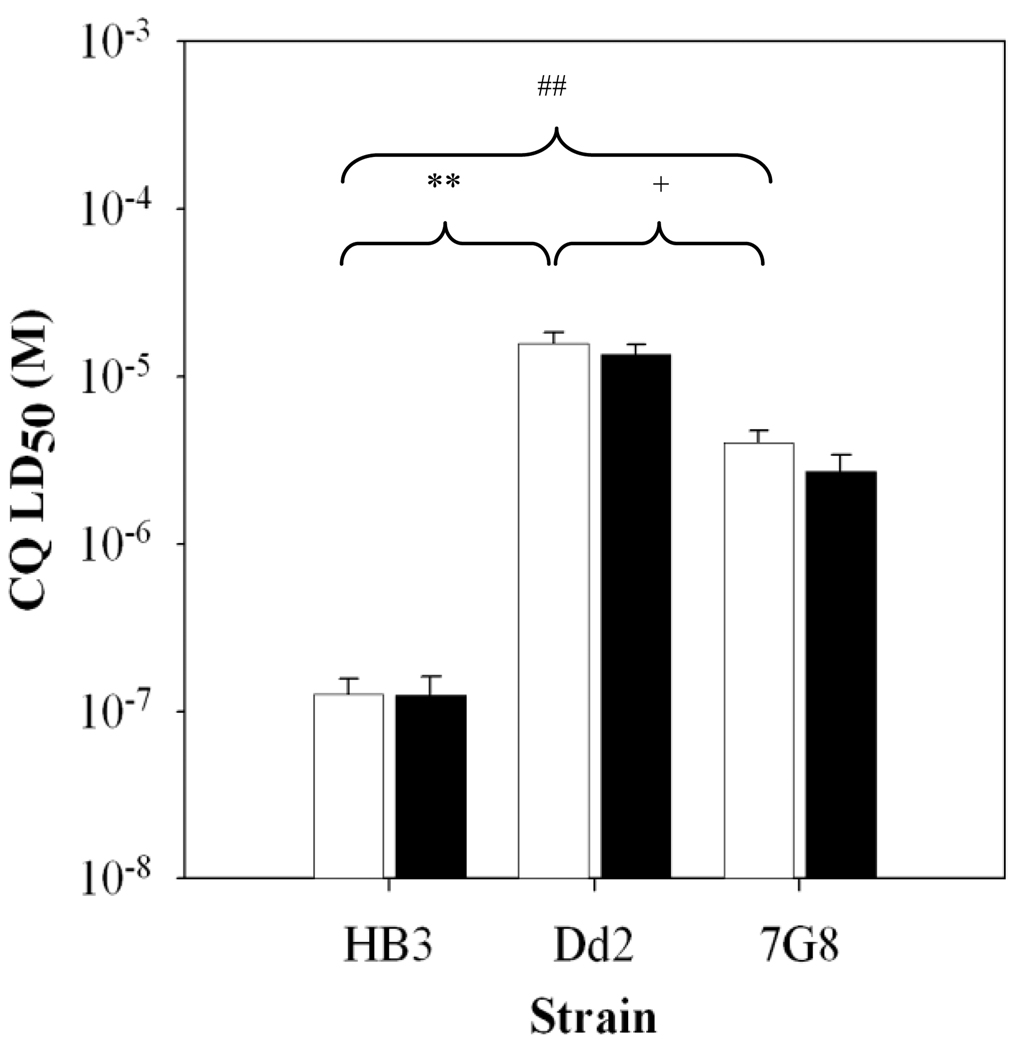
Another well known, established concept derived from inspection of antimalarial IC50 data is that VPL is known to reverse elevated IC50 for Dd2 parasites, hence the common description of VPL as an antimalarial “chemoreversal agent” [6]. VPL has a much lower chemoreversal effect for strain 7G8. This is consistent with recent data that shows VPL strongly inhibits CQ probe binding to the Dd2 PfCRT isoform, but not to the 7G8 PfCRT isoform [9]. Interestingly, a standard similar chemoreversal dose of VPL (~0.9 µM) administered in bolus fashion along with bolus CQ does not lower CQ LD50 for any of the three strains (not shown). However, we note that proper definition of a chemoreversal agent means that it is non-toxic when administered at the same dose alone. In our hands, 0.9 µM VPL is the highest dose for non-growth inhibitory administration of VPL in continuous culture, meaning, above this dose VPL is cytostatic. This means that “reversal” at dosages > 0.9 µM is actually combined growth inhibition by both CQ and VPL, not necessarily VPL chemoreversal of CQR. Since any cytocidal activity of VPL might be time-dependent as is the case with CQ, we titrated [VPL] vs. time of bolus treatment to determine the highest concentration that was non-cytocidal in a 6 hr pulse (data not shown) and found this to be 5 µM. Importantly then, we also find that VPL does not chemoreverse the cytocidal activity of CQ at the highest non toxic dose (Figure 2). Incubation of HB3, Dd2, or 7G8 parasites with CQ in the presence of 5 µM VPL (filled bars) yielded CQ LD50 values that were not significantly different from parasites treated with CQ alone (open bars).
FIG. 2.
Verapamil does not chemosensitize the CQR strains Dd2 and 7G8 to CQ as indicated by no statistically significant change in CQ LD50 (open bars) upon addition of 5 µM VPL (filled bars). CQ LD50 for HB3 was found to be significantly different from Dd2 (**, p < 0.001) and 7G8 (##, p <0.001). Dd2 and 7G8 CQ LD50 values were also significantly different from each other (+, p < 0.01), but there were no significant differences for either of the 3 strains +/− VPL (see also Table 1)
Discussion
Limitations in speed and sensitivity, as well as multiple sources of error in crude visual counting approaches reported earlier [14] enticed us to develop a simpler, more sensitive, and faster SYBR Green-based approach for quantifying malarial parasite LD50 vs diffusible antimalarial drugs. This assay is superior in most respects but we also note some potential complexities. First, cytocidal potency is defined indirectly in this new assay and assumes that those cells not killed by toxic levels of CQ grow at a similar rate relative to parasites not treated with drug. This may be an oversimplification, since it is possible some cells treated at ≥LD50 dosages might grow more slowly once the drug is removed. We currently have no evidence for this, and note that it is unlikely since LD50 quantified at 48 hrs vs 72 hrs are the same (data not shown). However, if some strains (or field isolates) of P. falciparum do behave in that manner, quantification of the number of parasites that have actually been killed might be slightly overestimated via the new assay (in contrast to under estimation that occurs via visual quantification, see below).
Regardless, importantly, we note that the LD50 we calculate are entirely consistent with clinical data. For instance, we would expect fast clearance rates for CQS parasites with LD50 near 200 nM, as has been observed in clinical studies, because plasma levels of CQ can range from 1–10 µM (e.g. [17]). LD50 approximately 4–15 µM for CQR parasites (Table 1), is similarly consistent with the responses that have been observed clinically for patients infected with CQR parasites. Fewer measurements of plasma levels of QN have been done, but published data show that plasma QN is ~ 2 – 20 – fold higher than plasma CQ [18,19], again consistent with measured LD50 for QNS (HB3) and QNR (Dd2) strain behavior. Interestingly, we note strain 7G8 is QNS via IC50 data, but QNR via LD50. This suggests that the intracellular targets for QN cytostatic vs cytocidal effects likely differ. Related to these points, we note that there is patient variability not just in peak plasma concentrations of quinoline antimalarials, but also in time above what is usually assumed to be maximum inhibitory concentration (MIC). The antimalarial potency of quinoline drugs is usually envisioned to be related to the time above MIC. However, data in this paper suggest that peak plasma concentration is also quite relevant. If so, perhaps patient variability in both peak plasma concentrations as well as time above MIC is relevant to emergence of chloroquine resistance phenomena.
Because of dissimilar stage-dependent behavior for CQS HB3 vs. CQR Dd2 parasites (Fig. 1B), data summarized in Table 1 are from experiments using asynchronous culture. Earlier attempts to measure CQ LD50 [14] for synchronized stages of HB3 and Dd2 cultures was performed by rather tedious visual counting of parasites. The CQ LD50 values obtained in the previous study are similar but not identical to those we report here. Differences and increased error in the previous study are due to several factors. For example, Gligorijevic et al. [14] primarily used bolus dose times of 2 hrs (as opposed to 6 hrs used in the present work) and visual counting of Giemsa-stained parasites instead of fluorescence quantification of new parasite nuclei to estimate survival. For strains grown under standard conditions, one parasite infected RBC in live culture that successfully proceeds through schizogony provides 3–6 new iRBC, yielding a 3–6 fold increase in calculated parasitemia. However, each of those 3–6 new parasites generates 12–28 new nuclei that can be stained by SYBR Green I. Therefore, visual counting provides only a 3–6 fold dynamic range for quantifying increases over time while SYBR Green I staining potentially provides 36–168 fold ([3–6] × [12–28]). Signal-to-noise ratio is thus much higher (and intrinsic error is thus lower) with the SYBR Green approach. Also, through the work presented in this paper, we now appreciate that many dead (pyknotic) parasites would still be visually counted via the earlier approach but would not proceed to schizogony nor fluoresce as do live cells in the current assay, which highlights another significant source of error in the earlier work [14]. Our earlier work also used an absolute time line for stage-dependent treatments with drug wherein survival for bolus dosed rings was visually counted 56 hrs after treatment but bolus dosed trophozoites were counted 46 hrs post treatment [14]. The different times were necessary because of the high volume of tedious visual counts that were required for that study. In contrast, measurements in the present work involving synchronized culture rings and trophozoites (Fig. 1A) were done at the same times post treatment for each stage.
Although this new assay is an improvement over methods that rely on visually counting parasites, additional improvements in high-throughput cytocidal assays are certainly possible. These would include definition of unambiguous, direct markers for parasite cell death that are also amenable to high throughput assay formats. What makes SYBR Green I and certain other DNA dyes so useful in malarial parasite assays is that the dye stains nuclei that are multiplied 8–36 fold in each division. In addition, SYBR Green I fluorescence quantum yield increases 7-fold upon binding to parasite nucleic acid [1]. These two features added together provide a very large dynamic range for detecting variance in parasite density via convenient fluorescence plate reader equipment, but also requires that surviving parasites proceed through one iRBC cycle after bolus treatment with drug (so that they produce new nuclei in amplified numbers). A more direct cytocidal assay would be able to detect dead parasites immediately after bolus drug treatment, but currently no reliable detection methods for this exist. In attempting to devise one that is amenable for high-throughput format, we have stained iRBC treated with CQ LD50 dosages and find in various experiments that 40–60 percent of the parasites will stain with the dead cell indicator trypan blue, but to variable intensities. Other fully toxic treatments, such as 5 mM azide for 60 min or 20 µM ionophore for 60 min, yield higher but still variable trypan blue positive parasites. Intensity of trypan blue staining is known to vary for dead cells, but we suspect trypan blue staining is also especially variable for parasites within iRBC because the RBC membrane makes uniform transport of (toxic) trypan blue difficult in short periods of time before the dye itself begins to kill parasites (data not shown). That is, concentrations of trypan blue needed for high contrast staining are in and of themselves toxic. Furthermore, the dynamic range in trypan blue signal is not amenable to the precision and accuracy required for a convenient high-throughput assay, because in this case the signal is much lower and what is measured is less specific chromophore absorbance, not nucleic acid-specific intense fluorescence. Other fluorescent stains that might in theory be used to directly detect parasite death include hydroxyethidine, carbocyanines, and others. Hydroxyethidine must be converted to a different form within the cell to yield live cell specific fluorescence, and carbocyanines require high membrane potentials in live cells to distinguish live vs. dead. Similar to trypan blue, we find that staining with these dyes is quite variable for CQ treated cultures and that the dynamic range for measuring live vs. dead is again quite low, making accurate quantification of LD50 difficult. This is likely the case because hydroxyethidine conversion in live cells is not uniform or as reliable as might be hoped [20] and because the low membrane potential for only a single mitochondrion in iRBC yields high background carbocyanine staining.
Our experiences with these dyes notwithstanding, this does not mean that other approaches cannot in theory be developed. Based on these initial provocative indications of a much wider than expected range of LD50 for CQR strains, non-reversal of strain Dd2 LD50 by VPL, and altered patterns of multidrug resistance relative to previous definitions that came from ranking IC50, we urge continued inspection of LD50 phenomena for malarial parasites. Standardized LD50 data provides important additional information in the development of new candidate antimalarial drugs.
We also suggest additional mapping of QTL and other genetic descriptions of malarial drug resistance phenomena are warranted using LD50 (not IC50) as the physiologic or phenotypic indicator. Although our results are only preliminary, in our initial scan through the progeny of the well known HB3xDd2 cross, we do not find the same relative ranking of the progeny using CQ LD50 vs. CQ IC50 [M.F.P., K.G.B. & P.D.R. unpublished]. This indicates to us that perhaps unknown genetic loci contribute to elevated LD50 phenomena, or that known genetic loci that dictate elevated IC50 have a different rank order of resistance conferring effects when cytocidal activity of CQ is quantified. Such a scenario appears likely since parasite CQ induced death vs growth inhibition likely involves different targets [8]. The dissection of these determinants could significantly reframe our reliance on IC50 data. Also, a wider distribution of LD50 values for the progeny (>100-fold resistances are seen via LD50 ratios), relative to IC50 values (only 10–20-fold resistances are seen), should make definition of any genetic features that complement pfcrt mutation in conferring resistance less difficult.
Finally, we note that since radiolabeled versions of other drugs used in this study are not available to us, it was not possible to verify complete removal of other quinoline drugs by three washing steps as described in Methods. But because these drugs are very similar to CQ in their chemical properties, and are not known be metabolized, activated or permanently covalently attached to intracellular targets under these conditions, it is most likely that they are completely removed by these washing steps, similar to CQ. Artemisinin-based drugs, on the contrary, are activated to highly reactive species that then irreversibly alkylate multiple targets and are hence not removed by washing steps, as is the case for quinoline drugs. Pharmacologic characteristics (and resistance phenomena) for this class of drugs will therefore of course differ significantly from what is observed for quinolines (e.g. see [21]). For highly reactive drugs that covalently attach to their target, LD50 and IC50 are expected to be similar, since even a short term bolus dose results in the continuous (intracellular) presence of the drug. However, more detailed transport studies with artemisinin – based drugs are needed to define concentration dependent saturation; if accumulation is not saturated in short periods of time, LD50 and IC50 might differ more than is expected for some artemisinin – based drugs.
Acknowledgements
This work was funded by NIH grants AI045957 and AI056312 (to PDR). We thank Drs. R. Cooper (Old Dominion), M. Ferdig (Notre Dame), and P. Rathod (Univ. Washington) for helpful discussions, Ms. K. Sherlach (Georgetown) for sharing trypan blue staining data and Ms. Emily Rubinson for technical help.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- 1.Bennett TN, Paguio M, Gligorijevic B, Seudieu C, Kosar AD, Davidson E, Roepe PD. Novel, rapid, and inexpensive cell-based quantification of antimalarial drug efficacy. Antimicrob Agents Chemother. 2004;48:1807–1810. doi: 10.1128/AAC.48.5.1807-1810.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Co E-MA, Dennull RA, Reinbold DD, Waters NC, Johnson JD. Assessment of malaria in vitro drug combination screening and mixed-strain infections using the malaria Sybr green I-based fluorescence assay. Antimicrob Agents Chemother. 2009;53:2557–2563. doi: 10.1128/AAC.01370-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferdig MT, Cooper RA, Mu J, Deng B, Joy DA, Su XZ, Wellems TE. Dissecting the loci of low-level quinine resistance in malaria parasites. Mol Microbiol. 2004;52:985–997. doi: 10.1111/j.1365-2958.2004.04035.x. [DOI] [PubMed] [Google Scholar]
- 4.Fidock DA, Nomura T, Talley AK, Cooper RA, Dzekunov SM, Ferdig MT, Ursos LM, Sidhu AB, Naudé B, Deitsch KW, Su XZ, Wootton JC, Roepe PD, Wellems TE. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol Cell. 2000;6:861–871. doi: 10.1016/s1097-2765(05)00077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ndiaye D, Patel V, Demas A, LeRoux M, Ndir O, Mboup S, Clardy J, Lakshmanan V, Daily JP, Wirth DF. A non-radioactive DAPI-based high-throughput in vitro assay to assess Plasmodium falciparum responsiveness to antimalarials--increased sensitivity of P. falciparum to chloroquine in Senegal. Am J Trop Med Hyg. 2010;82:228–230. doi: 10.4269/ajtmh.2010.09-0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patel JJ, Thacker D, Tan JC, Pleeter P, Checkley L, Gonzales JM, Deng B, Roepe PD, Cooper RA, Ferdig MT. Chloroquine susceptibility and reversibility in a Plasmodium falciparum genetic cross. Mol Microbiol. 2010;78:770–787. doi: 10.1111/j.1365-2958.2010.07366.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sá JM, Twu O, Hayton K, Reyes S, Fay MP, Ringwald P, Wellems TE. Geographic patterns of Plasmodium falciparum drug resistance distinguished by differential responses to amodiaquine and chloroquine. Proc Natl Acad Sci U S A. 2009;106:18883–18889. doi: 10.1073/pnas.0911317106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cabrera M, Paguio MF, Xie C, Roepe PD. Reduced digestive vacuolar accumulation of chloroquine is not linked to resistance to chloroquine toxicity. Biochemistry. 2009;48:11152–11154. doi: 10.1021/bi901765v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lekostaj JK, Natarajan JK, Paguio MF, Wolf C, Roepe PD. Photoaffinity labeling of the Plasmodium falciparum chloroquine resistance transporter with a novel perfluorophenylazido chloroquine. Biochemistry. 2008;47:10394–10406. doi: 10.1021/bi8010658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pleeter P, Lekostaj JK, Roepe PD. Purified Plasmodium falciparum multi-drug resistance protein (PfMDR 1) binds a high affinity chloroquine analogue. Mol Biochem Parasitol. 2010;173:158–161. doi: 10.1016/j.molbiopara.2010.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paguio MF, Cabrera M, Roepe PD. Chloroquine transport in Plasmodium falciparum. 2. Analysis of PfCRT-mediated drug transport using proteoliposomes and a fluorescent chloroquine probe. Biochemistry. 2009;48:9482–9491. doi: 10.1021/bi901035j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roepe PD. PfCRT – Mediated Drug Transport in Malarial Parasites. Biochemistry. 2011;50:163–171. doi: 10.1021/bi101638n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Young RD, Rathod PK. Clonal viability measurements on Plasmodium falciparum to assess in vitro schizonticidal activity of leupeptin, chloroquine, and 5-fluoroorotate. Antimicrob Agents Chemother. 1993;37:1102–1107. doi: 10.1128/aac.37.5.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gligorijevic B, Purdy K, Elliott DA, Cooper RA, Roepe PD. Stage independent chloroquine resistance and chloroquine toxicity revealed via spinning disk confocal microscopy. Mol Biochem Parasitol. 2008;159:7–23. doi: 10.1016/j.molbiopara.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trager W, Jensen JB. Human malaria parasites in continuous culture. Science. 1976;193:673–675. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- 16.Lambros C, Vanderberg JP. Synchronization of Plasmodium falciparum erythrocytic stages in culture. J Parasitol. 1979;65:418–420. [PubMed] [Google Scholar]
- 17.Krishna S, White NJ. Pharmscokinetics of Quinine, Chloroquine and Amodiaquine. Clinical Implications. Clin Pharmacokinet. 1996;30:263–299. doi: 10.2165/00003088-199630040-00002. [DOI] [PubMed] [Google Scholar]
- 18.Fernandes Vieira JL, Guimaraes Borges LM, Silva Nascimento MT, de LS, Gomes A. Quinine Levels in Patients with Uncomplicated falciparum Malaria in the Amazon Region of Brazil. The Brazilian J. of Infec. Diseases. 2008;12:353–354. doi: 10.1590/s1413-86702008000500001. [DOI] [PubMed] [Google Scholar]
- 19.Mirghani RA, Elagib I, Elghazali G, Hellgren U, Gustafsson LL. Effects of Plasmodium falciparum Infection on the Pharmacokinetics of Quinine and its Metabolites in Pregnant and Non – Pregnant Sudanese Women. Eur. J. Clin. Pharmacol. 2010;66:1229–1234. doi: 10.1007/s00228-010-0877-3. [DOI] [PubMed] [Google Scholar]
- 20.Zielonka J, Kalyanaraman B. Hydroethidine- and MitoSOX-derived red fluorescence is not a reliable indicator of intracellular superoxide formation: another inconvenient truth. Free Radic Biol Med. 2010;48:983–1001. doi: 10.1016/j.freeradbiomed.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Teuscher F, Gatton ML, Chen N, Peters J, Kyle D, Cheng Q. Artemisinin – Induced Dormancy in Plasmodium falciparum: Duration, Recovery Rates, and Implications in Treatment Failure. J. Infect. Disease. 2010;202:1362–1368. doi: 10.1086/656476. [DOI] [PMC free article] [PubMed] [Google Scholar]