Abstract
Fatty acid biosynthesis is a promising novel antibiotic target. Two inhibitors of fatty acid biosynthesis, platencin and platensimycin, were recently discovered and their molecular targets identified. Numerous structure-activity relationship studies for both platencin and platensimycin are currently being undertaken. We established a proteomic signature for fatty acid biosynthesis inhibition in Bacillus subtilis using platencin, platensimycin, cerulenin, and triclosan. The induced proteins, FabHA, FabHB, FabF, FabI, PlsX, and PanB, are enzymes involved in fatty acid biosynthesis and thus linked directly to the target pathway. The proteomic signature can now be used to assess the in vivo mechanisms of action of compounds derived from structure-activity relationship programs, as demonstrated for the platensimycin-inspired chromium bioorganometallic PM47. It will further serve as a reference signature for structurally novel natural and synthetic antimicrobial compounds with unknown mechanisms of action. In summary, we described a proteomic signature in B. subtilis consisting of six upregulated proteins that is diagnostic of fatty acid biosynthesis inhibition and thus can be applied to advance antibacterial drug discovery programs.
INTRODUCTION
Bacterial infections continue to be a challenge throughout the world, especially in light of increasing development and dissemination of multiresistant pathogens that are more and more difficult to treat (33). Therefore, prudent use of approved antibiotics and, more importantly, new antibiotics, preferably with new mechanisms of action and low resistance development rates, are urgently required to restrain infectious diseases. Two main strategies for antibiotic development are being pursued today: (i) identification of structurally novel antibiotics using screening approaches based on either natural or synthetic compounds and (ii) chemical modification of known antibiotics aiming at improving their antibacterial or pharmacological properties or at circumventing existing resistance mechanisms.
Since the recent discovery of platencin and platensimycin, two potent natural inhibitors of bacterial growth from Streptomyces platensis (15, 24, 31, 32), there is renewed interest in fatty acid biosynthesis as an antibacterial target. Platensimycin as well as cerulenin, discovered in the 1960s, inhibit the 3-oxoacyl-acyl carrier protein (ACP) synthase II FabF (21, 32), whereas platencin inhibits both FabF and 3-oxoacyl-ACP synthases III FabHA and FabHB (15, 31). These enzymes catalyze the initial condensation of acyl-ACPs and existing fatty acid chains with malonyl-ACP, respectively (9). In contrast, triclosan, another fatty acid biosynthesis inhibitor, discovered in the 1970s, targets the second reduction step in the fatty acid chain elongating biosynthesis cycle, inhibiting the enoyl-ACP reductase FabI (11) (see Fig. 1 for an overview). Neither triclosan nor cerulenin is used clinically as an antimicrobial agent. However, triclosan is used in consumer products, such as toothpaste (17, 29, 30), while cerulenin is currently being evaluated as an antitumor therapeutic in combination therapies (10). Platensimycin and platencin showed efficacy in a Staphylococcus aureus mouse infection model (31). A number of structure-activity relationship (SAR) studies are being performed to identify lead structures for further development (14, 16, 18, 20). In addition, the search for effective natural analogues continues (12, 35, 36).
Fig. 1.
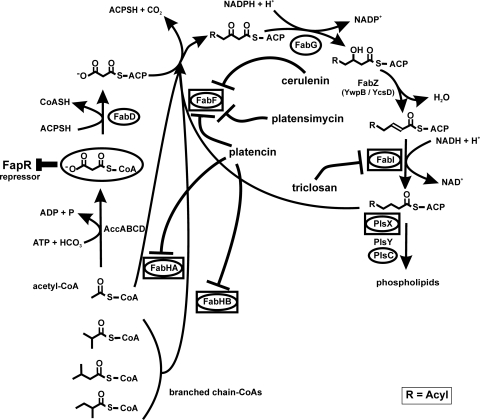
Fatty acid biosynthesis in B. subtilis. Cerulenin, platensimycin, and platencin inhibit the 3-oxoacyl-ACP synthase FabF. Platencin targets FabF and FabHA/HB. Triclosan inhibits the enoyl-ACP-reductase FabI. Proteins belonging to the FapR regulon are circled. Proteins induced by fatty acid biosynthesis inhibitors are placed inside boxes. Modified according to Fujita et al. (9) with permission of the publisher.
Proteomic profiling can support antibacterial drug discovery by contributing to target identification and mechanism-of-action studies (4, 7). We have previously established a comprehensive proteomic response reference compendium using the Gram-positive model organism Bacillus subtilis. It contains proteomic response patterns for over 40 antibacterial compounds (2). As the bacterial response to antibiotic treatment mirrors the inflicted damage, it is highly specific and closely linked to the antibiotic mechanism. Proteomic signatures indicative of the antibiotic mechanism of action can be established, if structurally different inhibitors of the same pathway are available (28). Once they are established, these signatures can aid in mechanism-of-action identification of structurally novel compounds. For instance, the proteomic signature for inhibition of translation identified peptidyltransferase inhibition as the mechanism of action of the structurally novel compound Bay 50-2369 (2). The signatures can also serve as references to confirm the in vivo mechanism of action of SAR-derived compounds, provided the modified compound still has the same mechanism as the lead structure.
In this study we investigated the bacterial response to fatty acid biosynthesis inhibitors triclosan, cerulenin, platensimycin, and platencin. On the basis of the proteomic response profiles of B. subtilis, we were able to establish a proteomic signature for fatty acid biosynthesis inhibition. We then applied the newly established signature to investigate the mechanism of action of the chromium bioorganometallic PM47, a compound inspired by platensimycin, which displayed low activity against Gram-positive bacteria (19). On the basis of the findings of proteome analysis, we could rule out fatty acid biosynthesis as its primary mechanism of action.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacillus subtilis 168 (trpC2) (1) was grown at 37°C under steady agitation in a defined medium previously described (25). Cerulenin and triclosan were purchased from Merck KGaA, Darmstadt, Germany; platencin was synthesized by D. Chen; and platensimycin was provided by Merck & Co., Inc., Rahway, NJ. All antibiotic stock solutions were prepared in dimethyl sulfoxide (DMSO). MICs were determined in a test tube assay as described previously (2). Two milliliters of defined medium was inoculated with 105 bacteria per ml, and the mixture was incubated at 37°C under agitation for 18 h. The MIC was defined as the lowest concentration inhibiting visible growth.
In growth experiments, bacterial cultures were exposed to antibiotics at different concentrations during early exponential growth phase after they reached an optical density at 500 nm (OD500) of 0.35. An antibiotic concentration leading to a reduction in growth rate of approximately 50 to 70% was chosen for proteomic profiling experiments.
Preparation of cytoplasmic l-[35S]methionine-labeled protein fractions.
For pulse-labeling experiments, 5 ml of a bacterial culture in early exponential growth phase was exposed to 0.5 μg/ml triclosan, 5 μg/ml cerulenin, 5 μg/ml platensimycin, 0.2 μg/ml platencin, or 25 μg/ml PM47 or was left untreated as a control. After 10 min of antibiotic treatment, cells were pulse-labeled radioactively with 1.8 MBq l-[35S]methionine (Hartmann Analytic, Braunschweig, Germany) for 5 min. Methionine incorporation was stopped by adding 1 mg/ml chloramphenicol and an excess of nonradioactive l-methionine (10 mM) and by immediately transferring samples onto ice. Cells were harvested by centrifugation and washed three times with 100 mM Tris–1 mM EDTA buffer, before disruption by ultrasonication in a VialTweeter instrument (Hielscher, Teltow, Germany) in 10 mM Tris buffer containing 1.39 mM phenylmethylsulfonyl fluoride. The soluble protein fraction was separated from cell debris by centrifugation at 16.1 × g for 20 min. Protein concentrations were estimated using a Bradford-based Roti NanoQuant assay (Roth, Karlsruhe, Germany).
2D-PAGE.
Unless otherwise noted, chemicals for two-dimensional (2D) gel electrophoresis were ordered from Sigma-Aldrich or Roth in electrophoresis-grade quality. Cytosolic proteins were solubilized in 400 μl buffer containing 7 M urea, 2 M thiourea, 6.5 mM 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate, 0.5% Triton X-100, 1.04% Pharmalyte 3-10 (GE Healthcare, Uppsala, Sweden), and 50 mM dithiothreitol (DTT). Fifty or 300 μg of protein for radioactive analytical gels and for nonradioactive preparative gels (for protein identification by mass spectrometry), respectively, was loaded onto 24-cm immobilized pH gradient (IPG) strips, pH 4 to 7 (GE Healthcare), by passive rehydration for 18 h. Isoelectric focusing was carried out using a Multiphor II apparatus (GE Healthcare), applying the following gradient: 0 to 500 V for 1 kVh, 500 V for 0.02 kVh, 500 to 3,500 V for 3 kVh, and 3,500 V for 57 kVh at 20°C. Prior to SDS-PAGE, proteins were reduced and subsequently alkylated for 20 min each in equilibration buffer (50 mM Tris, pH 8.8, 6 M urea, 30% glycerol, 2% SDS) supplemented with 1% DTT and 2.5% iodoacetamide, respectively. IPG strips were placed onto 12.5% SDS-polyacrylamide gels (acrylamide, bisacrylamide, 0.375 M Tris, 01.% SDS, 0.05% ammonium persulfate, 0.0138% N,N,N′,N′-tetramethylethylenediamine) and covered with 0.5% agarose in running buffer (25 mM tris, 192 mM glycine, 0.1% SDS). Electrophoresis was carried out using an Ettan DALTtwelve system from GE Healthcare at 0.5 W/gel for 1 h to allow transfer of proteins from the IPG strip into the polyacrylamide gel, followed by 10 W/gel for protein separation using the above-described running buffer (2× buffer in the upper chamber, 1× buffer in the lower chamber). Proteins were stained with 0.003% ruthenium(II) Tris(4,7-diphenyl-1,10-phenantrolin disulfonate) (RuBPs) (22) and imaged using a Typhoon Trio+ variable-mode imager (GE Healthcare) set at an excitation wavelength of 532 nm and using a 610-nm emission filter. Analytical gels were dried on Whatman paper and exposed to storage phosphor screens (GE Healthcare). Screens were scanned with the Typhoon Trio+ imager at a 633-nm excitation wavelength and using a 390-nm emission filter. Images were analyzed as described by Bandow et al. (3) using Decodon Delta 2D image analysis software (Decodon, Greifswald, Germany). Proteins found to be induced more than 2-fold in three independent biological replicates are reported as marker proteins.
Protein identification.
Protein spots were excised from preparative 2D gels and transferred into 96-well microtiter plates. Tryptic digestion with subsequent spotting on a matrix-assisted laser desorption ionization (MALDI) target was carried out automatically with an Ettan spot handling workstation (Amersham Biosciences, Uppsala, Sweden) as described by Eymann et al. (8).
MALDI-time of flight (TOF) measurements were carried out on a 4800 MALDI TOF/TOF analyzer (Applied Biosystems, Foster City, CA) designed for high-throughput measurements. The instrument allows automatic measurement of the samples, calibration of the spectra, and analysis of the data using 4000 Explorer software, version 3.5.3. Spectra were recorded in a mass range from 900 to 3,700 Da with a focus mass of 2,000 Da. For one main spectrum, 25 subspectra with 100 shots per subspectrum were accumulated using a random-search pattern. If the autolytical fragment of trypsin with the monoisotopic (M+H)+ m/z at 2211.104 reached a signal-to-noise (S/N) ratio of at least 10, internal calibration was automatically performed as one-point calibration using this peak. The standard mass deviation was less then 0.15 Da. If the automatic mode failed (for less than 1% of samples), the calibration was carried out manually. After calibration, the peak lists were created by using the “peak-to-mascot” script of the 4000 Explorer software, version 3.5.3, using the following settings: mass range from 900 to 3,700 Da, peak density of 50 peaks per 200 Da, minimal area of 100, and a maximum of 200 peaks per spot. The peak list was created for an S/N ratio of 6.
The MALDI-TOF-TOF measurements were also carried out on the 4800 MALDI TOF/TOF analyzer. The three strongest peaks of the TOF spectra were measured. For one main spectrum, 20 subspectra with 125 shots per subspectrum were accumulated using a random-search pattern. Internal calibration was automatically performed as one-point calibration, with the monoisotopic arginine (M+H)+ m/z at 175.119 or lysine (M+H)+ m/z at 147.107 reaching an S/N ratio of at least 5. The peak lists were created by using the script of the 4000 Explorer Software, version 3.5.3, with the following settings: mass range from 60 to precursor mass plus 20 Da, peak density of 5 peaks per 200 Da, minimal area of 100, and a maximum of 20 peaks per precursor. The peak list was created for an S/N ratio of 5. For database search, the Mascot search engine, version 2.1.04 (Matrix Science Ltd., London, United Kingdom), with a specific B. subtilis sequence database was used.
RESULTS
Proteomic signature of fatty acid biosynthesis inhibition.
Bacterial cells react quickly to subinhibitory concentrations of antibiotics by adjusting their protein synthesis (2). In many cases, translation capacity is allocated to production of proteins that counteract the loss of function or the damage inflicted by the antibiotic action. The cellular response is highly specific for each antibiotic compound, with some of the responder proteins reflecting the antibiotic mechanism of action and others being structure specific. Structurally different antibiotics that cause the same physiological condition in the cell induce the same responder proteins. Those responder proteins indicative of the antibiotic mechanism constitute the antibiotic proteomic signature.
To establish an antibiotic proteomic signature for fatty acid biosynthesis inhibition, the protein synthesis patterns of triclosan, cerulenin, platensimycin, and platencin were compared to identify common responder proteins. Reproducibility of the proteome response patterns for each of the antibiotics is absolutely critical and highly dependent on reproducibility of the cellular growth and antibiotic treatment conditions. Differences in antibiotic concentration, cell density, or incubation time may have a significant impact on protein synthesis profiles. This is of particular concern, when an antibiotic mode of action is known to shift in a concentration-dependent manner. Triclosan, e.g., inhibits fatty acid biosynthesis at low concentrations but at higher doses also damages multiple cytoplasmic and membrane targets (23, 26, 27).
We first determined the MICs of the four antibiotics against B. subtilis under growth conditions similar to those used for proteomic sample generation. The MICs in defined medium with cells aerated during overnight incubation were 0.1 μg/ml for triclosan, 0.2 μg/ml for platencin, 1 μg/ml for platensimycin, and 5 μg/ml for cerulenin. Growth experiments with different antibiotic concentrations added to the cultures in early exponential growth phase were then performed to identify a concentration that inhibited bacterial growth visibly without killing the cells. A growth rate reduction of 50 to 70% in the exponential growth phase compared to the untreated control growth had previously proven useful for proteome analysis (2). In order to generate the desired growth inhibition, concentrations of up to five times the MIC were used to treat the cultures. Representative growth curves of B. subtilis cultures exposed to fatty acid biosynthesis inhibitors at concentrations selected for proteome analyses are shown in Fig. 2.
Fig. 2.
Antibiotic concentrations leading to approximately 50% growth reduction. B. subtilis was grown in defined medium to exponential phase and treated with platensimycin (5 μg/ml) (A), platencin (0.2 μg/ml) (B), cerulenin (5 μg/ml) (C), and triclosan (0.5 μg/ml) (D). The time point of antibiotic addition is marked by arrowheads.
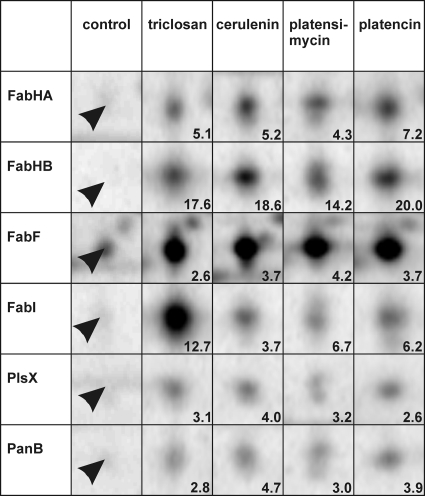
Once a suitable antibiotic concentration was found, pulse-labeling experiments with l-[35S]methionine were performed to specifically label those proteins newly synthesized during the 5-min pulse either after treatment with a fatty acid biosynthesis inhibitor or under control conditions. Highly reproducible protein expression profiles were obtained from three biological replicate experiments for each antibiotic following a standardized protocol for protein separation and gel image analysis. Marker proteins reproducibly induced at least 2-fold for each antibiotic are listed in Table 1. All fatty acid biosynthesis inhibitors tested induced six common marker proteins (Fig. 3). These six markers make up the proteomic signature for fatty acid biosynthesis inhibition. All of them are involved in fatty acid biosynthesis. Indirectly contributing to fatty acid or phospholipid biosynthesis is PanB, an enzyme of the pantothenate biosynthesis pathway. Pantothenate is a precursor of the cofactor of ACP and a precursor of coenzyme A (CoA). Coenzyme A is essential for fatty acid biosynthesis, as the condensation cycle always starts with acetyl-CoA or branched-chain CoAs (Fig. 1) and chain elongation requires malonyl-CoA. The other five marker proteins are involved directly in fatty acid biosynthesis. The 3-oxoacyl-ACP synthases FabHA, FabHB, and FabF initiate the condensation reaction with different substrate preferences, whereas enoyl-ACP reductase FabI catalyzes the second reduction step of the condensation cycle. PlsX starts the conversion of fatty acids to phospholipids. On the transcriptional level in B. subtilis, all five enzymes are negatively controlled by the FapR repressor and are therefore part of the same regulon (Fig. 1).
Table 1.
Marker proteins induced by fatty acid biosynthesis inhibitors
| Marker proteina | Induction factor |
Protein function | Pathway | |||
|---|---|---|---|---|---|---|
| Platensimycin | Platencin | Cerulenin | Triclosan | |||
| FabHA | 5.1 | 5.2 | 4.3 | 7.2 | 3-Oxoacyl-ACP synthase III | Fatty acid biosynthesis |
| FabHB | 17.6 | 18.6 | 14.2 | 20.0 | 3-Oxoacyl-ACP synthase III | Fatty acid biosynthesis |
| FabF | 2.6 | 3.7 | 4.2 | 3.7 | 3-Oxoacyl-ACP synthase II | Fatty acid biosynthesis |
| FabI | 12.7 | 3.7 | 6.7 | 6.2 | Enoyl-ACP reductase | Fatty acid biosynthesis |
| PlsX | 3.1 | 4.0 | 3.2 | 2.6 | Putative glycerol-3-phosphate acyltransferase PlsX | Phospholipid biosynthesis |
| PanB | 2.8 | 4.7 | 3.0 | 3.9 | 3-Methyl-2-oxobutanoate hydroxymethyltransferase | Pantothenate biosynthesis |
| CP_1 | 7.0 | 5.4 | Protein not identified | |||
| YkrS | 2.6 | Methylthioribose-1-phosphate isomerase | Amino acid metabolism | |||
| SerA | 3.5 | d-3-Phosphoglycerate dehydrogenase | Amino acid metabolism | |||
| Plc_3 | 3.1 | Protein not identified | ||||
| Plc_4 | 4.0 | Protein not identified | ||||
| Ald | 2.6 | Putative alanine dehydrogenase | Amino acid metabolism | |||
| Cer_1 | 4.4 | Protein not identified | ||||
| YurP | 2.8 | Similar to glutamine–fructose-6-phosphate transaminase | Amino acid metabolism | |||
| Cer_4 | 2.3 | Protein not identified | ||||
| PdhC | 4.3 | Pyruvate dehydrogenase subunit E2b | Acetyl-CoA, branched-chain CoA metabolism | |||
| Trc_2 | 3.9 | Protein not identified | ||||
| SucD | 2.3 | Succinyl-CoA synthetase subunit alpha | Citric acid cycle | |||
| Trc_4 | 2.6 | Protein not identified | ||||
Cer, unidentified protein induced by cerulenin; CP, unidentified protein induced by cerulenin and platencin; Plc, unidentified protein induced by platencin; Trc, unidentified protein induced by triclosan.
This subunit is also part of branched-chain alpha 2-oxoacid dehydrogenase.
Fig. 3.
Details of 2D gels depict the six marker proteins (marked by arrowheads) making up the proteomic signature of fatty acid biosynthesis inhibition under control conditions as well as triclosan, cerulenin, platensimycin, and platencin treatment. Induction factors displayed in the lower right reflect averages over three biological replicates.
Platensimycin exclusively induced the marker proteins of the proteomic signature for fatty acid biosynthesis inhibition. Triclosan, cerulenin, and platencin each induced some additional marker proteins specific for each compound (Table 1). Some of these proteins are related indirectly to fatty acid biosynthesis, as they are involved in acetyl-CoA production. Cerulenin-induced Ald is a putative dehydrogenase forming pyruvate from l-alanine, while triclosan-induced PdhC is a subunit of pyruvate dehydrogenase as well as branched-chain alpha 2-oxoacid dehydrogenase, which generate acetyl-CoA and branched-chain CoAs, respectively.
Some of the marker proteins, such as platencin-induced YkrS involved in the methionine salvage pathway, could not be connected directly to fatty acid biosynthesis. Others could not be identified by mass spectrometry due to a lack of protein accumulation. However, these protein signals defined by their coordinates on the 2D gel map still serve as antibiotic-specific markers, as they are reproducibly induced after antibiotic treatment.
Proteomic analysis of platensimycin analogue PM47.
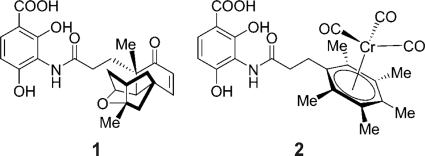
Patra et al. synthesized different organometallic platensimycin derivatives aiming to preserve the steric properties while at the same time simplifying synthesis and potentially improving antibacterial properties (19). One of these compounds is the chromium bioorganometallic PM47 (Fig. 4), which shows low activity against Gram-positive bacteria and has a MIC of 80 μg/ml against B. subtilis (19).
Fig. 4.
Structures of platensimycin (structure 1) and PM47 (structure 2). The tetracyclic cage of platensimycin was replaced by an organometallic core. Me, methyl.
The proteomic response of B. subtilis to 25 μg/ml PM47 was investigated to test whether PM47 still inhibits fatty acid biosynthesis or has another mechanism of action. Induction of the marker proteins belonging to the proteomic signature for fatty acid biosynthesis inhibition would be expected if the mechanism of action were preserved.
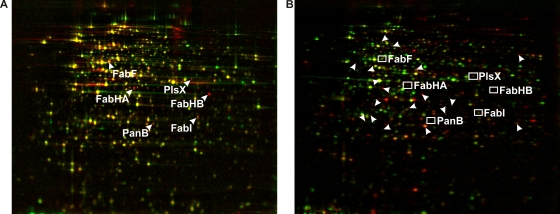
The proteomic responses to PM47 and platensimycin were compared, as the latter induced exclusively the proteins belonging to the fatty acid biosynthesis inhibition signature (Fig. 5). None of the six signature markers nor any of the compound-specific marker proteins were induced by PM47, indicating that PM47 does not inhibit fatty acid biosynthesis. Seventeen proteins were significantly upregulated upon PM47 treatment. The comparison of the proteomic response to PM47 with the proteomic profiles in the reference compendium (2) did not yield a close match, although some of the PM47 marker proteins are also induced by membrane-active antibiotics (data not shown). There was further evidence that platensimycin derivative PM47 has another mechanism of action than its lead compound. In contrast to bacteriostatic platensimycin, PM47 was shown to be bacteriolytic at higher doses.
Fig. 5.
The protein expression profiles of B. subtilis in response to treatment with 5 μg/ml platensimycin (A) and 25 μg/ml PM47 (B) do not share marker proteins. In panel B, unidentified marker proteins of PM47 are indicated by arrowheads, while boxes indicate the locations of fatty acid biosynthesis marker proteins. Pulse-labeled cytosolic proteins were separated by 2D PAGE. Red false-color images representing protein biosynthesis after antibiotic treatment were warped onto green false-color images showing protein synthesis under control conditions. Antibiotic-induced proteins appear red, repressed proteins appear green, and proteins synthesized at equal rates under both conditions appear yellow.
DISCUSSION
Bacteria respond to antibiotic treatment in a highly specific way. The proteome mirrors the physiological impairment caused by the antibiotic. Due to this tight connection between mechanism of action and proteomic response, we can use proteomics to support mechanism-of-action studies in antibacterial drug discovery programs (2, 5, 6). To this end, we established a reference compendium of proteome response profiles for over 40 antibiotics, including almost all clinically relevant antibiotic classes. Recently, the discovery of novel inhibitors with antibacterial activity sparked renewed interest in fatty acid biosynthesis as an antibiotic target (34). The comparison of proteomic responses to four different inhibitors of fatty acid biosynthesis allowed the description of a proteomic signature diagnostic of fatty acid biosynthesis inhibition consisting of six upregulated proteins connected to the target pathway. This proteomic signature, together with the signatures already available in the reference compendium, serves to classify structurally novel synthetic or natural compounds with regard to their mechanisms of action. Proteome analyses of the bacterial response to different fatty acid biosynthesis inhibitors revealed that five enzymes regulated by fatty acid biosynthesis repressor FapR are induced regardless of the molecular target in the pathway. They are directly involved in fatty acid or phospholipid biosynthesis (Fig. 1). For the four enzymes directly involved in fatty acid biosynthesis, FabI, FabF, FabHA, and FabHB, Hutter et al. previously reported induction of transcription by fatty acid biosynthesis inhibitors cerulenin and triclosan (13). Malonyl-CoA sterically inhibits FapR (9). It can be expected that this precursor of fatty acid biosynthesis accumulates upon fatty acid biosynthesis inhibition, causing derepression of the fatty acid biosynthesis enzymes. The sixth marker protein of the proteomic signature, PanB, is involved in the biosynthesis of coenzyme A, an essential cofactor for fatty acid biosynthesis, as the condensation reaction always starts with acetyl-CoA or branched-chain CoAs. None of the six signature proteins were induced by any of the antibiotics with mechanisms other than fatty acid biosynthesis inhibition tested so far.
Beside the six proteins induced by all inhibitors tested, some additional proteins are upregulated in response to one or two of the antibiotics. Some of these are connected to fatty acid biosynthesis as they contribute to the acetyl-CoA metabolism. Others may be induced as a result of compound-specific reactions.
The newly established proteomic signature for fatty acid biosynthesis inhibition was used to evaluate the mechanism of action of a compound derived from an SAR program around the lead structure of platensimycin. Chromium-containing organometallic analogue PM47 showed low antibacterial activity (19) against Gram-positive pathogens. PM47 treatment did not induce fatty acid biosynthesis enzymes, proving that the in vivo mechanism of action is not inhibition of fatty acid biosynthesis. This finding is interesting, as in silico docking studies indicated that the compound fits well into the same binding pocket as platensimycin (19). However, a different mechanism of action is in concordance with the observation that, unlike fatty acid biosynthesis inhibitors, PM47 is bacteriolytic at higher doses. Interestingly, the entire reference compendium does not contain a proteomic signature or protein response profile matching that of PM47, pointing to an unprecedented mechanism of action. There is some overlap in marker proteins with membrane-active antibiotics. Further studies would be necessary to elucidate the mechanism of action. However, PM47 shows cytotoxicity against mammalian cells at sub-MICs (19), suggesting a nonspecific, toxic mechanism.
ACKNOWLEDGMENTS
We thank Merck & Co., Inc., Rahway, NJ, for providing platensimycin. We further thank Elmar Langenfeld, formerly of the Medizinisches Proteom-Center, Ruhr University Bochum, for helping us with RuBP synthesis and Lars I. O. Leichert from the same institution for critically reading the manuscript. We are also grateful for the superb technical support by our colleagues at Rubion.
This work was financially supported by a startup grant from the Ruhr University Bochum and a grant from the state of North Rhine-Westphalia (NRW), Germany, and the European Union, European Regional Development Fund, Investing in your future, to J.E.B.
Footnotes
Published ahead of print on 7 March 2011.
REFERENCES
- 1. Agnostopoulos C., Spizizen J. 1961. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81:741–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bandow J. E., Brötz H., Leichert L. I., Labischinski H., Hecker M. 2003. Proteomic approach to understanding antibiotic action. Antimicrob. Agents Chemother. 47:948–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bandow J. E., et al. 2008. Improved image analysis workflow for 2D gels enables large-scale 2D gel-based proteomics studies—COPD biomarker discovery study. Proteomics 8:3030–3041 [DOI] [PubMed] [Google Scholar]
- 4. Bandow J. E., Hecker M. 2007. Proteomic profiling of cellular stresses in Bacillus subtilis reveals cellular networks and assists in elucidating antibiotic mechanisms of action. Prog. Drug Res. 64:79, 81-101 [DOI] [PubMed] [Google Scholar]
- 5. Beyer D., et al. 2004. New classes of bacterial phenylalanyl-tRNA synthetase inhibitors with high potency and broad-spectrum activity. Antimicrob. Agents Chemother. 48:525–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brötz-Oesterhelt H., et al. 2005. Dysregulation of bacterial proteolytic machinery by a new class of antibiotics. Nat. Med. 11:1082–1087 [DOI] [PubMed] [Google Scholar]
- 7. Brötz-Oesterhelt H., Bandow J. E., Labischinski H. 2005. Bacterial proteomics and its role in antibacterial drug discovery. Mass Spectrom. Rev. 24:549–565 [DOI] [PubMed] [Google Scholar]
- 8. Eymann C., et al. 2004. A comprehensive proteome map of growing Bacillus subtilis cells. Proteomics 4:2849–2876 [DOI] [PubMed] [Google Scholar]
- 9. Fujita Y., Matsuoka H., Hirooka K. 2007. Regulation of fatty acid metabolism in bacteria. Mol. Microbiol. 66:829–839 [DOI] [PubMed] [Google Scholar]
- 10. Haase D., et al. 2010. Fatty acid synthase as a novel target for meningioma therapy. Neuro. Oncol. 12:844–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Heath R. J., Yu Y. T., Shapiro M. A., Olson E., Rock C. O. 1998. Broad spectrum antimicrobial biocides target the FabI component of fatty acid synthesis. J. Biol. Chem. 273:30316–30320 [DOI] [PubMed] [Google Scholar]
- 12. Herath K. B., et al. 2008. Structure and semisynthesis of platensimide A, produced by Streptomyces platensis. Org. Lett. 10:1699–1702 [DOI] [PubMed] [Google Scholar]
- 13. Hutter B., et al. 2004. Prediction of mechanisms of action of antibacterial compounds by gene expression profiling. Antimicrob. Agents Chemother. 48:2838–2844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jang K. P., et al. 2009. Isoplatensimycin: synthesis and biological evaluation. Bioorg. Med. Chem. Lett. 19:4601–4602 [DOI] [PubMed] [Google Scholar]
- 15. Jayasuriya H., et al. 2007. Isolation and structure of platencin: a FabH and FabF dual inhibitor with potent broad-spectrum antibiotic activity. Angew. Chem. Int. ed. Engl. 46:4684–4688 [DOI] [PubMed] [Google Scholar]
- 16. Krauss J., Knorr V., Manhardt V., Scheffels S., Bracher F. 2008. Synthesis of platensimycin analogues and their antibiotic potency. Arch. Pharm. (Weinheim) 341:386–392 [DOI] [PubMed] [Google Scholar]
- 17. Moran J., Addy M., Newcombe R. G., Marlow I. 2001. A study to assess the plaque inhibitory action of a newly formulated triclosan toothpaste. J. Clin. Periodontol. 28:86–89 [DOI] [PubMed] [Google Scholar]
- 18. Nicolaou K. C., Tria G. S., Edmonds D. J., Kar M. 2009. Total syntheses of (+/−)-platencin and (−)-platencin. J. Am. Chem. Soc. 131:15909–15917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Patra M., et al. 2009. Synthesis and biological evaluation of chromium bioorganometallics based on the antibiotic platensimycin lead structure. ChemMedChem. 4:1930–1938 [DOI] [PubMed] [Google Scholar]
- 20. Patra M., et al. 2010. Synthesis and biological evaluation of ferrocene-containing bioorganometallics inspired by the platensimycin lead structure. Organometallics 29:4312–4319 [Google Scholar]
- 21. Price A. C., et al. 2001. Inhibition of beta-ketoacyl-acyl carrier protein synthases by thiolactomycin and cerulenin. Structure and mechanism. J. Biol. Chem. 276:6551–6559 [DOI] [PubMed] [Google Scholar]
- 22. Rabilloud T., Strub J. M., Luche S., van Dorsselaer A., Lunardi J. 2001. A comparison between SyproRuby and ruthenium II Tris (bathophenanthroline disulfonate) as fluorescent stains for protein detection in gels. Proteomics 1:699–704 [DOI] [PubMed] [Google Scholar]
- 23. Russell A. D. 2004. Whither triclosan? J. Antimicrob. Chemother. 53:693–695 [DOI] [PubMed] [Google Scholar]
- 24. Singh S. B., et al. 2006. Isolation, structure, and absolute stereochemistry of platensimycin, a broad spectrum antibiotic discovered using an antisense differential sensitivity strategy. J. Am. Chem. Soc. 128:11916–11920 [DOI] [PubMed] [Google Scholar]
- 25. Stülke J., Hanschke R., Hecker M. 1993. Temporal activation of beta-glucanase synthesis in Bacillus subtilis is mediated by the GTP pool. J. Gen. Microbiol. 139:2041–2045 [DOI] [PubMed] [Google Scholar]
- 26. Suller M. T. E., Russell A. D. 1999. Antibiotic and biocide resistance in methicillin-resistant Staphylococcus aureus and vancomycin-resistant enterococcus. J. Hosp. Infect. 43:281–291 [DOI] [PubMed] [Google Scholar]
- 27. Suller M. T. E., Russell A. D. 2000. Triclosan and antibiotic resistance in Staphylococcus aureus. J. Antimicrob. Chemother. 46:11–18 [DOI] [PubMed] [Google Scholar]
- 28. vanBogelen R. A., Schiller E. E., Thomas J. D., Neidhardt F. C. 1999. Diagnosis of cellular states of microbial organisms using proteomics. Electrophoresis 20:2149–2159 [DOI] [PubMed] [Google Scholar]
- 29. Van Loveren C., Bujis J. F., ten Cate J. M. 1999. Protection of dentin by triclosan toothpaste in a bacterial demineralization model. Eur. J. Oral Sci. 107:114–120 [DOI] [PubMed] [Google Scholar]
- 30. van Loveren C., Bujis J. F., ten Cate J. M. 2000. The effect of triclosan toothpaste on enamel demineralization in a bacterial demineralization model. J. Antimicrob. Chemother. 45:153–158 [DOI] [PubMed] [Google Scholar]
- 31. Wang J., et al. 2007. Discovery of platencin, a dual FabF and FabH inhibitor with in vivo antibiotic properties. Proc. Natl. Acad. Sci. U. S. A. 104:7612–7616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang J., et al. 2006. Platensimycin is a selective FabF inhibitor with potent antibiotic properties. Nature 441:358–361 [DOI] [PubMed] [Google Scholar]
- 33. World Health Organization 2005. Containing antimicrobial resistance. World Health Organization, Geneva, Switzerland [Google Scholar]
- 34. Wright H. T., Reynolds K. A. 2007. Antibacterial targets in fatty acid biosynthesis. Curr. Opin. Microbiol. 10:447–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang C., et al. 2010. Isolation, structure and biological activities of platencin A(2)-A(4) from Streptomyces platensis. Bioorg. Med. Chem. 18:2602–2610 [DOI] [PubMed] [Google Scholar]
- 36. Zhang C., et al. 2008. Isolation, structure and fatty acid synthesis inhibitory activities of platensimycin B1-B3 from Streptomyces platensis. Chem. Commun. (Camb.) 5034-5036 [DOI] [PubMed] [Google Scholar]