Abstract
Proteus mirabilis isolates obtained in 1999 to 2008 from three European countries were analyzed; all carried chromosomal AmpC-type cephalosporinase blaCMY genes from a Citrobacter freundii origin (blaCMY-2-like genes). Isolates from Poland harbored several blaCMY genes (blaCMY-4, blaCMY-12, blaCMY-14, blaCMY-15, and blaCMY-38 and the new gene blaCMY-45), while isolates from Italy and Greece harbored blaCMY-16 only. Earlier isolates with blaCMY-4 or blaCMY-12, recovered in France from Greek and Algerian patients, were also studied. All isolates showed striking similarities. Their blaCMY genes resided within ISEcp1 transposition modules, named Tn6093, characterized by a 110-bp distance between ISEcp1 and blaCMY, and identical fragments of both C. freundii DNA and a ColE1-type plasmid backbone. Moreover, these modules were inserted into the same chromosomal site, within the pepQ gene. Since ColE1 plasmids carrying ISEcp1 with similar C. freundii DNA fragments (Tn6114) had been identified earlier, it is likely that a similar molecule had mediated at some stage this DNA transfer between C. freundii and P. mirabilis. In addition, isolates with blaCMY-12, blaCMY-15, and blaCMY-38 genes harbored a second blaCMY copy within a shorter ISEcp1 module (Tn6113), always inserted downstream of the ppiD gene. Sequence analysis of all mobile blaCMY-2-like genes indicated that those integrated in the P. mirabilis chromosome form a distinct cluster that may have evolved by the stepwise accumulation of mutations. All of these observations, coupled to strain typing data, suggest that the blaCMY genes studied here may have originated from a single ISEcp1-mediated mobilization-transfer-integration process, followed by the spread and evolution of a P. mirabilis clone over time and a large geographic area.
INTRODUCTION
Acquired cephalosporinases of the AmpC type constitute a significant source of resistance to most of the newer-generation β-lactams in Enterobacteriaceae (3, 18, 29). Similar to their natural precursors in several Gram-negative species, e.g., Aeromonas spp., Enterobacter spp., Morganella morganii, or Citrobacter freundii, these enzymes hydrolyze penicillins, most cephalosporins, and aztreonam and are poorly inhibited by β-lactam inhibitors (5, 18). Usually, acquired AmpCs are expressed constitutively, conferring resistance to all of their substrates and inhibitor combinations, although resistance levels depend on the amounts of enzymes produced, as well as the presence of other resistance mechanisms (18, 29). The acquired ampC genes have escaped from the chromosome of some species to plasmids, following mobilizations mediated by such elements as IS26, ISEcp1, or ISCR1 (14, 21, 24, 27, 34). The identity of these elements, their location with respect to the ampC gene, and the size of the DNA fragment mobilized are diagnostic of specific escape events. Mobilizations of the C. freundii ampC have given rise to blaCMY genes observed in many species worldwide (1, 18, 29). With more than 40 genes identified thus far in this family, in which blaCMY-2 is the most common, this is the largest group of mobile ampCs (18, 29). blaCMY-2 may also be the precursor of other blaCMY genes through the accumulation of neutral mutations (1).
ISEcp1 has played an important role in mobilizing blaCMY-2-like genes, since it is often found at their 5′ flanks (14, 21, 23, 26, 32). It may transpose together with adjacent DNA fragments on the 3′ side, producing transposition modules of various sizes. This process utilizes sequences similar to the ISEcp1 inverted right repeat (IRR), and such alternative IRRs mark precisely the modules' 3′ ends (20). ISEcp1 has been identified 116 bp upstream from blaCMY-2, blaCMY-4, blaCMY-7, blaCMY-21, and blaCMY-23 genes in plasmids of the IncA/C and IncI1 groups disseminated worldwide among Escherichia coli, Klebsiella pneumoniae, Salmonella enterica, and Proteus mirabilis (14–16, 26, 32, 36, 37). It has also been found 110 bp upstream from blaCMY-5, blaCMY-31, and blaCMY-36 genes in highly similar ColE1-type plasmids in Klebsiella oxytoca (pTKH11), S. enterica serovar Newport (pA172), and K. pneumoniae (pH 205) from Sweden, the United States, and Greece, respectively (38, 39).
Chromosomal blaCMY genes (blaCMY-3, blaCMY-4, and blaCMY-12) in P. mirabilis were first observed sporadically in France, in patients with Greek and Algerian origins (4, 11). Later, P. mirabilis isolates with ISEcp1 close to blaCMY-2-like genes in their chromosome were also found in Poland (blaCMY-4, blaCMY-12, blaCMY-14, blaCMY-15, and blaCMY-38) (13, 21), Italy (blaCMY-16) (9, 23), and Greece (25). The present international molecular epidemiology study aimed to compare all of the above P. mirabilis isolates, in order to assess the clonality of blaCMY-2-like gene sequences, their transposition modules, and insertion regions, as well as of the bacterial chromosomes themselves.
MATERIALS AND METHODS
Bacterial strains.
Twenty-one CMY-producing P. mirabilis isolates were studied (Table 1). Eighteen had been recovered between 1999 and 2008 in hospitals of different cities of Poland, Greece, and Italy and were selected (six isolates per country) from larger groups of isolates partially described previously (9, 13, 21, 23, 25, 35). The Polish isolates belonged to various pulsed-field gel electrophoresis (PFGE) patterns and harbored all of the blaCMY genes identified in P. mirabilis in that country thus far; the Italian and Greek isolates represented different PFGE patterns and/or clinical centers where these organisms have been studied to date. The three remaining P. mirabilis strains with chromosomal blaCMYs, kindly provided by G. Arlet, were isolated in hospitals in Paris from patients originating either from either Greece or Algeria. Strains 22317 and PLAR (Greece) were among the earliest CMY-4 producers reported, while strain 34955 (Algeria) was the first CMY-12 producer ever identified (11). Seven AmpC-negative, epidemiologically unrelated P. mirabilis isolates were included for comparative typing; these had been collected from different Greek hospitals from 2006 to 2008.
Table 1.
P. mirabilis isolates: basic information from previous studies and the present study
| Strain | Yr of isolationa | Country | City | Specimen | CMY variant | Other β-lactamase(s) | ISEcp1 modules | Ribotype | Source or reference |
|---|---|---|---|---|---|---|---|---|---|
| PL 6735/99 | 1999 | Poland | Warsaw | Urine | CMY-14 | TEM-1 | Tn6093 | R5 | 21 |
| PL 1662/00 | 2000 | Poland | Warsaw | Pus | CMY-15 | TEM-2 | Tn6093, Tn6113 | R5 | 21 |
| PL 27/00 | 1999 | Poland | Grajewo | Urine | CMY-12 | TEM-2 | Tn6093, Tn6113 | R5 | 21 |
| PL 1376/00 | 2000 | Poland | Zielona Góra | Urine | CMY-45 | TEM-1 | Tn6093 | R6 | This study |
| PL 864/01 | 2001 | Poland | Lublin | Sputum | CMY-4 | TEM-1 | Tn6093 | R5 | 21 |
| PL 1455/04 | 2004 | Poland | Łódź | Urine | CMY-38 | TEM-2 | Tn6093, Tn6113 | R5 | 13 |
| IT NO-051/03 | 2003 | Italy | Novara | Cutaneous ulcer | CMY-16 | TEM-1 | Tn6093 | R5 | 9 |
| IT VA-1017/03 | 2003 | Italy | Varese | Urine | CMY-16 | TEM-1 | Tn6093 | R5 | 9 |
| IT VA-832/05 | 2005 | Italy | Varese | Urine | CMY-16 | TEM-1 | Tn6093 | R5 | 23 |
| IT VA-070/06 | 2006 | Italy | Varese | Urine | CMY-16 | TEM-1 | Tn6093 | R5 | 23 |
| IT VA-414/06 | 2006 | Italy | Varese | Urine | CMY-16 | TEM-1 | Tn6093 | R5 | 23 |
| IT LC-10/08 | 2008 | Italy | Lecco | Blood | CMY-16 | TEM-1 | Tn6093 | R5 | This study |
| GR 17/04 | 2004 | Greece | Thessaloniki | Urine | CMY-16 | VIM-1, TEM-1 | Tn6093 | R5 | 35 |
| GR 15184/05 | 2005 | Greece | Thessaloniki | Urine | CMY-16 | VIM-1, TEM-1 | Tn6093 | R5 | 35 |
| GR 15315/05 | 2005 | Greece | Thessaloniki | Urine | CMY-16 | VIM-1, TEM-1 | Tn6093 | R5 | 35 |
| GR 2506/07 | 2007 | Greece | Athens | Sputum | CMY-16 | VIM-1, TEM-1 | Tn6093 | R5 | This study |
| GR 2530/07 | 2007 | Greece | Athens | Urine | CMY-16 | VIM-1, TEM-1 | Tn6093 | R5 | This study |
| GR 2720/08 | 2008 | Greece | Athens | Sputum | CMY-16 | VIM-1, TEM-1 | Tn6093 | R5 | 25 |
| 22317 | – | France/Greece | Paris | Bile | CMY-4 | TEM-1 | Tn6093 | R7 | 11 |
| PLAR | – | France/Greece | Paris | Feces | CMY-4 | TEM-1 | Tn6093 | R7 | 11 |
| 34955 | – | France/Algeria | Paris | Urinary catheter | CMY-12 | TEM-2 | Tn6093, Tn6113 | R5 | 11 |
| GR-485-S | 2007 | Greece | Athens | Urine | No | None | No | R4 | This study |
| GR-20A-S | 2007 | Greece | Athens | Urine | No | None | No | NDb | This study |
| GR-99-S | 2007 | Greece | Athens | Pus | No | None | No | R1 | This study |
| GR-106-S | 2007 | Greece | Athens | Pus | No | None | No | R1 | This study |
| GR-20-S | 2008 | Greece | Athens | Pus | No | None | No | R2 | This study |
| GR-28-S | 2008 | Greece | Athens | Pus | No | None | No | R2 | This study |
| GR-59-S | 2008 | Greece | Athens | Sputum | No | None | No | R3 | This study |
–, The date of isolation was not provided in the reference paper (11).
ND, not determined.
Detection and sequencing of blaCMY genes.
The entire coding regions of blaCMY genes were amplified with the primers ampC1 and ampC2 (11) and directly sequenced by using several blaCMY-specific primers (Table 2). blaCMY sequences were compared for evolutionary relationships by using the MEGA v3.1 software (19). Subsequently, the sequences, prealigned in Mauve 2.3.0 (10), were analyzed by using CloneFrame 1.1 software (12), and the results were visualized by using SplitsTree 4.10 (17) as a consensus network. DNA sequencing in all experiments of the present study was performed either in-house, using the Applied Biosystems technology and equipment (Foster City, CA), or by an external facility (Macrogen, Inc., Seoul, Korea).
Table 2.
Oligonucleotides used in this study
| Primer | Gene | Oligonucleotide sequence (5′–3′) | Positiona | Purpose | Source or reference |
|---|---|---|---|---|---|
| ampC1 | blaCMY | ATG ATG AAA AAA TCG TTA TGC | 1–21* | PCR, sequencing | 11 |
| ampC2 | blaCMY | TTG CAG CTT TTC AAG AAT GCG C | 1122–1143* | PCR, sequencing | 11 |
| ampC5 | blaCMY | CAG CGT TTG CTG CGT G | 222–237* | PCR, sequencing | This study |
| E4/F | blaCMY | TGG GTT CAG GCC AAC ATG GAT GC | 757–779* | PCR mapping, sequencing | This study |
| E7/R | blaCMY | TGC CAG CAT CAC GAT GCC AAG G | 1059–1080* | PCR mapping, sequencing | This study |
| ecpF2 | 3′ end of ISEcp1 | GTT GCT CTG TGG ATA ACT TG | 2084–2102† | PCR, sequencing | 39 |
| ecpR1 | ISEcp1 | CCT AAA TTC CAC GTG TGT | 2247–2264† | PCR | This study |
| ISEcp1/F | tnpA of ISEcp1 | CAT GCT CTG CGG TCA CTT C | 959–977† | PCR mapping | This study |
| E8/F | Downstream of blaCMY | CCA GGA TAT TGG GCC TC | 3534–3550† | PCR mapping | This study |
| blc/R | blc | GAC AAC CAG GAA TGC AGC | 3647–3664† | PCR mapping | This study |
| sugE/R | sugE | GCC TGA TAT GTC CTG GAT CGT | 4448–4468† | PCR mapping | This study |
| ORF6/R | orf6 | CTT CAT CCC TAT CAT CGC CA | 5634–5653† | PCR mapping | This study |
| ORF6e/R | orf6 | AAT CAG CAA TAA CAT CAC CAT G | 5770–5791† | PCR mapping | This study |
| mobB/R | mobB | TAG AGC AGC AGA AGC CAG CT | 6205–6224† | PCR mapping | This study |
| RNAIIp/R | RNAII | TCA TTC CAC GCC TGA CAC TC | 6741–6760† | PCR mapping | This study |
| pepQ/F | pepQ | CAC CTG TTG ATT ATT GGC AT | 3872704–3872723‡ | PCR mapping, sequencing | This study |
| pepQ/R | pepQ | TCA GGC TAA ATG TAA GTC TC | 3873769–3873787‡ | PCR mapping, sequencing | This study |
| blc/F | blc | CTG TGG ATC CTT TCA CGG A | 4025–4043† | Inverse PCR | This study |
| C12-tnpA/r | tnpA of ISEcp1 | TAT TCT GAA GAG TCC AAG GAA | 1921–1901† | Inverse PCR | 6 |
| ppiD/F | Intergenic region PMI0120/ppiD | GCG ATT ACT GAA TGC CAT C | 150710–150692‡ | PCR mapping, sequencing | This study |
| ppiD/R | ppiD | CAA CGG CAG AAC AGC TTG | 150366–150383‡ | PCR mapping, sequencing | This study |
*, Position numbering is according to the first nucleotide of the coding sequence of the blaCMY gene; †, position numbering is according to the sequence of plasmid pH 205 (GenBank acc. no. EU331426) (39); ‡, position numbering is according to the sequence of the P. mirabilis HI4320 genome (GenBank accession no. AM942759) (28).
Detection of the ISEcp1 element.
The ISEcp1 element was detected upstream of blaCMY genes by PCR with the primers ecpF2 and ampC5 (Table 2), which were also used for sequencing the spacer region between ISEcp1 and the blaCMY genes.
Assays of blaCMY locus number and localization.
The number of loci containing blaCMY genes was assessed by hybridization of blaCMY and ISEcp1 probes with chromosomal DNA digested with EcoRI and HindIII restriction enzymes (MBI Fermentas, Vilnius, Lithuania), as described previously (21). The blaCMY (1,143-bp) and ISEcp1 (181-bp) probes were obtained by amplifying DNA from isolate PL 1662/00 with the primer pairs ampC1/ampC2 and ecpF2/ecpR1 (Table 2), respectively. Chromosomal versus plasmidic localization of blaCMY genes was assessed by I-CeuI analysis (22), as described previously (9).
Partial cloning and sequencing of ISEcp1-blaCMY modules.
DNA from all Polish and French isolates was digested with EcoRI and HindIII, and ligated into plasmid pHSG398 (33). E. coli DH5α electroporants were selected with 2 μg of cefotaxime/ml and 25 μg of chloramphenicol/ml (Sigma-Aldrich, St. Louis, MO). Recombinant plasmids were checked for inserts of ∼4.2 kb (“main blaCMY locus”) and ∼3.1 kb (“additional blaCMY locus”). Sequencing of the inserts was performed using primers specific for insert-flanking vector sequences, ISEcp1 and blaCMYs (Table 2), and other primers designed according to the accumulating sequence. Sequences were analyzed with the DNASTAR Lasergene software (Madison, WI).
Characterization of entire ISEcp1-blaCMY modules and their chromosomal insertion site: “main locus.”
The structure and site of integration of ISEcp1-blaCMY transposition modules within the P. mirabilis chromosome at the “main locus” was initially investigated by inverse PCR. Total DNA (1 μg) of isolate IT NO-051/03 was digested by AgeI (New England Biolabs, Inc., Beverly, MA). The digestion mixture, purified with the Wizard SV Gel and PCR Cleanup system (Promega, Madison, WI), was diluted 1:10 and then self-ligated by using T4 DNA ligase (Promega). The ligation mixture (2 μl) was then used in a PCR with the primers blc/F and C12-tnpA/r (7) (Table 2) to amplify the adjacent regions of the ISEcp1-blaCMY module. Sequences flanking the module were compared to the genomic sequence of P. mirabilis strain HI4320 (GenBank accession no. AM942759) (28), using the microbial BLAST tool (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi). For the remaining isolates, the module structure and the integration site were analyzed by PCR mapping using primers targeting various regions (Table 2 and Fig. 1), designed based on sequences of the inverse PCR product and the P. mirabilis HI4320 locus pepQ (positions 3872453 to 3873787) (28).
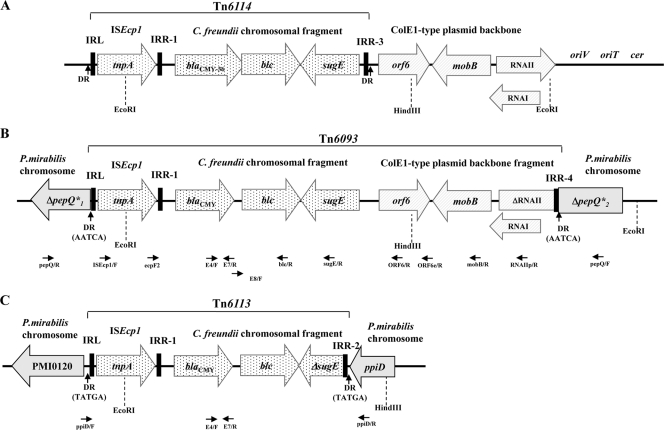
Fig. 1.
Schematic representation of ISEcp1 transposition modules containing the C. freundii-derived blaCMY genes, enzyme restriction sites, and primer hybridization sites used in the analysis of the Tn6093 and Tn6113 structures. (A) Tn6114 module present in the pH 205 plasmid; (B) Tn6093 module inserted into the P. mirabilis pepQ gene (“main locus”); (C) Tn6113 module inserted between ppiD and PMI0120 ORF in the P. mirabilis chromosome (“additional locus”). The schemes are aligned according to the ISEcp1 position. EcoRI and HindIII sites, used in cloning experiments, are indicated by dashed vertical lines. Black arrows below the Tn6093 and Tn6113 schemes represent primers used for PCR mapping and sequencing.
Analysis of the insertion site for ISEcp1-blaCMY modules: “additional locus.”
Sequences 3′ to the ISEcp1-blaCMY modules of the “additional locus,” determined by sequencing of cloned fragments, were compared to the genomic sequence of P. mirabilis HI4320 (28). The matching sequence of the ppiD region was then used to design primers for mapping the modules in the spacer between the ppiD gene (positions 148619 to 150496) and the PMI0120 open reading frame (ORF; 150751 to 151125) (Table 2 and Fig. 1) and for sequencing the 5′ junction.
Molecular typing.
Ribotyping was performed after HincII (New England Biolabs) digestion of genomic DNA, as described by Pignato et al. (30). For PFGE, total P. mirabilis DNA in agarose plugs was prepared as described previously (8), digested with NotI and SfiI (New England Biolabs), and electrophoresed using a CHEF III Bio-Rad apparatus (Hercules, CA). All electrophoretic patterns were analyzed using GelCompar v4.1 (Applied Maths NV, Sint-Martens-Latem, Belgium), using the Dice coefficient and clustering by UPGMA (unweighted pair-group method with arithmetic averages), with 1% tolerance in band position differences. For the NotI+SfiI PFGE patterns, a similarity cutoff of 80% was applied for discerning clusters of related isolates (31).
Antimicrobial susceptibility testing.
The MICs of β-lactams were evaluated by the agar dilution method according to Clinical and Laboratory Standards Institute criteria (6). For the isolates tested earlier, the methodology was described in previous reports (9, 11, 21, 23, 25, 35).
Nucleotide sequence accession numbers.
Sequences of the blaCMY-45 gene coding region and of the Tn6113 (blaCMY-12) and Tn6093 (blaCMY-16) modules will appear in the EMBL database under accession numbers FN546177, FR716828, and FM995219, respectively.
RESULTS
Sequence and location of the blaCMY genes in P. mirabilis isolates.
A total of 21 P. mirabilis isolates producing CMY-2-like β-lactamases from different countries and periods were investigated (Table 1). The sequences of the blaCMY genes were already known in five isolates from Poland (blaCMY-4, blaCMY-12, blaCMY-14, blaCMY-15, and blaCMY-38), five isolates from Italy (blaCMY-16), and three isolates from France (although from patients of Greek and Algerian origins [blaCMY-4 and blaCMY-12, respectively]) (9, 11, 13, 21, 23). PCR and sequencing showed that the remaining eight isolates carried either blaCMY-16 (the recent isolates from Greece and the IT LC-10/08 isolate from Italy) or a new allele (the PL 1376/00 isolate from Poland), named blaCMY-45, which differs from blaCMY-2 (2) by four point mutations: C418A (Gln140Lys), G511T (Ala171Ser), T661C (Trp221Arg), and G1019T (Gly340Val).
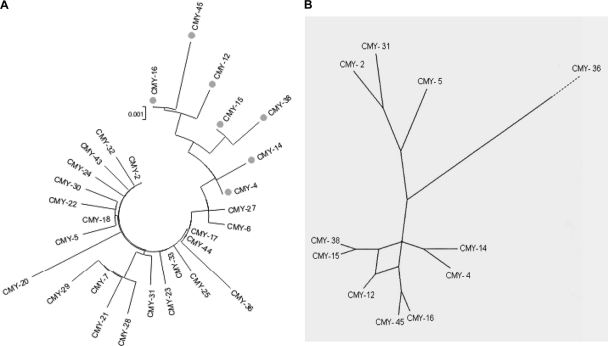
Comparison to other known blaCMY-2-like alleles indicated that those carried by our P. mirabilis isolates form a distinct cluster of more closely related sequences (Fig. 2). Sequence comparisons also suggested that the present study's alleles may have evolved from blaCMY-4 by stepwise mutations; blaCMY-12 was notable in that it shared, on the one hand, the G511T (Ala171Ser) mutation with blaCMY-16 and blaCMY-45, and, on the other, the G1088A (Ser363Asn) mutation with blaCMY-15 and blaCMY-38 (Fig. 2B).
Fig. 2.
Sequence comparison of blaCMY-2-like genes. (A) Dendrogram of all mobile blaCMY-2-like gene sequences obtained with the MEGA v3.1 software (19); (B) consensus network of the blaCMY-2-like genes located in ISEcp1 modules with a 110-bp distance between ISEcp1 and blaCMY (plasmidic Tn6114-like, and P. mirabilis Tn6093-like modules). The consensus network was generated with the SplitsTree 4.10 software (17), after prealignment in Mauve 2.3.0 (10) and analysis by the CloneFrame 1.1 software (12).
I-CeuI analysis proved (or confirmed) that the blaCMY genes were chromosome-borne in all studied isolates (data not shown). PCR and sequencing assays revealed (or confirmed) the presence of an ISEcp1 element inserted 110bp upstream of the blaCMY genes in all isolates (thus correcting the value of 106 bp previously published for blaCMY-4, blaCMY-12, blaCMY-14, and blaCMY-15 [21]). Southern blot experiments carried out with genomic DNAs digested with EcoRI (which cuts inside ISEcp1) and HindIII (which does not cut inside either ISEcp1 or any of the blaCMYs) and using either a blaCMY or a ISEcp1 probe yielded a hybridizing band of ∼4.2 kb with all isolates (“main blaCMY locus”) and another band of ∼3.1 kb with the isolates carrying blaCMY-12, blaCMY-15, and blaCMY-38 (“additional blaCMY locus”). These results revealed (or confirmed) that, while in most isolates the blaCMY gene was present in a single copy, two copies were present in isolates with blaCMY-12, blaCMY-15, and blaCMY-38 from Poland and France (Algeria) (21).
Genetic context of the blaCMY genes and their integration sites.
The genetic context of the blaCMY genes and their integration sites were investigated by a combination of cloning, inverse PCR, and PCR mapping and sequencing, as detailed in Materials and Methods. The results showed that all of the isolates carried variants of a 6,210-bp ISEcp1-blaCMY module, named Tn6093, integrated into the P. mirabilis pepQ gene coding for Xaa-Pro dipeptidase (28). In all isolates, Tn6093 was inserted in the same position within pepQ and was flanked by 5-bp direct repeats (AATCA), indicating that the ISEcp1-mobilized module had integrated by transposition into the P. mirabilis chromosome. Tn6093 contained ISEcp1, followed by the C. freundii-derived blaCMY, blc, and sugE genes, a part of the ColE1-type plasmid backbone (regions orf6, mobB, and ΔRNAII), and terminating with an ISEcp1 IRR-like sequence, named IRR-4 (Fig. 1). Tn6093 matched very well fragments of plasmids pTKH11, pA172, and pH 205 with ISEcp1 modules called Tn6114, carrying complete blaCMY, blc, and sugE genes and terminating with the IRR-like motif IRR-3 (Fig. 1) (38, 39). Compared to the homologous part of pH 205 (39), the Tn6093 variants had, apart from variable mutations within blaCMYs, eight stable single nucleotide polymorphisms: one between ISEcp1 and blaCMY and seven downstream of Tn6114. The Tn6093-containing locus gave rise to the ∼4.2-kb band (“main blaCMY locus”) recognized in Southern blot experiments with all P. mirabilis isolates.
In the isolates with two blaCMY copies, a second ISEcp1-blaCMY module, 3,783 bp long and named Tn6113, was inserted into the P. mirabilis intergenic spacer between ORF PMI0120 and the ppiD gene encoding peptidyl-prolyl cis-trans isomerase D (28). In all of these isolates, Tn6113 was inserted at the same position, 154 bp downstream of ppiD, and was flanked by 5-bp direct repeats (TATGA), which indicated that this module also had integrated into the chromosome by transposition. Tn6113 consisted of ISEcp1, followed by the C. freundii-like sequences blaCMY, blc, and ΔsugE, and terminating with an IRR-like motif inside sugE, named IRR-2 (Fig. 1). Therefore, Tn6113 was shorter by 247 bp than Tn6114. The Tn6113-containing locus gave rise to the ∼3.1-kb band (“additional blaCMY locus”) identified in Southern blot experiments with the P. mirabilis isolates carrying two blaCMY copies; the same blaCMY allele was always present in both Tn6093 and Tn6113 modules.
Clonality of the blaCMY-carrying P. mirabilis isolates.
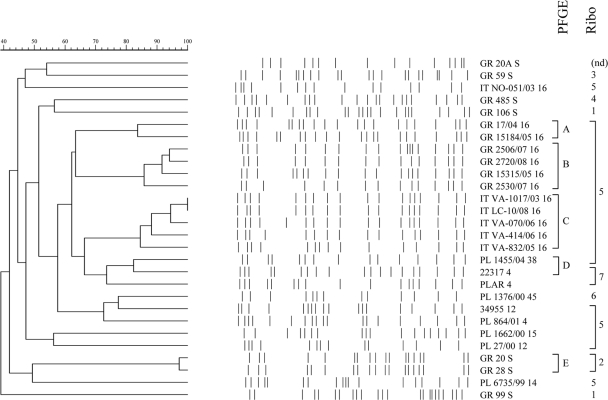
Given the identical structures and insertion sites of blaCMY-carrying elements, we investigated the possible clonality of P. mirabilis chromosomes using ribotyping and PFGE of genomic DNA after NotI-SfiI digestion. All blaCMY-carrying isolates belonged to three ribotypes. The main one, R5, grouped 18 isolates, while the remaining three isolates belonged to two other ribotypes (R6, n = 1; R7, n = 2). In contrast, the six blaCMY-negative control isolates tested, belonged to four distinct ribotypes, R1 to R4 (Fig. 3). As expected, PFGE profiling resulted in a somewhat higher discrimination, nevertheless confirming the considerable relatedness of blaCMY-positive isolates compared to that of blaCMY-negative ones (Fig. 3). Clonal relationships similar to those revealed by ribotyping were also obtained when the I-CeuI typing patterns were compared, as expected, given that this enzyme cuts within rrn loci (data not shown).
Fig. 3.
Dendrogram based on NotI+SfiI PFGE patterns, indicating clusters of the resulting types, as well as ribotypes. The bands arising after NotI+SfiI PFGE for each isolate are shown. The number next to the isolate code denotes the CMY variant. PFGE type clusters (>80% similarity, as per the percent scale at top left) are labeled A to E; ribotypes 1 to 7 are shown to the right of these.
Antimicrobial susceptibility testing.
The β-lactam resistance patterns of the study isolates were typical as for producers of CMY-type cephalosporinases (1, 21, 29), with high-level resistance to penicillins, cefoxitin, cefotaxime, and ceftazidime (Table 3). However, there were significant differences in resistance levels, even among the isolates that produced the same enzyme variant, and this variation concerned both the compounds that are substrates of CMY-like β-lactamases and those that are not (for imipenem, the presence of metallo-β-lactamase VIM-1 in the isolates from Greece was noteworthy). Although high, the MICs for the isolates with double blaCMY gene copies were not consistently higher than those for the isolates with single blaCMY copies.
Table 3.
β-Lactam susceptibility of the study P. mirabilis isolates
| Isolate | Major β-lactamase(s) | MIC (μg/ml)a |
Source or reference | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP/AMX | PIP | TZP | CTX | CAZ | FEP | FOX | ATM | IPM | |||
| Clinical P. mirabilis isolatesb | |||||||||||
| PL 6735/99 | CMY-14 | >512 | 128 | 8 | 64 | 128 | 32 | 128 | 16 | 2 | 21 |
| PL 1662/00 | CMY-15 | >512 | 256 | 32 | 128 | 128 | 8 | 512 | 8 | 16 | 21 |
| PL 27/00 | CMY-12 | >512 | 256 | 32 | 64 | 128 | 16 | 256 | 8 | 4 | 21 |
| PL 1376/00 | CMY-45 | >512 | 256 | 16 | 64 | 128 | 4 | 128 | 8 | 2 | This study |
| PL 864/01 | CMY-4 | >512 | 128 | 1 | 32 | 16 | 4 | 64 | 0.5 | 2 | 21 |
| PL 1455/04 | CMY-38 | >512 | 128 | 32 | >128 | 64 | 16 | 128 | 8 | 2 | This study |
| IT NO-051/03 | CMY-16 | >128 | 64 | 4 | 64 | 32 | 2 | 32 | 1 | 2 | 9 |
| IT VA-1017/03 | CMY-16 | >128 | 64 | 4 | >128 | 32 | 2 | 32 | 1 | 2 | 9 |
| IT VA-832/05 | CMY-16 | >128 | >128 | 4 | >128 | 32 | 2 | 64 | 2 | 4 | 23 |
| IT VA-070/06 | CMY-16 | >128 | >128 | 2 | 128 | 16 | 2 | 32 | 1 | 2 | 23 |
| IT VA-414/06 | CMY-16 | >128 | >128 | 2 | 128 | 16 | 2 | 32 | 1 | 2 | 23 |
| IT LC-10/08 | CMY-16 | >128 | >128 | 4 | 128 | 32 | 4 | 32 | 2 | 2 | This study |
| GR 17/04 | CMY-16 + VIM-1 | >128 | >128 | 16 | >128 | 32 | 4 | 32 | 2 | 2 | 35 |
| GR 15184/05 | CMY-16 + VIM-1 | >128 | >128 | 16 | 128 | 32 | 8 | >128 | 2 | 2 | 35 |
| GR 15315/05 | CMY-16 + VIM-1 | >128 | >128 | 32 | >128 | 32 | 16 | >128 | 2 | 4 | 35 |
| GR 2506/07 | CMY-16 + VIM-1 | >128 | >128 | 64 | >128 | 128 | 16 | >128 | 8 | 8 | This study |
| GR 2530/07 | CMY-16 + VIM-1 | >128 | >128 | 64 | 128 | 64 | 4 | >128 | 4 | 4 | This study |
| GR 2720/08 | CMY-16 + VIM-1 | >128 | >128 | 16 | >128 | 128 | 16 | >128 | 4 | 8 | 25 |
| 22317 | CMY-4 | >128 | 64 | 2 | 64 | 32 | 0.125 | 32 | 1 | 1 | 11 |
| PLAR | CMY-4 | >128 | 128 | 4 | 128 | 64 | 0.25 | 128 | 1 | 1 | 11 |
| 34955 | CMY-12 | >128 | 32 | 2 | >128 | >128 | 0.25 | 128 | 2 | 0.5 | 11 |
| ATCC isolates | |||||||||||
| E. coli ATCC 25922 | 4 | 1 | 0.5 | 0.03 | 0.25 | 0.125 | 8 | 0.125 | 0.06 | 21 | |
| P. mirabilis ATCC 7002 | 1 | ≤0.25 | ≤0.25 | ≤0.015 | 0.06 | 0.125 | 2 | ≤0.015 | 0.5 | 21 | |
Abbreviations: AMP, ampicillin; AMX, amoxicillin; ATM, aztreonam; CAZ, ceftazidime; CTX, cefotaxime; FEP, cefepime; FOX, cefoxitin; IPM, imipenem; PIP, piperacillin; TZP, piperacillin + tazobactam. For AMP/AMX, PIP, and FOX, different highest concentrations of the compounds were used in the MIC determinations in the three laboratories participating in the study (512 or 128 μg/ml). For TZP, the tazobactam concentration was 4 μg/ml.
Most of the isolates were obtained from Poland (PL), Italy (IT), and Greece (GR). Isolates 22317 and PLAR originated also from Greece. Isolate 34955 originated from Algeria.
DISCUSSION
This study follows several reports on the international presence of unusual P. mirabilis strains with chromosomal blaCMY-2-like genes. Such isolates were first recovered in the 1990s in France, some of them from Greek and Algerian patients (4, 11); further studies revealed their emergence and spread in Poland (13, 21) and, more recently, in Italy (9, 23) and Greece (25, 35). Comparative analysis of representative isolates from all of the above countries revealed striking similarities among them, regardless of geographic and temporal origins. First, their blaCMY genes resided in transposition modules of the same structure, called Tn6093, and composed of ISEcp1, the C. freundii chromosomal fragment and a part of the ColE1-type plasmid backbone. Second, Tn6093 elements in all isolates were integrated into the same site of the P. mirabilis chromosomal pepQ gene (28). These data are consistent with a single mobilization of these blaCMY genes' ancestor and identified its putative donor to the P. mirabilis chromosome. This was most probably a ColE-like plasmid similar to pTKH11, pA172, and pH 205, all of which carry an ISEcp1 module with blaCMY-2-like genes, named Tn6114 (38, 39). Tn6114-like elements, extended by plasmid scaffold sequences, formed the Tn6093 modules. During the putative mobilization-transfer-integration, ISEcp1 would have utilized different alternative IRRs while transposing from the C. freundii chromosome to the plasmid (IRR-3) and then from the plasmid to the P. mirabilis chromosome (IRR-4), thus giving rise to the Tn6114- and Tn6093-like structures, respectively.
In sequence comparisons, the P. mirabilis blaCMY alleles formed a distinct cluster among all blaCMY-2-like genes. This lends further support to the hypothesis of a single mobilization-transfer-integration event, as posited by the data just discussed: according to this, the seven blaCMY alleles studied here will have evolved by the stepwise acquisition of mutations after Tn6093 chromosomal integration. Allele blaCMY-12, since it shares polymorphisms specific to two separate gene sublineages, may have arisen from recombination between two different alleles. Most of the blaCMY genes identified in the P. mirabilis chromosome have only been observed in this genetic environment thus far. Their direct precursor is likely to have been blaCMY-4, identified in early isolates from France/Greece and Poland (although not in a plasmidic Tn6114-like module), rather than the blaCMYs (blaCMY-5, blaCMY-31, and blaCMY-36) of the ColE1-like plasmids described to date (38, 39), which all have higher numbers of nucleotide differences. Moreover, several other polymorphisms have been detected between Tn6093 and the homologous parts of these plasmids. As has been shown in earlier studies on recombinant E. coli expressing most of the blaCMY genes studied here, mutations acquired during these genes' evolution were neutral (9, 21); however, the clinical P. mirabilis strains themselves varied in resistance levels to β-lactams (9, 21, 23, 25, 35) (Table 3). Such variation could be due to other enzymes, as in the case of the Greek isolates, which produced VIM-1 metallo-β-lactamase (25, 35) (Table 1), or to nonenzymatic mechanisms, e.g., reduction in permeability, as previously proposed (21).
The presence of a second blaCMY module in a subset of our isolates was also intriguing. Previously, Polish isolates had been found to contain two copies of blaCMY-12 and blaCMY-15 (21). The present study, while confirming these observations, also showed that the French-Algerian isolate with blaCMY-12 (11) and the Polish one with blaCMY-38 (13) also had two gene copies. These “duplicated” alleles are related: within the blaCMY sequence cluster, blaCMY-15 and blaCMY-38 form a sublineage, to which blaCMY-12 is closely linked (Fig. 2). All “additional blaCMY loci” contained an identical ISEcp1 module, Tn6113, inserted into the same site, downstream of the ppiD gene (28). Most probably, this emerged from a transposition event in which ISEcp1 utilized yet another IRR-like motif (IRR-2); however, it is impossible to decide on whether Tn6113 resulted from a “main blaCMY locus” Tn6093 or a plasmidic Tn6114 transposition. At any rate, this integration also probably occurred once, at the origin of the specific sublineage of blaCMYs. Interestingly, the Tn6093 and Tn6113 found in the same chromosome always harbored identical blaCMY variants, possibly due to sequence homogenization by a gene-conversion-like mechanism. As reported previously, isolates with double blaCMY copies did not show consistently higher β-lactam resistance levels compared to single-copy isolates (21) (Table 3). This might again have resulted from the already mentioned contributions of β-lactamase- and non-β-lactamase-mediated mechanisms alike on overall resistance.
Addressing the possible clonality of the blaCMY-carrying isolates required a method able to assess phylogenetic similarity among organisms collected over a large geographic area and a long period of time. Therefore, in the absence of an MLST scheme for P. mirabilis, we used ribotyping. This revealed a significant relatedness among most of our isolates, especially compared to a group of random blaCMY-negative P. mirabilis strains. In general, these results were congruent with those obtained with the more discriminatory, PFGE-based methods (using either I-Ceu-I-digested or NotI-SfiI-digested genomic DNA).
The present study indicates a common origin of P. mirabilis strains with acquired and chromosomally inserted blaCMY-2-like genes, spreading in Europe since the 1990s. Their dissemination is probably quite efficient: in Poland, such isolates comprised ∼20% of the nosocomial P. mirabilis isolates in the mid-2000s (13), while in Italy their progressive spread has also been documented (9, 23). It is therefore likely that they represent a true international clonal group of P. mirabilis undergoing continuous diversification over time and place.
ACKNOWLEDGMENTS
We are grateful to Guillaume Arlet for the P. mirabilis strains 22317, PLAR, and 34955 and to Joanna Empel for helpful discussions.
This study was supported in part by grants from the European Commission (TROCAR contract HEALTH-F3-2008-223031 and TEMPOTest-QC contract HEALTH-F3-2009-241742) and by the grant SPUB MIKROBANK from the Polish Ministry of Science and Higher Education.
Footnotes
Published ahead of print on 14 March 2011.
REFERENCES
- 1. Barlow M., Hall B. G. 2002. Origin and evolution of the AmpC β-lactamases of Citrobacter freundii. Antimicrob. Agents Chemother. 46:1190–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bauernfeind A., Stemplinger I., Jungwirth R., Giamarellou H. 1996. Characterization of the plasmidic β-lactamase CMY-2, which is responsible for cephamycin resistance. Antimicrob. Agents Chemother. 40:221–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Beceiro A., Bou G. 2004. Class C β-lactamases: an increasing problem worldwide. Rev. Med. Microbiol. 15:141–152 [Google Scholar]
- 4. Bret L., et al. 1998. Chromosomally encoded AmpC-type β-lactamase in a clinical isolate of Proteus mirabilis. Antimicrob. Agents Chemother. 42:1110–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bush K., Jacoby G. A. 2010. Updated functional classification of β-lactamases. Antimicrob. Agents Chemother. 54:969–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Clinical and Laboratory Standards Institute 2010. Performance standards for antimicrobial susceptibility testing. CLSI M100-S20U. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 7. Colinon C., Miriagou V., Carattoli A., Luzzaro F., Rossolini G. M. 2007. Characterization of the IncA/C plasmid pCC416 encoding VIM-4 and CMY-4 β-lactamases. J. Antimicrob. Chemother. 60:258–262 [DOI] [PubMed] [Google Scholar]
- 8. Daikos G. L., et al. 2007. Enterobacteriaceae bloodstream infections: presence of integrons, risk factors, and outcome. Antimicrob. Agents Chemother. 51:2366–2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. D'Andrea M. M., et al. 2006. CMY-16, a novel acquired AmpC-type β-lactamase of the CMY/LAT lineage in multifocal monophyletic isolates of Proteus mirabilis from northern Italy. Antimicrob. Agents Chemother. 50:618–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Darling A. C. E., Mau B., Blattner F. R., Perna N. T. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14:1394–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Decré D., et al. 2002. Characterization of CMY-type β-lactamases in clinical strains of Proteus mirabilis and Klebsiella pneumoniae isolated in four hospitals in the Paris area. J. Antimicrob. Chemother. 50:681–688 [DOI] [PubMed] [Google Scholar]
- 12. Didelot X., Falush D. 2007. Inference of bacterial microevolution using multilocus sequence data. Genetics 175:1251–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Empel J., et al. 2008. Molecular survey of β-lactamases conferring resistance to newer β-lactams in Enterobacteriaceae isolates from Polish hospitals. Antimicrob. Agents Chemother. 52:2449–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Giles W. P., et al. 2004. DNA sequence analysis of regions surrounding blaCMY-2 from multiple Salmonella plasmid backbones. Antimicrob. Agents Chemother. 48:2845–2852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hopkins K. L., Batchelor M. J., Liebana E., Deheer-Graham A. P., Threlfall E. J. 2006. Characterisation of CTX-M and AmpC genes in human isolates of Escherichia coli identified between 1995 and 2003 in England and Wales. Int. J. Antimicrob. Agents 28:180–192 [DOI] [PubMed] [Google Scholar]
- 16. Hopkins K. L., et al. 2006. Replicon typing of plasmids carrying CTX-M or CMY β-lactamases circulating among Salmonella and Escherichia coli isolates. Antimicrob. Agents Chemother. 50:3203–3206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huson D. H., Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23:254–267 [DOI] [PubMed] [Google Scholar]
- 18. Jacoby G. A. 2009. AmpC β-lactamases. Clin. Microbiol. Rev. 22:161–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kumar S., Nei M., Dudley J., Tamura K. 2008. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 9:299–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lartigue M. F., Poirel L., Aubert D., Nordmann P. 2006. In vitro analysis of ISEcp1B-mediated mobilization of naturally occurring β-lactamase gene blaCTX-M of Kluyvera ascorbata. Antimicrob. Agents Chemother. 50:1282–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Literacka E., et al. 2004. Four variants of the Citrobacter freundii AmpC-type cephalosporinases, including two novel enzymes, CMY-14 and -15, in a Proteus mirabilis clone widespread in Poland. Antimicrob. Agents Chemother. 48:4136–4143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu S. L., Hessel A., Sanderson K. E. 1993. Genomic mapping with I-Ceu I, an intron-encoded endonuclease specific for genes for rRNA, in Salmonella spp., Escherichia coli, and other bacteria. Proc. Natl. Acad. Sci. U. S. A. 90:6874–6878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Luzzaro F., et al. 2009. Spread of multidrug-resistant Proteus mirabilis isolates producing an AmpC-type β-lactamase: epidemiology and clinical management Int. J. Antimicrob. Agents 33:328–333 [DOI] [PubMed] [Google Scholar]
- 24. Miriagou V., Carattoli A., Tzelepi E., Villa L., Tzouvelekis L. S. 2005. IS26-associated In4-type integrons forming multiresistance loci in enterobacterial plasmids. Antimicrob. Agents Chemother. 49:3541–3543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Miriagou V., et al. 2010. Detecting VIM-1 production in Proteus mirabilis by an imipenem-dipicolinic acid double disk synergy test. J. Clin. Microbiol. 48:667–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nakano R., Okamoto R., Nagano N., Inoue M. 2007. Resistance to gram-negative organisms due to high-level expression of plasmid-encoded ampC β-lactamase blaCMY-4 promoted by insertion sequence ISEcp1. J. Infect. Chemother. 13:18–23 [DOI] [PubMed] [Google Scholar]
- 27. Nakano R., et al. 2004. CFE-1, a novel plasmid-encoded AmpC β-lactamase with an ampR gene originating from Citrobacter freundii. Antimicrob. Agents Chemother. 48:1151–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pearson M. M., et al. 2008. Complete genome sequence of uropathogenic Proteus mirabilis, a master of both adherence and motility. J. Bacteriol. 190:4027–4037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Philippon A., Arlet G., Jacoby G. A. 2002. Plasmid-determined AmpC-type β-lactamases. Antimicrob. Agents Chemother. 46:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pignato S., Giammanco G. M., Grimont F., Grimont P. A., Giammanco G. 1999. Molecular characterization of the genera Proteus, Morganella, and Providencia by ribotyping. J. Clin. Microbiol. 37:2840–2847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Struelens M. J., Schwam V., Deplano A., Baran D. 1993. Genome macrorestriction analysis of diversity and variability of Pseudomonas aeruginosa strains infecting cystic fibrosis patients. J. Clin. Microbiol. 31:2320–2326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Su L. H., et al. 2006. Distribution of a transposon-like element carrying blaCMY-2 among Salmonella and other Enterobacteriaceae. J. Antimicrob. Chemother. 57:424–429 [DOI] [PubMed] [Google Scholar]
- 33. Takeshita S., Sato M., Toba M., Masahashi W., Hashimoto-Gotoh T. 1987. High-copy-number and low-copy-number plasmid vectors for lacZ α-complementation and chloramphenicol or kanamycin resistance selection. Gene 61:63–74 [DOI] [PubMed] [Google Scholar]
- 34. Verdet C., Arlet G., Barnaud G., Lagrange P. H., Phillippon A. 2000. A novel integron in Salmonella enterica serovar Enteritidis, carrying, the blaDHA-1 gene and its regulator gene ampR, originated from Morganella morganii. Antimicrob. Agents Chemother. 44:222–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vourli S., et al. 2006. Emergence of Proteus mirabilis carrying the bla metallo-β-lactamase gene. Clin. Microbiol. Infect. 12:691–694 [DOI] [PubMed] [Google Scholar]
- 36. Whichard J. M., et al. 2007. Human Salmonella and concurrent decreased susceptibility to quinolones and extended-spectrum cephalosporins. Emerg. Infect. Dis. 13:1681–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Woodford N., Reddy S., Fagan E. J., R. L., et al. 2007. Wide geographic spread of diverse acquired AmpC β-lactamases among Escherichia coli and Klebsiella spp. in the UK and Ireland. J. Antimicrob. Chemother. 59:102–105 [DOI] [PubMed] [Google Scholar]
- 38. Wu S. W., Dornbusch K., Kronvall G., Norgren M. 1999. Characterization and nucleotide sequence of a Klebsiella oxytoca cryptic plasmid encoding a CMY-type β-lactamase: confirmation that the plasmid-mediated cephamycinase originated from the Citrobacter freundii AmpC β-lactamase. Antimicrob. Agents Chemother. 43:1350–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zioga A., et al. 2009. CMY-31 and CMY-36 cephalosporinases encoded by ColE1-like plasmids. Antimicrob. Agents Chemother. 53:1256–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]