Abstract
Methicillin-resistant Staphylococcus aureus (MRSA) strains are major pathogens causing infections of the skin and soft tissues and more serious, life-threatening diseases, including sepsis and necrotizing pneumonia. The vraSR operon encodes the key regulatory system that modulates the stress response of S. aureus elicited upon exposure to cell wall antibiotics. Mutation of vraS and vraR results in decreased oxacillin resistance in vitro. We investigated the effect of oxacillin treatment in experimental models employing a clinical USA300 MRSA strain (strain 923) and an isogenic vraSR deletion mutant (strain 923-M23). In a murine model of S. aureus necrotizing pneumonia, animals were treated with oxacillin, beginning 15 min after inoculation. Among mice infected with mutant strain 923-M23, oxacillin treatment significantly improved survival compared with saline treatment, whereas oxacillin treatment had no effect in mice infected with strain 923. Similarly, treatment with oxacillin decreased the bacterial burden among animals infected with strain 923-M23 but not among animals infected with strain 923. In a murine skin infection model, oxacillin eliminated the development of dermonecrosis among 923-M23-infected mice and decreased the bacterial burden in the lesions, but not among strain 923-infected mice. We conclude that deletion of the vraSR operon allowed an oxacillin regimen to be effective in murine models of MRSA pneumonia and skin infection. These findings provide proof-of-principle for development of a new antibiotic that could restore the usefulness of oxacillin against MRSA by inhibiting VraS or VraR.
INTRODUCTION
Staphylococcus aureus is a major pathogen causing a variety of infections, including skin and soft tissue infections, necrotizing pneumonia, foreign body infections, septic arthritis, osteomyelitis, endovascular infections, endocarditis, and sepsis in adults and children (22). Since strains of S. aureus resistant to methicillin (MRSA) were first identified (15), they have increased in prevalence in both the healthcare- and community-associated settings (10, 11, 18, 27, 30). The emergence of these resistant isolates has limited the empirical use of β-lactams (9). As a consequence, development of new antimicrobial strategies against these resistant strains is imperative.
MRSA isolates are resistant to all available β-lactam antibiotics by producing a penicillin-binding protein (PBP), PBP2a, which has low affinity for β-lactams (14). The transcription of mecA is regulated by the variably present MecI/MecR1 and the homologous regulatory proteins of staphylococcal β-lactamase, BlaI/BlaR1 (7, 23).
In S. aureus, exposure to cell wall antibiotics induces a stress response resulting in transcriptional induction of over 100 genes that is referred to as the cell wall stimulon (31). Among these genes is a vancomycin- and β-lactam-inducible two-component regulatory system (VraSR) that regulates the transcription of genes involved in cell wall biosynthesis. The VraSR system consists of a sensor kinase, VraS, and a cytoplasmic response regulator, VraR. Their mutation in clinical strains increases in vitro susceptibility to β-lactams and/or vancomycin (6, 12, 20, 32). The basis for the decreased methicillin resistance phenotype of vraSR mutants is unknown, but the increased in vitro susceptibility to oxacillin in MRSA vraSR mutants is independent of oxacillin-induced mecA transcription and the production of its product, PBP2a (6). It is also independent of transcription of pbpB (6) that encodes the native PBP2, which is also involved in effecting methicillin resistance (28).
The effect of VraS and VraR on the methicillin resistance phenotype suggests that these genes and/or their products could be used as targets for antimicrobial therapy when administered together with oxacillin. To investigate this possibility, we studied whether treatment with oxacillin would be effective in experimental pneumonia and skin infection caused by a clinical MRSA isolate with a vraSR deletion.
MATERIALS AND METHODS
Construction of strain 923-M23.
The CA-MRSA USA300 clinical strain 923 has been previously described (6, 24). A vraSR deletion was constructed in this strain using a strategy that used the temperature-sensitive allelic replacement shuttle vector pMAD (1) (Table 1). The upstream and downstream DNA sequence fragments that flank the vraSR operon were each amplified from strain 923 by PCR. The primers used were Vrasr-up-f and Vrasr-up-r for the upstream fragment and Vrasr-down-f and Vrasr-down-r for the downstream fragment (Table 2). The upstream and downstream fragments were sequentially inserted in the pGEM-T vector (Promega). The chloramphenicol acetyltransferase gene fragment was PCR amplified from plasmid pLI50 by using the primers CmR-f and CmR-r (Table 2) and inserted into the XbaI site between the upstream and downstream fragments. The resulting chimeric fragment was ligated into pMAD (in the NcoI site), and transformed into E. coli. Plasmid DNA obtained from E. coli was then transformed into the intermediate S. aureus host strain, RN4220 (19), by electroporation (2). Plasmid DNA harvested from strain RN4220(pQ2820) was then transformed into MRSA strain 923. Blue transformants were selected at 30°C on tryptic soy agar (TSA) containing chloramphenicol or erythromycin (5 mg/liter) and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; 100 mg/liter).
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| 923 | USA 300 CA-MRSA | 6 |
| Q2820 | RN4220 derivative carrying the pMAD vraSR operon deletion construct | This study |
| 923-M23 | Strain 923 with the vraSR operon deleted | This study |
| Q2590 | RN4220 derivative carrying the vraSR operon cloned in pAW8 (pVRASR-2) | 6 |
| 923-M23(pVRASR-2) | Strain 923-M23 carrying pVRASR-2 | This study |
| Plasmids | ||
| pMAD | S. aureus-E. coli allelic replacement vector | 1 |
| pGEM-T vector | Vector used for cloning of DNA fragments in E. coli | Promega |
| pQ2820 | vraSR operon deletion construct cloned in pMAD | This study |
| pAW8 | S. aureus-E. coli shuttle vector | 16 |
| pVRASR-2 | vraSR operon cloned in pAW8 | 6 |
| pLI50 | S. aureus-E. coli shuttle vector containing chloramphenicol acetyltransferase gene | 21 |
Table 2.
Primers used in this study
| Primer | Sequence (5′–3′)a |
|---|---|
| Vrasr-up-f | AAAGGTACCTTGGATACATATGCGCTAAAGTGCGC |
| Vrasr-up-r | AAATCTAGAGAATAGGCTCCGGAACTAAATGATG |
| Vrasr-down-f | AAATCTAGACAATAGTTCGTATCGAATTAAG |
| Vrasr-down-r | AAACTGCAGAGGTTAAGGAGTGCACATGTCG |
| CmR-f | AAATCTAGACGCCGGCAATAGTTACCC |
| CmR-r | AAATCTAGAGGATATGGATCTGGAGCTG |
| SAV1889-f | CAGCTGTTAAAATGGCATAGATGGG |
| SAV1882-f | AATCCTGAATTAGAACGTTC |
Underlined sequences indicate restriction enzyme recognition sequences for KpnI (GGTACC), XbaI (TCTAGA), or PstI (CTGCAG).
To obtain the vraSR operon deletion mutant, strain 923 harboring pQ2820 was grown overnight in tryptic soy broth (TSB) containing erythromycin (5 mg/liter), diluted 1:100 in plain TSB and incubated for 2 h at 30°C, followed by 6 h at 42°C. Serial dilutions of this broth culture were spread onto TSA containing X-Gal and chloramphenicol and then incubated at 42°C overnight. A white colony was selected for further study (strain 923-M23). Deletion of the vraSR operon from the chromosome was confirmed by the absence of a vraSR-specific band in a Southern blot and by DNA sequencing of a PCR fragment from strain 923-M23 that was obtained using the primers Sav1889-f and Sav1882-f corresponding to the upstream and downstream sequences of the vraSR operon (Table 2).
To verify in vitro restoration of the resistant phenotype, we complemented strain 923-M23. To accomplish this, a PCR fragment containing the vraSR operon expressed from its own promoter was cloned in the low-copy-number plasmid pAW8 as described previously (6). The plasmid was introduced into 923-M23 to create strain 923-M23(pVRASR-2).
Broth MIC determinations.
MICs were determined according to Clinical and Laboratory Standards Institute (CLSI) guidelines (8) in 24-well culture plates (Corning, Inc., Corning, NY). Colonies grown on TSA overnight were diluted to a density equivalent to a 0.5 McFarland standard in 0.9% saline and then suspended in TSB containing 2% NaCl to yield a final concentration of 5 × 105 CFU/ml. Oxacillin was tested at 0, 2, 4, 6, 12, 16, 24, 32, 48, 64, 96, 128, and 256 mg/liter. Tetracycline (5 mg/liter) was used to test strains containing pVRASR2 to maintain the plasmid. Turbidity was evaluated after incubation at 37°C for 24 h.
Preparation of bacteria for inoculation and antibiotics for treatment.
Strains 923 and 923-M23 were each subcultured from a −80°C stock onto TSA and incubated at 37°C overnight. A single colony from each strain was inoculated into 5 ml of TSB and incubated at 37°C with shaking. The overnight culture was diluted 1:100 in TSB and grown to early stationary phase, ascertained from the optical density at 600 nm. The culture was then centrifuged at 3,700 × g for 10 min, washed, and resuspended in phosphate-buffered saline (PBS). Each bacterial suspension was diluted to the desired concentration and quantified by plating serial dilutions on TSA; the colonies were enumerated after 24 h of incubation at 37°C. Oxacillin (Sandoz GmbH, Kundl, Austria) was diluted in sterile saline (80 mg/ml). Normal saline of identical volume (0.1 ml) was used for the control treatment. Both solutions were stored at 4°C and used within 4 h following preparation.
Treatment of experimental pneumonia.
The experimental pneumonia model was a modification of that described previously (24, 26). In brief, 6-week-old C57BL/6 male mice (Harlan Laboratories, Indianapolis, IN) were anesthetized with ketamine and xylazine and inoculated intranasally. After inoculation, the mice were held upright for 20 s. Three doses of oxacillin (400 mg/kg) were injected intraperitoneally 1 h apart; the first dose was administered 10 to 15 min after the bacterial inoculation. All animals were allowed free access to chow and water.
To compare the effect of oxacillin treatment in mice infected with strains 923 and 923-M23, we used a high inoculum (4.5 × 108 to 5.5 × 108 CFU/mouse) and a low inoculum, (1.0 × 108 to 2.0 × 108 CFU/mouse). The high inoculum was used to assess mortality, and the low inoculum was used to quantify bacteria in the lung. For both inocula, animals were randomly assigned into four groups of eight animals, each consisting of mice infected with strain 923 or 923-M23 and treated with oxacillin or saline.
After infection with the high inoculum, survival was assessed for 72 h. Mice that received the low inoculum were euthanized with pentobarbital 24 h after inoculation for quantification of the bacterial burden in the lung. The left lung was harvested using aseptic technique, suspended in PBS, and homogenized. After homogenization, the mixture was serially diluted in PBS and plated on mannitol-salt agar. Colonies were enumerated 24 h after incubation at 37°C and expressed as the number of CFU per lung.
Treatment of experimental skin infection.
We modified a model of skin infection in mice that we described previously (25, 26). In brief, 6-week-old C57BL/6 male mice were anesthetized with ketamine and xylazine, shaved, and inoculated by subcutaneous injection in the right flank with 50 μl of a S. aureus suspension (1.4 × 107 CFU/mouse). Oxacillin was injected intraperitoneally at a dose of 400 mg/kg; the dose was administered 20 to 30 min after the bacterial inoculation. Thirty-two animals were randomly assigned into four groups of eight, each consisting of mice infected with strain 923 or strain 923-M23 and treated with oxacillin or saline. The presence and area of dermonecrosis were recorded over the course of 14 days. The area of dermonecrosis (in mm2) was calculated by using the formula l × w, where l is length (in mm) and w is width (in mm). To assess the bacterial burden in the skin lesions, eight mice from each group were sacrificed 3 days after subcutaneous infection with S. aureus (1.2 × 107 CFU/mouse). The homogenized skin lesion was serially diluted in PBS and plated on mannitol-salt agar. The colonies were enumerated 24 h after incubation at 37°C.
Animal procedures were performed in compliance with the University of Chicago Institutional Animal Care and Use Committee guidelines.
Oxacillin pharmacokinetics in mouse serum.
To measure the serum oxacillin concentration, we modified a previously described disc diffusion bioassay (3, 29) using Bacillus subtilis (ATCC 6633) as the indicator strain. Briefly, a standard curve was produced by applying 10-μl aliquots of various oxacillin concentrations (1, 2, 4, 6, 8, 10, 20, 40, 60, 80, 100, 200, 400, 600, 800, and 1,000 mg/liter) to sterile discs that were overlaid on TSA preinoculated with a lawn of B. subtilis. After overnight incubation, the magnitudes of zones of clearing were plotted against the corresponding oxacillin concentrations. A standard curve was determined by using an equation for nonlinear regression fit.
To determine unknown serum oxacillin concentrations among the treated animals, 10 μl of murine serum obtained by cardiac puncture was applied to a disc, and the diameter of the zone of inhibition was compared to that of the standard curve. Mice were sacrificed at multiple time points after dosing with oxacillin (5, 10, 15, 30, and 60 min after the first and second doses of oxacillin and 15, 30, 60, 120, 180, and 300 min after the third dose). After sacrifice, serum was stored at −80°C until testing. The assay was performed in quadruplicate for each time point.
Statistical analysis.
The bacterial densities in the tissue were compared by using a nonparametric Mann-Whitney test. Differences in survival were compared using the Mantel-Cox test. The prevalence of dermonecrosis was compared by the Fisher exact test, and the area of dermonecrosis was compared by using an unpaired t test. All statistical data were analyzed by using Prism 5 program (GraphPad Software, Inc., San Diego, CA). A P value of <0.05 was considered significant.
RESULTS
Deletion of vraSR increased susceptibility to oxacillin in vitro.
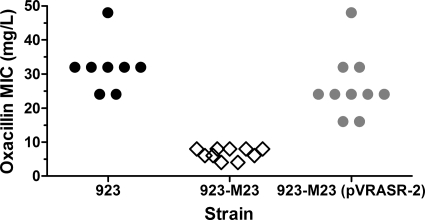
The mode MIC of oxacillin for the CA-MRSA USA300 clinical strain 923 was 32 mg/liter. Deleting the vraSR operon from strain 923 greatly increased susceptibility to oxacillin (mode MIC, 8 mg/liter) (Fig. 1). Complementation of the vraSR deletion strain, 923-M23, with a shuttle vector containing the vraSR operon restored resistance to oxacillin (mode MIC, 24 mg/liter) (Fig. 1). These data confirm conclusions from previous studies that the vraSR operon plays a key role in oxacillin resistance in vitro.
Fig. 1.
MIC of oxacillin of S. aureus strains. Each determination was performed eight to 10 times. Each symbol represents a single result.
Deletion of vraSR increased susceptibility to oxacillin in a mouse model of necrotizing pneumonia.
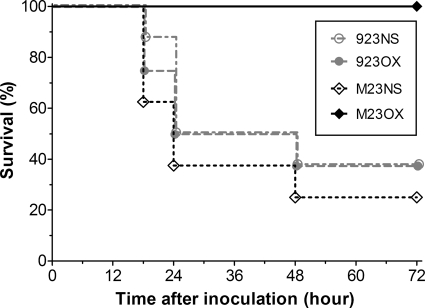
The survival rate of saline-treated animals infected with strain 923-M23 was somewhat lower than that found among the strain 923-infected animals (38% versus 50%, respectively, at 24 h and 25% versus 38%, respectively, at 72 h postinoculation) (Fig. 2). The difference, however, was not significant (P = 0.45). Thus, the vraSR operon deletion did not significantly affect the virulence of strain 923-M23.
Fig. 2.
Effect of oxacillin treatment on the survival of mice infected with wild-type CA-MRSA (strain 923) or its isogenic vraSR deletion mutant (strain 923-M23). The survival rates at 18, 24, 48, and 72 h after inoculation are depicted. 923 and M23 refer to strains 923 and 923-M23, respectively. NS, normal saline treated; OX, oxacillin treated.
Among strain 923-infected mice, treatment with oxacillin did not affect survival compared with saline-treated animals (P = 0.89). In contrast, all oxacillin-treated mice infected with strain 923-M23 survived the 72-h observation period and looked active and well. Thus, the survival rate of oxacillin-treated mice infected with strain 923-M23 was significantly greater than that of oxacillin-treated mice infected with strain 923 (P < 0.01). At 72 h, the survival rate of mice infected with strain 923-M23 and treated with oxacillin also significantly exceeded that found among mice infected with the same strain but treated with saline (100% versus 25%) (P < 0.01) (Fig. 2).
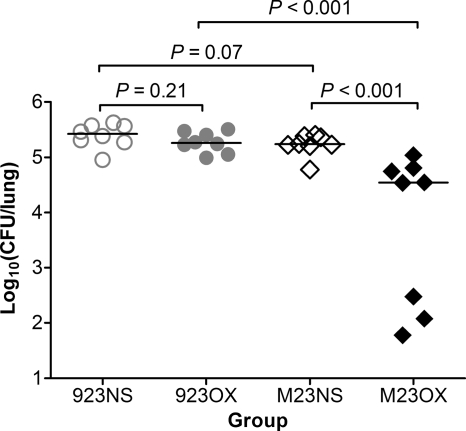
Oxacillin treatment decreased the bacterial burden in the lung following infection with a vraSR deletion mutant.
Among mice infected with strain 923-M23, oxacillin treatment significantly decreased the bacterial burden in the lungs compared with saline-treated animals inoculated with this strain (P < 0.001) or compared with oxacillin-treated mice inoculated with wild-type strain 923 (P < 0.001) (Fig. 3). In contrast, there was no significant difference in the lung bacterial burden comparing oxacillin- and saline-treated mice infected with strain 923 (P = 0.21) (Fig. 3).
Fig. 3.
Bacterial density in the lung 24 h after inoculation. Horizontal lines indicate the median CFU for each group. 923NS, group infected with strain 923 and treated with normal saline; 923OX, group infected with strain 923 and treated with oxacillin; M23NS, group infected with strain 923-M23 and treated with normal saline; M23OX, group infected with strain 923-M23 and treated with oxacillin.
Oxacillin treatment prevented dermonecrosis after infection with a vraSR deletion mutant.
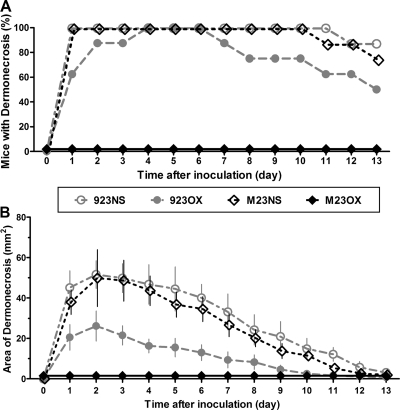
Saline-treated mice inoculated either with strain 923 or strain 923-M23 developed a dermonecrotic skin lesion (Fig. 4A) that reached a maximal size 2 days after inoculation and diminished thereafter (Fig. 4B) until disappearing at about day 11 or thereafter. The mean maximal area of dermonecrosis did not differ significantly among mice inoculated with either of these two strains (P = 0.91).
Fig. 4.
Dermonecrosis among mice subcutaneously inoculated with wild-type CA-MRSA (strain 923) or a strain with the vraSR operon deleted (strain 923-M23). (A) Percentage of animals per group that had dermonecrosis on each day in the 14-day period. (B) Area of dermonecrosis. The data are means ± the standard errors of the mean (SEM). 923NS, group infected with strain 923 and treated with normal saline; 923OX, group infected with strain 923 and treated with oxacillin; M23NS, group infected with strain 923-M23 and treated with normal saline; M23OX, group infected with strain 923-M23 and treated with oxacillin.
Oxacillin-treated mice inoculated with strain 923 uniformly had a dermonecrotic lesion that followed a time course similar to that observed among the saline-treated animals (Fig. 4B), except that the appearance of dermonecrosis was delayed, with onset on days 2 to 4 in three animals in this group (Fig. 4A); also, the mean maximal area of dermonecrosis was smaller than that occurring among saline-treated controls (P = 0.02).
In contrast, dermonecrosis did not occur in mice inoculated with strain 923-M23 that were treated with oxacillin compared with oxacillin-treated mice infected with strain 923 (P < 0.01) (Fig. 4). Only three of the eight 923-M23-inoculated, oxacillin-treated animals developed a subcutaneous abscess. The abscesses were small (diameter < 2 mm) and resolved within 7 days after inoculation.
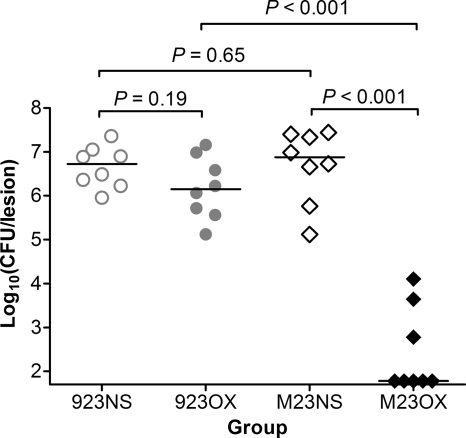
Oxacillin treatment decreased the bacterial burden in skin lesions following infection with a vraSR deletion mutant.
The bacterial burden in the skin lesions of saline-treated animals inoculated with strain 923 or strain 923-M23 did not differ significantly (P = 0.65). Similarly, there was no significant difference in CFU between the oxacillin- and saline-treated mice infected with strain 923 (P = 0.19).
However, among oxacillin-treated mice infected with strain 923-M23, the bacterial burden in the skin lesions was significantly lower than that among the saline-treated mice (P < 0.001) and oxacillin-treated mice infected with strain 923 (P < 0.001) (Fig. 5).
Fig. 5.
Bacterial density in the skin lesion 3 days after inoculation. Horizontal lines indicate the median CFU for each group. 923NS, group infected with strain 923 and treated with normal saline; 923OX, group infected with strain 923 and treated with oxacillin; M23NS, group infected with strain 923-M23 and treated with normal saline; M23OX, group infected with strain 923-M23 and treated with oxacillin.
Serum oxacillin pharmacokinetics.
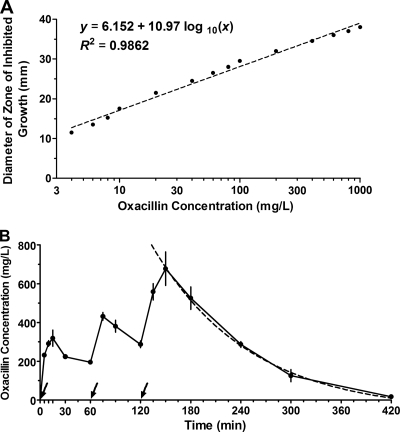
Measured zones of inhibition of growth ranged between 11 and 38 mm. The relationship between the diameter (y [mm]) and the oxacillin concentration (x [mg/liter]) was linear and described by a nonlinear regression fit of the equation: y = 6.152 × 10.97 log10(x), R2 = 0.9862 (Fig. 6A).
Fig. 6.
Oxacillin pharmacokinetics in mice. (A) Standard curve of the relationship between oxacillin concentration and diameter of zone of inhibited growth. Each standard concentration was tested in quadruplicate and each symbol represents the mean (dot) and the SEM (bar). (B) Serum oxacillin half-life in treated mice. Arrows indicate the times of the three doses administered in treatment of pneumonia. Each symbol represents the mean (dot) and the SEM (vertical line) for four serum samples at each time point. Each sample was from one 6-week-old C56BL/6 mouse and tested in quadruplicate. The half-life was determined by using a one compartment exponential decay model and the data points 180 to 420 min (dashed line).
The serum samples from mice treated with oxacillin produced zones of inhibition that ranged from 15 to 39 mm in size. Based on the standard curve, the serum oxacillin concentration peaked 30 min after the third injection, and the half-life was 80 min when calculated with a one-compartment exponential decay model (Fig. 6B).
DISCUSSION
Our data demonstrate that the decreased in vitro oxacillin resistance observed in an MRSA genetic background with a vraSR operon deletion is associated with improved outcome with oxacillin treatment in both experimental pneumonia and skin infection. The vraSR operon deletion was constructed in MRSA strain 923, which belongs to the USA300 lineage that is epidemic in the United States (10) and is highly virulent (24). In experimental pneumonia, the vraSR deletion eliminated mortality and lowered the CFU recovered from the lungs of animals treated with oxacillin. In experimental skin infections, the vraSR operon deletion eliminated dermonecrosis and decreased the number of bacteria present in lesions of animals treated with oxacillin. We did not explore whether the identical effect would have been achieved had we waited longer to initiate treatment after experimental inoculation.
We did not reproduce antimicrobial regimens that are typically used for patients. The half-life of oxacillin administered intravenously to patients is 20 to 30 min (4, 17), which is somewhat shorter than the 80-min half-life we calculated after 3 intraperitoneal administrations, 1 h apart. Nevertheless, the treatment conditions we used were effective in demonstrating a difference in susceptibility of the wild-type and vraSR mutant strains in vivo.
We followed inoculated animals for 72 hours. It is possible that relapse may have occurred after that time. The likelihood seems remote as surviving animals looked well at that time. The vraSR operon mutant strain, 923-M23, had a dramatically decreased MIC of oxacillin in vitro, confirming that vraSR is essential to expression of the methicillin resistance phenotype (20). However, the MIC was still in the resistant range according to CLSI guidelines. Nevertheless, the decrease in oxacillin MIC was sufficient to significantly improve the outcome of the oxacillin regimen that we used in the rodent models. We previously reported that insertional inactivation of the vraS and vraR genes produced a phenotype in vitro similar to that seen in the present study with deletion of the complete operon (6), and we hypothesize, therefore, that experimental infection with a strain with these two genes inactivated would produce similar results to those that we report here.
The mechanism by which increased susceptibility to oxacillin occurs in MRSA isolates with a vraSR mutation is unknown. The oxacillin-dependent transcriptional induction of many genes was shown to be dysregulated in S. aureus strains that had undergone allelic replacement in the vraSR locus (6, 20). Therefore, the implication of increased susceptibility to oxacillin in strain 923-M23 is that the oxacillin-mediated induction of one or more of those downstream genes is essential for the methicillin resistance phenotype. The decrease in the methicillin resistance phenotype occurred despite the maintenance of mecA (that resides in this strain on an SCCmec IV element), as well as the native pbpB gene; both are necessary for the methicillin resistance phenotype (28). Also, the observed increase in oxacillin susceptibility in strain 923-M23 is not likely to be related to mecA expression since it was paradoxically increased in vraSR mutants we have studied (6). In a similar vein, although β-lactam-mediated pbpB induction is lost in vraSR mutants, constitutive pbpB transcription and translation persists, irrespective of the presence of β-lactam antibiotics (5). Moreover, overexpression of pbpB in trans did not restore resistance to a vraS mutant (6). Thus, although mecA and pbpB play an important role in effecting the methicillin resistance phenotype (28), their behavior in vraSR mutants does not readily explain the decreased oxacillin resistance that we observed.
The vraSR operon consists of four ORFs with a single known transcription start site. The two distal genes, vraS and vraR, have characteristics of a typical two-component signal transduction system. The functions of the two proximal genes, orf1 and orf2, are not known. The stimuli for induction of the vraSR operon are incompletely understood. Induction of the operon occurs after exposure to a broad spectrum of antibiotics that work by interfering with cell wall synthesis including β-lactams, daptomycin, and cationic peptides (6, 20, 12). Antibiotics that work by inhibiting protein synthesis do not upregulate vraSR.
Our findings here have potential importance for new antimicrobial drug design. The Infectious Diseases Society of America has initiated a program called the “10 × 20 Initiative” (13). This program has grown out of concern that the development of new antibiotics has dramatically slowed in the last 20 years. New antimicrobials are urgently needed. Especially valuable in this regard is basic research to identify new antimicrobial targets and compounds that limit microbial activities other than synthesis and maintenance of cell walls. The identification of a cell signaling mechanism such as VraSR and the recognition of its central role in allowing S. aureus pathogenic isolates to express an antibiotic resistance phenotype is an example of a novel strategy by which antimicrobials could target microorganisms. For example, a β-lactam antimicrobial combined with a VraSR inhibitor might be an effective treatment and represents an antimicrobial directed against a novel target that can potentiate the effect of cell wall antimicrobials. In this regard, the present study provides the first proof of principle in vivo for targeting VraSR as a potentiator of oxacillin.
ACKNOWLEDGMENTS
This study was supported by the Research Funds of Chonbuk National University in 2009 (D.S.J.); the Pediatric Critical Care Scientist Development Program and National Institute of Allergy and Infectious Disease (1K08AI076596-01A1) (C.P.M.); and grants R56, A1040481-08 A1S1, and 1R01A1067 584-04 (R.S.D. and S.B.-V.).
R.S.D. served on paid advisory boards sponsored by Clorox (in 2007 and 2010), Pfizer (in 2007), Astellas (in 2007 and 2010), Merck (in 2008), GlaxoSmithKline (in 2009), Novartis (in 2009), and Glycovaxyn (in 2010). The other authors report no potential conflicts of interest.
Footnotes
Published ahead of print on 7 March 2011.
REFERENCES
- 1. Arnaud M., Chastanet A., Debarbouille M. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl. Environ. Microbiol. 70:6887–6891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Augustin J., Gotz F. 1990. Transformation of Staphylococcus epidermidis and other staphylococcal species with plasmid DNA by electroporation. FEMS Microbiol. Lett. 54:203–207 [DOI] [PubMed] [Google Scholar]
- 3. Axline S. G., Yaffe S. J., Simon H. J. 1967. Clinical pharmacology of antimicrobials in premature infants. II. Ampicillin, methicillin, oxacillin, neomycin, and colistin. Pediatrics 39:97–107 [PubMed] [Google Scholar]
- 4. Baxter Healthcare Corp. 3 November 2010, accession date Oxacillin (oxacillin sodium) injection, solution. Baxter Healthcare Corp., Deerfield, IL: http://dailymed.nlm.nih.gov/dailymed/drugInfo.cfm?id=4055 [Google Scholar]
- 5. Boyle-Vavra S., Yin S., Challapalli M., Daum R. S. 2003. Transcriptional induction of the penicillin-binding protein 2 gene in Staphylococcus aureus by cell wall-active antibiotics oxacillin and vancomycin. Antimicrob. Agents Chemother. 47:1028–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Boyle-Vavra S., Yin S., Daum R. S. 2006. The VraS/VraR two-component regulatory system required for oxacillin resistance in community-acquired methicillin-resistant Staphylococcus aureus. FEMS Microbiol. Lett. 262:163–171 [DOI] [PubMed] [Google Scholar]
- 7. Chambers H. F. 1997. Methicillin resistance in staphylococci: molecular and biochemical basis and clinical implications. Clin. Microbiol. Rev. 10:781–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Clinical and Laboratory Standards Institute 2007. Performance standards for antimicrobial susceptibility testing; 17th informational supplement. CLSI M100-S17. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 9. Daum R. S. 2007. Clinical practice: skin and soft-tissue infections caused by methicillin-resistant Staphylococcus aureus. N. Engl. J. Med. 357:380–390 [DOI] [PubMed] [Google Scholar]
- 10. David M. Z., Daum R. S. 2010. Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin. Microbiol. Rev. 23:616–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fridkin S. K., et al. 2005. Methicillin-resistant Staphylococcus aureus disease in three communities. N. Engl. J. Med. 352:1436–1444 [DOI] [PubMed] [Google Scholar]
- 12. Gardete S., Wu S. W., Gill S., Tomasz A. 2006. Role of VraSR in antibiotic resistance and antibiotic-induced stress response in Staphylococcus aureus. Antimicrob. Agents Chemother. 50:3424–3434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Infectious Diseases Society of America 2010. The 10 × '20 Initiative: pursuing a global commitment to develop 10 new antibacterial drugs by 2020. Clin. Infect. Dis. 50:1081–1083 [DOI] [PubMed] [Google Scholar]
- 14. Ito T., et al. 2001. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45:1323–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jevons M. P. 1961. “Celbenin”-resistant staphylococci. BMJ 1:124–125 [Google Scholar]
- 16. Katayama Y., Zhang H. Z., Hong D., Chambers H. F. 2003. Jumping the barrier to beta-lactam resistance in Staphylococcus aureus. J. Bacteriol. 185:5465–5472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kind A. C., Tupasi T. E., Standiford H. C., Kirby W. M. M. 1970. Mechanisms responsible for plasma levels of nafcillin lower than those of oxacillin. Arch. Intern. Med. 125:685–690 [PubMed] [Google Scholar]
- 18. Klevens R. M., et al. 2007. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 298:1763–1771 [DOI] [PubMed] [Google Scholar]
- 19. Kreiswirth B. N., et al. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709–712 [DOI] [PubMed] [Google Scholar]
- 20. Kuroda M., et al. 2003. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol. Microbiol. 49:807–821 [DOI] [PubMed] [Google Scholar]
- 21. Lee C. Y., Buranen S. L., Ye Z. H. 1991. Construction of single-copy integration vectors for Staphylococcus aureus. Gene 103:101–105 [DOI] [PubMed] [Google Scholar]
- 22. Lowy F. D. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520–532 [DOI] [PubMed] [Google Scholar]
- 23. McKinney T. K., Sharma V. K., Craig W. A., Archer G. L. 2001. Transcription of the gene mediating methicillin resistance in Staphylococcus aureus (mecA) is corepressed but not coinduced by cognate mecA and beta-lactamase regulators. J. Bacteriol. 183:6862–6868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Montgomery C. P., et al. 2008. Comparison of virulence in community-associated methicillin-resistant Staphylococcus aureus pulsotypes USA300 and USA400 in a rat model of pneumonia. J. Infect. Dis. 198:561–570 [DOI] [PubMed] [Google Scholar]
- 25. Montgomery C. P., Boyle-Vavra S., Daum R. S. 2009. The arginine catabolic mobile element is not associated with enhanced virulence in experimental invasive disease caused by the community-associated methicillin-resistant Staphylococcus aureus USA300 genetic background. Infect. Immun. 77:2650–2656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Montgomery C. P., Boyle-Vavra S., Daum R. S. 2010. Importance of the global regulators agr and saeRS in the pathogenesis of CA-MRSA USA300 infection. PLoS One 5:e15177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Moran G. J., et al. 2006. Methicillin-resistant S. aureus infections among patients in the emergency department. N. Engl. J. Med. 355:666–674 [DOI] [PubMed] [Google Scholar]
- 28. Pinho M. G., de Lencastre H., Tomasz A. 2001. An acquired and a native penicillin-binding protein cooperate in building the cell wall of drug-resistant staphylococci. Proc. Natl. Acad. Sci. U. S. A. 98:10886–10891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Simon H. J., Yin E. J. 1970. Microbioassay of antimicrobial agents. Appl. Microbiol. 19:573–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tiemersma E. W., et al. 2004. Methicillin-resistant Staphylococcus aureus in Europe, 1999–2002. Emerg. Infect. Dis. 10:1627–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Utaida S., et al. 2003. Genome-wide transcriptional profiling of the response of Staphylococcus aureus to cell-wall-active antibiotics reveals a cell-wall-stress stimulon. Microbiology 149:2719–2732 [DOI] [PubMed] [Google Scholar]
- 32. Yin S., Daum R. S., Boyle-Vavra S. 2006. VraSR two-component regulatory system and its role in induction of pbp2 and vraSR expression by cell wall antimicrobials in Staphylococcus aureus. Antimicrob. Agents Chemother. 50:336–343 [DOI] [PMC free article] [PubMed] [Google Scholar]