Abstract
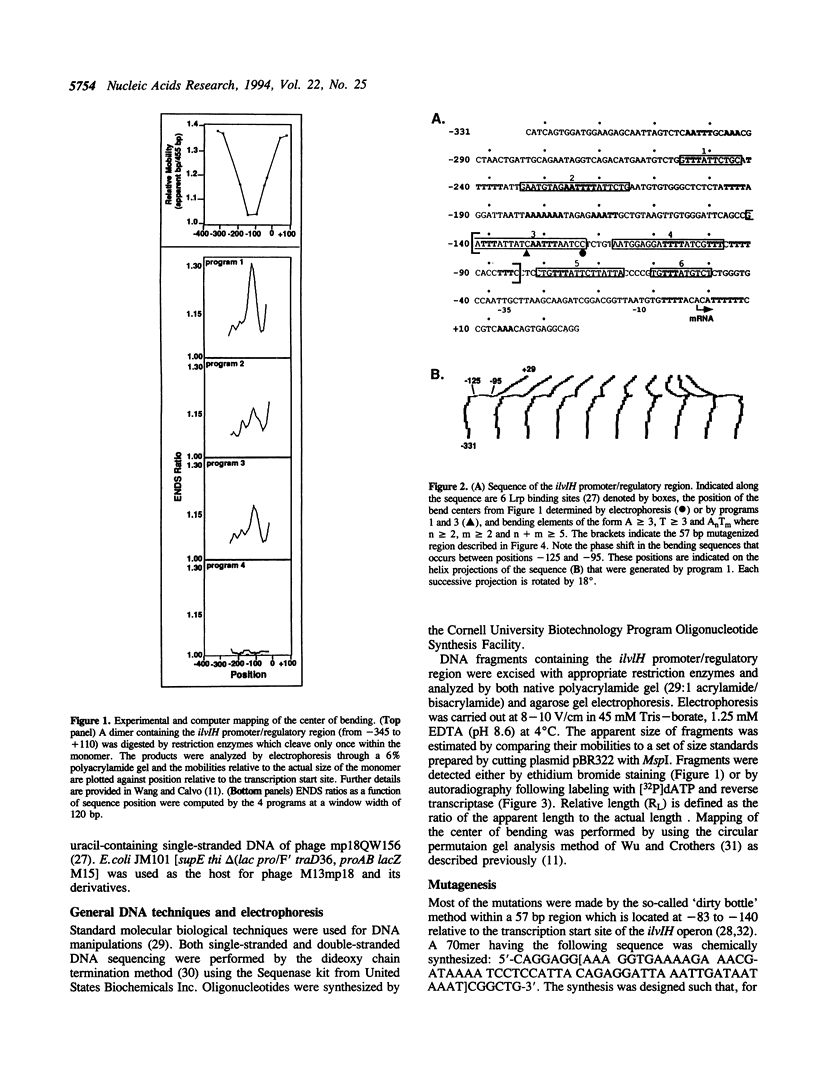
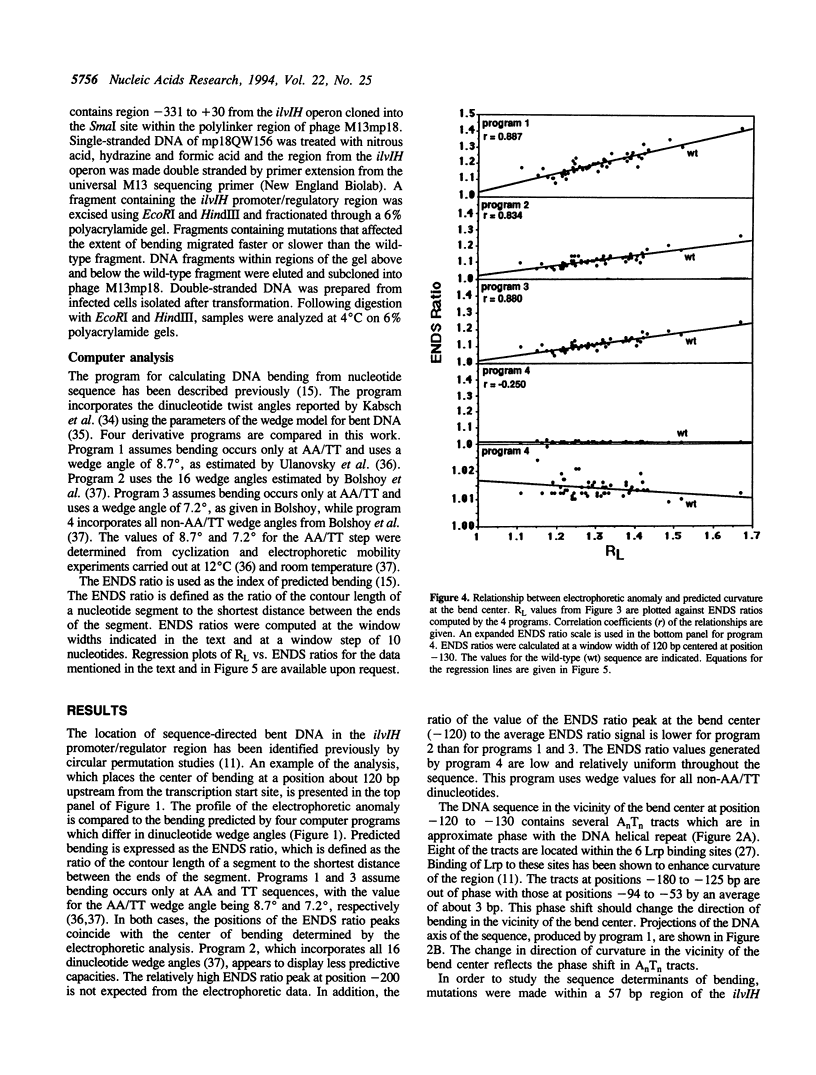
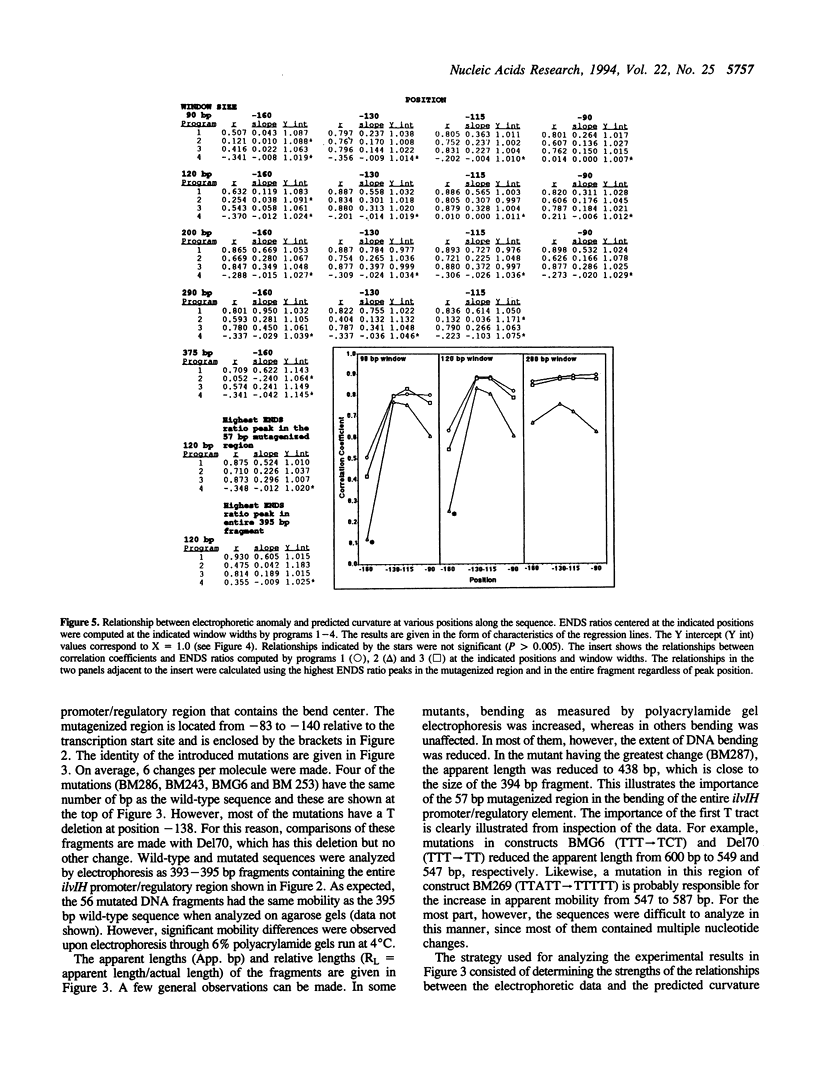
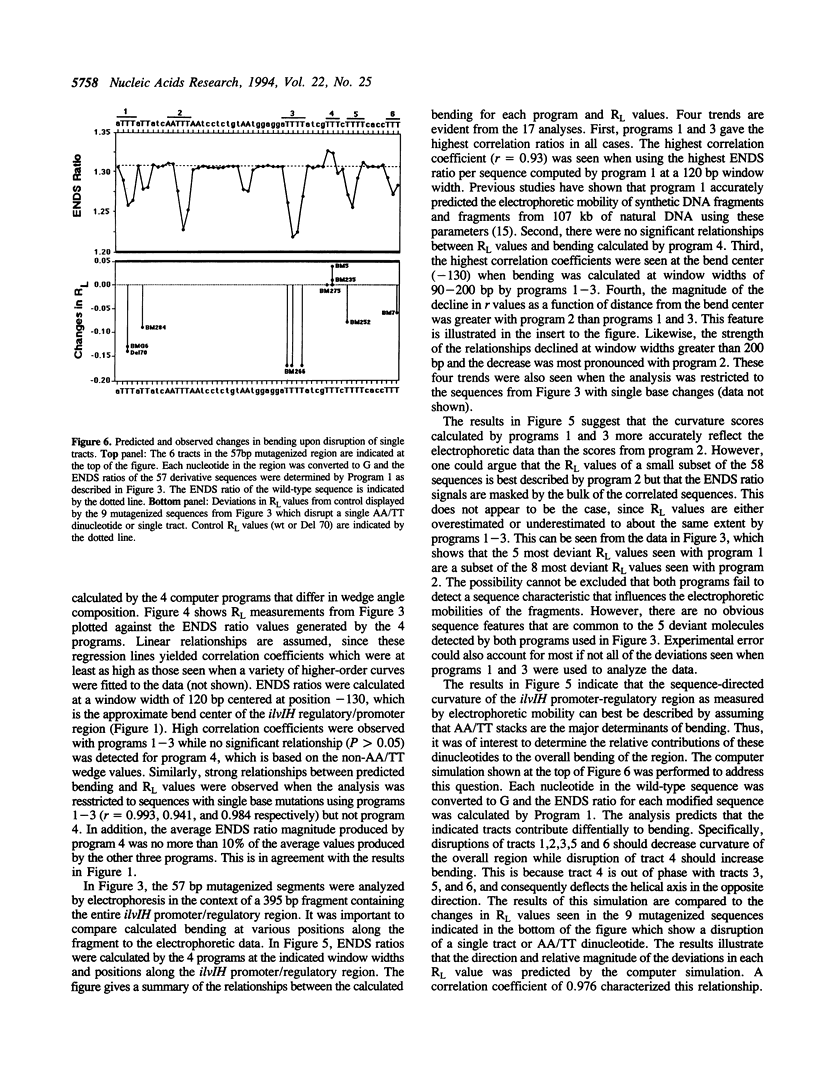
Previous studies have shown that the promoter/regulatory region of the ilvlH operon displays intrinsic curvature, with the bend center located at position -120 relative to the transcription start site. In this report, a 57 bp sequence spanning the bend center was mutagenized in vitro in order to study the relationship between nucleotide sequence and curvature measured by electrophoresis. The strategy used for analyzing the results consisted of determining the strengths of the relationships between electrophoretic anomaly and predicted curvature calculated by computer programs that differ in wedge angle composition. The results revealed that programs which assume that bending occurs only at AA/TT display good predictive value, with correlation coefficients between electrophoretic anomaly and predicted curvature as high as 0.93. In contrast, a program which assumes that bending occurs at all 16 dinucleotide steps exhibited lower predictive value, while there were no significant relationships between the experimental data and curvature calculated by a program that was based on all non-AA/TT wedge values. These results show that the complete wedge model which incorporates values for all dinucleotide steps does not adequately describe the electrophoretic data in this report.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson J. N. Detection, sequence patterns and function of unusual DNA structures. Nucleic Acids Res. 1986 Nov 11;14(21):8513–8533. doi: 10.1093/nar/14.21.8513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer B. F., Kar E. G., Elford R. M., Holmes W. M. Sequence determinants for promoter strength in the leuV operon of Escherichia coli. Gene. 1988;63(1):123–134. doi: 10.1016/0378-1119(88)90551-3. [DOI] [PubMed] [Google Scholar]
- Bolshoy A., McNamara P., Harrington R. E., Trifonov E. N. Curved DNA without A-A: experimental estimation of all 16 DNA wedge angles. Proc Natl Acad Sci U S A. 1991 Mar 15;88(6):2312–2316. doi: 10.1073/pnas.88.6.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossi L., Smith D. M. Conformational change in the DNA associated with an unusual promoter mutation in a tRNA operon of Salmonella. Cell. 1984 Dec;39(3 Pt 2):643–652. doi: 10.1016/0092-8674(84)90471-9. [DOI] [PubMed] [Google Scholar]
- Bracco L., Kotlarz D., Kolb A., Diekmann S., Buc H. Synthetic curved DNA sequences can act as transcriptional activators in Escherichia coli. EMBO J. 1989 Dec 20;8(13):4289–4296. doi: 10.1002/j.1460-2075.1989.tb08615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crothers D. M., Haran T. E., Nadeau J. G. Intrinsically bent DNA. J Biol Chem. 1990 May 5;265(13):7093–7096. [PubMed] [Google Scholar]
- Eckdahl T. T., Anderson J. N. Computer modelling of DNA structures involved in chromosome maintenance. Nucleic Acids Res. 1987 Oct 26;15(20):8531–8545. doi: 10.1093/nar/15.20.8531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckdahl T. T., Anderson J. N. Conserved DNA structures in origins of replication. Nucleic Acids Res. 1990 Mar 25;18(6):1609–1612. doi: 10.1093/nar/18.6.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald D. J., Dryden G. L., Bronson E. C., Williams J. S., Anderson J. N. Conserved patterns of bending in satellite and nucleosome positioning DNA. J Biol Chem. 1994 Aug 19;269(33):21303–21314. [PubMed] [Google Scholar]
- Gartenberg M. R., Crothers D. M. Synthetic DNA bending sequences increase the rate of in vitro transcription initiation at the Escherichia coli lac promoter. J Mol Biol. 1991 May 20;219(2):217–230. doi: 10.1016/0022-2836(91)90563-l. [DOI] [PubMed] [Google Scholar]
- Goodman S. D., Nash H. A. Functional replacement of a protein-induced bend in a DNA recombination site. Nature. 1989 Sep 21;341(6239):251–254. doi: 10.1038/341251a0. [DOI] [PubMed] [Google Scholar]
- Hagerman P. J. Sequence-directed curvature of DNA. Annu Rev Biochem. 1990;59:755–781. doi: 10.1146/annurev.bi.59.070190.003543. [DOI] [PubMed] [Google Scholar]
- Haughn G. W., Squires C. H., De Felice M., Largo C. T., Calvo J. M. Unusual organization of the ilvIH promoter of Escherichia coli. J Bacteriol. 1985 Jul;163(1):186–198. doi: 10.1128/jb.163.1.186-198.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W., Sander C., Trifonov E. N. The ten helical twist angles of B-DNA. Nucleic Acids Res. 1982 Feb 11;10(3):1097–1104. doi: 10.1093/nar/10.3.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H. S., Wu H. M., Crothers D. M. DNA bending at adenine . thymine tracts. Nature. 1986 Apr 10;320(6062):501–506. doi: 10.1038/320501a0. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Lavigne M., Herbert M., Kolb A., Buc H. Upstream curved sequences influence the initiation of transcription at the Escherichia coli galactose operon. J Mol Biol. 1992 Mar 20;224(2):293–306. doi: 10.1016/0022-2836(92)90995-v. [DOI] [PubMed] [Google Scholar]
- Lyubchenko Y. L., Shlyakhtenko L. S., Appella E., Harrington R. E. CA runs increase DNA flexibility in the complex of lambda Cro protein with the OR3 site. Biochemistry. 1993 Apr 20;32(15):4121–4127. doi: 10.1021/bi00066a038. [DOI] [PubMed] [Google Scholar]
- Marini J. C., Levene S. D., Crothers D. M., Englund P. T. Bent helical structure in kinetoplast DNA. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7664–7668. doi: 10.1073/pnas.79.24.7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister C. F., Achberger E. C. Rotational orientation of upstream curved DNA affects promoter function in Bacillus subtilis. J Biol Chem. 1989 Jun 25;264(18):10451–10456. [PubMed] [Google Scholar]
- McNamara P. T., Bolshoy A., Trifonov E. N., Harrington R. E. Sequence-dependent kinks induced in curved DNA. J Biomol Struct Dyn. 1990 Dec;8(3):529–538. doi: 10.1080/07391102.1990.10507827. [DOI] [PubMed] [Google Scholar]
- McNamara P. T., Harrington R. E. Characterization of inherent curvature in DNA lacking polyadenine runs. J Biol Chem. 1991 Jul 5;266(19):12548–12554. [PubMed] [Google Scholar]
- Mizuno T. Static bend of DNA helix at the activator recognition site of the ompF promoter in Escherichia coli. Gene. 1987;54(1):57–64. doi: 10.1016/0378-1119(87)90347-7. [DOI] [PubMed] [Google Scholar]
- Myers R. M., Lerman L. S., Maniatis T. A general method for saturation mutagenesis of cloned DNA fragments. Science. 1985 Jul 19;229(4710):242–247. doi: 10.1126/science.2990046. [DOI] [PubMed] [Google Scholar]
- Plaskon R. R., Wartell R. M. Sequence distributions associated with DNA curvature are found upstream of strong E. coli promoters. Nucleic Acids Res. 1987 Jan 26;15(2):785–796. doi: 10.1093/nar/15.2.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross W., Shulman M., Landy A. Biochemical analysis of att-defective mutants of the phage lambda site-specific recombination system. J Mol Biol. 1982 Apr 15;156(3):505–522. doi: 10.1016/0022-2836(82)90263-7. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satchwell S. C., Drew H. R., Travers A. A. Sequence periodicities in chicken nucleosome core DNA. J Mol Biol. 1986 Oct 20;191(4):659–675. doi: 10.1016/0022-2836(86)90452-3. [DOI] [PubMed] [Google Scholar]
- Snyder U. K., Thompson J. F., Landy A. Phasing of protein-induced DNA bends in a recombination complex. Nature. 1989 Sep 21;341(6239):255–257. doi: 10.1038/341255a0. [DOI] [PubMed] [Google Scholar]
- Squires C. H., De Felice M., Wessler S. R., Calvo J. M. Physical characterization of the ilvHI operon of Escherichia coli K-12. J Bacteriol. 1981 Sep;147(3):797–804. doi: 10.1128/jb.147.3.797-804.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers A. A., Klug A. The bending of DNA in nucleosomes and its wider implications. Philos Trans R Soc Lond B Biol Sci. 1987 Dec 15;317(1187):537–561. doi: 10.1098/rstb.1987.0080. [DOI] [PubMed] [Google Scholar]
- Trifonov E. N. DNA in profile. Trends Biochem Sci. 1991 Dec;16(12):467–470. doi: 10.1016/0968-0004(91)90181-t. [DOI] [PubMed] [Google Scholar]
- Trifonov E. N., Sussman J. L. The pitch of chromatin DNA is reflected in its nucleotide sequence. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3816–3820. doi: 10.1073/pnas.77.7.3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulanovsky L. E., Trifonov E. N. Estimation of wedge components in curved DNA. Nature. 1987 Apr 16;326(6114):720–722. doi: 10.1038/326720a0. [DOI] [PubMed] [Google Scholar]
- Ulanovsky L., Bodner M., Trifonov E. N., Choder M. Curved DNA: design, synthesis, and circularization. Proc Natl Acad Sci U S A. 1986 Feb;83(4):862–866. doi: 10.1073/pnas.83.4.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanWye J. D., Bronson E. C., Anderson J. N. Species-specific patterns of DNA bending and sequence. Nucleic Acids Res. 1991 Oct 11;19(19):5253–5261. doi: 10.1093/nar/19.19.5253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Calvo J. M. Lrp, a global regulatory protein of Escherichia coli, binds co-operatively to multiple sites and activates transcription of ilvIH. J Mol Biol. 1993 Jan 20;229(2):306–318. doi: 10.1006/jmbi.1993.1036. [DOI] [PubMed] [Google Scholar]
- Wang Q., Calvo J. M. Lrp, a major regulatory protein in Escherichia coli, bends DNA and can organize the assembly of a higher-order nucleoprotein structure. EMBO J. 1993 Jun;12(6):2495–2501. doi: 10.1002/j.1460-2075.1993.tb05904.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Sacco M., Ricca E., Lago C. T., De Felice M., Calvo J. M. Organization of Lrp-binding sites upstream of ilvIH in Salmonella typhimurium. Mol Microbiol. 1993 Mar;7(6):883–891. doi: 10.1111/j.1365-2958.1993.tb01179.x. [DOI] [PubMed] [Google Scholar]
- Williams J. S., Eckdahl T. T., Anderson J. N. Bent DNA functions as a replication enhancer in Saccharomyces cerevisiae. Mol Cell Biol. 1988 Jul;8(7):2763–2769. doi: 10.1128/mcb.8.7.2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willins D. A., Calvo J. M. In vitro transcription from the Escherichia coli ilvIH promoter. J Bacteriol. 1992 Dec;174(23):7648–7655. doi: 10.1128/jb.174.23.7648-7655.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. M., Crothers D. M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984 Apr 5;308(5959):509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- Zahn K., Blattner F. R. Direct evidence for DNA bending at the lambda replication origin. Science. 1987 Apr 24;236(4800):416–422. doi: 10.1126/science.2951850. [DOI] [PubMed] [Google Scholar]
- Zahn K., Blattner F. R. Sequence-induced DNA curvature at the bacteriophage lambda origin of replication. Nature. 1985 Oct 3;317(6036):451–453. doi: 10.1038/317451a0. [DOI] [PubMed] [Google Scholar]
- de Felice M., Lago C. T., Squires C. H., Calvo J. M. Acetohydroxy acid synthase isoenzymes of Escherichia coli K12 and Salmonella typhimurium. Ann Microbiol (Paris) 1982 Mar-Apr;133(2):251–256. [PubMed] [Google Scholar]



