Abstract
Technology has advanced to where it is possible to design and grow—with predefined geometry and surprisingly good fidelity—living networks of neurons in culture dishes. Here we overview the elements of design, emphasizing the lithographic techniques that alter the cell culture surface which in turn influences the attachment and growth of the neural networks. Advanced capability in this area makes it possible to design networks of desired complexity. Other issues addressed include the influence of glial cells and media on activity and the potential for extending the designs into three dimensions. Investigators are advancing the art and science of analyzing and controlling through stimulation the function of the neural networks, including the ability to take advantage of their geometric form in order to influence functional properties.
Keywords: Brain on chip, cellular lithography, neural culture, neural engineering, patterning
I. INTRODUCTION
Building a brain on a chip has caught the imagination of a growing number of researchers. The origins of the idea date to pioneering work of Thomas [1], Pine [2], and Gross [3] who showed 30 years ago the feasibility of recording from cultured neurons, myocytes and isolated ganglia with planar electrode arrays. A related technique, the recording and stimulation of activity from brain slices, has progressed from its initial demonstrations [4]–[6] to its current state where the use of multielectrode arrays (MEAs) is so widespread as to support multiple commercial vendors. Common to these efforts is the idea that stimulation and recording of these networks with large numbers of electrodes would lead to better understanding of the basic neuroscience of learning and memory, neural coding, and properties of signal propagation in neural networks. The technology also provides unique approaches to the understanding of disease states such as epilepsy and stroke, and has potential for screening for neuroactivity of drugs in development [7]. That the brain shows both strong structure as well as local randomness has enticed a number of investigators to pursue the means to grow neurons in patterns so as to influence their functional behavior.
The thesis of this paper is that technology has developed to the point where it is beginning to be appropriate to talk seriously about designing a brain on a chip, not just investigating the properties of neural cell culture or brain slices. Most notably, the geometric pattern of growth can be controlled in dramatic fashion. However, there are a number of other design choices that are increasingly selectable by the neuroengineer. These start with the nature of the tissue (dissociated primary neurons vs. cell line vs. brain slice vs. ganglion) and its identity, and include the density of the cells, the composition of the media, the duration of the experiment, and the stimulation and recording protocols. It is even possible to construct three-dimensional culture systems, including recording and stimulation devices. This paper discusses these issues, with an emphasis on the lithographic techniques and the resulting neuron growth patterns.
Although the work described below is dominantly from our laboratories, there are researchers around the world involved in similar or complementary studies. The reader is referred to the MEA Conference Proceedings for a much more thorough list [8]. A preliminary version of this paper appeared as a conference abstract [9].
II. CELLULAR LITHOGRAPHY
Although patterning substrates to control cellular growth in culture has long roots [10]–[12] the field accelerated after the publication by Kleinfeld of the use of photoresist technology to pattern hydrophobic and hydrophilic materials to control neuronal cell attachment [13], as well as the introduction of UV photoablation [14]. The introduction of variety of techniques, including UV techniques [15], [16], photoresist patterning [17]–[19], microcontact printing [20]–[25], microfluidic deposition [26], [27], and micro-machined surfaces [28]–[30]. Materials patterned include hydrophobic alkyl- and hydrophilic amino-silanes on glass (and their thiol equivalents on gold), protein resistant polyethylene glycol, proteins, biological macromolecules, and critical peptide sequences. Linkers, including epoxy-silane [31], enhance the effectiveness of the surface chemistry. Substrates have included insulators glass, silicon, and various plastic polymers.
We have begun to understand the principles that control in vitro cell patterning. Initial observations are that hydrophilic materials are more conducive to cell attachment and growth (“cytophilic”), than “cytophobic” hydrophobic materials; positively charged materials are preferred to negatively charged molecules [13], [14]; growth is correlated with amine group density [32]. More complex materials, such as laminin and fibronectin, are known to control attachment. We have found a protein resistant material (polyethylene glycol) to be very successful at restricting neurite outgrowth [33], [34]. Some materials enhance axonal outgrowth, including mixtures of laminin and polylysine [35], [36] and the laminin derived peptide sequence P20 [37]. Topology is also important [28], [29], [38], [39].
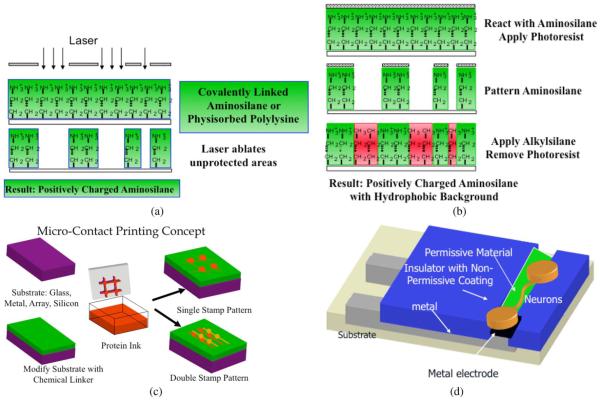
Fig. 1 illustrates the patterning techniques and their use with electrode arrays. Most of the materials are compatible with multiple patterning techniques, including microcontact printing, photoresist patterning, laser ablation, microfluidic deposition, and microchannel deposition. However, to our knowledge only printing permits multiple different biomolecular cues to be applied to the surface. The surfaces may be any of the popular electrode array metals (gold, platinum, indium tin oxide, titanium nitride) and insulators (silicon nitride, silicon dioxide, glass, polyimide, PDMS). The materials deposited may be permissive to cell growth, including polylysine, laminin, or various aminosilanes, or they may be nonpermissive, including polyethylene glycol, albumin, and chondroitin sulfate. Of great interest, but still only recently exploited is the use of neural growth factors such as BDNF to provide precision guidance of neurons [40]. The biomolecules may be covalently linked through a variety of surface chemistry linkers, or they may be physisorbed onto the surface. Taken collectively, the experimenter has a very rich toolbox from which to choose.
Fig. 1.
Micropatterning technologies commonly used for cellular lithography. (a) Laser ablation: a permissive coating is ablated through a mask. (b) Photoresist processing: here a permissive material is protected in desired areas by photoresist; unprotected areas are chemically etched and then reacted with a non-permissive coating. (c) Micro-contact printing: the surface is prepared with a chemical linker, the stamp is inked with the permissive material and the surface stamped, perhaps with multiple inks, and perhaps reacted with a nonpermissive material in the final step. (d) Electrode arrays with metal conductors have their insulating surfaces modified to lead neurons to grow over the top of electrodes.
Confinement may be with physical channels or posts [41], [42] or wells [43] that restrict the movement of cell bodies while permitting axonal extension. Tunnels under PDMS [44] and agarose [45] have worked to provide pathways for networked neurons. As discussed below, the recent development of microtunnel/chamber structures show promise for isolating pure populations of neurons while permitting axonal communication [46], [47].
III. GEOMETRIC DESIGN CHOICES
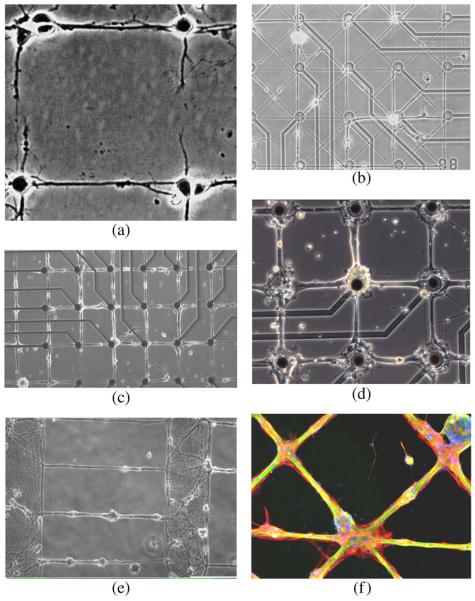
The availability of working lithographic technology implies that the engineer must design the network. We are gaining experience in the ability to control growth patterns. Fig. 2 shows images of networks grown in our laboratories. These indicate that the choices include line width, node size, and geometric pattern. The range is from an approximation not too far from connecting individual neurons [Fig. 2(a), (b)] to using “bundles” of varying width [Fig. 2(c), (d)] to creating neuropil-like structure with connecting lines [Fig. 2(e)]. The crossing pattern [Fig. 2(f)] has dimensions comparable to Fig. 2(d).
Fig. 2.
Geometric patterns of rat hippocampal neuron growth in culture. (a) Network of individual neurons (laser patterned, Reproduced by permission of Wiley & Sons [14]. (b) Thin line network on 3 μm lines at 14 days in culture. (c) 10 μm wide line network at 55 days in culture. (d) 30 μm lines and 80 μm square nodes at 21 days in culture. (e) Neuropil structure separated by 500 μm with 3 μm wide lines. (f) Cross pattern of 80 μm nodes and 30 μm lines, stained for neurons (green), astroglia (red), and nuclei (blue) (figure was used with permission of the first author for the cover on the Journal of Neural Engineering). In (c), (d) epoxy-silane served as linker for stamped poly-D-lysine and as cytophobic background [31].
Networks of single or limited numbers of neurons may be too fragile for practical experimental use. Rutten’s group has studied the dependence of neural activity on size of clusters of neurons, emphasizing that a critical population size of neurons is needed to support both activity and survivability. They report delayed onset of activity and gradual breakdown of pattern (e.g., one month) [48], [49]. Anecdotally we have found that lines that are too thin provide too little substrate for robust attachment and survival of neurons. In one study, we observed migration of neural cell bodies to the larger nodes and measured compliance to pattern [14]. In another we found that isolated lines thinner than 25 μm wide resulted in clumping of neurons and insufficient electrical spontaneous or evoked activity to permit usage [50], [51]. A third study reported that when local cell densities on patterns exceeded 250 cells/mm2, there was substantial activity [52]. With these results one of our design choices is to utilize 80 μm square nodes and 25 μm wide lines, as shown in Fig. 2(d), (f). However, an investigation with 4 μm wide lines and 10 μm diameter nodes, in conjunction with patch clamping, was quite successful for identifying circuit connections of individual cells [53]. For reference, the networks in Fig. 2(c), (d), (f) are representative of networks for which recording of signals is likely, while recording signals is unlikely in Fig. 2(a), (b), (e).
IV. MANIPULATING FIRING RATES WITH ASTROGLIA AND OPTIMIZED MEDIA
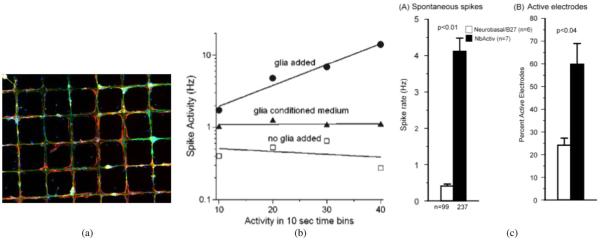
Although our cultures are virtually free of astroglia when initially plated [54], astroglia grow rapidly thereafter [55]. By our counting at one week the ratio of glia to neural cells is one after one week, increasing to a small multiple after several weeks, mimicking natural conditions [56]. If the neurons follow patterns, then the astroglia follow the neurons [Fig. 3(a)] [57], even though astroglia are much less likely to follow surface chemical cues than are neurons. We find that the emergence of widespread spontaneous electrical activity (one to two weeks) is correlated with the emergence of astroglia. We note that glia are known to be beneficial to this activity in vivo [58], [59].
Fig. 3.
Designing with astroglia and defined media. (a) Astroglia develop after neurons in culture, but much prefer neurons to cytophobic substrates, adding stability to a complex network (26 days in culture. Red astroglia, green neurons; reproduced with permission of Koninklijke Brill NV [57]). (b) Deliberately adding astroglia increases firing rates (reproduced with permission of Cambridge University Press [60].) (c) Deliberately fine tuning the media also increases neural activity. (reproduced with permission of Elsevier [61].)
From a signaling perspective, neural cultures have limited action potential firing rates, making it difficult to detect down-regulation of firing as part of a coding strategy. Adding extra glia to the cultures increases firing rates [Fig. 3(b)], prevents desensitization due to glutamate, causes less release from inhibition due to bicuculline, and likely increases the size of the inhibitory population [60]. Activity can also be increased by manipulation of the composition of the cell culture medium [61]. These cultures also exhibit considerable bursting that is suggestive of seizure activity. Stimulating the culture at moderate rates (e.g., 20 Hz) disrupts bursting phenomena and permits use of a wider range of stimuli for probing the network’s properties [62]. Hence the neuroengineer has several tools—glial addition, changes in media composition, or stimulation—with which to manipulate baseline activity level.
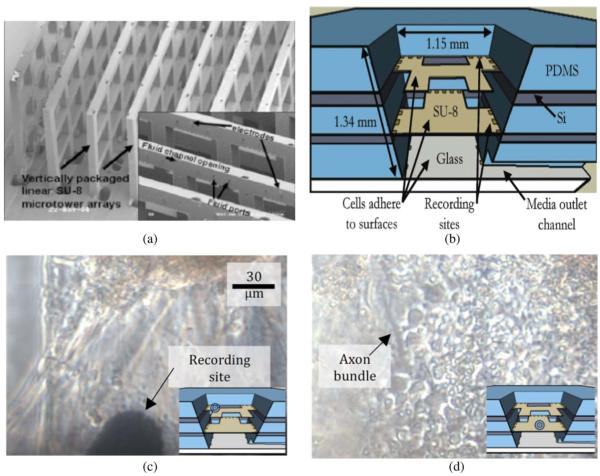
V. MICROTUNNELS—MEMS CAMPENOT CHAMBERS
Campenot chambers, reported in 1977 [63], consist of two neural growth compartments separated by a septum under which needle scratches defined paths for axonal extension from one compartment to the other. The modern MEMS equivalent was reported by Jeon’s group in 2005 [46] consisting of microfabricated tunnels in PDMS which is bonded to a substrate. In time the neurons in one compartment extend their axons through the tunnels, providing pure axonal material on which to do biomolecular analyses [64]–[66].
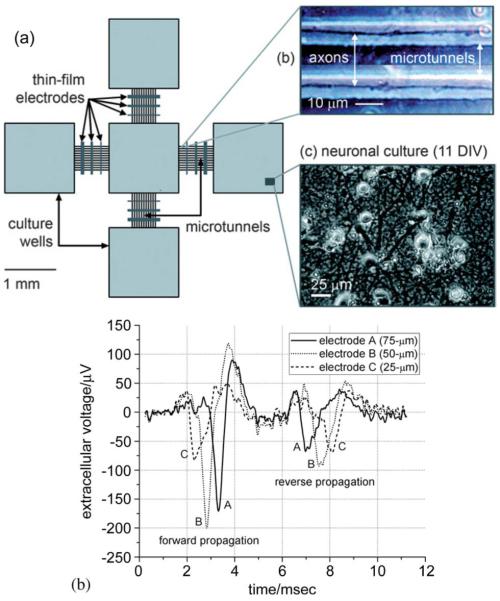
When combined with electrode arrays, the approach offers many substantial advantages. As shown in our work [47], the microtunnel PDMS structure is compatible with electrode arrays. Fig. 4 illustrates the concept. As shown in Fig. 4(b), the signals recordable from the axons in the tunnels are much larger (~100 μV) than in the open chambers where they are often so small as to be in the noise. This is due to the large series resistance of the narrow tunnel (~ 16 MΩ).
Fig. 4.
Microtunnel electrode array. (a) Schematic diagram showing four culture wells, each connected by a series of microtunnels to a central well. There are stimulating/recording electrodes in the wells and crossing the tunnels. The tunnels are 3 μm high by 10 μm wide by 750 μm long. (b) The amplitudes of the axonal potentials are much higher than normal due to the restricted and hence high resistance space in which the axons grow. Direction of propagation can be determined by the temporal order in which spikes occur on the electrodes place along the tunnels. (Reproduced by permission of The Royal Society of Chemistry (RSC) [47].)
The construct offers tremendous advantages for brain on a chip design. Each chamber can be filled with a different population of neurons (e.g., granule or pyramidal cells) or of muscle fibers. Unidirectional transmission can be created through timed growth of the neurons. Various logical geometries of network can be constructed. Separate fluidic treatment of the different populations should be possible. The signals from within the tunnels provide a unique opportunity to sample action potential signals from which one can separately monitor the activity of the source, the target, and the communication channel between them.
VI. BRAIN IN A CHIP
The future will include three-dimensional constructs. There has been progress in developing culture techniques to support neural growth in three-dimensional hydrogels [67]–[69], with experiments showing the critical importance of fluid flow within the hydrogels to enhance cell survival [70]. We and others [71], [72] have printed or photolinked neural guidance molecules onto deformable substrates. These suggest that it will be possible to create three-dimensional substrates in which neurons grow in controlled geometric patterns.
Recording signals from three-dimensional constructs will be challenging. Already two designs have appeared in the literature (Fig. 5; [73], [74]). Both offer multiple electrode contacts and perfusion ports for either maintenance of tissue or the application of drugs for testing. The layered structure proved especially compatible with existing commercial hardware as electrode contacts could easily be made to match the footprint of commercial amplifiers. Demonstration experiments showed that neural cultures could be kept alive for weeks, that they developed correlated spontaneous activity and could be stimulated. Further fluidic perfusion and drug application were successfully performed. Hence these offer models for further development of “braininachip” technologies.
Fig. 5.
3-D microelectrode arrays for neural culture. (a) Picket fence style 3-D electrode array with integrated electrodes and fluidics [74]). (b) Layered array used successfully to culture neurons, record correlated activity on different layers, and deliver drug to alter activity [73]). (c), (d) views of neurons growing on the top and middle locations in the chamber. [All figures reproduced by permission of The Royal Society of Chemistry (RSC).]
VII. CODING AND PLASTICITY
There is a substantial literature investigating how information may be represented in patterns of action potential activity recorded from MEA, and only a suggestion as to the breadth of the studies is possible here. Strong model formation has come from physicists [75], including modeling bursts as avalanches [76], the use of information theory [75] and affinity and field theory measures [77], [78], and engineers with state-space [79], clustering and multidimensional scaling approaches. Characteristic bursting events [80]–[82]} are often the focus of the analysis, including analyses of burst propagation [83]. However, much is to be done because of the high dimensionality of the data (often 60 or more channels of activity) and wide spread in the time scales of importance (submillisecond for synaptic phenomena, weeks for developmental phenomena).
Progress is being made in developing learning and memory models. Plasticity in burst response has been quantified [84]. The work of Jimbo et al. [85], recently repeated with Grainger causality analysis [86], showed widespread but highly variable potentiation and depression of the pathways connecting stimulating electrode to recorded neuron. Ruaro et al. showed that elementary pattern recognition and signal processing functions could be impressed on a cultured network [87]. Marom’s group showed differential learning of rare and frequent stimuli [88], which formed the basis for DeMarse’s demonstration of control of a simulated device [89]. Feedback control of network behavior has been demonstrated [90].
VIII. FUNCTION FOLLOWS FORM
We are seeing the beginning of the exploration of how functional properties are affected by network geometry. We see increases in neural activity with chronic stimulation of cultured networks [91] and increases in activity with synapse density [92]. These predict enhanced development over weeks of networks geometrically patterned to have greater connectivity, for which we have preliminary evidence [93]. Different sized networks show different statistical properties [94]. A more dramatic example of how geometrically designed networks can have designed function is the narrow line (150 μm) network of Jacobi and Moses [95] wherein propagation of electrical activity is a function of cell density. More recently Feinerman et al. showed how to design the functional equivalents of delay lines, diodes, and and gates with patterned neurons [96].
IX. DISCUSSION AND SUMMARY
The goal of creating a brain on a chip has certainly not been met. However, the progress toward controlling and understanding neural activity in a culture dish has come a long way in the thirty years since first started, and the less than twenty years since the first serious attempts to create patterning technologies. As highlighted above, there are many encouraging reports of progress toward not only understanding the behaviors of these in vitro networks, but of being able to customized their functional properties so that they may be more valuable in the pursuit of basic and applied science applications.
The report above does not highlight the difficulties in performing the work. While culturing, recording and stimulating neurons without patterns is routine in many laboratories around the world, the addition of patterning greatly increases the complexity and risk and reduces the likelihood of any one culture being successful. Still, in our hands networks similar to those in Fig. 2(c), (d) maintain reasonable fidelity to their patterns for a month often enough for experimentation; some have survived for several months with relatively good fidelity [51].
The retreat from a focus on networks of single neurons (from, e.g., [97], [98]) appears largely due to these difficulties, especially for mammalian preparations, leading to “spoke-and-cluster” or “street-plan” constructs for which activity is more robust. Migration to Campenot chamber type recordings with distinct populations of cells in the different wells will be a natural response to searching for a more robust experimental platform. The addition of glia and change of media represent attempts to address the issue of robust electrical activity.
A fortunate aspect of this work has been the tremendous advance in instrumentation and signal processing available from commercial vendors and from software freely available on the web. The front end processing—detecting signals, averaging, performing pairwise correlations, and extracting basic signal properties—is easily accomplished. (We note, however, that there is growing development of integrated circuit microelectrodes with thousands of channels for stimulation and recording, promising a substantial shift in how researchers approach neural culture studies. [99]–[101]) The current technology has enabled investigators to pursue sophisticated statistical approaches to understanding the underlying activity. Much remains to be done, however, as the dimensions of the data set and the inherent nonstationarity of the preparation provide great challenges.
X. EXPERIMENTAL OVERVIEW
Our work has involved mostly primary rat hippocampal or cortical cells taken from embryonic day 18 rat pups. They are cultured according to standard protocols that may be found in the referenced papers. Briefly, cells are taken from the embryonic brains, dissociated, and allowed to settle onto the electrode array or culture dish surfaces, attaching as shown in multiple figures in this paper. They can be maintained for several months if not longer. All procedures were approved by animal use protocols at the University of Illinois and at the Southern Illinois University School of Medicine.
Acknowledgment
The authors acknowledge the tremendous accomplishments of students and staff in their laboratories, including (at Illinois) Joe Corey (original patterning work), Darren Branch (developed microstamping), John Chang (microstamping, cell density, and glial measures), Yoonkey Nam (flexible MEMS, glial patterning, neural activity), David Khatami (optimal patterns for neural activity), Betty Ujehlji [cell culture and Fig. 2(f)], Mauricio Vieira (3-D culturing), Kate Musick (3-D array), Rudi Scharnweber (microtunnel cultures), Brad Dworak (microtunnel array). At Southern Illinois University, Mike Boehler (development of optimal media formulation and addition of glia to enhance activity.) Dr. Brewer acknowledges financial interest in Brain Bits, LLC, which sells neurons and NBActive4, the media formulation optimized to enhance spiking activity.
This work was supported by U.S. taxpayers via multiple grants from National Institutes of Health, most importantly R01 NS 052233 and R01 EB000786, and the National Science Foundation EIA 0130828, as well as Illinois and now Florida taxpayers through support for the investigators and students.
Biography

Bruce C. Wheeler (Fellow, IEEE) received the B.S. degree from the Massachusetts Institute of Technology, Cambridge, and the M.S. and Ph.D. in electrical engineering from Cornell University, Ithaca, NY.
He was at the University of Illinois for 28 years, including service as Founding and Interim Head of the Bioengineering Department, Associate Head of Electrical and Computer Engineering, and Chair of the Neuroscience Program. He was Professor of BioE, ECE, and the Beckman Institute. He recently moved to the University of Florida and was appointed Interim Chair of the J. Crayton Pruitt Family Department of Biomedical Engineering. His research interests lie in the application of electrical engineering methodologies to neuroscience, including neural spike sorting technologies, microelectrode arrays for use with brain slices and cell culture, lithography to the geometric patterns of neural growth in culture.
Prof. Wheeler is a Fellow of the AIMBE. He is the Editor in Chief of the IEEE Transactions on Biomedical Engineering.

Gregory J. Brewer received the B.S. degree from California Institute of Technology, Pasadena, and the Ph.D. degree in biology from the University of California, San Diego.
He holds the Kenneth Stark Endowed Chair in Alzheimer’s Research and is Professor of Neurology and Medical Microbiology, Immunology and Cell Biology at Southern Illinois University School of Medicine, where he has been for 29 years. His research interests lie in the application of engineering and biochemistry principles to neuron cell culture, development and live neural networks. He also has projects on the neuroscience of aging and is founder and President of BrainBits LLC, which supplies live brain tissue to neuroscientists.
Contributor Information
Bruce C. Wheeler, Pruitt Family Department of Biomedical Engineering, University of Florida, Gainesville, FL 32611 USA. Departments of Bioengineering and Electrical and Computer Engineering, Neuroscience Program and Beckman Institute, University of Illinois, Urbana, IL 61801 USA (bwheeler@ufl.edu)..
Gregory J. Brewer, Departments of Neurology and Medical Microbiology, Immunology and Cell Biology, Southern Illinois University School of Medicine, Springfield, IL 62794 USA (gbrewer@siumed.edu).
REFERENCES
- [1].Thomas CA, Springer PA, Okun LM, Berwald Y, Loeb GE. Miniature microelectrode array to monitor bioelectric activity of cultured cells. Exp. Cell Res. 1972;vol. 74(no. 1):61–66. doi: 10.1016/0014-4827(72)90481-8. [DOI] [PubMed] [Google Scholar]
- [2].Pine J. Recording action-potentials from cultured neurons with extracellular micro-circuit electrodes. J. Neurosci. Methods. 1980;vol. 2(no. 1):19–31. doi: 10.1016/0165-0270(80)90042-4. [DOI] [PubMed] [Google Scholar]
- [3].Gross GW. Simultaneous single unit recording invitro with a photoetched laser deinsulated gold multi-micro-electrode surface. IEEE Trans. Biomed. Eng. 1979 May;vol. BME-26(no. 5):273–279. doi: 10.1109/tbme.1979.326402. [DOI] [PubMed] [Google Scholar]
- [4].Jobling DT, Smith JG, Wheal HV. Active microelectrode array to record from the mammalian central nervous system in vitro. Med. Biol. Eng. Comput. 1981;vol. 19(no. 5):553–560. doi: 10.1007/BF02442768. [DOI] [PubMed] [Google Scholar]
- [5].Novak JL, Wheeler BC. Multisite hippocampal slice recording and stimulation using a 32 element microelectrode array. J. Neurosci. Methods. 1988;vol. 23(no. 2):149–159. doi: 10.1016/0165-0270(88)90187-2. [DOI] [PubMed] [Google Scholar]
- [6].Wheeler BC, Novak JL. Current source density-estimation using microelectrode array data from the hippocampal slice preparation. IEEE Trans. Biomed. Eng. 1986 Dec;vol. BME-33(no. 12):1204–1212. doi: 10.1109/TBME.1986.325701. [DOI] [PubMed] [Google Scholar]
- [7].Gross GW, Harsch A, Rhoades BK, Gopel W. Odor, drug and toxin analysis with neuronal networks in vitro: Extracellular array recording of network responses. Biosens. Bioelectron. 1997;vol. 12(no. 5):373–393. doi: 10.1016/s0956-5663(97)00012-2. [DOI] [PubMed] [Google Scholar]
- [8].Proceedings of the MEA Meeting; Stuttgart, Germany: BioPro Baden Wuerttemburg GmbH; 2008. 2008. [Google Scholar]
- [9].Wheeler BC. Building a brain on a chip. Proc. EMBS 2008; Vancouver: BC, Canada. Aug. 20–25, 2008. [Google Scholar]
- [10].Carter SB. Principles of cell motility—Direction of cell movement and cancer invasion. Nature. 1965;vol. 208(no. 5016):1183–1187. doi: 10.1038/2081183a0. [DOI] [PubMed] [Google Scholar]
- [11].Ivanova OY, Margolis LB. Use of phospholipid film for shaping cell-cultures. Nature. 1973;vol. 242(no. 5394):200–201. doi: 10.1038/242200a0. [DOI] [PubMed] [Google Scholar]
- [12].Letourneau PC. Cell-to-substratum adhesion and guidance of axonal elongation. Dev. Biol. 1975;vol. 44(no. 1):92–101. doi: 10.1016/0012-1606(75)90379-6. [DOI] [PubMed] [Google Scholar]
- [13].Kleinfeld D, Kahler KH, Hockberger PE. Controlled outgrowth of dissociated neurons on patterned substrates. J. Neurosci. 1988;vol. 8(no. 11):4098–4120. doi: 10.1523/JNEUROSCI.08-11-04098.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Corey JM, Wheeler BC, Brewer GJ. Compliance of hippocampal-neurons to patterned substrate networks. J. Neurosci. Res. 1991;vol. 30(no. 2):300–307. doi: 10.1002/jnr.490300204. [DOI] [PubMed] [Google Scholar]
- [15].Dulcey CS, Georger JH, Krauthamer V, Stenger DA, Fare TL, Calvert JM. Deep UV photochemistry of chemisorbed monolayers—Patterned coplanar molecular assemblies. Science. 1991;vol. 252(no. 5005):551–554. doi: 10.1126/science.2020853. [DOI] [PubMed] [Google Scholar]
- [16].Hickman JJ, Bhatia SK, Quong JN, Schoen P, Stenger DA, Pike CJ, Cotman CW. Rational pattern design for in-vitro cellular networks using surface photochemistry. J. Vac. Sci. Technol. A, Vac. Surf. Films. 1994;vol. 12(no. 3):607–616. [Google Scholar]
- [17].Corey JM, Wheeler BC, Brewer GJ. Micrometer resolution silane-based patterning of hippocampal neurons: Critical variables in photoresist and laser ablation processes for substrate fabrication. IEEE Trans. Biomed. Eng. 1996 Sep;vol. 43(no. 9):944–955. doi: 10.1109/10.532129. [DOI] [PubMed] [Google Scholar]
- [18].Lom B, Healy KE, Hockberger PE. A versatile technique for patterning biomolecules onto glass coverslips. J. Neurosci. Methods. 1993;vol. 50(no. 3):385–397. doi: 10.1016/0165-0270(93)90044-r. [DOI] [PubMed] [Google Scholar]
- [19].Wyart C, Ybert C, Bourdieu L, Herr C, Prinz C, Chatenay D. Constrained synaptic connectivity in functional mammalian neuronal networks grown on patterned surfaces. J. Neurosci. Methods. 2002;vol. 117(no. 2):123–131. doi: 10.1016/s0165-0270(02)00077-8. [DOI] [PubMed] [Google Scholar]
- [20].Branch DW, Corey JM, Weyhenmeyer JA, Brewer GJ, Wheeler BC. Microstamp patterns of biomolecules for high-resolution neuronal networks. Med. Biol. Eng. Comput. 1998;vol. 36(no. 1):135–141. doi: 10.1007/BF02522871. [DOI] [PubMed] [Google Scholar]
- [21].Chang JC, Brewer GJ, Wheeler BC. A modified microstamping technique enhances polylysine transfer and neuronal cell patterning. Biomaterials. 2003;vol. 24(no. 17):2863–2870. doi: 10.1016/s0142-9612(03)00116-9. [DOI] [PubMed] [Google Scholar]
- [22].Lopez GP, Dimilla PA, Kumar A, Harter R, Whitesides GM. Use of self-assembled monolayers of alkanethiolates on gold for the study and manipulation of interactions of proteins and cells with solid-surfaces. J. Cell. Biochem. 1994:250. [Google Scholar]
- [23].Singhvi R, Kumar A, Lopez GP, Stephanopoulos GN, Wang DIC, Whitesides GM, Ingber DE. Engineering cell-shape and function. Science. 1994;vol. 264(no. 5159):696–698. doi: 10.1126/science.8171320. [DOI] [PubMed] [Google Scholar]
- [24].StJohn PM, Kam L, Turner SW, Craighead HG, Issacson M, Turner JN, Shain W. Preferential glial cell attachment to microcontact printed surfaces. J. Neurosci. Methods. 1997;vol. 75(no. 2):171–177. doi: 10.1016/s0165-0270(97)00069-1. [DOI] [PubMed] [Google Scholar]
- [25].Vogt AK, Wrobel G, Meyer W, Knoll W, Offenhausser A. Synaptic plasticity in micropatterned neuronal networks. Biomaterials. 2005;vol. 26(no. 15):2549–2557. doi: 10.1016/j.biomaterials.2004.07.031. [DOI] [PubMed] [Google Scholar]
- [26].Chiu DT, Jeon NL, Huang S, Kane RS, Wargo CJ, Choi IS, Ingber DE, Whitesides GM. Patterned deposition of cells and proteins onto surfaces by using three-dimensional microfluidic systems. Proc. Natl. Acad. Sci. USA. 2000;vol. 97(no. 6):2408–2413. doi: 10.1073/pnas.040562297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Vielmetter J, Stolze B, Bonhoeffer F, Stuermer CAO. Invitro assay to test differential substrate affinities of growing axons and migratory cells. Exp. Brain Res. 1990;vol. 81(no. 2):283–287. doi: 10.1007/BF00228117. [DOI] [PubMed] [Google Scholar]
- [28].Clark P, Connolly P, Curtis ASG, Dow JAT, Wilkinson CDW. Topographical control of cell behavior.2. Multiple grooved substrata. Development. 1990;vol. 108(no. 4):635–644. doi: 10.1242/dev.108.4.635. [DOI] [PubMed] [Google Scholar]
- [29].Curtis ASG, Clark P. The effects of topographic and mechanical-properties of materials on cell behavior. Crit. Rev. Biocompatibility. 1990;vol. 5(no. 4):343–362. [Google Scholar]
- [30].Suzuki I, Sugio Y, Jimbo Y, Yasuda K. Stepwise pattern modification of neuronal network in photo-thermally-etched agarose architecture on multi-electrode array chip for individual-cell-based electrophysiological measurement. Lab Chip. 2005;vol. 5(no. 3):241–247. doi: 10.1039/b406885h. [DOI] [PubMed] [Google Scholar]
- [31].Nam Y, Branch DW, Wheeler BC. Epoxy-silane linking of biomolecules is simple and effective for patterning neuronal cultures. Biosens. Bioelectron. 2006;vol. 22(no. 5):589–597. doi: 10.1016/j.bios.2006.01.027. [DOI] [PubMed] [Google Scholar]
- [32].Stenger DA, Pike CJ, Hickman JJ, Cotman CW. Surface determinants of neuronal survival and growth on self-assembled monolayers in culture. Brain Res. 1993;vol. 630(no. 1–2):136–147. doi: 10.1016/0006-8993(93)90651-3. [DOI] [PubMed] [Google Scholar]
- [33].Branch DW, Wheeler BC, Brewer GJ, Leckband DE. Long-term maintenance of patterns of hippocampal pyramidal cells on substrates of polyethylene glycol and microstamped polylysine. IEEE Trans. Biomed. Eng. 2000 Mar;vol. 47(no. 3):290–300. doi: 10.1109/10.827289. [DOI] [PubMed] [Google Scholar]
- [34].Branch DW, Wheeler BC, Brewer GJ, Leckband DE. Long-term stability of grafted polyethylene glycol surfaces for use with microstamped substrates in neuronal cell culture. Biomaterials. 2001;vol. 22(no. 10):1035–1047. doi: 10.1016/s0142-9612(00)00343-4. [DOI] [PubMed] [Google Scholar]
- [35].Esch T, Lemmon V, Banker G. Local presentation of substrate molecules directs axon specification by cultured hippocampal neurons. J. Neurosci. 1999;vol. 19(no. 15):6417–6426. doi: 10.1523/JNEUROSCI.19-15-06417.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Oliva AA, James CD, Kingman CE, Craighead HG, Banker GA. Patterning axonal guidance molecules using a novel strategy for microcontact printing. Neurochem. Res. 2003;vol. 28(no. 11):1639–1648. doi: 10.1023/a:1026052820129. [DOI] [PubMed] [Google Scholar]
- [37].Matsuzawa M, Tabata T, Knoll W, Kano M. Formation of hippocampal synapses on patterned substrates of a laminin-derived synthetic peptide. Eur. J. Neurosci. 2000;vol. 12(no. 3):903–910. doi: 10.1046/j.1460-9568.2000.00977.x. [DOI] [PubMed] [Google Scholar]
- [38].Li NZ, Folch A. Integration of topographical and biochemical cues by axons during growth on microfabricated 3-D substrates. Exp. Cell Res. 2005;vol. 311(no. 2):307–316. doi: 10.1016/j.yexcr.2005.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Goldner JS, Bruder JM, Li G, Gazzola D, Hoffman-Kim D. Neurite bridging across micropatterned grooves. Biomaterials. 2006;vol. 27(no. 3):460–472. doi: 10.1016/j.biomaterials.2005.06.035. [DOI] [PubMed] [Google Scholar]
- [40].Mai J, Fok L, Gao HF, Zhang X, Poo MM. Axon initiation and growth cone turning on bound protein gradients. J. Neurosci. 2009;vol. 29(no. 23):7450–7458. doi: 10.1523/JNEUROSCI.1121-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Zeck G, Fromherz P. Noninvasive neuroelectronic interfacing with synaptically connected snail neurons immobilized on a semiconductor chip. Proc. Nat. Acad. Sci. USA. 2001;vol. 98(no. 18):10 457–10 462. doi: 10.1073/pnas.181348698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Fromherz P, Bainbridge WS, Roco MC, editors. Progress in Convergence: Technologies for Human Wellbeing. Blackwell; Oxford, U.K.: 2006. Three levels of neuroelectronic interfacing—Silicon chips with ion channels, nerve cells, and brain tissue; pp. 143–160. [DOI] [PubMed] [Google Scholar]
- [43].Erickson J, Tooker A, Tai YC, Pine J. Caged neuron MEA: A system for long-term investigation of cultured neural network connectivity. J. Neurosci. Methods. 2008;vol. 175(no. 1):1–16. doi: 10.1016/j.jneumeth.2008.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Morin F, Nishimura N, Griscom L, LePioufle B, Fujita H, Takamura Y, Tamiya E. Constraining the connectivity of neuronal networks cultured on microelectrode arrays with microfluidic techniques: A step towards neuron-based functional chips. Biosens. Bioelectron. 2006;vol. 21(no. 7):1093–1100. doi: 10.1016/j.bios.2005.04.020. [DOI] [PubMed] [Google Scholar]
- [45].Suzuki I, Sugio Y, Jimbo Y, Yasuda K. Individual-cell-based electrophysiological measurement of a topographically controlled neuronal network pattern using agarose architecture with a multi-electrode array. Cell Struct. Funct. 2004;vol. 29:59. [Google Scholar]
- [46].Taylor AM, Blurton-Jones M, Rhee SW, Cribbs DH, Cotman CW, Jeon NL. Nat. Methods. 2005;vol. 2(no. 8):599–605. doi: 10.1038/nmeth777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Dworak BJ, Wheeler BC. Novel MEA platform with PDMS microtunnels enables the detection of action potential propagation from isolated axons in culture. Lab Chip. 2009;vol. 9(no. 3):404–410. doi: 10.1039/b806689b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Ruardij TG, van den Boogaart MAF, Rutten WLC. Adhesion and growth of electrically-active cortical neurons on polyethyleneimine patterns microprinted on PEO-PPO-PEO triblockcopolymer-coated hydrophobic surfaces. IEEE Trans. Nanobioscience. 2002 Mar;vol. 1(no. 1):1–8. doi: 10.1109/tnb.2002.806921. [DOI] [PubMed] [Google Scholar]
- [49].Rutten WLC, Ruardij TG, Marani E, Roelfsema PR. Cultured neural networks: Optimization of patterned network adhesiveness and characterization of their neural activity. Appl. Bionics Biomech. 2006;vol. 3(no. 1):1–7. [Google Scholar]
- [50].Khatami D. M.S. thesis. Univ. Illinois; Urbana: 2004. Effects of spatial confinement on the spontaneous and evoked activity of linearly patterned biological neural networks. [Google Scholar]
- [51].Khatami D. Ph.D. thesis. Univ. Illinois; Urbana: 2008. Considerations for electrical stimulation and functional connectivity analysis of neuronal networks patterned on microelectrode arrays. [Google Scholar]
- [52].Chang JC, Brewer GJ, Wheeler BC. Modulation of neural network activity by patterning. Biosens. Bioelectron. 2001;vol. 16(no. 7–8):527–533. doi: 10.1016/s0956-5663(01)00166-x. [DOI] [PubMed] [Google Scholar]
- [53].Vogt AK, Brewer GJ, Offenhausser A. Connectivity patterns in neuronal networks of experimentally defined geometry. Tissue Eng. 2005;vol. 11(no. 11–12):1757–1767. doi: 10.1089/ten.2005.11.1757. [DOI] [PubMed] [Google Scholar]
- [54].Brewer GJ, Torricelli JR, Evege EK, Price PJ. Optimized survival of hippocampal-neurons in B 27-supplemented Neurobasal(TM), a new serum-free medium combination. J. Neurosci. Res. 1993;vol. 35(no. 5):567–576. doi: 10.1002/jnr.490350513. [DOI] [PubMed] [Google Scholar]
- [55].Nam Y, Chang J, Khatami D, Wheeler BC. Patterning to enhance activity of cultured neuronal networks. IEE Proc. Nanobiotechnol. 2004;vol. 151(no. 3):109–115. doi: 10.1049/ip-nbt:20040706. [DOI] [PubMed] [Google Scholar]
- [56].Chang JC, Brewer GJ, Wheeler BC. Neuronal network structuring induces greater neuronal activity through enhanced astroglial development. J. Neural Eng. 2006;vol. 3(no. 3):217–226. doi: 10.1088/1741-2560/3/3/004. [DOI] [PubMed] [Google Scholar]
- [57].Nam Y, Brewer GJ, Wheeler BC. Development of astroglial cells in patterned neuronal cultures. J. Biomater. Sci.-Polym. Ed. 2007;vol. 18(no. 8):1091–1100. doi: 10.1163/156856207781494430. [DOI] [PubMed] [Google Scholar]
- [58].Pfrieger FW, Barres BA. Synaptic efficacy enhanced by glial cells in vitro. Science. 1997;vol. 277(no. 5332):1684–1687. doi: 10.1126/science.277.5332.1684. [DOI] [PubMed] [Google Scholar]
- [59].Ullian EM, Sapperstein SK, Christopherson KS, Barres BA. Control of synapse number by glia. Science. 2001;vol. 291(no. 5504):657–661. doi: 10.1126/science.291.5504.657. [DOI] [PubMed] [Google Scholar]
- [60].Boehler MD, Wheeler BC, Brewer GJ. Added astroglia promote greater synapse density and higher activity in neuronal networks. Neuron Glia Biol. 2007;vol. 3:127–140. doi: 10.1017/S1740925X07000440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Brewer GJ, Boehler MD, Jones TT, Wheeler BC. NbActiv4 medium improvement to Neurobasal/B27 increases neuron synapse densities and network spike rates on multielectrode arrays. J. Neurosci. Methods. 2008;vol. 170(no. 2):181–187. doi: 10.1016/j.jneumeth.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Wagenaar DA, Madhavan R, Pine J, Potter SM. Controlling bursting in cortical cultures with closed-loop multi-electrode stimulation. J. Neurosci. 2005;vol. 25(no. 3):680–688. doi: 10.1523/JNEUROSCI.4209-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Campenot RB. Local control of neurite development by nerve growth-factor. Proc. Natl. Acad. Sci. USA. 1977;vol. 74(no. 10):4516–4519. doi: 10.1073/pnas.74.10.4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Park JW, Vahidi B, Kim HJ, Rhee SW, Jeon NL. Quantitative analysis of CNS axon regeneration using a microfluidic neuron culture device. BioChip J. 2008;vol. 2(no. 1):44–51. [Google Scholar]
- [65].Park JW, Vahidi B, Taylor AM, Rhee SW, Jeon NL. Microfluidic culture platform for neuroscience research. Nat. Protoc. 2006;vol. 1(no. 4):2128–2136. doi: 10.1038/nprot.2006.316. [DOI] [PubMed] [Google Scholar]
- [66].Scharnweber R. Ph.D. thesis. Univ. Illinois; Urbana: 2009. Neuronal polarization and axonal mRNA localization on microfabricated substrates. [Google Scholar]
- [67].Edelman DB, Keefer EW. A cultural renaissance: In vitro cell biology embraces three-dimensional context. Exp. Neurol. 2005;vol. 192(no. 1):1–6. doi: 10.1016/j.expneurol.2004.10.005. [DOI] [PubMed] [Google Scholar]
- [68].Irons HR, Cullen DK, Shapiro NP, Lambert NA, Lee RH, LaPlaca MC. Three-dimensional neural constructs: A novel platform for neurophysiological investigation. J. Neural Eng. 2008;vol. 5(no. 3):333–341. doi: 10.1088/1741-2560/5/3/006. [DOI] [PubMed] [Google Scholar]
- [69].Ma W, Fitzgerald W, Liu QY, O’Shaughnessy TJ, Maric D, Lin HJ, Alkon DL, Barker JL. CNS stem and progenitor cell differentiation into functional neuronal circuits in three-dimensional collagen gels. Exp. Neurol. 2004;vol. 190(no. 2):276–288. doi: 10.1016/j.expneurol.2003.10.016. [DOI] [PubMed] [Google Scholar]
- [70].Cullen DK, Vukasinovic J, Glezer A, LaPlaca MC. Microfluidic engineered high cell density three-dimensional neural cultures. J. Neural Eng. 2007;vol. 4(no. 2):159–172. doi: 10.1088/1741-2560/4/2/015. [DOI] [PubMed] [Google Scholar]
- [71].Hynd MR, Frampton JP, Dowell-Mesfin N, Turner JN, Shain W. Directed cell growth on protein-functionalized hydrogel surfaces. J. Neurosci. Methods. 2007;vol. 162(no. 1–2):255–263. doi: 10.1016/j.jneumeth.2007.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Luo Y, Shoichet MS. A photolabile hydrogel for guided three-dimensional cell growth and migration. Nat. Mater. 2004;vol. 3(no. 4):249–253. doi: 10.1038/nmat1092. [DOI] [PubMed] [Google Scholar]
- [73].Musick K, Khatami D, Wheeler BC. Three-dimensional micro-electrode array for recording dissociated neuronal cultures. Lab Chip. 2009;vol. 9(no. 14):2036–2042. doi: 10.1039/b820596e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Rowe L, Almasri M, Lee K, Fogleman N, Brewer GJ, Nam Y, Wheeler BC, Vukasinovic J, Glezer A, Frazier AB. Active 3-D microscaffold system with fluid perfusion for culturing in vitro neuronal networks. Lab Chip. 2007;vol. 7(no. 4):475–482. doi: 10.1039/b700795g. [DOI] [PubMed] [Google Scholar]
- [75].Bettencourt LMA, Stephens GJ, Ham MI, Gross GW. Functional structure of cortical neuronal networks grown in vitro. Phys. Rev. E. 2007;vol. 75(no. 2):10. doi: 10.1103/PhysRevE.75.021915. [DOI] [PubMed] [Google Scholar]
- [76].Beggs JM, Plenz D. Neuronal avalanches are diverse and precise activity patterns that are stable for many hours in cortical slice cultures. J. Neurosci. 2004;vol. 24(no. 22):5216–5229. doi: 10.1523/JNEUROSCI.0540-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Segev R, Benveniste M, Hulata E, Cohen N, Palevski A, Kapon E, Shapira Y, Ben-Jacob E. Long term behavior of lithographically prepared in vitro neuronal networks. Phys. Rev. Lett. 2002;vol. 88(no. 11):4. doi: 10.1103/PhysRevLett.88.118102. [DOI] [PubMed] [Google Scholar]
- [78].Segev R, Baruchi I, Hulata E, Ben-Jacob E. Hidden neuronal correlations in cultured networks. Phys. Rev. Lett. 2004;vol. 92(no. 11):4. doi: 10.1103/PhysRevLett.92.118102. [DOI] [PubMed] [Google Scholar]
- [79].Dimoka A, Courellis SH, Gholmieh GI, Marmarelis VZ, Berger TW. Modeling the nonlinear properties of the in vitro hippocampal perforant path-dentate system using multielectrode array technology. IEEE Trans. Biomed. Eng. 2008 Feb;vol. 55(no. 2):693–702. doi: 10.1109/TBME.2007.908075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].van Pelt J, Wolters PS, Corner MA, Rutten WLC, Ramakers GJA. Long-term characterization of firing dynamics of spontaneous bursts in cultured neural networks. IEEE Trans. Biomed. Eng. 2004 Nov;vol. 51(no. 11):2051–2062. doi: 10.1109/TBME.2004.827936. [DOI] [PubMed] [Google Scholar]
- [81].Wagenaar DA, Pine J, Potter SM. An extremely rich repertoire of bursting patterns during the development of cortical cultures. BMC Neurosci. 2006;vol. 7:17. doi: 10.1186/1471-2202-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Stegenga J, Le Feber J, Marani E, Rutten WLC. Analysis of cultured neuronal networks using intraburst firing characteristics. IEEE Trans. Biomed. Eng. 2008 Apr;vol. 55(no. 4):1382–1390. doi: 10.1109/TBME.2007.913987. [DOI] [PubMed] [Google Scholar]
- [83].Eckmann JP, Jacobi S, Marom S, Moses E, Zbinden C. Leader neurons in population bursts of 2D living neural networks. New J. Phys. 2008;vol. 10:19. [Google Scholar]
- [84].Stegenga J, Le Feber J, Marani E, Rutten WLC. The effect of learning on bursting. IEEE Trans. Biomed. Eng. 2009 Apr;vol. 56(no. 4):1220–1227. doi: 10.1109/TBME.2008.2006856. [DOI] [PubMed] [Google Scholar]
- [85].Jimbo Y, Tateno T, Robinson HPC. Simultaneous induction of pathway-specific potentiation and depression in networks of cortical neurons. Biophys. J. 1999;vol. 76(no. 2):670–678. doi: 10.1016/S0006-3495(99)77234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Cadotte AJ, DeMarse TB, He P, Ding M. Causal measures of structure and plasticity in simulated and living neural networks. PLoS ONE. 2008;vol. 3(no. 10):e3355. doi: 10.1371/journal.pone.0003355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Ruaro ME, Bonifazi P, Torre V. Toward the neurocomputer: Image processing and pattern recognition with neuronal cultures. IEEE Trans. Biomed. Eng. 2005 Mar;vol. 52(no. 3):371–383. doi: 10.1109/TBME.2004.842975. [DOI] [PubMed] [Google Scholar]
- [88].Eytan D, Brenner N, Marom S. Selective adaptation in networks of cortical neurons. J. Neurosci. 2003;vol. 23(no. 28):9349–9356. doi: 10.1523/JNEUROSCI.23-28-09349.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].DeMarse TB, Dockendorf KP. Adaptive flight control with living neuronal networks on microelectrode arrays. Proc. Int. Joint Conf. Comput. and Neural Netw..2005. pp. 1548–1551. [Google Scholar]
- [90].Bakkum DJ, Chao ZC, Potter SM. Spatio-temporal electrical stimuli shape behavior of an embodied cortical network in a goal-directed learning task. J. Neural Eng. 2008;vol. 5(no. 3):310–323. doi: 10.1088/1741-2560/5/3/004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Brewer GJ, Boehler MD, Ide AN, Wheeler BC. Chronic electrical stimulation of cultured hippocampal networks increases spontaneous spike rates. J. Neurosci. Methods. 2009;vol. 184(no. 1):104–109. doi: 10.1016/j.jneumeth.2009.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Brewer GJ, Boehler MD, Pearson RA, DeMaris AA, Ide AN, Wheeler BC. Neuron network activity scales exponentially with synapse density. J. Neural Eng. 2009;vol. 6(no. 1):7. doi: 10.1088/1741-2560/6/1/014001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Khatami D, Brewer GJ, Wheeler BC. Effect of short- and long-term electrical stimulation on network behavior. Proc. MEA 2008 (6th Int. Mtg., Substrate-Integrated Microelectrode Arrays); Reutlingen, Germany. 2008. pp. 57–58. [Google Scholar]
- [94].Volman V, Baruchi I, Ben-Jacob E. Manifestation of function-follow-form in cultured neuronal networks. Phys. Biol. 2005;vol. 2(no. 2):98–110. doi: 10.1088/1478-3975/2/2/003. [DOI] [PubMed] [Google Scholar]
- [95].Jacobi S, Moses E. Variability and corresponding amplitude-velocity relation of activity propagating in one-dimensional neural cultures. J. Neurophysiol. 2007;vol. 97(no. 5):3597–3606. doi: 10.1152/jn.00608.2006. [DOI] [PubMed] [Google Scholar]
- [96].Feinerman O, Rotem A, Moses E. Reliable neuronal logic devices from patterned hippocampal cultures. Nat. Phys. 2008;vol. 4(no. 12):967–973. [Google Scholar]
- [97].Maher MP, Pine J, Wright J, Tai YC. The neurochip: A new multielectrode device for stimulating and recording from cultured neurons. J. Neurosci. Methods. 1999;vol. 87(no. 1):45–56. doi: 10.1016/s0165-0270(98)00156-3. [DOI] [PubMed] [Google Scholar]
- [98].Merz M, Fromherz P. Polyester microstructures for topographical control of outgrowth and synapse formation of snail neurons. Adv. Mater. 2002;vol. 14(no. 2):141–144. [Google Scholar]
- [99].Eversmann B, Jenkner M, Hofmann F, Paulus C, Brederlow R, Holzapfl B, Fromherz P, Merz M, Brenner M, Schreiter M, Gabl R, Plehnert K, Steinhauser M, Eckstein G, Schmitt-Landsiedel D, Thewes R. A 128 × 128 CMOS biosensor array for extracellular recording of neural activity. IEEE J. Solid-State Circuits. 2003 Dec;vol. 38(no. 12):2306–2317. [Google Scholar]
- [100].Berdondini L, van der Wal PD, Guenat O, de Rooij NF, Koudelka-Hep M, Seitz P, Kaufmann R, Metzler P, Blanc N, Rohr S. High-density electrode array for imaging in vitro electrophysiological activity. Biosens. Bioelectron. 2005;vol. 21(no. 1):167–174. doi: 10.1016/j.bios.2004.08.011. [DOI] [PubMed] [Google Scholar]
- [101].Frey U, Egert U, Heer F, Hafizovic S, Hierlemann A. Microelectronic system for high-resolution mapping of extracellular electric fields applied to brain slices. Biosens. Bioelectron. 2009;vol. 24(no. 7):2191–2198. doi: 10.1016/j.bios.2008.11.028. [DOI] [PubMed] [Google Scholar]