Abstract
207Pb NMR spectroscopy can be used to monitor the binding of Pb(II) to thiol rich biological small molecules such as glutathione and to zinc finger proteins. The UV/visible (UV/Vis) absorption band centered at 334 nM and the observed 207Pb-signal in 207Pb NMR (δ ~ 5750 ppm) indicate that glutathione binds Pb(II) in a trigonal pyramidal geometry (PbS3) at pH 7.5 or higher with a 1:3 molar ratio of Pb(II) to GSH. While previous studies using UV/Vis and extended X-ray absorption fine structure (EXAFS) spectroscopy were interpreted to show that the zinc binding domain from HIV nucleocapsid protein (HIV-CCHC) binds Pb(II) in a single PbS3 environment, the more sensitive 207Pb NMR spectra (at pH 7.0, 1:1 molar ratio) provide compelling evidence for the presence of two PbS3 structures (δ = 5790 and 5744 ppm), one of which is more stable at high temperatures. It has previously been proposed that the HIV-CCHH peptide does not fold properly to afford a PbS2N motif, because histidine does not bind to Pb(II). These predictions are confirmed by the present studies. These results demonstrate the applicability of 207Pb NMR to biomolecular structure determination in proteins with cysteine binding sites for the first time.
Keywords: Glutathione, 207Pb-NMR Spectroscopy, heavy metal toxicity, zinc fingers
1. Introduction
Heavy metal (Pb, Hg, As, Cd) toxicity is a worldwide concern. Plants and animals, including humans, have been exposed to toxic metal ions through soil, contaminated food, drinking water, and the inhalation of polluted air. Heavy metals can cause cellular damage via diverse mechanisms such as interrupting essential metal ions homeostatsis, perturbing membrane potentials and through inhibition of metalloenzymes active sites [1]. Pb(II) targets several zinc enzymes and proteins such as δ -aminolevulinic acid dehydratase (ALAD) [2], carbonic anhydrase, acetylcoline esterase and zinc-finger proteins (Cys4, Cys3His and Cys2His2). In addition, the similarity of Pb(II) to Ca(II) in higher coordination numbers and with oxygen and nitrogen ligands makes calcium binding proteins another potential target [2,3]. To protect the human body against heavy metal toxicity, cells synthesize thiol-rich small molecules such as glutathione and metallothionein which can sequester toxic metal ions before they can cause deleterious effects. Glutathione (GSH), a tripeptide containing glutamic acid, cysteine and glycine (γ-Glu-Cys-Gly) residues is present in millimolar concentrations (0.1 to 10 mM) in erythrocytes [4], and is the most abundant nonprotein thiol in cellular systems. GSH plays multiple roles in cells. As an antioxidant, GSH protects cells from reactive oxygen species that can damage DNA and RNA. Heavy metals are quite thiophilic and show a high affinity for the thiol of cysteine, allowing GSH to serve as a chelater for heavy metals in cellular compartments [5]. Thus, defining the chemistry of glutathione with Pb(II) is important for understanding the toxicity of this element.
Heavy metals also target gene regulatory proteins such as zinc fingers. Several zinc-dependent enzymes and proteins have been identified in which zinc functions either catalytically or structurally [6]. The most common and best studied structural zinc dependent proteins are the zinc fingers. Based on the ligand sets present in the metal binding sites, zinc fingers are classified into 3 types [7]: (i) Cys2His2 or the cellular/transcription factor type (e.g. Xenopus TFIIIA, Sp1), (ii) Cys2HisCys or the retroviral type, represented by the retroviral nucleocapsid proteins; and (iii) Cys4 or the steroid/thyroid hormone receptor type. It has been shown that As(III), Cd(II), Hg(II) and Pb(II) inhibit the DNA repair process due to the displacement of zinc ion in zinc finger structures of DNA repair proteins [8,9]. Based on sequence alignment of 131 sequences of transcription factor proteins, Berg and coworkers designed a consensus zinc finger peptide that contains 26 amino acid residues [10,11]. Recently, Godwin and coworkers reported detailed lead binding studies on a series of zinc binding domains of consensus peptides (CP-CCHC, CP-CCHH and CP-CCCC) and the HIV nucleocapsid protein (HIV-CCHC) and suggested that they are potential targets for Pb(II) [12,13]. Zawia and his coworkers have suggested that Pb(II) can target the traditional Cys2His2 type of zinc fingers by displacing Zn(II), altering the structure of the zinc finger motifs and consequently altering the regulation of gene expression [14]. In contrast, Godwin’s studies on model peptides with Cys3 and Cys4 motifs show that Pb(II) has higher binding affinity than Zn(II); but Pb(II) cannot compete with Zn(II) for binding to the His2Cys2 site [13]. This is probably due to the improper folding of the peptide and that histidine may not coordinate Pb(II) ion tightly.
A variety of spectroscopic techniques including 113Cd and 199Hg NMR spectroscopy have been used to study the binding mode of these heavy metal ions to the peptides or proteins. However, 207Pb NMR spectroscopy has not been used to study Pb(II) binding to thiol rich centers in natural or synthetic biomolecules until very recently. We reported the use of 207Pb NMR spectroscopy to probe a PbS3 environment in designed three stranded coiled coil peptides, demonstrating that 207Pb NMR could now be fruitfully applied to understanding the molecular mechanism of Pb(II) toxicity in thiol rich systems [15]. Resonances within the 207Pb NMR chemical shift range from 5600 to 5800 ppm were observed for PbS3 complexes in this homoleptic peptide environment.
Despite the great importance of Pb-toxicity, there are no examples of 207Pb NMR spectroscopy being used to study Pb-binding to biomolecules such as glutathione and zinc finger proteins. The binding of Pb(II) to glutathione [16] and zinc finger proteins [13,17] has been studied in detail using UV/Vis, CD, EXAFS, and 1H-NMR spectroscopy, but these techniques have not given complete insight into the bonding geometry of the Pb(II) ion. To obtain a better understanding of how Pb(II) is bound to sulfur rich biomolecules, we have used 207Pb NMR spectroscopy to study the binding of Pb(II) to GSH under physiological concentrations (5 mM [Pb(GSH)3]− rather than the 90 mM [Pb(GSH)3]− that was previously studied) [16] and zinc finger peptides. 207Pb NMR spectroscopy has yet to be used to study the binding of Pb(II) with these biologically important biomolecules. We believe this new application of 207Pb NMR spectroscopy will bring the powerful structural potential of NMR to clarify Pb(II) toxicity, and potentially opens the door for probing Pb(II) toxicity in Pb(II) poisoned human blood samples.
2. Experimental Section
2.1. Materials
N-α-(9-Fluorenyl methyloxycarbonyl) (Fmoc) protected amino acids, MBHA Rink Amide resin, N-hydroxybenzotriazole (HOBt), and 2-(1H-benzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HBTU) were purchased from AnaSpec. Reduced glutathione, diisopropylethylamine (DIPEA), acetic anhydride, and pyridine were purchased from Aldrich. N-methylpyrrolidinone (NMP) was purchased from Fisher Scientific. The N-terminus of all amino acids were protected by Fmoc whereas side groups were protected as triphenylmethyl (Trt) and t-butyl carbonate (Boc) as follows: Cys(Trt), His(Trt), Glu(OtBu), Lys(Boc). There were no side chain protecting groups for Ala and Ilu. Enriched Pb(NO3)2 (92.4%) was purchased from Oak Ridge National Laboratory, TN. Dithiothreitol (DTT) was purchased from Ultra Pure. All chemicals were used as received without any further purification.
2.2. Peptide Synthesis and Purification
All peptides were synthesized on an Applied Biosystems 433A peptide synthesizer using standard Fmoc/tBu-based protection strategies on Rink Amide Methylbenzhydrylamine (MBHA) resin (0.25 mmole scale) with HBTU/HOBt/DIEPA coupling method, and purified and characterized as described previously [15]. The crude peptides were reduced by dithiothreitol (DTT) before their purification on HPLC. The purity of the reduced peptides was assessed by electrospray ionization (ESI)-mass spectrometry and reverse phase HPLC. The purity of the peptides was over 90%. The N-terminus of the peptides were acylated whereas the C-terminus was amidated to avoid the coordination of Pb(II) by these two ends. The sequence for the zinc binding domain from HIV-1 nucleocapsid proteins are: (HIV-CCHC = AcHN-VKCFNCGKEGHIARNCRA-CONH2; HIV-CCGC = AcHN-VKCFNCGKEGGIARNCRA- CONH2, and HIV-CCHH = AcHN-VKCFNCGKEGHIARNHRA-CONH2) [13].
2.3. UV/Vis Spectroscopy
Electronic absorption experiments were carried out on a Cary 100 spectrometer using quartz cuvettes (1 cm path lengths) in 100 mM Tris.HCl buffer at pH 7.5–9.5. All reagents or buffer solutions were degassed with argon for at least 30 min before the preparation of any solution. The stock solution concentrations of GSH and peptides were determined by using Ellman's test [18]. A 20 µM peptide solution or 60 µM GSH was prepared and 1.0 equiv of Pb(NO3)2 per peptide (or 1/3 of total GSH concentration) was added to the peptide or GSH solution at pH 7.5. The pH of the metallopeptides was readjusted by the addition of small aliquots of dil. HCl or NaOH. The spectra were recorded in the range of 200 nm to 450 nm.
2.4. 207Pb NMR Spectroscopy
All NMR samples were prepared by dissolving ~ 5 mg of GSH or ~ 10–12 mg of the pure lyophilized and degassed peptides in 500–600 µL of 10% D2O (in 90% H2O, v/v) under a nitrogen atmosphere. The GSH and peptide concentrations were determined by using Ellman’s test [18]. Pb(II)-GSH complex was prepared by the addition of a calculated amount of isotopically enriched 207Pb(NO3)2 (92.4%, Oak Ridge National Laboratory, TN) so as to obtain a 3:1 GSH to Pb(II) stoichiometry and diluted in a 10% D2O to get a final concentration of 5–6 mM complex. Similarly, zinc finger peptides and Pb(II) complexes were prepared to get a molar ratios of 1:1 (5–6 mM). The pH of the metallopeptide solutions were adjusted by the addition of KOH/D2O and HCl/D2O. All 207Pb NMR spectra were recorded at a frequency of 104.31 MHz on a Varian 500 MHz spectrometer at room temperature (25°C) using 60° pulses, a 0.02 s relaxation delay, and a 0.02 s acquisition time (spectral width 166.6 KHz). A linear prediction was performed to remove the noise, and the real FID was determined before the data processing. After zero-filling, the data (16 K data points) were processed with an exponential line broadening of 200–250 Hz using the software MestRe-C [19]. The 207Pb NMR chemical shifts are reported downfield from tetramethyllead (δ = 0 ppm; toluene) using 1.0 M Pb(NO3)2 salt (natural) as an external standard (δ = −2990 ppm, D2O, 25°C; relative to PbMe4). Therefore, the reported 207Pb signals are the results of observed signal plus 2990 ppm.
3. Results and Discussion
3.1. Pb-glutathione studies
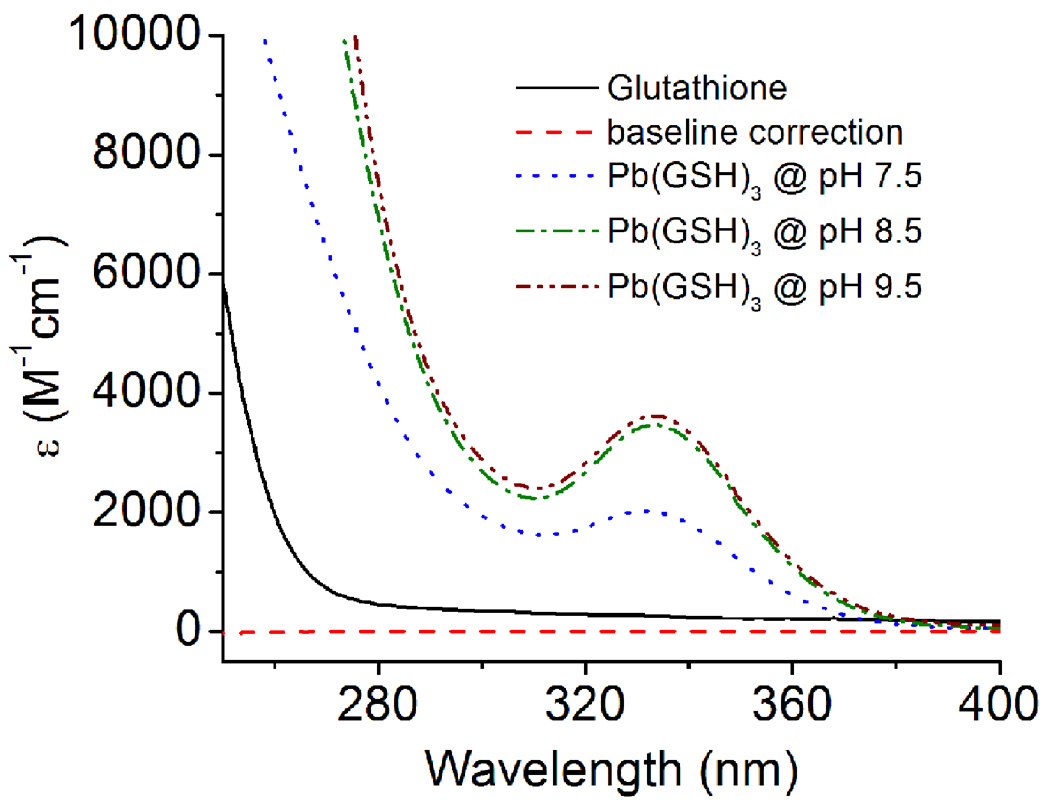
The formation of Hg(II)-GSH and Cd(II)-GSH complexes has extensively been studied using 199Hg- and 113Cd-NMR spectroscopy and X-ray absorption spectroscopy [20,21]. A three coordinate [Hg(GS)3]− and a four coordinate [Cd(GS)4]2− have been reported [21–23]. Similarly, the complexation of As(III) with glutathione is proposed as a trigonal pyramidal structure, As(GS)3, at physiological pH [24]. Pb(II) has a versatile nature of coordination behavior. It can be complexed by a combination of S, O, N, and P-containing ligands with a coordination number ranging from 2–9. However, the most preferred geometry for Pb(II) in a thiol rich environment is trigonal pyramidal with a lone pair occupying the apical position (hemidirected) [17]. Recently Polec-Pawlak and coworkers have used size exclusion chromatography coupled to ICP-MS and ESI to probe the speciation of Pb(II) ions with glutathione [25]. A variety of species were observed including [Pb(GS)3]−. Despite the fact that ESI is a soft ionization technique, fragmentation of major species can occur in the gas phase before reaching the analyzer [26]. A similar study by Mah et al. did not observe species corresponding to Pb(II) ions bound to more than three GSH [16]. Microscopic acid dissociation constants of GSH (pk) have been determined from 1H NMR chemical shift data, pH titration, and by the spectrophotometric method and found to be 2.06, 3.50, 8.69 and 9.62 for the Glu-COOH, Gly-COOH, Cys-SH and Glu-NH3+, respectively [27]. Although glutathione has a number of potential metal binding sites, the Glu-COOH (pKa 2.06), the Gly-COOH (pKa 3.50) and the Cys-SH (pKa 8.69), but not Glu-NH3+ (pKa 9.62), would be deprotonated in the presence of Pb(II) at the pH used in this study [28]. Upon the addition of Pb(II), the lowering of pKa of cysteine thiol is probably due to the coordination of Pb(II) to sulfur atom which can ease the deprotonation. Due to the high enthalpy of Pb-S bond formation [29], it is believed that the coordination of thiolate moeities with Pb(II) is thermodynamically more favorable than Pb(II)-carboxylate complexation. The formation of a PbS3 complex is supported by the appearance of a ligand to metal charge transfer (LMCT) band in UV/Vis absorption spectra at 334 nm (ε = ~ 3500 M−1cm−1) (Fig. 1). The absorption wavelength and extinction coefficients of [Pb(GS)3]− obtained in our studies are very close to the values reported in the literature [30].
Fig. 1.
UV/Vis spectra of Pb(II) bound glutathione (GSH) at different pH values. The spectrum at pH 7.5 (blue), pH 8.5 (green) and pH 9.5 (brown) were the difference spectra after the subtraction of GSH absorbance. The spectra were recorded in TrisHCl buffer (100 mM) at 25°C.
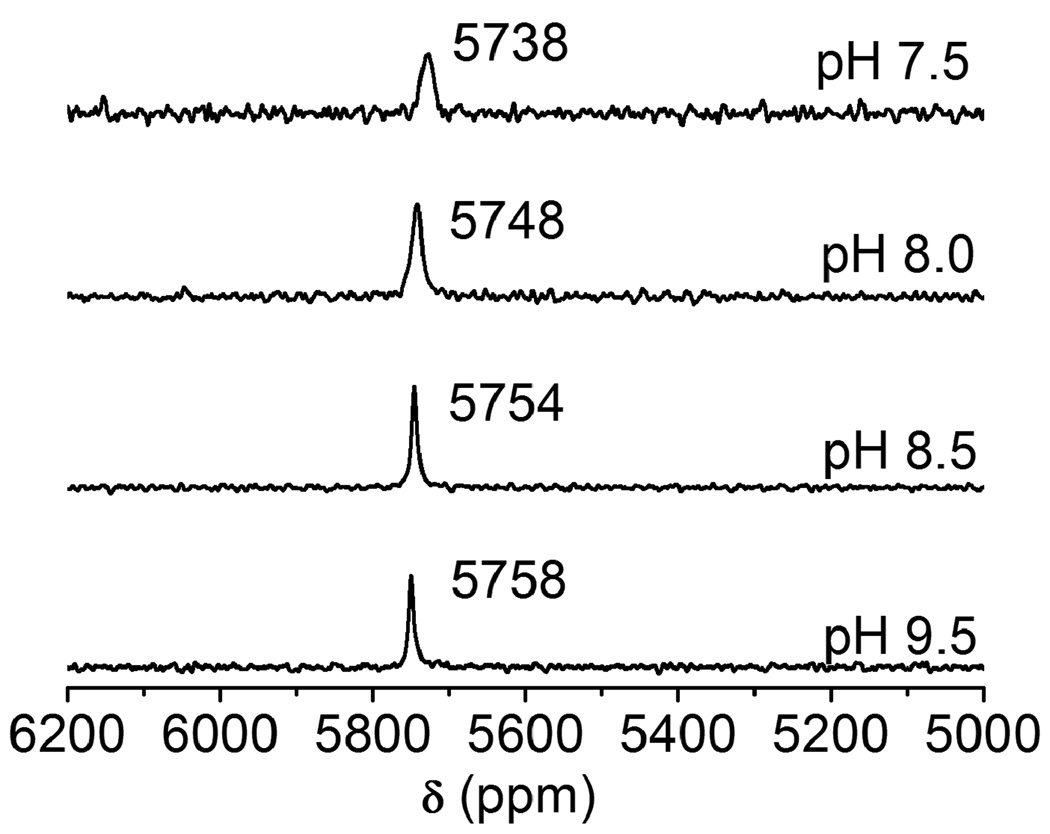
We recently used 207Pb NMR spectroscopy to investigate the coordination environment of Pb(II) in the thiol rich sites of three stranded coiled coil peptides which directly mimic the structural site of a zinc enzyme, ALAD [15]. Now, we are interested utilizing this technique on real biological molecules and have started with the simple, but thoroughly studied, biomolecules glutathione and zinc finger proteins. Fig. 2 shows the 207Pb NMR spectra of Pb-GSH complexes (1:3 ratios) at different pH conditions. The observed 207Pb signal at 5758 ppm (at pH 9.5) is in between the signals obtained for the a site and d site of three stranded coiled coil peptides in a PbS3 coordination environment [15]. This suggests that the major species present in the solution is [Pb(GS)3]− where Pb(II) is coordinated in a trigonal pyramidal geometry (hemidirected) with a lone pair occupying the apical position (Scheme 1). At high pH, where all cysteine thiolates are deprotonated and able to bind Pb(II), sharp signals with relatively narrow band widths (pH 9.5: δ = 5758 ppm, w1/2 = 7 ppm; pH 8.5: δ = 5754 ppm, w1/2 = 7 ppm) are observed. The signal broadens and shifts slightly when the pH is lowered to 8.0 (δ = 5748 ppm, w1/2 = 14 ppm) and 7.5 (δ = 5738 ppm, w1/2 = 25 ppm). This may be due to the slow exchange of bound and unbound GSH in solution, to a decrease in the number of [Pb(GS)3]− species in the solution or a small conformational change of the chelate complex as other amino acid residues are protonated. It should be remembered that the 207Pb NMR chemical shift range is enormous (over 16000 ppm), so that a shift of 16 ppm is negligible with respect to the first coordination sphere ligands. Thus, we can conclude that under these conditions, Pb(II) binds to glutathione as a PbS3 structure.
Fig. 2.
207Pb NMR spectra of Pb(II)-bound reduced glutathione (GSH) in a molar ratio of 1:3 (5 mM Pb(II): 15 mM GSH) at different pHs. All spectra were recorded for 2 h using enriched 207Pb(NO3)2 (207Pb = 92.4%) at 25°C.
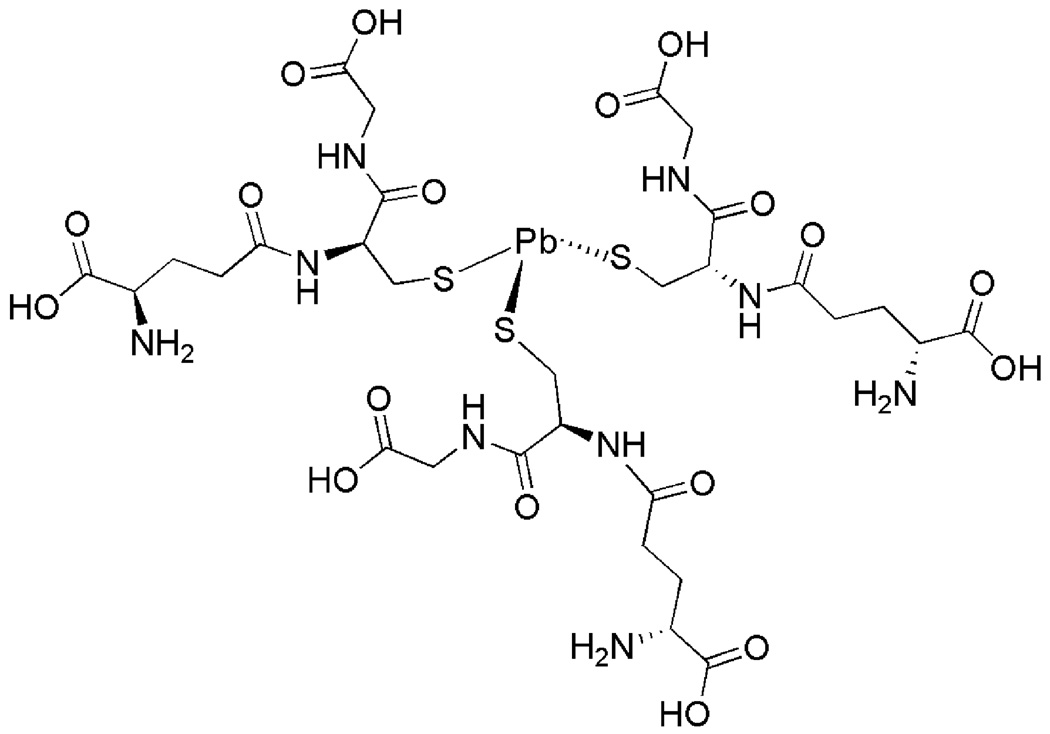
Scheme 1.
Proposed structure of Pb(II) bound Glutathione at physiological pH.
The electronic absorption spectra of [Pb(GS)3]− at different pH values suggests that the decrease in intensity of the Pb-signal is due to the incomplete deprotonation of cysteine thiolates. The UV/Vis spectrum at pH 7.0 clearly shows that the coordination of all cysteine thiolates to Pb(II) is not complete. When the pH is raised above 8.5, the LMCT band centered at 334 nm becomes more prominent and the observed extinction coefficient (ε334 = 3500±100 M−1cm−1) is very close with that obtained for PbS3 in homoleptic three stranded coiled coil peptides (Fig. 1) [31]. Temperature dependent 207Pb NMR spectra obtained at pH 8.0 shows that the intensity of the signal decreases as the temperature is elevated (Fig. 3 and 4). When the temperature is raised to 35°C, the intensity of the signal begins to decrease and it completely disappears at 65°C. However, the line broadening at 25°C and 35°C are similar. When the sample was cooled back to 25°C the original spectrum with similar line widths and intensities was obtained (spectrum not shown). This may be due to either the complex undergoing fast exchange or the decomposition of Pb-GSH complex. If decomposition occurs this may result from a decrease in pH of the solution caused by the change in temperature or to the thermodynamic instability with increased temperature. To probe whether the decrease of NMR signal was due to the complex decomposition, we examined the temperature dependence of the UV/Vis spectra since electronic and NMR spectroscopies have drastically different timescales. The results show that the absorbance at 334 nm gradually decreases with the rise in temperature of the solution and the original absorption spectrum can be reproduced upon cooling (Fig. S1). Therefore, the loss of 207Pb NMR signal upon the temperature increase is best explained by complex dissociation at higher temperatures. Furthermore, to establish whether the complex instability was a consequence of a temperature dependent pH change, we monitored, and then corrected, the solution pH. Once again the signal was lost at high temperature and returned to the PbS3 spectrum at room temperature, indicating that the complex dissociation is not due to a change in solution pH, which is further supported by 207Pb NMR. At pH 9.5, a Pb-signal at 5758 ppm was observed for the [Pb(GS)3]− complex at 25°C. When the temperature was raised to 65°C (where the pH should not be lowered below 8.0 and we should be able to observe the PbS3-signal), we did not observe 5758 ppm resonance. Instead, the solution became turbid due to the decomposion of the complex and formation of Pb(OH)2. Most important, we are able to detect the Pb-signal at a temperature close to the physiological temperature; however, these observations suggest that glutathione may become a less effective chelating agent for Pb(II). We were unable to detect the Pb-signal for a 1:2 stoichiometry of Pb:GSH, nor did we observe any significant change in the 207Pb NMR signal when the stoichiometry of Pb:GSH was increased to 1:6 (Fig. S2).
Fig. 3.
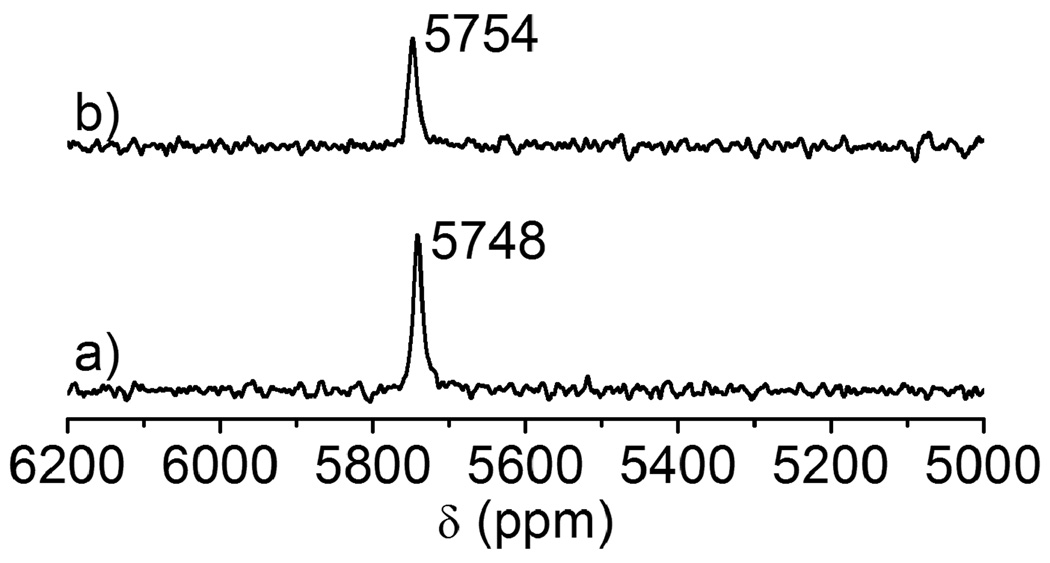
207Pb NMR spectra of Pb(II) bound reduced glutathione (GSH) in a molar ratio of 1:3 (5 mM Pb(II): 15 mM GSH) at two different temperature at pH 8.0: (a) 25°C, (b) 35°C. Both spectra were recorded for 2 h using enriched 207Pb(NO3)2 (207Pb = 92.4%).
Fig. 4.
207Pb NMR spectra of Pb(II) bound reduced glutathione (GSH) in a molar ratio of 1:6 (5 mM Pb(II): 30 mM GSH) at two different temperature at pH 8.0: (a) 25°C, (b) 35°C. Both spectra were recorded for 2 h using enriched 207Pb(NO3)2 (207Pb = 92.4%).
3.2. Zn-Finger studies
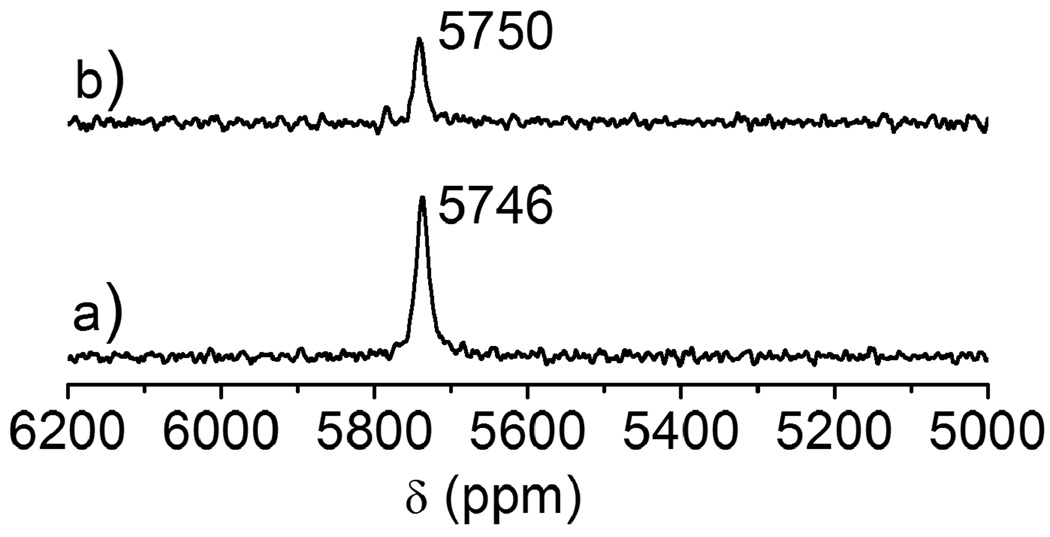
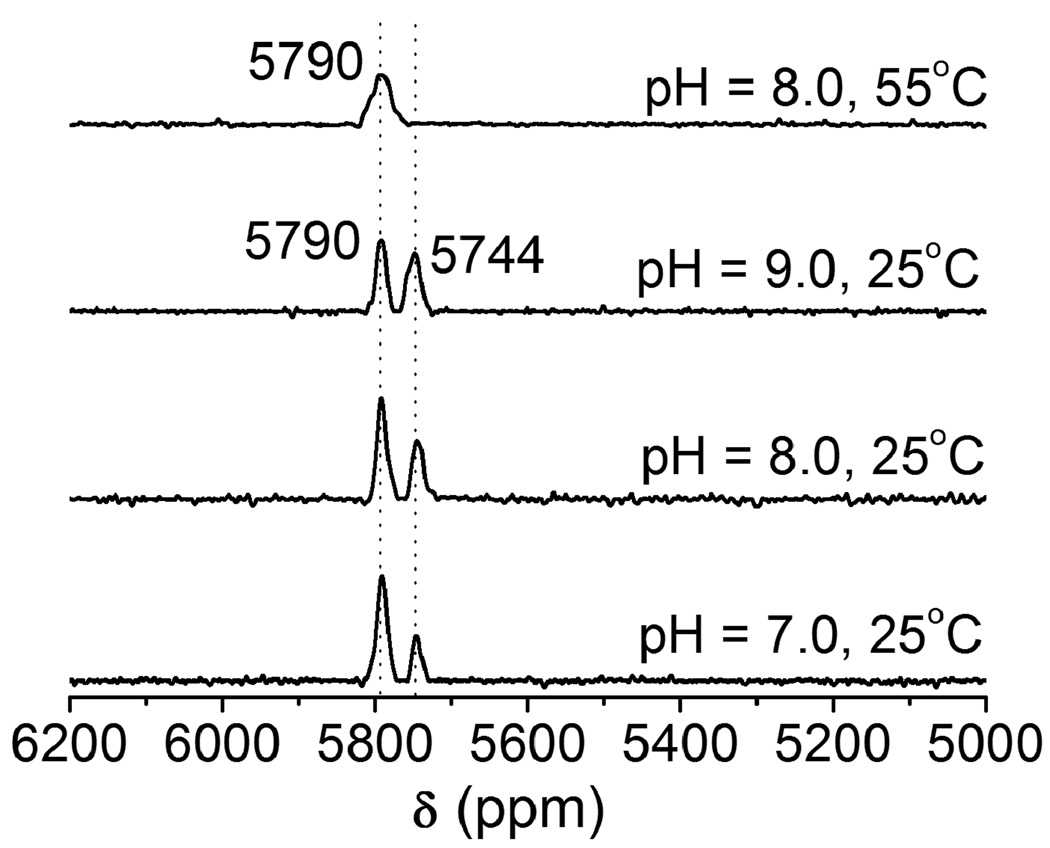
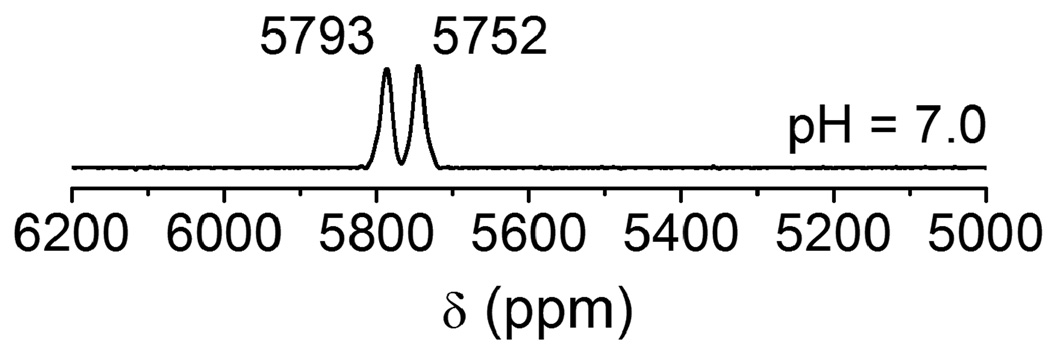
Zinc fingers are thiol rich proteins that are essential for gene regulation, transcriptional activation and development. A number of studies show that heavy metal ions such as Pb(II) can replace the zinc ion in various types of zinc fingers (Sp1 and TFIIIA) and perturb or inhibit their functions [14,32]. Zawia and coworker have utilized a synthetic Cys2His2 zinc finger peptide that is found in a transcription factor Sp1 protein and studied the DNA-binding inhibition by a variety of metal ions. They observed that heavy metal ions such as Hg(II), Cd(II), and Pb(II) inhibit the zinc finger from binding DNA, whereas other metals such as Ca(II), Ba(II), and Sn(II) neither bind to the zinc finger nor inhibit the peptide from binding DNA [14]. Recent lead binding studies to consensus peptides of zinc fingers (CP-CCHH, CP-CCHC and CP-CCCC) and the zinc binding domain from HIV nucleocapsid protein (HIV-CCHC) by the Godwin group have suggested that Pb(II) preferentially binds in a PbS3 coordination environment, while Zn(II) binds in a tetrahedral four coordinate mode. Consequently, Pb(II) does not stabilize the proper folding of the zinc binding domain as Zn(II) does [13]. The binding affinity of Pb(II) depends on the number of cysteine residues in the metal binding sites and progressively decreases as cysteine residues are replaced with histidine residues. Despite the detailed Pb(II) binding studies using these peptides, no 207Pb NMR spectroscopy has been accomplished to probe the thiolate-rich biomolecule scaffolds. We chose HIV-CCHC as a model peptide to study the binding mode of Pb(II) using 207Pb NMR spectroscopy. Recently we used this technique to study the complexation of Pb(II) to cysteine side chains inside designed three stranded coiled coil peptides (a vs d site) [15] and now hope to extend the applicability to native protein folds in systems implicated in lead toxicity. The electronic absorption spectra of Pb(HIV-CCHC) and Pb(HIV-CCGC) (at pH 7.0) with a single potential binding domain display a LMCT band at ~ 334 nm (spectrum not shown) which indicates that both zinc finger peptides have similar PbS3 coordination environments at the Pb(II) center. The absorption bands and extinction coefficients are very similar to those observed by Godwin and coworkers [13], however, the 207Pb NMR spectrum (Fig. 5(a)) displays two Pb signals (δ = 5790 ppm, ω1/2 = 13 ppm; and 5744 ppm, ω1/2 = 8 ppm) with different intensities. This observation of two species immediately underscores the enhanced sensitivity for structure assessment using 207Pb NMR. Possibilities which would give two chemical shifts could be the combination of PbS3 and PbS3N, PbS2N, PbS3O or PbS2O2. The chemical shift region from 5600 ppm to 5800 ppm has been assigned to PbS3 species with trigonal pyramidal geometry and homoleptic thiol sites in peptides [15] and in small synthetic molecules [33–36]. A huge upfield chemical shift has been observed for PbS2O2 (4100–4500 ppm) [37] and PbN2S (5318 ppm) complexes [29] suggesting that both conformations retain a PbS3 structure. Another possible explanation of the signal at 5744 ppm could be the weak coordination of a histidine residue to Pb(II) to give PbS3N. If this is the case, the peak at 5744 ppm would not be present if the histidine residue were not present. To test this, a peptide in which the His is substituted with the non-coordinating Gly residue, HIV-CCGC, was prepared and reacted with Pb(NO3)2. The spectrum of this metalloprotein is shown in Fig. 6. Two peaks of equal intensity are observed at 5793 ppm and 5752 ppm which are very close to those observed for Pb(HIV-CCHC). Therefore, we can rule out the coordination of nitrogen (from imidazole of His) or oxygen from water molecules. Interestingly, the signals are of equal relative intensities (and thus two conformations are equally populated) when the substitution of histidine by glycine was made. Thus, a long distance/weak coordination of histidine to Pb(II) can be ruled out. These results are in-line with those obtained by Godwin’s group using EXAFS to study consensus zinc finger peptides [17] and computational studies performed by Jarzecki [38].
Fig. 5.
207Pb NMR spectra of Pb(HIV-CCHC) in a 1:1 molar ratio of Pb(II):peptide at three different pH. The spectra (a)–(c) were recorded at 25°C and spectrum (d) was recorded at 55°C. All spectra were recorded for 2 h using enriched 207Pb(NO3)2 (207Pb = 92.4%).
Fig. 6.
207Pb NMR spectra of Pb(HIV-CCGC) in a 1:1 molar ratio of Pb(II):peptide at pH 7.0. The spectrum was recorded at 25°C for 2 h using enriched 207Pb(NO3)2 (207Pb = 92.4%).
A more likely scenario is that the observed peaks for Pb(HIV-CCHC) are due to the formation of PbS3 motifs in two different conformations. As the pH of the solution is increased, the position of each peak remains unshifted but the intensity and line width of the peak at 5744 ppm increases and the peak eventually disappears at 55°C (at pH 8.0). This suggests that one conformation is more stable than the other. Thus, not only has 207Pb-NMR identified a new species present in this system, it can differentiate the relative thermodynamic stabilities of these two structures. To test whether buffer may lead to a proper folding of the peptides and would give a single conformation, we recorded the Pb-NMR spectra of Pb(HIV-CCHC) and Pb(HIV-CCGC) in 100 mM Tris buffer/10% D2O (pH 7.0) at room temperature. The results show that there is no significant change in chemical shifts and intensities of Pb-signals with and without buffer (Fig. S3). Similar results were also observed in the presence of 150 mM NaCl (data are not shown). This suggests that one of the two Pb-signals (upfield signals: 5744 ppm for Pb(HIV-CCHC) and 5752 for Pb(HIV-CCGC)) is not due to the folding intermediate in which no thermodynamic minimum was reached.
Finally, we tried to observe the effect of His-Pb coordination on the 207Pb NMR chemical shift and quantify that upfield shift using the HIV-CCHH peptide. In this case, we expected to observe a PbS2N species. However, Pb(II) did not bind the HIV-CCHH peptide (in H2O or in 100 mM TrisHCl buffer, pH 7.5) and Pb(II) precipitated from solution when the pH was increased to 7.0 or higher. This further suggests that Pb(II) prefers the PbS3 coordination environment and that the peptide may not be folded properly so that His is not accessible for coordination (sequence preference), which may in part explain why Pb(II) precipitates with the HIV-CCHH peptide.
4. Conclusions
Heavy metal ions can target thiol rich molecules such as glutathione and zinc fingers in cells. We studied the interaction of glutathione and zinc finger peptides with Pb(II) by using 207Pb NMR spectroscopy. The study shows that glutathione is preferentially bound in a PbS3 coordination environment in the pH range from 7.0 to 9.5. We were unable to detect the presence of any PbS2O/N species, probably due to the fast exchange of GSH on the NMR time scale. We also observed that the glutathione complex is unstable at higher temperature but the effect is reversible upon returning to room temperature. Like glutathione, zinc finger peptides from HIV retrovirus nucleocapsid protein (HIV-CCHC) also bind Pb(II) in a PbS3 environment; however, we can now demonstrate for the first time that two conformations appear to exist for the CCHC variant, one of which is more stable at high temperatures. Substitution of the His residue in the protein with a Gly residue (HIV-CCGC) does not affect the observed 207Pb-chemical shifts but two conformers are equally populated. This suggests that the His is not coordinating to Pb(II), however it may affect the folding of the peptide around the Pb(II) center.
Supplementary Material
Acknowledgments
This work was supported by a grant from the National Institutes of Health (R01 ES0 12236).
Abbreviations
- ALAD
δ-aminolevulinic acid dehydratase
- GSH
Reduced glutathione
- EXAFS
Extended X-ray absorption fine structure
- ESI
Electrospray Ionization
- 207Pb-NMR
207Pb Nuclear Magnetic Resonance Spectroscopy
- HIV-CCHC
AcHN-VKCFNCGKEGHIARNCRA-CONH2
- HIV-CCGC
AcHN-VKCFNCGKEGGIARNCRA-CONH2
- HIV-CCHH
AcHN-VKCFNCGKEGHIARNHRA-CONH2
- HBTU
O-Benzotriazole-N,N,N′,N′-tetramethyl-uronium-hexafluoro-phosphate
- HOBt
N-Hydroxybenzotriazole
- DIEPA
N,N-Diisopropylethylamine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Patrick L. Altern. Med. Rev. 2006;11:114–127. [PubMed] [Google Scholar]
- 2.Simons TJB. Eur. J. Biochem. 1995;234:178–183. doi: 10.1111/j.1432-1033.1995.178_c.x. [DOI] [PubMed] [Google Scholar]
- 3.Zawia NH, Crumpton T, Brydie M, Reddy GR, Razmiafshari M. Neurotoxicology. 2000;21:1069–1080. [PubMed] [Google Scholar]
- 4.Sen C. J. Nutr. Biochem. 1997;8:660–672. [Google Scholar]
- 5.Kromidas L, Trombetta LD, Jamall IS. Toxicol. Lett. 1990;51:67–80. doi: 10.1016/0378-4274(90)90226-c. [DOI] [PubMed] [Google Scholar]
- 6.Coleman JE. Annu. Rev. Biochem. 1992;61:897–946. doi: 10.1146/annurev.bi.61.070192.004341. [DOI] [PubMed] [Google Scholar]
- 7.Kaptein R. Curr. Opin. Strc. Biol. 1991;1:63–70. [Google Scholar]
- 8.Huang M, Krepkiy D, Hu W, Petering DH. J. Inorg. Biochem. 2004;98:775–785. doi: 10.1016/j.jinorgbio.2004.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hartwig A. Antioxid. Redox Signal. 2001;3:625–634. doi: 10.1089/15230860152542970. [DOI] [PubMed] [Google Scholar]
- 10.Krizek BA, Amann BT, Kilfoil VJ, Merkle DL, Berg JM. J. Am. Chem. Soc. 1991;113:4518–4523. [Google Scholar]
- 11.Krizek BA, Merkle DL, Berg JM. Inorg. Chem. 1993;32:937–940. [Google Scholar]
- 12.Ghering AB, Jenkins LMM, Schenck BL, Deo S, Mayer RA, Pikaart MJ, Omichinski JG, Godwin HA. J. Am. Chem. Soc. 2005;127:3751–3759. doi: 10.1021/ja0464544. [DOI] [PubMed] [Google Scholar]
- 13.Payne JC, ter Horst MA, Godwin HA. J. Am. Chem. Soc. 1999;121:6850–6855. [Google Scholar]
- 14.Razmiafshari M, Zawia NH. Toxicol. Appl. Pharmacol. 2000;166:1–12. doi: 10.1006/taap.2000.8950. [DOI] [PubMed] [Google Scholar]
- 15.Neupane KP, Pecoraro VL. Angew. Chem. Int. Ed. 2010;49:8177–8180. doi: 10.1002/anie.201004429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mah V. PhD thesis. Albarta: University of Calgary; 2009. pp. 185–225. [Google Scholar]
- 17.Magyar JS, Weng TC, Stern CM, Dye DF, Rous BW, Payne JC, Bridgewater BM, Mijovilovich A, Parkin G, Zaleski JM, Penner-Hahn JE, Godwin HA. J. Am. Chem. Soc. 2005;127:9495–9505. doi: 10.1021/ja0424530. [DOI] [PubMed] [Google Scholar]
- 18.Ellman GL. Arch. Biochem. Biophys. 1958;74:443–450. doi: 10.1016/0003-9861(58)90014-6. [DOI] [PubMed] [Google Scholar]
- 19.Cobas C, Cruces J, Sardina FJ. Spain: Universidad de Santiago de Compostela; 2000. [Google Scholar]
- 20.Mah V, Jalilehvand F. J. Biol. Inorg. Chem. 2008;13:541–553. doi: 10.1007/s00775-008-0342-2. [DOI] [PubMed] [Google Scholar]
- 21.Mah V, Jalilehvand F. J. Biol. Inorg. Chem. 2010;15:441–458. doi: 10.1007/s00775-009-0616-3. [DOI] [PubMed] [Google Scholar]
- 22.Cheesman BV, Arnold AP, Rabenstein DL. J. Am. Chem. Soc. 1988;110:6359–6364. [Google Scholar]
- 23.Shoukry MM, Cheesman BV, Rabenstein DL. Can. J. Chem. 1988;66:3184–3189. [Google Scholar]
- 24.Rey NA, Howarth OW, Pereira-Maia EC. J. Inorg. Biochem. 2004;98:1151–1159. doi: 10.1016/j.jinorgbio.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 25.Polec-Pawlak K, Ruzika R, Lipiec E. Talanta. 2007;72:1564–1572. doi: 10.1016/j.talanta.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 26.Di Marco VB, Bombi GG. Mass Spectrom. Rev. 2006;25:347–379. doi: 10.1002/mas.20070. [DOI] [PubMed] [Google Scholar]
- 27.Rabenstein DL. In: Glutathione: Chemical, Biochemical, and Medical aspects. Dolphin D, Poulson R, Avramovic O, editors. vol. 3. New York: Wiley-Interscience; 1989. pp. 147–186. [Google Scholar]
- 28.Oram PD, Fang X, Fernando Q, Letkeman D. Chem. Res. Toxicol. 1996;9:709–712. doi: 10.1021/tx9501896. [DOI] [PubMed] [Google Scholar]
- 29.Andersen RJ, diTargiani RC, Hancock RD, Stern CL, Goldberg DP, Godwin HA. Inorg. Chem. 2006;45:6574–6576. doi: 10.1021/ic060497z. [DOI] [PubMed] [Google Scholar]
- 30.Bae W, Mehra RK. J. Inorg. Biochem. 1997;68:201–210. [Google Scholar]
- 31.Matzapetakis M, Ghosh D, Weng TC, Penner-Hahn JE, Pecoraro VL. J. Biol. Inorg. Chem. 2006;11:876–890. doi: 10.1007/s00775-006-0140-7. [DOI] [PubMed] [Google Scholar]
- 32.Zawia NH, Sharan R, Brydie M, Oyama T, Crumpton T. Dev. Brain Res. 1999;107:291–298. doi: 10.1016/s0165-3806(98)00023-6. [DOI] [PubMed] [Google Scholar]
- 33.Christou G, Folting K, Huffman JC. Polyhedron. 1984;3:1247–1253. [Google Scholar]
- 34.Arsenault JJI, Dean PAW. Can. J. Chem. 1983;61:1516–1523. [Google Scholar]
- 35.Claudio ES, Godwin HA, Magyar JS. Prog. Inorg. Chem. 2003;51:1–144. [Google Scholar]
- 36.Dean PW, Vittal JJ, Payne NC. Inorg. Chem. 1984;23:4232–4236. [Google Scholar]
- 37.Rupprecht S, Franklin SJ, Raymond KN. Inorg. Chim. Acta. 1995;235:185–194. [Google Scholar]
- 38.Jarzecki AA. Inorg. Chem. 2007;46:7509–7521. doi: 10.1021/ic700731d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.