Abstract
PSI-352938 is a novel cyclic phosphate prodrug of β-d-2′-deoxy-2′-α-fluoro-2′-β-C-methylguanosine 5′-monophosphate that has potent activity against hepatitis C virus (HCV) in vitro. The studies described here characterize the in vitro anti-HCV activity of PSI-352938, alone and in combination with other inhibitors of HCV, and the cross-resistance profile of PSI-352938. The effective concentration required to achieve 50% inhibition for PSI-352938, determined using genotype 1a-, 1b-, and 2a-derived replicons stably expressed in the Lunet cell line, were 0.20, 0.13, and 0.14 μM, respectively. The active 5′-triphosphate metabolite, PSI-352666, inhibited recombinant NS5B polymerase from genotypes 1 to 4 with comparable 50% inhibitory concentrations. In contrast, PSI-352938 did not inhibit the replication of hepatitis B virus or human immunodeficiency virus in vitro. PSI-352666 did not significantly affect the activity of human DNA and RNA polymerases. PSI-352938 and its cyclic phosphate metabolites did not affect the cyclic GMP-mediated activation of protein kinase G. Clearance studies using replicon cells demonstrated that PSI-352938 cleared cells of HCV replicon RNA and prevented replicon rebound. An additive to synergistic effect was observed when PSI-352938 was combined with other classes of HCV inhibitors, including alpha interferon, ribavirin, NS3/4A inhibitors, an NS5A inhibitor, and nucleoside/nucleotide and nonnucleoside inhibitors. Cross-resistance studies showed that PSI-352938 remained fully active against replicons containing the S282T or the S96T/N142T amino acid alteration. Replicons that contain mutations conferring resistance to various classes of nonnucleoside inhibitors also remained sensitive to inhibition by PSI-352938. PSI-352938 is currently being evaluated in a phase I clinical study in genotype 1-infected individuals.
INTRODUCTION
Hepatitis C virus (HCV), an important member of the Flaviviridae family of viruses, is a major health problem affecting approximately 170 million people worldwide. In the United States, where it is the leading cause of liver transplantation, nearly 2% of the U.S. population are HCV carriers (2). The current standard of care (SOC), which consists of pegylated alpha interferon (IFN-α) and ribavirin, results in only about a 50% sustained virological response in patients infected with genotype 1 HCV, the predominant genotype in the United States and Europe (13, 18). The limited cure rate and potential side effects associated with interferon and ribavirin (10, 11) prompted the development of direct-acting antiviral agents (DAAs) in order to improve clinical efficacy and tolerability.
Among the DAAs currently in clinical development, nucleoside/nucleotide analogs as inhibitors of HCV replication have the advantage of broad genotype coverage and a high barrier to resistance (14). The active 5′-triphosphates of nucleoside/nucleotide analogs target the HCV NS5B RNA-dependent RNA polymerase (Pol) by serving as alternative substrate inhibitors during RNA synthesis (6). Both of the two resistance mutations identified so far, S96T and S282T, are highly conserved residues and unfit for HCV replicon replication (33). Among the nucleoside/nucleotide analogs, RG7128 (the prodrug of PSI-6130, β-d-2′-fluoro-2′-C-methylcytidine) is the most advanced anti-HCV nucleoside and is currently in phase IIb clinical studies. So far, results from the clinical studies showed that RG7128 is generally safe and effective in reducing the viral loads for genotype 1, 2, 3, and 4 HCV-infected patients when it is combined with SOC, with no resistance-related breakthrough (16, 21, 27, 37).
More recently, we reported results of in vitro studies characterizing PSI-7851 and PSI-7977 (the single Sp isomer of PSI-7851), phosphoramidate prodrugs of β-d-2′-fluoro-2′-C-methyluridine 5′-monophosphate (30, 45, 48). While RG7128 was designed to improve the pharmacokinetic profile of PSI-6130, PSI-7851 and PSI-7977 were synthesized to bypass the nonproductive first phosphorylation step, as the corresponding nucleoside analog was incapable of being converted to the monophosphate form. In a 3-day phase I monotherapy study, a 1.95-log viral load reduction was observed in HCV-infected patients administered 400 mg of PSI-7851 once a day (32). In a phase IIa clinical study of PSI-7977 in combination with SOC, rapid virological response (RVR) rates of 88 to 94% were observed in HCV-infected patients, whereas the RVR was 21% in patients receiving placebo and SOC (31). PSI-7977 is currently being evaluated in genotype 2/3-infected patients in a phase IIb clinical trial with promising interim results: in all HCV-infected patients treated with PSI-7977 and SOC, there was a rapid viral suppression and the level of virus remained below the limit of detection through the end of treatment at week 12 (28).
PSI-352938, a cyclic phosphate prodrug of β-d-2′-deoxy-2′-α-fluoro-2′-β-C-methylguanosine-5′-monophosphate, is currently being evaluated in a phase I clinical trial. PSI-352938 was synthesized as a nucleotide prodrug to bypass the rate-limiting first phosphorylation step in the metabolism of the parent nucleoside to the active triphosphate, β-d-2′-deoxy-2′-α-fluoro-2′-β-C-methylguanosine-5′-triphosphate (PSI-352666) (47). Structure-activity relationship (SAR) studies showed that PSI-352938 was about 60-fold more active than its corresponding nucleoside analog, with no significant cellular and mitochondrial toxicity, and produced high levels of triphosphate in both primary human hepatocytes and the liver of rats administered a single dose of PSI-352938 of 50 mg/kg of body weight (47). Herein we report on the activity of PSI-352938 and its triphosphate metabolite in a panel of cell-based HCV and NS5B polymerase assays demonstrating that PSI-352938 has pan-genotype anti-HCV activity. Replicon clearance studies were performed to determine the ability of PSI-352938 to clear the HCV replicon and prevent replicon RNA rebound. We also evaluated the anti-HCV effect of combining PSI-352938 with other classes of HCV inhibitors, including IFN-α, ribavirin, and compounds that target the NS3 protease, NS4A cofactor, NS5A phosphoprotein, and NS5B polymerase. Finally, we evaluated the activity of PSI-352938 against replicons containing mutations in the NS5B polymerase that confer resistance to nucleoside and nonnucleoside inhibitors.
MATERIALS AND METHODS
Compounds.
PSI-352938 (CAS registry number 199809-32-9), PSI-354091 {(4aR,6R,7R,7aR)-6-(2-amino-6-ethoxy-purin-9-yl)-7-fluoro-7-methyl-2-oxo-tetrahydro-2λ5-furo[3,2-d][1,3,2]dioxaphosphinin-2-ol}, PSI-354519 {2-amino-9-[(4aR,6R,7R,7aR)-7-fluoro-2-hydroxy-7-methyl-2-oxo-tetrahydro-2λ5-furo[3,2-d][1,3,2]dioxaphosphinin-6-yl]-9H-purin-6-ol}, PSI-7851 (CAS registry number 1064684-44-1), d-ddFCTP (β-d-2′,3′-dideoxy-5-fluorocytidine 5′-triphosphate), R1479 (4′-azidocytidine), INX-08189, ITMN-191, BMS-790052, ACH-806, and the nonnucleoside NS5B inhibitors (NNIs) that include the benzofuran compound 5-cyclopropyl-2-(4-fluoro-phenyl)-6-[(2-hydroxy-ethyl)-methanesulfonyl-amino]-benzofuran-3-carboxylic acid methylamide (HCV-796), thiophene compound 3-[(1R,4R)-N-isopropyl-4-methylcyclohexanecarboxamido]-5-phenylthiophene-2-carboxylic acid, benzothiadiazine compound N-{3-[4-hydroxy-1-(3-methyl-butyl)-2-oxo-1,2-dihydro-pyrrolo[1,2-b]pyridazin-3-yl]-1,1-dioxo-1,4-dihydro-1λ6-benzo[1,2,4]thiadiazin-7-yl}-N-methyl-methanesulfonamide, and the 2-phenol indole compound 5a-amino-12-cyclohexyl-N-(N,N-dimethylsulfamoyl)-3-methoxy-4b,5,5a,6-tetrahydrobenzo [3,4] cyclopropa [5,6] azepino [1,2-a] indole-9-carboxamide were synthesized at Pharmasset, Inc. PSI-352666 (2′-fluoro-2′-C-methylguanosine-5′-triphosphate) was synthesized by NuBlocks (Vista, CA). [α-32P]dCTP, [α-32P]UTP, and [α-32P]GTP were purchased from Perkin Elmer (Waltham, MA). 3′-dCTP was purchased from Trilink Biotechnology (San Diego, CA). Aphidicolin and α-amanitin were purchased from Sigma (St. Louis, MO).
Cells and viruses.
Clone A (Apath LLC, Brooklyn, NY) and Huh 5-2 cells (kindly provided by R. Bartenschlager, University of Heidelberg, Heidelberg, Germany), which contain a genotype 1b replicon derived from the Con1 strain (GenBank accession no. AJ238799.1), were maintained in medium containing 0.25 mg/ml G418 as described previously (30, 51). The genotype 1b-derived ET replicon (Con1 strain) with adaptative mutations E1202G, T1280I, and K1846T (38) and the Lunet cell line (25) were kindly provided by R. Bartenschlager (University of Heidelberg, Heidelberg, Germany). Genotype 1a-derived H77 (NCBI reference sequence NC_004102.1) with adaptive mutations P1496L and S2204I (4) and genotype 2a-derived J6/JFH-1 replicon-containing plasmids were licensed from Apath. The J6/JFH-1 replicon encodes a partial core (first 19 amino acids) and the 3′ nontranslated region (NTR) from the J6 strain (GenBank accession no. AF177036) and the 5′ NTR and NS3 to NS5B region from the JFH-1 strain (GenBank accession no. AB047639). To generate the ET, H77, and J6/JFH-1 replicon cell lines, plasmid DNA containing ET, H77, and J6/JFH-1 replicons were linearized with ScaI, HpaI, and XbaI, respectively. After in vitro transcription, each replicon RNA was transfected into the highly permissive Lunet cells and selected in medium containing 0.75 mg/ml G418 (1a replicon cells) or 0.25 mg/ml G418 (1b and 2a replicon cells) (30). The Huh 5-2 and ET-Lunet cells contained replicon that encodes the Firefly luciferase reporter gene, while the J6/JFH-1 replicon contains the Renilla luciferase reporter gene. HepAD38 hepatitis B virus (HBV) cells (kindly provided by B. Korba, Georgetown University, Washington, DC) and the P4- or human immunodeficiency virus type 1 (HIV-1)-infectible HeLa cell line (kindly provided by P. Charneau, Institut Pasteur, France) were maintained as described previously (30, 51). Primary human mononuclear (PBM) cells were prepared from buffy coat of blood from healthy donors (Biological Specialty Corporation, Colmar, PA).
HCV, HBV, and HIV inhibition assays.
HCV replicon assays using clone A, Huh 5-2, ET-Lunet, H77-Lunet, and J6/JFH-1-Lunet cells were performed as described previously (30, 52). Briefly, replicon-containing cells (1,500 to 3,000 cells/well in a 96-well plate) were incubated for 4 days with serially diluted test compounds. Inhibition of HCV RNA replication was determined by real-time PCR (RT-PCR) using primers that anneal to the 5′ NTR or by measuring the levels of luminescence expressed via the Firefly or Renilla luciferase reporter gene using Bright-Glo and Renilla-Glo reagents, respectively (Promega, Madison, WI). The EC50 and EC90 values, the concentrations at which 50% inhibition and 90% inhibition were achieved, were determined using GraphPad Prism software (San Diego, CA). Assays against the H77sV2 and JFH-1 infectious viruses were performed as previously described (55). Briefly, fresh compound-containing medium was replaced every day for 3 days, after which cells were fixed and incubated with a primary HCV-core mouse monoclonal IgG1 antigen (Affinity BioReagents, Golden, CO), which was reacted with a fluorescein isothiocyanate (FITC)-labeled secondary antibody to mouse IgG (Kirkegaard & Perry Laboratories, Gaithersburg, MD). Clusters of infected cells were considered to constitute a single infectious focus. Focus counts were analyzed with a Zeiss LSM 510 laser scanning confocal microscope, and EC50 and EC90 values were determined using GraphPad Prism software. Assays against HBV were performed using HepAD38 cells, which replicate HBV under conditions that can be regulated with tetracycline, as previously described (19, 50). Assays against HIV were performed using P4 cells or PBM cells (7).
Polymerase assays.
Cloning, expression, and purification of genotype 1b, 2a, 3a, and 4a recombinant HCV NS5B polymerase (NS5BΔ21) have been described previously (30, 52). The polymerase assay reactions were performed as described previously (30). Briefly, purified NS5BΔ21 polymerase (40 ng/μl) was incubated with genotype 1b internal ribosome entry site (IRES) RNA template (minus strand) (20 ng/μl), all four natural ribonucleotides (1 μM), [α-32P]UTP, and various concentrations of PSI-352666. The radioactive RNA products were quantified on a Hybond N+ membrane (GE Healthcare) using a phosphorimager (Perkin Elmer) (44). Reactions for genotype 3a and 4a NS5BΔ21 polymerases were allowed to proceed for 3 h, while the reaction mixtures for genotype 1b and 2a NS5BΔ21 polymerases were incubated for 30 min. The activity of human RNA polymerase II was followed in an in vitro transcription reaction initiated by HeLaScribe nuclear extract (Promega) (30). Briefly, reactions with mixtures containing PSI-352666 (500 μM) or α-amanitin (1 μM) incubated with cytomegalovirus immediate-early promoter DNA, all four ribonucleotides, [α-32P]GTP, and HeLaScribe nuclear extract were allowed to proceed for 60 min at 30°C. RNA polymerase II synthesized products were resolved on a 6% polyacrylamide sequencing gel and quantified by using a phosphorimager. Human DNA polymerase α, β, or γ (CHIMRx, Milwaukee, WI) was tested in the presence of increasing concentrations of PSI-352666 and the appropriate positive controls (30). Briefly, increasing concentrations of PSI-352666, d-ddFCTP, or aphidicolin were incubated in reaction mixtures containing activated calf thymus DNA (GE Healthcare), DNA polymerase α, β, or γ, all four deoxynucleoside triphosphates, and [α-32P]dCTP for 30 min at 37°C. The radiolabeled products were quantified using the Whatman DE81 paper binding assay as described previously (44). A nonlinear fit was performed to determine the 50% inhibitory concentration (IC50) using the GraphFit program (Erithacus Software, Horley, Surrey, United Kingdom).
Human bone marrow toxicity assays.
The bone marrow stem cell cytotoxicity assays using human erythroid and myeloid hematopoietic progenitor cells derived from normal bone marrow (Lonza, Walkersville, MD) were performed by Reachbio (Seattle, WA) using a semisolid methylcellulose-based medium (R&D Systems, Minneapolis, MN) containing recombinant human (rh) stem cell factor (50 ng/ml), rh-interleukin-3 (10 ng/ml), rh-granulocyte/monocyte colony-stimulating factor (10 ng/ml), and rh-erythropoietin (3 U/ml). Increasing concentrations of PSI-352938 (up to 50 μM) diluted in dimethyl sulfoxide (DMSO) or DMSO was added to progenitor cells seeded at 2 × 104 cells per culture in the methylcellulose-based medium so that the final DMSO concentration was 0.1%. 5-Fluorouracil was used as a positive control at concentrations of 0.01, 0.1, and 1.0 μg/ml. The cultures were set up in triplicate at 2 × 104 cells per culture. Following 14 days of incubation, the number of colonies was assessed and scored on the basis of size and morphology to determine the 50% inhibition concentration as described previously (49).
PKG studies.
Human recombinant protein kinase G 1β (PKG), cyclic GMP (cGMP), and the substrate peptide (Arg-Lys-Arg-Ser-Arg-Ala-Glu) were purchased from Sigma. PKG activation reactions were performed in 50 μl, and each reaction mixture contained 0.1, 1.0, or 10 μM cGMP; PSI-352938, PSI-354091, or PSI-354519; 10 ng/μl of peptide substrate; 50 μM [γ-32P]ATP (0.1 μCi/μl); 0.5 ng/μl PKG; and 10 mM MgCl2 in 20 mM Tris HCl, pH 7.5, buffer. The inhibition of cGMP-mediated activation of PKG was assayed in the presence of 10 μM cGMP and 1, 10, or 100 μM PSI-352938, PSI-354091, or PSI-354519 in the same buffer. The reactions were performed at 30°C, and an 8-μl aliquot was taken and mixed with 2 μl of 0.5 M EDTA at the 0-, 10-, 20-, 30-, and 40-min time points. A 5-μl aliquot of the quenched reaction mix was spotted on a P81 phosphocellulose paper disk (Whatman). The disks were washed three times with 75 mM phosphoric acid for 10 min. After the disks were dried, radioactivity was counted by a scintillation counter. The reaction rates were determined by fitting the data to linear regression using the GraphFit program.
Rebound and clearance studies.
ET-Lunet replicon cells were seeded at 1 × 105 cells per well in a 6-well plate in culture medium without G418. PSI-352938 or other test compounds were dissolved in DMSO and added to cells at concentrations 0-, 1-, 10-, and 20-fold over their approximate EC50 (0.1 μM for PSI-352938, PSI-7851, and the benzothiadiazine NNI; 0.6 nM for ITMN-191). The final DMSO concentration in the cultures was 0.5%. When the cells reached approximately 90% confluence, they were passaged at ratios of 1:4 or 1:5 every 3 to 4 days for 2 weeks and replenished with fresh medium containing the appropriate amount of compounds. At each passage, an aliquot of cells was harvested for RNA analysis. After 2 weeks, cells were passaged into culture medium containing 0.25 mg/ml G418 without inhibitor and cultured for an additional 2 weeks. At the end of the experiment, total RNA was extracted from all cell samples using an RNeasy-96 kit (Qiagen). Levels of HCV RNA and rRNA were determined using RT-PCR as described above. To determine relative log HCV RNA reduction, HCV RNA was normalized to rRNA by calculating the change in the threshold cycle (ΔCT; HCV CT − rRNA CT). ΔΔCT was calculated by subtracting the average value for the DMSO controls from each ΔCT value. The level of HCV RNA was calculated as log(2−ΔΔCT). Results were expressed as log change in HCV RNA levels between compound-treated cells and DMSO-treated control cells. The effect of compound treatment was also examined by colony formation. At the end of the 4-week experiment, cells were washed with phosphate-buffered saline, fixed, and stained using a solution of 0.5% crystal violet in 20% methanol for 30 min at room temperature and then washed with water.
Combination studies.
PSI-352938 was evaluated in combination with IFN-α, ribavirin, an NS3 protease inhibitor (ITMN-191), an NS3/4A inhibitor (ACH-806), an NS5A inhibitor (BMS-790052), a nonnucleoside inhibitor (a benzothiadiazine compound), a nucleoside inhibitor (PSI-6130), or a nucleotide inhibitor (PSI-7851). Twofold serial dilutions were made such that the final range for the combinations covered ratios corresponding to the in vitro EC90s, EC75s, and EC50s for the individual compounds. The corresponding monotherapy concentrations were assayed in parallel. Compounds were combined in a checkerboard cross in an opaque 96-well plate where 25 μl of PSI-352938 (4× concentration) dilutions was added horizontally and 25 μl of another compound (4× concentration) was added vertically. Huh 5-2 cells or ET-Lunet cells were added to the compound plates at 3,000 cells/well in 50 μl to make the final 1× drug concentration. The plates were incubated at 37°C in a humidified 5% CO2 atmosphere for 4 days prior to measurement of luciferase activity. Percent inhibition was calculated as the amount of luciferase activity detected in the presence of a single compound or combination of compounds relative to that for the no-drug control. At least two separate experiments were performed for each combination. The effects of drug-drug combinations were evaluated using the CalcuSyn program (Biosoft, Fergusen, MO). Combination indices (CIs) were calculated at the EC50, EC75, and EC90 values of the combinations. A CI value of less than 1 indicates synergy, and a CI value equal to 1 indicates additivity, while a CI value more than 1 indicates antagonism.
Cross-resistance studies.
HCV clone A cells containing the NS5B S282T replicon mutant were established previously by selecting the cells with 2′-C-methyladenosine (51). The NS5B S96T/N142T mutant replicon cell line was generated as described previously (30). Resistance mutations (C316Y, M414T, M423T, and P495L) associated with nonnucleoside inhibitors were generated in the ET replicon using a QuikChange II XL site-directed mutagenesis kit (Stratagene, Inc.) with the following primers and their complements: C316Y (5′-CTG CAC GAT GCT CGT ATA CGG AGA CGA CCT TGT CG), M414T (5′-CTA GGC AAC ATC ATC ACG TAT GCG CCC ACC), M423T (5′-CCT TGT GGG CAA GGA CGA TCC TGA TGA CTC ATT TC), and P495L (5′-GGA AAC TTG GGG TAC TGC CCT TGC GAG TC) (Integrated DNA Technologies, Coralville, IA). All mutations were confirmed by sequencing analysis (Cogenics, Houston, TX). Plasmids containing the mutated replicon were in vitro transcribed into RNA using a Ribomax large-scale RNA production system as recommended by the manufacturer (Promega). RNA (5 μg) was electroporated into the highly permissive Lunet cells using a BTX ElectoSquare Porator apparatus (Harvard Apparatus) as previously described (29). Cells containing replicon mutants were selected with 0.25 mg/ml G418. Anti-HCV replicon activity in wild-type or mutated replicon cells was evaluated using either RT-PCR or a luciferase-based replicon assay.
RESULTS
Antiviral activity of PSI-352938 and activity of PSI-352666 against the NS5B polymerase.
In order to determine the breadth of anti-HCV genotype activity of PSI-352938, we performed in vitro studies using genotype 1a (H77)-, 1b (Con1)-, and 2a (J6/JFH-1)-derived replicon cell lines and genotype 1a (H77) and 2a (JFH-1) infectious virus, as well as enzymatic assays using recombinant NS5B polymerases from genotypes 1 to 4. Results showed that PSI-352938 had similar activity against genotype 1a, 1b, and 2a replicons, with EC50s ranging from 0.13 to 0.20 μM and EC90 values ranging from 0.35 to 0.74 μM (Table 1). PSI-352938 also effectively inhibited HCV replication in the infectious virus assays: the EC50 and EC90 values were 0.28 ± 0.083 μM and 0.63 ± 0.018 μM, respectively, against the H77 infectious virus and 0.39 ± 0.31 μM and 1.16 ± 0.64 μM, respectively, against the JFH-1 infectious virus (Table 1). In contrast, PSI-352938 was not active against HBV or HIV up to the highest concentration tested (EC50 > 100 μM).
Table 1.
Activity of PSI-352938 in genotype 1a, 1b, and 2a subgenomic replicon cells and genotype 1a and 2a infectious virus system
| Cell-based assay and genotype | EC50 (μM) | EC90 (μM) |
|---|---|---|
| HCV replicona | ||
| 1b | 0.13 ± 0.076 | 0.51 ± 0.34 |
| 1a | 0.20 ± 0.12 | 0.74 ± 0.30 |
| 2a | 0.14 ± 0.040 | 0.35 ± 0.075 |
| Infectious HCVb | ||
| 1a | 0.28 ± 0.083 | 0.63 ± 0.018 |
| 2a | 0.39 ± 0.31 | 1.16 ± 0.64 |
Cells were treated with PSI-352938 for 4 days prior to determination of HCV inhibition. Quantitative real-time PCR or a luciferase-based reporter assay was used to quantify levels of HCV inhibition. Values are reported as averages of at least three independent experiments ± standard deviations.
Infectious foci were quantified using a primary HCV-core mouse monoclonal antibody reacted with a FITC-labeled secondary antibody. Values are reported as averages ± standard deviations of two independent experiments performed in duplicate.
PSI-352666, the active 5′-triphosphate metabolite of PSI-352938, was evaluated for its ability to inhibit recombinant HCV NS5B polymerase isolated from genotypes 1 to 4. Increasing concentrations of PSI-352666 were titrated into reaction mixtures containing HCV NS5B, and results showed that all four polymerases were inhibited in a dose-dependent manner. As summarized in Table 2, the IC50s for PSI-352666 against genotype 1b, 2a, 3a, and 4a NS5B polymerases were determined to be 1.0 ± 0.2, 4.7 ± 0.6, 1.3 ± 0.5, and 4.2 ± 0.8 μM, respectively.
Table 2.
Activity of PSI-352666 against genotype 1b, 2a, 3a, and 4a recombinant NS5BΔ21 polymerases and human RNA polymerase II and DNA polymerases α, β, and γ
| Enzyme-based assay and genotype or enzyme | IC50 (μM) |
|
|---|---|---|
| PSI-352666a | Positive controlb | |
| HCV NS5B | ||
| 1b | 1.0 ± 0.2 | 0.1 ± 0.04 |
| 2a | 4.7 ± 0.6 | 0.1 ± 0.02 |
| 3a | 1.3 ± 0.5 | 0.03 ± 0.004 |
| 4a | 4.2 ± 0.8 | 2.9 ± 0.6 |
| Human polymerase | ||
| DNA Pol α | 900 ± 140 | 16 ± 2 |
| DNA Pol β | >1,000 | 9.0 ± 0.4 |
| DNA Pol γ | >1,000 | 79 ± 4 |
| RNA Pol II | >500 | <1 |
HCV NS5B polymerase activity in the presence of PSI-352666 was determined by measuring the incorporation of [α-32P]UTP into the RNA product using an HCV IRES template. The amount of product was quantified by phosphorimaging. Human DNA polymerase α, β, and γ activities were quantified by measuring the synthesized radiolabeled products with a filter binding assay. Human RNA polymerase II synthesized RNA products from in vitro transcription using HeLa nuclear extract were analyzed using a 6% polyacrylamide sequencing gel and phosphorimaging. Results were obtained from at least two independent experiments in duplicate and are reported as means ± standard deviations.
The positive control for HCV NS5B was 3′-dCTP, that for DNA Pol α was aphidicolin, that for DNA Pols β and γ was d-ddFCTP, and that for RNA Pol II was α-amanitin.
In vitro safety assessment of PSI-352938 and metabolites.
PSI-352938 was previously reported not to be cytotoxic (50% cytotoxic concentrations [CC50s] > 100 μM) or toxic to mitochondria (CC50s > 100 μM) in various cell types (47). Because bone marrow toxicity has been associated with a number of nucleoside antiviral analogs (49), we evaluated the effect of PSI-352938 on the proliferation of human bone marrow progenitor cells. No significant inhibition by PSI-352938 was observed in human erythroid (CFU-E and BFU-E) or myeloid progenitor (CFU-GM) cells in this 14- to 16-day assay (CC50s >50 μM). At 1 μg/ml (4 μM) the positive control, 5-fluorouracil, inhibited CFU-E progenitor cell colony formation by >80% and completely inhibited BFU-E and CFU-GM cell colony formation.
To further evaluate the in vitro specificity profile of PSI-352938, we tested the ability of PSI-352666, the active triphosphate metabolite, to inhibit the activity of human DNA polymerases α, β, and γ and human RNA polymerase II (Table 2). PSI-352666 was a very weak inhibitor of DNA polymerase α, with an IC50 of 900 μM, and showed no significant inhibition of polymerases β and γ at concentrations up to 1 mM, the highest concentration tested. The effect of PSI-352666 on human RNA Pol II was assessed using an in vitro transcription assay. No significant inhibition was observed at concentrations of PSI-352666 up to 500 μM, the highest concentration tested, whereas α-amanitin, a known RNA Pol II inhibitor, almost completely inhibited the reaction at 1 μM.
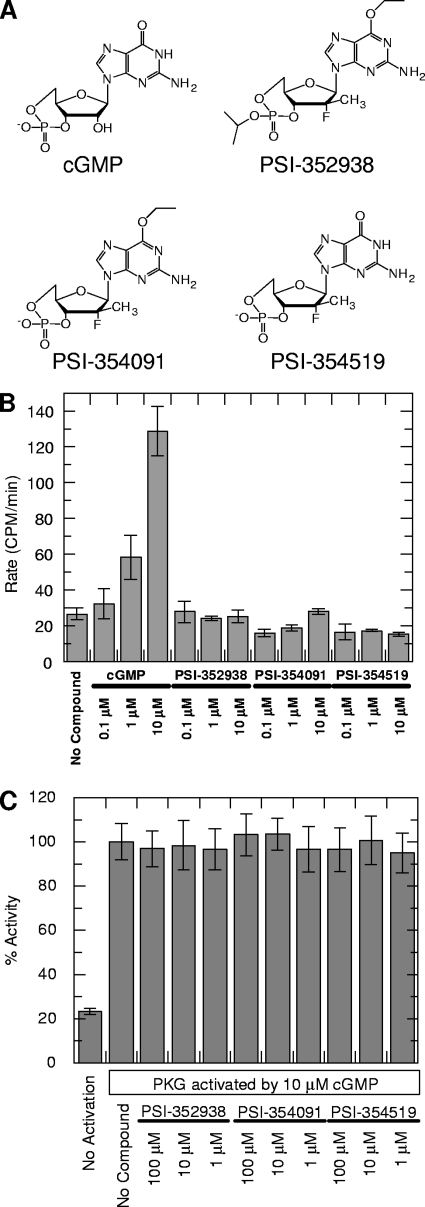
cGMP plays an important role as an intracellular secondary messenger in regulating many signaling transduction pathways, especially in the cardiovascular system (3). One of its major targets is cGMP-dependent PKG, which functions to phosphorylate other cellular proteins upon cGMP activation (39, 54). Because PSI-352938 and its metabolites PSI-354091 and PSI-354519 share a structural similarity with cGMP (see chemical structures in Fig. 1A), we evaluated the ability of PSI-352938, PSI-354091, and PSI-354519 to activate PKG. As a positive control, assays were also performed with the natural activator cGMP. Results showed that only cGMP activated PKG in a dose-dependent manner, with no significant PKG activation observed with PSI-352938, PSI-354091, or PSI-354519 (Fig. 1B). The ability of PSI-352938, PSI-354091, and PSI-354519 to inhibit the cGMP-mediated activation of PKG was also studied. Results showed that almost the same levels of activation were observed in the presence or absence of PSI-352938 and its metabolites (Fig. 1C). Taken together, our data indicate that PSI-352938, PSI-354091, and PSI-354519 do not affect the activity or the cGMP-mediated activation of PKG.
Fig. 1.
Effect of PSI-352938 and its potential cyclic metabolites on PKG activity. (A) Chemical structures of cyclic cGMP, PSI-352938, PSI-354091, and PSI-354519; (B) effect of PSI-352938, PSI-354091, and PSI-354519 on the activation of PKG compared to that of cGMP; (C) effect of PSI-352938, PSI-354091, and PSI-354519 on the cGMP-mediated PKG activation compared to that of the no-compound control.
Rebound and clearance studies.
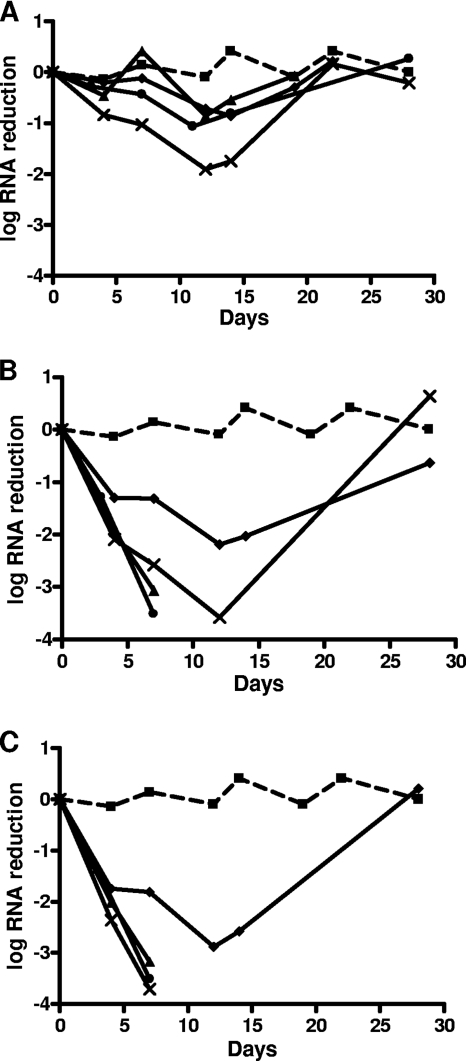
Using the replicon system, PSI-352938 was evaluated for its ability to clear HCV replicon RNA in ET-Lunet cells. PSI-7851 (phosphoramidate prodrug of 2′-α-fluoro-2′-β-C-methyluridine), a benzothiadiazine compound (NS5B NNI), and ITMN-191 (an NS3 protease inhibitor) were also included for comparison. The first part of the study involved treating replicon cells with each compound for 14 days in the absence of G418 at approximately 1, 10, and 20 times the approximate EC50. During this time, cells were passaged twice a week with replenishment of fresh medium and compounds, and an aliquot was removed to analyze for HCV replicon RNA levels. As shown in Fig. 2, control cells incubated in the absence of compound maintained a stable level of replicon RNA throughout the course of the experiment. At the end of the 2-week compound incubation period, cells treated with 1× EC50 of ITMN-191 achieved 1.75 log reduction, while cells treated with 1× EC50 of PSI-7851, PSI-352938, and the NNI achieved reductions of HCV RNA levels of 0.54, 0.81, and 0.86 log, respectively (Fig. 2A). At 10× EC50s, the levels of HCV RNA were below the limit of detection for cells treated with PSI-7851 or PSI-352938 by day 12, while the NNI and ITMN-191 reduced HCV RNA levels by 2.2 and 3.6 log, respectively (Fig. 2B). At 20× EC50s, the HCV RNA levels were below the limit of detection for cells treated with PSI-7851, PSI-352938, or ITMN-191 by the end of the 2-week treatment, while HCV replicon RNA was still detectable in cells treated with the NNI (Fig. 2C).
Fig. 2.
Rebound and clearance study of HCV replicon RNA. Replicon cells were treated with PSI-352938, PSI-7851, a benzothiadiazine NNI, or ITMN-191 for 2 weeks in the absence of G418, followed by removal of compounds and G418 selection for an additional 2 weeks. An aliquot of cells at each passage was harvested for RNA analysis. Level of HCV replicon RNA change in the presence of DMSO (■), PSI-352938 (●), PSI-7851 (▲), a benzothiadiazine NNI (♦) or ITMN-191 (×) at their approximate 1× EC50s (A), 10× EC50s (B), or 20× EC50s (C).
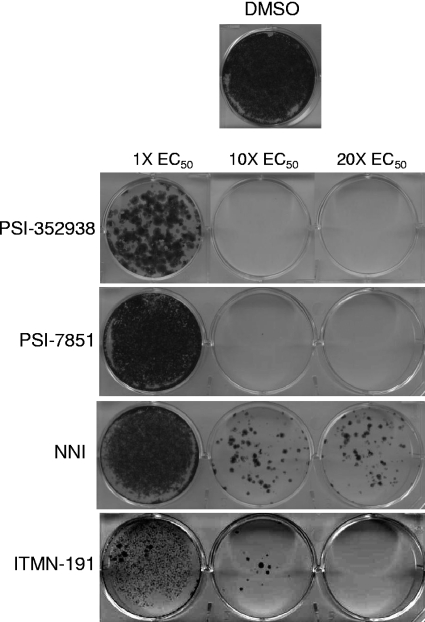
To determine whether clearance of the HCV replicon occurred after 14 days of treatment, each compound was withdrawn and the cells were incubated for an additional 14 days in the presence of G418. Cells that grew in the presence of G418 contained replicon RNA, whereas cells that were cleared of the replicon did not survive G418 treatment. In addition to quantifying the HCV replicon RNA levels, colony formation assays were also performed to evaluate the ability of each compound in preventing rebound of replicon cells. By the end of the 4-week study, the levels of all cells treated with compounds at 1× their EC50s rebounded to levels similar to that of the no-drug control cells (Fig. 2A and 3). At 10× EC50s, the level of HCV replicon RNA was below the limit of detection in the RT-PCR assay for cells treated with PSI-352938 or PSI-7851, while rebound of HCV replicon was observed in cells treated with the NNI or ITMN-191 (Fig. 2B). The colony formation study showed only one very small colony at 10× EC50 of PSI-352938 or PSI-7851, while significantly more colonies were observed for cells treated with the NNI or ITMN-191 at 10× their EC50s (Fig. 3). Complete clearance was observed in cells treated with 20× the EC50s of the two nucleotide inhibitors and ITMN-191, while replicon cells cultured in the presence of the NNI rebounded even when they were treated at 20× its EC50 (Fig. 2C and 3).
Fig. 3.
Colony formation analysis from the rebound and clearance study using replicon cells treated with 1×, 10×, or 20× EC50s of PSI-352938, PSI-7851, a benzothiadiazine NNI, or ITMN-191. Cells were incubated with compounds for 2 weeks in the absence of G418, with replenishment of each compound with fresh medium at every passage, followed by removal of compounds and G418 selection for an additional 2 weeks. Cells remaining at the end of the experiment were fixed and stained with crystal violet.
Combination studies.
The anti-HCV effect of combining PSI-352938 with other known HCV inhibitors was evaluated using the replicon cell system. Each of the combination studies was performed by combining PSI-352938, which was added horizontally (10 different concentrations), and another HCV inhibitor, which was added vertically (8 different concentrations) in a checkerboard cross in a 96-well plate. Four different molar ratios, each containing at least six different combinations of PSI-352938 and another test compound, were used for data analysis to allow variability in the estimates of relative activity. The CI analysis method, which determined CI values at concentrations of compounds that achieve 50, 75, or 90% inhibition, was based on that described by Chou and Talalay (8). Generally, a CI value of 1 indicates additivity, a CI value of less than 1 indicates synergism, and a CI value of greater than 1 indicates an antagonistic effect. The linear correlation coefficient value, r, represents the goodness of fit for the data to the dose-effect equation.
We first combined PSI-352938 with IFN-α and ribavirin. As defined by the CalcuSyn analysis, combining PSI-352938 with IFN-α appears to have an additive to moderate synergistic effect, while combining PSI-352938 with ribavirin appears to be synergistic (Table 3). We also combined PSI-352938 with other small-molecule HCV inhibitors targeting the NS3 protease (ITMN-191), NS4A cofactor (ACH-806), NS5A phosphoprotein (BMS-790052), or NS5B polymerase (a benzothiadiazine-derived NS5B NNI, the nucleoside/nucleotide analogs PSI-6130 and PSI-7851) (see Table S1 in the supplemental material for compound structures). As summarized in Table 4, the combination of PSI-352938 with ITMN-191 resulted in an additive to moderate synergistic effect, the combination of PSI-352938 with ACH-806, the benzothiadiazine NNI, or PSI-6130 was synergistic, and the combination of PSI-352938 with BMS-790052 or PSI-7851 resulted in an overall additive effect on inhibition of HCV RNA replication in the replicon cells.
Table 3.
CalcuSyn analysis of PSI-352938 combination studies with IFN-α or ribavirin
| Combination and ratioa | Combination indexb |
rc | ||
|---|---|---|---|---|
| EC50 | EC75 | EC90 | ||
| PSI-352938 and IFN-α | ||||
| 1:40 | 0.74 | 0.71 | 0.69 | 0.99 |
| 1:20 | 0.68 | 0.71 | 0.74 | 0.97 |
| 1:10 | 0.88 | 0.76 | 0.65 | 0.97 |
| 1:5 | 1.02 | 0.87 | 0.74 | 0.98 |
| Ribavirin and PSI-352938 | ||||
| 400:1 | 0.59 | 0.6 | 0.62 | 0.98 |
| 200:1 | 0.63 | 0.67 | 0.72 | 0.99 |
| 100:1 | 0.44 | 0.52 | 0.60 | 0.95 |
| 50:1 | 0.36 | 0.41 | 0.46 | 0.91 |
Compounds were combined in a checkerboard format, where 10 different concentrations of PSI-352938 were added horizontally and 8 different concentrations of IFN-α or ribavirin were added vertically to a 96-well plate. For each ratio indicated, at least six different combinations of PSI-352938 and IFN-α or ribavirin were included for calculations of the combination index value.
Combination index values at 50%, 75%, and 90% inhibition were calculated using the CalcuSyn program.
The r value is the linear correlation coefficient that indicates the goodness of fit for the data.
Table 4.
CalcuSyn analysis of PSI-352938 combination studies with ITMN-191, ACH-806, BMS-790052, a benzothiadiazine NNI, PSI-6130, or PSI-7851
| Combination and ratioa | Combination indexb |
rd | ||
|---|---|---|---|---|
| EC50 | EC75 | EC90 | ||
| PSI-352938 and ITMN-191 | ||||
| 20:1 | 0.64 | 0.68 | 0.73 | 0.97 |
| 40:1 | 0.72 | 0.73 | 0.74 | 0.96 |
| 80:1 | 0.93 | 0.83 | 0.73 | 0.99 |
| 160:1 | 1.13 | 1.12 | 1.10 | 0.98 |
| PSI-352938 and ACH-806 | ||||
| 1:5 | 0.67 | 0.68 | 0.69 | 0.97 |
| 1:2.5 | 0.63 | 0.64 | 0.66 | 0.95 |
| 1:1.25 | 0.78 | 0.76 | 0.73 | 0.98 |
| 1:0.63 | 0.76 | 0.76 | 0.76 | 0.99 |
| PSI-352938 and BMS-790052 | ||||
| 12,500:1 | 1.01 | 0.92 | 0.85 | 0.97 |
| 25,000:1 | 1.09 | 1.11 | 1.14 | 0.98 |
| 50,000:1 | 0.91 | 0.89 | 0.88 | 0.93 |
| 100,000:1 | 0.92 | 0.9 | 0.88 | 0.93 |
| PSI-352938 and NNIc | ||||
| 0.8:1 | 0.60 | 0.61 | 0.63 | 0.99 |
| 1.6:1 | 0.80 | 0.70 | 0.62 | 0.95 |
| 3.2:1 | 0.70 | 0.63 | 0.58 | 0.99 |
| 6.4:1 | 0.77 | 0.77 | 0.78 | 0.92 |
| PSI-352938 and PSI-6130 | ||||
| 1:20 | 0.51 | 0.6 | 0.71 | 0.94 |
| 1:10 | 0.51 | 0.63 | 0.78 | 0.98 |
| 1:5 | 0.62 | 0.66 | 0.71 | 0.98 |
| 1:2.5 | 0.58 | 0.62 | 0.66 | 0.93 |
| PSI-352938 and PSI-7851 | ||||
| 1:1.25 | 0.80 | 0.76 | 0.71 | 0.99 |
| 1:0.63 | 1.06 | 1.03 | 1.00 | 0.98 |
| 1:0.31 | 1.02 | 0.91 | 0.81 | 0.99 |
| 1:0.16 | 1.17 | 1.15 | 1.13 | 0.98 |
Compounds were combined in a checkerboard format, where 10 different concentrations of PSI-352938 were added horizontally and 8 different concentrations of another HCV inhibitor were added vertically to a 96-well plate. For each ratio indicated, at least six different combinations of PSI-352938 and the other anti-HCV compound were included for calculation of the combination index value.
Combination index values at 50%, 75%, and 90% inhibition were calculated using the CalcuSyn program.
The NS5B NNI used here was a benzothiadiazine-derived compound targeting an allosteric site within the palm domain of NS5B.
The r value is the linear correlation coefficient that indicates the goodness of fit for the data.
Cross-resistance studies.
Investigators previously demonstrated that the NS5B S282T mutant replicon was less susceptible to inhibition by 2′-modified nucleosides/nucleotides, including 2′-C-methyladenosine, 2′-C-methylguanosine, 2′-C-methylcytidine, 2′-fluoro-2′-C-methycytidine (PSI-6130), and 2′-fluoro-2′-C-methyluridine prodrugs (PSI-7851 and PSI-7977) (1, 30, 33, 43, 48). Because PSI-352938 was also modified at the 2′ position of the ribose ring, it was anticipated that the compound would show reduced activity against a replicon that contained the S282T mutation. To determine if the S282T mutation conferred resistance to PSI-352938, the activity of the compound was tested against cells that contained the S282T replicon mutant. Results showed that the S282T replicon remained fully susceptible to PSI-352938 (Table 5). We also included INX-08189 (prodrug of 2′-C-methylguanosine) and R1479 (4′-azidocytidine) as positive and negative controls, respectively (see Table S1 in the supplemental material for compound structures). As shown in Table 5, R1479 retained its activity in the S282T replicon cells, while INX-08189 was 6.1-fold less active against the S282T replicon than the wild-type replicon.
Table 5.
Cross-resistance studies of PSI-352938 using replicon mutants resistant to nucleoside/nucleotide analogs and nonnucleoside inhibitors
| NS5B resistance mutation | HCV inhibitor | EC90 (μM)a |
EC90 fold changeb | |
|---|---|---|---|---|
| WT replicon | Mutant replicon | |||
| S282T | R1479 | 22.35 ± 4.58 | 12.53 ± 3.78 | 0.56 |
| INX-08189 | 0.018 ± 0.001 | 0.11 ± 0.004 | 6.11 | |
| PSI-352938 | 1.38 ± 0.67 | 1.61 ± 0.19 | 1.17 | |
| S96T/N142T | R1479 | 15.76 ± 3.67 | 74.12 ± 12.12 | 4.70 |
| INX-08189 | 0.0054 ± 0.0036 | 0.0059 ± 0.0027 | 1.09 | |
| PSI-352938 | 0.59 ± 0.34 | 0.53 ± 0.28 | 0.89 | |
| C316Y | HCV-796 | 0.026 ± 0.017 | 1.92 ± 0.77 | 73.85 |
| PSI-352938 | 0.88 ± 0.24 | 0.95 ± 0.30 | 1.08 | |
| M414T | Benzothiadiazine compound | 1.23 ± 0.55 | 28.39 ± 13.70 | 23.08 |
| PSI-352938 | 0.88 ± 0.24 | 0.93 ± 0.38 | 1.06 | |
| M423T | Thiophene compound | 3.65 ± 0.21 | 69.52 ± 9.24 | 19.05 |
| PSI-352938 | 0.88 ± 0.24 | 0.76 ± 0.20 | 0.86 | |
| P495L | 2-Phenol indole compound | 1.17 ± 0.46 | 60.75 ± 19.90 | 51.92 |
| PSI-352938 | 0.88 ± 0.24 | 0.65 ± 0.19 | 0.74 | |
The wild-type (WT) and S282T replicon cells were derived from the clone A cell line. EC90s were determined using the RT-PCR assay. The S96T/N142T, C316Y, M414T, M423T, and P495L replicons were derived from the ET construct transfected into Lunet cells. EC90s were determined using the Firefly luciferase assay and compared to those generated from the wild-type ET-Lunet replicon cells. All values are reported as the means ± standard deviations from at least three independent experiments performed in duplicate.
Fold change in EC90 was calculated by normalizing the EC90 value for the test compound in the mutant replicon cells with that of the wild type.
The S96T/N142T mutation within the NS5B polymerase was selected by R1479 in the replicon cells and was subsequently shown to confer resistance to this inhibitor (33). The S96T/N142T mutant replicon was not associated with any cross-resistance to PSI-352938 and also remained fully susceptible to inhibition by INX-08189. As expected, S96T/N142T replicon RNA synthesis was less susceptible to inhibition by R1479: the EC90 value for R1479 in the S96T/N142T cells was 74.1 μM, which was 4.7-fold higher than that achieved in the wild-type cells.
We also tested PSI-352938 against replicons containing mutations that confer resistance to nonnucleoside inhibitors of NS5B. These included C316Y, a palm 2 site mutation that confers resistance to HCV-796; M414T, a palm 1 site mutation that confers resistance to benzothiadiazine-derived compounds; M423T, a thumb 2 site mutation that confers resistance to thiophene-derived compounds; and P495L, a thumb 1 site mutation that confers resistance to 2-phenol indole- and benzimidazole-derived compounds (20, 34, 46, 53). Results showed that these NNI-resistant replicons were also fully susceptible to inhibition by PSI-352938 (Table 5). In contrast, each mutant conferred resistance to the corresponding nonnucleoside inhibitor (see Table S1 in the supplemental material for compound structures), with the reduction in activity ranging from about 20- to 70-fold.
DISCUSSION
As a cyclic monophosphate prodrug, PSI-352938 is currently the first of its class to enter clinical trials for the treatment of hepatitis C virus infection. Modified 2′-C-methyl nucleosides with a cyclic phosphate moiety have been explored previously as inhibitors of HCV using the replicon system (17, 42). Meppen et al. examined the SAR of cyclic phosphoramidate prodrugs of 2′-C-methylcytidine. Those compounds showed relatively weak or no improvement in activity compared to the corresponding nucleoside analog (42). In contrast, cyclic monophosphate prodrugs of 2′-C-methyl-substituted purine nucleoside analogs showed significant enhancement of activity in the replicon assay (17). These compounds contained various phosphate ester moieties, such as pivaloyloxymethyl, S-acetyl-2-thioethyl, and carbonate, on the monophosphate and either an adenine or guanine base with modification at C-6 (17). More recently, we described an analysis of the SARs of a panel of 2′-fluoro-2′-C-methyguanosine cyclic monophosphate prodrugs, which identified PSI-352938 for further development because of its overall favorable activity in the replicon assay, metabolism to the active triphosphate (PSI-352666), lack of cytotoxicity, and stability profiles (47). Results from the HCV and NS5B inhibition studies presented here further demonstrate that PSI-352938 and PSI-352666 possessed broad genotype coverage, having similar activities against genotype 1 and 2 HCV replicon RNA synthesis and infectious virus replication and against genotype 1 to 4 recombinant NS5B polymerases.
In addition to the anti-HCV effect, the potential of PSI-352666, PSI-352938, and its metabolites PSI-354091 and PSI-354519 to target cellular proteins was evaluated. Our human polymerase studies showed that PSI-352666 (triphosphate form of 2′-fluoro-2′-C-methylguanosine) did not significantly inhibit RNA polymerase II or DNA polymerase α, β, or γ, indicating that the 2′-fluoro-2′-C-methyl modification at the sugar moiety made it a poor inhibitor for human RNA polymerase II and DNA polymerases α, β, and γ. Another potential cellular target that we examined was the cGMP-dependent PKG, since PSI-352938 was a guanosine cyclic phosphate analog and its cyclic phosphate metabolites PSI-354091 and PSI-354519 shared structural similarities with cGMP. cGMP, an important secondary messenger, triggers signaling cascades that contribute to the function of several vascular cell types (3, 54). Results from the cGMP-dependent PKG studies confirmed that PSI-352938 and its cyclic phosphate metabolites neither activate nor inhibit the cGMP-mediated activation of PKG. Among the three cyclic phosphate analogs, PSI-354519 most resembled cGMP, differing only at the 2′ position of the ribose moiety, further validating that modification at this position prevented these compounds from targeting cellular proteins, thus reducing the potential for off-target side effects. Indeed, lack of cytotoxicity was demonstrated for PSI-352938 in a number of different cell lines, including Huh7 and HepG2 hepatoma cells, pancreatic BxPC3 cells, CEM lymphocytes, and human bone marrow progenitor cells (47).
In order to cure HCV infection, it is critical for antiviral compounds to clear the virus and prevent viral RNA rebound. The cell-based rebound and clearance studies showed that PSI-352938 cleared the HCV replicon more effectively than the NS3 protease inhibitor ITMN-191 at concentrations corresponding to 10× their EC50s. The benzothiadiazine NNI studied here did not clear HCV replicon RNA even at 20× its EC50, which may be related to the mechanism of action of NNIs targeting allosteric regions on the NS5B polymerase. Our in vitro clearance and rebound study was a first step toward demonstrating that PSI-352938 was capable of clearing HCV RNA. It is possible that the outcome from an in vivo situation could be different from the results of in vitro studies, especially since HCV could infect organs other than the liver, such as the lymphatic system (24).
Migliaccio et al. demonstrated that a single mutation within the NS5B active site, S282T, was sufficient to confer resistance to 2′-C-methyladenosine and the corresponding guanosine ribonucleoside (43). Further cross-resistance and selection studies with other 2′-modified nucleosides/nucleotides, including the prodrugs of 2′-C-methylguanosine (IDX184 and INX-08189), 2′-C-methylcytidine and its prodrug (NM283), 2′-fluoro-2′-C-methylcytidine (PSI-6130) and its prodrug (RG7128), and 2′-fluoro-2′-C-methyluridine prodrugs (PSI-7851/PSI-7977), showed reduced activity against the S282T-mutated replicon (1, 23, 30, 33, 40, 48, 51). It was previously suggested that resistance due to the S282T mutation was directed toward the ribose moiety: incorporation of the 2′-C-methyl-modified nucleotide in the primer position could result in steric hindrance between the methyl group and the ribose of the incoming nucleoside triphosphate (43). Our data showing that PSI-352938 retained its activity against the S282T replicon were therefore quite surprising. First, similar to 2′-C-methylguanosine and its prodrugs (IDX184 and INX-08189), guanine is the base for the active 5′-triphosphate of PSI-352938. However, unlike these compounds, PSI-352938 contains a fluorine at the 2′ α position, in addition to the 2′-C-methyl substitution at the β position. Second, the S282T mutation conferred resistance to the 2′-α-fluoro-2′-β-C-methyl-substituted pyrimidine analogs PSI-6130, PSI-7851, and PSI-7977. Together, our results suggest that both the base and the 2′-fluoro-2′-C-methyl substitutions play a role in the activity of PSI-352938 against the S282T replicon cells.
In addition to the S282T mutation, we also tested the activity of PSI-352938 against a panel of resistant replicons each containing representative mutations selected by other classes of nucleoside and nonnucleoside inhibitors. Among the mutations (C316Y, M414T, M423T, and P495L) selected by the NNIs, we were particularly interested in the P495L mutation, which was located near the alternative GTP binding site on the thumb domain. It has previously been reported that the binding of GTP in this allosteric region was important for initiating RNA synthesis, with P495 providing a hydrophobic platform for the guanine base (5). Nonnucleoside inhibitors, including the benzimidazoles and 2-phenol indole-derived compounds, binding to this region have been shown to block NS5B-mediated RNA synthesis by interfering with the formation of the polymerase-RNA complex (53). Our results showed that the P495L replicon cells were fully susceptible to the inhibition by PSI-352938. Similarly, PSI-352938 remained active against the other replicon mutants. Together with the results from the S282T and S96T/N142T replicon cells, the cross-resistance studies using various NS5B replicon mutants showed that the active triphosphate of PSI-352938 likely interacts with NS5B in a unique way.
To date we have not been able to select genotype 1a- or 1b-resistant replicons using the conventional methodology in which replicon cells were incubated in the presence of increasing concentrations of PSI-352938 and G418. We observed that either PSI-352938 would totally clear the replicon from the cells, which led to cell death, or under situations in which cells did survive, no consistent mutation that conferred resistance to PSI-352938 was selected. We were, however, able to raise resistance using a genotype 2a replicon cell line. Studies are under way to confirm the amino acid changes to define the resistance profile of these replicon cells for PSI-352938.
Combinations of inhibitors of different mechanisms are likely to become the essential components of future anti-HCV therapies in order to maximize antiviral efficacy and prevent the emergence of resistance. Emergence of resistance mutations remains to be a major challenge for DAAs currently in development: as in the case of NS3 protease, NS5A and NS5B nonnucleoside inhibitors which rapidly select for resistance in cell culture and in the clinic (12, 22). It has already been observed that the rate of naturally existing polymorphisms was higher for NNIs and NS3 protease inhibitors, and therefore, they would be more likely to select for preexisting resistance mutations (26, 36). In contrast to these inhibitors, the nucleoside/nucleotide analogs are known to have a high barrier to resistance, which may be due to low viral fitness of mutants selected with nucleoside analogs (35, 41). The INFORM-1 study, which was a phase IIA clinical trial in HCV genotype 1-infected patients that combined RG7227 (ITMN-191) and RG7128, has already shown the proof of concept for significantly reducing the viral load in HCV-infected patients in the absence of SOC (15). The in vitro combination studies that we performed here with PSI-352938 and regimens from SOC or other classes of HCV inhibitors targeting NS3, NS4A, NS5A, or NS5B showed favorable antiviral effects. Nucleoside analogs already have a proven track record as therapies for viral infections caused by herpesviruses, HIV, and hepatitis B virus (9). With the pan-genotype activity and high barrier to resistance, nucleotide analogs are promising candidates as HCV inhibitors to serve as the backbone of future combination anti-HCV therapies. The complementary resistance profile of PSI-352938 further positions it to be an ideal compound for combination therapy with other HCV inhibitors, including other classes of NS5B nucleoside/nucleotide analogs.
Supplementary Material
ACKNOWLEDGMENTS
The work performed at the University of Texas Medical Branch was supported in part by a contract (AI-25488) from the National Institute of Allergy and Infectious Diseases.
We thank David Frick (University of Wisconsin, Milwaukee, WI) and Julie Heck (New York Medical College, Valhalla, NY) for providing the recombinant NS5B polymerase derived from the genotype 2a JFH-1 strain. We also thank Emer Clarke at ReachBio LLC (Seattle, WA) for performing human bone marrow progenitor cell toxicity assays.
Footnotes
Supplemental material for this article may be found at http://aac.asm.org/.
Published ahead of print on 28 March 2011.
REFERENCES
- 1. Ali S., et al. 2008. Selected replicon variants with low-level in vitro resistance to the hepatitis C virus NS5B polymerase inhibitor PSI-6130 lack cross-resistance with R1479. Antimicrob. Agents Chemother. 52:4356–4369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alter H. J. 2005. HCV natural history: the retrospective and prospective in perspective. J. Hepatol. 43:550–552 [DOI] [PubMed] [Google Scholar]
- 3. Birschmann I., Walter U. 2004. Physiology and pathophysiology of vascular signaling controlled by guanosine 3′,5′-cyclic monophosphate-dependent protein kinase. Acta Biochim. Pol. 51:397–404 [PubMed] [Google Scholar]
- 4. Blight K. J., McKeating J. A., Marcotrigiano J., Rice C. M. 2003. Efficient replication of hepatitis C virus genotype 1a RNAs in cell culture. J. Virol. 77:3181–3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bressanelli S., Tomei L., Rey F. A., De Francesco R. 2002. Structural analysis of the hepatitis C virus RNA polymerase in complex with ribonucleotides. J. Virol. 76:3482–3492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carroll S. S., Olsen D. B. 2006. Nucleoside analog inhibitors of hepatitis C virus replication. Infect. Disord. Drug Targets 6:17–29 [DOI] [PubMed] [Google Scholar]
- 7. Charneau P., Alizon M., Clavel F. 1992. A second origin of DNA plus-strand synthesis is required for optimal human immunodeficiency virus replication. J. Virol. 66:2814–2820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chou T. C., Talalay P. 1984. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv. Enzyme Regul. 22:27–55 [DOI] [PubMed] [Google Scholar]
- 9. De Clercq E. 2004. Antiviral drugs in current clinical use. J. Clin. Virol. 30:115–133 [DOI] [PubMed] [Google Scholar]
- 10. Di Bisceglie A. M., Hoofnagle J. H. 2002. Optimal therapy of hepatitis C. Hepatology 36:S121–S127 [DOI] [PubMed] [Google Scholar]
- 11. Foster G., Mathurin P. 2008. Hepatitis C virus therapy to date. Antivir. Ther. 13(Suppl. 1):3–8 [PubMed] [Google Scholar]
- 12. Fridell R. A., Qiu D., Wang C., Valera L., Gao M. 2010. Resistance analysis of the hepatitis C virus NS5A inhibitor BMS-790052 in an in vitro replicon system. Antimicrob. Agents Chemother. 54:3641–3650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fried M. W., et al. 2002. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N. Engl. J. Med. 347:975–982 [DOI] [PubMed] [Google Scholar]
- 14. Furman P. A., Lam A. M., Murakami E. 2009. Nucleoside analog inhibitors of hepatitis C viral replication: recent advances, challenges and trends. Future Med. Chem. 1:1429–1452 [DOI] [PubMed] [Google Scholar]
- 15. Gane E., et al. 2009. Combination therapy with a nucleoside polymerase (R7128) and protease (R7227/ITMN-191) inhibitor in HCV: safety, pharmacokinetics, and virologic results from INFORM-1, abstr. 193. Abstr. 60th Annu. Meet. Am. Assoc. Study Liver Dis [Google Scholar]
- 16. Gane E. J., et al. 2010. Sustained virologic response following RG7128 1500 mg BID PEG-IFN/RBV for 28 days in HCV genotype 2/3 prior non-responders, abstr. 37. Abstr. 45th Annu. Meet. Eur. Assoc. Study Liver [Google Scholar]
- 17. Gunic E., et al. 2007. Cyclic monophosphate prodrugs of base-modified 2′-C-methyl ribonucleosides as potent inhibitors of hepatitis C virus RNA replication. Bioorg. Med. Chem. Lett. 17:2452–2455 [DOI] [PubMed] [Google Scholar]
- 18. Hadziyannis S. J., et al. 2004. Peginterferon-alpha2a and ribavirin combination therapy in chronic hepatitis C: a randomized study of treatment duration and ribavirin dose. Ann. Intern. Med. 140:346–355 [DOI] [PubMed] [Google Scholar]
- 19. Hassan A. E., et al. 2003. Synthesis and antiviral evaluation of 2′,3′-dideoxy-2′-fluoro-3′-C-hydroxymethyl-beta-d-arabinofuranosyl pyrimidine nucleosides. Nucleosides Nucleotides Nucleic Acids 22:891–894 [DOI] [PubMed] [Google Scholar]
- 20. Howe A. Y., et al. 2008. Molecular mechanism of hepatitis C virus replicon variants with reduced susceptibility to a benzofuran inhibitor, HCV-796. Antimicrob. Agents Chemother. 52:3327–3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jensen D. M., et al. 2010. High rates of early viral response, promising safety profile and lack of resistance-related breakthrough in HCV GT 1/4 patients treated with RG7128 plus PEG-IFN alfa-2a (40KD)/RBV: planned week 12 interim analysis from the Propel study, abstr. 81. Abstr. 61st Annu. Meet. Am. Assoc. Study Liver Dis [Google Scholar]
- 22. Kim A. Y., Timm J. 2008. Resistance mechanisms in HCV: from evolution to intervention. Expert Rev. Anti Infect. Ther. 6:463–478 [DOI] [PubMed] [Google Scholar]
- 23. Kolykhalov A., et al. 2009. In vitro characterization of INX-189, a highly potent phosphoramidate nucleoside analogue inhibitor of HCV, abstr. 101. Abstr. HEPDART: Frontiers in Drug Development for Viral Hepatitis. [Google Scholar]
- 24. Kondo Y., et al. 2010. Lymphotropic HCV strain can infect human primary naive CD4(+) cells and affect their proliferation and IFN-gamma secretion activity. J. Gastroenterol. 46:232–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Koutsoudakis G., Herrmann E., Kallis S., Bartenschlager R., Pietschmann T. 2007. The level of CD81 cell surface expression is a key determinant for productive entry of hepatitis C virus into host cells. J. Virol. 81:588–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kuntzen T., et al. 2008. Naturally occurring dominant resistance mutations to hepatitis C virus protease and polymerase inhibitors in treatment-naive patients. Hepatology 48:1769–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lalezari J., et al. 2008. Potent antiviral activity of the HCV nucleoside polymerase inhibitor R7128 with PEG-IFN and ribavirin: interim results of R7128 500mg BID for 28 days, abstr. 66. Abstr. 43rd Annu. Meet. Eur. Assoc. Study Liver [Google Scholar]
- 28. Lalezari J., et al. 2011. Once daily PSI-7977 plus PegIFN/RBV in a phase 2B trail: rapid virologic suppression in treatment-naive patients with HCV GT2/GT3, abstr. 61. Abstr. 46th Annu. Meet. Eur. Assoc. Study Liver [Google Scholar]
- 29. Lam A. M., Frick D. N. 2006. Hepatitis C virus subgenomic replicon requires an active NS3 RNA helicase. J. Virol. 80:404–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lam A. M., et al. 2010. PSI-7851, a pronucleotide of beta-d-2′-deoxy-2′-fluoro-2′-C-methyluridine monophosphate, is a potent and pan-genotype inhibitor of hepatitis C virus replication. Antimicrob. Agents Chemother. 54:3187–3196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lawitz E., et al. 2010. High rapid virologic response (RVR) with PSI-7977 daily dosing plus PEG-IFN/RBV in a 28-day phase 2a trial, abstr. 806. Abstr. 61st Am. Assoc. Study Liver Dis. [Google Scholar]
- 32. Lawitz E., et al. 2009. Potent antiviral activity observed with PSI-7851, a novel nucleotide polymerase inhibitor for HCV, following multiple ascending oral doses for 3 days in patients with chronic HCV infection, abstr. 102. Abstr. HepDart. [Google Scholar]
- 33. Le Pogam S., et al. 2006. In vitro selected Con1 subgenomic replicons resistant to 2′-C-methyl-cytidine or to R1479 show lack of cross resistance. Virology 351:349–359 [DOI] [PubMed] [Google Scholar]
- 34. Le Pogam S., et al. 2006. Selection and characterization of replicon variants dually resistant to thumb- and palm-binding nonnucleoside polymerase inhibitors of the hepatitis C virus. J. Virol. 80:6146–6154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Le Pogam S., et al. 2008. Low level of resistance and low viral fitness in vitro and absence of resistance mutations in baseline quasispecies may contribute to high barrier to R1626 resistance in vivo, abstr. 6. Abstr. 3rd Int. Workshop Hepatitis C-Resist. New Compounds. [Google Scholar]
- 36. Le Pogam S., et al. 2008. Existence of hepatitis C virus NS5B variants naturally resistant to non-nucleoside, but not to nucleoside, polymerase inhibitors among untreated patients. J. Antimicrob. Chemother. 61:1205–1216 [DOI] [PubMed] [Google Scholar]
- 37. Le Pogam S., et al. 2009. No evidence of R7128 drug resistance after up to 4 weeks treatment of GT1,2 and 3 hepatitis c virus infected individuals. J. Hepatol. 50S:348 [Google Scholar]
- 38. Lohmann V., Hoffmann S., Herian U., Penin F., Bartenschlager R. 2003. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J. Virol. 77:3007–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lucas K. A., et al. 2000. Guanylyl cyclases and signaling by cyclic GMP. Pharmacol. Rev. 52:375–414 [PubMed] [Google Scholar]
- 40. McCarville J. F., et al. 2010. In vitro and in vivo resistance profile of IDX184, a novel nucleotide analog for the treatment of HCV infection, abstr. 9. Abstr. 5th Int. Workshop Hepatitis C-Resist. New Compounds. [Google Scholar]
- 41. McCown M. F., et al. 2008. The hepatitis C virus replicon presents a higher barrier to resistance to nucleoside analogs than to nonnucleoside polymerase or protease inhibitors. Antimicrob. Agents Chemother. 52:1604–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Meppen M., et al. 2009. Cyclic phosphoramidates as prodrugs of 2′-C-methylcytidine. Eur. J. Med. Chem. 44:3765–3770 [DOI] [PubMed] [Google Scholar]
- 43. Migliaccio G., et al. 2003. Characterization of resistance to non-obligate chain-terminating ribonucleoside analogs that inhibit hepatitis C virus replication in vitro. J. Biol. Chem. 278:49164–49170 [DOI] [PubMed] [Google Scholar]
- 44. Murakami E., et al. 2007. Mechanism of activation of beta-d-2′-deoxy-2′-fluoro-2′-C-methylcytidine and inhibition of hepatitis C virus NS5B RNA polymerase. Antimicrob. Agents Chemother. 51:503–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Murakami E., et al. 2010. Mechanism of activation of PSI-7851 and its diastereoisomer PSI-7977. J. Biol. Chem. 285:34337–34347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nguyen T. T., et al. 2003. Resistance profile of a hepatitis C virus RNA-dependent RNA polymerase benzothiadiazine inhibitor. Antimicrob. Agents Chemother. 47:3525–3530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Reddy P. G., et al. 2010. 2′-Deoxy-2′-alpha-fluoro-2′-beta-C-methyl 3′,5′-cyclic phosphate nucleotide prodrug analogs as inhibitors of HCV NS5B polymerase: discovery of PSI-352938. Bioorg. Med. Chem. Lett. 20:7376–7380 [DOI] [PubMed] [Google Scholar]
- 48. Sofia M. J., et al. 2010. Discovery of a beta-d-2′-deoxy-2′-alpha-fluoro-2′-beta-C-methyluridine nucleotide prodrug (PSI-7977) for the treatment of hepatitis C virus. J. Med. Chem. 53:7202–7218 [DOI] [PubMed] [Google Scholar]
- 49. Sommadossi J. P., Schinazi R. F., Chu C. K., Xie M. Y. 1992. Comparison of cytotoxicity of the (−)- and (+)-enantiomer of 2′,3′-dideoxy-3′-thiacytidine in normal human bone marrow progenitor cells. Biochem. Pharmacol. 44:1921–1925 [DOI] [PubMed] [Google Scholar]
- 50. Stuyver L. J., et al. 2002. Antiviral activities and cellular toxicities of modified 2′,3′-dideoxy-2′,3′-didehydrocytidine analogues. Antimicrob. Agents Chemother. 46:3854–3860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Stuyver L. J., et al. 2006. Inhibition of hepatitis C replicon RNA synthesis by beta-d-2′-deoxy-2′-fluoro-2′-C-methylcytidine: a specific inhibitor of hepatitis C virus replication. Antivir. Chem. Chemother. 17:79–87 [DOI] [PubMed] [Google Scholar]
- 52. Stuyver L. J., et al. 2003. Dynamics of subgenomic hepatitis C virus replicon RNA levels in Huh-7 cells after exposure to nucleoside antimetabolites. J. Virol. 77:10689–10694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tomei L., et al. 2003. Mechanism of action and antiviral activity of benzimidazole-based allosteric inhibitors of the hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 77:13225–13231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Vaandrager A. B., de Jonge H. R. 1996. Signalling by cGMP-dependent protein kinases. Mol. Cell. Biochem. 157:23–30 [DOI] [PubMed] [Google Scholar]
- 55. Yi M., Villanueva R. A., Thomas D. L., Wakita T., Lemon S. M. 2006. Production of infectious genotype 1a hepatitis C virus (Hutchinson strain) in cultured human hepatoma cells. Proc. Natl. Acad. Sci. U. S. A. 103:2310–2315 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.