Abstract
MX-2401 is a semisynthetic calcium-dependent lipopeptide antibiotic (analogue of amphomycin) in preclinical development for the treatment of serious Gram-positive infections. In vitro and in vivo, MX-2401 demonstrates broad-spectrum bactericidal activity against Gram-positive organisms, including antibiotic-resistant strains. The objective of this study was to investigate the mechanism of action of MX-2401 and compare it with that of the lipopeptide daptomycin. The results indicated that although both daptomycin and MX-2401 are in the structural class of Ca2+-dependent lipopeptide antibiotics, the latter has a different mechanism of action. Specifically, MX-2401 inhibits peptidoglycan synthesis by binding to the substrate undecaprenylphosphate (C55-P), the universal carbohydrate carrier involved in several biosynthetic pathways. This interaction resulted in inhibition, in a dose-dependent manner, of the biosynthesis of the cell wall precursors lipids I and II and the wall teichoic acid precursor lipid III, while daptomycin had no significant effect on these processes. MX-2401 induced very slow membrane depolarization that was observed only at high concentrations. Unlike daptomycin, membrane depolarization by MX-2401 did not correlate with its bactericidal activity and did not affect general membrane permeability. In contrast to daptomycin, MX-2401 had no effect on lipid flip-flop, calcein release, or membrane fusion with 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC)/1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (sodium salt) (POPG) liposomes. MX-2401 adopts a more defined structure than daptomycin, presumably to facilitate interaction with C55-P. Mutants resistant to MX-2401 demonstrated low cross-resistance to other antibiotics. Overall, these results provided strong evidence that the mode of action of MX-2401 is unique and different from that of any of the approved antibiotics, including daptomycin.
INTRODUCTION
MX-2401 is a semisynthetic calcium (Ca2+)-dependent lipopeptide antibiotic in preclinical development for the treatment of serious Gram-positive infections. An analogue of amphomycin, MX-2401, demonstrates broad-spectrum bactericidal activity against Gram-positive organisms, including strains resistant to vancomycin, macrolides, penicillin, methicillin, gentamicin, and other marketed antimicrobials (12, 15). In animal models of infection, MX-2401 exhibits potent dose-dependent activity against Gram-positive organisms, including resistant strains, with an excellent ability to kill bacteria in neutropenic mice (9).
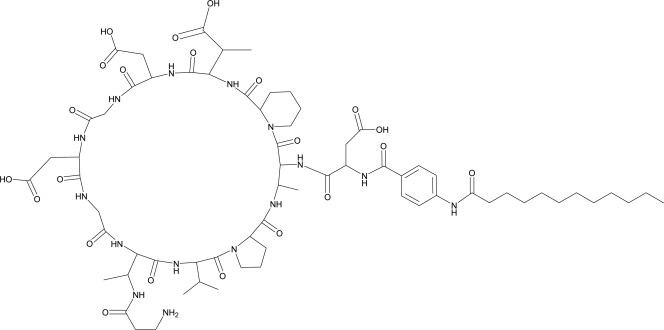
MX-2401 (Fig. 1) is chemically related to a number of antimicrobial lipopeptides, such as amphomycin, friulimicin, and daptomycin (7, 11, 25, 46). Specifically, amphomycin, friulimicin, and MX-2401 share the same cyclopeptide ring core comprising 10 amino acids and differ only in their exocylic amino acids, with asparagine in friulimicin and aspartic acid in MX-2401 and amphomycin (25, 46). MX-2401 differs from amphomycin in the side chain of residue 9 (1). In contrast, daptomycin has a different core structure in that it shares only the general lipopeptide framework with a cyclic decapeptide core and a decanoyl fatty acid tail interlinked by 3 extracyclic amino acids (11). Daptomycin is further classified as a lipodepsipeptide with the decapetide core cyclized by an ester bond.
Fig. 1.
MX-2401 structure.
Daptomycin, a first-in-class lipopeptide antibiotic, has been approved for the treatment of complicated skin and skin structure infections, endocarditis, and bacteremia. Daptomycin has been proposed to kill bacteria by interacting with the cytoplasmic membrane (19) and causing membrane depolarization or perturbation (35, 36). It was demonstrated that in the presence of Ca2+ daptomycin interacts with phosphatidylglycerol in the bacterial membrane and perturbs the membrane bilayer (18, 19). In model membranes containing phosphatidylglycerol, daptomycin also causes membrane fusion in the presence of Ca2+ (18). The perturbation of the bacterial cell membrane and its depolarization are proposed to lead to intracellular ion leakage and rapid cell death (36). Daptomycin adopts similar structures in the apo form, in the presence of one or more equivalents of Ca2+, and in the presence of phosphatidylcholine/Ca2+ (14, 32). It is thought that the highly dynamic nature of daptomycin enables it to adapt to these different environments and to effectively interact with the bacterial cell membrane. Once daptomycin has permeated the membrane, however, it is still possible that it interacts with other intracellular targets (19, 31). This fact may account for the high potency of daptomycin.
In contrast, lipopeptide antibiotics, such as amphomycin and friulimicin, function by inhibiting the biosynthesis of the membrane-bound peptidoglycan (component) precursor of Gram-positive cell walls (28, 39, 41). Specifically, earlier studies with amphomycin indicated the inhibition of bacterial peptidoglycan biosynthesis, probably acting at the level of phospho-N-acetylmuramoyl-pentapeptide-transferase (MraY). This enzyme links the final soluble cytoplasmic cell wall building block, N-acetylmuramyl pentapeptide (UDP-MurNAc-pp) to the membrane carrier lipid undecaprenyl-phosphate (bactoprenol-phosphate [C55-P]). However, the molecular details of the amphomycin mode of action have not been determined (39, 40, 41). It was further shown that amphomycin inhibits the synthesis of dolichol-linked saccharides, which are essential in the glycosylation of glycoproteins in eukaryotes (20). Since dolichyl-phosphate and undecaprenyl-phosphate differ simply in the length of the carrier lipid, i.e., 12 isoprenoid units rather than 11 (60 carbon atoms versus 55 in bactoprenol), this finding suggests that the bactericidal action of amphomycin may be achieved through interaction with the lipid carrier C55-P. Similarly, friulimicin was shown to form a Ca2+-dependent complex with C55-P, resulting in inhibition of cell wall biosynthesis (28).
At present, little is known about the mechanism of action of the novel semisynthetic amphomycin analogue MX-2401. The purpose of the current study was to investigate the mechanism of action of MX-2401 and to compare it with that of amphomycin and daptomycin. Our data indicate that its action is completely distinct from that of daptomycin. Resistance training studies indicated that its mechanism is also very different from that of other common antibiotics.
MATERIALS AND METHODS
Organisms.
Staphylococcus simulans 22 (clinical isolate), Staphylococcus aureus ATCC 29213, Enterococcus faecalis ATCC 29212, and Staphylococcus epidermidis ATCC 12228 were used in these studies.
Antibiotics.
Daptomycin was obtained from Cubist Pharmaceuticals, Inc., Lexington, MA. Vancomycin, gentamicin, trimethoprim, ampicillin, ceftriaxone, erythromycin, and tetracycline were purchased from Sigma Chemical Co., St. Louis, MO, and ciprofloxacin from BioChemika Fluka. MX-2401 and amphomycin were produced by BioWest Therapeutics Inc. (formerly Migenix Inc.) with assistance from BioSource Pharm Inc.
Susceptibility testing.
Minimal growth-inhibitory concentrations (MICs) of amphomycin, daptomycin, and MX-2401 were determined by the standard broth microdilution method based on the most recent Clinical and Laboratory Standards Institute guidelines (8) using cation-adjusted Mueller-Hinton broth supplemented with 50 μg/ml Ca2+ (denoted CAMHBc). The organisms in the exponential growth phase were diluted to a final inoculum of 1 × 105 to 5 × 105 CFU/ml. MICs were read after 16 to 20 h of incubation at 37°C.
Serial passaging of S. aureus and E. faecalis in MX-2401 and daptomycin.
To generate mutants of S. aureus (ATCC 29213) and E. faecalis (ATCC 29212) with decreased susceptibility to MX-2401 and daptomycin, the strains were serially passaged in the presence of sub- to supra-MIC concentrations of MX-2401 or daptomycin. For each passage, MICs were determined using the CLSI broth microdilution method (47). Briefly, 90 μl/well of bacterial suspension at 5 × 105 CFU/ml in CAMHBc was added to a 96-well plate, along with 10 μl of serially diluted lipopeptide solution. After the plate was incubated for 18 h at 37°C, the MIC was taken as the concentration at which no growth was visibly observed (as determined by visual inspection). The well containing bacterial suspension at the lipopeptide concentration corresponding to half the MIC was used as the inoculum for the second passage. This mixture was diluted by a factor of 1/200 in CAMHBc, and 90 μl/well was added to a new 96-well plate, along with 10 μl serially diluted lipopeptides. Five concentrations of the lipopeptides were tested for each serial passage, the half MIC and two doubling dilutions below and two doubling dilutions above the half MIC. This procedure consisted of one serial passage. The passages were serially repeated 27 times in all.
Cross-resistance testing.
Cross-resistance was investigated by doing CLSI MIC tests using various antibiotics on the S. aureus and E. faecalis serial-passage mutants with decreased susceptibility to MX-2401 and daptomycin. MIC testing (47) was performed by using CAMHBc for MICs of daptomycin and MX-2401, while CAMHB was used for all the other antimicrobials.
Intracellular accumulation of the final soluble cell wall precursor, UDP-N-acetylmuramyl pentapeptide.
For analysis of the cytoplasmic peptidoglycan nucleotide precursor pool, we followed the protocol of Kohlrausch and Höltje (21), elaborated for Bacillus subtilis, with slight modifications. S. simulans 22 cells were grown in 20 ml of half-concentrated Mueller-Hinton broth containing 1.25 mM Ca2+ to an optical density at 600 nm (OD600) of 0.6 and then supplemented with 130 μg/ml of chloramphenicol and incubated for 15 min. Chloramphenicol is necessary to prevent induction of an autolytic process and de novo synthesis of enzymes hydrolyzing the nucleotide-activated sugars interfering with determination of the soluble precursor, under the impact of the antibiotic under investigation (10). Then, lipopeptides were added at 10× MIC as determined under the standard conditions described above and incubated for another 45 min. Subsequently, the cells were rapidly cooled on ice and spun down (15,000 × g; 5 min; 4°C), resuspended in cold water, and, under stirring, treated with 2 volumes of boiling water. Cell debris was removed (48,000 × g; 30 min), and the supernatant was lyophilized. For C-18 reverse-phase high-performance liquid chromatography (HPLC), lyophilisates were dissolved in water and acidified to pH 2 with 20% (vol/vol) phosphoric acid; insoluble material was removed (15,000 × g; 5 min); and aliquots of the supernatants, adjusted to identical cell wet weights for the differently treated cultures, were applied to the column. Separation was achieved under isocratic conditions with 50 mM sodium phosphate, pH 5.2, as a solvent. UDP-linked cell wall precursors were confirmed by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry.
Cloning and purification of recombinant proteins.
Cloning, expression, and purification of the recombinant S. aureus NCTC MraY enzyme were performed as described previously (28). TagO (Llm) of S. aureus N315 was cloned, expressed, and purified according to the protocol elaborated for MraY. The TagO gene, llm, was amplified by PCR using the primer pair llm1-for and llm2-rev (Table 1). The S. aureus murA to murF genes were amplified using forward and reverse primers listed in Table 1 and cloned into a pET21b vector (Novagen) using NheI or NdeI and XhoI restriction sites to generate C-terminal His6 fusion proteins. Escherichia coli BL21(DE3) (Promega) cells transformed with the appropriate recombinant plasmid were grown in LB medium (Becton Dickinson) at 30°C. At an OD600 of 0.6, IPTG (isopropyl-β-d-thiogalactopyranoside) was added at a concentration of 0.5 mM to induce expression of the recombinant proteins. After 3 h, cells were harvested and resuspended in lysis buffer (50 mM NaH2PO4, pH 7.8, 300 mM NaCl, 10 mM imidazole). Aliquots of 200 mg/ml lysozyme, 100 mg/ml DNase, and 10 mg/ml RNase were added, and the cells were incubated for 30 min on ice and sonicated. The cell debris was spun down, and the supernatant was applied to Ni-nitrilotriacetic acid (NTA)-agarose slurry (Qiagen). This mixture was gently stirred at 4°C for 1 h and then loaded onto a column support. After being washed with lysis buffer, weakly bound material was removed with 50 mM NaH2PO4, pH 7.8, 300 mM NaCl, and 20 mM imidazole. His-tagged MurA-MurF proteins eluted with buffer containing 50 mM NaH2PO4, pH 7.8, 300 mM NaCl, and 200 mM imidazole. Three fractions each were collected and stored in 50% glycerol at −20°C. Purity was controlled by SDS-PAGE.
Table 1.
Primers used in this study
| Primer | Sequence (5′–3′)a |
|---|---|
| llm5-for | AAGGATCCGTTACATTATTACTAGTTGCAG |
| llm5-rev | ATTCTCGAGTTCCTCTTTATGAGATGAC |
| murA-for | TTTGCTAGCGATAAAATAGTAATCAAAGGTG |
| murA-rev | ACTCTCGAGATCGTTAATACGTTCAAT |
| murB-for | TGAGCTAGCATAAATAAAGACATCTATCAAG |
| murB-rev | CTTCTCGAGCGATTCCTTTGGATGTT |
| murC-for | TATGCTAGCACACACTATCATTTTGTC |
| murC-rev | AACCTCGAGAAACGCATTTTTCATGC |
| murE-for | TTCGCTAGCGATGCAAGTACGTTGT |
| murE-rev | AATCTCGAGTTGATCAACAGGGCCA |
| murF-for | GTTGCTAGCATTAATGTTACATTAAAG |
| murF-rev | TAACTCGAGTGAAATTAAAGCATTTAC |
| ddlA-for | AACGCTAGCACAAAAGAAAATATTTGT |
| ddlA-rev | CCTCTCGAGGTCAATTTTGTATTTA |
Restriction sites are underlined.
In vitro lipid II synthesis reaction using membrane preparations of Micrococcus luteus.
In vitro lipid II synthesis was performed using membranes of Micrococcus luteus as described previously (5, 30). In short, membrane preparations (200 μg protein) were incubated in the presence of purified substrates, 5 nmol C55-P, 50 nmol UDP-N-acetylmuramic acid pentapeptide (UDP-MurNAc-pp), and 50 nmol [14C]UDP-GlcNAc (UDP-activated N-acetyl-glucosamine) in 60 mM Tris-HCl, 5 mM MgCl2, pH 7.5, 1.25 mM Ca2+, and 0.5% (wt/vol) Triton X-100 in a total volume of 50 μl for 1 h at 30°C. Bactoprenol-containing products were extracted with the same volume of butanol-pyridine acetate (2:1 [vol:vol]; pH 4.2) and analyzed by thin-layer chromatography (TLC) using phosphomolybdic acid (PMA) staining (30). Quantification was carried out using a storage phosphor screen to visualize radioactivity in a Storm imaging system (GE Healthcare). For testing the impacts of amphomycin and MX-2401, the lipopeptides were added in molar ratios of 0.25 to 2 with respect to C55-P.
Synthesis of [14C]UDP-MurNAc-pentapeptide by S. aureus MurA-MurF and DdlA enzymes.
[14C]UDP-MurNAc-pentapeptide was synthesized on the basis of the protocol of Wong et al. (48) with modifications. UDP-GlcNAc (100 nmol) was incubated in the presence of 15 μg MurA-MurF and DdlA protein in 50 mM bis-Tris-propane, pH 8, 25 mM (NH4)2SO4, 5 mM MgCl2, 5 mM KCl, 0.5 mM dithiothreitol (DTT), 2 mM ATP, 2 mM phosphoenolpyruvate (PEP), 2 mM NADPH, 1 mM each amino acid (14C-l-Lys, d-Glu, l-Ala, and d-Ala), and 10% dimethyl sulfoxide (DMSO) in a total volume of 125 μl for 60 min at 30°C; 31.25 μl of the reaction mixture, corresponding to 25 nmol [14C]UDP-MurNAc-pentapeptide, was added to the MraY synthesis assay without further purification.
In vitro lipid I synthesis reaction using purified MraY.
To determine the impacts of amphomycin and MX-2401 on the MraY-catalyzed lipid I synthesis, the assay was carried out in a total volume of 60 μl containing 2.5 nmol C55-P, 25 nmol of [14C]UDP-MurNAc-pentapeptide in 100 mM Tris-HCl, 30 mM MgCl2, pH 7.5, 10% (vol/vol) DMSO, and 10 mM N-lauroyl sarcosine in the presence of 1.25 mM CaCl2. The reaction was initiated by the addition of 7.5 μg of the enzyme and incubated for 2 h at 30°C. Amphomycin and MX-2401 were added in molar ratios ranging from 0.25 to 2 with respect to the amount of C55-P. Synthesized lipid I was extracted from the reaction mixtures with n-butanol/pyridine acetate, pH 4.2 (1:1 [vol/vol]), analyzed by TLC, and quantified by phosphorimaging.
In vitro lipid III synthesis reaction using purified TagO.
The enzymatic activity of the TagO-catalyzed lipid III (undecaprenol-PP-GlcNAc)-synthesis was determined using purified recombinant TagO protein incubated in the presence of 5 nmol C55-P, 50 nmol [14C]UDP-GlcNAc, 80 mM Tris-HCl, pH 8, 6 mM MgCl2, 1.25 mM CaCl2, 10% DMSO (vol/vol), and 10 mM N-lauroyl sarcosine in a final volume of 60 μl. Amphomycin and MX-2401 were added in molar ratios ranging from 0.25 to 2 with respect to the amount of C55-P. The reaction mixture was incubated for 90 min at 30°C. Analysis and quantification were carried out as described for MraY.
Membrane depolarization and cell viability assays.
Membrane depolarization was determined by a fluorescence assay based on the method described previously (36), with some modifications. An inoculum of ∼107 CFU/ml of S. epidermidis at exponential phase was incubated with MX-2401, daptomycin, or amphomycin at various concentrations in CAMHBc for 15 to 240 min. A membrane potential-sensitive dye, 3,3′-dipropylthiadicarbocyanine iodide [DiSC3(5); Invitrogen, CA] was added to the cells. To compare the membrane depolarization, the relative fluorescence change (in relative fluorescence units [RFU]) over 120 s was measured for each treatment group following addition of the dye and was normalized against its corresponding control. An aliquot of cells was removed from each sample before the addition of the fluorescent dye to determine cell viability/bactericidal activity by counting CFU.
Bacterial membrane permeability assay.
Changes in bacterial membrane permeability following antimicrobial treatment were analyzed using a modified LIVE/DEAD BacLight Bacterial Viability Kit (Invitrogen, CA) as described by Singh (37). Briefly, an exponential-phase S. epidermidis strain (ATCC 12228) was harvested by centrifugation and resuspended in 2% CAMHBc to an OD600 of 0.2 (cells with intact membranes). The cells with damaged membranes were generated by incubating a portion of this bacterial suspension with 70% propanol for 1 h and then resuspended in 2% CAMHBc to the same OD600. The intact and damaged cells were mixed to obtain cell suspensions containing five different ratios, i.e., 100:0, 75:25, 50:50, 25:75, and 0:100 (percent), of intact and damaged cells for generating a standard curve. For antibiotic treatment, the intact cells were incubated with MX-2401 or daptomycin at 35°C for 60 min with vigorous shaking. At the end of the drug treatment period, 100 μl of the treated cells or the standard samples was mixed with 100 μl of the 1× BacLight dye mixture (24 μl each of components A and B in 16 ml of deionized water) and incubated for 15 min in the dark. The fluorescence intensities of the samples and standards were measured using a SpectroMax M5 (Molecular Devices, CA) with an excitation wavelength of 485 nm and dual emissions at 535 and 615 nm for green (G) and red (R) fluorescence, respectively. The G/R fluorescence ratio, obtained by dividing the green and red fluorescence intensities, was plotted against cell suspensions with the known ratio of cells with intact or damaged membranes to generate a standard curve. The percentage of intact cells in the antibiotic-treated samples was calculated based on the standard curve.
Lipid flip-flop assay.
Lipid flip-flop experiments were done using unilamellar liposomes that were made up of an equimolar mixture of 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (sodium salt) (POPG) and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) lipids asymmetrically labeled with 0.5 mol% 1-palmitoyl-2-{6-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]hexanoyl}-sn-glycero-3-phosphocholine (C6-NBD-PC) lipids (Avanti Polar Lipids Inc.) as previously described (19, 49). Briefly, the liposomes were prepared by dissolving the lipid mixture in chloroform and drying it under a stream of nitrogen gas, followed by 2 h of vacuum drying, resulting in a thin lipid film. The lipid film was rehydrated with TSE buffer (10 mM Tris, 150 mM NaCl, 1 mM EDTA [disodium salt], pH 7.5) for 1 h on a shaker at 37°C and then freeze-thawed five times. Using a Lipex Extruder (Northern Lipids Inc.), the lipid solution was extruded 10 times through two stacked polycarbonate filters with a pore size of 100 nm, producing symmetrically labeled liposomes. To produce asymmetrically labeled liposomes, the lipid solution was incubated with 1 M sodium dithionite in 1 M Tris-HCl, pH 7.5 (26 mM lipids-38 mM Na2S2O4) for 15 min at room temperature, quenching the NBD groups in the outer leaflet of the liposome bilayers. The dithionite was separated out by filtration through a column (0.6 by 6 cm) containing Sephadex (G-50; fine), using TSE as the running buffer. On the fluorescence spectrometer (Perkin Elmer; catalog number LS50B), the emission wavelength was set to 530 nm and the excitation wavelength was set to 460 nm with a slit width of 8 nm. The baseline fluorescence for TSE containing asymmetrically labeled liposomes, at a lipid concentration of 180 μM, was established for approximately 100 s before the treatments were added, along with 5 mM CaCl2 and 10 mM Na2S2O4. The fluorescence was recorded 10 min after treatment. Triton X-100 (0.1%) and gramicidin S (GMS) were used as positive controls. The percent lipid flip-flop was calculated relative to 0.1% Triton X-100, which was set to 100% using the following equation: percent flip-flop = 100 × (F0 − FS)/(F0 − FT), where F0, FS, and FT represent the fluorescence intensity in asymmetrically labeled liposomes without treatment, with treatment, and with Triton X-100, respectively.
Calcein release.
Calcein release experiments were done using unilamellar liposomes that were made up of an equimolar mixture of POPG and POPC lipids entrapped with calcein as previously described (18, 49), similar to the method described for the lipid flip-flop assay. The lipid film was rehydrated in 5 mM HEPES, pH 7.4, containing 100 nM calcein. The lipid-calcein mixture was freeze-thawed and extruded as described above. The free calcein was removed by gel filtration through a column (0.6 by 6 cm) containing Sephadex (G-50; fine), using 20 mM HEPES, 150 mM NaCl, 1 mM EDTA (disodium salt), pH 7.4, as the running buffer. Calcein-free liposomes were also made in order to adjust the final liposome concentration for the calcein release assay. These liposomes were made the same way as the calcein-entrapped liposomes, except they were rehydrated in running buffer and there was no need to run them on a column. The calcein release assay was performed on the same fluorescence spectrometer as the lipid flip-flop assay with the emission wavelength set to 520 nm and the excitation wavelength set to 490 nm, with the slit width set to 2.5 nm. The baseline fluorescence for the running buffer containing 120 μM calcein-free liposomes and 60 μM calcein-entrapped liposomes was established for 100 s before the treatments were added, along with 5 mM CaCl2. The fluorescence was recorded 5 min after treatment; 0.1% Triton X-100 and GMS were used as positive controls. The percent calcein release was calculated relative to 0.1% Triton X-100, which was set to 100% using the following equation: percent calcein release = 100 × (FS − F0)/(FT − F0), where F0, FS, and FT represent the fluorescence intensity of the mixture of calcein-entrapped and calcein-free liposomes without treatment, with treatment, and with Triton X-100, respectively.
Membrane fusion.
POPC, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), and POPG were purchased from Avanti Polar Lipids (Alabaster, AL). Liposomes were prepared by the extrusion method using a small-volume extrusion apparatus as previously described (18). In summary, lipids were dissolved in CHCl3. The CHCl3 was evaporated off with N2, and the lipids were further dried under vacuum overnight. The dried lipid films were resuspended with 20 mM HEPES buffer, pH 7.4, and freeze-thawed five times. The resulting lipid suspensions were extruded 10 times through two 0.1-mm filters (AMD Manufacturing Inc., Mississauga, ON, Canada).
The size of a population of unilamellar liposomes consisting of POPC, POPC-POPG, or POPE-POPG with a molar ratio of 1:1 was determined using a Beckman Coulter (Mississauga, ON, Canada) N4 Plus photon correlation spectrometer. Liposome samples containing either 5 mM CaCl2 alone or with added daptomycin or MX-2401 were incubated at 23°C for 15 min prior to the measurement. The concentration of daptomycin or MX-2401 was 8 μg/ml. The measurements were made at 23°C using a 600-nm laser at a 90° angle. The resulting data were analyzed using a unimodal distribution curve.
NMR structure of MX-2401 in SDS.
MX-2401 (1 mM) in 100 mM deuterated SDS-d25 was prepared in 500 μl 100 mM phosphate buffer, 7% D2O, pH 7.8 (adjusted with a pH meter). Two-dimensional total correlation spectroscopy (TOCSY) (4) and nuclear Overhauser effect spectroscopy (NOESY) (17) 1H nuclear magnetic resonance (NMR) spectra were recorded at 298 K on a 700-MHz Bruker Avance spectrometer (operating at a proton frequency of 700.13 MHz), with a TXI SB 5-mm probe with z gradients. In phase-sensitive mode, time-proportional phase incrementation (TPPI) (22) in the indirect dimension was applied. The TOCSY experiment used the DIPSI2 sequence for mixing (mixing time = 70 ms) and excitation sculpting with gradients for water suppression (16). The two-dimensional (2D) data set consisted of 2,048 data points in t2 and 512 points in t1 (where t1 and t2 are the time intervals before and after mixing) and was processed to yield a 1,000 by 1,000 matrix. The NOESY experiment was acquired with a mixing time of 150 ms and also used excitation sculpting for water suppression. The data size for this data set was the same as for the TOCSY spectrum. The spectra were assigned using the Bruker software TOPSPIN. Volumes were extracted using the same software and calibrated to the distance between the two protons in the aromatic linker joining Asp1 to the acyl chain (2.46 Å).
The GROMOS96 biomolecular simulation package and the 43A1 force field were used (33, 44). All simulations were performed in explicit methanol under rectangular periodic boundary conditions in the normal pressure, P, and temperature, T (NPT) ensemble (T = 300 K; P = 1 atm) imposed by the Berendsen weak-coupling methods (2). Covalent bonds were constrained using the SHAKE method (27) with a relative geometric tolerance of 10−4. A reaction field long-range correction (42) to the truncated Coulomb potential was applied. For the initial coordinates, residues were attached to one another to form an extended chain in CHIMERA (26), which was then minimized within GROMOS in a vacuum to obtain a cyclic peptide. This structure was solvated and minimized in methanol, after which the system was rendered neutral by the addition of 3 Na+ ions. After a second minimization step, initial atomic velocities were generated from a Maxwell-Boltzmann distribution at 300 K, and the system was equilibrated for 5 ns without positional restraints.
A refinement simulation 26 ns in length was performed, starting from this equilibrated system, without any positional restraints for data collection. The experimentally determined NOE distance restraint data were introduced into the simulation using a time-averaged potential energy function (43) with a relaxation time (τdr) of 10 ps and a force constant (Kdr) of 5 kJ · mol−1 · nm−2 (24).
RESULTS
Intracellular accumulation of UDP-N-acetylmuramyl pentapeptide in S. simulans 22.
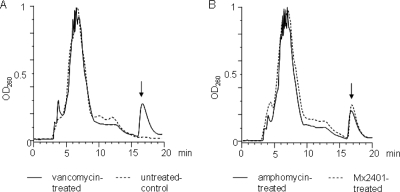
Antibiotics, such as vancomycin and β-lactams, that interfere with the late membrane-bound stages of peptidoglycan biosynthesis trigger the accumulation of the final soluble peptidoglycan precursor UDP-MurNAc-pp in the cytoplasm of growing bacterial cells (21). We therefore treated S. simulans 22 with 10× MIC of amphomycin and MX-2401 and found both lipopeptides caused the accumulation of UDP-MurNAc-pp (Fig. 2B) to an extent comparable to that with the vancomycin-treated positive control (Fig. 2A). The results indicated that (i) UDP-MurNAc-pentapeptide, the final soluble cell wall precursor, was correctly formed in the presence of the lipopeptides and (ii) one of the subsequent membrane-associated steps of cell wall biosynthesis was blocked. The accumulation of the soluble cell wall precursor also indicated that neither peptide impaired or depolarized the cytoplasmic membrane, since the precursor was retained in the cytoplasm and did not leak from treated cells within the 45 min of treatment. The results also indicated the increased potency of MX-2401 over amphomycin, since similar levels of precursor accumulation were achieved with 5 μg/ml and 20 μg/ml, respectively.
Fig. 2.
Intracellular accumulation of UDP-N-acetylmuramyl pentapeptide in untreated and vancomycin-treated (A) and amphomycin-treated and MX-2401-treated (B) S. simulans 22 cells. The experiment was performed with 10× MIC of the respective antibiotic compounds for 45 min. Subsequently, cells were extracted with boiling water, and the intracellular nucleotide pool was analyzed by reverse-phase HPLC. The identity of UDP-MurNAc-pentapeptide was confirmed by MALDI-TOF mass spectrometry, yielding a molecular mass of 1,148.72 Da for the indicated peaks (arrows).
Lipid II synthesis by cytoplasmic membrane preparations.
The late membrane-associated peptidoglycan biosynthesis steps leading to the formation of lipid II and the subsequent incorporation of this central cell wall building block into the growing peptidoglycan network involve a series of enzymatic reactions (6). The phospho-MurNAc-pentapeptide translocase MraY catalyzes the first lipid-linked step, which transfers the soluble cell wall precursor UDP-MurNAc-pentapeptide to the bactoprenol carrier (C55-P), yielding lipid I. MurG subsequently adds UDP-activated N-acetyl-glucosamine (UDP-GlcNAc) to form lipid II (undecaprenylphosphate-GlcNAc-MurNAc-pentapeptide).
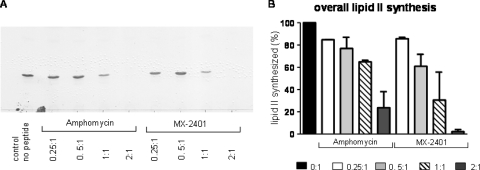
Cytoplasmic membrane preparations of M. luteus contain sufficient MraY and MurG activity for the synthesis of lipid I and lipid II in vitro (28, 29). In this test system, using isolated cytoplasmic membranes supplemented with defined amounts of C55-P and UDP-activated sugars, the addition of amphomycin and MX-2401 inhibited the synthesis of lipid II compared to the untreated negative control, in which an almost complete conversion of bactoprenol-phosphate to lipid II was achieved (Fig. 3). Increasing concentrations of amphomycin and MX-2401 led to enhanced inhibition of lipid II synthesis, as demonstrated by quantitative analysis using radiolabeled [14C]UDP-GlcNAc (Fig. 3). To achieve complete inhibition of the synthesis, antibiotics had to be added in at least 2-fold molar excess over C55-P (Fig. 3), suggesting that both compounds stoichiometrically bind to the substrate C55-P, similar to what has been shown for friulimicin (28), rather than inhibiting the enzyme.
Fig. 3.
Impacts of amphomycin and MX-2401 on the overall in vitro lipid II synthesis by cytoplasmic membrane preparations of M. luteus. (A) The synthesis of lipid II in the presence of increasing concentrations of amphomycin and MX2401 was qualitatively analyzed using TLC and PMA staining. (B) Quantitative analysis was carried out using [14C]UDP-GlcNAc as described in Materials and Methods. Peptides were added at molar ratios of 0.25 to 2 with respect to the amount of the substrate C55-P. The reaction product of the untreated control was taken as 100%. The error bars indicate standard deviations.
Bactoprenol-phosphate-consuming reactions.
The impacts of amphomycin and MX-2401 on the individual bactoprenol-phosphate-consuming reactions were investigated in more detail. To this end, we set up in vitro assays using purified recombinant proteins (MraY and TagO) and radiolabeled substrates, i.e., UDP-MurNAc-(14C-Lys)-pentapeptide and [14C]UDP-GlcNAc, respectively. UDP-MurNAc-pentapeptide is synthesized by a cascade of ligase reactions, which catalyze the assembly of the stem peptide moiety by sequential addition of d- and l-amino acids (45). The reconstitution of this cytoplasmic pathway using purified S. aureus MurA-MurF and DdlA (d-Ala–d-Ala) proteins and radiolabeled amino acids (i.e., 14C-Lys) allowed the quantification of the MraY synthesis product lipid I in vitro.
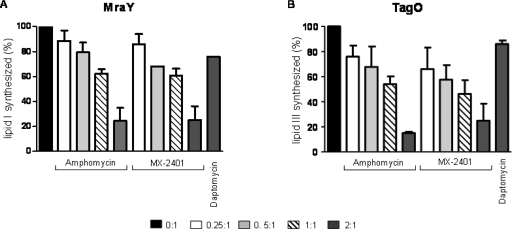
In the presence of C55-P and radiolabeled UDP-MurNAc-pentapeptide, MraY-His6 synthesized 14C-lipid I (Fig. 4A, lane 1). Amphomycin and MX-2401 inhibited the synthesis of lipid I in a dose-dependent fashion (Fig. 4A) analogous to what was observed in the overall lipid II synthesis assay (Fig. 3). When peptides were added in equimolar concentrations, the formation of lipid I was inhibited by about 60%, while almost complete inhibition was achieved at a 2:1 (peptide/C55-P) molar ratio compared to the control, where no antibiotic was added (Fig. 4A, lanes 5 and 9).
Fig. 4.
Impacts of amphomycin and MX-2401 on bactoprenol-phosphate-consuming reactions catalyzed by MraY (A) and TagO (B). The incorporation of [14C]UDP-MurNAc-pentapeptide into lipid I (A) and [14C]UDP-GlcNAc into lipid III (B) was analyzed in the presence of increasing peptide concentrations. Peptides were added in molar ratios of 0.25 to 2 with respect to the substrate C55-P. Daptomycin was added in 2-fold molar excess with respect to C55-P. Quantification was carried out using phosphorimaging. The error bars indicate standard deviations.
C55-P serves as a universal carbohydrate carrier involved in several biosynthetic pathways. To address the effect on other C55-P-consuming biosynthesis pathways, we investigated the impacts of the lipopeptides on the first lipid-linked step of wall teichoic acid biosynthesis catalyzed by TagO (phospho-GlcNAc-translocase). As shown in Fig. 4B, lipid III (bactoprenol-PP-GlcNAc) was synthesized by S. aureus TagO-His6 in the presence of 14C-UDP-GlcNAc and C55-P. The formation of lipid III was clearly inhibited in a concentration-dependent manner by the addition of amphomycin and MX-2401 (Fig. 4B). When both lipopeptides were added at a molar ratio of 2:1 with respect to the precursor C55-P, only about 20% of the total amount of lipid III was synthesized, confirming that the mode of action of amphomycin and MX-2401 relied on complex formation with C55-P.
Daptomycin, in contrast, showed only a minor effect on both lipid I and lipid III synthesis in vitro when added in 2-fold molar excess over the substrate, C55-P (Fig. 4).
Bacterial membrane depolarization and permeabilization.
The antimicrobial mechanism of daptomycin has been proposed to be direct insertion into bacterial membranes, which results in rapid loss of membrane potential, cessation of macromolecule synthesis, and cell death (19, 35, 36). To determine if MX-2401 shares this mechanism of action with daptomycin, we evaluated the effect of MX-2401 on depolarization and permeabilization of the bacterial membrane of S. epidermidis and determined if those effects correlated with the bactericidal activity of MX-2401.
To establish appropriate concentration ranges to be tested in the membrane depolarization assay, the MICs were first determined. In the CAMHBc medium, MX-2401 had an MIC of 2 μg/ml, while daptomycin and amphomycin had MICs of 0.5 μg/ml and 4 μg/ml, respectively. To determine the effect of MX-2401 on bacterial membrane potential, we adapted the DiSC3(5) assay (36) to exponential-growth-phase S. epidermidis. In this assay, the degree of membrane depolarization is estimated by the kinetics of the fluorescence increase as the dye is released from the cell and dequenches.
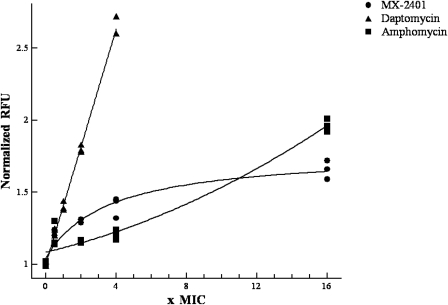
The depolarization of S. epidermidis was examined following incubation with MX-2401, daptomycin, or amphomycin for various periods. The results indicated that all compounds caused concentration-dependent depolarization; however, daptomycin was significantly more efficient: only 1 μg/ml (2× MIC) was needed to depolarize the cells to the same extent as 32 μg/ml MX-2401 (16× MIC) and 64 μg/ml of amphomycin (16× MIC) (Fig. 5). The results observed with daptomycin were consistent with previous studies (36). Depolarization induced by both MX-2401 and daptomycin appeared to be time dependent; however, MX-2401 induced depolarization at a significantly lower rate than daptomycin. For example, 8 μg/ml of daptomycin (16× MIC) effectively depolarized S. epidermidis following 15 min of incubation (data not shown). In contrast, prolonged exposure (>30 min) to MX-2401 was required to reduce the bacterial membrane potential at 16× MIC, the highest concentration tested (data not shown).
Fig. 5.
Dose-dependent membrane depolarization by MX-2401, daptomycin, and amphomycin. Cells were incubated with antibiotics for 60 min before depolarization was measured.
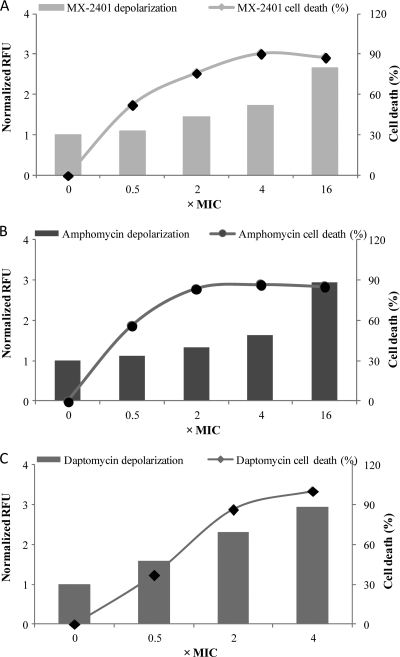
In light of these findings that MX-2401 can gradually depolarize cell membranes, we investigated whether the membrane depolarization was associated with the bactericidal activity of MX-2401. Therefore, we quantified bacterial viability in parallel with the membrane potential change. When S. epidermidis cells were incubated with 2 to 16 μg/ml of MX-2401 for 90 min, their membrane potential was reduced, as demonstrated by the increase of fluorescent dye release (Fig. 6A). However, there was no correlation between the changes in the membrane potential and cell death. For example, MX-2401 at 1 μg/ml did not cause significant changes in the membrane potential compared to the control level; however, more than 50% cell death was observed at this concentration. At higher concentrations (8 and 32 μg/ml), the bactericidal effect had already reached a maximum level (∼90% death) while the membrane permeabilization continued to increase. These findings suggest that membrane depolarization is not the mechanism by which MX-2401 executes its bactericidal activity. Similarly, the bactericidal activity of amphomycin was not associated with its depolarization effect (Fig. 6B). In contrast, in cells treated with daptomycin, cell viability decreased in parallel with the reductions in membrane potential (Fig. 6C).
Fig. 6.
Membrane depolarization and cell death in S. epidermidis following incubation with sub- and supra-MIC concentrations of antibiotics in the presence or absence of surfactant. (A) Cells incubated with MX-2401 for 90 min. (B) Cells incubated with amphomycin for 90 min. (C) Cells incubated with daptomycin for 60 min.
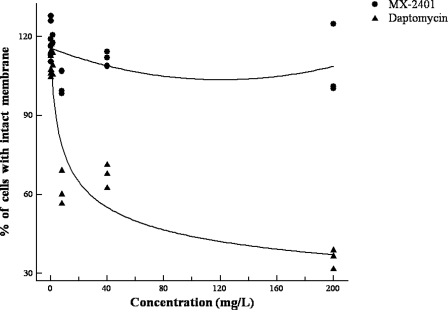
To further explore the effect of MX-2401 on bacterial membrane integrity, we examined bacterial membrane permeability following MX-2401 exposure, using a BacLight Bacterial Viability Kit. This method enables differentiation between cells with intact membranes and cells with damaged membranes based on their cytoplasmic membrane permeability to propidium iodide (3). Bacterial killing by antibiotics, such as vancomycin and penicillin, that do not permeabilize bacterial membranes cannot be detected by this method (29). In the cells incubated with MX-2401 for 60 min, no membrane damage was observed with MX-2401 concentrations up to 200 μg/ml (100× MIC), indicating that MX-2401 had no significant effect on membrane permeability (Fig. 7). In contrast, daptomycin caused concentration-dependent permeabilization of the cells, which was in line with its proposed mechanism of action (36).
Fig. 7.
Effects of MX-2401 and daptomycin on bacterial membrane permeability.
Model membrane studies.
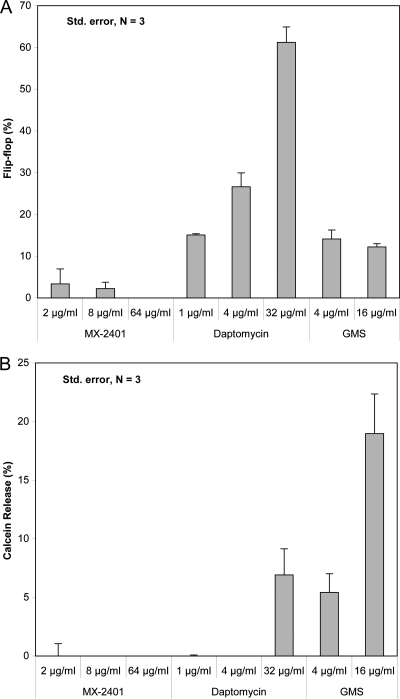
To further assess the potential mechanism of action of MX-2401, we utilized a series of assays that were previously used to demonstrate the intrinsic activity of daptomycin on lipid membranes. The effects of MX-2401 on lipid flip-flop and calcein release were determined in unilamellar liposomes. Lipid flip-flop (the movement of lipid molecules between the outer and inner monolayers of lipid bilayers) occurs at very low frequency in unperturbed liposomes but is a highly sensitive indicator of membrane perturbation with cationic peptides and daptomycin. It was monitored with a fluorescent lipid probe, C6-NBD-PC, using asymmetrically labeled POPC/POPG liposomes treated with MX-2401, daptomycin, and GMS in the presence of 5 mM CaCl2 (Fig. 8A). The results indicated that MX-2401 had no effect on lipid flip-flop at all concentrations tested, including those significantly exceeding its MIC levels. In contrast, daptomycin induced substantial lipid flip-flop in a concentration-dependent manner, in agreement with data reported earlier (19). A positive control, the cationic peptide gramicidin S, also induced lipid flip-flop.
Fig. 8.
Ability of MX-2401 to interact with model membranes. (A) Dose-dependent lipid flip-flop of C6-NBD-PC asymmetrically labeled POPC/POPG liposomes treated with MX-2401, daptomycin, and GMS in the presence of 5 mM CaCl2. (B) Dose-dependent calcein release from POPC/POPG liposomes treated with MX-2401, daptomycin, and GMS in the presence of 5 mM CaCl2. The error bars indicate standard deviations.
We also determined whether MX-2401 has the ability to induce model membrane leakage by measuring the release of an encapsulated dye, calcein, from unilamellar liposomes made up of an equimolar mixture of POPG and POPC lipids. No calcein release was observed in the presence of MX-2401, while calcein leakage was induced by a high daptomycin concentration (Fig. 8B).
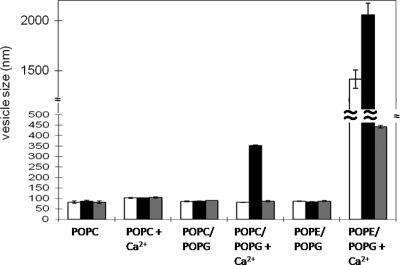
Daptomycin can cause fusion of lipids by entering membranes and bridging between adjacent liposomes to cause membrane fusion (18). Control experiments were conducted on the appropriate lipid mixture in buffer only. The results show an increase in vesicle size only when daptomycin was mixed with POPC, POPG, and Ca2+, as previously reported (18). MX-2401 did not cause membrane fusion under these conditions. Indeed MX-2401 appeared to have the reverse effect by reducing the modest Ca2+-dependent fusion in POPE/POPG plus Ca2+ vesicles, which tend to aggregate on their own when calcium is added (Fig. 9).
Fig. 9.
Effect of MX-2401 and daptomycin on membrane fusion as determined by light scattering. Daptomycin (black bars) induces fusion in the presence of POPC, POPG, and Ca2+ as observed by the increase in vesicle size, whereas MX-2401 (white bars) does not, similarly to the control (gray bars; lipid only). The error bars represent the error associated with three repeats.
NMR structure.
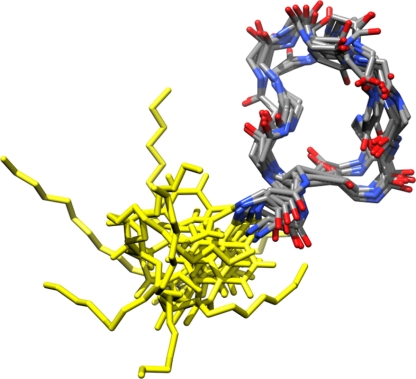
In order to further understand the mechanism of action of MX-2401, its three-dimensional structure was determined. One hundred seventy-nine NOE-derived restraints were applied to the starting structure. Over the course of the refinement, 29% of the NOEs were violated, which is comparable to what was found for daptomycin (32). Additional structural statistics are summarized in Table 2. From the refinement simulations, 2,600 configurations chosen at 10-ps time intervals were selected and subjected to cluster analysis, as previously described (34). The eight representative structures obtained from the clustering analysis are shown in Fig. 10. The backbone trace of the cyclic moiety shows two distinct conformations, which are well defined.
Table 2.
Statistical analysis of NMR-derived structures of MX-2401 in SDS micelles
| Parameter | Value |
|---|---|
| No. of NOE restraints | 179 |
| No. of intraresidue restraints | 90 |
| No. of interresidue restraints | 89 |
| Total no. of NOE restraints violateda | 52 |
| Total % NOE violations | 29.1 |
| Total no. of intraresidue restraints violated | 10 |
| Total no. of interresidue restraints violated | 42 |
| Avg RMSDb (Cα − residuesc) | 1.0 ± 0.5 Å |
| Avg no. of relative NOE violations | 0.081 |
A restraint is considered to be violated if the relative average violation is larger than 0.1. The average is over the 26 ns of simulation trajectory (the first 5 ns of equilibration were not used for the analysis).
RMSD, root mean square deviation, calculated by comparing the RMSD values between the representative structures shown in Fig. 10.
Cα − residues are the alpha carbon residues.
Fig. 10.
Representative structures of MX-2401 as obtained from the refinement simulations. The backbone trace on the right represents residues Asp1 → Pro11 (carbon, gray; nitrogen, blue; and oxygen, red), while the traces on the left (yellow) represent the acyl chain and linker connected to the N-terminal residue Asp1. The cyclic part is well defined (relative to daptomycin) and appears to adopt two major conformations. The image was created using UCSF CHIMERA (26).
Serial passaging and cross-resistance study.
A cross-resistance study was conducted with MX-2401 and daptomycin-resistant S. aureus and E. faecalis to evaluate the potential for resistance and possible mechanistic overlaps with other classes of antibiotics. Cross-resistance is expected to be observed if agents share some aspects of their mechanisms of action and the resulting resistance arises through a common genetic mutation. No mutants were obtained by plating 1010 S. aureus cells on selective medium. Serial passaging of S. aureus and E. faecalis with MX-2401 and daptomycin showed that resistance developed in an unstable manner, slowly peaking after 27 passages, and only 2- to 8-fold changes in the MIC were determined after 27 passages. The results of the cross-resistance study are presented in Table 3, and in only one circumstance was cross-resistance observed between the two lipopeptide antibiotics, whereby the MX-2401-trained mutant in S. aureus had a 4-fold increase in the daptomycin MIC. Daptomycin led to no other cross-resistances. With respect to other antibiotics, MX-2401 led to only one other significant cross-resistance, namely, to trimethoprim in S. aureus. On the other hand, MX-2401 training led to significantly increased susceptibility to ceftriaxone in E. faecalis and to gentamicin and ampicillin in S. aureus, while daptomycin training led only to enhanced susceptibility to tetracycline in E. faecalis. Such very distinct cross-resistance/susceptibility patterns are consistent with the above data that indicate that daptomycin and MX-2401 work through different mechanisms and that they are discrete from the mechanisms of other antibiotics.
Table 3.
Cross-resistance of MX-2401 and daptomycin-resistant mutants of S. aureus and E. faecalis to various antibiotics
| Bacteriuma | Cross-resistant strain | MIC (μg/ml) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Vancomycin | Gentamicin | Trimethoprim | Ampicillin | Ceftriaxone | Ciprofloxacin | Erythromycin | Tetracycline | MX-2401 | Daptomycin | ||
| E. faecalis ATCC 29212 | Wild-type | 2 | 16 | 0.25 | 1 | 256 | 1 | 4 | 32 | 4 | 4 |
| MX-2401 mutant | 1 | 8 | 0.25 | 1 | 64 | 1 | 4 | 16 | 16 | 4 | |
| Daptomycin mutant | 1 | 16 | 0.25 | 1 | 256 | 0.5 | 2 | 8 | 8 | 8 | |
| S. aureus ATCC 29213 | Wild-type | 1 | 1 | 4 | 2 | 4 | 0.5 | 0.5 | 0.5 | 2 | 1 |
| MX-2401 mutant | 2 | 0.25 | 16 | 0.125 | 2 | 0.25 | 0.5 | 0.5 | 16 | 4 | |
| Daptomycin mutant | 1 | 1 | 2 | 2 | 2 | 0.5 | 0.25 | 0.5 | 4 | 4 | |
The bacteria were taken from passage 27 grown in 2 and 4 μg/ml daptomycin for S. aureus and E. faecalis, respectively, and in 8 μg/ml MX-2401 for both organisms. The MICs are the modes of five biological replicates.
DISCUSSION
The emergence of resistant organisms in both hospital and community settings continues to escalate, with pathogen resistance to more than one class of agents becoming increasingly common and with resistance to recently introduced antibiotics emerging rapidly. Therefore, there is a continuous need for the development of novel antibiotics to treat infections caused by Gram-positive pathogens.
Lipopeptides have proven to be a promising class of novel antibacterial agents, with the first drug, daptomycin, already approved for the treatment of skin and skin structure infections, endocarditis, and bacteremia. The mode of action of daptomycin is not yet fully understood, but it is believed that daptomycin interacts with the bacterial membrane and causes rapid depolarization in a manner similar to that of the cationic peptides (15, 18, 32, 36, 38). Functional and structural studies suggest that this process is Ca2+ dependent, with Ca2+ and daptomycin interacting to form micelles that may help to deliver daptomycin at high concentrations to bacterial membranes (14). Recent work, which made use of fluorescently tagged daptomycin, has challenged the view that daptomycin forms oligomers in the presence of Ca2+ when the former is at concentrations close to the MIC (23). Nevertheless, both proposed mechanisms (14, 23, 38) indicate that daptomycin inserts into the membrane through a process that is also potentiated by Ca2+ through bridging between daptomycin and phosphatidylglycerol in the bacterial membranes. Downstream events result in membrane depolarization, leakage, and rapid cell death.
The research reported here indicates that although both daptomycin and MX-2401 are in the structural class of Ca2+-dependent lipopeptide antibiotics, the latter has a different mechanism of action. Specifically, MX-2401 and its parent compound, amphomycin, inhibit peptidoglycan synthesis by binding to the substrate C55-P, the universal carbohydrate carrier involved in several biosynthetic pathways. This interaction results in inhibition, in a dose-dependent manner, of the biosynthesis of the cell wall precursors lipids I and II and the wall teichoic acid precursor lipid III, while daptomycin had no significant effect on these processes. These results are in agreement with our earlier studies, in which daptomycin was shown not to promote precursor accumulation (28), pointing toward fundamental differences in the modes of action of these lipopeptides. MX-2401 showed optimal inhibition at a 2:1 (MX-2401/C55-P) molar ratio. This 2:1 stoichiometry of binding suggests dimerization of the lipopeptide, as has been described for the structurally related tsushimycin (7). As seen from the NMR structure, the relative rigidity of MX-2401 compared to daptomycin most likely favors interaction with C55-P, which has many unsaturated carbons. Rigidity is probably important for favorable packing between MX-2401 and C55-P (50).
With respect to membrane depolarization, MX-2401 induced only very slow membrane depolarization that was observed at concentrations significantly above MIC levels. Unlike daptomycin, membrane depolarization by MX-2401 did not correlate with its bactericidal activity and did not affect general membrane permeability. The delay between bactericidal activity and the start of membrane depolarization indicates that the limited observed depolarization may be a consequence, rather than the cause, of cell death. This is further supported by the data showing that, in contrast to daptomycin, MX-2401 had no effect on lipid flip-flop, calcein release, or membrane fusion with POPC/POPG liposomes in the presence of Ca2+. Overall, these results provided strong evidence that the antimicrobial effect of MX-2401 is not associated with any effects on the bacterial membrane potential and that MX-2401 had a mechanism different from that of daptomycin.
Perhaps as a consequence of its mechanism of action, which reflects its ability to directly insert into the bacterial cytoplasmic membrane, daptomycin is inactivated by pulmonary surfactant. In mammalian species, pulmonary surfactant consists of ∼80% dipalmitoylphosphatidylcholine and ∼10% phosphatidylglycerol; the latter is also a prominent component of the bacterial cytoplasmic membrane (35). In the presence of surfactant, daptomycin demonstrates Ca2+-dependent and concentration-dependent insertion into the surfactant lipid that results in loss of its ability to depolarize and/or permeabilize the bacterial membrane. Due to inhibition by the surfactant, daptomycin failed in clinical trials for the treatment of pneumonia (35). In contrast to the lipopeptide daptomycin, the activity of MX-2401 was not inhibited in the presence of lung surfactant, and the drug shows promising activity in a bronchial-alveolar pneumonia model in which daptomycin was not active (13, 35). This advantage in pharmacological activity is a direct result of the different mechanism of action of MX-2401.
Another positive implication of the current studies is the very low cross-resistance to other antibiotics of mutants resistant to MX-2401 and the observation of increased susceptibility to certain antibiotics among the trained mutants. This provides strong hope that MX-2401 will not suffer from rapid resistance development and consequent lack of therapeutic options, analogous to the situation with daptomycin. When combined with other characteristics, such as broad-spectrum bactericidal activity against Gram-positive organisms (including resistant strains) that have been demonstrated in both in vitro and in vivo studies (9, 12, 15) and a beneficial pharmacokinetic profile (25), our present results showing its mechanism of action and low potential for serious resistance development suggest that the compound may fulfill a serious medical need for the treatment of life-threatening infections by Gram-positive bacteria.
ACKNOWLEDGMENTS
We acknowledge Ray Sui and Jeremy Finn for their help with the experiments. S.K.S. thanks Paulo Sgarbi for useful discussions.
S.K.S. acknowledges funding from NSERC (Discovery grant) and the Michael Smith Foundation for Health Research (Career Investigator Scholar). H.G.S. thanks the German Research Foundation for funding (DFG Sa 292/13-1) and the BONFOR program of the Medical Faculty, University of Bonn, Bonn, Germany, for general support. R.E.W.H. holds a Canada Research Chair and acknowledges additional support from the Canadian Institutes for Health Research.
Footnotes
Published ahead of print on 4 April 2011.
REFERENCES
- 1. Baltz R. H., Miao V., Wrigley S. K. 2005. Natural products to drugs: daptomycin and related lipopeptide antibiotics. Nat. Prod. Rep. 22:717–741 [DOI] [PubMed] [Google Scholar]
- 2. Berendsen H. J. C., Postma J. P. M., van Gunsteren W. F., Dinola A., Haak J. R. 1984. Molecular-dynamics with coupling to an external bath. J. Chem. Phys. 81:3684–3690 [Google Scholar]
- 3. Berney M., Hammes F., Bosshard F., Weilenmann H. U., Egli T. 2007. Assessment and interpretation of bacterial viability by using the LIVE/DEAD BacLight Kit in combination with flow cytometry. Appl. Environ. Microbiol. 73:3283–3290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Braunschweiler L., Ernst R. R. 1983. Coherence transfer by isotropic mixing: application to proton correlation spectroscopy. J. Magn. Reson. 53:521–528 [Google Scholar]
- 5. Brötz H., et al. 1998. Role of lipid-bound peptidoglycan precursors in the formation of pores by nisin, epidermin and other lantibiotics. Mol. Microbiol. 30:317–327 [DOI] [PubMed] [Google Scholar]
- 6. Brown S., Zhang Y. H., Walker S. 2008. Revised pathway proposed for Staphylococcus aureus wall teichoic acid biosynthesis based on in vitro reconstitution of the intracellular steps. Chem. Biol. 15:12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bunkóczi G., Vertesy L., Sheldrick G. M. 2005. Structure of the lipopeptide antibiotic tsushimycin. Acta Crystallogr. D Biol. Crystallogr. 61:1160–1164 [DOI] [PubMed] [Google Scholar]
- 8. Clinical and Laboratory Standards Institute 2007. Methods for antimicrobial susceptibility testing of anaerobic bacteria; approved standard, 7th ed CLSI document M11-A7. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 9. Craig W. A., Andes D., Satmstad T. 2010. In vivo pharmacodynamics of a new lipopeptide MX-2401. Antimicrob. Agents Chemother. 54:5092–5098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dai D., Ishiguro E. E. 1988. murH, a new genetic locus in Escherichia coli involved in cell-wall peptidoglycan biosynthesis. J. Bacteriol. 170:2197–2201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Debono M., et al. 1988. Enzymatic and chemical modifications of lipopeptide antibiotic A21978C: the synthesis and evaluation of daptomycin (LY146032). J. Antibiot. 41:1093–1105 [DOI] [PubMed] [Google Scholar]
- 12. Dugourd D., et al. 2006. In vitro characterization of MX-2401—a novel amphomycin derivative active against gram positive bacteria, abstr. F1-1879. Abstr. 46th Intersci. Conf. Antimicrob. Agents Chemother [Google Scholar]
- 13. Dugourd D., Yang H., Rubinchik E. 2009. MX-2401: a novel lipopeptide active in the presence of lung surfactant and in Streptococcus pneumoniae bronchial-alveolar pneumonia model, abstr. F1-2026. Abstr. 49th Intersci. Conf. Antimicrob. Agents Chemother [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ho S. W., et al. 2008. Effect of divalent cations on the structure of the antibiotic daptomycin. Eur. Biophys. J. 37:421–433 [DOI] [PubMed] [Google Scholar]
- 15. Hoban D. J., Weshnoweski B., Vashisht R., Zhanel G. G., Dugourd D. In vitro activity of MX-2401, a novel lipopeptide against multi-drug resistant (MDR) Staphylococcus aureus (SA), abstr. F1-363. Abstr. 48th Intersci. Conf. Antimicrob. Agents Chemother [Google Scholar]
- 16. Hwang T. L., Shaka A. J. 1995. Water suppression that works—excitation sculpting using arbitrary wave-forms and pulsed-field gradients. J. Magn. Reson. Ser. A 112:275–279 [Google Scholar]
- 17. Jeener J., Meier B. H., Bachmann P., Ernst R. R. 1979. Investigation of exchange processes by two-dimensional NMR spectroscopy. J. Chem. Phys. 71:4546–4553 [Google Scholar]
- 18. Jung D., Powers J. P., Straus S. K., Hancock R. E. W. 2008. Lipid-specific binding of the calcium-dependent antibiotic daptomycin leads to changes in lipid polymorphism of model membranes. Chem. Phys. Lipids 154:120–128 [DOI] [PubMed] [Google Scholar]
- 19. Jung D., Rozek A., Okon M., Hancock R. E. W. 2004. Structural transitions as determinants of the action of the calcium-dependent antibiotic daptomycin. Chem. Biol. 11:949–957 [DOI] [PubMed] [Google Scholar]
- 20. Kang M. S., Spencer J. P., Elbein A. D. 1978. Amphomycin inhibition of mannose and GlcNAc incorporation into lipid-linked saccharides. J. Biol. Chem. 253:8860–8866 [PubMed] [Google Scholar]
- 21. Kohlrausch U., Holtje J. V. 1991. Analysis of murein and murein precursors during antibiotic-induced lysis of Escherichia coli. J. Bacteriol. 173:3425–3431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Marion D., Wuthrich K. 1983. Application of phase sensitive two-dimensional correlated spectroscopy (COSY) for measurements of 1H-1H spin-spin coupling constants in proteins. Biochem. Biophys. Res. Commun. 113:967–974 [DOI] [PubMed] [Google Scholar]
- 23. Muraih J. K., Pearson A., Silverman J., Palmer M. 2011. Oligomerization of daptomycin on membranes. Biochim. Biophys. Acta 1808:1154–1160 [DOI] [PubMed] [Google Scholar]
- 24. Nanzer A. P., van Gunsteren W. F., Torda A. E. 1995. Parametrization of time-averaged distance restraints in Md simulations. J. Biomol. NMR 6:313–320 [DOI] [PubMed] [Google Scholar]
- 25. Pasetka C. J., Erfle D. J., Cameron D. R., Clement J. J., Rubinchik E. 2010. Novel antimicrobial lipopeptides with long in vivo half-lives. Int. J. Antimicrob. Agents 35:182–185 [DOI] [PubMed] [Google Scholar]
- 26. Pettersen E. F., et al. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612 [DOI] [PubMed] [Google Scholar]
- 27. Ryckaert J. P., Ciccotti G., Berendsen H. J. C. 1977. Numerical-integration of Cartesian equations of motion of a system with constraints—molecular-dynamics of N-alkanes. J. Comput. Phys. 23:327–341 [Google Scholar]
- 28. Schneider T., et al. 2009. The lipopeptide antibiotic Friulimicin B inhibits cell wall biosynthesis through complex formation with bactoprenol phosphate. Antimicrob. Agents Chemother. 53:1610–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schneider T., et al. 2010. Plectasin, a fungal defensin, targets the bacterial cell wall precursor Lipid II. Science 328:1168–1172 [DOI] [PubMed] [Google Scholar]
- 30. Schneider T., et al. 2004. In vitro assembly of a complete, pentaglycine interpeptide bridge containing cell wall precursor (lipid II-Gly5) of Staphylococcus aureus. Mol. Microbiol. 53:675–685 [DOI] [PubMed] [Google Scholar]
- 31. Schneider T., Sahl H. G. 2010. An oldie but a goodie—cell wall biosynthesis as antibiotic target pathway. Int. J. Med. Microbiol. 300:161–169 [DOI] [PubMed] [Google Scholar]
- 32. Scott W. R. P., Baek S. B., Jung D., Hancock R. E. W., Straus S. K. 2007. NMR structural studies of the antibiotic lipopeptide daptomycin in DHPC micelles. Biochim. Biophys. Acta 1768:3116–3126 [DOI] [PubMed] [Google Scholar]
- 33. Scott W. R. P., et al. 1999. The GROMOS biomolecular simulation program package. J. Phys. Chem. A 103:3596–3607 [Google Scholar]
- 34. Scott W. R., et al. 2006. Characterization of de novo four-helix bundles by molecular dynamics simulations. Proteins 64:719–729 [DOI] [PubMed] [Google Scholar]
- 35. Silverman J. A., Mortin L. I., VanPraagh A. D. G., Li T., Alder J. 2005. Inhibition of daptomycin by pulmonary surfactant: in vitro modeling and clinical impact. J. Infect. Dis. 191:2149–2152 [DOI] [PubMed] [Google Scholar]
- 36. Silverman J. A., Perlmutter N. G., Shapiro H. M. 2003. Correlation of daptomycin bactericidal activity and membrane depolarization in Staphylococcus aureus. Antimicrob. Agents Chemother. 47:2538–2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Singh M. P. 2006. Rapid test for distinguishing membrane-active antibacterial agents. J. Microbiol. Methods 67:125–130 [DOI] [PubMed] [Google Scholar]
- 38. Straus S. K., Hancock R. E. W. 2006. Mode of action of the new antibiotic for Gram-positive pathogens, daptomycin: comparison with cationic antimicrobial peptides and lipopeptides. Biochim. Biophys. Acta 1758:1215–1223 [DOI] [PubMed] [Google Scholar]
- 39. Tanaka H., Iwai Y., Oiwa R., Shinohara S., Shimizu S., Oka T., Omura S. 1977. Studies on bacterial cell wall inhibitors. II. Inhibition of peptidoglycan synthesis in vivo and in vitro by amphomycin. Biochim. Biophys. Acta 497:633–640 [DOI] [PubMed] [Google Scholar]
- 40. Tanaka H., Oiwa R., Matsukura S., Inokoshi J., Omura S. 1982. Studies on bacterial cell wall inhibitors. X. Properties of phospho-N-acetylmuramoyl pentapeptidetransferase in peptidoglycan synthesis of Bacillus megaterium and its inhibition by amphomycin. J. Antibiot. 35:1216–1221 [DOI] [PubMed] [Google Scholar]
- 41. Tanaka H., Oiwa R., Matsukura S., Omura S. 1979. Amphomycin inhibits phospho-N-acetylmuramyl-pentapeptide translocase in peptidoglycan synthesis of Bacillus. Biochem. Biophys. Res. Commun. 86:902–908 [DOI] [PubMed] [Google Scholar]
- 42. Tironi I. G., Sperb R., Smith P. E., van Gunsteren W. F. 1995. A generalized reaction field method for molecular-dynamics simulations. J. Chem. Phys. 102:5451–5459 [Google Scholar]
- 43. Torda A. E., Scheek R. M., van Gunsteren W. F. 1989. Time-dependent distance restraints in molecular-dynamics simulations. Chem. Phys. Lett. 157:289–294 [Google Scholar]
- 44. van Gunsteren W. F., et al. 1996. Biomolecular simulation: the GROMOS96 manual and user guide. VdF, Hochschulverlag AG an der ETH Zurich BIOMOS b.v., Zurich, Switzerland [Google Scholar]
- 45. van Heijenoort J. 2001. Formation of the glycan chains in the synthesis of bacterial peptidoglycan. Glycobiology 11:25R–36R [DOI] [PubMed] [Google Scholar]
- 46. Vértesy L., et al. 2000. Friulimicins: novel lipopeptide antibiotics with peptidoglycan synthesis inhibiting activity from Actinoplanes friuliensis sp. nov. J. Antibiot. 53:816–827 [DOI] [PubMed] [Google Scholar]
- 47. Wiegand I., Hilpert K. K., Hancock R. E. W. 2008. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 3:163–175 [DOI] [PubMed] [Google Scholar]
- 48. Wong K. K., et al. 1998. Engineering a cell-free murein biosynthetic pathway: combinatorial enzymology in drug discovery. J. Am. Chem. Soc. 120:13527–13528 [Google Scholar]
- 49. Zhang L., Rozek A., Hancock R. E. W. 2001. Interaction of cationic antimicrobial peptides with model membranes. J. Biol. Chem. 276:35714–35722 [DOI] [PubMed] [Google Scholar]
- 50. Zhou G.-P., Troy A. F., II 2005. NMR study of the preferred membrane orientation of polyisoprenols (Dolichol) and the impact of their complex with polyisoprenyl recognition sequence peptides on membrane structure. Glycobiology 15:347–359 [DOI] [PubMed] [Google Scholar]