Abstract
Antibiotic resistance and tolerance are increasing threats to global health as antibiotic-resistant bacteria can cause severe morbidity and mortality and can increase treatment cost 10-fold. Although several genes contributing to antibiotic tolerance among pneumococci have been identified, we report here that ClpL, a major heat shock protein, could modulate cell wall biosynthetic enzymes and lead to decreased penicillin susceptibility. On capsular type 1, 2, and 19 genetic backgrounds, mutants lacking ClpL were more susceptible to penicillin and had thinner cell walls than the parental strains, whereas a ClpL-overexpressing strain showed a higher resistance to penicillin and a thicker cell wall. Although exposure of Streptococcus pneumoniae D39 to penicillin inhibited expression of the major cell wall synthesis gene pbp2x, heat shock induced a ClpL-dependent increase in the mRNA levels and protein synthesized by pbp2x. Inducible ClpL expression correlated with PBP2x expression and penicillin susceptibility. Fractionation and electron micrograph data revealed that ClpL induced by heat shock is localized at the cell wall, and the ΔclpL showed significantly reduced net translocation of PBP2x into the cell wall. Moreover, coimmunoprecipitation with either ClpL or PBP2x antibody followed by reprobing with ClpL or PBP2x antibody showed an interaction between ClpL and PBP2x after heat stress. This interaction was confirmed by His tag pulldown assay with either ClpLHis6 or PBP2xHis6. Thus, ClpL stabilized pbp2x expression, interacted with PBP2x, and facilitated translocation of PBP2x, a key protein of cell wall synthesis process, contributing to the decrease of antibiotic susceptibility in S. pneumoniae.
INTRODUCTION
Streptococcus pneumoniae is the major cause of community-associated pneumonia, otitis media, septicemia, and meningitis (36). Pneumonia caused by S. pneumoniae infections has one of the highest morbidity and mortality rates (≥2 million deaths every year) (53) and is the sixth-leading cause of death in the United States (5). β-Lactam antibiotics are the most common treatment for pneumococcal pneumonia (48), and resistance to β-lactams is associated with changes in the penicillin binding proteins (PBPs) (17), which catalyze the final transpeptidase reaction in cell wall peptidoglycan synthesis (48). Since antibiotic resistance increases morbidity, mortality, length of hospitalization, and medical costs (12), the elucidation of factors modulating antibiotic resistance and tolerance is clinically important.
Several stresses, including antibiotics, DNA damage, oxidative stress, and high osmolarity, induce heat shock proteins (HSPs) in Escherichia coli and Bacillus subtilis (43). However, these stresses failed to induce major HSPs such as GroEL and DnaK in pneumococci (11). HSPs in E. coli decline rapidly to steady-state levels after the stimulus is removed (43), whereas pneumococcal HSPs are transiently induced by heat shock but can remain present at 1 h after return to normal temperature (27). Therefore, pneumococcal HSPs may have different fates upon exposure to stresses than those of E. coli.
In the Gram-negative organism E. coli, GroEL is essential for the expression of the cell wall synthesis enzyme dihydropicolinate synthase (DapA), which is the first enzyme in the synthesis pathway of the cell wall precursor diaminopimelic acid (DAP) (40). Similarly, the Coxiella burnetii DnaK homologue is localized to the cell wall and the cytoplasm (35). Consistent with this, in Gram-positive bacteria, DnaK is associated with the cell wall in Lactobacillus salivarius (24), Lactobacillus plantarum (23), and Listeria monocytogenes (55). In Staphylococcus aureus, DnaK is a member of the cell wall stress stimulon and induced by inhibition of peptidoglycan biosynthesis (47). The dnaK mutant of S. aureus is highly susceptible to cell wall-active antibiotic stress conditions (56). Moreover, both GroEL and DnaK are found on the bacterial surface in Mycobacterium tuberculosis (21) and L. plantarum (23). Thus, GroEL or DnaK facilitate protein refolding on the cell wall or cell surface. Moreover, treatment of S. aureus infections with cefoxitin, an expanded-spectrum cephalosporin, induces DnaK, GroES, and Clp ATPase subunit (ClpB and ClpL) levels (25). It has been reported that the bacterial stress response may affect antibiotic resistance (31). Therefore, HSPs could be involved in cell wall synthesis. However, no bacterial stress proteins in S. pneumoniae involved in antibiotic resistance have been identified.
ClpL is a member of the HSP100/Clp (caseinolytic protease) chaperone family found mainly in Gram-positive bacteria (http://www.ncbi.nlm.nih.gov/sites/entrez). It has been found that ATP-dependent proteolytic activity is increased in extracts from E. coli cells overexpressing the clpL gene (22). It is also involved in virulence modulation in S. pneumoniae (31, 59). HSPs interact transiently with hydrophobic residues, refold unfolded polypeptides, and translocate them across the membrane (14). HSPs induced by antibiotic stress might facilitate translocation of proteins into the membrane/cell wall, where they could affect cell wall synthesis and antibiotic resistance. In this study, we examined this hypothesis and obtained direct evidence that higher ClpL levels correlated with decreased penicillin susceptibility and increased pbp2x mRNA and cell wall thickness. Also, heat shock-induced ClpL interacted with PBP2x and colocalized with PBP2x at the cell wall.
(A part of this study's data was presented at the 7th International Symposium on Tonsils and Mucosal Barriers of the Upper Airways, Asahikawa, Japan, 2010.)
MATERIALS AND METHODS
Bacterial strains, culture, and transformation.
The bacterial strains and plasmids used in this study are shown in Table 1. Encapsulated S. pneumoniae D39 (serotype 2) and its isogenic ΔclpL strain (HYK304), which was created with the ermB cassette inserted in the opposite orientation relative to that of the gene to exclude polar effects (27), were used. The D39 ClpL-overexpressing strain (RT172), which has a recombinant pMV158 plasmid with the functional clpL gene in the D39 ΔclpL background, was described previously (59). The clpL-overexpressing strain produces ClpL from a plasmid that is not regulated by the normal bacterial regulators. This strain constitutively overexpressed ClpL regardless of heat shock. All pneumococci were cultured in Todd-Hewitt broth containing yeast extract (THY broth), as described previously (59). Nonencapsulated S. pneumoniae CP1200, a derivative of Rx1, and its isogenic ΔclpL strain (HYK1) were described previously (27) and grown in Casitone-tryptone-based medium (11). Competence was controlled by addition of competence-stimulating peptide and quantitated as antibiotic-resistant transformants obtained after exposure of cells to DNA in culture medium, as described previously (6). Erythromycin (2.5 μg/ml) or tetracycline (1 μg/ml) was used to screen positive transformants. Escherichia coli culture and transformation were as described previously (28).
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Antibiotic resistancea | Reference or source |
|---|---|---|---|
| S. pneumoniae strains | |||
| CP1200 | Nonencapsulated derivative of Rx1 malM511 str-1 | 11 | |
| HYK1 | CP1200 ΔclpL::ermB | ERY | 27 |
| D39 | Encapsulated, type 2 | 2 | |
| HYK304 | D39 ΔclpL::ermB | ERY | 27 |
| RT172 | D39 ΔclpL::ermB containing pMV158(clpL) | ERY, TET | 59 |
| TDHT01 | D39 containing pMV158 | TET | This study |
| TDHT02 | D39 ΔclpL::ermB containing pMV158 | ERY, TET | This study |
| ATCC 6301 | Type 1 | ATCC | |
| KEH 6301 | ATCC 6301 ΔclpL::ermB | ERY | This study |
| TDHT 6301 | ATCC 6301 ΔclpL::ermB containing pMV158(clpL) | ERY, TET | This study |
| ATCC 6319 | Type 19 | ATCC | |
| KEH 6319 | ATCC 6319 ΔclpL::ermB | ERY | This study |
| TDHT 6319 | ATCC 6319 ΔclpL::ermB containing pMV158(clpL) | ERY, TET | This study |
| D39 F-clpL | D39 PfcsK::clpL | ERY | This study |
| E. coli strains | |||
| BL21(DE3) | gal (λ cI ts857 ind1 Sam7 nin5 lacUV5-T7 gene 1) | Novagen | |
| DH5α | supE44 ΔlacU169 (φ80 lacZΔM15) | Gibco BRL | |
| Plasmids | |||
| pET32b(+) | 5,899 bp | AMP | Novagen |
| pKHY004 | 7,500 bp, His-tagged clpL in pET30(a) | KAN | 27 |
| pTDHT001 | 8,178 bp, His-tagged pbp2x in pET32b(+) | AMP | This study |
| pTDHT002 | 6,556 bp, His-tagged vncR in pET32b(+) | AMP | This study |
| pTDHT003 | 7,228 bp, His-tagged vncS in pET32b(+) | AMP | This study |
| pMV158 | 5,300 bp, streptococcal plasmid | TET | 7 |
| pLNT001 | 7,400 bp, clpL in pMV158 | TET | 59 |
ERY, erythromycin; TET, tetracycline; AMP, ampicillin; KAN, kanamycin.
Construction of clpL deletion mutants.
ΔclpL of serotypes 1 (ATCC 6301) and 19 (ATCC 6319) were constructed using the same primers described previously (27). To create an insertion-deletion mutation of clpL (ΔclpL::ermB) in S. pneumoniae, an 860-bp ermB cassette was amplified with primers prs3 and prs4 (Table 2) from erythromycin-resistant E. coli chromosomal DNA and used to disrupt clpL. A 410-bp fragment (clpLup) containing part of both clpL and the 5′ end of ermB was amplified with primers hlp3 and hlp1 (Table 2) from CP1200 DNA. A 300-bp fragment (clpLdown) containing part of both the downstream clpL sequence and the 3′ terminus of ermB was amplified with primers hlp2 and hlp4 (Table 2) from CP1200 DNA.
Table 2.
Primers used in this study
| Name | Primer sequences | Reference or source |
|---|---|---|
| pbp2x | Forward: TGC AGA TGC CAC GAT TCG | This study |
| Reverse: CAT CAT TCT GCC ACC AGT CAA | ||
| murM | Forward: TGG TAT GAA ACG GCT CGC TAT | This study |
| Reverse: TTT TCA ACA CCA CCT AAA TTT TGC | ||
| 16S RNA | Forward: CCC CTT ATG ACC TGG GCT ACA | This study |
| Reverse: CGG CTT GCG ACT CGT TGT | ||
| clpL | Forward: CGC CCA ATC GGC AAC TT | This study |
| Reverse: GCT TAG CAA GCT CCG TCT TAC C | ||
| PBP2x | Forward: GGC CGC CCA TGG CGA TGA AGT GGA CAA AAA GAG TAA TC | This study |
| Reverse: CCT CCC CTC GAG TTA GTC TCC TAA AGT TAA TG | ||
| VncR | Forward: GGC CGC CCA TGG GCA TGA AAA TTT TAA TTG TAG | This study |
| Reverse: CCT CCC CTC GAG TCA TTT TCG CTC CAA TTT ATA ACC | ||
| VncS | Forward: GGG CCC GGA TCC GAT GAA ACG AAC AGG TTT ATT TGC | This study |
| Reverse: GGC CCG CTC GAG CTA GTC TTG GAC GAC TTT TGG | ||
| prs3 | CCG GGC CCA AAA TTT GTT TGA T | 27 |
| prs4 | AGT CGG CAG CGA CTC ATA GAA T | 27 |
| hlp3 | CGG TAC CAT GAA CAA TAA TTT TAA C | 27 |
| hlp1 | ATC AAA CAA ATT TTG GGC CCG GTC AGA TGT TTC TTG AAT TTC C | 27 |
| hlp2 | ATT CTA TGA GTC GCT GCC GAC TGT TCT AGA TGA TGG TCG TTT G | 27 |
| hlp4 | GGC CGA GCT CTT AGA CTT TCT CAC GAA TAA C | 27 |
| clpL1 | TTT TTC GGT AGG CAG TCC TAC CGT GGC TTA CCG TTC GTA TAG | This study |
| clpL2 | TTT TCT TCT CTC TTC GTC CTT GA | This study |
| clpL3 | CCA TCA GTG CTG GAA TTG TG | This study |
| clpL4 | TAG GAC TGC CTA CCG AAA AAT TAC ATC AAA TAC AAA ATT GC | This study |
| clpL5 | GGA CGA AGA GAG AAG AAA AAT GAA CAA CAA TTT TAA TAA | This study |
| clpL6 | CTG GAT GTT GAG CCG CCA AG | This study |
The three PCR products were used as a mixed template for PCR with primers hlp3 and hlp4 (Table 2) to produce a 1.6-kb fragment with a 1,300-bp deletion of clpL that was replaced by the ermB gene. The tripartite 1.6-kb fragment was subsequently introduced into either S. pneumoniae type 1 or type 19 strains by transformation, and recipient bacteria that had integrated the recombinant fragment into the chromosome by homologous recombination were selected by resistance to erythromycin. Transformants were screened for the correct deletion by PCR and Western blot analysis. Type 1 (KEH 6301) and 19 (KEH 6319) ΔclpL strains containing the correct deletion within clpL were used for further studies.
Construction of ClpL-overexpressing strain.
The recombinant pMV158 plasmid with the clpL gene (pLNT001) was purified from the D39 ClpL-overexpressing strain (RT172) as described previously (59) and transformed into the type 1 (KEH 6301) or type 19 (KEH 6319) ΔclpL strain. Subsequently, the tetracycline-resistant transformants (TDHT 6301 and TDHT 6319, respectively) were selected and confirmed by colony PCR and Western blotting.
Construction of clpL-regulatable strain.
An S. pneumoniae D39 clpL-regulatable strain (D39 F-clpL) was constructed by placing clpL under the control of the PfcsK inducible promoter in the chromosome of S. pneumoniae by using a three-piece PCR amplification with overlapping primers (Table 2) (8, 63; this study). First, a promoter replacement cassette containing the PfcsK promoter, transcriptional terminators (t1, t2) located at the 5′ end of the PfcsK promoter, and an independent erythromycin resistance marker (ermAM) was amplified from the Cheshire cassette, which was provided by D. A. Morrison (63), using primers clpL1 and clpL2 (Table 2). This piece was flanked by DNA sequences of the gene immediately upstream of clpL (mraY) and the start of the clpL open reading frame, which were amplified from D39 genomic DNA by primers clp3 and clpL4 and primers clpL5 and clpL6, respectively (Table 2). The triple-joining construct, amplified by primers clpL1 and clpL6, was integrated into S. pneumoniae D39 at the clpL locus by transformation. The transformant was selected by 0.1 μg/ml erythromycin, and transformation was confirmed by sequencing.
Cloning and purification of recombinant ClpLHis6, PBP2xHis6, VncRHis6, and VncSHis6 in E. coli.
The pbp2X, vncR, and vncS genes were amplified by PCR using D39 genomic DNA as a template with primers PBP2x, VncR, and VncS (Table 2) that incorporated NcoI and XhoI (PBP2x and VncR) and BamHI and XhoI (VncS) restriction enzyme sites. The PCR products were digested with the NcoI and XhoI or the BamHI and XhoI enzymes (New England BioLabs) and cloned into the corresponding restriction sites in pET32b(+) (Novagen) to generate plasmids pTDHT001, pTDHT002, and pTDHT003 (Table 1). After transformation into E. coli DH5α, transformants were selected by resistance to 100 μg/ml of ampicillin. Recombinant plasmids were confirmed by NcoI-XhoI or BamHI-XhoI double digestion and sequencing.
In order to purify the ClpLHis6, PBP2xHis6, VncRHis6, and VncSHis6 proteins, recombinant plasmids pKHY004, pTDHT001, pTDHT002, and pTDHT003, respectively (Table 1), were transformed into E. coli BL21(DE3) (Table 1). Proteins were then induced by 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and purified using a nickel-nitrilotriacetic acid column (Ni-NTA; Probond; Invitrogen), according to the manufacturer's suggestion, and dialyzed against 50 mM Tris buffer (pH 7.5).
Antisera, gel electrophoresis, and Western blotting.
Antisera against ClpL, PBP2x, VncR, and VncS were prepared as described previously (28). Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) (10% polyacrylamide gel) was performed as described previously (29) using 10 μg of proteins. A bacterial pellet was collected by centrifugation at 6,000 rpm for 10 min and washed once with phosphate-buffered saline (PBS; pH 7.4), if required. The pellet was then resuspended in lysis buffer (50 mM Tris-HCl, pH 8.0, 1 mM dithiothreitol, 0.1% Triton X-100), followed by 15 min freezing at −80°C and 10 min thawing at 37°C (repeated 3 times). Then, the lysate was sonicated for 10 s and cell debris was removed by centrifugation at 13,000 rpm for 5 min. The supernatant was collected and used for further studies. The proteins (10 μg) were electroblotted onto polyvinylidene difluoride membranes, blocked with 3% skim milk (Difco), and then probed with a 1:500 dilution of polyclonal mouse sera raised against ClpL, PBP2x, VncR, and VncS. The secondary antibody was a 1:5,000 dilution of goat anti-mouse immunoglobulin G conjugated to horseradish peroxidase (HRP; Promega). Both chemiluminescence and colorimetry were used to detect HRP-conjugated secondary antibody used in Western blots. The relative amount of protein was determined by measuring histogram using the Photoshop program.
Determination of MIC values.
MICs were determined as described previously (1), with some modifications. Bacteria were cultured in THY broth until the optical density at 550 nm (OD550) was 0.3 and were serially diluted to 10−3, 10−4, and 10−5. One microliter of each diluted culture (3 drops/plate) was dropped onto a plate containing 0.01, 0.02, 0.04, 0.08, 0.16, 0.32, 0.64, 1.28, or 2.56 μg/ml penicillin and incubated overnight at 37°C. A series of experiments with doubling increments of the penicillin concentration were repeatedly performed to define a MIC value close to the real resistance level. The MIC value was defined as the lowest concentration of penicillin that killed all bacteria at all dilution levels after overnight incubation.
Microscopy of morphology.
Pneumococcus grown in THY broth until mid-exponential phase (OD550 = 0.3) was fixed in 5% Formalin (Sigma) for 15 min, centrifuged, and washed 3 times in distilled water. Bacteria were resuspended in 100 μl of distilled water. Twenty microliters of that was dropped onto a slide, air dried at room temperature, and covered with 20 μl of 50% glycerol. A coverslip was applied and sealed with nail polish. The morphology and chain length were observed by a microscope (Olympus) at ×100 magnification.
TEM and ClpL localization by TEM.
Pneumococci cultured on THY blood agar overnight were inoculated into THY broth and cultured until early exponential phase (OD 550 nm = 0.1). Transmission electron microscopy (TEM) was performed as described previously (30). The cell pellets were fixed with modified Karnovsky's fixative consisting of 2% glutaraldehyde and 2% paraformaldehyde in 0.05 M sodium cacodylate buffer (pH 7.2) at 4°C overnight. They were then washed 3 times with the same buffer for 10 min each time. The specimens were postfixed with 1% (wt/vol) osmium tetroxide in the same buffer at 4°C for 2 h and washed briefly with distilled water twice. The postfixed specimens were stained en bloc with 0.5% uranyl acetate at 4°C overnight, dehydrated in a graded ethanol series (30, 50, 70, and 80%, each for 10 min), and embedded in LR white resin. Ultrathin sections (ca. 60 nm thick) were cut with a diamond knife using an ultramicrotome (MT-X; RMC, Tucson, AZ) and then mounted on bare copper grids. They were stained with 2% uranyl acetate and Reynolds' lead citrate each for 7 min and examined with an energy-filtering transmission electron microscope (LIBRA 120; Carl Zeiss, Oberkochen, Germany) operated at an accelerating voltage of 120 kV.
For localization of ClpL, pneumococci were heat shocked at 42°C for 30 min and collected by centrifugation, followed by immediate fixation for electron microscopy. Thin sections were treated with normal serum or anti-ClpL antibodies as primary antibody, followed by anti-rabit IgG conjugated to colloidal gold.
qRT-PCR.
Bacterial RNA was isolated using the hot phenol method described previously (27). One microgram of bacterial RNA was reverse transcribed into cDNA using a random primer (Takara). Quantitative reverse transcriptional PCR (qRT-PCR) was performed according to the manufacturer's instructions (Applied Biosystems). The reaction mixture contained 1.8 μl of 10 pmol each appropriate primer, 10 μl of 2× real-time PCR mixtures (Applied Biosystems), and 5 μl of 10- to 50-fold-diluted cDNA in a 0.2-μl fast reaction tube (Applied Biosystems). Nuclease-free water (Ambion) was added to 20 μl. A standard run of the real-time PCR program was as follows: hold (95°C for 10 min), cycling (95°C for 15 s, 55°C for 30 s, 72°C for 30 s) for 40 cycles, and melt curve (95°C for 15 s, 60°C for 1 min, 95°C for 15 s). The collection data point was set at the last step of the melt curve. Each condition was analyzed in quadruplicate. The expression data were analyzed by the Sigma plot program.
Localization of ClpL and PBP2x by subcellular fractionation.
Exponentially growing nonencapsulated pneumococci were harvested by centrifugation, and sucrose-induced protoplast formation was performed as described previously (62). Briefly, cells were converted to protoplasts by incubation at 30°C for 1 h with 1 M sucrose buffer (1 M sucrose, 100 mM Tris-HCl, pH 7.6, 2 mM MgCl2, and 1 mM phenylmethylsulfonyl fluoride [PMSF]). Centrifugation at 13,000 × g for 20 min separated the cell wall fraction (supernatant) from the protoplasts (pellet). The protoplasts were subjected to osmotic lysis by dilution of 19 volumes of hypotonic buffer (100 mM Tris-HCl, pH 7.6, 1 mM PMSF, 1 mM EDTA). Lysates were centrifuged initially at 5,000 × g for 5 min to remove unlysed cells and then at 70,000 rpm for 30 min to obtain the cytoplasmic fraction (supernatant) and the membrane fraction (pellet). The localization of ClpL and PBP2x was detected by Western blotting. VncR was used as a cytosolic marker.
Coimmunoprecipitation using anti-ClpL or anti-PBP2x antibody.
Pneumococcal lysates were precleared with 50 μl of mouse normal serum for 1 h on ice, and the mixture was gently agitated in the presence of 100 μl of protein A-anchored agarose bead slurry (Sigma) for 30 min at 4°C. Subsequently, 200 μg of precleared protein lysates was incubated with either anti-ClpL or anti-PBP2x antibody at 4°C overnight with gentle agitation. Preimmune serum (normal serum) was used as a negative control. The mixture was mixed with 100 μl of protein A-anchored agarose beads at 4°C for 4 h with gentle agitation. The beads were collected and washed 3 times with lysis buffer. Immunoprecipitated proteins were detached from the beads by adding 50 μl of 2× SDS-PAGE sample buffer and boiling at 100°C for 5 min. The supernatant was analyzed by Western blotting with both anti-ClpL and anti-PBP2x antibodies.
His tag pulldown assay.
His tag pulldown assay was carried out as described previously (18), with some modifications. Briefly, wild-type (WT) strain D39 was cultured in THY broth at 30°C until mid-log phase (OD550 = 0.3) and then heat shocked at 42°C for 30 min. One hundred micrograms of each cell extract was incubated with 10 μg of either ClpLHis6 or PBP2xHis6 for 4 h at 4°C with gentle shaking. Ni-NTA agarose beads (0.1 ml; Qiagen), prewashed three times in lysis buffer, were added into the mixture, and the mixture was further incubated overnight at 4°C with gentle shaking. The beads were transferred to a new 1.5-ml tube and washed three times with 1 ml lysis buffer (0.5 M NaCl, 50 mM NaH2PO4, 20 mM imidazole, 1% Triton X-100, and 1 mM PMSF) and three times with 1 ml of the same buffer but with 20 mM imidazole. Bound proteins were eluted by boiling the beads with 2× SDS-PAGE sample buffer in lysis buffer containing 1 M imidazole for 20 min at 100°C. Samples were analyzed by Western blotting using anti-ClpL and anti-PBP2x antibodies.
Statistical analysis.
Statistical differences between groups were analyzed by either the Student t test, one-way analysis of variance (ANOVA; Tukey method), or nonparametric test (Kruskal-Wallis method), depending on the data. A probability level of 0.05 was considered significant.
RESULTS
ClpL protein increases resistance to DOC-, Triton X-100-, and penicillin-triggered lysis.
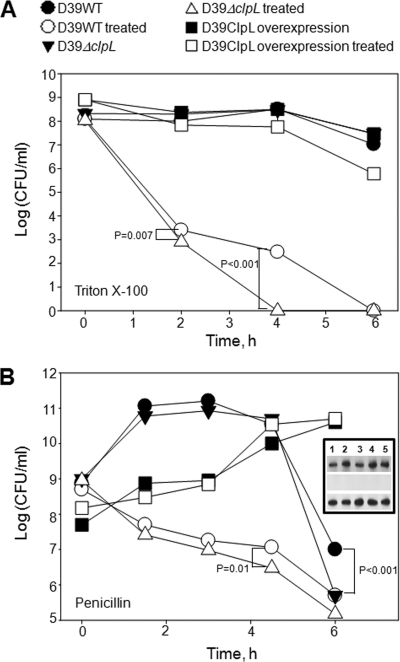
We previously demonstrated that ClpL has chaperone activity (27) and that the ClpL chaperone represses adherence of S. pneumoniae to host cells (59). Although the ClpL-overexpressing strain (RT172) showed a similar adherence phenotype as the wild type (59), it was more resistant to lysis by Triton X-100, penicillin, or deoxycholate (DOC) (see Fig. S1A in the supplemental material), an autolysis inducer in pneumococci. Triton X-100 killed all D39 WT and ΔclpL (HYK304) bacteria within 4 h and 6 h, respectively (P < 0.001), and the D39 WT was more resistant to Triton X-100 than the ΔclpL strain (HYK304) after 4 h treatment (P < 0.001). In contrast, the D39 ClpL-overexpressing strain (RT172) was only marginally affected (Fig. 1A; see Fig. S1B in the supplemental material).
Fig. 1.
Increased resistance of pneumococcus to Triton X-100 and penicillin-triggered lysis by ClpL overexpression. Pneumococci were cultured in THY broth until the OD550 was 0.2. Then, Triton X-100 (A) or penicillin (B) was added to the culture at final concentrations of 0.05% and 0.1 μg/ml, respectively, followed by counting of viable cells. Each data point is the mean value from three independent experiments. (Inset) Expression level of ClpL in D39 WT (row 1), D39 ΔclpL (row 2), and the D39 ClpL-overexpressing strain (row 3) after exposure to 0.1 μg/ml penicillin for 0, 1.5, 3, 4.5, and 6 h (lanes 1, 2, 3, 4, and 5, respectively). Statistical differences were analyzed by Student's t test. Standard deviations are smaller than the symbols. The D39 ClpL-overexpressing strain was only marginally affected by Triton X-100 (A) and was not affected by penicillin (B).
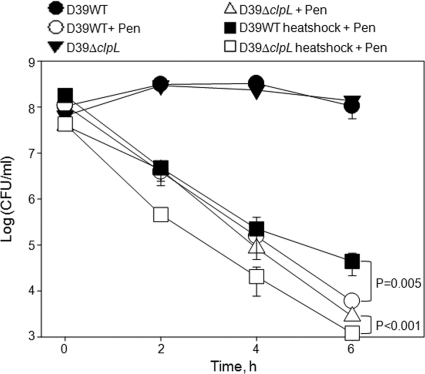
Penicillin binding to PBPs inhibits peptidoglycan synthesis and results in bacterial lysis (16). Exposure of the D39 WT and ΔclpL strain (HYK304) to penicillin significantly decreased viability but did not affect the D39 ClpL-overexpressing strain (RT172) (Fig. 1B; see Fig. S1C in the supplemental material). Moreover, the number of viable D39 WT bacteria was higher than that of the ΔclpL strain (HYK304) during the penicillin incubation period and was significantly higher at the end of stationary phase, which was at 4.5 h of incubation (P = 0.01) (Fig. 1B). The ΔclpL strain (HYK304) containing the pMV158 vector only did not show increased penicillin resistance (see Fig. S1D in the supplemental material). However, heat shock pretreatment to induce ClpL increased resistance of the D39 WT 9-fold against a 6-h penicillin treatment (P = 0.005), whereas heat shock made the ΔclpL strain (HYK304) 5-fold more susceptible to penicillin treatment (P < 0.001) at all time points (Fig. 2; see Fig. S1E in the supplemental material) and heat shock itself did not affect the growth of either D39 WT or the D39 ΔclpL strain (HYK304) (see Fig. S2 in the supplemental material). Although only 20% of D39 ClpL-overexpressing bacteria were lysed, ClpL protein in the D39 ClpL-overexpressing strain (RT172) was constantly overexpressed and its level was significantly higher than that in the D39 WT even in the presence or absence of penicillin in a time-dependent manner. However, the amount of ClpL protein in D39 WT was induced 3-fold after exposure to penicillin for 1.5 h. In contrast, no ClpL was detected in D39 ΔclpL (Fig. 1B, inset). Thus, ClpL plays a role in pneumococci resistance to DOC and Triton X-100 and penicillin-triggered lysis.
Fig. 2.
Heat shock-induced ClpL increases penicillin resistance. Pneumococci were cultured in THY broth at 30°C until the OD550 was 0.2 and then heat shocked at 42°C for 30 min. After heat shock, penicillin (Pen) was added to the culture at a final concentration of 0.2 μg/ml, followed by viable cell counting. The figure shows the standard deviation from five independent experiments. Statistical differences were analyzed by Student's t test. In some cases, standard deviations are smaller than the symbol.
Overexpression of ClpL protein increases resistance to penicillin.
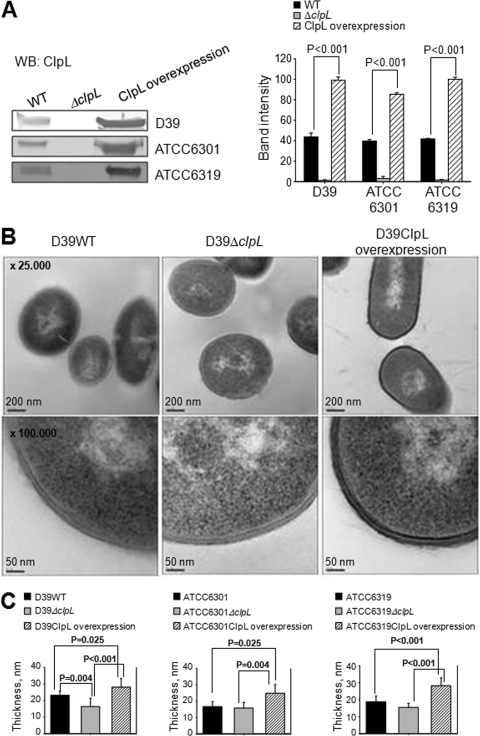
We next compared the MIC of penicillin of all three ClpL strains. The MIC for the D39 ClpL-overexpressing strain (RT172) was 0.16 μg/ml, but the MICs were only 0.04 and 0.01 μg/ml for the D39 WT and the ΔclpL strain (HYK304), respectively (see Table S1 in the supplemental material). Consistent with penicillin resistance level, the ClpL level of the ClpL-overexpressing strain of all three tested strains (RT172, TDHT 6301, and TDHT 6319) was 2.5-fold (P < 0.001) higher than that of the respective WT (Fig. 3A).
Fig. 3.
Thicker cell wall by ClpL overexpression. (A) Amount of ClpL in WT, ΔclpL, and ClpL-overexpressing type 2, 1, and 19 strains. WT, ΔclpL, and ClpL-overexpressing strains were grown in THY broth at 37°C until the OD550 was 0.3. Ten micrograms of the cell lysates was applied for Western blot (WB) analysis to determine ClpL level. Colorimetry was used to detect HRP-conjugated secondary antibody used in Western blots. Band density was analyzed by Photoshop. The figure shows representative results of three independent experiments. (B and C) ClpL overexpression increased cell wall thickness. Pneumococci were grown in THY broth until mid-exponential phase (OD550 = 0.3). The bacterial pellet was fixed and used to analyze cell wall thickness by TEM at ×25,000 magnification (upper panels) or ×100,000 magnification (lower panels). TEM images of serotypes 1 and 19 are not shown (B). The figure shows representative results of three independent experiments. Cell wall thickness was determined from 10 random bacterial cells for each strain (C). Significant differences were analyzed by one-way ANOVA.
ClpL-overexpressing strains have a thicker cell wall, mucoid colony morphology, longer chain length, and a longer generation time.
We next analyzed cell wall thickness with TEM. The D39 ClpL-overexpressing strain (RT172) was more electron dense and the cell wall was 126% of the thickness of the D39 WT cell wall (P = 0.025), whereas the ΔclpL strain (HYK304) was less electron dense and its cell wall was only 73% of the thickness of the D39 WT cell wall (P = 0.004) (Fig. 3B and C). The cell wall thicknesses of serotypes 1 and 19 were similarly affected by ClpL overexpression (Fig. 3C). The immunostaining data showed that D39 WT and the ΔclpL strain (HYK304) retain the ability to produce capsule, while the overexpressing strain (RT172) was a mix of capsule-producing and unencapsulated cells since the overexpression strain showed a mixture of bright fluorescence and nonfluorescent pneumococci (see Fig. S3 in the supplemental material).
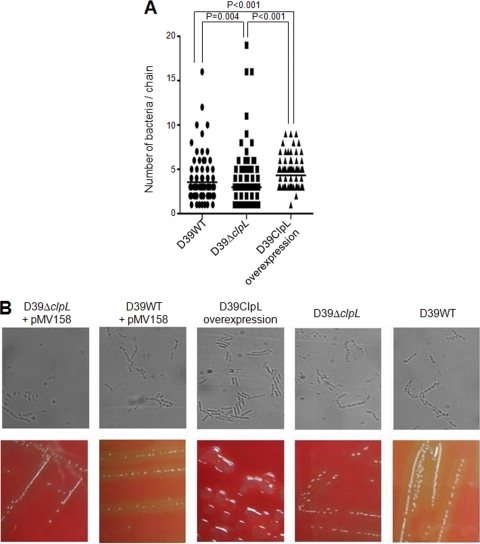
The ClpL-overexpressing strain (RT172) had a generation time of 300 min, versus 48 min for the D39 WT and the ΔclpL (HYK304), when it was grown in bacterial broth. Heat shock did not affect the generation time of either the D39 WT or ΔclpL strain (HYK304) (see Fig. S2 in the supplemental material). Pneumococci grow in chains in liquid media, and the ClpL-overexpressing strain (RT172) produced 4 bacterial cells/chain (median), compared to 3 bacterial cells/chain for D39 WT (P < 0.001), whereas the ΔclpL strain (HYK304) produced 2 bacterial cells/chain (P = 0.004) (Fig. 4A). Inserting the vector used to overexpress ClpL in the D39 WT or ΔclpL strain (HYK304) did not affect chain length (Fig. 4B, upper panels). Also, the D39 ClpL-overexpressing strain (RT172) formed colonies that were more mucoid than those of D39 WT or the ΔclpL strain (HYK304) (Fig. 4B, lower panels).
Fig. 4.
Longer chains and mucoid colonies by ClpL overexpression. Pneumococcal strains at mid-exponential phase (OD550 = 0.3) were fixed onto a glass slide (B, upper panels), and the average chain length was analyzed by microscopy at ×100 magnification from 100 random samples (A and B); or the strains were streaked onto a THY blood agar plate without antibiotic to observe colony morphology (B, lower panels). The figure shows representative results of three independent experiments. Significant differences were analyzed by one-way ANOVA.
Exposure to penicillin increases expression of ClpL and PBP2x in D39 WT.
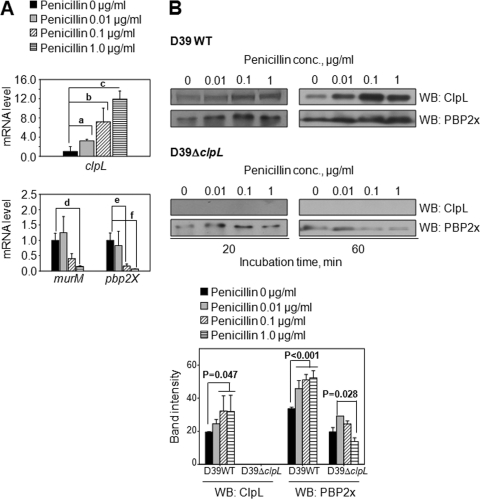
The pbp2b, pbp2x, and murM mutations in pneumococci can affect penicillin resistance (48, 44). Moreover, antibiotic resistance and autolysis could also be affected by the capsule (15) and ciaRH (18). We therefore measured mRNA levels of penicillin resistance-related genes (pbp1A, pbp2b, pbp2x, murM, cps2A, ciaR, and ciaH) in D39 WT with and without penicillin treatment (see Fig. S4 in the supplemental material). Although both PBP2b and PBP2x, the targets of penicillin, were decreased by exposure to penicillin, so far, the PBP2x mutation has been found in all S. pneumoniae isolates with β-lactam resistance, but PBP2b has not (44). Moreover, for structural and functional analyses of PBPs, PBP2x has been used as a model for investigation of penicillin resistance evolution and has been well characterized in many penicillin resistance-related studies (44). Therefore, PBP2x was chosen for further studies. The 3 most significantly changed genes, pbp2x, murM, and clpL, were confirmed by qRT-PCR (Fig. 5A). clpL mRNA levels increased 3.2-fold (P = 0.005), 7.1-fold (P < 0.001), and 11.9-fold (P < 0.001) with increasing doses relative to the levels for the nontreated controls, whereas penicillin decreased mRNA levels of murM at 1 μg/ml (P = 0.005) and pbp2x at 0.1 μg/ml (P = 0.0015) and 1 μg/ml (P < 0.001) (Fig. 5A). To corroborate these data, protein levels of ClpL, PBP2x, and MurM after exposure of D39 to penicillin were analyzed by Western blotting. The protein level of ClpL was increased 1.5-fold (P = 0.047) after D39 WT was exposed to 0.1 μg/ml or 1 μg/ml (Fig. 5B) after 20 min incubation. Surprisingly, however, the protein level of PBP2x was also increased 1.5-fold after D39 WT was exposed to penicillin (P < 0.001) (Fig. 5B, upper panels). The increase of ClpL and PBP2x protein levels was even more significant after exposure of D39 WT to penicillin for 1 h (Fig. 5B, right panels). In contrast, when D39 ΔclpL (HYK304) was exposed to penicillin, the PBP2x protein level was significantly lower (2- to 10-fold, dose dependently) than that of the D39 WT, and protein was even faintly detected at 0.1 and 1 μg/ml of penicillin after 1 h of incubation (Fig. 5B, lower panels). The protein level of MurM was not analyzed because it was not affected by heat shock or the presence of ClpL (Fig. 6B). These data suggested that the presence of ClpL is required for stabilizing the protein expression of PBP2x after exposure to penicillin.
Fig. 5.
Induction of ClpL by penicillin stress and stabilization of PBP2x expression by ClpL. (A) After exposure of D39 WT to penicillin for 1 h, bacterial RNA was isolated and mRNA levels of clpL, murM, and pbp2x were analyzed by quantitative RT-PCR. Each sample was tested in quadruplicate. The figure shows the standard deviation from three independent experiments. Significant differences were analyzed by one-way ANOVA. (B) D39 WT (upper panels) and D39 ΔclpL (lower panels) at the mid-exponential phase (OD550 = 0.3) were exposed to penicillin for 20 and 60 min. Ten micrograms of bacterial lysate was used for Western blotting (WB) with either anti-ClpL or anti-PBP2x antibody. Both chemiluminescence (for PBP2x) and colorimetry (for ClpL) were used to detect HRP-conjugated secondary antibody used in Western blots. The figure shows results representative of three independent experiments. Band density at 20 min was analyzed by Photoshop. The figure shows standard deviations from three independent experiments.
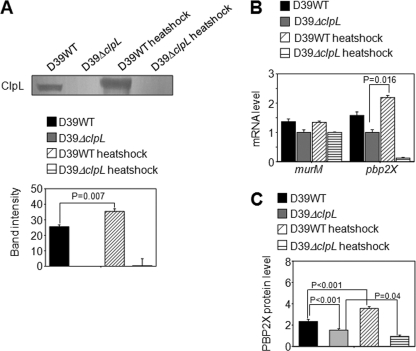
Fig. 6.
Induction of ClpL and PBP2x by heat shock. (A) D39 WT and D39 ΔclpL at the mid-exponential phase (OD550 = 0.3) were heat shocked at 42°C for 30 min. Ten micrograms of bacterial lysate was use to analyze ClpL protein level by Western blot. Colorimetry was used to detect HRP-conjugated secondary antibody used in Western blots. The figure shows representative results of three independent experiments. Band density was analyzed by Photoshop. The figure shows standard deviations from three independent experiments. (B and C) After heat shock, mRNA levels were determined by quantitative RT-PCR (B), and PBP2x protein levels were analyzed by enzyme-linked immunosorbent assay (C) from five (B) and six (C) independent experiments. Significant differences were analyzed by one-way ANOVA.
Heat-induced ClpL increases pbp2x but not murM expression.
mRNA levels could not be determined in the ClpL-overexpressing strain (RT172) because it was very hard to lyse and obtain intact RNA. As a result, the direct effect of ClpL on the expression of murM and pbp2x was further examined in D39 WT and the ΔclpL strain (HYK304) by heat shock. As expected, the level of ClpL protein was increased 1.5-fold (P = 0.007) after heat shock (Fig. 6A), which was similar to the data detected for the D39 ClpL-overexpressing strain (Fig. 3A). In D39 WT, heat shock increased pbp2x mRNA and PBP2x protein levels 1.5-fold (P = 0.016) and 1.5-fold (P < 0.001), respectively. In contrast, in the ΔclpL strain (HYK304), both pbp2x mRNA and PBP2x protein levels were significantly decreased after heat shock (Fig. 6B and C). Moreover, the PBP2x protein level was 175% higher in D39 WT than in the ΔclpL strain (HYK304) (P < 0.001) (Fig. 6C) under the normal condition. Heat shock did not affect murM expression in either D39 WT or the ΔclpL strain (HYK304) (Fig. 6B). These results demonstrated that ClpL could contribute to the modulation of PBP2x expression at both the transcription and translation levels under the heat stress condition.
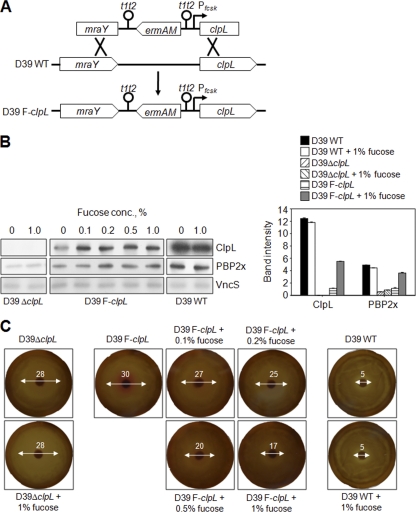
Inducible ClpL expression correlated with PBP2x expression and penicillin susceptibility.
In order to demonstrate a direct effect of ClpL on PBP2x expression and penicillin susceptibility, a D39 clpL-regulatable strain (D39 F-clpL) was constructed by placing clpL under the control of the PfcsK inducible promoter in the chromosome of S. pneumoniae D39 (Fig. 7A). ClpL expression was induced 5-fold in the presence of 0.1 to 1% l-fucose, which was consistent with a previous report (8). Moreover, the expression of PBP2x was also increased 3-fold when D39 F-clpL was exposed to 0.2 to 1% l-fucose (Fig. 7B). However, the maximum amount of ClpL in fucose-induced D39 F-clpL was only half of that in D39 WT. Consistently, the maximum amount of PBP2x also could not reach that of D39 WT. In contrast, the presence of l-fucose in D39 WT and D39 ΔclpL did not affect expression of ClpL or other proteins in S. pneumoniae type 2, including VncS sensor kinase (Fig. 7B). These results suggested that ClpL could directly modulate the expression of PBP2x in vivo.
Fig. 7.
Increase of ClpL expression correlated with PBP2x expression and penicillin susceptibility. (A) An S. pneumoniae D39 clpL-regulated strain (D39 F-clpL) was constructed by placing clpL under the control of the PfcsK inducible promoter in the chromosome of S. pneumoniae. (B) Bacteria were cultured in THY broth until the OD550 was 0.3 and then exposed to 0, 0.1, 0.2, 0.5, and 1% l-fucose for 4 h. Ten micrograms of bacterial lysate was used to analyze the protein expression level of ClpL, PBP2x, and VncS by Western blotting. Band density was analyzed by Photoshop. The figure shows representative results of two independent experiments. (C) D39 WT, D39 ΔclpL, and D39 F-clpL were cultured in THY broth until the OD550 was 0.3. An aliquot of 50 μl of each culture was spread onto a THY agar plate containing 5% sheep blood and 0, 0.1, 0.2, 0.5, or 1% l-fucose. Penicillin (0.02 μg) was placed on sterile filter paper disks, and plates were then incubated at 37°C for 1 day.
To investigate the effect of ClpL on penicillin susceptibility, the inhibition zone diameter around the penicillin-containing disk was measured. In the presence of l-fucose, the inhibition zone diameter was decreased dose dependently (Fig. 7C), suggesting that increase of ClpL expression decreased the penicillin susceptibility of D39 F-clpL. Moreover, the inhibition zone diameter of D39 F-clpL at 1% l-fucose was 3-fold bigger than that of D39 WT, which was consistent with the expression levels of ClpL and PBP2x shown previously (Fig. 7B). These results demonstrated that ClpL was specifically induced after addition of l-fucose and the induction of ClpL directly increased PBP2x expression, which resulted in a decrease of penicillin susceptibility.
Although both ClpL and PBP2x protein levels were significantly different between D39 WT and D39 ΔclpL (Fig. 5), the difference of cell wall thickness between them could not be observed even after TEM analysis (Fig. 3). Moreover, both ClpL and PBP2x protein levels of D39 F-clpL were intermediate between those of D39 WT and D39 ΔclpL (Fig. 7). Therefore, cell wall thickness of D39 F-clpL in the presence of various fucose concentrations was not analyzed.
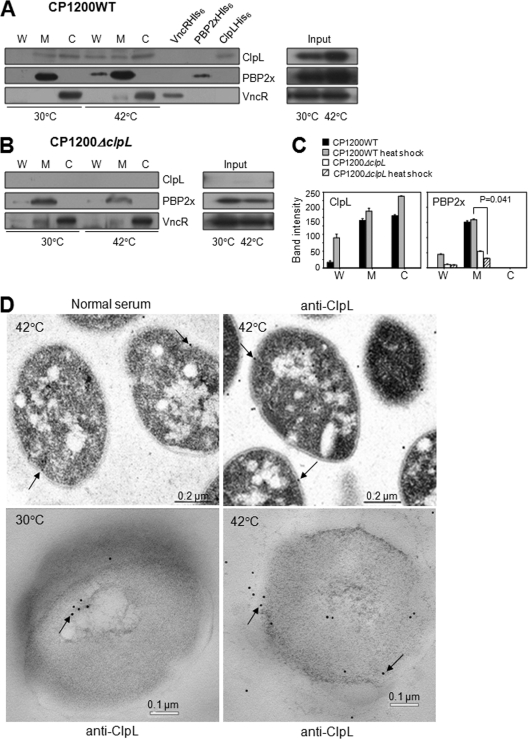
ClpL cotranslocates with PBP2x into cell wall after heat shock.
HSP chaperones refold proteins containing hydrophobic residues and translocate them across membranes (14). Therefore, a ClpL chaperone might facilitate translocation of PBPs into cell wall. To check this possibility, the nonencapsulated wild-type pneumococcus (CP1200) and the isogenic ΔclpL strain (HYK1) were heat shocked and fractionated into cytosol, membrane, and cell wall, followed by Western blot analysis using anti-ClpL and anti-PBP2x antibodies. In the wild-type bacteria at 30°C, ClpL was localized in the membrane and cytosol fractions, while PBP2x was localized predominantly in the membrane fraction. However, neither of them was detected at the cell wall (Fig. 8A). VncR served as a cytosol marker and was detected mostly (>95%) in the cytosol fraction at both 30°C and 42°C (Fig. 8A). After heat shock, small amounts of ClpL and PBP2x were found in the cell wall fraction and cell membrane fractions, suggesting that in wild-type bacteria ClpL and PBP2x were translocated into the cell wall after heat shock (Fig. 8A and C). In contrast to the results with wild-type bacteria, when the ΔclpL strain (HYK1) was grown at 30°C, PBP2x was detected in the membrane fraction; but after heat shock, the PBP2x concentration in the cell membrane decreased (P = 0.041) and became undetectable in the cell wall fractions (P = 0.102) (Fig. 8B and C), suggesting that ClpL is required for both PBP2x stabilization and net translocation into the cell wall. TEM of thin-sectioned samples consistently showed that ClpL was mainly present in the cytosol under normal growth conditions, whereas it was consistently in the cell wall after heat shock (Fig. 8D).
Fig. 8.
Colocalization of ClpL with PBP2x at the cell wall after heat shock. (A to C) After heat shock, the cell wall (W), cell membrane (M), and cytosol (C) of D39 WT (A) and D39 ΔclpL (B) were fractionated. Western blotting using anti-ClpL, anti-PBP2x, and anti-VncR (cytosole marker) antibodies was carried out to localize ClpL and PBP2x. Purified VncR (VncRHis6), PBP2x (PBP2xHis6), and ClpL (ClpLHis6) were added as positive controls. Total cell lysate was used as input. Both chemiluminescence (for ClpL and VncR) and colorimetry (for PBP2x) were used to detect HRP-conjugated secondary antibody used in Western blots. Band density was analyzed by Photoshop. The figure shows the standard deviations from two independent experiments (C). Significant differences were analyzed by ANOVA. (D) Pneumococci cultured at 30°C were heat shocked at 42°C for 30 min. Thin-sectioned samples were treated with preimmune rabbit serum as primary antibody (left and upper panel) or anti-ClpL antibodies, followed by anti-rabbit IgG conjugated to colloidal gold (right, upper and lower panels). The colloidal gold is seen as electron-dense particles (black arrows). The figure shows representative data from three different sections.
ClpL interacts with PBP2x.
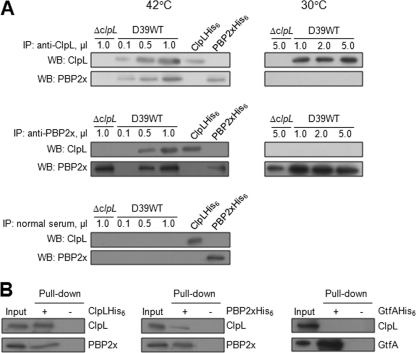
To test for an interaction between ClpL and PBP2x in vivo, cell lysates of the D39 WT and the ΔclpL strain were immunoprecipitated with either anti-ClpL antibody or anti-PBP2x and reprobed with either anti-ClpL or anti-PBP2x antibody. When anti-ClpL was used, PBP2x coimmunoprecipitated with ClpL and the amount of ClpL precipitated increased in proportion to the amount of antibody added (Fig. 9A, upper panels). Also, when anti-PBP2x antibody was used for immunoprecipitation, ClpL coimmunoprecipitated with PBP2x and the amount of PBP2x precipitated increased in proportion to the amount of antibody added (Fig. 9A, middle panels). However, under the non-heat shock condition (30°C), ClpL and PBP2x were not coimmunoprecipitated with PBP2x and ClpL, respectively (Fig. 9A, right panels), suggesting that ClpL did not interact with PBP2x under nonstress conditions. Neither PBP2x nor ClpL was detected after immunoprecipitation with preimmune serum (Fig. 9A, lower panel). To corroborate this result in vitro, a His tag pulldown assay was carried out using either purified ClpLHis6 or PBP2xHis6. The result revealed that PBP2x was pulled down in the presence of ClpL but not in the absence of ClpL (Fig. 9B, left panel) and vice versa (Fig. 9B, right panel). When GtfA was used as a negative control, ClpL was not pulled down (Fig. 9B). Taken together, these data indicate that after heat shock ClpL interacts with PBP2x to promote cell wall synthesis.
Fig. 9.
Interaction of ClpL with PBP2x in vivo and in vitro. (A) Pneumococci cultured at 30°C were heat shocked at 42°C for 30 min. After heat shock, 200 μg of cell lysates of D39 WT was immunoprecipitated (IP) using 0.1 μl (lane 2), 0.5 μl (lane 3), and 1.0 μl (lane 4) of either anti-ClpL (upper panels) or anti-PBP2x (middle panels) antibody. Lysate of the ΔclpL strain was used as a control. The immunoprecipitated products were used for Western blotting (WB) using either anti-ClpL or anti-PBP2x antibody. As a control, preimmune (normal) serum was used instead of anti-ClpL or anti-PBP2x antibody (lower panel), followed by ClpL and PBP2x detection. Purified ClpL and PBP2x were used as positive controls. For the non-heat shock condition (right panels), 1 μl, 2 μl, and 5 μl of either anti-ClpL (upper panels) or anti-PBP2x (lower panels) were used for immunoprecipitation. The figure shows representative results of two independent experiments. (B) Interaction of ClpL and PBP2x in vitro. D39 WT was heat shocked, and the lysate was used for His tag pulldown assay using either ClpLHis6 (left panel) or PBP2xHis6 (right panel). After the membrane was probed with anti-ClpL antibody, the membrane was reprobed with anti-PBP2x antibody, and vice versa. The figure shows representative results of two independent experiments. Chemiluminescence was used to detect HRP-conjugated secondary antibody used in Western blots. GtfAHis6 was used as a negative control.
DISCUSSION
The emergence of high-level resistance to antimicrobials is an increasing threat to global health (33, 39), and even a small increase in antibiotic-refractory bacterial subpopulations or MIC could herald the emergence of higher-level resistance (39, 3, 4). Moreover, the phenomena of bacterial persistence in vitro and antibiotic tolerance in vivo are well established (13). Therefore, any factor contributing to an increase of antibiotic resistance is critically important. Since HSPs have been reported to be induced by antibiotics (43), they could contribute to in vivo antibiotic tolerance. Our finding that penicillin-triggered lysis could be partially prevented by heat shock pretreatment makes it clear that in vivo stresses, such as inflammation, respiratory bursts in phagocytes, and temperature upshift, may induce higher ClpL levels and increase resistance to penicillin.
Penicillin dose-dependently increased clpL levels but decreased pbp2x levels. Because ClpL is induced by heat shock and other stresses (11, 27), pneumococcus survival seems to depend on competition between the amounts of ClpL and PBP2x. Low-level stress may induce ClpL and increase cell resistance to penicillin compared to normal cells. Therefore, low-level stress could improve pneumococcus survival. Roger et al. (52) reported that after exposure to penicillin, transcriptome analysis of S. pneumoniae D39 showed significant increases in the levels of luxS and ctsR genes, which are involved in quorum sensing (61) and regulation of Clp ATP-dependent protease (10), respectively; however, expression of clpL or pbp2x was not observed. Since luxS is located next to the clpL and the CtsR repressor binding sequence is present upstream of clpL in the D39 genome (40), clpL expression could have been indirectly affected in their study. Moreover, in this study, expression of clpL and pbp2x was significantly affected at 0.1 μg/ml of penicillin, but not at 0.01 μg/ml (Fig. 5A), which was consistent with the results obtained at 0.03 μg/ml of penicillin in the study of Roger et al. (52). Thus, the difference in gene expression levels in our result from the previous one seems to be due to different penicillin concentrations.
The proteins induced by antibiotic exposure may affect susceptibility to these antibiotics (64). In E. coli, disruption of the heat shock proteins DnaK and GroEL, and particularly the ATP-dependent protease, Lon, increases susceptibility to fluoroquinolones (64). The increased susceptibility of the dnaK, groEL, and lon mutants can partly be explained by the disruption of the quality control system in the E. coli cytosol (64). In addition, HSPs increased resistance of E. coli to lysis by cephaloridine and cefsulodin, which could be abolished by a mutation in any of five heat shock genes (dnaK, dnaJ, grpE, groES, or groEL) (49). S. pneumoniae with mutations of HSP ClpL showed increased susceptibility to penicillin-induced lysis (59). We also found that heat shock pretreatment significantly increased resistance of the wild-type pneumococci to penicillin, whereas heat shock made the ΔclpL strain (HYK304) 5-fold more susceptible (Fig. 2). This susceptibility could be due to a disrupted quality control system in the ΔclpL strain (HYK304), thereby abolishing the resistance-increasing effect of ClpL by heat shock. Moreover, since the ΔclpL strain (HYK304) was in chains that are half as long (Fig. 4 in this study), only two cocci, on average, have to be killed to get rid of a ΔclpL CFU compared to WT CFU, where an average of 4 cocci have to be killed. That could be the explanation for why the ΔclpL strain (HYK304) is easier to kill than the WT strain or why the ΔclpL strain (HYK304) was more susceptible than the WT.
In S. pneumoniae, deoxycholate induces autolysis by triggering autolysin (42). The major autolysin, LytA, digests cell wall peptidoglycans (54) and results in release of intracellular and cell wall components (34). LytA is encoded by the cinA-recA-dinF-lytA operon, which is regulated by ComDE, a two-component system composed of the histidine kinase ComD and its cognate response regulator, ComE. Therefore, LytA is under the control of competence (41), and competence induction triggers autolysis in S. pneumoniae (57). CiaRH, which is involved in the early control of competence induction, prevents autolysis induced by treatments with cell wall inhibitors and by mutations in pbp2x (38). A nonencapsulated R6 strain with a vncS mutation is tolerant to β-lactams, cephalosporins, and vancomycin (45). However, loss of function of VncRS or Pep27 (an effector molecule encoded upstream of the vncRS locus) does not alter autolysis (20, 51), suggesting that the connection between penicillin resistance and VncRS operon function is still unclear. Subinhibitory concentrations of penicillin (0.5× MIC) induced ciaRH and ctsR but repressed the competence genes comD and comE in S. pneumoniae (52). Therefore, ClpL may be modulated by factors such as CtsR, as ciaRH is not modulated by ClpL and lytA mRNA levels were similar in the ΔclpL (HYK304) and the D39 WT strains after 30 min heat shock (data not shown).
The ClpL-overexpressing strain (RT172) had a generation time of 300 min, versus 48 min for the D39 WT and the ΔclpL strain (HYK304), when it was grown in bacterial broth. ClpL is a nonessential gene that could be disrupted in S. pneumoniae R6 and D39 (40, 50). Also in Lactobacillus gasseri, the ΔclpL strain had growth characteristics that were indistinguishable from those of the wild type under several stress conditions, except a lethal temperature (58). Consistently, in our study, although the ClpL-overexpressing strain (RT172) had a significantly lengthened generation time, the ΔclpL strain (HYK304) showed growth similar to that of the D39 WT. Such a long generation time could be one mechanism postulated to account for the declining sensitivity and survival of bacteria confronted with bactericidal antibiotics (32).
In S. aureus, β-lactam and vancomycin treatment induces the VraSR (vancomycin resistance-associated sensor/regulator) system, which is associated with cell wall synthesis, such as PBP 2, MurZ, and SgtB (26). VraS kinase senses damage of the cell wall structure or inhibition of cell wall synthesis (26), and vraSR mutants show less resistance against β-lactam, teicoplanin, bacitracin, and fosfomycin, indicating that VraSR is a positive regulator of peptidoglycan synthesis (26). In contrast, S. pneumoniae VncRS, which is equivalent to the VraSR system in S. aureus, did not change after β-lactam and vancomycin treatment, and vncR, vncS, and pep27 mutants did not affect vancomycin resistance (20, 51).
In S. pneumoniae, the ΔclpL strain is sensitive to high temperature (27) and adhered more efficiently to A549 human type II alveolar cells (59). In this study, the ΔclpL strain (HK304) was more sensitive to penicillin (Fig. 1B) and had a shorter chain length (Fig. 4A). In contrast, the clpC mutant is resistant to high temperature and penicillin and deficient in adhering to A549 cells (9). Moreover, the clpC mutant formed long chains and is not lysed by treatment with penicillin or vancomycin (9). These results suggest that ClpC and ClpL might have different functions upon induction by the stress response, although both are induced by high temperature.
Because ClpL homologs are found in pathogens such as Staphylococcus aureus, Streptococcus spp., including Streptococcus pyogenes, and Enterococcus spp., as well as lactic acid bacteria (Lactococcus, Lactobacillus, and Leuconostoc) (http://www.ncbi.nlm.nih.gov/sites/entrez), stresses such as DNA damage, viral infection, starvation, and other stressful insults, including hyperthermia and ethanol, may induce antibiotic resistance and tolerance.
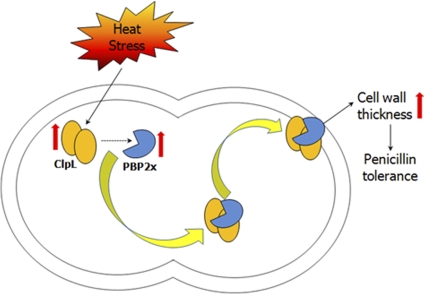
ClpL is induced by heat shock in S. pneumoniae (11, 27) (Fig. 6) and was colocalized with PBP2x at the cell wall after heat shock (Fig. 8A). Penicillin binding proteins, Clp proteases, and two-component sensor regulators, as well as ABC transporters responsible for the translocation of the RTX class of bacterial toxins, are known to be exported or membrane-associated proteins (46). In this study, ClpL was found to interact with PBP2x in the cytoplasm (Fig. 9A) and to bind to denatured PBP2x (data not shown). However, ClpL did not associate with PBP2x under the nonstress condition (Fig. 9), suggesting that ClpL might be required only to stabilize and reactivate PBP2x under stress conditions. Since HSP chaperones refold proteins containing hydrophobic residues and translocate them across membranes (14), our results suggest that PBP could be a substrate for ClpL. Therefore, it seems that ClpL could facilitate translocation of PBP2x into the cell wall and subsequently increase cell wall thickness and penicillin resistance (Fig. 10). However, the mechanism of how a membrane-tethered protein like PBP2x is facilitated to move from the membrane to the cell wall requires further study.
Fig. 10.
Model of how ClpL could increase penicillin resistance. Under heat shock condition, ClpL protein expression is induced, which contributes to the stabilization and increase of PBP2x protein level. As a molecular chaperone, ClpL then interacts with PBP2x in the cytosol and facilitates translocation of PBP2x into the cell wall, which results in a PBP-mediated increase in cell wall thickness and penicillin resistance.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a South Korea Science and Engineering Foundation (KOSEF) grant funded by the South Korean government (MEST) (WCU R33-10045 and R01-2006-000-10504-0).
Footnotes
Supplemental material for this article may be found at http://aac.asm.org/.
Published ahead of print on 21 March 2011.
REFERENCES
- 1. Andrews J. M. 2001. Determination of minimum inhibitory concentrations. J. Antimicrob. Chemother. 48(Suppl 1):5–16 [DOI] [PubMed] [Google Scholar]
- 2. Avery O. T., MacLeod C. M., McCarty M. 1944. Studies on the chemical nature of the substance inducing transformation of pneumococcal types. Induction of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type III. J. Exp. Med. 79:137–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Balaban N. Q., Merrin J., Chait R., Kowalik L., Leibler S. 2004. Bacterial persistence as a phenotypic switch. Science 305:1622–1625 [DOI] [PubMed] [Google Scholar]
- 4. Barclay M. L., Begg E. J., Chambers S. T. 1992. Adaptive resistance following single doses of gentamicin in a dynamic in vitro model. Antimicrob. Agents Chemother. 36:1951–1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bartlett J. G., et al. 2000. Practice guidelines for the management of community-acquired pneumonia in adults. Clin. Infect. Dis. 31:347–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bricker A. L., Camilli A. 1999. Transformation of a type 4 encapsulated strain of Streptococcus pneumoniae. FEMS Microbiol. Lett. 172:131–135 [DOI] [PubMed] [Google Scholar]
- 7. Burdett V. 1980. Identification of tetracycline-resistant R-plasmids in Streptococcus agalactiae (group B). Antimicrob. Agents Chemother. 18:753–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chan P. F., et al. 2003. Characterization of a novel fucose-regulated promoter (PfcsK) suitable for gene essentiality and antibacterial mode-of-action studies in Streptococcus pneumoniae. J. Bacteriol. 185:2051–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Charpentier E., Novak R., Tuomanen E. 2000. Regulation of growth inhibition at high temperature, autolysis, transformation, and adherence in Streptococcus pneumoniae. Mol. Microbiol. 37:717–726 [DOI] [PubMed] [Google Scholar]
- 10. Chastanet A., Prudhomme M., Claverys J. P., Msadek T. 2001. Regulation of Streptococcus pneumoniae clp genes and their role in competence development and stress survival. J. Bacteriol. 183:7295–7307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Choi I. H., et al. 1999. Limited stress response in Streptococcus pneumoniae. Microbiol. Immunol. 43:807–812 [DOI] [PubMed] [Google Scholar]
- 12. Cosgrove S. E. 2006. The relationship between antimicrobial resistance and patient outcomes: mortality, length of hospital stay, and health care costs. Clin. Infect. Dis. 42(Suppl. 2):S82–S89 [DOI] [PubMed] [Google Scholar]
- 13. Eagle H., Fleischman R., Musselman A. D. 1950. The bactericidal action of penicillin in vivo: the participation of the host, and the slow recovery of the surviving organisms. Ann. Intern. Med. 33:544–571 [DOI] [PubMed] [Google Scholar]
- 14. Feldman D. E., Frydman J. 2000. Protein folding in vivo: the importance of molecular chaperones. Curr. Opin. Struct. Biol. 10:26–33 [DOI] [PubMed] [Google Scholar]
- 15. Fernebro J., et al. 2004. Capsular expression in Streptococcus pneumoniae negatively affects spontaneous and antibiotic-induced lysis and contributes to antibiotic tolerance. J. Infect. Dis. 189:328–338 [DOI] [PubMed] [Google Scholar]
- 16. Ghooi R. B., Thatte S. M. 1995. Inhibition of cell wall synthesis—is this the mechanism of action of penicillins? Med. Hypotheses 44:127–131 [DOI] [PubMed] [Google Scholar]
- 17. Goffin C., Ghuysen J. M. 1998. Multimodular penicillin-binding proteins: an enigmatic family of orthologs and paralogs. Microbiol. Mol. Biol. Rev. 62:1079–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gong F., Fahy D., Smerdon M. J. 2006. Rad4-Rad23 interaction with SWI/SNF links ATP-dependent chromatin remodeling with nucleotide excision repair. Nat. Struct. Mol. Biol. 13:902–907 [DOI] [PubMed] [Google Scholar]
- 19. Reference deleted.
- 20. Haas W., Sublett J., Kaushal D., Tuomanen E. I. 2004. Revising the role of the pneumococcal vex-vncRS locus in vancomycin tolerance. J. Bacteriol. 186:8463–8476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hickey T. B., Thorson L. M., Speert D. P., Daffe M., Stokes R. W. 2009. Mycobacterium tuberculosis Cpn60.2 and DnaK are located on the bacterial surface, where Cpn60.2 facilitates efficient bacterial association with macrophages. Infect. Immun. 77:3389–3401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huang D. C., Huang X. F., Novel G., Novel M. 1993. Two genes present on a transposon-like structure in Lactococcus lactis are involved in a Clp-family proteolytic activity. Mol. Microbiol. 7:957–965 [DOI] [PubMed] [Google Scholar]
- 23. Izquierdo E., et al. 2009. 2-DE and MS analysis of key proteins in the adhesion of Lactobacillus plantarum, a first step toward early selection of probiotics based on bacterial biomarkers. Electrophoresis 30:949–956 [DOI] [PubMed] [Google Scholar]
- 24. Kelly P., et al. 2005. Correlation of probiotic Lactobacillus salivarius growth phase with its cell wall-associated proteome. FEMS Microbiol. Lett. 252:153–159 [DOI] [PubMed] [Google Scholar]
- 25. Kuroda H., Kuroda M., Cui L., Hiramatsu K. 2007. Subinhibitory concentrations of beta-lactam induce haemolytic activity in Staphylococcus aureus through the SaeRS two-component system. FEMS Microbiol. Lett. 268:98–105 [DOI] [PubMed] [Google Scholar]
- 26. Kuroda M., et al. 2003. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol. Microbiol. 49:807–821 [DOI] [PubMed] [Google Scholar]
- 27. Kwon H. Y., et al. 2003. Effect of heat shock and mutations in ClpL and ClpP on virulence gene expression in Streptococcus pneumoniae. Infect. Immun. 71:3757–3765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kwon H. Y., et al. 2004. The ClpP protease of Streptococcus pneumoniae modulates virulence gene expression and protects against fatal pneumococcal challenge. Infect. Immun. 72:5646–5653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Laemmli U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 30. Lee I. J., Kim K. W., Hyun J. W., Lee Y. H., Park E. W. 2009. Comparative ultrastructure of nonwounded Mexican lime and Yuzu leaves infected with the citrus canker bacterium Xanthomonas citri pv. citri. Microsc. Res. Tech. 72:507–516 [DOI] [PubMed] [Google Scholar]
- 31. Lee S., et al. 2009. Targeting a bacterial stress response to enhance antibiotic action. Proc. Natl. Acad. Sci. U. S. A. 106:14570–14575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Levin B. R. 2004. Noninherited resistance to antibiotics. Science 305:1578–1579 [DOI] [PubMed] [Google Scholar]
- 33. Levy S. B., Marshall B. 2004. Antibacterial resistance worldwide: causes, challenges and responses. Nat. Med. 10:S122–S129 [DOI] [PubMed] [Google Scholar]
- 34. Lopez R., Garcia E., Garcia P., Garcia J. L. 1997. The pneumococcal cell wall degrading enzymes: a modular design to create new lysins? Microb. Drug Resist. 3:199–211 [DOI] [PubMed] [Google Scholar]
- 35. Macellaro A., Tujulin E., Hjalmarsson K., Norlander L. 1998. Identification of a 71-kilodalton surface-associated Hsp70 homologue in Coxiella burnetii. Infect. Immun. 66:5882–5888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mandell L. A., et al. 2007. Infectious Diseases Society of America/American Thoracic Society consensus guidelines on the management of community-acquired pneumonia in adults. Clin. Infect. Dis. 44(Suppl. 2):S27–S72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Reference deleted.
- 38. Mascher T., Heintz M., Zähner D., Merai M., Hakenbeck R. 2006. The CiaRH system of Streptococcus pneumoniae prevents lysis during stress induced by treatment with cell wall inhibitors and by mutations in pbp2x involved in beta-lactam resistance. J. Bacteriol. 188:1959–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Massey R. C., Buckling A., Peacock S. J. 2001. Phenotypic switching of antibiotic resistance circumvents permanent costs in Staphylococcus aureus. Curr. Biol. 11:1810–1814 [DOI] [PubMed] [Google Scholar]
- 40. McLennan N., Masters M. 1998. GroE is vital for cell-wall synthesis. Nature 392:139. [DOI] [PubMed] [Google Scholar]
- 41. Mortier-Barriere I., de Saizieu A., Claverys J. P., Martin B. 1998. Competence-specific induction of recA is required for full recombination proficiency during transformation in Streptococcus pneumoniae. Mol. Microbiol. 27:159–170 [DOI] [PubMed] [Google Scholar]
- 42. Mosser J. L., Tomasz A. 1970. Choline-containing teichoic acid as a structural component of pneumococcal cell wall and its role in sensitivity to lysis by an autolytic enzyme. J. Biol. Chem. 245:287–298 [PubMed] [Google Scholar]
- 43. Neidhardt F. C., VanBogelen R. A. 1987. Heat shock response, p. 1334–1345 In Neidhardt F. C. (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology. American Society for Microbiology, Washington, DC [Google Scholar]
- 44. Normark B. H. 2007. Molecular epidemiology and mechanisms for antibiotic resistance in Streptococcus pneumoniae, p. 269–290 In Hakenbeck R., Chhatwal S. (ed.), Molecular biology of streptococci. Horizon Bioscience, United Kingdom [Google Scholar]
- 45. Novak R., Henriques B., Charpentier E., Normark S., Tuomanen E. 1999. Emergence of vancomycin tolerance in Streptococcus pneumoniae. Nature 399:590–593 [DOI] [PubMed] [Google Scholar]
- 46. Pearce B. J., Yin Y. B., Masure H. R. 1993. Genetic identification of exported proteins in Streptococcus pneumoniae. Mol. Microbiol. 9:1037–1050 [DOI] [PubMed] [Google Scholar]
- 47. Pechous R., Ledala N., Wilkinson B. J., Jayaswal R. K. 2004. Regulation of the expression of cell wall stress stimulon member gene msrA1 in methicillin-susceptible or -resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 48:3057–3063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pernot L., et al. 2004. A PBP2x from a clinical isolate of Streptococcus pneumoniae exhibits an alternative mechanism for reduction of susceptibility to beta-lactam antibiotics. J. Biol. Chem. 279:16463–16470 [DOI] [PubMed] [Google Scholar]
- 49. Powell J. K., Young K. D. 1991. Lysis of Escherichia coli by beta-lactams which bind penicillin-binding proteins 1a and 1b: inhibition by heat shock proteins. J. Bacteriol. 173:4021–4026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Robertson G. T., Ng W. L., Gilmour R., Winkler M. E. 2003. Essentiality of clpX, but not clpP, clpL, clpC, or clpE, in Streptococcus pneumoniae R6. J. Bacteriol. 185:2961–2966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Robertson G. T., et al. 2002. Vancomycin tolerance induced by erythromycin but not loss of vncRS, vex3, or pep27 function in Streptococcus pneumoniae. J. Bacteriol. 184:6987–7000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rogers P. D., et al. 2007. Gene expression profiling of the response of Streptococcus pneumoniae to penicillin. J. Antimicrob. Chemother. 59:616–626 [DOI] [PubMed] [Google Scholar]
- 53. Rudan I., Boschi-Pinto C., Biloglav Z., Mulholland K., Campbell H. 2008. Epidemiology and etiology of childhood pneumonia. Bull. World Health Organ. 86:408–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sanchez-Puelles J. M., et al. 1986. Searching for autolysin functions. Characterization of a pneumococcal mutant deleted in the lytA gene. Eur. J. Biochem. 158:289–293 [DOI] [PubMed] [Google Scholar]
- 55. Schaumburg J., et al. 2004. The cell wall subproteome of Listeria monocytogenes. Proteomics 4:2991–3006 [DOI] [PubMed] [Google Scholar]
- 56. Singh V. K., et al. 2007. Role for dnaK locus in tolerance of multiple stresses in Staphylococcus aureus. Microbiology 153:3162–3173 [DOI] [PubMed] [Google Scholar]
- 57. Steinmoen H., Knutsen E., Havarstein L. S. 2002. Induction of natural competence in Streptococcus pneumoniae triggers lysis and DNA release from a subfraction of the cell population. Proc. Natl. Acad. Sci. U. S. A. 99:7681–7686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Suokko A., Poutanen M., Savijoki K., Kalkkinen N., Varmanen P. 2008. ClpL is essential for induction of thermotolerance and is potentially part of the HrcA regulon in Lactobacillus gasseri. Proteomics 8:1029–1041 [DOI] [PubMed] [Google Scholar]
- 59. Tu le N., et al. 2007. Modulation of adherence, invasion, and tumor necrosis factor alpha secretion during the early stages of infection by Streptococcus pneumoniae ClpL. Infect. Immun. 75:2996–3005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Reference deleted.
- 61. Vendeville A., Winzed K., Heurlier K., Tang C. M., Hardie K. R. 2005. Making ‘sense’ of metabolism: autoinducer-2, LUXS and pathogenic bacteria. Nat. Rev. Microbiol. 3:383–396 [DOI] [PubMed] [Google Scholar]
- 62. Vijayakumar M. N., Morrison D. A. 1986. Localization of competence-induced proteins in Streptococcus pneumoniae. J. Bacteriol. 165:689–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Weng L., Biswas I., Morrison D. A. 2009. A self-deleting Cre-lox-ermAM cassette, Cheshire, for marker-less gene deletion in Streptococcus pneumoniae. J. Microbiol. Methods 79:353–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yamaguchi Y., Tomoyasu T., Takaya A., Morioka M., Yamamoto T. 2003. Effects of disruption of heat shock genes on susceptibility of Escherichia coli to fluoroquinolones. BMC Microbiol. 3:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.