Abstract
With the spread of chloroquine (CQ)-resistant malaria in India, sulfadoxine-pyrimethamine (SP) alone or in combination with artesunate is used as an alternative antimalarial drug. Due to continuous drug pressure, the Plasmodium falciparum parasite is exhibiting resistance to antifolates because of mutations in candidate genes dihydrofolate reductase (dhfr) and dihydropteroate synthetase (dhps). Our earlier study on flanking microsatellite markers of dhfr mutant alleles from India had shown a single origin of the pyrimethamine resistance and some minor haplotypes which shared haplotypes with Southeast Asian (Thailand) strains. In the present study, we have analyzed 193 of these Indian P. falciparum isolates for 15 microsatellite loci around dhps to investigate the genetic lineages of the mutant dhps alleles in different parts of the country. Eighty-one of these samples had mutant dhps alleles, of which 62 were from Andaman and Nicobar Islands and the remaining 19 were from mainland India. Of 112 isolates with a wild-type dhps allele, 109 were from mainland India and only 3 were from Andaman and Nicobar Islands. Consistent with the model of selection, the mean expected heterozygosity (He) around mutant dhps alleles (He = 0.55; n = 81) associated with sulfadoxine resistance was lower (P ≤ 0.05) than the mean He around the wild-type dhps allele (He = 0.80; n = 112). There was more genetic diversity in flanking microsatellites of dhps than dhfr among these isolates, which confirms the assertion that dhps mutations are at a very early stage of fixation in the parasite population. Microsatellite haplotypes around various mutant dhps alleles suggest that the resistant dhps alleles have multiple independent origins in India, especially in Andaman and Nicobar Islands. Determining the genetic lineages of the resistant dhps alleles on Andaman and Nicobar Islands and mainland India is significant, given the role of Asia in the intercontinental spread of chloroquine- and pyrimethamine-resistant parasites in the past.
INTRODUCTION
The molecular basis of sulfadoxine-pyrimethamine (SP) resistance has been well documented (6, 26, 30–32). This drug combination mainly targets the folate biosynthetic pathway of the malarial parasite, which arrests the nucleic acid biosynthesis and hence causes parasite death. Sulfadoxine is the competitive inhibitor of the dihydropteroate synthetase (DHPS) enzyme, and pyrimethamine targets dihydrofolate reductase (DHFR) of Plasmodium falciparum. Mutations at five amino acid positions of P. falciparum DHPS cause alterations in the sulfadoxine binding site of the enzyme. A change from alanine to glycine at codon 437 (A437G) is the first step to resistance to sulfa drugs, followed by sequential mutations at codons 436 (S436A), 540 (K540E), 581 (A581G), and 613 (A613S/T), which cause a further increase in drug resistance (32).
The extent of sulfadoxine resistance in P. falciparum isolates differs in different parts of the world due to the presence of different P. falciparum dhps (pfdhps) mutant alleles. Double mutant allele SGEAA (mutated amino acids are boldfaced and underlined; amino acids shown here correspond to positions 436, 437, 540, 581, and 613, respectively) is prevalent in East Africa (25), whereas AGKAA is prevalent in West and Central Africa (15). In South America, there is a prevalence of the triple mutant SGEGA allele (16). Triple mutant alleles AGEAA and SGEGA, along with novel triple mutant allele SGNGA, are prevalent in Thailand and Cambodia (4, 33). In India, the prevalence of different mutant dhps alleles varies from region to region (1, 3, 11), due to different rates of malaria transmission and level of drug resistance (10). Wild-type dhps allele SAKAA predominates among isolates from mainland India (1, 3). There is a prevalence of double and triple mutant dhps alleles (AGKAA, SGEGA, and AGEAA) in Andaman and Nicobar Islands (3). Novel triple mutant allele AGNAA was observed only in Andaman and Nicobar Islands of India (11). These triple mutant alleles, AGEAA and SGEGA, were usually associated with quadruple mutant dhfr allele AIRNL (the positions of these amino acids are 16, 51, 59, 108, and 164, respectively) among isolates from Andaman and Nicobar Islands, whereas in Assam, these mutations were associated with the double mutant dhfr ANRNI allele (1, 3). The novel mutant allele AGNAA was predominantly associated with double mutant dhfr allele ANRNI (11).
Microsatellite analysis around dhfr suggested that there are limited numbers of origins of high-level pyrimethamine resistance dhfr alleles (those with 3 or more mutations) worldwide. In Southeast Asia, dhfr alleles with a single mutation have multiple independent origins (21, 22), whereas alleles with 2 or more mutations have a single common origin (12, 21, 22). The Southeast Asian triple mutant dhfr allele AIRNI later spread to Africa, where this is in abundance (7, 14, 24, 28, 29). However, there is evidence of indigenous origins of triple mutant dhfr allele AIRNI in several African countries like Kenya, Ghana, and Cameroon (18, 19). Two distinct lineages of triple mutant dhfr alleles (RICNI and CICNL) have been reported from South America (8, 17, 27). In India, all dhfr alleles with multiple mutations (2 or more) had identical or closely related microsatellite haplotypes, suggesting their common genetic backgrounds. They were also closely related to the Thai type, which confirmed that the highly resistant dhfr alleles in India have come from Southeast Asia (12).
Although the origin of dhfr mutant alleles in malarial parasites is well documented, there is limited information on the origin of mutant dhps alleles from the world over (4, 25, 33). Recent reports of sulfadoxine resistance revealed that there are multiple origins of dhps mutant alleles from Africa and from Southeast Asia (4, 25, 33). In South America, there is a single origin of dhps mutant alleles, with some independent origins being found (5, 16). Here, we report the multiple origins and ongoing selection events of dhps alleles in the Indian subcontinent, where mutant dhfr alleles have already shown fixation in these isolates (12).
MATERIALS AND METHODS
PCR amplification and size scoring of microsatellite loci around dhps.
Two hundred microliters of heparinized blood samples was collected from malaria patients infected with P. falciparum from different parts of India (Uttar Pradesh [U.P.; Aligarh and Ghaziabad], Assam [Kamrup]), Andaman and Nicobar Islands [Car Nicobar], Orissa [Ganjam and Jagatsingh Pur], and Madhya Pradesh [M.P.; Jabalpur]) according to institutional ethical guidelines. These were the same samples which were used for the previous study to analyze the microsatellite markers around dhfr (12). Parasite DNA was extracted from a Bioneer Corporation (South Korea) genomic DNA extraction kit according to the manufacturer's protocol and used for PCR amplification. The msp1 gene and two neutral microsatellite loci on chromosomes 2 and 3 were amplified to exclude samples with multiple infections (see Table S1 in the supplemental material). For the remaining samples with single clonal infections, PCR amplification of the dhps gene (3) and its flanking microsatellite was done. The DNA was subjected to two rounds of PCR according to a primary nested strategy. The details of the primers and the cycling conditions are provided in Table S1 in the supplemental material. The PCR products were resolved on a 1.8% agarose gel, and the products were then purified and eluted using an Accuprep Gel purification kit (Bioneer Corporation, South Korea) according to the manufacturer's protocol. The amplified products were then sequenced for dhps mutation detection at codons 436, 437, 540, 581, and 613. The purified products of microsatellite loci were diluted in a ratio of 1:10 and then separated on an ABI 3130xl genetic analyzer and analyzed using GeneMapper software (version 3.7; Applied Biosystems, Foster City, CA).
Microsatellite polymorphism.
A total of 15 microsatellite loci flanking dhps were scored. Eight microsatellite loci (−30, −22.7, −8.9, −7.5, −3.43, −2.9, −1.5, −0.13) were located upstream and seven (0.005, 1.37, 3.9, 6.7, 8.97, 24.6, 28.6) were located downstream of the dhps gene. To estimate the genetic variation in these microsatellite loci, expected heterozygosity values (Hes) were calculated for each locus using the formula [n/(n − 1)] [1 − Σpi2], where n is the number of isolates and pi is the frequency of the ith allele. The Microsatellite Excel tool kit was used for calculating He values and allele frequencies (23). Various (unique) haplotypes of the microsatellite loci along with the dhps genotype were constructed using Arlequin software (version 3.0) (9).
RESULTS
Mutations in dhps.
A total of 193 single clonal infections from five different geographical areas, viz., Andaman and Nicobar Islands (islands close to Southeast Asia, n = 65), Madhya Pradesh (central India, n = 30), Orissa (eastern region of India, n = 35), Assam (northeast region of India, n = 30), and Uttar Pradesh (northern India, n = 33), with different malarial transmission rates and endemicities (10) were analyzed for their dhps sequences at five different codons (codons 436, 437, 540, 581, and 613) and 15 microsatellite loci. The dhps sequences at these codons have previously been reported for 92 of these 193 isolates (3, 11), and the remaining 101 isolates were sequenced in the present study. The wild-type dhps allele was predominant (58.0%) among these 193 isolates. It was also predominant in all the regions except Andaman and Nicobar Islands, where it was present in only 4.7% (n = 65) of the isolates (Table 1). Parasite populations from U.P., M.P., and Orissa had only two types of dhps alleles, compared to four and eight different types of dhps alleles from Assam and Andaman and Nicobar Islands, respectively. Isolates with certain genotypes, viz., AGKAA, SGKGA, AGEAA, SGEGA, AGNAA, and AGEGA, were present in Andaman and Nicobar Islands and were not found in other regions (Table 1). These results indicate the existence of higher levels of sulfadoxine resistance-associated mutations among parasite isolates from Andaman and Nicobar Islands than isolates from mainland India (Table 1).
Table 1.
Geographic distribution of dhps alleles among P. falciparum isolates from India
| Serial no. | dhps allelea | No. (%) of isolatesb |
|||||
|---|---|---|---|---|---|---|---|
| A & N (n = 65) | Assam (n = 30) | M.P. (n = 30) | Orissa (n = 35) | U.P. (n = 33) | Total (n = 193) | ||
| 1 | SAKAA | 3 (4.6) | 22 (73.3) | 29 (96.7) | 31 (88.6) | 27 (81.8) | 112 (58.0) |
| 2 | SGKAA | 1 (1.5) | 3 (10.0) | 1 (3.3) | 0 | 6 (18.2) | 11 (5.7) |
| 3 | AGKAA | 9 (13.8) | 1 (3.3) | 0 | 0 | 0 | 10 (5.1) |
| 4 | SGKGA | 10 (15.4) | 2 (6.7) | 0 | 0 | 0 | 12 (6.2) |
| 5 | SGEGA | 8 (12.3) | 0 | 0 | 0 | 8 (4.1) | |
| 6 | AGEAA | 13 (20.0) | 2 (6.7) | 0 | 4 (11.4) | 0 | 19 (9.8) |
| 7 | AGNAA | 19 (29.2) | 0 | 0 | 0 | 19 (9.8) | |
| 8 | AGEGA | 2 (3.1) | 0 | 0 | 0 | 0 | 2 (1.0) |
Mutated amino acids are boldfaced and underlined. Positions of amino acids shown are 436, 437, 540, 581, and 613, respectively.
n, number of isolates; A & N, Andaman and Nicobar Islands; M.P., Madhya Pradesh; U.P., Uttar Pradesh.
Genetic diversity at microsatellite loci.
All 193 isolates were analyzed for 15 microsatellite loci flanking the dhps gene. The genetic diversity in terms of He was calculated for each locus for all dhps alleles. The wild type showed more heterozygosity (mean He = 0.80 ± 0.14) than mutant dhps alleles (mean He = 0.55 ± 0.19) (P ≤ 0.05). The genotypes of two neutral microsatellite loci analyzed here showed higher He values (mean He = 0.76 ± 0.07) than the genotypes of the microsatellites surrounding the mutant dhps alleles (mean He = 0.55 ± 0.19).
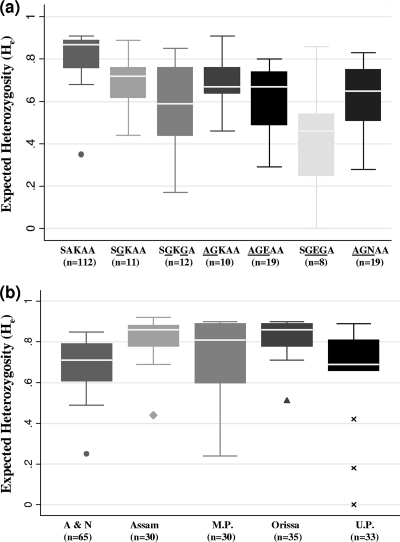
There were two types of double mutant dhps alleles in the population. Parasites with the SGKGA allele showed slightly lower (P = 1.00) heterozygosity (mean He = 0.58 ± 0.2) than those that showed the AGKAA allele (mean He = 0.69 ± 0.1) (Fig. 1a). When the heterozygosity values of triple mutant dhps alleles were compared with the value for the wild type, there was preferred selection for the SGEGA allele (mean He = 0.44 ± 0.2; P ≤ 0.05) over the AGEAA allele (mean He = 0.61 ± 0.16; P = 0.067). Genetic diversity in terms of He values for novel mutant allele AGNAA (mean He = 0.63 ± 0.14) was insignificantly lower than that for the wild-type allele (P = 0.18). Also, the selective sweep was wider and more symmetrical for the SGEGA allele than the AGEAA and AGNAA dhps alleles (see Table S2 in the supplemental material).
Fig. 1.
Microsatellite genetic diversity around different dhps alleles among all Indian P. falciparum isolates tested (a) and isolates from different geographical regions of India (b). The box for dhps allele AGEGA is not shown, as the number of isolates bearing this genotype was only two. Mutated amino acids are underlined. The expected heterozygosity around 15 loci flanking the dhps gene was calculated, and box plots show median, minimum, and maximum values and interquartile ranges. n, number of isolates; A & N, Andaman and Nicobar Islands; M.P., Madhya Pradesh; U.P., Uttar Pradesh.
Regional variation in genetic diversity around dhps.
There were regional differences in the genetic diversity around dhps alleles among Indian P. falciparum isolates (Fig. 1b; see Table S3 in the supplemental material). Genetic diversity around dhps alleles in terms of He values for isolates from Andaman and Nicobar Islands (mean He = 0.67 ± 0.15) was less than that for isolates from Assam (mean He = 0.80 ± 0.12; P = 0.0034), M.P. (mean He = 0.74 ± 0.19; P = 0.087), and Orissa (mean He = 0.82 ± 0.1; P = 0.0013). This could be expected because isolates with higher levels of sulfadoxine resistance-associated mutant alleles were present in Andaman and Nicobar Islands (Table 1). Furthermore, the majority of isolates from mainland India contained wild-type dhps and thus higher He values.
Distinct haplotypes.
Eleven loci were considered for constructing the haplotypes (±8.9 kb) of the dhps gene. We have observed 75 distinct haplotypes among mutant dhps alleles. Single mutant dhps allele SGKAA showed a decrease in heterozygosity and an increase in linkage among the nearest loci to dhps. There are two different lineages to double mutant dhps alleles AGKAA and SGKGA, as there is a lack of sharing of loci among the samples harboring double mutant dhps alleles (see Table S4 in the supplemental material). Certain microsatellite alleles were shared between double mutant allele SGKGA and triple mutant allele SGEGA, as well as quadruple mutant allele AGEGA (see Table S4 in the supplemental material). Haplotype H17 was the only common haplotype shared between SGKGA and SGEGA. We also observed microsatellite alleles shared between double mutant allele AGKAA and triple mutant allele AGEAA. Thus, haplotype construction suggests the multiple and distinct origins of mutant dhps alleles, with few alleles being shared in India (see Table S4 in the supplemental material).
DISCUSSION
Favorable mutated alleles get selected when drug pressure exists in the parasite population. This leads to hitchhiking of the markers flanking the gene and an increase in linkage disequilibrium. Genetic hitchhiking across the dhfr alleles had previously been reported from several countries (7, 12, 14, 16, 21, 22, 24, 28, 29), in contrast to fewer reports on dhps alleles (4, 25, 28, 33). Earlier, we have reported that antimalarial drug resistance is widespread in the Indian P. falciparum population (1–3, 20). We also reported that there is a selective sweep around dhfr mutant alleles among isolates in the Indian P. falciparum population (12). For example, quadruple mutant allele AIRNL showed a greater strength of hitchhiking than triple AIRNI and double ANRNI dhfr mutant alleles (12). The predominant haplotype of the mutant dhfr alleles was similar to that of isolates from Thailand, indicating the probable gene flow from the area of proximity (12). On the contrary, when these samples were analyzed in the present study for genetic hitchhiking around dhps mutant alleles, they showed greater genetic variability in the flanking microsatellites (see Tables S2 and S4 in the supplemental material). This indicates that dhps mutations are at a very early stage of fixation in the parasite population.
As the first mutation occurs at codon 437 (A437G), there is a decrease in He values and allelic skewing. A further decrease in heterozygosity occurs with an increased number of mutations at the dhps locus (Fig. 1a). These data are consistent with the observation that with the increased number of mutations in dhps, the 50% inhibitory concentration (IC50) of sulfadoxine against the parasite also increases proportionately (32). Further, there are significant differences in the extent of genetic diversity among some of the mutant dhps alleles. For example, triple mutant allele SGEGA shows greater fixation than double mutant dhps allele AGKAA (P = 0.04). Similarly, single mutant dhps allele SGKAA shows more variation than triple mutant allele SGEGA (P = 0.002). The He values observed around double and triple mutant dhps alleles in Indian isolates were higher than the He values reported from Southeast Asia (33). This indicates that, unlike Southeast Asia, there is a lack of a distinct selective sweep around mutant dhps alleles among Indian isolates (33).
There are 75 unique haplotypes observed among the mutant dhps alleles (see Table S4 in the supplemental material). On the basis of the results, it is quite clear that the resistance-associated dhps alleles in India have evolved on multiple genetic backgrounds, which is consistent with results of previous studies from South America (5, 16), Southeast Asia (4, 33), and Africa (25). The SGKGA and AGKAA double mutants originated independently on SGKAA genetic backgrounds (see Table S4 in the supplemental material). There are two major and several minor lineages of the double mutant alleles. The majority of the SGEGA triple mutant alleles and AGEGA quadruple mutants shared identical or closely related haplotypes with SGKGA, suggesting their common ancestry. Though most of AGEAA triple mutants were related to AGKAA double mutant backgrounds, there were also a number of isolates with unique microsatellite haplotypes. These different haplotypes could have arisen either independently or through recombination and mutation events in the limited number of ancestors. Thus, there are two major and independent origins for these two triple mutant alleles in India. The AGNAA allele, however, has a quite diverse microsatellite background, which is suggestive of the multiple origins for this triple mutant. We compared our data for 3 microsatellite loci located upstream (−0.3 kb, −1.5 kb, and −2.9 kb) with the published data from Thailand and Cambodia (4, 33) and found similar haplotypes, which probably suggests that these alleles might have originated from a common progenitor.
Sulfadoxine resistance-associated dhps mutant alleles are mainly concentrated in Andaman and Nicobar Islands and are least present in mainland India (3, 11). Indeed, parasite isolates with the wild-type dhps allele predominate in mainland India (Table 1) and show complete linkage equilibrium. Isolates from Andaman and Nicobar Islands have more triple mutant dhps alleles (AGEAA, SGEGA, and novel triple mutant AGNAA) than isolates from mainland India (11). Therefore, the genetic diversity is expectedly low on these islands compared to that in other geographical regions of the country (Fig. 1b). This leads to a regional bias in the genetic diversity values among flanking microsatellites of mutant dhps alleles, where minimum He values were observed in Andaman and Nicobar Islands (Fig. 1b). The regional variations in genetic diversity could also be explained because of differences in malarial transmission intensity (10), which contribute to differences in the distribution of the dhps alleles.
The novel mutation K540N in dhps seen among isolates from Andaman and Nicobar Islands (11) had also been reported from Cambodia and Thailand, but with a different genotype, i.e., SGNGA instead of the AGNAA genotype (4, 33).The contemporary mutation at K540E in triple mutant dhps allele AGEAA shows greater fixation (see Table S2 in the supplemental material) than AGNAA, suggesting that there could be a recent selection of the novel mutant, which may have a lower level of resistance than its contemporary partner. This is supported by computer modeling data for the DHPS protein, which suggest that a change to K540E causes a greater alteration in binding of sulfadoxine to its cleft than a change to K540N (11), and also by cell-based inhibition studies, which show slightly lower IC50s for the K540N mutant than the K540E mutant (13). Hence, K540N causes a moderate decrease in resistance to sulfadoxine compared to that caused by K540E.
A single origin of dhps alleles among South American isolates with some independent origins (5, 16) and multiple origins among African and Southeast Asian isolates have been reported (4, 24, 33). Our results are consistent with those of the previous studies, as we also observed multiple origins of dhps alleles in Indian P. falciparum isolates. Also, we did not observe any of the strong selective sweeps around dhps alleles observed for dhfr alleles in the same samples (12). This is in agreement with the assertion that under SP pressure dhfr mutations are fixed first in the parasite population, followed by dhps mutations. Therefore, the reason for this difference in selective sweeps could be that we have analyzed the flanking microsatellite markers for the dhps allele at a very early stage of their fixation, which otherwise, with the course of time, may lead to strong selective signatures. Furthermore, gene flow may be playing an important role in the population dynamics of the parasite, besides the possibility that recombination events break the linkage of flanking microsatellite markers with dhps alleles. Therefore, a strong surveillance system to check the migration of parasite populations across countries is strongly recommended. In conclusion, we observed multiple origins of dhps alleles and single/shared origins of dhfr alleles in India.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by financial assistance from the Department of Biotechnology (Government of India) and the Indian Council of Medical Research. V.L. received a senior research fellowship from the Council for Scientific and Industrial Research.
We are grateful to R. M. Pandey and Amit Srivastava for their help with statistical analysis of the data. The facility of the Bio-Technology Information System (BTIS) of the Biotechnology Department is gratefully acknowledged. We thank Shalini Narang for preparing the manuscript.
Footnotes
Supplemental material for this article may be found at http://aac.asm.org/.
Published ahead of print on 21 March 2011.
REFERENCES
- 1. Ahmed A., et al. 2004. Plasmodium falciparum isolates in India exhibit a progressive increase in mutations associated with sulfadoxine-pyrimethamine resistance. Antimicrob. Agents Chemother. 48:879–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ahmed A., Das M. K., Dev V., Saifi M. A., Wajihullah, Sharma Y. D. 2006. Quadruple mutations in dihydrofolate reductase of Plasmodium falciparum isolates from Car Nicobar Island, India. Antimicrob. Agents Chemother. 50:1546–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ahmed A., Lumb V., Das M. K., Dev V., Wajihullah, Sharma Y. D. 2006. Prevalence of mutations associated with higher levels of sulfadoxine-pyrimethamine resistance in Plasmodium falciparum isolates from Car Nicobar Island and Assam, India. Antimicrob. Agents Chemother. 50:3934–3938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Alam M. T., et al. 2011. Tracking origins and spread of sulfadoxine-resistant Plasmodium falciparum dhps alleles in Thailand. Antimicrob. Agents Chemother. 55:155–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bacon D. J., et al. 2009. Dynamics of malaria drug resistance patterns in the Amazon Basin region following changes in Peruvian national treatment policy for uncomplicated malaria. Antimicrob. Agents Chemother. 53:2042–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brooks D. R., et al. 1994. Sequence variation of the hydroxymethyldihydropterin pyrophosphokinase: dihydropteroate synthase gene in lines of the human malaria parasite, Plasmodium falciparum, with differing resistance to sulfadoxine. Eur. J. Biochem. 224:397–405 [DOI] [PubMed] [Google Scholar]
- 7. Certain L. K., et al. 2008. Characteristics of Plasmodium falciparum dhfr haplotypes that confer pyrimethamine resistance, Kilifi, Kenya, 1987-2006. J. Infect. Dis. 197:1743–1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cortese J. F., Caraballo A., Contreras C. E., Plowe C. V. 2002. Origin and dissemination of Plasmodium falciparum drug-resistance mutations in South America. J. Infect. Dis. 186:999–1006 [DOI] [PubMed] [Google Scholar]
- 9. Excoffier L., Laval G., Schneider S. 2005, posting date Arlequin version 3.0: an integrated software package for population genetics data analysis. http//cmpg.unibe.ch/software/arlequin3/ Zoological Institute, University of Bern, Bern, Switzerland: [PMC free article] [PubMed] [Google Scholar]
- 10. Kumar A., Valecha N., Jain T., Dash A. P. 2007. Burden of malaria in India: retrospective and prospective view. Am. J. Trop. Med. Hyg. 77:69–78 [PubMed] [Google Scholar]
- 11. Lumb V., et al. 2009. Emergence of an unusual sulfadoxine-pyrimethamine resistance pattern and a novel K540N mutation in dihydropteroate synthetase in Plasmodium falciparum isolates obtained from Car Nicobar Island, India, after the 2004 tsunami. J. Infect. Dis. 199:1064–1073 [DOI] [PubMed] [Google Scholar]
- 12. Lumb V., Das M. K., Singh N., Dev V., Wajihullah, Sharma Y. D. 2009. Characteristics of genetic hitchhiking around dihydrofolate reductase gene associated with pyrimethamine resistance in Plasmodium falciparum isolates from India. Antimicrob. Agents Chemother. 53:5173–5180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lumb V., Sharma Y. D. 22 February 2011, posting date Novel K540N mutation in Plasmodium falciparum dihydropteroate synthetase confers a lower level of sulfa drugs resistance than K540E mutation. Antimicrob. Agents and Chemother. [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Maiga O., et al. 2007. A shared Asian origin of the triple-mutant dhfr allele in Plasmodium falciparum from sites across Africa. J. Infect. Dis. 196:165–172 [DOI] [PubMed] [Google Scholar]
- 15. McCollum A. M., Basco L. K., Tahar R., Udhayakumar V., Escalante A. A. 2008. Hitchhiking and selective sweeps of Plasmodium falciparum sulfadoxine and pyrimethamine resistance alleles in a population from central Africa. Antimicrob. Agents Chemother. 52:4089–4097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McCollum A. M., Mueller K., Villegas L., Udhayakumar V., Escalante A. A. 2007. Common origin and fixation of Plasmodium falciparum dhfr and dhps mutations associated with sulfadoxine-pyrimethamine resistance in a low-transmission area in South America. Antimicrob. Agents Chemother. 51:2085–2091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McCollum A. M., et al. 2006. Antifolate resistance in Plasmodium falciparum: multiple origins and identification of novel dhfr alleles. J. Infect. Dis. 194:189–197 [DOI] [PubMed] [Google Scholar]
- 18. Mita T., et al. 2009. Indigenous evolution of Plasmodium falciparum pyrimethamine resistance multiple times in Africa. J. Antimicrob. Chemother. 63:252–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mita T., et al. 2007. Independent evolution of pyrimethamine resistance in Plasmodium falciparum isolates in Melanesia. Antimicrob. Agents Chemother. 51:1071–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mittra P., et al. 2006. Progressive increase in point mutations associated with chloroquine resistance in Plasmodium falciparum isolates from India. J. Infect. Dis. 193:1304–1312 [DOI] [PubMed] [Google Scholar]
- 21. Nair S., et al. 2003. A selective sweep driven by pyrimethamine treatment in Southeast Asian malaria parasites. Mol. Biol. Evol. 20:1526–1536 [DOI] [PubMed] [Google Scholar]
- 22. Nash D., et al. 2005. Selection strength and hitchhiking around two anti-malarial resistance genes. Proc. Biol. Sci. 272:1153–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Park S. D. E. 2001. Trypanotolerance in West African cattle and the population genetic effects of selection. Ph.D. thesis University of Dublin, Dublin, Ireland [Google Scholar]
- 24. Pearce R., et al. 2005. Reduced variation around drug-resistant dhfr alleles in African Plasmodium falciparum. Mol. Biol. Evol. 22:1834–1844 [DOI] [PubMed] [Google Scholar]
- 25. Pearce R. J., et al. 2009. Multiple origins and regional dispersal of resistant dhps in African Plasmodium falciparum malaria. PLoS Med. 6:e1000055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Peterson D. S., Walliker D., Wellems T. E. 1988. Evidence that a point mutation in dihydrofolate reductase-thymidylate synthase confers resistance to pyrimethamine in falciparum malaria. Proc. Natl. Acad. Sci. U. S. A. 85:9114–9118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Plowe C. V., et al. 1997. Mutations in Plasmodium falciparum dihydrofolate reductase and dihydropteroate synthase and epidemiologic patterns of pyrimethamine-sulfadoxine use and resistance. J. Infect. Dis. 176:1590–1596 [DOI] [PubMed] [Google Scholar]
- 28. Roper C., et al. 2003. Antifolate antimalarial resistance in southeast Africa: a population-based analysis. Lancet 361:1174–1181 [DOI] [PubMed] [Google Scholar]
- 29. Roper C., et al. 2004. Intercontinental spread of pyrimethamine-resistant malaria. Science 305:1124. [DOI] [PubMed] [Google Scholar]
- 30. Sirawaraporn W., Sirawaraporn R., Cowman A. F., Yuthavong Y., Santi D. V. 1990. Heterologous expression of active thymidylate synthase-dihydrofolate reductase from Plasmodium falciparum. Biochemistry 29:10779–10785 [DOI] [PubMed] [Google Scholar]
- 31. Triglia T., Cowman A. F. 1994. Primary structure and expression of the dihydropteroate synthetase gene of Plasmodium falciparum. Proc. Natl. Acad. Sci. U. S. A. 91:7149–7153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Triglia T., Menting J. G., Wilson C., Cowman A. F. 1997. Mutations in dihydropteroate synthase are responsible for sulfone and sulfonamide resistance in Plasmodium falciparum. Proc. Natl. Acad. Sci. U. S. A. 94:13944–13949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vinayak S., et al. Origin and evolution of sulfadoxine resistant Plasmodium falciparum. PLoS Pathog. 6:e1000830. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.