Abstract
A lack of patient response to alpha interferon (α-IFN) plus ribavirin (RBV) treatment is a major problem in eliminating hepatitis C virus (HCV). We screened chemical libraries for compounds that enhanced cellular responses to α-IFN and identified a triterpenoid, toosendanin (TSN). Here, we studied the effects and mechanisms of action of TSN on HCV replication and its effect on α-IFN signaling. We treated HCV genotype 1b replicon-expressing cells and HCV-J6/JFH-infected cells with TSN, with or without α-IFN, and the level of HCV replication was quantified. To study the effects of TSN on α-IFN signaling, we detected components of the interferon-stimulated gene factor 3 (ISGF3), phosphorylated signal transducer and activator of transcription 1 (STAT1), and STAT2 by Western blotting analysis; expression levels of mRNA of interferon regulatory factor 9 using real-time reverse transcription-PCR (RT-PCR); and interferon-stimulated response element reporter activity and measured the expression levels of interferon-inducible genes for 2′,5′-oligoadenylate synthetase, MxA, protein kinase R, and p56 using real-time RT-PCR. TSN alone specifically inhibited expression of the HCV replicon (50% effective concentration = 20.6 nM, 50% cytotoxic concentration > 3 μM, selectivity index > 146). Pretreatment with TSN prior to α-IFN treatment was more effective in suppressing HCV replication than treatment with either drug alone. Although TSN alone did not activate the α-IFN pathway, it significantly enhanced the α-IFN-induced increase of phosphorylated STATs, interferon-stimulated response element activation, and interferon-stimulated gene expression. TSN significantly increased baseline expression of interferon regulatory factor 9, a component of interferon-stimulated gene factor 3. Antiviral effects of treatment with α-IFN can be enhanced by pretreatment with TSN. Its mechanisms of action could potentially be important to identify novel molecular targets to treat HCV infection.
INTRODUCTION
Hepatitis C virus (HCV) is one of the most important pathogens causing acute and chronic hepatitis, liver cirrhosis, and hepatocellular malignancies (29). Alpha interferon (α-IFN) combined with ribavirin (RBV) is the standard treatment for HCV infection (6, 10). However, virus elimination rates are about 50% among treated patients, and therapy is often accompanied by substantial side effects (6, 44). It was recently reported that genetic polymorphisms of the IL28B gene, which codes for lambda IFN, are critical for predicting responses to α-IFN plus RBV therapy (8, 35, 38). Patients with minor variants of IL28B, who comprise ∼50% of Caucasian, 25% of Asian, and ∼70% of African populations, showed poor responses to α-IFN treatment. Although new specific anti-HCV drugs are under development, many of them require combined use with α-IFN and RBV (26). Taken together, current difficulties in eliminating HCV are mostly attributable to the limited treatment options and to the limited activity of α-IFN against the virus. For this reason, the development of safe and effective agents that enhance antiviral actions against HCV has been a strong motivation in academia and industry.
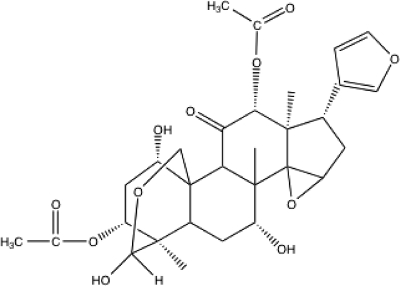
To search for a new agent which enhances the effect of α-IFN, we used interferon-stimulated response element (ISRE) reporter screening. We screened a chemical library (60,500 compounds) for compounds that enhance ISRE activity when they are used in combination with α-IFN, using ISRE reporter screening, and identified several compounds that increased the ISRE reporter activities when they are used in combination with α-IFN and that did not show cytotoxicity. Among the hit compounds, toosendanin (TSN; C30H38O11; molecular weight = 574) (Fig. 1), which is a triterpenoid derivative extracted from the bark of Melia toosendan Sieb et Zucc, was the strongest in enhancing α-IFN-induced ISRE reporter activation and the expression of interferon-stimulated genes (ISGs). TSN has been used as an anthelmintic vermifuge against ascaris (31). Although TSN has some other biological effects against toxin-producing anaerobic bacteria and against carcinoma cells (32, 45), antiviral activity has not been reported.
Fig. 1.
Chemical structure of toosendanin.
In this study, we showed, using an HCV replicon system, that TSN, with or without α-IFN, inhibits HCV replication in a cultured human hepatoma Huh7 cell line and that the combination of TSN and α-IFN shows synergistic effects on viral replication. We have investigated the mechanisms of action of TSN further and show that TSN induced activation of a component of interferon-stimulated gene factor 3 (ISGF3).
MATERIALS AND METHODS
Reagents.
Alpha interferon was from Otsuka (Tokushima, Japan). TSN was from APIN Chemicals (Oxon, United Kingdom). Purity was over 77.32%. The designated concentration was achieved through dilution with cell culture medium (the final concentration of dimethyl sulfoxide [DMSO] in the medium was less than 0.3%). Beta-mercaptoethanol was from Wako (Osaka, Japan). The TSN used in this study was solubilized in DMSO.
Cells and cell culture.
The human hepatoma cell line Huh7 was maintained in Dulbecco's modified Eagle's medium (Sigma, St. Louis, MO) supplemented with 10% fetal bovine serum at 37°C under 5% CO2. To maintain cell lines carrying an HCV subgenomic replicon (Huh7/Rep-Feo), G418 (Nakalai Tesque, Kyoto, Japan) was added to the culture medium at a final concentration of 500 μg/ml.
HCV subgenomic replicon construct.
The HCV subgenomic replicon plasmid pRep-Feo expresses a fusion gene comprising the firefly luciferase and neomycin phosphotransferase (37, 43). RNA was synthesized in vitro from the plasmid and transfected into Huh7 cells. After culture in the presence of G418, cell lines stably expressing the replicon were established.
Reporter constructs.
We analyzed the effects of TSN, with or without α-IFN, on signal transduction of ISRE and nuclear factor-kappaB (NF-kappaB). A plasmid, pCIneo-Rluc-IRES-Fluc, was constructed to analyze HCV internal ribosome entry site (IRES)-mediated translation efficiency (23). Plasmids pISRE-TA-Luc and pNF-kappaB-Luc (Clontech Laboratories, Franklin Lakes, NJ) contained consensus motifs upstream of the firefly luciferase gene. A plasmid, pTA-Luc (Clontech), which lacks the enhancer element, was used to determine the background. Plasmid pRL-CMV (Promega, Madison, WI), which expresses the Renilla luciferase protein, was used for normalization of transfection efficiency (17).
ISRE reporter screening.
Huh7 cells were seeded in 384-well plates at a density of 3.0 × 103 cells/well. An ISRE-responsive firefly luciferase reporter was introduced using Lipofectamine 2000 (Invitrogen). Five hours after transfection, the cells were treated with 60,500 compounds from chemical libraries at a concentration of 3 μg/ml for 24 h and then treated with α-IFN at a concentration of 3 IU/ml. Six hours later, cells were lysed, and luciferase activities were quantified using a Steady Glo luciferase assay kit (Promega). The compounds were stored in 100% DMSO, and thus, the final concentration of DMSO was 0.3%. Z′ factors were calculated as reported previously (46).
Luciferase assays and measurements of antiviral activity.
Huh7/Rep-Feo cells were cultured with various concentrations of compound, such that the final DMSO concentration was 0.1%. Levels of HCV replication were quantified by internal luciferase assay after 48 h of culture. Luciferase activities were quantified using a luminometer (Promega) and the Bright-Glo luciferase assay system (Promega). Assays were performed in triplicate, and the results are expressed as mean percentage of the controls ± standard deviation (SD). The 50% effective concentration (EC50) values were calculated using the probit method (2, 33). The determination of EC50s was performed three times, and EC50s are presented as means ± SDs for each compound.
MTS assays.
To evaluate cell viability, dimethylthiazol carboxymethoxyphenyl sulfophenyl tetrazolium (MTS) assays were performed using a Cell Titer 96 Aqueous One Solution cell proliferation assay (Promega) as previously reported (18, 22). Huh7/Rep-Feo cells and HCV-J6/JFH1-infected Huh7 cells were seeded in 96-well plates at a density of 8.0 × 103 cells/well. After treatment, to analyze the therapeutic index with the same concentration of the drug and administration time, 20 μl/well of Cell Titer 96 Aqueous One Solution reagent was added to the cells cultured in a 96-well plate, the plate was incubated at 37°C for 60 min, and then the absorbance at 490 nm was recorded with a 96-well plate reader. The cells were analyzed when the growth became confluent. Cell viability was expressed as the concentration required for 50% cytotoxicity (CC50). The drug selectivity index was calculated as CC50/EC50. All experiments were performed in triplicate.
Analyses of drug synergism.
The effects of treatment of Huh7/Rep-Feo cells with α-IFN, alone and in combination with TSN, were analyzed by using isobologram analysis as described previously (27, 37). Dose inhibition curves of α-IFN and TSN were drawn with the two drugs used alone or in combination. In each drug combination, EC50s for α-IFN and TSN were plotted against the fractional concentration of α-IFN and TSN on the x and y axes, respectively. A theoretical line of additivity is drawn between plots of the EC50 for either drug that was used alone. The combined effects of the two drugs were considered additive, synergistic, or antagonistic if the plots of the drug combination were located on the line, below, or above the line of additivity, respectively.
HCV-J6/JFH1 cell culture.
HCV-J6/JFH1 (21), which is a recombinant of HCV-JFH1 (42), was used. In vitro-synthesized HCV-J6/JFH1 RNA was transfected into naïve Huh7 cells (48), and the cells were cultured in the presence of drugs (34). Cellular viral RNA expression levels were measured using a real-time reverse transcription-PCR (RT-PCR) system.
Real-time RT-PCR analysis.
Real-time RT-PCR was carried out as described previously (7). Total cellular RNA was extracted from cultured cells using Isogen (Nippon Gene, Tokyo, Japan), reverse transcribed, and subjected to real-time RT-PCR analyses. Expression of mRNA was quantified using TaqMan Universal PCR master mix, an ABI 7500 real-time PCR system (Applied Biosystems, CA), and a QuantiTect SYBR green PCR kit (Qiagen, CA). Some primers have been described elsewhere (30, 34). The primers used were -S (5′-TTT GAA ACA TCA AAG TTT TTC ACA GAC CTA-3′), -AS (5′-CAC AGT CAA GGT CCT TAG TAT TTC AGA TGT-3′), p56-S (5′-ACT TCG GAG AAA GGC ATT AGA TCT GGA AAG-3′), p56-AS (5′-TAA GGA CCT TGT CTC ACA GAG TTC TCA AAG-3′), Viperin-S (5′-GCT ACC AAG AGG AGA AAG CA-3′), Viperin-AS (5′-TTG ATC TTC TCC ATA CCA GC-3′), ISG20-S (5′-CTA CGA CAC GTC CAC TGA CAG G-3′), ISG20-AS (5′-CAT CGT TGC CCT CGC ATC TTC-3′), IRF9-S (5′-GCA GCA GCA GCC CTG AGC CAC AGG AAG TTA-3′), IRF9-AS (5′-TTA CCT GGA ACT TCG GTG GGG GGC CCA GGC-3′), IFNAR1-S (5′-CTT TCA AGT TCA GTG GCT CC-3′), IFNAR1-AS (5′-CAT CAG ATG CTT GTA CGC GGA G-3′), IFNAR2-S (5′-GCC AGA ATG CCT TCA TCG TCA G-3′), and IFNAR2-AS (5′-GTG AGT TGG TAC AAT GGA GTG G-3′).
Western blotting.
Twenty micrograms of total cell lysate was separated by SDS-PAGE and blotted onto a polyvinylidene fluoride Western blotting membrane. The membrane was incubated with the primary antibodies, followed by incubation with a peroxidase-labeled anti-IgG antibody, and were visualized by chemiluminescence using an enhance chemiluminescence Western blotting analysis system (Amersham Biosciences, Buckinghamshire, United Kingdom). The antibodies used were mouse anti-NS5A (BioDesign, ME), rabbit anti-signal transducer and activator of transcription 1 (anti-STAT1) p84/p91, rabbit anti-phospho-STAT1 (Tyr 701), rabbit anti-STAT2, rabbit anti-phospho-STAT2 (Tyr 690) (Santa Cruz, CA), and anti-beta-actin antibody (Sigma). NIH image software was used to analyze the densitometry of the Western blot analysis. Quantification of STAT phosphorylation was done using NIH image software, and the results correspond to the ratio between the phosphorylated STAT1 (p-STAT1) or p-STAT2 amount and the STAT1 or STAT2 amount normalized to the amount for the control without α-IFN and TSN. The results correspond to the ratio between the NS5A amount and the beta-actin amount normalized to the amount for the control without α-IFN and TSN.
Statistical analyses.
Statistical analyses were performed using Student's t test. P values of less than 0.05 were considered statistically significant.
RESULTS
ISRE reporter screening.
At the primary screening (n = 1), we defined a 1.5-fold induction in response to α-IFN to be a hit compound, and the hit rate was about 1%. At the secondary screening (n = 4), we selected the compound whose cps were 2 SDs larger than that for the drug used as a negative control, and the hit compound rate was 0.2% of the original library. Both assays were highly reproducible, and reflecting this, the Z′ factor (46) for the ISRE reporter screen was 0.97.
TSN has activity against HCV RNA replication.
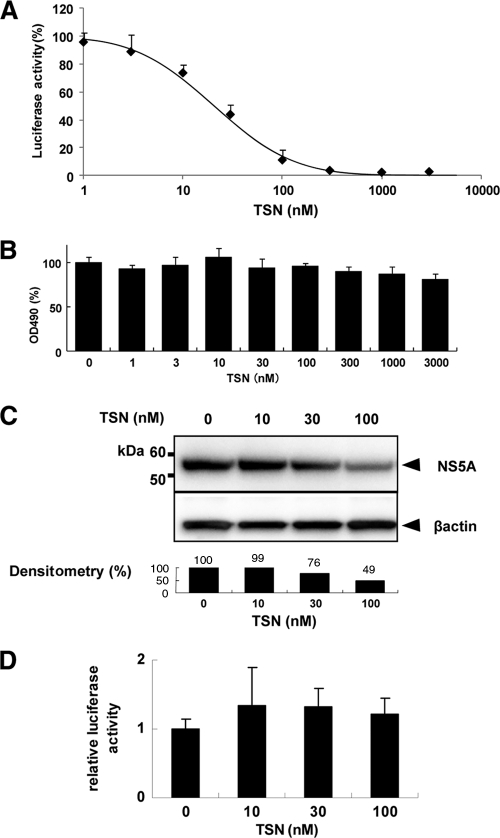
Huh7/Rep-Feo cells were cultured with various concentrations of TSN, and the effect was measured using a luciferase assay. TSN caused a marked suppression of HCV RNA replication in a dose-dependent manner (Fig. 2A). The EC50 of TSN was 20.6 nM. In contrast, MTS assays showed that treatment with TSN had little effect on cellular viability and replication, with a CC50 of over 3 μM and a selectivity index of more than 146. These results indicated that TSN had an effect against HCV RNA replication when it was used alone and that the effect was specific for HCV replication and not attributable to nonspecific cytotoxicity (Fig. 2B). Similarly, by Western blotting (Fig. 2C), the expression of HCV NS5A protein was shown to be reduced by corresponding amounts following treatment with TSN. To determine whether TSN suppresses HCV IRES-dependent translation, we used a Huh7 cell line that had been stably transfected with pCIneo-Rluc-IRES-Fluc (Fig. 2D). Treatment of these cells with TSN resulted in no significant change of the internal luciferase activities at concentrations of TSN that suppressed expression of the HCV replicon.
Fig. 2.
Effect of TSN on expression of HCV replicon. (A) HCV replicon cells were treated with various concentrations of TSN for 48 h. Replication levels of HCV RNA were analyzed by luciferase assay. Bars indicate luciferase activities relative to that of the drug-negative control. (B) Cell viability was determined by MTS assay. Bars indicate the value relative to that of the drug-negative control. (C) Western blotting analyses. The expression of NS5A and beta-actin was detected using anti-NS5A and anti-beta-actin antibodies. Densitometry of NS5A protein was performed, and the result is indicated as a percentage of the result for the drug-negative control. The assay was repeated three times, and a representative result is shown. (D) A bicistronic reporter gene plasmid, pCIneo-Rluc-IRES-Fluc, was transfected into Huh7 cells. The cells were cultured with TSN at the concentrations indicated, and dual luciferase activities were measured after 24 h of treatment. Values are displayed as ratios of Fluc to Rluc. In panels A, B, and D, the assays were done in triplicate and repeated three times. Error bars indicate means ± SDs.
TSN increased ISRE reporter activity with α-IFN.
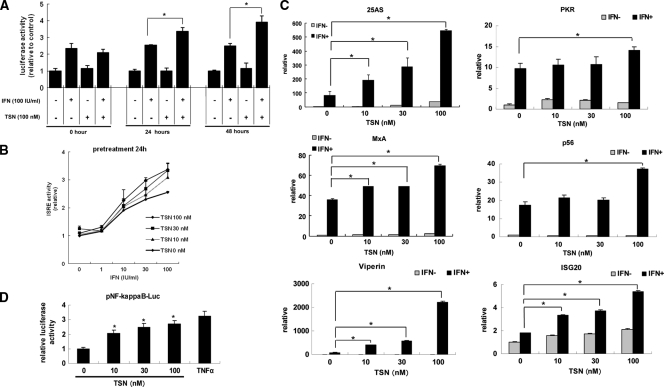
Because we identified TSN originally through ISRE reporter-based drug screening, we analyzed the effects of TSN on the cellular responses to α-IFN following pretreatment with TSN. First, we treated ISRE-TA-Luc-transfected Huh7 cells with TSN and α-IFN simultaneously or pretreated the cells with 10 to 100 nM TSN at 24 or 48 h prior to α-IFN treatment. Luciferase assays were performed 6 h after addition of α-IFN at concentrations of 0.1 to 100 IU/ml (Fig. 3A and B). Treatment with TSN alone did not increase ISRE reporter activity. Similarly, simultaneous treatment with TSN and α-IFN did not enhance α-IFN-induced ISRE reporter activation more than treatment with α-IFN alone. In contrast, pretreatment with TSN 24 or 48 h before addition of α-IFN significantly increased ISRE activation compared to that achieved by treatment with α-IFN alone (Fig. 3A). On the basis of these results, we performed the subsequent experiments with addition of TSN 24 h before α-IFN treatment.
Fig. 3.
ISRE reporter screening and aberrant pathway of α-IFN. (A) Pretreatment with TSN. Huh7 cells transfected with a reporter gene (pISRE-Luc and pRL-CMV) were pretreated with TSN (0 or 100 nM) for 0, 24, or 48 h, followed by treatment with α-IFN (0 or 100 IU/ml). Six hours later, the relative ISRE-luciferase activity (n = 4) was determined as described in Materials and Methods. The data are expressed as means ± SDs and are a representative example of the data from three similar experiments. (B) Pretreatment with TSN at the concentrations indicated for 24 h, followed by treatment with α-IFN (0 to 100 IU/ml). The ISRE reporter assay was performed as described for panel A. (C) Type I IFN-induced antiviral ISG expression in Huh7 cells. Huh7 cells were treated with TSN for 24 h, followed by treatment with α-IFN at 100 IU/ml for 24 h. The total cellular RNA was then isolated for real-time RT-PCR analysis of the mRNAs of 25AS, MxA, p56, viperin, and ISG20. Beta-actin was used as a control. The data are expressed as means ± SDs and are a representative example of data from three similar experiments. *, P < 0.05. (D) Analysis of aberrant pathways of α-IFN signaling under the influence of TSN. Promoter activities of NF-kappaB were analyzed by luciferase reporter assays. These cells were transfected with pNF-kappaB-TA-Luc, pTA-Luc which lacks the enhancer element and which was used as a negative control, and pRL-CMV to normalize transfection efficiency. At 24 h after transfection with these reporters, treatment with TSN (0, 10, 30, 100 nM) was carried out. After 24 h, the relative levels of induction of NF-kappaB activity for each treatment were calculated. TNF-α (50 ng/ml), which was used as a positive control for NF-kappaB, was added 6 h before analysis. The assays were done in triplicate and repeated three times. Error bars indicate means ± SDs. *, P < 0.05.
We next quantified the expression levels of ISGs, including those for 2′,5′-oligoadenylate synthetase (25AS), MxA, protein kinase R, p56, viperin, and ISG20, which encode proteins with direct antiviral activity (14, 15, 25). Naïve Huh7 cells were treated with TSN for 24 h, followed by treatment with 100 IU/ml α-IFN for 24 h. The expression of each ISG was significantly elevated in a dose-dependent manner following pretreatment with TSN and α-IFN stimulation (Fig. 3C). These results indicated that TSN pretreatment significantly enhanced the cellular response to α-IFN-induced, ISRE-regulated expression of ISGs.
It has been reported that α-IFN receptor-mediated signaling cross talks with several alternative pathways, including the NF-kappaB, gamma IFN, phosphatidylinositol 3-kinase (PI3K), and mitogen-activated protein kinase (MAPK) pathways (9, 16, 24, 28). Therefore, we analyzed the effect of TSN on the signaling pathways indicated above. Cells were transfected with various reporter plasmids, including NF-kappaB, gamma interferon activation site (GAS), or activator protein 1 (AP1) Fluc plasmids. The reporter activities were measured after culture with or without TSN. As shown in Fig. 3D, there was no significant effect of TSN on GAS or AP1 reporter activities (data not shown). In contrast, NF-kappaB reporter activity was significantly elevated by TSN in a dose-dependent fashion.
Synergistic inhibitory effects of TSN and α-IFN on the replicon.
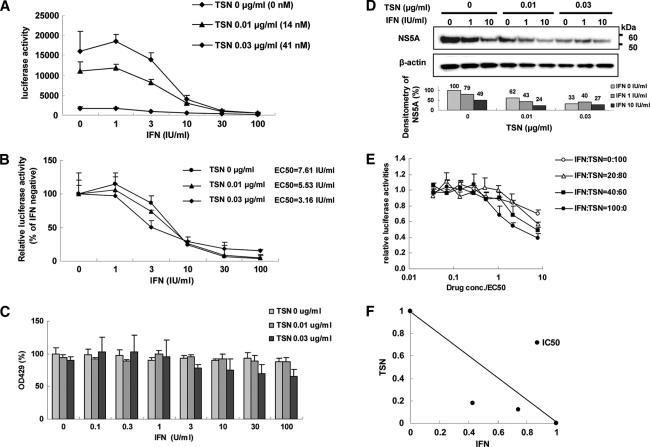
We next assessed the effects of TSN combination with α-IFN on the intracellular replication of the HCV genome. Huh7/Rep-Feo cells were treated with various concentrations of TSN (0, 0.01, and 0.03 μg/ml) and α-IFN (0 to 100 IU/ml). Replication of the HCV replicon was suppressed by pretreatment with TSN, followed by treatment with α-IFN, in a dose-dependent manner (Fig. 4A; see Fig. S1 in the supplemental material). The EC50 of α-IFN in the absence of TSN was 7.61 IU/ml, while that after pretreatment with 0.03 μg/ml (41 nM) TSN was 3.16 IU/ml. These results indicated that pretreatment with TSN before α-IFN treatment is more effective in inhibiting HCV replication than treatment with α-IFN alone.
Fig. 4.
Suppression of HCV RNA replication by TSN combined with α-IFN. (A and B) Luciferase activity (A, absolute value; B, relative value). Huh7/Rep-Feo cells, which constitutively express an HCV replicon, enable the quantification of replication levels through the measurement of luciferase activity. Absolute and relative dose-response curves in the presence of 24 h of pretreatment of various concentrations of TSN (0, 0.01, 0.03 μg/ml) and α-IFN (0, 100 IU/ml). (A) Bars indicate luciferase activities. (B) Bars indicate luciferase activities relative to the activity of each α-IFN-negative control. Luciferase assays were performed in triplicate. Error bars indicate means ± SDs. (C) MTS assay of Huh7/Rep-Feo cells cultured with the indicated concentrations of TSN and α-IFN. The assays were done in triplicate and repeated three times. Error bars indicate means ± SDs. (D) Western blotting. Ten micrograms of total cellular protein was separated by polyacrylamide gel electrophoresis and transferred onto the membrane. Monoclonal anti-NS5A antibody or an anti-beta-actin antibody was used as the primary antibody. Densitometry of NS5A or beta-actin protein was performed and the result is indicated as a percentage of that for the drug-negative control. The assay was repeated three times, and representative results are shown. (E) Dose-inhibition curves of α-IFN and TSN when they were combined at the indicated ratios, adjusted by the EC50 of the individual drug. Assays were done in triplicate, and mean values were plotted and indicated as means ± SDs. (F) Graphical representation of the isobologram analysis. For each drug combination in panel E, the EC50s of α-IFN and TSN for inhibition of HCV replication were plotted against the fractional concentrations of α-IFN and TSN, which are indicated on the x and y axes, respectively. A theoretical line of additivity is drawn between the EC50 for each drug alone. All of the fractional EC50 plots for the TSN and α-IFN combinations fell below the line of additivity, indicating synergy.
Subsequently, we conducted the following assay to determine whether TSN and α-IFN have a synergistic inhibitory effect on the replicon. The relative dose-inhibition curves of α-IFN were plotted for several concentrations of TSN and α-IFN. The curves shifted to the left with increasing concentrations of TSN (Fig. 4B), demonstrating that HCV replication was considerably reduced by the combination compared with that by either TSN or α-IFN alone. An MTS-based cell viability assay did not show significant cytotoxicity from TSN (Fig. 4C). Western blot analysis and densitometry of each blot showed results essentially identical to those from the luciferase assay (Fig. 4D).
We used isobologram analysis to determine whether the anti-HCV effect of TSN is synergistic with that of α-IFN (27, 37). Huh7/Rep-Feo cells were treated with a combination of α-IFN and TSN at an EC50 ratio of 1:0, 2:3, 1:4, or 0:1, and the dose-effect plots were drawn (Fig. 4E). The fractional EC50s for α-IFN and TSN were plotted on the x and y axes, respectively, to generate an isobologram (Fig. 4F). Each plot showing the fractional EC50 of each drug ratio fell below the line showing additivity, indicating that the effect of the drug combination on intracellular HCV RNA replication was synergistic. The MTS values at the drug concentrations used in this isobologram analysis did not show any significant decrease, suggesting that the synergistic action of α-IFN and TSN on HCV replication is through their pharmacological effects and is not due to augmentation of cytotoxicity.
Suppression of HCV-J6/JFH1 infection by pretreatment of TSN with α-IFN.
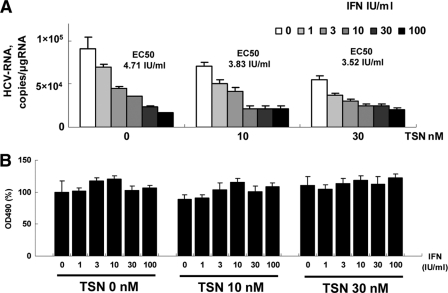
The inhibitory effects of pretreatment with TSN prior to α-IFN treatment demonstrated on HCV subgenomic replication were validated further using HCV-J6/JFH1 cell culture (21, 42). Various concentrations of TSN and α-IFN were added to HCV-J6/JFH1-infected Huh7 cells, and intracellular HCV RNA was quantified after 48 h of incubation. As shown in Fig. 5A, TSN with or without α-IFN suppressed expression of intracellular HCV RNA in a dose-dependent manner. The EC50s of α-IFN with TSN at 0, 10, and 30 nM were 4.71 IU/ml, 3.83 IU/ml, and 3.52 IU/ml, respectively. An MTS-based cell viability assay did not show significant cytotoxicity from TSN (Fig. 5B). These data indicate that pretreatment with TSN also augmented the α-IFN effect on the JFH1 system.
Fig. 5.
Suppression of full HCV-J6/JFH1 replication by pretreatment of TSN with α-IFN. Ten micrograms of HCV-J6/JFH1 RNA was transfected into Huh7 cells. At 48 h after transfection, cells were pretreated with TSN for 24 h, followed by treatment with α-IFN (0, 1, 3, 10, 30, 100 IU/ml). At 48 h after α-IFN addition, cells were harvested. (A) Real-time RT-PCR analysis; (B) effect of pretreatment TSN with α-IFN on cell viability. MTS assays were performed 48 h after culture in the presence of pretreatment TSN with α-IFN. Bars indicate values relative to that of the drug-negative control. In panels A and B, the assays were done in triplicate and repeated three times. Error bars indicate means ± SDs.
TSN upregulates ISGF3 in combination with α-IFN.
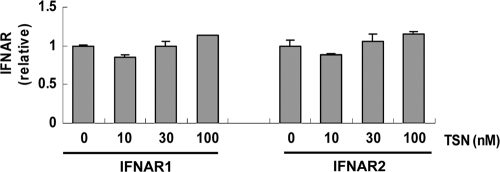
Subsequently, we performed experiments to investigate the mechanisms of action of TSN. First, we quantified expression of alpha/beta IFN receptor subunit (IFNAR) 1 and IFNAR2 and the effect of TSN. Real-time RT-PCR analysis showed no change in levels of IFNAR1 and IFNAR2 mRNA expression with or without TSN (Fig. 6).
Fig. 6.
IFNAR expression. Huh7 cells were pretreated with TSN for 24 h, followed by treatment with 100 IU/ml α-IFN for 6 h. The total cellular RNA was then isolated for real-time RT-PCR analysis of the mRNAs of IFNAR1 and IFNAR2. The assays were done in triplicate and repeated three times. The data are shown as means ± SDs.
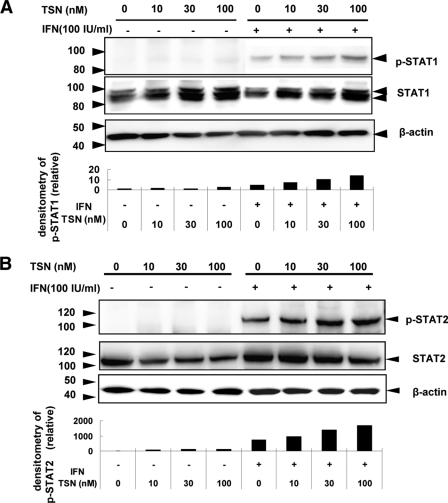
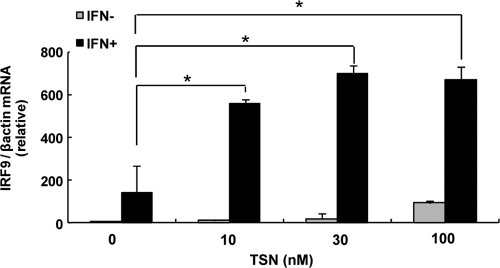
Next, we investigated the ISGF3 components, STAT1, and STAT2, using Western blotting, and interferon regulatory factor 9 (IRF9), using real-time RT-PCR. Huh7 cells were treated with various concentrations of TSN or 0.01% DMSO. Twenty-four hours after TSN treatment, 100 IU/ml of α-IFN was added, and STATs and IRF9 were detected. Western blot analysis demonstrated that phosphorylated STAT1 and STAT2 levels were increased more by treatment with α-IFN and TSN than by α-IFN treatment alone (Fig. 7A and B). In addition, IRF9 mRNA expression was significantly higher following pretreatment with TSN prior to α-IFN therapy than by α-IFN monotherapy (Fig. 8). These findings are consistent with the hypothesis that TSN activates ISGF3 components in combination with α-IFN.
Fig. 7.
TSN with α-IFN treatment of Huh7 cells increases phosphorylation of STAT1 and STAT2. (A) Western blotting. Alteration in the distribution of α-IFN-induced phosphorylation of STAT1 and STAT2 by TSN. Huh7 cells were treated with TSN or 0.01% DMSO for 24 h. After that, the cells were stimulated by 100 IU/ml α-IFN for 30 min. Cells were harvested, and the resulting lysates were analyzed for phosphorylated and total STAT1 or STAT2. The relative amounts of phosphorylated STAT1 or STAT2 were normalized to the amount of total STAT1 or STAT2 and expressed relative to the amount for the drug-negative control. The assay was repeated three times, and a representative result is shown.
Fig. 8.
IRF9 mRNA expression after combination treatment with TSN and α-IFN. Real-time RT-PCR analysis. Huh7 cells were treated with TSN for 24 h. After 6 h, the cells were stimulated by α-IFN (100 IU/ml). We used the method described in the legend to Fig. 6 to analyze the mRNA of IRF9. The assays were done in triplicate and repeated three times. Error bars indicate means ± SDs. *, P < 0.05.
DISCUSSION
In this study, we investigated the molecular actions of TSN on HCV replication and on α-IFN-mediated cellular antiviral responses. Treatment of cells expressing an HCV subgenomic replicon with TSN alone specifically inhibited HCV replication with a selectivity index of more than 146 (Fig. 2). In addition, pretreatment of cells with TSN prior to addition of α-IFN augmented α-IFN receptor-mediated, ISRE-regulated gene expression (Fig. 3). Consistent with these findings, TSN pretreatment significantly enhanced the suppressive effects of α-IFN on the HCV replicon and HCV cell culture (Fig. 4 and 5). Finally, we demonstrated that the α-IFN-enhancing effects of TSN are through increased transcriptional activation of a component of ISGF3 (Fig. 7 and 8). Taken together, out results demonstrate that TSN is potentially an effective antiviral agent when it is used alone and especially when it is used in combination with α-IFN and that screening for such α-IFN-enhancing agents may identify promising antiviral therapeutics. Because TSN treatment alone or simultaneous treatment with TSN and α-IFN did not increase ISRE activity or augment α-IFN-mediated ISRE activation, TSN may affect α-IFN sensitivity by upregulating molecules that affect α-IFN receptor-mediated signaling without activating ISRE signaling directly.
Type I interferon plays a central role in eliminating viruses through its innate antiviral activity or following therapeutic application. Binding of α-IFNs to their receptors activates the Jak-STAT pathway to form a complex with ISGF3, which translocates to the nucleus, binds the ISRE located in the promoter/enhancer region of the ISGs, and activates expression of ISGs (28, 39, 40). In this study, we demonstrated that TSN enhanced α-IFN effects by upregulating ISGF3, which may cancel the suppressive effect of HCV gene products on the α-IFN signaling pathway.
In our study, it was not proved that increasing ISRE activities had direct relevance to inhibition of HCV replication. In Fig. 3C, we showed that TSN with α-IFN treatment had elevated the level of expression of mRNA of ISGs. Previous studies suggested that overexpression of known ISGs inhibited HCV replication in HCV replicon-containing Huh7 cells (13, 14). These findings may support the possibility that TSN had the potential to augment the α-IFN effect.
Other than the canonical Jak/STAT-mediated α-IFN signaling pathway, several alternative α-IFN pathways have been reported, including the NF-kappaB, gamma IFN, PI3K, and MAPK pathways (9, 16, 24, 28). We carried out reporter assays using NF-kappaB, AP1, and GAS reporter plasmid constructs and treated the cells with TSN. As shown in Fig. 3D, TSN activated NF-kappaB-regulated gene expression significantly. NF-kappaB is a sequence-specific transcription factor which regulates the expression of numerous cellular and viral genes and plays important roles in inflammation, innate immune responses, tumorigenesis, and cell survival (3, 19). Activation of NF-kappaB is principally regulated by tumor necrosis factor alpha (TNF-α), Toll-like receptors (TLRs), and RIG-I, which may possibly be associated with the molecular mechanisms of TSN monotherapy. Horsmans et al. (12) and Agrawal and Kandimalla (1) reported that TLR7, -8, and -9 agonists have the ability to modulate TLR-mediated immune responses in targeting a broad range of disease vectors, including HCV, alone or in combination with other therapeutic agents. These reports support the hypothesis that activation of NF-kappaB may be one of the mechanisms of action of TSN.
It has been reported that TSN exhibits cytotoxic/antiproliferative potential at high concentrations (36, 47). In our study, the selectivity index of TSN against HCV was sufficient to ascertain that the antiviral effects are not simply due to the cytotoxicity of TSN. A recent study showed that a triterpenoid compound, dammarenolic acid, inhibits retrovirus, human immunodeficiency virus, simian immunodeficiency virus, murine leukemia virus, and respiratory syncytial virus infections in vitro (4) (5). We have analyzed the effects of dammarenolic acid on antiviral actions on Huh7/Rep-Feo cells, cytotoxicity, and ISRE reporter activation. However, dammarenolic acid did not inhibit HCV replication or enhanced α-IFN-induced ISRE activity (data not shown). These findings suggest that the anti-HCV and α-IFN enhancer effects are distinctive features of TSN among triterpenoid compounds. Hiasa et al. have reported that ME3738, a triterpenoid saponin, suppressed HCV replication through production of endogenous beta interferon (11). ME3738 is now in clinical trials for treatment of HCV-infected patients. Taking these findings together, despite reports on the cell-suppressive effect of triterpenoids, properly selected or designed compounds might be used as drugs against HCV infection.
Because the mechanisms of action of these triterpenoid compounds against these viruses are poorly understood, further investigation of the mechanism of action of TSN on HCV may be valuable to implement antiviral strategies against other viruses. It would be important to assess drug resistance after continuous treatment with TSN. There is no in vitro or in vivo report on resistance of TSN or cellular attenuation of responses to TSN. Such information, if any is found, would help elucidate the mechanism of action of TSN.
Given the current situation of limited therapeutic options against HCV, the search for more potent and less toxic antiviral drugs is needed to improve clinical anti-HCV chemotherapeutics. Several direct antiviral agents with activity against HCV are currently undergoing clinical trials. These include NS3 protease inhibitors and NS5B polymerase inhibitors (41). However, the frequent emergence of drug-resistant viruses is a major weakness of such agents (20). Our results indicate that TSN is also effective at suppressing HCV infection and replication. Future studies with TSN, its derivatives, and other chemicals that target the α-IFN pathway could be directed toward developing a new class of antiviral treatment regimens and drugs.
Supplementary Material
ACKNOWLEDGMENTS
We thank Frank Chisari for providing Huh7.5.1 cells, Charles Rice for providing plasmid pJ6/JFH1full, and Takaji Wakita for providing plasmid pJFH1full.
This study was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan, the Japan Society for the Promotion of Science, the Ministry of Health, Labor and Welfare of Japan, the Japan Health Sciences Foundation, the National Institute of Biomedical Innovation, and the Miyakawa Memorial Research Foundation.
Footnotes
Supplemental material for this article may be found at http://aac.asm.org/.
Published ahead of print on 28 March 2011.
REFERENCES
- 1. Agrawal S., Kandimalla E. R. 2007. Synthetic agonists of Toll-like receptors 7, 8 and 9. Biochem. Soc. Trans. 35:1461–1467 [DOI] [PubMed] [Google Scholar]
- 2. Bailey M., Williams N. A., Wilson A. D., Stokes C. R. 1992. PROBIT: weighted probit regression analysis for estimation of biological activity. J. Immunol. Methods 153:261–262 [DOI] [PubMed] [Google Scholar]
- 3. Baldwin A. S., Jr 2001. Series introduction: the transcription factor NF-kappaB and human disease. J. Clin. Invest. 107:3–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Esimone C. O., et al. 2008. Potential anti-respiratory syncytial virus lead compounds from Aglaia species. Pharmazie 63:768–773 [PubMed] [Google Scholar]
- 5. Esimone C. O., et al. Dammarenolic acid, a secodammarane triterpenoid from Aglaia sp. shows potent anti-retroviral activity in vitro. Phytomedicine 17:540-547 [DOI] [PubMed] [Google Scholar]
- 6. Fried M. W., et al. 2002. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N. Engl. J. Med. 347:975–982 [DOI] [PubMed] [Google Scholar]
- 7. Funaoka Y., et al. Analysis of interferon signaling by infectious hepatitis C virus clones with substitutions of core amino acids 70 and 91. J. Virol., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ge D., et al. 2009. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature 461:399–401 [DOI] [PubMed] [Google Scholar]
- 9. Goodbourn S., Didcock L., Randall R. E. 2000. Interferons: cell signalling, immune modulation, antiviral response and virus countermeasures. J. Gen. Virol. 81:2341–2364 [DOI] [PubMed] [Google Scholar]
- 10. Hadziyannis S. J., et al. 2004. Peginterferon-alpha2a and ribavirin combination therapy in chronic hepatitis C: a randomized study of treatment duration and ribavirin dose. Ann. Intern. Med. 140:346–355 [DOI] [PubMed] [Google Scholar]
- 11. Hiasa Y., et al. 2008. Hepatitis C virus replication is inhibited by 22beta-methoxyolean-12-ene-3beta, 24(4beta)-diol (ME3738) through enhancing interferon-beta. Hepatology 48:59–69 [DOI] [PubMed] [Google Scholar]
- 12. Horsmans Y., et al. 2005. Isatoribine, an agonist of TLR7, reduces plasma virus concentration in chronic hepatitis C infection. Hepatology 42:724–731 [DOI] [PubMed] [Google Scholar]
- 13. Itsui Y., et al. 2009. Antiviral effects of the interferon-induced protein guanylate binding protein 1 and its interaction with the hepatitis C virus NS5B protein. Hepatology 50:1727–1737 [DOI] [PubMed] [Google Scholar]
- 14. Itsui Y., et al. 2006. Expressional screening of interferon-stimulated genes for antiviral activity against hepatitis C virus replication. J. Viral Hepat. 13:690–700 [DOI] [PubMed] [Google Scholar]
- 15. Jiang D., et al. 2008. Identification of three interferon-inducible cellular enzymes that inhibit the replication of hepatitis C virus. J. Virol. 82:1665–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kalvakolanu D. V. 2003. Alternate interferon signaling pathways. Pharmacol. Ther. 100:1–29 [DOI] [PubMed] [Google Scholar]
- 17. Kanazawa N., et al. 2004. Regulation of hepatitis C virus replication by interferon regulatory factor 1. J. Virol. 78:9713–9720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Karakama Y., et al. 2010. Inhibition of hepatitis C virus replication by a specific inhibitor of serine-arginine-rich protein kinase. Antimicrob. Agents Chemother. 54:3179–3186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Karin M., Lin A. 2002. NF-kappaB at the crossroads of life and death. Nat. Immunol. 3:221–227 [DOI] [PubMed] [Google Scholar]
- 20. Kuntzen T., et al. 2008. Naturally occurring dominant resistance mutations to hepatitis C virus protease and polymerase inhibitors in treatment-naive patients. Hepatology 48:1769–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lindenbach B. D., et al. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623–626 [DOI] [PubMed] [Google Scholar]
- 22. Mishima K., et al. 2010. Cell culture and in vivo analyses of cytopathic hepatitis C virus mutants. Virology 405:361–369 [DOI] [PubMed] [Google Scholar]
- 23. Nakagawa M., et al. 2005. Suppression of hepatitis C virus replication by cyclosporin a is mediated by blockade of cyclophilins. Gastroenterology 129:1031–1041 [DOI] [PubMed] [Google Scholar]
- 24. Randall R. E., Goodbourn S. 2008. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 89:1–47 [DOI] [PubMed] [Google Scholar]
- 25. Sadler A. J., Williams B. R. 2008. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 8:559–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sakamoto N., Watanabe M. 2009. New therapeutic approaches to hepatitis C virus. J. Gastroenterol. 44:643–649 [DOI] [PubMed] [Google Scholar]
- 27. Sakamoto N., et al. 2007. Bone morphogenetic protein-7 and interferon-alpha synergistically suppress hepatitis C virus replicon. Biochem. Biophys. Res. Commun. 357:467–473 [DOI] [PubMed] [Google Scholar]
- 28. Samuel C. 2001. Antiviral actions of interferons. Clin. Microbiol. Rev. 14:778–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sangiovanni A., et al. 2006. The natural history of compensated cirrhosis due to hepatitis C virus: a 17-year cohort study of 214 patients. Hepatology 43:1303–1310 [DOI] [PubMed] [Google Scholar]
- 30. Sekine-Osajima Y., et al. 2008. Development of plaque assays for hepatitis C virus-JFH1 strain and isolation of mutants with enhanced cytopathogenicity and replication capacity. Virology 371:71–85 [DOI] [PubMed] [Google Scholar]
- 31. Shi Y. L., Li M. F. 2007. Biological effects of toosendanin, a triterpenoid extracted from Chinese traditional medicine. Prog. Neurobiol. 82:1–10 [DOI] [PubMed] [Google Scholar]
- 32. Shi Y. L., Wang Z. F. 2004. Cure of experimental botulism and antibotulismic effect of toosendanin. Acta Pharmacol. Sin. 25:839–848 [PubMed] [Google Scholar]
- 33. Soothill J. S., Ward R., Girling A. J. 1992. The IC50: an exactly defined measure of antibiotic sensitivity. J. Antimicrob. Chemother. 29:137–139 [DOI] [PubMed] [Google Scholar]
- 34. Suda G., et al. IL-6-mediated intersubgenotypic variation of interferon sensitivity in hepatitis C virus genotype 2a/2b chimeric clones. Virology 407:80–90 [DOI] [PubMed] [Google Scholar]
- 35. Suppiah V., et al. 2009. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat. Genet. 41:1100–1104 [DOI] [PubMed] [Google Scholar]
- 36. Tada K., Takido M., Kitanaka S. 1999. Limonoids from fruit of Melia toosendan and their cytotoxic activity. Phytochemistry 51:787–791 [DOI] [PubMed] [Google Scholar]
- 37. Tanabe Y., et al. 2004. Synergistic inhibition of intracellular hepatitis C virus replication by combination of ribavirin and interferon-alpha. J. Infect. Dis. 189:1129–1139 [DOI] [PubMed] [Google Scholar]
- 38. Tanaka Y., Nishida N., Sugiyama M., Tokunaga K., Mizokami M. Lambda-interferons and the single nucleotide polymorphisms: a milestone to tailor-made therapy for chronic hepatitis C. Hepatol. Res. 40:449–460 [DOI] [PubMed] [Google Scholar]
- 39. Taniguchi T., Ogasawara K., Takaoka A., Tanaka N. 2001. IRF family of transcription factors as regulators of host defense. Annu. Rev. Immunol. 19:623–655 [DOI] [PubMed] [Google Scholar]
- 40. Taniguchi T., Takaoka A. 2002. The interferon-alpha/beta system in antiviral responses: a multimodal machinery of gene regulation by the IRF family of transcription factors. Curr. Opin. Immunol. 14:111–116 [DOI] [PubMed] [Google Scholar]
- 41. Thompson A. J., McHutchison J. G. 2009. Antiviral resistance and specifically targeted therapy for HCV (STAT-C). J. Viral Hepat. 16:377–387 [DOI] [PubMed] [Google Scholar]
- 42. Wakita T., et al. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yokota T., et al. 2003. Inhibition of intracellular hepatitis C virus replication by synthetic and vector-derived small interfering RNAs. EMBO Rep. 4:602–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zeuzem S., et al. 2000. Peginterferon alfa-2a in patients with chronic hepatitis C. N. Engl. J. Med. 343:1666–1672 [DOI] [PubMed] [Google Scholar]
- 45. Zhang B., Wang Z. F., Tang M. Z., Shi Y. L. 2005. Growth inhibition and apoptosis-induced effect on human cancer cells of toosendanin, a triterpenoid derivative from Chinese traditional medicine. Invest. New Drugs 23:547–553 [DOI] [PubMed] [Google Scholar]
- 46. Zhang J. H., Chung T. D., Oldenburg K. R. 1999. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 4:67–73 [DOI] [PubMed] [Google Scholar]
- 47. Zhang Y., et al. 2008. Roles of reactive oxygen species and MAP kinases in the primary rat hepatocytes death induced by toosendanin. Toxicology 249:62–68 [DOI] [PubMed] [Google Scholar]
- 48. Zhong J., et al. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. U. S. A. 102:9294–9299 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.