Abstract
ACH-702, a novel isothiazoloquinolone (ITQ), was assessed for antibacterial activity against a panel of Gram-positive and Gram-negative clinical isolates and found to possess broad-spectrum activity, especially against antibiotic-resistant Gram-positive strains, including methicillin-resistant Staphylococcus aureus (MRSA). For Gram-negative bacteria, ACH-702 showed exceptional potency against Haemophilus influenzae, Moraxella catarrhalis, and a Neisseria sp. but was less active against members of the Enterobacteriaceae. Good antibacterial activity was also evident against several anaerobes as well as Legionella pneumophila and Mycoplasma pneumoniae. Excellent bactericidal activity was observed for ACH-702 against several bacterial pathogens in time-kill assays, and postantibiotic effects (PAEs) of >1 h were evident with both laboratory and clinical strains of staphylococci at 10× MIC and similar in most cases to those observed for moxifloxacin at the same MIC multiple. In vivo efficacy was demonstrated against S. aureus with murine sepsis and thigh infection models, with decreases in the number of CFU/thigh equal to or greater than those observed after vancomycin treatment. Macromolecular synthesis assays showed specific dose-dependent inhibition of DNA replication in staphylococci, and biochemical analyses indicated potent dual inhibition of two essential DNA replication enzymes: DNA gyrase and topoisomerase IV. Additional biological data in support of an effective dual targeting mechanism of action include the following: low MIC values (≤0.25 μg/ml) against staphylococcal strains with single mutations in both gyrA and grlA (parC), retention of good antibacterial activity (MICs of ≤0.5 μg/ml) against staphylococcal strains with two mutations in both gyrA and grlA, and low frequencies for the selection of higher-level resistance (<10−10). These promising initial data support further study of isothiazoloquinolones as potential clinical candidates.
INTRODUCTION
Antibiotic resistance creates significant treatment issues for the worldwide health care community in regard to serious bacterial infections (3). Current pathogens of concern include methicillin-resistant Staphylococcus aureus (MRSA) (35), penicillin-resistant Streptococcus pneumoniae (PRSP) (22, 36), vancomycin-resistant enterococci (VRE) (15), extended-spectrum β-lactamase (ESBL) Gram-negative bacteria (17, 29, 31, 47), and multidrug-resistant Pseudomonas aeruginosa (17, 31). Staphylococci, particularly MRSA but also including coagulase-negative strains, have posed a challenge in hospital settings, resulting in substantial morbidity and mortality. Vancomycin is often used to treat MRSA infections, but in recent years there have been reports of vancomycin-nonsusceptible isolates and the reduced effectiveness of this drug (2, 6, 7, 18, 30). In addition, community-acquired MRSA accounts for an increasing number of serious infections (8). Despite the growing unmet medical need, few new antibacterial agents have been introduced in recent years that are effective against many of these often highly resistant clinical isolates (5).
One of our most important classes of antibiotics has been the fluoroquinolones; however, resistance to these drugs has also increased over time. In particular, most MRSA clinical isolates became resistant to fluoroquinolones within 5 years of their introduction for clinical use (1). Previously, we described a class of compounds with structural similarities to quinolones, the isothiazoloquinolones (ITQs; subset of heteroaryl isothiazolones), which displayed potent and broad-spectrum antibacterial activity against a variety of important pathogens, including fluoroquinolone-resistant isolates (32, 43, 44, 45). Prototype representatives from this class have been synthesized previously (10, 11), but none to date have been successfully developed as antibacterial drugs, for reasons unknown. These compounds were found to be excellent inhibitors of both bacterial DNA gyrase and topoisomerase IV, essential enzymes involved in DNA replication. This potent dual targeting of both enzymes probably accounts for the retention of good antibacterial activity against quinolone-resistant strains with multiple target mutations and the difficulty in obtaining mutants by selection with ITQs (9).
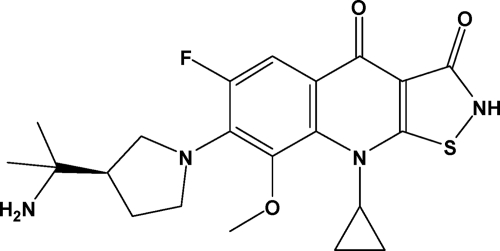
In this work, we describe our optimized lead compound, ACH-702 (Fig. 1), which had the best overall in vitro profile in our ITQ analog library in regard to antibacterial, protein binding and target inhibition properties (33, 41). The effectiveness of this compound against Gram-positive isolates, particularly MRSA and including quinolone-resistant strains, is especially attractive. In addition, ACH-702 also exhibits antibacterial activity against many Gram-negative strains, a property that is lacking among most currently marketed drugs used to treat MRSA infections. The mechanism of action involves potent inhibition of two clinically validated bacterial targets, DNA gyrase and topoisomerase IV, and thus suggests a more difficult path for resistance emergence. Therefore, this bactericidal compound offers the potential for further development as a new antibacterial agent, particularly against antibiotic-resistant Gram-positive pathogens.
Fig. 1.
Chemical structure of ACH-702.
MATERIALS AND METHODS
Bacterial strains.
Specific strains used in this work are listed in Table 1. All clinical isolates used and listed in Tables 2, 3, and 4, including MRSA strain ACH-0231, were from the strain collection at Eurofins Medinet, Chantilly, VA. Isolates were selected to include important emerging resistance phenotypes. For Staphylococcus aureus, this included methicillin-, vancomycin-, and quinolone-resistant, vancomycin intermediate-resistant, and linezolid-nonsusceptible isolates. For the streptococci, macrolide- and penicillin-resistant isolates were included. All other organisms were randomly selected from the Eurofins Medinet collection that contains isolates from clinical sites around the world. MRSA strains NY2746, BSA643, and BSA678 were obtained from Donald Low, Department of Microbiology, Toronto Medical Laboratories and Mount Sinai Hospital, Toronto, Canada (23). MRSA strain BK2384 was obtained from Barry Kreiswirth at the Public Health Research Institute/UMDNJ.
Table 1.
Bacterial strains and relevant genotypesa
| Organism | Strain | Genotypeb or description | Source and/or reference |
|---|---|---|---|
| S. aureus | ATCC 29213 | CLSI QC reference strain; MSSA | ATCC |
| ATCC 13709 | Laboratory standard strain; strain Smith | ATCC | |
| ATCC 33591 | Laboratory standard strain; MRSA | ATCC | |
| ACH-0204 | ATCC 29213 gyrA(Ser84Leu) | 9 | |
| ACH-0216 | ATCC 29213 grlA(Ser80Phe) | 9 | |
| ACH-0126 | ATCC 29213 gyrA(Ser84Leu) grlA(Ser80Phe) | 9 | |
| ACH-0130 | ATCC 29213 gyrA(Ser84Leu Glu88Val) grlA(Ser80Phe Ala116Val) | 9 | |
| COL | MRSA clinical isolate | 26 | |
| BK2384 | Cipr, Tmpr, Tetr, Eryr; HA-MRSA gyrA(Ser84Leu) grlA(Ser80Phe) | B. Kreiswirth, PHRI/UMDNJ | |
| ATCC 700699 | mecA+gyrA(Ser84Leu Glu409Lys) | ATCC (18) | |
| grlA(Ser80Phe); Mu50; VISA; MRSA | |||
| NY2746 | mecA+gyrA(Ser84Leu) | 23 | |
| grlA(Ser80Phe); MRSA | |||
| BSA643 | mecA+gyrA(Ser84Leu) | 23 | |
| grlA(Ser80Tyr Glu84Gly); MRSA | |||
| BSA678 | mecA+gyrA(Ser84Leu Ser85Pro) | 23 | |
| grlA(Ser80Phe Glu84Lys); MRSA | |||
| ACH-0231 | mecA+gyrA(Ser84Leu Glu88Lys) | Eurofins Medinet, Chantilly, VA | |
| grlA(Ser80Tyr Glu84Gly); MRSA | |||
| E. faecalis | ATCC 29212 | CLSI QC reference strain | ATCC |
| E. coli | ATCC 25922 | CLSI QC reference strain | ATCC |
| P. aeruginosa | ATCC 27853 | CLSI QC reference strain | ATCC |
Abbreviations: Cipr, ciprofloxacin resistant; Tmpr, trimethoprim resistant; Tetr resistant, tetracycline; Eryr, erythromycin resistant; CLSI, Clinical and Laboratory Standards Institute, Wayne, PA; ATCC, American Type Culture Collection, Manassas, VA; HA-MRSA, hospital-associated MRSA.
mecA is the gene encoding PBP2a; gyrA and grlA are the genes encoding the A subunit of gyrase and topoisomerase IV, respectively.
Table 2.
ACH-702 antibacterial activity against Gram-positive clinical isolates
| Organism (no. of isolates)b | Antimicrobial | MIC (μg/ml)a |
||
|---|---|---|---|---|
| Range | MIC50 | MIC90 | ||
| Staphylococcus aureus (82) | ||||
| MSSA, all isolates (12) | ACH-702 | 0.015–1 | 0.03 | 0.25 |
| Levofloxacin | 0.12–>16 | 0.25 | >16 | |
| Oxacillin | 0.25–0.5 | 0.5 | 0.5 | |
| Linezolid | 2–2 | 2 | 2 | |
| Vancomycin | 1–2 | 1 | 2 | |
| MRSA, all isolates (70) | ACH-702 | 0.008–0.5 | 0.06 | 0.25 |
| Levofloxacin | 0.12–>16 | 16 | >16 | |
| Oxacillin | 4–>16 | >16 | >16 | |
| Linezolid | 1–2 | 1 | 2 | |
| Vancomycin | 0.5–2 | 1 | 1 | |
| MRSA, FQNS (49) | ACH-702 | 0.03–0.5 | 0.06 | 0.25 |
| Levofloxacin | 2–>16 | 16 | >16 | |
| Oxacillin | 4–>16 | >16 | >16 | |
| Linezolid | 1–2 | 2 | 2 | |
| Vancomycin | 0.5–2 | 1 | 1 | |
| Staphylococcus epidermidis | ||||
| All isolates (21) | ACH-702 | 0.015–0.25 | 0.03 | 0.25 |
| Levofloxacin | 0.12–>16 | 0.25 | >16 | |
| Oxacillin | 0.25–>16 | 1 | >16 | |
| Linezolid | 0.5–1 | 1 | 1 | |
| Vancomycin | 1–2 | 2 | 2 | |
| FQNS (9) | ACH-702 | 0.12–0.25 | 0.12 | 0.25 |
| Levofloxacin | 8–>16 | 16 | >16 | |
| Oxacillin | 0.25–>16 | 8 | >16 | |
| Linezolid | 0.5–1 | 1 | 1 | |
| Vancomycin | 1–2 | 2 | 2 | |
| Streptococcus pneumoniae | ||||
| All isolates (31) | ACH-702 | 0.015–0.06 | 0.03 | 0.06 |
| Levofloxacin | 0.5–2 | 1 | 1 | |
| Ceftriaxone | 0.015–8 | 0.5 | 1 | |
| Linezolid | 0.5–2 | 1 | 1 | |
| Penicillin | <0.015–8 | 1 | 4 | |
| PENNS (18) | ACH-702 | 0.03–0.06 | 0.03 | 0.06 |
| Levofloxacin | 0.5–2 | 1 | 2 | |
| Ceftriaxone | 0.5–8 | 1 | 2 | |
| Linezolid | 0.5–2 | 1 | 1 | |
| Penicillin | 0.25–8 | 4 | 4 | |
| Enterococcus faecalis | ||||
| All isolates (15) | ACH-702 | 0.06–0.5 | 0.25 | 0.5 |
| Levofloxacin | 0.5–>16 | >16 | >16 | |
| Linezolid | 0.5–>16 | 2 | >16 | |
| Vancomycin | 0.25–>32 | 1 | >32 | |
| FQNS (9) | ACH-702 | 1–2 | 1 | 1 |
| Levofloxacin | 16–>16 | >16 | >16 | |
| Linezolid | 0.5–>16 | 2 | >16 | |
| Vancomycin | 1–>32 | 1 | >32 | |
| Enterococcus faecium (14)c | ACH-702 | 0.25–4 | 1 | 2 |
| Levofloxacin | 4–>16 | >16 | >16 | |
| Linezolid | 0.5–>16 | 2 | 2 | |
| Vancomycin | 0.25–>32 | >32 | >32 | |
For current CLSI breakpoints for comparators, see reference 14.
FQNS, fluoroquinolone-nonsusceptible isolates as indicated by levofloxacin MICs relative to CLSI breakpoints; PENNS, penicillin-nonsusceptible isolates, including intermediate (I) and resistant (R) isolates.
All E. faecium isolates tested were fluoroquinolone resistant.
Table 3.
ACH-702 antibacterial activity against Gram-negative clinical isolates
| Organism (no. of isolates)b | Antimicrobial | MIC (μg/ml)a |
||
|---|---|---|---|---|
| Range | MIC50 | MIC90 | ||
| Enterobacteriaceae, all isolates (120) | ||||
| Escherichia coli (30) | ACH-702 | 0.06–16 | 0.12 | 8 |
| Ciprofloxacin | 0.015–>64 | 0.03 | 32 | |
| Moxifloxacin | 0.06–32 | 0.06 | 16 | |
| Ceftazidime | ≤0.03–>16 | 0.25 | 0.25 | |
| Imipenem | 0.12–0.25 | 0.25 | 0.25 | |
| Gentamicin | ≤0.12–>8 | 0.5 | >8 | |
| Enterobacter cloacae (30) | ACH-702 | 0.06–32 | 0.25 | 8 |
| Ciprofloxacin | 0.015–>64 | 0.03 | 16 | |
| Moxifloxacin | 0.06–>64 | 0.12 | 16 | |
| Ceftazidime | ≤0.03–>16 | 0.25 | >16 | |
| Imipenem | 0.06–2 | 0.5 | 1 | |
| Gentamicin | ≤0.12–>8 | 0.25 | >8 | |
| Klebsiella pneumoniae (30) | ACH-702 | 0.06–64 | 0.25 | 2 |
| Ciprofloxacin | 0.015–>64 | 0.06 | 1 | |
| Moxifloxacin | 0.03–>64 | 0.12 | 2 | |
| Ceftazidime | ≤0.03–>16 | 0.12 | >16 | |
| Imipenem | 0.12–8 | 0.25 | 0.5 | |
| Gentamicin | ≤0.12–>8 | 0.25 | 0.5 | |
| Proteus mirabilis (30) | ACH-702 | 0.06–16 | 0.25 | 4 |
| Ciprofloxacin | 0.03–>64 | 0.06 | 32 | |
| Moxifloxacin | 0.25–>64 | 0.5 | 64 | |
| Ceftazidime | ≤0.03–0.12 | 0.06 | 0.06 | |
| Imipenem | 0.06–4 | 2 | 4 | |
| Gentamicin | 0.5–>8 | 1 | >8 | |
| Enterobacteriaceae FQNS (22) | ACH-702 | 0.5–64 | 8 | 16 |
| Ciprofloxacin | 2–>64 | 32 | >64 | |
| Moxifloxacin | 4–>64 | 16 | >64 | |
| Ceftazidime | 0.06–>16 | 16 | >16 | |
| Imipenem | 0.06–8 | 0.5 | 4 | |
| Gentamicin | 0.25–>8 | 1 | >8 | |
| Nonfermenters, all isolates (60) | ||||
| Acinetobacter baumannii (30) | ACH-702 | 0.06–8 | 1 | 4 |
| Ciprofloxacin | 0.15–>64 | 32 | >64 | |
| Moxifloxacin | 0.03–64 | 4 | 32 | |
| Ceftazidime | 0.25–>16 | 4 | >16 | |
| Imipenem | 0.12–>8 | 0.5 | >8 | |
| Gentamicin | ≤0.12–>8 | 1 | >8 | |
| A. baumannii FQNS (16) | ACH-702 | 1–8 | 2 | 8 |
| Ciprofloxacin | 32–>64 | 64 | >64 | |
| Moxifloxacin | 4–64 | 16 | 32 | |
| Ceftazidime | 2–>16 | >16 | >16 | |
| Imipenem | 0.12–>8 | 8 | >8 | |
| Gentamicin | 0.25–>8 | >8 | >8 | |
| Pseudomonas aeruginosa (30) | ACH-702 | 0.12–32 | 1 | 8 |
| Ciprofloxacin | 0.03–32 | 0.25 | 8 | |
| Moxifloxacin | 0.12–64 | 2 | 64 | |
| Ceftazidime | ≤0.03–>16 | 2 | 8 | |
| Imipenem | 0.5–>8 | 2 | 4 | |
| Gentamicin | 0.25–>8 | 1 | 2 | |
| P. aeruginosa FQNS (11) | ACH-702 | 1–32 | 8 | 32 |
| Ciprofloxacin | 2–32 | 8 | 32 | |
| Moxifloxacin | 4–64 | 64 | 64 | |
| Ceftazidime | 0.5–>16 | 2 | 16 | |
| Imipenem | 0.5–>8 | 2 | 8 | |
| Gentamicin | 0.25–>8 | 2 | 8 | |
| Respiratory Gram-negatives (23) | ||||
| Haemophilus influenzae (10) | ACH-702 | 0.06–0.12 | 0.06 | 0.12 |
| Levofloxacin | 0.015–0.06 | 0.015 | 0.03 | |
| Ampicillin | 0.12–>16 | >16 | >16 | |
| Ceftriaxone | 0.015–0.03 | 0.015 | 0.015 | |
| Moraxella catarrhalis (13) | ACH-702 | 0.03–0.12 | 0.06 | 0.06 |
| Levofloxacin | 0.03–0.06 | 0.03 | 0.06 | |
| Ampicillin | 1–16 | 4 | 8 | |
| Ceftriaxone | 0.03–2 | 1 | 2 | |
For current CLSI breakpoints for comparators, see reference 14.
FQNS, fluoroquinolone-nonsusceptible isolates as defined by ciprofloxacin MICs relative to CLSI breakpoints.
Table 4.
ACH-702 antibacterial activity against anaerobic clinical isolates
| Organism (no. of isolates) | Antimicrobial | MIC (μg/ml)a |
||
|---|---|---|---|---|
| Range | MIC50 | MIC90 | ||
| Bacteroides fragilis (10) | ACH-702 | 0.06–0.12 | 0.12 | 0.12 |
| Clindamycin | 0.25–4 | 1 | 4 | |
| Imipenem | 0.25–0.5 | 0.25 | 0.25 | |
| Penicillin | 0.5–32 | 16 | 16 | |
| Clostridium difficile (10) | ACH-702 | 0.25–4 | 0.25 | 4 |
| Clindamycin | 1–>32 | 2 | >32 | |
| Imipenem | 4–16 | 4 | 4 | |
| Penicillin | 0.5–4 | 1 | 1 | |
| Propionibacterium acnes (10) | ACH-702 | 0.03–0.06 | 0.06 | 0.06 |
| Clindamycin | ≤0.03–0.06 | 0.06 | 0.06 | |
| Imipenem | 0.03–0.12 | 0.06 | 0.06 | |
| Penicillin | ≤0.12–≤0.12 | ≤0.12 | ≤0.12 | |
| Peptostreptococcus sp. (10) | ACH-702 | 0.008–0.12 | 0.015 | 0.06 |
| Clindamycin | 0.12–4 | 0.25 | 1 | |
| Imipenem | 0.03–0.5 | 0.06 | 0.25 | |
| Penicillin | ≤0.25–0.5 | 0.25 | 0.5 | |
For current CLSI breakpoints for comparators, see reference 14.
In vitro susceptibility testing.
All susceptibility testing was done either at Achillion Pharmaceuticals or Eurofins Medinet in cation-adjusted Mueller-Hinton II broth (CAMHB; media obtained from BD, Sparks, MD, unless otherwise indicated). Streptococci were supplemented with 2 to 5% lysed horse blood, and Haemophilus influenzae was tested in Haemophilus test medium. Aerobic isolates were tested by broth microdilution using CLSI standards (12), with final inoculum sizes of approximately 5 × 105 CFU/ml for most strains. Inoculated plates were incubated aerobically at 35 to 37°C for 24 h, and the MIC was defined as the minimum concentration of compound that resulted in no visible growth after 24 h at 35 to 37°C. Anaerobic organisms were tested by agar dilution as recommended by CLSI (13) using Brucella agar supplemented with 5 mg/ml hemin, 1 mg/ml vitamin K, and 5% laked sheep blood. Inocula consisted of colonies taken from 24-h growth on enriched blood agar and resuspended to approximate the turbidity of a 0.5 McFarland standard in Brucella broth. Plates were incubated under anaerobic conditions at 35 to 37°C for 48 h. The MIC50 and MIC90 values were calculated by standard methods at Eurofins Medinet.
Time-kill studies.
Time-kill studies were done as described previously (4, 32). Briefly, all strains were cultured overnight at 35 to 37°C in CAMHB, and cells were diluted into fresh medium, grown to exponential phase, and then diluted again in medium to adjust cell densities to approximately 107 CFU/ml. Compounds were then added at concentration multiples of the MIC. Rates of killing were determined by measuring the reduction in viable bacteria (log10 CFU/ml) at 0, 1, 2, 4, 6, and 24 h at fixed concentrations of compound. Experiments were performed in duplicate. If plates contained fewer than 10 CFU/ml, the number of colonies was considered to be below the limit of quantitation. Samples of culture containing compound were diluted at least 10-fold to minimize drug carryover to the CAMHB plates.
PAE.
The postantibiotic effect (PAE) was determined following treatment of S. aureus with compound at 10× MIC for 1 h (16). Prewarmed 10-ml CAMHB cultures with or without antibiotic were inoculated with approximately 2 to 4 × 107 CFU/ml S. aureus cells in logarithmic growth phase, and then grown with shaking at 35°C to 37°C for 1 h. After this exposure period, cultures were diluted 1:1,000 in prewarmed CAMHB to initiate recovery. Viable counts were measured prior to exposure, immediately after dilution, and hourly for 4 to 8 h of recovery, by plating appropriate dilutions onto CAMHB agar. An additional control with untreated bacteria was diluted in media plus compound at 0.01× MIC to establish that the residual compound during recovery had no effect on growth and viability (data not shown). The PAE was defined as T − C, where T is the time required for viability counts of an antibiotic-exposed culture to increase 1 log10 above the counts present immediately after dilution and C is the corresponding time for the growth control without antibiotic (28).
Macromolecular synthesis assay.
The effects of ACH-702 and control compounds ciprofloxacin (DNA synthesis), rifampin (RNA synthesis), and chloramphenicol (protein synthesis) on DNA, RNA, and protein synthesis in bacteria were measured using radiolabeled precursors [3H]thymidine (DNA), [3H]uracil (RNA), and [3H]leucine (protein) in mid-exponential-phase cultures (108 CFU/ml) of S. aureus ATCC 29213 in chemically defined medium (40) as previously described (27). Final concentrations of 5 μCi/ml for thymidine and 2.5 μCi/ml for each of the other precursors were added to cultures immediately before the addition of antibiotics (10× MIC). Negative controls for the macromolecular assays consisted of all reaction materials with no antibiotics added. The resulting counts were used as the 100% values. After an additional 20-min incubation at 37°C in the presence of antibiotics, samples were removed for trichloroacetic acid precipitation and subsequent scintillation counter analyses for the determination of radioactive incorporation into DNA, RNA, or protein, and the data were expressed as the percent inhibition of incorporation into a drug-free control.
Selection of resistant mutants.
S. aureus parent strains, ATCC 29213 or ATCC 33591, were cultured in brain heart infusion (BHI) broth overnight and concentrated in a 1/10 volume of phosphate-buffered saline, and approximately 1 × 1010 CFU were plated onto BHI agar plates containing 1×, 2×, 4× and 8× MIC concentrations of compounds and incubated at 37°C for 48 to 72 h as described previously (9). Selection frequencies of resistant mutants were calculated as the ratios of resistant mutants to untreated control cells. All resistant mutants were confirmed by MIC and DNA sequencing of the quinolone resistance determining regions (QRDRs) (46) of gyrA and grlA.
DNA sequencing.
Template genomic DNAs were prepared by boiling one fresh S. aureus colony in 50 μl of distilled water and were subjected to PCR amplification. ABI 3100 DNA sequencers used PCR DNAs as templates for automated sequencing at W. M. Keck Facility, Yale University, New Haven, CT.
Cloning, protein expression, and purification of S. aureus enzymes.
Expression of wild-type proteins GyrA, GyrB, GrlA (ParC), and GrlB (ParE) and mutant proteins GyrA(Ser84Leu), GrlA(Ser80Phe), GyrA(Ser84Leu Glu88Lys), and GrlA(Ser80Phe Glu84Lys) was performed as previously described (9, 39), with S. aureus COL as the source for wild-type genes (26). Briefly, cloning was done using pET vector overexpression constructs with 6His tags (Novagen, EMD, La Jolla, CA). The GyrA constructs additionally encoded the cleavage site for tobacco etch virus (TEV) protease downstream of the thioredoxin gene for enhanced solubility (39). All mutant constructs were made by site-directed mutagenesis using the QuikChange II XL kit (Stragagene, Santa Clara, CA) according to the manufacturer's instructions. All proteins were expressed in Escherichia coli BL21(DE3) pLysS cells and purified individually to greater than 95% homogeneity from the supernatant of lysed cells by nickel affinity based on the ProBond protocol from Invitrogen followed by size exclusion column chromatography. GyrA protein was cleaved from thioredoxin with recombinant TEV protease (Invitrogen, Carlsbad, CA) during purification.
S. aureus topoisomerase IV and DNA gyrase assays.
Topoisomerase IV proteins were reconstituted in vitro as wild-type enzyme (GrlA[wild type] plus GrlB) or mutant enzyme (GrlA[mutant] plus GrlB) at subunit molar ratios of 1:1. Topoisomerase IV activity was measured by a decatenation assay that monitored the ATP-dependent unlinking of DNA minicircles from kinetoplast DNA (Topogen, Inc., Port Orange, FL). DNA gyrase proteins were reconstituted in vitro as wild-type enzyme (GyrA[wild type] plus GyrB) or mutant enzyme (GyrA[mutant] plus GyrB) at subunit molar ratios of 1:1. Gyrase activity was measured by a supercoiling assay that monitored the ATP-dependent conversion of relaxed pBR322 DNA to the supercoiled form (Topogen). The products of both enzymes were quantified by ethidium bromide fluorescence following agarose gel electrophoresis. IC50s for both enzymes were defined as the concentration of compound that reduced product formation to 50% of the uninhibited control. All values were the result of at least two independent experiments. Compound potencies were compared with values determined for quinolones such as moxifloxacin.
In vivo efficacy against experimental infections. (i) Mouse thigh muscle infection.
Adult female CD-1 mice (19 to 28 g; Charles River Laboratories, Wilmington, MA) were made neutropenic with two intraperitoneal injections of cyclophosphamide (Sigma; 150 mg/kg of body weight in 10 ml/kg) at 4 days and 1 day before infection. Groups of three mice each were infected intramuscularly with approximately 105 CFU of MRSA ATCC 33591 and were treated with a single subcutaneous dose of compound at 2 h postinfection. A control group of infected animals was not treated. Thighs from three animals were harvested at 2, 4, 6, or 24 h after treatment initiation. The thighs were processed by aseptic removal from each animal, dilution into sterile phosphate-buffered saline, and homogenization with a tissue homogenizer. Serial dilutions of the tissue homogenates were spread on Trypticase-soy agar (TSA; BD) plates, and the plates were incubated at 37°C overnight. The CFU/thigh values were determined from colony counts and compared with the control number of CFU/thigh at the time of treatment.
(ii) Mouse systemic infection.
Adult female CD-1 mice (20 to 29 g; Charles River Laboratories, Wilmington, MA) were inoculated intraperitoneally with a sufficient number of pathogens to kill 100% of the untreated animals within 5 days. Each mouse received ∼3.9 × 107 CFU/ml of S. aureus 13709 suspended in 5% (wt/vol) sterile hog gastric mucin in a volume of 0.5 ml. Test articles were administered at 1 h after pathogen inoculation. The number of mice that survived in each experimental group was monitored up to 5 days after pathogen inoculation, and the doses for 50% of the population of drug-treated animals that survived (50% protective doses [PD50s]) were determined. Each experimental group consisted of six animals, and a minimum of four different concentrations of compound was evaluated. The range of doses was 0.03 to 2 mg/kg of body weight.
Pharmacokinetics in mice.
Pharmacokinetic parameters were obtained as previously described (32). Briefly, plasma samples were collected at various time points, and the levels of ACH-702 were quantified by liquid chromatography-mass spectrometry (LC-MS/MS) using an internal standard. Resulting mean plasma concentrations were analyzed by noncompartmental methods with WinNonlin (Pharsight, Mountain View, CA). The area under the concentration (AUC)-time curve from time zero to the time of the last quantifiable plasma concentration (AUClast) was calculated using the linear trapezoidal rule. Plasma half life (t1/2) and time at which maximum concentration occurs after dosing (tmax) were calculated with WinNonlin.
RESULTS
Antibacterial activities against clinical isolates.
Table 2 compares the antibacterial activities of ACH-702 against Gram-positive clinical isolates to those of comparator drugs. Against methicillin-susceptible S. aureus (MSSA; 12 strains), ACH-702 possessed an MIC50 of 0.03 μg/ml and an MIC90 of 0.25 μg/ml. For MRSA (70 strains), good antibacterial activity was retained, with an MIC50 of 0.06 and an MIC90 of 0.25, while the comparator quinolone, levofloxacin, was essentially ineffective, as 49/70 strains (70%) had MICs of >2 μg/ml. The MIC90s of all currently marketed fluoroquinolones for quinolone-resistant MRSA are >4 μg/ml (4). Against those MRSA strains with reduced vancomycin susceptibility, including both intermediate-resistant (VISA) and resistant (VRSA) strains (n = 8), ACH-702 also displayed activity essentially equivalent to that against vancomycin-sensitive strains (data not shown). In addition, much greater activity was observed for ACH-702 than for comparator drugs against Staphylococcus epidermidis (Table 2), particularly against quinolone-nonsusceptible isolates.
The MIC50s and MIC90s for ACH-702 against Streptococcus pneumoniae (n = 31; Table 2), including penicillin-resistant isolates, were excellent, with no significant differences observed between penicillin-sensitive (PSSP; 13 strains), penicillin-intermediate (PISP; 3 strains), and penicillin-resistant (PRSP; 15 strains) phenotypes. The MICs for ACH-702 against penicillin nonsusceptible strains were lower than for penicillin and ceftriaxone, as the MIC90 remained at 0.06 μg/ml versus 1 and 2 μg/ml for ceftriaxone and penicillin, respectively. Against enterococcal strains, ACH-702 displayed MIC50 and MIC90 values of 0.25 and 0.5 μg/ml for Enterococcus faecalis. These values were lower than those for all comparators tested for E. faecalis. ACH-702 MIC50 and MIC90 values were 1 μg/ml for the quinolone-nonsusceptible E. faecalis isolates (9 strains), contrasting sharply with those for levofloxacin, for which all MICs for quinolone-nonsusceptible E. faecalis isolates were ≥16 μg/ml. However, ACH-702 was less active against E. faecium, with an MIC50 of 1 and an MIC90 of 2 μg/ml, similar to those for the linezolid comparator. Against additional streptococci, including Staphylococcus pyogenes and viridans group streptococci, ACH-702 was also quite active, with all MIC50s and MIC90s ≤0.03 μg/ml (data not shown).
Table 3 compares the antibacterial activities of ACH-702 against Gram-negative clinical isolates to those of several comparator drugs. Although MICs were not as impressive as those seen for Gram-positive strains, antibacterial activity comparable to those of ciprofloxacin and moxifloxacin was found for four Enterobacteriaceae: E. coli, Enterobacter cloacae, Klebsiella pneumoniae, and Proteus mirabilis, as well as P. aeruginosa. For Acinetobacter baumannii, however, ACH-702 was much more active than the two fluoroquinolone comparators, with MIC50 and MIC90 values of 1 and 4 μg/ml, respectively. However, against quinolone-nonsusceptible isolates of these species, ACH-702 possessed lower MIC50s and MIC90s than ciprofloxacin and moxifloxacin except for P. aeruginosa, for which ciprofloxacin was slightly more active (Table 3). For Haemophilus influenzae and Moraxella catarrhalis, ACH-702 was quite active, with MIC90s ranging from 0.06 to 0.12 μg/ml, including several ampicillin-resistant strains (Table 3). In addition, ACH-702 was tested against both Neisseria gonorrhea and Neisseria meningitidis, with MICs of ≤0.015 μg/ml for all isolates tested (n = 10 for each; data not shown).
MIC values for ACH-702 against four anaerobes are listed in Table 4. Against Bacteroides fragilis, MICs ranged from 0.06 to 0.12 μg/ml for all 10 strains tested. These values are slightly lower than those for imipenem and substantially lower than those for penicillin and clindamycin. For Clostridium difficile, the MIC50 and MIC90 were 0.25 and 4 μg/ml, respectively. MIC90 values were 0.06 μg/ml against both Propionibacterium acnes and Peptostreptococcus sp., equal to or better than those for the tested comparator drugs. ACH-702 also displayed good activity against Legionella pneumophila and Mycoplasma pneumoniae, with MIC90s of 0.015 μg/ml for both organisms (data not shown).
Bactericidal activity.
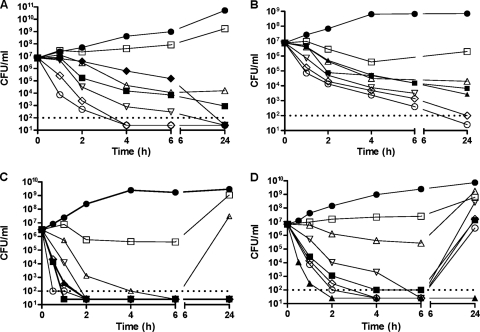
Bactericidal activity for ACH-702 was assessed using two Gram-positive and two Gram-negative strains treated in exponential growth phase. Figure 2A shows representative time-kill curves against an MRSA strain, S. aureus ATCC 33591, with vancomycin and moxifloxacin as comparators. Maximum killing was observed at concentrations of 4× MIC and higher, with a 3-log drop in the number of CFU/ml occurring by 2 h after compound addition. In comparison, killing was less rapid with 4× MIC concentrations of moxifloxacin and, especially, vancomycin. Killing was also less rapid at 2× MIC of ACH-702, for which about 4 h was required to cause a 3-log drop in the number of CFU/ml, suggesting a dose-dependent effect on staphylococcal killing up to a concentration equal to 4× MIC. Figure 2B shows bactericidal activity against E. faecalis ATCC 29212, for which again a 3-log drop in the number of CFU/ml was observed in approximately 2 h at ≥4× MIC, in contrast to the 6 h required by moxifloxacin and ciprofloxacin at 4× MIC. Against E. coli ATCC 25922, a 3-log drop in the number of CFU/ml was observed in less than 2 h at doses of ACH-702 ≥1× MIC as well as for ciprofloxacin and moxifloxacin at 4× MIC (Fig. 2C). For Pseudomonas aeruginosa ATCC 27853, a 3-log drop in the number of CFU/ml was observed for ACH-702 within 2 h at doses of ≥2× MIC (Fig. 2D). This was true for moxifloxacin and ciprofloxacin at 4× MIC. However, unexpectedly, a “rebound” effect was observed for moxifloxacin at 4× MIC and at all doses of ACH-702, including 8× MIC, at which cultures recovered to initial densities by 24 h. A rebound effect was also observed for E. coli at or below the MIC (Fig. 2C).
Fig. 2.
Time-kill curves for bacterial pathogens. Compounds were added to cultures at time zero, and samples were processed as described in Materials and Methods. Bacterial strains: S. aureus ATCC 33591 (A), E. faecalis ATCC 29212 (B), E. coli ATCC 25922 (C), and P. aeruginosa ATCC 27853 (D). Symbols: filled circles, untreated control; filled diamonds, vancomycin (4× MIC); filled squares, moxifloxacin (4× MIC); filled triangles, ciprofloxacin (4× MIC); open squares, ACH-702 (0.5× MIC); open triangles, ACH-702 (1× MIC); open inverted triangles, ACH-702 (2× MIC); open diamonds, ACH-702 (4× MIC); open circles, ACH-702 (8× MIC). The dotted lines indicate the lower limit of quantification; points below the dotted lines could not be assigned precise values for the number of CFU/ml.
PAEs.
The postantibiotic effect (PAE) is a measure of the effects of an antibiotic on the cellular viability and growth of bacteria after exposure to that antibiotic is terminated. The PAE of ACH-702 for staphylococci was assessed with four MRSA strains, ATCC 33591, ATCC 700699, BSA678, and BK2384, by measuring the time required for recovery after a defined treatment period (16). PAE results for these strains at 10× MIC were 1.2 to 4.6 h, similar to values obtained with moxifloxacin (data not shown).
Antibacterial activity against quinolone-resistant MRSA clinical isolates and laboratory-selected mutant strains.
The antibacterial activity of ACH-702 was determined against six quinolone-resistant MRSA clinical isolates with characterized target mutations (Table 5). The three marketed quinolones tested as comparators, moxifloxacin, gemifloxacin, and ciprofloxacin, were minimally active or inactive against the six strains that possessed two to four mutations in the QRDRs of gyrA and grlA. Interestingly, all six strains, including four with possible efflux contributions as indicated by ethidium bromide resistance, were susceptible to ACH-702, as the MICs remained at ≤0.5 μg/ml. Particularly noteworthy, ACH-702 retained good antibacterial activity against strains BSA678 and ACH-0231, for which quinolone MICs were all ≥64 μg/ml. Against quinolone-resistant laboratory mutants (9), differences in MIC values for ACH-702 versus moxifloxacin, gemifloxacin, and ciprofloxacin for strains possessing mutations in gyrA and/or grlA were evident, with ACH-702 retaining low MICs for strains with multiple QRDR mutations (Table 5).
Table 5.
MICs and target genotypes for laboratory strains and clinical isolates of S. aureus
| Isolate | ACH-702 MIC (μg/ml) | MIC (μg/ml) |
QRDR mutationsa |
||||
|---|---|---|---|---|---|---|---|
| MXFb | GEM | CIP | EtBr | gyrA | grlA | ||
| Clinical | |||||||
| ATCC 29213 | 0.004 | 0.06 | 0.03 | 0.25 | 4 | None | None |
| BK2384 | 0.08 | 2 | 1 | 64 | ND | Ser84Leu | Ser80Phe |
| NY2746 | 0.25 | 8 | 32 | 256 | 16 | Ser84Leu | Ser80Phe |
| ATCC 700699 | 0.125 | 4 | 4 | 64 | >64 | Ser84Leu Glu409Lys | Ser80Phe |
| BSA643 | 0.25 | 16 | 32 | 256 | 16 | Ser84Leu | Ser80Tyr Glu84Gly |
| BSA678 | 0.25 | >64 | 64 | 128 | 4 | Ser84Leu Ser85Pro | Ser80Phe Glu84Lys |
| ACH-0231 | 0.5 | 64 | >64 | >64 | 16 | Ser84Leu Glu88Lys | Ser80Tyr Glu84Gly |
| Laboratoryc | |||||||
| ACH-0204 | 0.016 | 0.125 | 0.125 | 0.5 | 4 | Ser84Leu | None |
| ACH-0216 | 0.008 | 0.25 | 0.125 | 2 | 4 | None | Ser80Phe |
| ACH-0126 | 0.125 | 4 | 4 | 64 | 4 | Ser84Leu | Ser80Phe |
| ACH-0130 | 0.5 | 32 | 32 | 256 | 32 | Ser84Leu Glu88Val | Ser80Phe Ala116Val |
QRDR, quinolone resistance determining region. No mutations found in gyrB or grlB.
Abbreviations: MXF, moxifloxacin; GEM, gemifloxacin; CIP, ciprofloxacin; EtBr, ethidium bromide; ND, not determined.
Parent strain is S. aureus ATCC 29213.
Activities of ACH-702 against in vitro-purified wild-type, single mutant, and double mutant DNA topoisomerase IV and gyrase.
To examine bacterial target enzyme inhibition by ACH-702, staphylococcal gyrase and topoisomerase IV enzymes were cloned, expressed, and purified for in vitro biochemical studies. In addition, mutant enzyme proteins were engineered via site-directed mutagenesis to introduce the GyrA(Ser84Leu) single and GyrA(Ser84Leu Glu88Lys) double mutations and the GrlA(Ser80Phe) single and GrlA(Ser80Phe Glu84Lys) double mutations that result in increased resistance to fluoroquinolones (19). Enzyme inhibition assays showed that IC50s of the comparator quinolones, moxifloxacin, gemifloxacin, and ciprofloxacin, for wild-type topoisomerase IV decatenation activity were 1.0, 0.4, and 3.0 μM, respectively. The IC50s were consistent with previous reports that quinolones have high binding affinities and thus potently inhibit topoisomerase IV (20, 21). The IC50 of ACH-702 was 0.12 μM, more than 3-fold lower than that of gemifloxacin at 0.4 μM, which was reported as dual targeting and one of the most potent quinolones against topoisomerase IV in staphylococci (21). The IC50s of the comparator quinolones for wild-type gyrase supercoiling activity were 7.7, 5.6, and 62 μM for moxifloxacin, gemifloxacin, and ciprofloxacin, respectively, while the IC50 of ACH-702 was 0.68 μM, more than 8-fold lower than that of gemifloxacin, the most potent marketed quinolone inhibitor of staphylococcal gyrase to date (21). These data demonstrate that ACH-702 significantly inhibits both topoisomerases in wild-type S. aureus, and this balanced activity, with lower IC50s against both enzymes compared with that of gemifloxacin, could account for the lower MIC of ACH-702 (0.004 μg/ml) against wild-type S. aureus than that obtained with gemifloxacin (0.03 μg/ml).
A similar analysis was made using gyrase and topoisomerase enzymes with single amino acid substitutions that result in decreased inhibition by quinolones. The IC50s of ACH-702 and comparator quinolones were obtained against mutant topoisomerase IV containing the GrlA(Ser80Phe) subunit and against mutant gyrase containing the GyrA(Ser84Leu) subunit. As shown in Table 6, moxifloxacin, gemifloxacin, and ciprofloxacin each lost between 12- and 25-fold activity against the mutant topoisomerase IV (IC50 = 11.9, 9.8, and 57 μM, respectively). In comparison, the IC50 of ACH-702 was 0.6 μM, which is only 5-fold greater than that against the wild-type enzyme and is 16- to 19-fold lower than that with moxifloxacin and gemifloxacin and in the same range as those displayed by the two quinolones against wild-type topoisomerase IV (1.0 and 0.4 μM, respectively). Results were even more pronounced for the mutant gyrase, as the comparator quinolones moxifloxacin, gemifloxacin, and ciprofloxacin showed complete inactivity in the assay (IC50s of >300 μM), while ACH-702 had an IC50 of 4.2 μM, which is only 6-fold greater than that against wild-type gyrase and in the same range as those of moxifloxacin and gemifloxacin against wild-type gyrase (7.7 and 5.6 μM, respectively).
Table 6.
Inhibition of wild-type (WT) and mutant S. aureus gyrase and topoisomerase IV catalytic activities by ACH-702 and quinolonesc
| Antimicrobial | IC50 (μM) |
|||||
|---|---|---|---|---|---|---|
| Topoisomerase IVa |
Gyraseb |
|||||
| WT | Ser80Phe | Ser80Phe Glu84Lys | WT | Ser84Leu | Ser84Leu Glu88Lys | |
| ACH-702 | 0.12 | 0.6 | 32 | 0.68 | 4.2 | 70 |
| Moxifloxacin | 1.0 | 11.9 | >200 | 7.7 | >300 | >500 |
| Gemifloxacin | 0.4 | 9.8 | >200 | 5.6 | >300 | >500 |
| Ciprofloxacin | 3.0 | 57 | >200 | 62 | >300 | ND |
Topoisomerase IV decatenation activity.
Gyrase supercoiling activity. ND, not determined.
Mutant enzymes made via site-directed mutagenesis of WT gene.
Additionally, the activity of ACH-702 and comparators were assessed against enzymes with double mutations that resulted in further resistance to inhibition by quinolones. The IC50s of ACH-702 and comparator quinolones against double mutant topoisomerase IV composed of GrlA(Ser80Phe Glu84Lys) and GrlB(wild type) subunits and against mutant gyrase composed of GyrA(Ser84Leu Glu88Lys) and GyrB(wild type) subunits were obtained (Table 6). Moxifloxacin and gemifloxacin were essentially inactive against both, with IC50 values of >200 and >500 μM against double mutant topoisomerase IV and gyrase, respectively. In comparison, the IC50s of ACH-702 were 32 and 70 μM, a range similar to that obtained for ciprofloxacin against wild-type gyrase.
Selection of resistant mutants.
The frequency of selecting resistant mutants of S. aureus ATCC 29213 by ACH-702 at 4× MIC after 48 h of incubation at 37°C was approximately 9 × 10−11, which was approximately 1 to 2 logs lower than that by ciprofloxacin (1 × 10−9 to 1.1 × 10−8) at 4× MIC (not shown); this value for ciprofloxacin was in agreement with previously published values (38). For MRSA strains, the frequency of mutant selection by ACH-702 at 4× MIC was <1 × 10−10 for ATCC 33591 and <4 × 10−10 for BK2384. These frequencies were ≥2 logs lower than those observed (approximately 1 × 10−8) using ciprofloxacin selection at 4× MIC for the two strains. Additional incubation of plates to 72 h yielded tiny ACH-702-resistant colonies at approximately 10-fold higher frequencies than those at 48 h, suggesting a fitness cost to these cells.
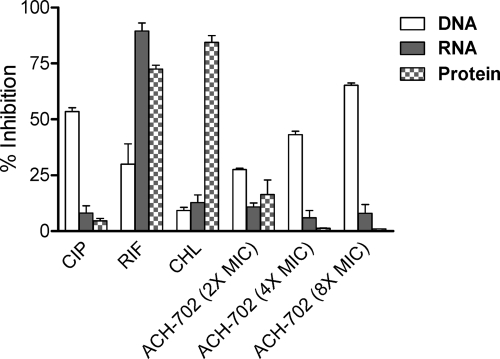
Macromolecular synthesis inhibition.
From the above enzyme inhibition data, it would follow that ACH-702 is a selective inhibitor of DNA synthesis in bacteria. To verify this assumption, three macromolecular synthesis inhibition assays were performed with ACH-702 and control antibiotics to evaluate [3H]thymidine, [3H]uridine, and [3H]leucine incorporation into DNA, RNA, and protein, respectively, after a short exposure to the compounds. The results of these assays are summarized in Fig. 3. Three antibacterial agents with validated mechanisms of action were tested as controls. As expected, ACH-702 selectively inhibited DNA synthesis in a dose-dependent manner. The control antibiotic inhibitors, ciprofloxacin, rifampin, and chloramphenicol, showed the predicted inhibition patterns for DNA, RNA, and protein synthesis, respectively.
Fig. 3.
Macromolecular synthesis inhibition in Staphylococcus aureus. The assays measured incorporation of radiolabeled precursors into DNA, RNA, or protein as described in Materials and Methods. The percent inhibition represents decrease from untreated controls. Positive control compounds: DNA, ciprofloxacin (CIP); RNA, rifampin (RIF); protein, chloramphenicol (CHL).
Therapeutic efficacy and pharmacokinetics in mice.
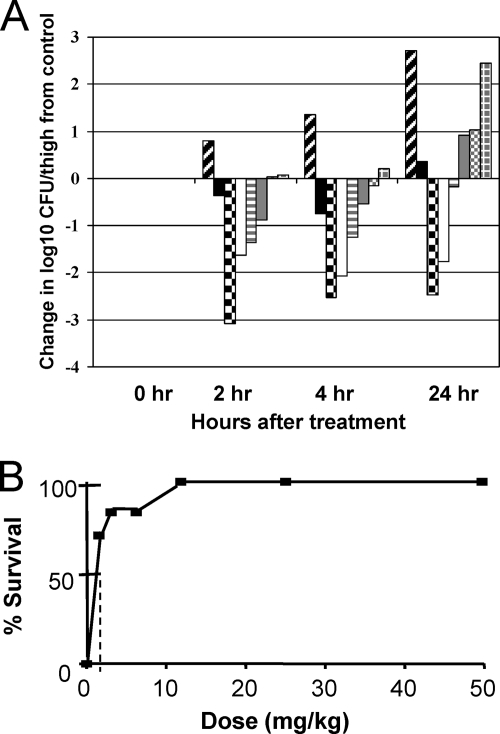
Therapeutic efficacy was demonstrated in a mouse thigh infection model using immunocompromised mice. The S. aureus ATCC 33591 (MRSA) inoculum concentration was 2.2 × 106 CFU/ml with 0.1 ml injected per thigh. The mean density of bacteria in control thighs at the time of antibiotic treatment (2 h postinfection) was 8.17 × 104 CFU/ml. The number of untreated control CFU/ml increased over 2.7 logs by 24 h postinfection (Fig. 4A). A standard vancomycin subcutaneous treatment at 10 mg/kg resulted in reductions in bacterial populations of 0.37 and 0.74 and an increase of 0.36 logs at 2, 4, and 24 h after antibiotic treatment, respectively. Subcutaneous administration of ACH-702 displayed a dose-dependent effect on the MRSA infection with a 24-h static dose of approximately 5 mg/kg (decrease of 0.18 logs). A 2.47-log reduction of the number of CFU/ml was observed at 24 h at a dose level of 20 mg/kg. The two lowest dose levels, 0.625 and 1.25 mg/kg, resulted in only minimal decreases in the number of CFU/ml compared with untreated control numbers. Efficacy was also observed with a mouse sepsis model using immunocompetent mice and S. aureus ATCC 13709, with a 50% protective dose (PD50) of approximately 0.5 mg/kg after subcutaneous administration of ACH-702, using a single dose at each concentration (Fig. 4B).
Fig. 4.
In vivo efficacy in two mouse infection models. (A) Mouse thigh infection. ACH-702 and vancomycin were dosed by subcutaneous injection at time zero, which is 2 h postinfection, with MRSA strain ATCC 33591. The numbers of CFU per thigh were calculated at the indicated time points; the graph depicts the change at the indicated time compared to the untreated control at time zero. Symbols: diagonal lines, untreated controls; black filled, vancomycin (10 mg/kg); checkered, ACH-702 (20 mg/kg); white filled, ACH-702 (10 mg/kg); horizontal lines, ACH-702 (5 mg/kg); gray filled, ACH-702 (2.5 mg/kg); gray dotted, ACH-702 (1.25 mg/kg); gray crosshatch, ACH-702 (0.625 mg/kg). (B) Mouse sepsis. ACH-702 was dosed at the indicated dose levels by subcutaneous injection 1 h after infection with S. aureus ATCC 13709 (see Materials and Methods). The y axis indicates survival after 5 days. The dotted line indicates the approximate PD50.
Pharmacokinetic parameters for ACH-702 in the plasma of treated mice were determined (Table 7). Mice were given a single subcutaneous dose at each concentration. At 10 mg/kg, the AUC from 0 to 24 h (AUC0–24) for ACH-702 was 0.94 μg · h/ml, which increased to 5.73 μg · h/ml at 40 mg/kg. The maximum concentrations of drug in serum (Cmaxs) were 0.45 and 1.41 μg/ml, respectively, at the two dosing concentrations. These values were lower than the exposures seen for ciprofloxacin at 20 mg/kg, for which the AUC0–24 and Cmax were 5.95 μg · h/ml and 3.36 μg/ml, respectively (32). This decreased exposure relative to that for ciprofloxacin was subsequently discovered to be the result of a more extensive glucuronidation of ACH-702 that reduced the levels of the parent compound (data not shown).
Table 7.
Plasma pharmacokinetic parameters after subcutaneous administration to micea
| Drug | Dose (mg/kg) | Cmax (μg/ml) | AUC0–24 (μg/ml) | t1/2 (h) | tmax (h) |
|---|---|---|---|---|---|
| ACH-702 | 10 | 0.45 | 0.94 | ND | 1.0 |
| 40 | 1.41 | 5.73 | 3.1 | 1.0 | |
| Ciprofloxacin | 20 | 3.36 | 5.95 | 1.07 | 0.5 |
| 50 | 7.42 | 14.43 | 1.17 | 0.5 |
Definitions of pharmacokinetic parameters can be found in reference 25. ND, not determined.
DISCUSSION
ACH-702, an isothiazoloquinolone (ITQ), represents an optimized analog of earlier compounds that were previously reported (9, 32, 43, 44, 45) to have reduced protein binding, enhanced inhibition of target enzymes, and improved in vitro and in vivo antibacterial activity. This compound displays potent in vitro antibacterial activity against both Gram-positive and Gram-negative bacteria (Tables 2 to 4), with particular effectiveness against resistant Gram-positive strains, including MRSA (Tables 2 and 5). The MICs against Gram-positive bacteria were generally superior to those of the comparators tested, including vancomycin, linezolid, and levofloxacin. The improved antibacterial activity of ACH-702 was particularly evident against fluoroquinolone-resistant isolates. Good activity (MICs of <1 μg/ml) was also observed against Haemophilus, Moraxella, and Neisseria strains as well as anaerobes, Legionella, and Mycoplasma clinical isolates, with the possible exception of C. difficile, for which the MIC90 was 4 μg/ml when tested against only 10 isolates. Activity against other Gram-negative bacteria was somewhat more limited but still was present for the majority of tested strains, as MIC50 values were ≤1 μg/ml (Table 3). Some improved antibacterial activity was seen against fluoroquinolone-resistant Enterobacteriaceae and A. baumannii compared with levofloxacin and ciprofloxacin. Good in vitro antibacterial activity was also demonstrated against Mycobacterium and Nocardia sp. (24, 34, 42).
A possible reason for the exceptional antibacterial activity of ACH-702 against Gram-positive bacteria is the potent dual targeting of both gyrase and topoisomerase IV enzymes, including those from quinolone-resistant strains, as demonstrated by the staphylococcal enzyme assays (Table 6). This is further supported by the observation that the effects of the quinolone-resistant phenotypes of the Gram-positive clinical isolates on the ACH-702 MIC90 values were not as significant as the major effects observed for quinolone MICs, which elevated many of them beyond their breakpoints. Also, laboratory-selected quinolone-resistant mutants were found to retain susceptibility to ACH-702 (Table 5). The effective targeting of two bacterial enzymes simultaneously may also account for the slow selection of resistant mutants and could prolong the clinical effectiveness of a drug with such a profile (37). The reduced antibacterial activity of ACH-702 against some Gram-negative strains could be the result of reduced permeability, more effective efflux of the compound, or a combination of both. This has not been further explored at present. As might be expected for a compound with potent activity on two targets, the frequencies of selection of spontaneous resistant mutants are quite low. In fact, mutants were difficult to obtain under previously reported conditions used for quinolones at 4× MIC for 48 h at 37°C (38). We did observe a 10-fold or greater increase in selection frequency if plates were allowed to incubate for an additional 24 h or longer (total of ≥72 h). However, the colonies were tiny and grew much more slowly, indicating potential fitness costs to the bacteria. How this would translate to infection conditions in the host is unclear.
Similar to quinolones, ACH-702 was found to be rapidly bactericidal against representative Gram-positive (S. aureus and E. faecalis) and Gram-negative (E. coli and P. aeruginosa) bacteria (Fig. 2), which may be a desirable property when treating a serious bacterial infection. At concentrations above 4× MIC, no further increases in the rates of killing were observed. Against the two Gram-positive strains, ACH-702 displayed greater bactericidal activity than both moxifloxacin and vancomycin, with greater killing evident at earlier times of exposure. Against the two Gram-negative strains, ACH-702 was comparable to ciprofloxacin and moxifloxacin in regard to both rate and extent of bacterial killing. Of note, a “rebound” effect was sometimes seen for the Gram-negative strains, especially with P. aeruginosa, where numbers of CFU/ml increased to nearly untreated control levels after 24 h. This rebound was observed for both ACH-702 and moxifloxacin against P. aeruginosa. The cause of this rebound effect is unknown. ACH-702 also demonstrated a postantibiotic effect (PAE) comparable to that of moxifloxacin in several S. aureus strains. This is another potential advantage during treatment of infections, as it could enhance the effective exposure of drug in a patient and contribute positively to successful clinical outcomes.
Therapeutic efficacy for ACH-702 was demonstrated in a mouse thigh infection model utilizing immunocompromised animals (Fig. 4A) and in a mouse sepsis model using immunocompetent animals (Fig. 4B). The 24-h static dose in the mouse thigh model against S. aureus was 5 mg/kg. The in vivo effectiveness of this compound is likely due to a combination of a low MIC for the infecting bacterial strains and relatively good pharmacokinetics in mice, although the plasma exposure was less than that of ciprofloxacin (Table 7). Efficacy was also observed with a mouse sepsis model with a PD50 value of 0.5 mg/kg and in a mouse lung infection model (Streptococcus pneumoniae) (M. Pucci, unpublished data). These studies were done with single subcutaneous doses, and improved results might be possible with multiple doses to compensate for the relatively short pharmacokinetic half-life in rodents. If exposure parameters track with ciprofloxacin, there is a reasonable expectation that half-lives and exposures will improve in other mammalian species, including humans. Preliminary data suggest that the ratio of the AUC to the MIC appears to be the most important pharmacokinetic/pharmacodynamic (PK/PD) driver for ACH-702, as with fluoroquinolones (W. A. Craig, unpublished data). We believe that this compound has good tissue distribution, as exposure levels determined from thigh tissue were similar to plasma levels in mice. Clinical observations of all animals treated with ACH-702 in these studies were normal at the doses tested.
ACH-702 demonstrates excellent Gram-positive antibacterial activity, including against MRSA strains, using both in vitro and in vivo assays. In addition, unlike most current antibiotics used to treat MRSA infections, it possesses Gram-negative antibacterial activity. Future efforts might generate further optimized derivatives that display improved potency against these pathogens. These data along with impressive bactericidal activity and postantibiotic effects suggested that ACH-702 might work well in animal infection models. Indeed, the compound was effective in treating animal bacterial infections, as efficacy was observed with murine sepsis, lung, and thigh infection models. However, pharmacokinetic analyses revealed that there were significantly lower exposures in mice than with ciprofloxacin. This was found to be due to rapid metabolism of the parent compound via glucuronidation, adversely affecting intravenous and especially oral administration of this compound. Other nonsystemic indications for ACH-702 are under investigation. We are currently working to identify more metabolically stable derivatives, as we believe that the overall profile of isothiazoloquinolones and the potential to treat serious antibiotic-resistant pathogens warrant further study of this class.
ACKNOWLEDGMENTS
We acknowledge Jason Wiles, Akihiro Hashimoto, Godwin Pais, Qiuping Wang, Edlaine Lucien, and Yongsheng Song for synthetic chemistry efforts and Jijun Cheng, Christy Thoma, Emily Ye, and Gohar Mushtaq for technical support. We thank Eurofins Medinet, especially Parveen Grover and Nina Brown for MIC testing of compounds and Chris Pillar and Dan Sahm for helpful discussions. We thank Ann O'Leary and colleagues at Ricerca for running mouse infection studies and William Craig for PK/PD analysis and mouse plasma samples. We also thank Donald Low, Darrin Bast, Barry Kreiswirth, and Eurofins Medinet for supplying MRSA clinical isolates.
Footnotes
Published ahead of print on 4 April 2011.
REFERENCES
- 1. Acar J. F., Goldstein F. W. 1997. Trends in bacterial resistance to fluoroquinolones. Clin. Infect. Dis. 24(Suppl. 1):S67–S73 [DOI] [PubMed] [Google Scholar]
- 2. Appelbaum P. C. 2006. The emergence of vancomycin-intermediate and vancomycin-resistant Staphylococcus aureus. Clin. Microbiol. Infect. 12(Suppl. 1):16–23 [DOI] [PubMed] [Google Scholar]
- 3. Arias C., Murray B. 2009. Antibiotic-resistant bugs in the 21st century—a clinical super-challenge. N. Engl. J. Med. 360:439–443 [DOI] [PubMed] [Google Scholar]
- 4. Bogdanovich T., et al. 2005. Antistaphylococcal activity of DX-619, a new des-F(6)-quinolone, compared to those of other agents. Antimicrob. Agents Chemother. 49:3325–3333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boucher H. W., et al. 2009. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 48:1–12 [DOI] [PubMed] [Google Scholar]
- 6. Centers for Disease Control and Prevention 1997. Update: Staphylococcus aureus with reduced susceptibility to vancomycin—United States, 1997. JAMA 278:1145–1146 [PubMed] [Google Scholar]
- 7. Centers for Disease Control and Prevention 2002. Vancomycin resistant Staphylococcus aureus—Pennsylvania, 2002. JAMA 288:2116. [PubMed] [Google Scholar]
- 8. Chambers H. F. 2005. Community-associated MRSA—resistance and virulence converge. N. Engl. J. Med. 352:2485–2487 [DOI] [PubMed] [Google Scholar]
- 9. Cheng J., et al. 2007. Dual targeting of DNA gyrase and topoisomerase IV: target interactions of heteroaryl isothiazolones in Staphylococcus aureus. Antimicrob. Agents Chemother. 51:2445–2453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chu D. T., Fernandes P. B., Claiborne A. K., Shen L., Pernet A. G. 1988. Structure-activity relationships in quinolone antibacterials: design, synthesis, and biological activities of novel isothiazoloquinolones. Drugs Exp. Clin. Res. 14:379–383 [PubMed] [Google Scholar]
- 11. Chu D. T., Lico I. M., Claiborne A. K., Plattner J. J., Pernet A. G. 1990. Structure-activity relationship of quinolone antibacterial agents: the effects of C-2 substitution. Drugs Exp. Clin. Res. 16:215–224 [PubMed] [Google Scholar]
- 12. CLSI 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, vol. 26, no. 2; approved standard, 7th ed CLSI document M7-A7. CLSI, Wayne, PA [Google Scholar]
- 13. CLSI 2006. Methods for antimicrobial susceptibility testing of anaerobic bacteria, vol. 27, no. 2; approved standard, 7th ed CLSI document M11-A7. CLSI, Wayne, PA [Google Scholar]
- 14. CLSI 2009. Performance standards for antimicrobial susceptibility testing; eighteenth informational supplement. CLSI document M100-S19. CLSI, Wayne, PA [Google Scholar]
- 15. Courvalin P. 2006. Vancomycin resistance in gram-positive cocci. Clin. Infect. Dis. 42(Suppl. 1):S25–S34 [DOI] [PubMed] [Google Scholar]
- 16. Craig W. A., Gudmundsson S. 1996. Postantibiotic effect, p. 296–329 In Lorian V. (ed.), Antibiotics in laboratory medicine. Williams and Wilkins Co., Baltimore, MD [Google Scholar]
- 17. Giske C. G., Monnet D. L., Cars O., Carmeli Y. on behalf of ReACt-Action on Antibiotic Resistance 2008. Clinical and economic impact of common multidrug-resistant gram-negative bacilli. Antimicrob. Agents Chemother. 52:813–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hiramatsu K., et al. 1997. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J. Antimicrob. Chemother. 40:135–136 [DOI] [PubMed] [Google Scholar]
- 19. Hooper D. C. 2002. Fluoroquinolone resistance among Gram-positive cocci. Lancet Infect. Dis. 2:530–538 [DOI] [PubMed] [Google Scholar]
- 20. Ince D., Zhang X., Hooper D. C. 2003. Activity of and resistance to moxifloxacin in Staphylococcus aureus. Antimicrob. Agents Chemother. 47:1410–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ince D., Zhang X., Silver L. C., Hooper D. C. 2003. Topoisomerase targeting with and resistance to gemifloxacin in Staphylococcus aureus. Antimicrob. Agents Chemother. 47:274–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jones M. E., Blosser-Middleton R. S., Thornsberry C., Karlowsky J. A., Sahm D. F. 2003. The activity of levofloxacin and other antimicrobials against clinical isolates of Streptococcus pneumoniae collected worldwide during 1999–2002. Diagn. Microbiol. Infect. Dis. 47:579–586 [DOI] [PubMed] [Google Scholar]
- 23. Low D. E., et al. 2002. Activity of BMS-284756, a novel des-fluoro(6) quinolone, against Staphylococcus aureus, including contributions of mutations to quinolone resistance. Antimicrob. Agents Chemother. 46:1119–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Molina-Torres C. A., Ocampo-Candiani J., Rendón A., Pucci M. J., Vera-Cabrera L. 2010. In vitro activity of a new isothiazoloquinolone, ACH-702, against Mycobacterium tuberculosis and other mycobacteria. Antimicrob. Agents Chemother. 54:2188–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mouton J. W., Dudley M. N., Cars O., Derendorf H., Drusano G. L. 2005. Standardization of pharmacokinetic/pharmacodynamic (PK/PD) terminology for anti-infective drugs: an update. J. Antimicrob. Chemother. 55:601–607 [DOI] [PubMed] [Google Scholar]
- 26. Murakami K., Tomasz A. 1989. Involvement of multiple genetic determinants in high-level methicillin resistance in Staphylococcus aureus. J. Bacteriol. 171:874–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mychajlonka M., McDowell T. D., Shockman G. D. 1980. Inhibition of peptidoglycan, ribonucleic acid, and protein synthesis in tolerant strains of Streptococcus mutans. Antimicrob. Agents Chemother. 17:572–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pankuch G. A., Appelbaum P. C. 2005. Postantibiotic effect of DX-619 against 16 gram-positive organisms. Antimicrob. Agents Chemother. 49:3963–3965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Paterson D. L., Bonomo R. A. 2005. Extended-spectrum beta-lactamases: a clinical update. Clin. Microbiol. Rev. 18:657–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Périchon B., Courvalin P. 2009. VanA-type vancomycin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 53:4580–4587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Poole K. 2004. Resistance to beta-lactam antibiotics. Cell. Mol. Life Sci. 61:2200–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pucci M. J., et al. 2007. In vitro and in vivo antibacterial activities of heteroaryl isothiazolones against resistant gram-positive pathogens. Antimicrob. Agents Chemother. 51:1259–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pucci M. J., et al. Abstr. 48th Intersci. Conf. Antimicrob. Agents Chemother., abstr. F1-2021.2008. [Google Scholar]
- 34. Pucci M. J., Ackerman M., Thanassi J. A., Shoen C. M., Cynamon M. H. 2010. In vitro antituberculosis activities of ACH-702, a novel isothiazoloquinolone, against quinolone-susceptible and quinolone-resistant isolates. Antimicrob. Agents Chemother. 54:3478–3480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ratnaraja N. V., Hawkey P. M. 2008. Current challenges in treating MRSA: what are the options? Expert Rev. Anti Infect. Ther. 6:601–618 [DOI] [PubMed] [Google Scholar]
- 36. Reinert R. R. 2009. The antimicrobial resistance profile of Streptococcus pneumoniae. Clin. Microbiol. Infect. 15(Suppl.3):7–11 [DOI] [PubMed] [Google Scholar]
- 37. Silver L. L. 2007. Multi-targeting by monotherapeutic antibacterials. Nat. Rev. Drug Discov. 6:41–55 [DOI] [PubMed] [Google Scholar]
- 38. Strahilevitz J., Truong-Bolduc Q. C., Hooper D. C. 2005. DX-619, a novel des-fluoro(6) quinolone manifesting low frequency of selection of resistant Staphylococcus aureus mutants: quinolone resistance beyond modification of type II topoisomerases. Antimicrob. Agents Chemother. 49:5051–5057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Strahilevitz J., Onodera Y., Hooper D. C. 2006. An improved expression plasmid for affinity purification of Staphylococcus aureus gyrase A subunit. Protein Expr. Purif. 47:10–15 [DOI] [PubMed] [Google Scholar]
- 40. Terleckyj B., Willett N. P., Shockman G. D. 1975. Growth of several strains of oral streptococci in a chemically defined medium. Infect. Immun. 11:649–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thanassi J. A., et al. Abstr. 48th Intersci. Conf. Antimicrob. Agents Chemother., abstr. F1-2022.2008. [Google Scholar]
- 42. Vera-Cabrera L., et al. 2010. In vitro activity of ACH-702, a new isothiazoloquinolone, against Nocardia brasiliensis compared with econazole and the carbapenems imipenem and meropenem alone or in combination with clavulanic acid. Antimicrob. Agents Chemother. 54:2191–2193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang Q., et al. 2007. Isothiazoloquinolones with enhanced antistaphylococcal activities against multidrug-resistant strains: effects of structural modifications at the 6-, 7-, and 8-positions. J. Med. Chem. 50:199–210 [DOI] [PubMed] [Google Scholar]
- 44. Wiles J. A., et al. 2006. Isothiazoloquinolones containing functionalized aromatic hydrocarbons at the 7-position: synthesis and in vitro activity of a series of potent antibacterial agents with diminished cytotoxicity in human cells. Bioorg. Med. Chem. Lett. 16:1272–1276 [DOI] [PubMed] [Google Scholar]
- 45. Wiles J. A., et al. 2006. Biological evaluation of isothiazoloquinolones containing aromatic heterocycles at the 7-position: in vitro activity of a series of potent antibacterial agents that are effective against methicillin-resistant Staphylococcus aureus. Bioorg. Med. Chem. Lett. 16:1277–1281 [DOI] [PubMed] [Google Scholar]
- 46. Yoshida H., Bogaki M., Nakamura M., Nakamura S. 1990. Quinolone resistance-determining region in the DNA gyrA gene of Escherichia coli. Antimicrob. Agents Chemother. 34:1271–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zahar J. R., et al. 2009. Addressing the challenge of extended-spectrum beta-lactamases. Curr. Opin. Investig. Drugs 10:172–180 [PubMed] [Google Scholar]