Abstract
The structures of staphylococcal cassette chromosome mec (SCCmec) elements carried by 31 clonal complex 398 (CC398) methicillin-resistant Staphylococcus aureus (MRSA) strains isolated from the participants at a conference were analyzed. The SCCmecs were classified into novel types, namely, IX, X, V(5C2&5) subtype c, and IVa. Type V(5C2&5) subtype c, IX, and X SCCmecs carried genes conferring resistance to metals. The structures of SCCmecs from CC398 strains were distinct from those normally found in humans, adding to the evidence that humans are not the original host for CC398.
TEXT
Recent reports of methicillin-resistant Staphylococcus aureus (MRSA) in livestock, particularly pigs, and in individuals who have contact with livestock provided evidence of the existence of a true livestock-associated MRSA reservoir (1, 13, 16, 18–19). Livestock-associated MRSA strains are nontypeable by pulsed-field gel electrophoresis using SmaI (2) and belong to specific spa types that group into multilocus sequence types (MLST) of clonal complex 398 (CC398), and the vast majority of strains are resistant to tetracycline (6, 12).
Although CC398 MRSA strains have been studied extensively (e.g., the whole genome sequence has been determined [15]), structures of staphylococcal cassette chromosome mec (SCCmec) elements carried by these strains are not yet fully understood. This study was undertaken to clarify the structures of SCCmec elements carried by CC398 MRSA strains in order to gain more understanding of their genetic background.
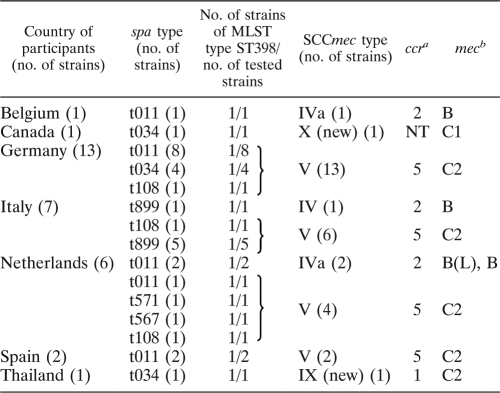
A total of 31 MRSA strains (Table 1) were collected from participants at the 19th International Pig Veterinary Conference in Denmark, 2006, including participants from Europe (n = 29), Thailand (n = 1), and Canada (n = 1); these strains were originally found to belong to spa types associated with CC398 in a previous study (20), which was confirmed by multilocus sequence typing (MLST) of 15 representative isolates (Table 1) (7).
Table 1.
Characteristics of 31 MRSA strains isolated from the participants of an international conference
aNT, nontypeable. A weak band was observed in some cases.
bB(L) signifies that the size of the DNA fragment amplified by a primer pair identifying the mecA gene lineage as IS1272 was longer than that of standard strains carrying the class B mec gene complex. A long-range PCR using primers mA7 and IS7 for the class B mec gene complex was performed, which produced a 4,159-bp amplicon, compared to a 2,827-bp amplicon produced by the standard strain, CA05. A 4,159-bp amplicon (GenBank accession no. HQ157182) containing a free copy of IS256 was inserted 45 bp downstream of the mecRI start codon on the opposite strand, which generated an 8-bp target site duplication (CATTGCTC) upon transposition.
The SCCmec elements were typed using a multiplex-PCR (M-PCR) assay described by Kondo et al. (14). Among 29 strains recovered from European participants, 25 (86.2%) carried type V SCCmec and 4 (13.8%) carried type IVa SCCmec (Table 1). Type IVa SCCmec of strain JCSC7158 contained a larger-than-normal class B mec gene complex [termed B(L) in Table 1]. Sequence analysis showed that a free copy of IS256 was inserted 45 bp downstream of the mecRI start codon. In contrast, strains JCSC6943 and JCSC6945 recovered from a Thai and a Canadian participant, respectively, carried SCCmecs that could not be classified into extant types. JCSC6943 was shown to carry a novel combination of the ccr gene complex (type 1) and mec gene complex (class C2), whereas neither ccr nor mec gene complexes in strain JCSC6945 could be amplified. The nucleotide sequences of the untypeable SCCmec elements carried by strains JCSC6943 and JCSC6945, as well as by a representative of the predominant type V SCCmec carried by strain JCSC6944, were determined using amplified DNA fragments with long-range PCR and fosmid clones that were prepared as described previously (3) and chosen by PCRs with pairs of primers identifying genes in SCCmecs (mecA, copB) and the chromosomal regions flanking SCCmecs. Open reading frames (ORFs) (≥100 bp) were identified with the In-silico Molecular Cloning genomics edition program (in silico biology, inc., Yokohama, Japan) and were compared to nucleotide sequences in the databases at the National Center for Biotechnology Information (Bethesda, MD) using BLAST for identifying homologues and predicting functions.
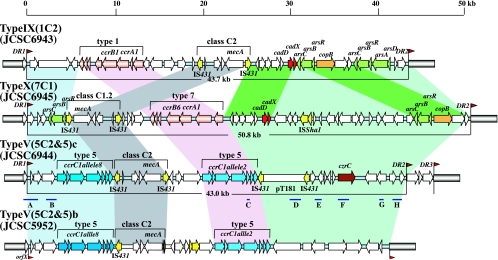
The structures of the SCCmec elements are illustrated in Fig. 1. The nucleotide sequences at the left and right chromosomal junctions of all three SCCmecs were similar to those of extant SCCmecs; they were demarcated by direct repeats containing the integration site sequence of the SCC (ISS), carried characteristic inverted repeats at both ends, and were integrated downstream of orfX.
Fig. 1.
Structures of three SCCmec elements identified in CC398 MRSA strains. The structures of type V (5C2&5)c, IX (1C2), and X (7C1) SCCmec elements from MRSA CC398 strains JCSC6944, JCSC6943, and JCSC6945, respectively, and type V(5C2&5)b SCCmec from strain JCSC5952 are illustrated based on the nucleotide sequences deposited in the DDBJ/EMBL/GenBank databases under accession no. AB505628 to AB505630 and AB478780. Blue bars indicate structures (A to H) distributed over the entire JCSC6944 SCCmec element, which were detected by a PCR-based approach (see the text). Red arrowheads indicate the locations of integration site sequences (ISS) for the SCC. Insertion sequences are indicated in yellow. Genes conferring resistance to metals are colored as follows: cadDX (red), arsRBC and arsDARBC (green), copB (orange), and czrC (brown). The ccr gene complexes, mec gene complexes, J3 regions, and J1 regions are shaded in pink, gray, blue, and light green, respectively. Structures of the J1 region that are conserved between JCSC6943's SCCmec and JCSC6945's SCCmec are shaded in deep green. The primers used for the amplification of DNA fragments, screening of fosmid clones, and structures A to H in JCSC6944's SCCmec are listed in Table S1 in the supplemental material. DR, direct repeat.
JCSC6943's SCCmec (43,675 bp) carried a type 1 ccr gene complex and class type C2 mec gene complex, although the nucleotide identities of JCSC6943's ccrA1 and ccrB1 genes to those of NCTC10442's SCCmec (type I) are 94.1% and 92.2%, respectively, and six ORFs surrounding the ccr genes showed 58.3 to 65.9% nucleotide identities to those of NCTC10442's SCCmec. The ccr gene complex was located between orfX and the class C2 mec gene complex, which is similar to the structure of JCSC6082's SCCmec (3). Based upon the novel combination of ccr and mec gene complexes (1C2), JCSC6943's SCCmec was classified as novel type IX.
JCSC6945's SCCmec (50,803 bp) carried a ccrA1 gene with 94.1% nucleotide identity to that of NCTC10442's SCCmec and a ccrB gene; ccrB was phylogenetically closer to ccrB6 of Staphylococcus saprophyticus ATCC 15305 than to ccrB1 of NCTC10442, with nucleotide identities of 89.1% and 87.0%, respectively (Fig. 2). Nucleotide identities of ORFs surrounding the ccr genes to those of SCCmec in NCTC10442 were 59.9 to 85.5%. Based on the novel combination of ccrA1 and ccrB6, the ccr gene complex was classified as a novel type, type 7. SCCmec in JCSC6945 carried a novel class C1-like mec gene complex (6,422 bp), which was distinct from the class C1 mec gene complex (7,212 bp) carried by type VII SCCmec in a Swedish community-associated MRSA strain, JCSC6082, in two respects: (i) SCCmec in JCSC6945 and SCCmec in JCSC6082 carry different direct repeat unit (dru) types (14f and 10a, respectively); IS431 is inserted into the mecR1 gene at different positions (17 bp and 968 bp downstream of the start codon, respectively). Furthermore, the orientation of the class C1 mec gene complex of JCSC6945 is opposite to that of mec gene complexes carried by type I to VII SCCmec elements. These data indicated that the class C1 mec gene complexes of SCCmec in JCSC6082 and of SCCmec in JCSC6945 have evolved independently. SCCmec in JCSC6945 was classified as a novel type, type X, which carries the novel combination of a type 7 ccr gene complex and a class C1 mec gene complex.
Fig. 2.
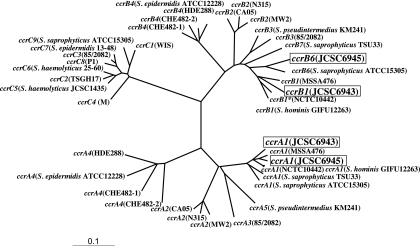
Phylogenetic relations of ccr genes. The ccrA and ccrB genes from the following strains were used. DDBJ/EMBL/GenBank database accession numbers are indicated in parentheses, and species are not indicated in the cases of S. aureus strains. ccrA1 and ccrB1, NCTC10442 (AB033763), MSSA476 (BX571857), and GIFU12263 (AB063171); ccrA2 and ccrB2, N315 (D86934), CA05 (AB063172), and MW2 (NC003923); ccrA3 and ccrB3, 85/2082 (AB037671); ccrA4 and ccrB4, HDE288 (AF411935), ATCC 12228 (AE015929), CHE482-1 (EF126185), and CHE482-2 (EF126186); ccrA1 and ccrB6, ATCC 15305 (NC007350); ccrA1 and ccrB7, TSU33 (AB353724); ccrA5 and ccrB3, KM241 (AM904731); and ccrC1, WIS (AB121219), TSGH17 (AY894416), 85/2082 (AB037671), M (U10927), S. haemolyticus JCSC1435 (AP006716), S. haemolyticus 25-60 (EF190467), S. epidermidis 13-48 (EF190468), P1 (AB656125), and S. saprophyticus ATCC 15305. The nucleotide sequences of 14 ccrA genes, 14 ccrB genes, and 9 ccrC genes were aligned by using the ClustalX program. The phylogenetic tree was generated by the neighbor-joining method by creating 2,000 bootstrap replicates. The tree was visualized with TreeView software, which was obtained from the website http://taxonomy.zoology.gla.ac.uk/ROD/treeview.html. The branch length indicates the distance, which is expressed as the number of substitutions per 100 bases.
SCCmec in JCSC6944 (43,381 bp) was a composite of an SCC carrying ccrC1 allele 2 (10 kb) and a type V SCCmec carrying ccrC1 allele 8 and a class C2 mec gene complex (17 kb). It was nearly identical to type V(5C2&5) SCCmec of the genome-sequenced ST398 MRSA strain S0385 (15). However, SCCmec in JCSC6944 carried an integrated plasmid, pT181, and the mec gene complexes of JCSC6944's SCCmec and S0385's SCCmec differed in their dru types (11a and 10a, respectively).
The joining (J) regions (J1, J2, and J3) constitute SCCmec components other than mec and ccr gene complexes in SCCmecs, and structural differences between the J regions within the same SCCmec type are used for defining subtypes (10). Whereas the first 27 ORFs of SCCmec in JCSC6944 showed very high similarity to the type V(5C2&5) SCCmecs carried by strains TSGH17 and PM1 (Taiwanese community-associated MRSA [CA-MRSA] isolates) and JCSC5952 (a Japanese CA-MRSA isolate), the last 18 ORFs, corresponding to the entire J1 region, were unique to SCCmec in JCSC6944 (Fig. 1). To distinguish them, we tentatively express the differences in J1 regions by using small letters while we await the decision of the International Working Group on the Classification of SCCmec Elements (IWG-SCC) (10): type V(5C2)a for the type V SCCmec identified in Australian CA-MRSA WIS; type V(5C2&5)b for the SCCmec elements identified in strains TSGH17, PM1, and JCSC5952; and type V(5C2&5)c for the SCCmec identified in JCSC6944.
Interestingly, the J regions of type V(5C2&5)c, IX, and X SCCmec elements carried genes related to the detoxification of heavy metals, such as cadmium, copper, zinc, and arsenate (Fig. 1). The J1 region (18 kb) of type IX SCCmec contained a cadDX operon, a copB gene, and two arsenate resistance operons, arsRBC and arsDARBC. All 20 ORFs in the J1 region were highly homologous to the ΨSCCmec elements in Staphylococcus haemolyticus JCSC1435, with nucleotide identities of 83.8 to 100%. The type X SCCmec elements contained a cadDX operon, a copB gene, and an arsRBC operon in the J1 region (28 kb) and an arsRBC operon in the J3 region.
In type X SCCmec, a 6-kb region containing the cadDX operon and a 7-kb region containing the copB gene and one of the arsRBC operons were highly homologous to the J1 region of type IX SCCmec, with nucleotide identities of 98.9% and 94.5%, respectively (indicated in dark green in Fig. 1). In addition, ISSha1, an insertion sequence typically identified in S. haemolyticus (17), was present in the J1 region of type X SCCmec.
The J1 region of type V(5C2&5)c SCCmec (17 kb) carried a czrC (cadmium zinc resistance C) gene, which is responsible for cadmium and zinc resistance (4), as well as an integrated plasmid, pT181, bearing tet(K). Notably, this plasmid was present as a free plasmid in the genome-sequenced MRSA ST398 strain S0385 but absent from SCCmec of S0385 (15).
To investigate whether the remaining type V SCCmec element identified in this study was identical or closely similar to JCSC6944's SCCmec, we designed a PCR assay to detect structures (A to H) distributed over JCSC6944's entire SCCmec (Fig. 1). A total of 18 strains (72.2%; 18/25), including strain JCSC6944, contained all eight structures (A to H), whereas three strains were negative for pT181/tetK (structure D), two strains were negative for czrC (structure F), and two strains were negative for ORF42 (structure H). These data support the idea that the majority of type V SCCmecs are highly similar and that the observed structural differences in a subset of strains are confined to the J1 region.
The J regions of type II and III SCCmec elements carried by hospital-associated MRSA strains also encode additional resistance determinants (11), which may cause a fitness advantage in the presence of antibiotic selection pressures. Given that antibiotics and metals are used worldwide for growth promotion in animal agriculture (8), it is likely that CC398 MRSA strains are under similar selection pressures and that the spread and persistence of CC398 MRSA strains are results of coselection by metal resistance-encoding genes in the J regions of type V(5C2&5)c, IX, and X SCCmec elements.
The majority of type V(5C2&5)c SCCmec elements in the collection of MRSA CC398 strains also contained the tetracycline resistance gene tet(K). However, the role of the tet(K) gene in the coselection of methicillin resistance remains unclear, given that another tetracycline resistance gene, tet(M), is widely distributed among CC398 strains (6, 12). The czrC gene carried by type V(5C2&5)c SCCmecs is closely related to those identified in the type VIII SCCmecs of a Canadian epidemic MRSA strain, C10682, and in SCCpbp4 of Staphylococcus epidermidis ATCC 12228. In addition, the copB genes carried by type IX SCCmecs and type X SCCmecs are closely related to each other and phylogenetically related to those identified in SCC elements of S. epidermidis ATCC 12228 and S. haemolyticus JCSC1435 as well as on the chromosome of S. aureus strains, e.g., FPR3757. These data indicate that genes conferring resistance to metals are disseminated widely among staphylococci and suggest that the genes might have been acquired from other species, e.g., S. haemolyticus, by horizontal gene transfer. This hypothesis is further supported by the presence of the S. haemolyticus insertion sequence ISSha1 in the J1 region of type X SCCmec.
Three SCCmec elements sequenced in this study are clearly distinct from SCCmecs previously identified in human isolates. The oppositely oriented class C1-like mec gene complex (although it should be classified as a subclass of C1), was identified in type X SCCmec. It suggests that other novel mec gene complexes will be generated in the future and adds to the evidence that SCCmec is formed by the acquisition of the mec gene complex by SCC. In contrast to the well-conserved mecA gene and its flanking region, the ccr gene complex and J regions are very diverse (Fig. 2). Although JCSC6943's SCCmec was judged to carry a type 1 ccr gene complex by PCR and nucleotide sequence comparisons, ccrA1 and ccrB1 in JCSC6943's SCCmec differed from prototypic ccrA1 and ccrB1 genes identified in NCTC10442's SCCmec (type I). JCSC6945's SCCmec carried ccrA1, which has 95% nucleotide identity to that of NCTC10442's SCCmec. Furthermore, JCSC6945's ccrB6 and other constituents of the ccr gene complex were more homologous to those carried by S. saprophyticus TSU33 and ATCC 15305, with 84 to 97% nucleotide identities, respectively, than to those in NCTC10442's SCCmec (9), suggesting that the ccr gene complex with ccrA1-ccrB6 might have evolved by undergoing recombination, although we cannot predict where such recombination has occurred. Such a combination of distinct allotypes of ccr, ccrA3, and ccrB5 has been reported in Staphylococcus pseudintermedius (5). S. pseudintermedius strains are carried mostly by dogs, and two kinds of SCCmec elements were identified in the species, the ones identical to those found in human strains and the very diverged-structured SCCmecs unique to this species. These data suggested that staphylococcal strains related to animals might carry SCCs or SCCmecs that might have evolved in a niche distinct from that of humans. All type V(5C2&5) SCCmec strains were from European attendees, while the novel SCCmec types were from Canadian and Thai attendees, pointing to a possibly geography-dependent epidemiologic evolution.
In conclusion, we identified two novel SCCmec types, IX and X, and present evidence that MRSA CC398 carrying an SCCmec of type V(5C2&5)c has been established as the predominant livestock-associated MRSA clone in Europe. How metal and antibiotic resistance genes are linked to SCC or SCCmec in animal agriculture, as well as the roles of SCCs and SCCmecs in the dissemination and persistence of MRSA strains, are the next questions to be answered.
Nucleotide sequence accession numbers.
The nucleotide sequences of SCCmec elements from MRSA CC398 strains JCSC6944, JCSC6943, and JCSC6945 have been deposited in the DDBJ/EMBL/GenBank databases under accession no. AB505628 to AB505630. The nucleotide sequence of the mec gene complex of JCSC7158 is deposited in GenBank under accession no. HQ157182.
Supplementary Material
Acknowledgments
We thank Makoto Onishi and Hidemasa Izumitani, National Institute of Health, Japan, for their kind help with analyzing dru types by using the BioNumerics software for sequence typing of polymorphic variable-number tandem repeats.
This work was supported by a Grant-in-Aid from the 21st Century Center of Excellence (COE) Program, a Grant-in-Aid for Scientific Research C19590456, and a Grant-in-Aid (S0991013) for the Foundation of Strategic Research Projects in Private Universities, all from the Ministry of Education, Science, Sports, Culture, and Technology of Japan.
Footnotes
Supplemental material for this article may be found at http://aac.asm.org/.
Published ahead of print on 21 March 2011.
REFERENCES
- 1. Armand-Lefevre L., Ruimy R., Andremont A. 2005. Clonal comparison of Staphylococcus aureus isolates from healthy pig farmers, human controls, and pigs. Emerg. Infect. Dis. 11:711–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bens C. C., Voss A., Klaassen C. H. 2006. Presence of a novel DNA methylation enzyme in methicillin-resistant Staphylococcus aureus isolates associated with pig farming leads to uninterpretable results in standard pulsed-field gel electrophoresis analysis. J. Clin. Microbiol. 44:1875–1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Berglund C., et al. 2008. Novel type of staphylococcal cassette chromosome mec in a methicillin-resistant Staphylococcus aureus strain isolated in Sweden. Antimicrob. Agents Chemother. 52:3512–3516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cavaco L. M., et al. 2010. Cloning and occurrence of czrC, a gene conferring cadmium and zinc resistance in methicillin-resistant Staphylococcus aureus CC398 isolates. Antimicrob. Agents Chemother. 54:3605–3608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Descloux S., Rossano A., Perreten V. 2008. Characterization of new staphylococcal cassette chromosome mec (SCCmec) and topoisomerase genes in fluoroquinolone- and methicillin-resistant Staphylococcus pseudintermedius. J. Clin. Microbiol. 46:1818–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. de Vries L. E., Christensen H., Skov R. L., Aarestrup F. M., Agerso Y. 2009. Diversity of the tetracycline resistance gene tet(M) and identification of Tn916- and Tn5801-like (Tn6014) transposons in Staphylococcus aureus from humans and animals. J. Antimicrob. Chemother. 64:490–500 [DOI] [PubMed] [Google Scholar]
- 7. Enright M. C., Day N. P., Davies C. E., Peacock S. J., Spratt B. G. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hasman H., Franke S., Rensing C. 2006. Resistance to metals used for agricultural production, p. 99–114 In Aarentrub E. M., Wegener H. C. (ed.), Antimicrobial resistance in bacteria of animal origin. ASM Press, Washington, DC [Google Scholar]
- 9. Higashide M., et al. 2008. Methicillin-resistant Staphylococcus saprophyticus isolates carrying staphylococcal cassette chromosome mec have emerged in urogenital tract infections. Antimicrob. Agents Chemother. 52:2061–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements (IWG-SCC) 2009. Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob. Agents Chemother. 53:4961–4967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ito T., et al. 2001. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45:1323–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kadlec K., et al. 2009. Diversity of antimicrobial resistance pheno- and genotypes of methicillin-resistant Staphylococcus aureus ST398 from diseased swine. J. Antimicrob. Chemother. 64:1156–1164 [DOI] [PubMed] [Google Scholar]
- 13. Khanna T., Friendship R., Dewey C., Weese J. S. 2008. Methicillin resistant Staphylococcus aureus colonization in pigs and pig farmers. Vet. Microbiol. 128:298–303 [DOI] [PubMed] [Google Scholar]
- 14. Kondo Y., et al. 2007. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob. Agents Chemother. 51:264–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schijffelen M. J. B. C., van Strijp J. A., Fluit A. C. 2010. Whole genome analysis of a livestock-associated methicillin-resistant Staphylococcus aureus ST398 isolate from a case of human endocarditis. BMC Genomics 11:376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Smith T. C., et al. 2009. Methicillin-resistant Staphylococcus aureus (MRSA) strain ST398 is present in midwestern U.S. swine and swine workers. PLoS One 4:e4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Takeuchi F., et al. 2005. Whole-genome sequencing of Staphylococcus haemolyticus uncovers the extreme plasticity of its genome and the evolution of human-colonizing staphylococcal species. J. Bacteriol. 187:7292–7308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. van Belkum A., et al. 2008. Methicillin-resistant and -susceptible Staphylococcus aureus sequence type 398 in pigs and humans. Emerg. Infect. Dis. 14:479–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Voss A., Loeffen F., Bakker J., Klaassen C., Wulf M. 2005. Methicillin-resistant Staphylococcus aureus in pig farming. Emerg. Infect. Dis. 11:1965–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wulf M. W., et al. 2008. Prevalence of methicillin-resistant Staphylococcus aureus among veterinarians: an international study. Clin. Microbiol. Infect. 14:29–34 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.