Abstract
DNA-binding proteins that control expression of drug efflux pump genes have been termed “local regulators” as their encoding gene is often located adjacent to the gene(s) that they regulate. However, results from recent studies indicate that they can control genes outside efflux pump-encoding loci, which we term as being “off target.” For example, the MtrR repressor was initially recognized for its ability to repress transcription of the mtrCDE-encoded efflux pump operon in the strict human pathogen Neisseria gonorrhoeae, but recent results from genetic and microarray studies have shown that it can control expression of nearly 70 genes scattered throughout the chromosome. One of the off-target MtrR-repressed genes is glnA, which encodes glutamine synthetase. Herein, we confirm the capacity of MtrR to repress glnA expression and provide evidence that such repression is due to its ability to negatively influence the binding of a second DNA-binding protein (FarR), which activates glnA. FarR was previously recognized as a transcriptional repressor of the farAB-encoded efflux pump operon. Thus, two DNA-binding proteins previously characterized as repressors of genes encoding efflux pumps that contribute to gonococcal resistance to antimicrobials can act in an opposing manner to modulate expression of a gene involved in basic metabolism.
INTRODUCTION
Over the past 2 decades, considerable progress has been made in characterizing the biochemistry, genetics, regulation, and function of efflux pump systems that export antimicrobials and contribute to bacterial resistance to antibiotics (1, 2, 8, 18, 20–22). Prior to the availability of genome sequence information, many of these efflux pumps were identified by the isolation of laboratory-derived mutants that expressed decreased susceptibility to antibiotics or other antimicrobials (18). These mutations frequently mapped to genes encoding DNA-binding proteins that normally function to dampen transcription of efflux pump protein-encoding genes. In this respect, the identification of the mtr (multiple transferable resistance) system of Neisseria gonorrhoeae by Pan and Spratt (19) was made possible by their cloning and sequencing of DNA fragments harboring mutant alleles of the mtrR gene, which encodes a repressor (MtrR) of the transcriptionally divergent mtrCDE efflux pump operon (10, 11, 19). When they are introduced into a wild-type strain of gonococci, mutant mtrR alleles can confer resistance to structurally diverse hydrophobic antimicrobial agents (9, 10, 19, 26, 28).
MtrR is a member of the TetR/QacR family of repressors (reviewed in reference 8), which are known to perform important roles in controlling expression of a number of bacterial efflux pump-encoding genes (21). With respect to repression of mtrCDE, two homodimers of MtrR bind the DNA sequence that overlaps the promoter used for mtrCDE transcription (12, 16), and a pseudo-direct repeat element (CCGTGCA and TCGTGTA), separated by a single nucleotide, within this binding site is important for MtrR recognition (12). Due to its ability to modulate levels of antimicrobial resistance and the frequent occurrence of mtrR mutants in clinical isolates (4, 25, 30, 31), we have been interested in how MtrR controls gene expression in gonococci (5–7, 15). Although repressors like MtrR have been thought of as local regulators (17) that control adjacent efflux pump genes, it is likely that they have the capacity to regulate additional genes outside those involved in drug efflux; we term these genes as being “off target.” Since it is unclear as to whether the model (12, 16) developed for MtrR repression of target genes (e.g., mtrCDE) would hold true for off-target genes, we examined its ability to control expression of glnA, which was identified in a recent microarray study (7) as being MtrR repressed. We were especially interested in MtrR regulation of glnA because glnA in Corynebacterium glutamicum has been shown (3) to be negatively controlled by another member of the TetR/QacR family (AmtR), and we thought that studies with MtrR would, in general, provide insights regarding the mechanism by which members of the TetR/QacR family regulate genes outside those encoding drug efflux proteins. We now report evidence that MtrR represses glnA in gonococci by negatively influencing the binding of a second DNA-binding protein (FarR), also involved in repressing expression of an antimicrobial efflux pump (15), which activates glnA expression. Taken together, our results indicate that DNA-binding proteins recognized for their ability to control expression of drug efflux pump genes and modulate levels of bacterial susceptibility to antimicrobials can have alternative targets and modes of regulation.
MATERIALS AND METHODS
Strains used and growth conditions.
All of the strains of N. gonorrhoeae employed (Table 1) are genetic derivatives of strain FA19 and were constructed in previous investigations (5–7, 15) or in this study. Gonococci were routinely cultured as nonpiliated, opacity-negative (P− Opa−) variants on GCB agar with defined supplements I and II (23) at 37°C under 3.8% (vol/vol) CO2; transformation experiments used piliated (P+) Opa− variants. Gonococci were also cultured in GCB broth (Difco Laboratories, Detroit, MI) with supplements I and II and 0.048% (wt/vol) sodium bicarbonate with shaking at 37°C. Escherichia coli strain DH5α mcr was routinely cultured in Luria-Bertani (LB) broth or on LB agar (Difco Laboratories) with antibiotics (100 μg/ml of ampicillin or kanamycin); antibiotics were purchased from Sigma Chemical Corporation (St. Louis, MO).
Table 1.
Gonococcal strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or remarks | Source or reference |
|---|---|---|
| Strains | ||
| FA19 | Wild type | P. F. Sparling |
| EL24 | As FA19 but farR::kan | 15 |
| EL28 | As EL24 but farR+ | 15 |
| JF1 | As FA19 but ΔmtrR | 5 |
| JF6 | As JF1 but mtrR+ | 7 |
| PJ9 | As FA19 but glnA::lacZ | This study |
| PJ10 | As JF1 but glnA::lacZ | This study |
| PJ11 | As EL24 but glnA::lacZ | This study |
| PJ12 | As JF6 but glnA::lacZ | This study |
| PJ13 | As PJ10 but farR::kan | This study |
| PJ14 | As EL28 but glnA::lacZ | This study |
| PJ15 | As PJ14 but farR+ | This study |
| PJ16 | As PJ12 but farR::kan | This study |
| PJ22 | As PJ13 but mtrR+ | This study |
| Plasmids | ||
| pLES94 | Cloning vector containing promoterless lacZ for insertion of gonococcal genes between proA and proB | V. Clark |
| pJF3 | As pLES94 but glnA::lacZ | This study |
| pGCC3 | NICS vector used for insertion of gonococcal genes between lctP and aspC | H. Seifert |
qRT-PCR.
The essentials of the quantitative real-time reverse transcription (RT)-PCR (qRT-PCR) protocol employed have been described previously (13). In order to confirm the microarray data reported by Folster et al. (7), a portion of each of the three RNA samples from parent strain FA19 and strain JF1 (as FA19 but ΔmtrR) used in that study was employed in qRT-PCR; these RNA samples were kindly provided by L. Jackson and D. Dyer (University of Oklahoma Health Sciences Center, Oklahoma City, OK). cDNA from each sample was synthesized in three independent RT reactions with random hexamer primers and Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA). The specific primers for glnA and 16S RNA used in qRT-PCR are listed in Table 2. qRT-PCR was performed on an iCycler iQ real-time PCR detection system (Bio-Rad Laboratories, Hercules, CA). iQSYBR Green Supermix (Bio-Rad Laboratories) was employed in a reaction volume of 25 μl with 200 nM 5′ and 3′ primers and 5-fold dilutions of RT reaction mixtures. 16S RNA cDNA was employed as the internal control. The results for changes in glnA expression due to loss of mtrR are reported (see below) as an average value (± standard deviation).
Table 2.
Oligonucleotides used in this study
| Oligonucleotide useda | Sequence (5′ → 3′) |
|---|---|
| 5′pglnA | GGGGATCCCATAAAGGCGGGGCGGTC |
| 3′pglnA | GGGGATCCCGGGACATCTTCAGCTCCTGAA |
| glnA1 | GCAACCGCCTGTTTCAAAAAATG |
| glnA2 | GGACATCTTCAGCTCCTGAAAAAG |
| glnAsec1F | TAACGTTTGCCCCGCAACC |
| glnAsec1R | CCCCCGCTACGCCGTTTTC |
| glnAsec2F | GGGCGTGCATAGTCATATTC |
| glnAsec2R | GCGTCAAATTCCAAGCCGG |
| glnAsec3F | AAACCGGTTTCAGACGGCAT |
| glnAsec3R | GGACATCTTCAGCTCCTGAA |
| glnAtrunckf | CATGGATCCGATGAATCTGCGGCGATTTG |
| rmpF | CATGTTTCTACAGCGGCCTG |
| rmpR | CGGCAAGATATTACCTAGCCT |
| 16SRTR | CATCGGTATTCCTCCACATCTC |
| 16SRTF | GTAGGGTGCGAGCGTTAATC |
| glnARTR | GGCTTTGGCGTGTTTGATG |
| glnARTF | TCCGATACCGCGCTCTACTAC |
Primers with ending letters RTR or RTF were used in qRT-PCR for the 16S rRNA and glnA transcripts.
Strain construction and β-Gal assays.
Translational lacZ fusions were constructed using pLES94 (27) by transformation into strains FA19, JF1, JF6, and EL24 or their derivatives. The strains bearing translational fusions are described in Table 1. β-Galactosidase (β-Gal) assays were performed as described by Folster and Shafer (5). Plasmid pJF3, which contained a glnA-lacZ fusion, was prepared essentially as described by Folster et al. (7). Briefly, the promoter sequence of glnA (summarized in Fig. 1) was amplified by PCR from strain FA19 using primers 5′pglnA and 3′pglnA (Table 2). Following amplification by PCR, the DNA sequence was cloned into the BamHI cloning site of pLES94 (27). The resulting plasmid construct was then transformed into E. coli DH5α mcr and transformants were recovered by ampicillin selection (100 μg/ml). The cloned fragment in the resulting transformant was identified by PCR analysis and DNA sequencing. This plasmid (pJF3) was then transformed (5) into strains FA19, JF1, JF6, and EL24, with insertion occurring at the nonessential proAB locus (27) of the recipient strains. Transformants were selected on GCB agar containing 1 μg/ml of chloramphenicol. The resulting transformants (Table 1) were then used in β-Gal assays as previously described (7).
Fig. 1.
Nucleotide sequence upstream of glnA. The nucleotide sequence of the DNA upstream of glnA is shown, including the first two codons encoding methionine (M) and serine (S); the translational start codon is identified by the bent arrow. This 506-bp sequence is divided into three sections that were used in EMSA experiments, with section I shown in green, section II in yellow, and section III in blue. The −10 and −35 hexamer sequences of the glnA promoter (section III) are identified with the −10 and −35 notation as well as a line under the sequences. The start point of transcription is shown by the † symbol and was determined by primer extension analysis as described previously (10). The MtrR-binding site in section I that is boxed in red was predicted on the basis of similarity to the binding site upstream of mtrCDE (12, 16). DNase I-hypersensitive sites observed in protection assays with MtrR are shown by asterisks. The FarR-binding site predicted by sequence similarity with that upstream of farAB (15) is shown in the black boxed area in section II, and the binding sites identified by DNase I protection (Fig. 4) are noted by the dotted line above the sense strand and below the antisense strand.
Protein purification and DNA-binding studies.
The production and purification of the maltose-binding protein (MBP)-MtrR and the histidine (His)-tagged FarR fusion proteins used in this study have been described previously (6, 15). These proteins were used in electrophoretic mobility shift assays (EMSAs) and DNase I protection studies using previously described methods (7, 15). The DNA probes for EMSAs consisted of PCR products that were obtained using sets of oligonucleotide primers (Table 2); the generation of specific PCR products is described in the relevant figure legend. These PCR products were end labeled with [γ-32P]dATP using T4 polynucleotide kinase (New England BioLabs, Beverly, MA) as described previously (15). The labeled PCR-generated products were incubated with purified MtrR-MBP or FarR-His, or both, in a final reaction volume of 30 μl consisting of the reaction buffer [10 mM Tris-HCl (pH 7.5), 0.5 mM dithiothreitol, 0.5 mM EDTA, 4% (vol/vol) glycerol, 1 mM MgCl2, 50 mM NaCl, poly(dI-dC) (0.05 μg/ml)] and distilled H2O at 25°C for 30 min. Following incubation, the reaction mixtures were subjected to gel electrophoresis utilizing a 6% (wt/vol) polyacrylamide gel at 4°C and dried, and autoradiography was performed for visualization. Competitive EMSAs were performed in the same manner, but with the addition of unlabeled specific competitor, generated from the same sequence as the target, or nonspecific competitor, generated from the rmp gene using primers rmpF and rmpR (Table 2). DNase I protection assays were performed essentially as described by Folster et al. (7). PCR-generated target DNA sequences were synthesized using primers glnAsec1F, glnAsec1R, glnAsec2F, or glnAsec2R and were labeled at the 5′ end of one strand as described for EMSA. Purified MtrR-MBP or FarR-His was then incubated with the target DNA sequence under the same binding conditions used in the EMSA for 15 min at 37°C. Following the addition of MgCl2 (5 mM) and CaCl2 (2.5 mM), DNase I (Promega, Madison, WI) was added and the reaction mixtures were incubated at 37°C for 1 min. The reactions were stopped with DNase I stop buffer (95% ethanol, 7.5 mM ammonium acetate, and nuclease-free H2O), snap-frozen on dry ice for 15 min, and then precipitated at −80°C overnight. The pellet was then washed in 70% (vol/vol) ethanol, dried, and resuspended in gel loading buffer (Epicentre, Madison, WI). Reaction mixtures were then loaded on 8% denaturing polyacrylamide gel, subjected to gel electrophoresis, and dried, and autoradiography was performed for visualization as described previously (6, 7).
RESULTS AND DISCUSSION
MtrR repression of off-target gene, glnA.
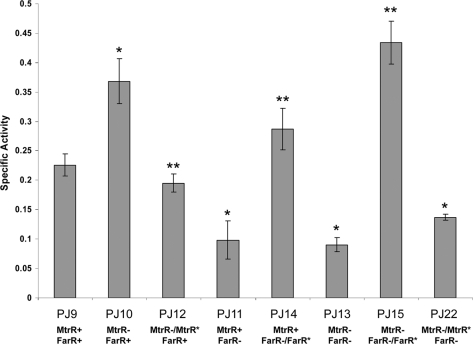
We first sought to confirm the earlier microarray data of Folster et al. (7), which revealed the capacity of MtrR to negatively control glnA expression by 1.6-fold (P < 0.05). Results from qRT-PCR experiments, which used total RNA isolated from isogenic strains FA19 and JF1 (as FA19 but ΔmtrR; Table 1), confirmed that MtrR could dampen (1.9- ± 0.23-fold; P < 0.05) glnA expression. As additional confirmation, we employed a translational promoterless lacZ expression system that measures gene expression and regulation from an ectopic site in gonococci (27). The glnA-lacZ translational fusion employed in this study consisted of 500 bp of DNA upstream and the first two codons of glnA (Fig. 1). Using this system, we found (Fig. 2) that glnA-lacZ expression in isogenic MtrR-positive and -negative genetic derivatives differed and was significantly (P = 0.005) elevated in MtrR-negative strain PJ10 compared to MtrR-positive strain PJ9. Importantly, this increase in glnA-lacZ expression in the MtrR-negative strain was reversed (P < 0.005) to wild-type levels when the mutation was complemented (see strain PJ12) with an ectopically expressed wild-type mtrR gene (Fig. 2).
Fig. 2.
MtrR and FarR regulation of glnA expression. The specific activity of β-Gal (expressed as nanomoles of o-nitrophenyl-β-d-galactopyranoside hydrolyzed per mg of protein) in strains PJ9 (FA19 glnA::lacZ), PJ10 (JF1 glnA::lacZ), PJ12 (JF6 glnA::lacZ), PJ11 (EL24 glnA::lacZ), PJ14 (EL28 glnA::lacZ), PJ13 (PJ10 farR::kn glnA lacZ), PJ15 (PJ14 farR+), and PJ22 (PJ13 mtrR+). The results are shown as an average value (± standard deviation) from three independent experiments. The single asterisk above a bar denotes a significant (P < 0.01) difference between the indicated strain and wild-type strain PJ9, while the double asterisk denotes a significant difference (P < 0.01) between a complemented strain and its respective mutant (comparisons of PJ12 versus PJ10, PJ14 versus PJ11, and PJ15 versus PJ13); the specific P values are provided in the text.
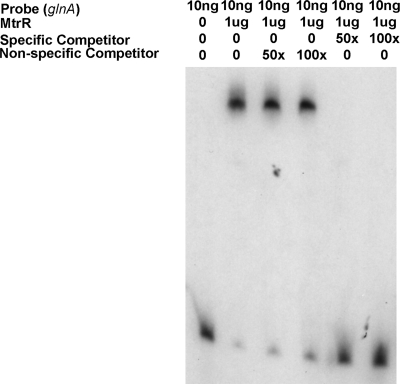
Since the qRT-PCR and lacZ translational fusion data confirmed the microarray data of Folster et al. (7) that glnA is subject to MtrR repression in gonococci, we tested if MtrR could bind the DNA sequence upstream of glnA. In preliminary EMSA experiments, we found that an MBP-MtrR fusion protein could bind in a specific manner (data not presented) to a labeled PCR probe that consisted of the DNA upstream of glnA shown in Fig. 1. In order to localize the site of specific DNA binding by MtrR, we prepared three probes (termed sections I, II, and III in Fig. 1) that consisted of truncated sequences upstream of glnA. Using EMSA, we found that MtrR bound only section I in a specific manner (data not presented and Fig. 3). On the basis of the MtrR-binding site sequence upstream of mtrCDE (12, 16), we identified a potential MtrR-binding site upstream of glnA within section I, which is shown in the red box in Fig. 1. However, when we used DNase I footprinting to identify potential sites for MtrR binding within section I, we could not reproducibly detect defined regions of protection. Interestingly, on the coding strand, at least three DNase I-hypersusceptible sites were evident (summarized in Fig. 1), suggesting the occurrence of MtrR-DNA interactions. Two of these hypersensitive sites were within the predicted MtrR-binding site (12, 16), while the third was 12 nucleotides upstream of this region.
Fig. 3.
Specific binding of MtrR upstream of glnA. The area of MtrR-specific binding was identified using the three sections of the full-length DNA sequence described in Fig. 1. The full-length probe was PCR amplified from DNA of strain FA19 using primers 5′pglnA and 3′pglnA (Table 2), while binding site I was amplified with primers glnAsec1F and glnAsec1R. The nonspecific probe was prepared using primers rmpF and rmpR. MtrR was found to specifically bind only (data not presented) to section I since specific competitor DNA, but not nonspecific competitor (up to 100 times), was able to abrogate the shifting of section I glnA DNA in EMSA; this result is shown in the figure.
FarR activates glnA expression.
Although MtrR can directly regulate certain genes in gonococci, it can also indirectly regulate others (7, 15). For instance, expression of the farAB efflux pump-encoding operon (15) is increased in an mtrR-null mutant because MtrR can repress farR, which encodes a direct repressor of the farAB-encoded efflux pump operon (15). Accordingly, we tested if MtrR regulation of glnA also involves FarR. For this purpose we used the translational glnA fusion system described above and monitored its expression in strains containing a farR-null mutation with and without a coresident mtrR-null mutation. Evidence for FarR regulation of glnA was obtained (Fig. 2) when the glnA-lacZ translational fusion was expressed in strain FA19 bearing a wild-type (strain PJ9) or an insertionally inactivated (strain PJ11) farR gene. With these strains we found (Fig. 2) that expression of glnA was significantly (P = 0.007) higher in FarR-positive strain PJ9.
As this result suggested that FarR is a positive regulator of glnA expression, we tested if complementation of the farR-null mutant with the wild-type farR gene expressed ectopically (strain PJ14) would restore levels of glnA expression and found this to be the case (Fig. 2); the difference in glnA expression between strains PJ11 and PJ14 was significant (P = 0.002). Due to these results, we next examined glnA expression in an MtrR− FarR− double mutant (strain PJ13). In double mutant strain PJ13, glnA expression mimicked (Fig. 2) that of the single mutant that only lacked FarR (strain PJ11). Moreover, complementation of the double mutant with a wild-type farR gene (strain PJ15) but not mtrR (strain PJ22) expressed ectopically enhanced glnA expression to a level that resembled that in MtrR− FarR+ strain PJ10 (Fig. 2); the difference in glnA expression between strains PJ13 and PJ15 was significant (P < 0.0001).
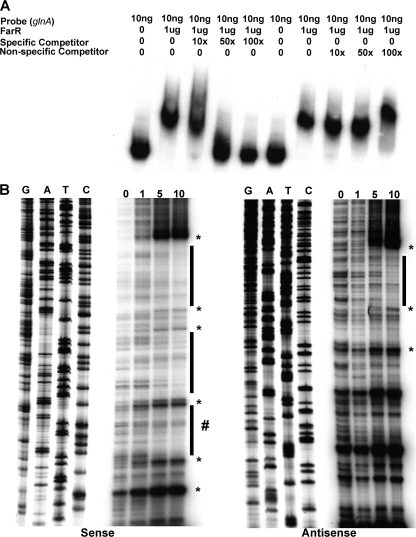
Evidence that FarR can bind upstream of glnA was obtained by both EMSA and DNase I protection. In EMSA experiments that monitored specific binding of FarR to sections I, II, and III (Fig. 1), we found that it bound only section II in a specific manner (data not presented and Fig. 4A). Using DNase I protection, we identified (Fig. 4B) three sites protected by FarR on the sense strand, all three of which had adjacent DNase I-hypersensitive nucleotides. Importantly, only one of the FarR-protected sites was within section II (shown by the # sign next to the vertical bar in Fig. 4B), which bound FarR in a specific manner (Fig. 4A). On the antisense strand, only a single predominant region gave evidence of protection. This protected region was complementary to a 28-nucleotide stretch on the sense strand in section II that was also protected by FarR (summarized in Fig. 1). Overall, this area of protection is located nearly 200 bp upstream of the previously annotated glnA promoter, which we verified (data not presented) by primer extension analysis.
Fig. 4.
Identification of the FarR-binding site upstream of the glnA promoter. (A) The area of FarR-specific binding was first identified using the subsections of the full-length glnA sequence shown in Fig. 1. The full-length probe was PCR amplified using primers 5′pglnA and 3′pglnA, while binding site II was amplified with primers glnAsec2F and glnAsec2R. The nonspecific probe was prepared using primers rmpF and rmpR. FarR was found (data not presented) to bind only section II in a specific manner, as determined by competitive EMSA (shown in the figure). (B) The FarR-binding sites on the sense and antisense strands were identified by DNase I protection assays that employed increasing amounts of purified FarR-His (0, 1, 5, and 10 μg). The protected regions on each probe are identified by the black bars, with the protected region on the sense strand within section II being denoted by a # sign next to the bar to distinguish it from the other two protected regions that lie within section I. DNase I-hypersensitive sites are shown by an asterisk. The sequencing reactions for each probe are adjacent to the DNase I protection reactions and oriented G, A, T, C.
MtrR binding to glnA negatively influences FarR binding.
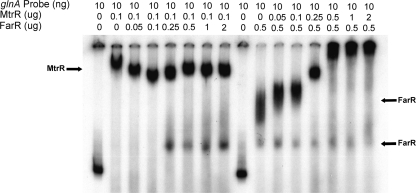
We hypothesized that the ability of MtrR to repress glnA expression could be due to its ability to impact FarR binding to a target DNA sequence upstream of glnA. In order to test if FarR::DNA complexes can be influenced by the presence of MtrR, the target DNA was preincubated with a fixed concentration of one protein and then incubated with increasing concentrations of the second protein. The results (Fig. 5) showed that in the absence of competing protein, MtrR and FarR gave distinct shifts of the probe. However, as the amount of FarR was increased after the DNA had been preincubated with a fixed amount of MtrR (0.1 μg), only the lower of the two FarR::DNA complexes appeared at ≥0.25 μg of competing FarR. Importantly, the electrophoretic mobility of the preformed MtrR::DNA complex showed only minor changes in mobility. In sharp contrast to these results, the pattern of the FarR-specific shifts was significantly changed by the addition of increasing amounts of MtrR. Specifically, the slower-migrating FarR-specific shift was lost and higher-molecular-weight complexes became evident and seemed to predominate, especially at amounts of MtrR of ≥0.25 μg; one of these complexes comigrated with the MtrR-specific shift (see the lane with 0.25 μg of competing MtrR in Fig. 5). The complexes that had a slower electrophoretic mobility than that of the MtrR-specific shift likely represent multimers of MtrR bound to the target DNA, as they were observed (data not presented) in other EMSAs that used large amounts of MtrR (≥0.5 μg) but not FarR.
Fig. 5.
MtrR influences FarR::DNA complexes. Shown are the results from an EMSA experiment that evaluated the binding of MtrR and FarR to the 32P-labeled full-length glnA probe (Fig. 1) that was prepared as described in the legend to Fig. 3. The lane assignments are as follows, from left to right: probe alone; probe plus 0.1 μg of MtrR-MBP; probe plus 0.1 μg of MtrR-MBP and increasing amounts of FarR-His (0.05, 0.1, 0.25, 0.5, 1.0, and 2.0 μg); probe alone; and probe plus 0.5 μg of FarR-His with increasing amounts of MtrR-MBP (0.05, 0.1, 0.25, 0.5, 1.0, and 2.0 μg). The positions of MtrR and FarR shifted bands in the absence of competing protein are shown.
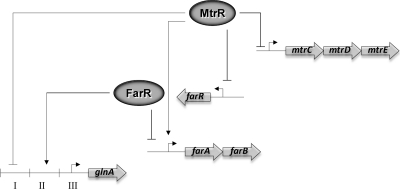
DNA-binding proteins that control efflux gene expression in bacteria have been termed “local regulators” (17), as their encoding gene is frequently located adjacent to their respective target genes that encode efflux pump proteins. While the ability of such regulators to control expression of efflux pump genes is certainly important in modulating levels of bacterial susceptibility to antimicrobials, they may have, on the basis of our work with MtrR described herein and elsewhere (6, 7, 15), as well as work with Salmonella enterica serovar Typhimurium (2), a more global action and control expression of genes, which we term off-target, that contribute to important physiological processes. The question addressed herein was whether the model developed for MtrR repression of the mtrCDE efflux pump locus, which involves DNA binding to the promoter region (12, 16), would be similar for a model off-target and repressed gene. In sharp contrast to MtrR repression of mtrCDE (12, 16), the results presented herein suggest that MtrR represses glnA both by its ability to reduce expression of farR (15) and by negatively influencing binding of FarR to its target site upstream of glnA or decreasing stability of such complexes (Fig. 5), which would normally result in activation of glnA. Thus, as summarized in Fig. 6, MtrR can directly repress its target sequence (the mtrCDE efflux pump operon [12, 16]) as well as off-target sequences upstream of farR (21) and glnA (Fig. 2). Conversely, transcriptional activation of glnA is mediated by FarR, which directly represses the farAB efflux operon (21), by its binding upstream of glnA (section II shown in Fig. 6). However, the binding of FarR to this target can be negatively influenced by binding of MtrR upstream of the FarR-binding site (Fig. 5).
Fig. 6.
Model of glnA regulation by MtrR and FarR. Direct repression and activation of genes is shown by the solid-barred and arrowed lines, respectively, while indirect activation (i.e., MtrR activation of farAB) is show by the dashed, arrowed line. The binding of proteins is shown adjacent to promoter regions (bent arrow). With respect to glnA, the binding of MtrR is shown within section I and the binding of FarR is shown within section II of the DNA sequence shown in Fig. 1. The ability of MtrR to repress glnA is proposed to be due to its capacity to repress farR expression (15) and its ability to diminish FarR binding within section II (Fig. 5).
The ability of a transcriptional regulator in the TetR/QacR family to repress glnA is not without precedent, as AmtR of Corynebacterium glutamicum was recently shown to negatively control glnA expression (3) by a yet-to-be-defined mechanism. Although measurements of glutamine synthetase activity in isogenic variants of strain FA19 bearing wild-type or mutant alleles of mtrR or farR did not reveal significant differences when such strains were grown in GCB broth lacking glutamine (P. J. T. Johnson and W. M. Shafer, unpublished observations), levels of glutamine at mucosal surfaces and within phagolysosomes (14), two important sites for harboring gonococci during infection (24), are scarce. We hypothesize that regulatory systems involving MtrR which modulate expression of glnA and other genes may be important for optimal growth and fitness of gonococci during infection. Accordingly, we are now using a murine model of vaginal infection, which previously revealed the importance of MtrR in determining levels of in vivo fitness of gonococci (29), to test the importance of off-target genes regulated by MtrR during infection.
ACKNOWLEDGMENTS
We thank L. Jackson and D. Dyer for RNA samples, C. Moran and Y. Zalucki for critical reading of the manuscript prior to submission, J. Folster and V. Dhulipala for preparing pJF3, and L. Pucko for help with manuscript preparation.
This work was supported by NIH grant R37 AI021150-25 and a VA Merit Award from the U.S. Department of Veterans Affairs, both to W.M.S. W.M.S. is the recipient of a Senior Research Career Scientist award from the U.S. Department of Veterans Affairs.
Footnotes
Published ahead of print on 21 March 2011.
REFERENCES
- 1. Ahmed M., Borsch C. M., Taylor S. S., Vazquez-Laslop N., Neyfakh A. A. 1994. A protein that activates expression of a multidrug efflux transporter upon binding the transporter substrates. J. Biol. Chem. 269:28506–28513 [PubMed] [Google Scholar]
- 2. Bailey A. M., et al. 2010. RamA, a member of the AraC/XylS family, influences both virulence and efflux in Salmonella enterica serovar Typhimurium. J. Bacteriol. 192:1607–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Buchinger S., et al. 2009. A combination of metabolome and transcriptome analyses reveals new targets of the Corynebacterium glutamicum nitrogen regulator AmtR. J. Biotechnol. 140:68–74 [DOI] [PubMed] [Google Scholar]
- 4. Eisenstein B. I., Sparling P. F. 1978. Mutations to increased antibiotic sensitivity in naturally-occurring gonococci. Nature 271:242–244 [DOI] [PubMed] [Google Scholar]
- 5. Folster J. P., Shafer W. M. 2005. Regulation of mtrF expression in Neisseria gonorrhoeae and its role in high-level antimicrobial resistance. J. Bacteriol. 187:3713–3720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Folster J. P., Dhulipala V., Nicholas R. A., Shafer W. M. 2007. Differential regulation of ponA and pilMNOPQ expression by the MtrR transcriptional regulatory protein in Neisseria gonorrhoeae. J. Bacteriol. 189:4569–4577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Folster J. P., et al. 2009. MtrR modulates rpoH expression and levels of antimicrobial resistance in Neisseria gonorrhoeae. J. Bacteriol. 191:287–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Grkovic S., Brown M. H., Skurray R. A. 2002. Regulation of bacterial drug export systems. Microbiol. Mol. Biol. Rev. 66:671–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hagman K. E., et al. 1995. Resistance of Neisseria gonorrhoeae to antimicrobial hydrophobic agents is modulated by the mtrRCDE efflux system. Microbiology 141:611–622 [DOI] [PubMed] [Google Scholar]
- 10. Hagman K. E., Shafer W. M. 1995. Transcriptional control of the mtr efflux system of Neisseria gonorrhoeae. J. Bacteriol. 177:4162–4165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hagman K. E., et al. 1997. The MtrD protein of Neisseria gonorrhoeae is a member of the resistance/nodulation/division protein family constituting part of an efflux system. Microbiology 143:2117–2125 [DOI] [PubMed] [Google Scholar]
- 12. Hoffmann K. M., Williams D., Shafer W. M., Brennan R. G. 2005. Characterization of the multiple transferable resistance repressor, MtrR, from Neisseria gonorrhoeae. J. Bacteriol. 187:5008–5012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Katzif S., Lee E.-H., Law A. B., Tzeng Y.-L., Shafer W. M. 2005. CspA regulates pigment production in Staphylococcus aureus through a SigB-dependent mechanism. J. Bacteriol. 187:8181–8184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Klose K. E., Mekalanos J. J. 1997. Simultaneous prevention of glutamine synthesis and high-affinity transport attenuates Salmonella typhimurium virulence. Infect. Immun. 65:587–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee E. H., Rouquette-Loughlin C., Folster J. P., Shafer W. M. 2003. FarR regulates the farAB-encoded efflux pump of Neisseria gonorrhoeae via an MtrR regulatory mechanism. J. Bacteriol. 185:7145–7152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lucas C. E., Balthazar J. T., Hagman K. E., Shafer W. M. 1997. The MtrR repressor binds the DNA sequence between the mtrR and mtrC genes of Neisseria gonorrhoeae. J. Bacteriol. 179:4123–4128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ma D., et al. 1995. Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli. Mol. Microbiol. 16:45–55 [DOI] [PubMed] [Google Scholar]
- 18. Nikaido H. 1996. Multidrug efflux pumps of gram-negative bacteria. J. Bacteriol. 178:5853–5859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pan W., Spratt B. G. 1994. Regulation of the permeability of the gonococcal cell envelope by the mtr system. Mol. Microbiol. 11:769–775 [DOI] [PubMed] [Google Scholar]
- 20. Paulsen I. T., Brown M. H., Skurray R. A. 1996. Proton-dependent multidrug efflux systems. Microbiol. Rev. 60:575–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ramos J. L., et al. 2005. The TetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. 69:326–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rouquette C., Harmon J. B., Shafer W. M. 1999. Induction of the mtrCDE-encoded efflux pump system of Neisseria gonorrhoeae requires MtrA, an AraC-like protein. Mol. Microbiol. 33:651–658 [DOI] [PubMed] [Google Scholar]
- 23. Shafer W. M., Guymon L. F., Lind I., Sparling P. F. 1984. Identification of an envelope mutation (env-10) resulting in increased antibiotic susceptibility and pyocin resistance in a clinical isolate of Neisseria gonorrhoeae. Antimicrob. Agents Chemother. 25:767–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shafer W. M., Rest R. F. 1989. Interactions of gonococci with phagocytic cells. Annu. Rev. Microbiol. 43:121–145 [DOI] [PubMed] [Google Scholar]
- 25. Shafer W. M., Balthazar J. T., Hagman K. E., Morse S. A. 1995. Missense mutations that alter the DNA-binding domain of the MtrR protein occur frequently in rectal isolates of Neisseria gonorrhoeae that are resistant to faecal lipids. Microbiology 141:907–911 [DOI] [PubMed] [Google Scholar]
- 26. Shafer W. M., Qu X., Waring A. J., Lehrer R. I. 1998. Modulation of Neisseria gonorrhoeae susceptibility to vertebrate antibacterial peptides due to a member of the resistance/nodulation/division efflux pump family. Proc. Natl. Acad. Sci. U. S. A. 95:1829–1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Silver L. E., Clark V. L. 1995. Construction of a translational lacZ fusion system to study gene regulation in Neisseria gonorrhoeae. Gene 166:101–104 [DOI] [PubMed] [Google Scholar]
- 28. Veal W. L., Nicholas R. A., Shafer W. M. 2002. Overexpression of the MtrC-MtrD-MtrE efflux pump due to an mtrR mutation is required for chromosomally mediated penicillin resistance in Neisseria gonorrhoeae. J. Bacteriol. 184:5619–5624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Warner D. M., Shafer W. M., Jerse A. E. 2008. Clinically relevant mutations that cause derepression of the Neisseria gonorrhoeae MtrC-MtrD-MtrE efflux pump system confer different levels of antimicrobial resistance and in vivo fitness. Mol. Microbiol. 70:462–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xia M., Whittington W. L., Shafer W. M., Holmes K. K. 2000. Gonorrhea among men who have sex with men: outbreak caused by a single genotype of erythromycin-resistant Neisseria gonorrhoeae with a single-base pair deletion in the mtrR promoter region. J. Infect. Dis. 181:2080–2082 [DOI] [PubMed] [Google Scholar]
- 31. Zarantonelli L., Borthagaray G., Lee E. H., Shafer W. M. 1999. Decreased azithromycin susceptibility of Neisseria gonorrhoeae due to mtrR mutations. Antimicrob. Agents Chemother. 43:2468–2472 [DOI] [PMC free article] [PubMed] [Google Scholar]