Abstract
Penicillin-binding protein 2a (PBP2a), the molecular determinant for high-level β-lactam resistance in methicillin-resistant Staphylococcus aureus (MRSA), is intrinsically resistant to most β-lactam antibiotics. The development and characterization of new inhibitors targeting PBP2a would benefit from an effective and convenient assay for inhibitor binding. This study was directed toward the development of a fluorescently detected β-lactam binding assay for PBP2a from MRSA. Biotinylated ampicillin and biotinylated cephalexin were tested as tagging reagents for fluorescence detection by using a streptavidin-horseradish peroxidase conjugate. Both bound surprisingly well to PBP2a, with binding constants of 1.6 ± 0.4 μM and 13.6 ± 0.8 μM, respectively. Two forms of the assay were developed, a one-step direct competition form of the assay and a two-step indirect competition form of the assay, and both forms of the assay gave comparable results. This assay was then used to characterize PBP2a binding to ceftobiprole, which gave results consistent with previous studies of ceftobiprole-PBP2a binding. This assay was also demonstrated for screening for PBP2a inhibitors by screening a set of 13 randomly selected β-lactams for PBP2a inhibition at 750 μM. Meropenem was observed to give substantial inhibition in this screen, and a follow-up titration experiment determined its apparent Ki to be 480 ± 70 μM. The availability of convenient and sensitive microtiter-plate based assays for the screening and characterization of PBP2a inhibitors is expected to facilitate the discovery and development of new PBP2a inhibitors for use in combating the serious public health problem posed by MRSA.
INTRODUCTION
Bacterial infections were the major cause of death and morbidity prior to the development of modern antibiotics, and the increasing resistance of pathogenic bacteria to commonly used antibacterial agents is of major public health concern. One organism of particular concern is methicillin-resistant Staphylococcus aureus (MRSA) (1, 7, 11). The high level of β-lactam resistance seen in MRSA, compared to methicillin-sensitive S. aureus (MSSA), is due to the presence of a novel acquired penicillin-binding protein (PBP): PBP2a (10; recently reviewed in references 13, 14, and 19). PBP2a is a high-molecular-mass (HMM) PBP that is intrinsically resistant to most β-lactam antibiotics. Assay methods for inhibitor binding to PBP2a have been described based on radiolabeled β-lactam binding (17) or on nitrocefin (9) (a chromogenic cephalosporin derivative) or BOCILIN-FL (a fluorescently tagged penicillin derivative) binding (8, 20). However, these assays are incompatible with a microtiter plate format required for efficient high-throughput inhibitor screening and characterization. Given the high intrinsic resistance of PBP2a to β-lactams, it was uncertain whether a microtiter plate β-lactam binding assay of the type we have described recently for other HMM PBPs (16), e.g., based on biotinylated β-lactams, would work with PBP2a. In the study reported here, we investigate the use of biotinylated β-lactams in microtiter plate assays for PBP2a inhibitor screening and characterization.
A common approach to determining PBP binding affinities is with a two-step assay, where the β-lactam test agent is preincubated with the PBP for a short period (10 to 30 min) to allow complex formation, followed by the addition of a saturating concentration of a β-lactam probe agent (such as a radiolabeled, fluorescently labeled, or biotin-labeled β-lactam) for a second short period (10 to 30 min), which reacts with and labels the uncomplexed PBP (see references 3, 4, and 15). This approach is based on relatively slow β-lactam release kinetics from the PBP target, so that the addition of probe cannot shift the equilibrium between the test agent-PBP complex substantially in the incubation time used. The slow off-rates observed for β-lactam probes bound to PBP2a indicates that such a kinetic approach is appropriate for measuring test agent β-lactam binding to PBP2a (9). To provide a basis for comparing the one-step steady-state approach we described previously (16) to a classic two-step kinetic approach, a microtiter plate-based two-step assay was also implemented.
MATERIALS AND METHODS
Cloning of a truncated mecA gene.
Chromosomal DNA of MRSA (ATCC 3300) was used as a template for PCR. Primers were designed based on the published mecA sequence from the National Center for Biotechnology Information and the primers being a forward primer 5′-PBP2a-EcoRI,BamHI (5′-GGATCCGAATTCCTGGAAGTTCTGTTCCAGGGGCCCATGGCTTCAAAAGATAAA-3′) and a reverse primer 3′-PBP2a-XhoI,HindIII (5′-AAGCTTCTCGAGTTATTCATCTATATCGTA-3′). The primers were designed so that the first 23 amino acids at the N terminus were deleted. The resulting DNA fragment (∼2 kb) was gel purified and then extracted by using a gel purification kit (Invitrogen) according to the manufacturer's protocol. The gene was ligated using T4 ligase into the pGEM-T vector (Promega, Madison, WI), and transformed into competent XL1-Blue cells. The mecA gene in the pGEM-T vector was sequenced by using T7 and SP6 promoter primers. The verified insert DNA and the pGEX-4T1 vector (GE Healthcare, Piscataway, NJ) were both digested with the same restriction enzymes (EcoRI and XhoI) and then ligated together to give the expression vector pGEX-PBP2a.
PBP2a expression.
The recombinant vector pGEX-PBP2a was transformed into Escherichia coli BL21(DE3) cells (Invitrogen) for protein expression. Cells were grown in Luria-Bertani broth containing 100 μg/ml of ampicillin at 37°C, until the culture reached an optical density at 600 nm of 0.6. The culture was chilled in an ice bath for 10 min and then placed in a shaker at 18°C, and protein expression was induced by adding 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Cells were then grown overnight (16 h) at 18°C with shaking and then harvested by centrifugation at 4°C. For large-scale production, three 1-liter flasks each containing 350 ml of culture were used.
GST-PBP2a purification.
All purification steps were performed at 4°C. The bacterial cell pellet was resuspended in 60 ml of cold lysis buffer (40 mM Na2HPO4, 10 mM KH2PO4, 300 mM NaCl [pH 7.4]). Bacterial cells were lysed using a Microfluidizer (model M100L; Microfluidics, Newton, MA). The bacterial extract was centrifuged for 30 min at 30,000 × g, the supernatant was collected and spun for an additional 20 min, and the supernatant was again collected and stored frozen at −80°C. The GST-PBP2a fusion protein was purified on GST resin (GenScript catalog no. L00206) according to the manufacturer's instructions.
Cleavage of the GST tag.
Thrombin (GE Healthcare, catalog no. 27-0846-01) was reconstituted with phosphate-buffered saline (PBS) to give a final solution of 1 U/μl. Small aliquots were stored at −80°C. To cleave the glutathione S-transferase (GST) tag from purified GST-PBP2a, 5 mg of GST-PBP2a was treated with 70 U of thrombin at 4°C for overnight. SDS-PAGE was used to confirm cleavage. The free GST tag and uncleaved GST-PBP2a were removed from the cleaved product PBP2a by passage over the GST resin as described above. Purified untagged PBP2a passed though the column in the flowthrough, whereas GST and GST-PBP2a were retained. Fractions were collected, analyzed for purity by SDS-PAGE and for protein concentration by Bradford assay, and then concentrated.
BIO-AMP and BIO-CEPH preparation.
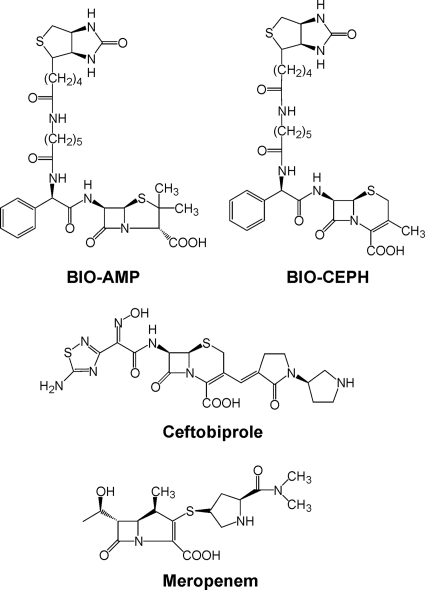
Biotinylated ampicillin (BIO-AMP) (Fig. 1) was prepared by a modification of the method of Dargis and Malouin (4) as described previously (16). Biotinylated cephalexin (BIO-CEPH) (Fig. 1) was prepared using the same procedure except that the final stock concentration was 4.2 mM due to the lower solubility of BIO-CEPH.
Fig. 1.
Structures of key β-lactams used in the present study.
General procedure for GST-PBP2a loading onto microtiter plates, labeling, and detection.
The protocol was similar to that described previously (16). Briefly, for GST-PBP2a attachment, black-walled microtiter plate wells (Costar, product no. 3631) were treated with 0.5 to 1 μg of GST-PBP2a in 50 μl of PBS–20% glycerol at 25°C for 30 min with gentle rocking, followed by treatment (three times) with 150 μl of blocking buffer (PBS–0.2% Tween 20)/well, and then washed three times with 200 μl of washing buffer (PBS–0.05% Tween 20)/well. To label GST-PBP2a in initial proof-of-principle experiments, 50 μl of 100 μM BIO-AMP or BIO-CEPH in PBS was added to the wells. After 15 min, the biotin-labeled GST-PBP2a was denatured with heating at 80°C for 3 min in a water bath, followed by quick cooling on ice. The plates were then washed three times with washing buffer (250 μl of PBS–0.05% Tween 20). Streptavidin-horseradish peroxidase (HRP) conjugate (Pierce, catalog no. 21126) (50 μl of a 0.4-μg/ml concentration) was then added to each well. After 30 min, the wells were washed three times with washing buffer, and 100 μl of a fluorescent HRP substrate mixture (1 mM H2O2 and 20 μM Amplex Red [Molecular Probes] in 100 mM Tris [pH 8.5]) was added to each well. After 30 to 60 min, the fluorescence signal was read (excitation, 546 nm; emission, 595 nm) in a Tecan Spectrafluor Plus microtiter plate reader. An identical protocol was also followed using untagged PBP2a for comparison.
Determination of BIO-AMP and BIO-CEPH Kmapp values for binding to PBP2a.
To assess BIO-AMP and BIO-CEPH binding to PBP2a, microtiter plate bound PBP2a was treated with serially diluted (two steps) concentrations of BIO-AMP or BIO-CEPH (each concentration tested in triplicate), and the remaining steps of the assay were performed as described above. To assess the effect of the labeling reaction incubation time (in the GST-PBP2a + BIO-AMP reaction) on the detected signal, an alternative incubation time of 30 min for labeling was also tested. The data (relative fluorescence units [RFU]) were analyzed for the Kmapp (i.e., the apparent steady-state binding constant to PBP2a, since PBPs turn over β-lactams, albeit generally very slowly) by fitting the data to equation 1 (16). No protein blanks were also included in these experiments.
| (1) |
Solution-phase SDS-PAGE-based BIO-AMP binding assay to confirm untagged PBP2a BIO-AMP binding activity and to provide a solution-phase Kmapp for BIO-AMP.
As described further below, GST-PBP2a gave good signals in the above-described assay, but untagged PBP2a failed to give any detectible signal, even when the amount of PBP2a loaded into each well was increased up to 8 μg/well. This observation suggested that either untagged PBP2a was unable to bind BIO-AMP (e.g., it was inactive) or that untagged PBP2a was unable to bind to the microtiter plate wells. It was also desirable to determine whether the affinity of GST-PBP2a for BIO-AMP in solution was the same as in the microtiter plate-based assay and also whether GST-PBP2a (in solution) had the same affinity as PBP2a (in solution, and also if active). To address these issues, GST-PBP2a and untagged PBP2a were labeled in solution with BIO-AMP, the complex denatured and resolved by SDS-PAGE, and BIO-AMP-labeled proteins were detected using a procedure similar to that described by Dargis and Malouin (4). A range of concentrations of BIO-AMP were used in the labeling reaction to assess affinity (0 × Kmapp, 1/8 × Kmapp, 1/2 × Kmapp, 2 × Kmapp, and 8 × Kmapp).
Characterization of inhibition of PBP2a using a one-step approach.
For inhibitor screening and characterization, the concentration of BIO-AMP was used at a fixed concentration equal to its determined Kmapp for PBP2a (1.6 μM). This was selected since it is high enough to give one-half the maximum possible assay signal and low enough to still allow inhibition to be readily detected. GST-PBP2a was first attached to the wells of a microtiter plate as described above. Serially (two steps) diluted solutions of the prospective inhibitor, plus BIO-AMP at a fixed concentration of 1.6 μM, in 100 μl of PBS were added to the wells, with each concentration tested in triplicate. After 15 min, the binding reactions were stopped, and the plates were developed as described above. With [BIO-AMP] = Kmapp and taking into account the background (blank) fluorescence, the competitive (with BIO-AMP) inhibitor binding isotherm will be described by equation 2 (16):
| (2) |
Inhibitor binding data were plotted, and the saturation curves were analyzed for the Kiapp of binding by fitting the data to equation 2.
Application to screening of potential PBP2a inhibitors using a one-step approach.
To demonstrate the potential of this assay for inhibitor screening and characterization, a demonstration screening of 13 randomly selected β-lactam antibiotics (Table 1) against GST-PBP2a was performed. Screening was done with a high concentration of each antibiotic (750 μM), with the BIO-AMP concentration fixed at its Kmapp. Each determination was made in triplicate.
Table 1.
Screening results in terms of the percent inhibition of BIO-AMP binding in order of increasing potency
| β-Lactam (750 μM) | % Inhibition (mean ± SE) |
|---|---|
| Penicillin G | 0 ± 2 |
| Cefoxitin | 0 ± 10 |
| Cefuroxime | 0 ± 8 |
| Carbenicillin | 0 ± 5 |
| Cephaloridine | 0 ± 2 |
| Cefotaxime | 0 ± 7 |
| Aztreonam | 0 ± 6 |
| Ceftriaxone | 0 ± 6 |
| Ampicillin | 8 ± 2 |
| Cephalexin | 15 ± 1 |
| Cefoperazone | 39 ± 1 |
| Amoxicillin | 48 ± 4 |
| Meropenem | 80 ± 2 |
Characterization of inhibition of PBP2a using a two-step assay approach.
GST-PBP was bound to microtiter plate wells as described above. In the first step, various concentrations of ceftobiprole (Fig. 1) in 50 μl of PBS were added to the wells. For the second step, after 15 min 50 μl of BIO-AMP at 16 times the Kmapp for BIO-AMP (25.6 μM) (final concentration of 8 times the Kmapp) was added to each well. After an additional 15 min, the reactions were stopped by heat denaturation, and the plates were developed as described above. Taking into account the background (blank) fluorescence, the inhibitor binding isotherm in this experimental design will be described by equation 3, which can be derived following the same general procedure as previously described for equation 2 (16).
| (3) |
Data collected using the two-step assay were analyzed by fitting with equation 3.
RESULTS AND DISCUSSION
Assay development.
Given the intrinsic resistance of PBP2a to most β-lactam antibiotics, it was uncertain at the outset whether BIO-AMP and/or BIO-CEPH would bind to PBP2a. PBP2a was therefore overexpressed as a GST fusion protein, and the potential of a BIO-AMP- or BIO-CEPH-based assay approach tested. In initial tests, strong signals were obtained with either 100 μM BIO-AMP or BIO-CEPH as the detection reagent, demonstrating the feasibility of this approach.
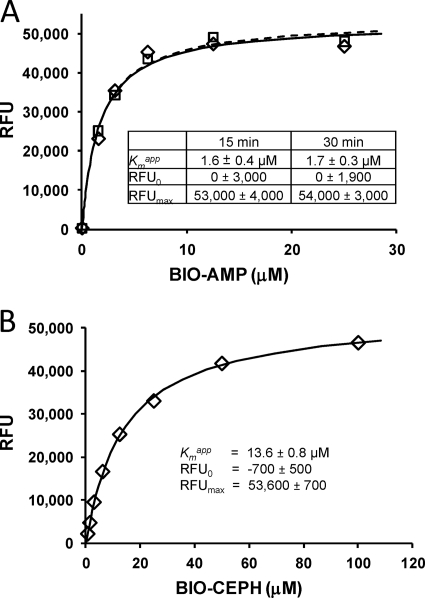
GST-PBP2a was next titrated with both BIO-AMP and BIO-CEPH to determine their respective apparent binding constants (Kmapp values). Both gave classic saturation binding curves (Fig. 2). Analysis of these curves demonstrated binding constants of 1.6 ± 0.4 μM for BIO-AMP and 13.6 ± 0.8 μM for BIO-CEPH (± the standard errors). These are both surprisingly good binding constants and demonstrate that the biotinyl group can apparently form favorable interactions within the active site of PBP2a. That these two biotinylated β-lactams can effectively bind to PBP2a provides a foundation for their use in characterizing other inhibitors of PBP2a in competitive binding experiments.
Fig. 2.
(A) Data, best-fit curves, and best-fit parameter values for titration of GST-PBP2a with BIO-AMP with 15- and 30-min incubations. The data points obtained after 15-min incubations are denoted by diamonds (♢) and those obtained after 30-min incubations are denoted by boxes (□). The best-fit curves through data points obtained after 15 min of incubation are denoted by a solid line, and the curve obtained after 30 min of incubation is denoted by a dashed line. (B) Data and results for BIO-CEPH with a 15-min incubation. For all values, means ± the standard errors are given.
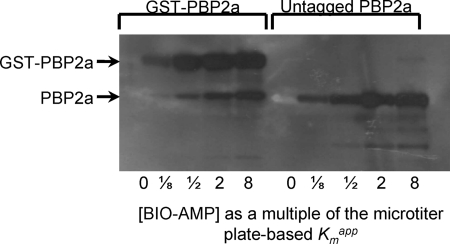
The same effort using untagged PBP2a (obtained by cleaving the GST tag using thrombin) gave no signals. At first it was suspected that the untagged PBP2a sample had lost its ability to bind BIO-AMP. However, a follow-up solution-phase binding experiment using the BIO-AMP reagent, followed by SDS-PAGE, blotting, and detection, revealed that both the GST-PBP2a and the untagged PBP were capable of binding to BIO-AMP (Fig. 3). This experimental observation indicates that GST-PBP2a can bind efficiently to microtiter plates, whereas untagged PBP2a appears unable to bind to microtiter plates under the assay conditions used here. As also illustrated in Fig. 3, the midpoint for the affinity of BIO-AMP to GST-PBP2a and untagged PBP2a in solution is similar to the Kmapp as determined for GST-PBP2a as determined in the microtiter plate-based assay (Fig. 2A).
Fig. 3.
SDS-PAGE gel of BIO-AMP-labeled GST-PBP2a (left) and untagged PBP2a (right), as a function of the BIO-AMP concentration relative to the Kmapp for BIO-AMP (as determined against GST-PBP2a in the microtiter plate-based assay). Some untagged PBP2a was present in the GST-PBP2a preparation due to the presence of endogenous proteases during GST-PBP2a purification.
The one-step assay is very precise. For the BIO-AMP titration at 15 min of incubation (Fig. 2A), the standard deviation for a set of replicate (n = 3) samples averaged 4% of the total signal change over the titration (a standard deviation of 2,004 RFU on average for each set of replicates, and with a total RFU change over the entire BIO-AMP titration RFU change of 53,000 RFU).
Steady-state binding isotherm for BIO-AMP and lack of time dependence.
To determine whether the reaction of BIO-AMP with PBP2a was reaching steady-state under the conditions used here, the results of BIO-AMP + PBP2a incubations for 15 and 30 min were determined and compared, and the data and analysis results are shown in Fig. 2A. It is clear that both the 15- and 30-min incubations gave virtually identical results and demonstrate that the reaction of BIO-AMP and PBP2a reached steady-state within 15 min. This is consistent with the time course for approach to steady-state for β-lactam binding in solution phase as reported by Graves-Woodward and Pratt (9). The excellent Michaelis-Menten saturation binding curves observed for both BIO-AMP and BIO-CEPH (Fig. 2) are also consistent with a steady-state reaction system.
Competitive one-step binding assay for characterization of ceftobiprole binding to PBP2a.
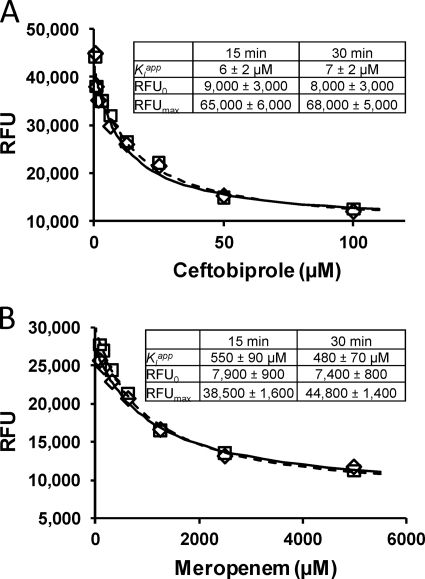
To demonstrate the utility of the one-step microtiter plate-based assay described here for the characterization of inhibitors of PBP2a, it was used to characterize PBP2a inhibition by ceftobiprole (Fig. 4), a new β-lactam designed to inhibit PBP2a and provide a β-lactam-based treatment option for MRSA infections (2, 12, 18). The results from the assay of variable concentrations of ceftobiprole versus a fixed concentration of BIO-AMP are shown in Fig. 4A. Both 15- and 30-min incubations were used to determine whether equilibrium was being achieved. Analysis of this competitive binding data by fitting with equation 2 gave a Kiapp of 6 ± 2 μM after 15 min of incubation and 7 ± 2 μM after 30 min of incubation. The close similarity in Kiapp values at the two different incubation times indicates that steady-state was reached within 15 min. Also, the competitive binding data fit very well to the competitive binding equation (equation 2), further supporting this conclusion.
Fig. 4.
Competitive titration of GST-PBP2a by ceftobiprole (A) and meropenem (B) in the presence of a fixed concentration (Kmapp = 1.6 μM) of BIO-AMP, including data points, error bars, best-fit lines, and best-fit parameter values. The data points obtained after 15 min of incubation are denoted by diamonds (♢), and those obtained after 30 min of incubation are denoted by boxes (□). Best-fit curves through data points obtained after 15 min of incubation are denoted by a solid line, and that obtained after 30 min of incubation is denoted by a dashed line. For all values, means ± the standard errors are given.
This Kiapp of 6 μM is greater than the MIC of ceftobiprole against MRSA of 1.9 μM (1 μg/ml) (reviewed in reference 18). This value is also slightly higher than range of 50% inhibitory concentrations (IC50s) reported for ceftobiprole binding to PBP2a in membrane extracts that ranged from 0.6 to 1.7 μM (5, 6).
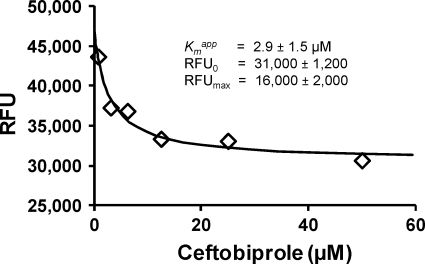
Determination of ceftobiprole binding to PBP2a using a two-step assay for comparison with one-step assay results.
In order to compare the results from a microtiter plate-based two-step assay for characterizing ceftobiprole binding to PBP2a to those from the one-step binding assay described above (Fig. 4A), a microtiter plate-based two-step assay was performed. The results of this two-step microtiter plate-based assay are shown in Fig. 5. Several features are notable. First, the Kiapp is lower than that determined using the one-step assay, but the observed difference is not significant at the P < 0.05 level. However, the lower Kiapp obtained with the two-step assay is more consistent with the MIC of ceftobiprole against MRSA of 1 μg/ml (1.9 μM; reviewed in reference 18). This value is also more consistent with the range of IC50s reported for ceftobiprole binding to PBP2a in membrane extracts ranging from 0.6 to 1.7 μM (5, 6).
Fig. 5.
Results from two-step binding assay result for ceftobiprole versus GST-PBP2a.
Demonstration inhibitor screening experiment and follow-up inhibitor characterization.
This assay (one step) was then tested for its utility for screening for PBP2a inhibitors. A random set of β-lactams was selected and screened for inhibition of PBP2a at 750 μM. This provided a ranked list of inhibitors (Table 1). As expected given PBP2a's intrinsic resistance to β-lactams, most of these β-lactams did not inhibit even at the high concentration of 750 μM; only meropenem (Fig. 1) yielded greater than 50% inhibition (Table 1). As a follow-up, the Kiapp for meropenem inhibition was determined using the one-step assay as described above for ceftobiprole, including with both 15- and 30-min incubations, which gave Kiapp values of 550 ± 90 (15-min incubation) and 480 ± 70 μM (30-min incubation) for meropenem (Fig. 4B). The value for meropenem is approximately twice the IC50 of 260 μM determined in membrane extracts (17), an ∼2-fold difference.
In conclusion, in the present study we developed and demonstrated a sensitive and convenient microtiter plate-based assay approach for the screening and characterization of inhibitors for PBP2a. The primary focus was on using a single-step assay with a relatively short incubation time of 15 min. This approach was validated against a solution phase β-lactam assay detected by SDS-PAGE, which demonstrated that GST-tagged PBP2a, necessary for binding to microtiter plates, bound BIO-AMP with the same affinity as untagged PBP2a and that the solution phase and microtiter plate assays gave the same apparent binding constants within the resolution of these different approaches. The one-step assay was further validated by using a longer (30-min) incubation time to demonstrate that the results using this assay were the same with the 15-min incubation time. Finally, a two-step assay protocol was also implemented in the microtiter plate format. The Kiapp for ceftobiprole with the two-step approach (2.9 μM) was lower that with the one-step approach (6 μM) but not by a statistically significant factor. The values for ceftobiprole binding obtained with the two-step microtiter plate-based approach were very similar to those of other studies using a two-step SDS-PAGE-based approach as cited in the text. The one-step approach gave more precise results than the two-step approach, but the two-step approach may provide a more accurate estimate, or at least an estimate more consistent with previous studies. For inhibitors (β-lactams) that reveal particularly slow acylation kinetics, a two-step assay with inhibitor binding in the absence of competing probe may be preferable. For rapid screening and characterization of prospective new inhibitors, the one-step protocol will likely be preferred. Also, for the discovery and characterization of novel noncovalent PBP2a inhibitors, a slow off rate is not expected, and the one-step assay will be required for such efforts. Given that PBP2a is a key molecular determinant for high-level β-lactam resistance in MRSA and that new inhibitors for PBP2a could provide new agents effective against MRSA, the microtiter plate-based assay approach described here is expected to facilitate the discovery, development, and characterization of new inhibitors against this important drug target.
ACKNOWLEDGMENTS
This study was supported by the American Heart Association (grant 0650179Z [WGG]) and by funds from the University of Missouri (University Research Board Grant K2303015 [W.G.G.]).
We thank Marilyn Yoder of the School of Biological Sciences at the University of Missouri–Kansas City for valuable advice on expressing GST-PBP2a in a soluble form by chilling the bacterial culture before induction, and we thank Anne Marie Queenan of Johnson and Johnson Pharmaceutical Research and Development, Raritan, NJ, for the generous gift of ceftobiprole.
Footnotes
Published ahead of print on 14 March 2011.
REFERENCES
- 1. Appelbaum P. C. 2007. Reduced glycopeptide susceptibility in methicillin-resistant Staphylococcus aureus (MRSA). Int. J. Antimicrob. Agents 30:398–408 [DOI] [PubMed] [Google Scholar]
- 2. Bazan J. A., Martin S. I., Kaye K. M. 2009. Newer beta-lactam antibiotics: doripenem, ceftobiprole, ceftaroline, and cefepime. Infect. Dis. Clin. N. Am. 23:983–996, ix [DOI] [PubMed] [Google Scholar]
- 3. Bush K., Smith S. A., Ohringer S., Tanaka S. K., Bonner D. P. 1987. Improved sensitivity in assays for binding of novel beta-lactam antibiotics to penicillin-binding proteins of Escherichia coli. Antimicrob. Agents Chemother. 31:1271–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dargis M., Malouin F. 1994. Use of biotinylated beta-lactams and chemiluminescence for study and purification of penicillin-binding proteins in bacteria. Antimicrob. Agents Chemother. 38:973–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Davies T. A., et al. 2007. Binding of ceftobiprole and comparators to the penicillin-binding proteins of Escherichia coli, Pseudomonas aeruginosa, Staphylococcus aureus, and Streptococcus pneumoniae. Antimicrob. Agents Chemother. 51:2621–2624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Entenza J. M., Hohl P., Heinze-Krauss I., Glauser M. P., Moreillon P. 2002. BAL9141, a novel extended-spectrum cephalosporin active against methicillin-resistant Staphylococcus aureus in treatment of experimental endocarditis. Antimicrob. Agents Chemother. 46:171–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fischbach M. A., Walsh C. T. 2009. Antibiotics for emerging pathogens. Science 325:1089–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fuda C., Suvorov M., Vakulenko S. B., Mobashery S. 2004. The basis for resistance to beta-lactam antibiotics by penicillin-binding protein 2a of methicillin-resistant Staphylococcus aureus. J. Biol. Chem. 279:40802–40806 [DOI] [PubMed] [Google Scholar]
- 9. Graves-Woodward K., Pratt R. F. 1998. Reaction of soluble penicillin-binding protein 2a of methicillin-resistant Staphylococcus aureus with beta-lactams and acyclic substrates: kinetics in homogeneous solution. Biochem. J. 332:755–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hartman B. J., Tomasz A. 1984. Low-affinity penicillin-binding protein associated with beta-lactam resistance in Staphylococcus aureus. J. Bacteriol. 158:513–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kluytmans J., Struelens M. 2009. Methicillin-resistant Staphylococcus aureus in the hospital. BMJ 338:b364. [DOI] [PubMed] [Google Scholar]
- 12. Lodise T. P., et al. 2008. Pharmacokinetic and pharmacodynamic profile of ceftobiprole. Diagn. Microbiol. Infect. Dis. 61:96–102 [DOI] [PubMed] [Google Scholar]
- 13. Macheboeuf P., Contreras-Martel C., Job V., Dideberg O., Dessen A. 2006. Penicillin-binding proteins: key players in bacterial cell cycle and drug resistance processes. FEMS Microbiol. Rev. 30:673–691 [DOI] [PubMed] [Google Scholar]
- 14. Mainardi J. L., Villet R., Bugg T. D., Mayer C., Arthur M. 2008. Evolution of peptidoglycan biosynthesis under the selective pressure of antibiotics in Gram-positive bacteria. FEMS Microbiol. Rev. 32:386–408 [DOI] [PubMed] [Google Scholar]
- 15. Spratt B. G. 1977. Properties of the penicillin-binding proteins of Escherichia coli K-12. Eur. J. Biochem. 72:341–352 [DOI] [PubMed] [Google Scholar]
- 16. Stefanova M., Bobba S., Gutheil W. G. 2010. A microtiter plate-based beta-lactam binding assay for inhibitors of high-molecular-mass penicillin-binding proteins. Anal. Biochem. 396:164–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sumita Y., Fukasawa M., Mitsuhashi S., Inoue M. 1995. Binding affinities of beta-lactam antibodies for penicillin-binding protein 2′ in methicillin-resistant staphylococcus aureus. J. Antimicrob. Chemother. 35:473–481 [DOI] [PubMed] [Google Scholar]
- 18. Vidaillac C., Rybak M. J. 2009. Ceftobiprole: first cephalosporin with activity against methicillin-resistant Staphylococcus aureus. Pharmacotherapy 29:511–525 [DOI] [PubMed] [Google Scholar]
- 19. Zapun A., Contreras-Martel C., Vernet T. 2008. Penicillin-binding proteins and beta-lactam resistance. FEMS Microbiol. Rev. 32:361–385 [DOI] [PubMed] [Google Scholar]
- 20. Zhao G., Meier T. I., Kahl S. D., Gee K. R., Blaszczak L. C. 1999. BOCILLIN FL, a sensitive and commercially available reagent for detection of penicillin-binding proteins. Antimicrob. Agents Chemother. 43:1124–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]