Abstract
A mathematical model of biofilm dynamics was used to investigate the protection from antibiotic killing that can be afforded to microorganisms in biofilms based on a mechanism of localized nutrient limitation and slow growth. The model assumed that the rate of killing by the antibiotic was directly proportional to the local growth rate. Growth rates in the biofilm were calculated by using the local concentration of a single growth-limiting substrate with Monod kinetics. The concentration profile of this metabolic substrate was calculated by solving a reaction-diffusion problem. The model predicted the following features: stratified patterns of growth with zones of no growth in the biofilm interior, slow killing of biofilm microorganisms that was further retarded as the initial biofilm thickness increased, nonuniform spatial patterns of killing inside the biofilm, biofilm killing rates that decrease in a nonlinear way as the concentration of the growth-limiting substrate feeding the biofilm is decreased, and heightened tolerance when external mass transfer resistance is manifested. This modeling study also provides motivation for further investigation of a hypothetical cell state in which damaged cells score as nonviable but continue to consume substrate. The existence of such a cell state can further retard biofilm killing, according to the simulations. The results support the important contributions of nutrient limitation and slow growth to the antibiotic tolerance of microorganisms in biofilms.
Bacteria and yeast that grow in biofilms are responsible for diverse persistent infections (2, 4). The tenacity of such infections is attributed, at least in part, to the reduced susceptibilities of microorganisms growing in biofilms to antimicrobial chemotherapy (12). One of the long-standing hypotheses to explain the poor killing of biofilm cells by antibiotics is that the biofilm contains slowly growing or nongrowing microorganisms that are protected by virtue of their inactivity (1, 18). Nutrient limitation and slow growth are known to be common features of the biofilm mode of growth. Experimental tests of the slow-growth mechanism of biofilm protection are generally consistent with the idea that this mechanism provides at least a partial explanation for the recalcitrance of biofilms to chemotherapy (5).
There is a reason to be skeptical of a biofilm defense based on nutrient limitation and slow growth. One would expect that as the growing cells in a biofilm are killed, nutrients should penetrate the biofilm, which would then feed the more deeply embedded cells and render them susceptible. The protection afforded by this mechanism would be only transient by this reasoning. This paradox is the motivation for the modeling study reported in this article. The interaction of microbial growth, nutrient utilization, nutrient diffusion, and antibiotic killing is complex and nonintuitive. The purpose of this study was to investigate, using a computer model of biofilm dynamics, the degree of protection that can be anticipated in a biofilm exposed to a growth-dependent antibiotic.
MATERIALS AND METHODS
The basic mathematical model used in this study has been described in detail elsewhere (11, 14). The model was based on the conceptual and mathematical formulation derived by Wanner and Reichert (16). The model described the growth of a uniformly thick biofilm in a continuous-flow stirred tank reactor, i.e., a chemostat with wall growth. Biologically, the system was conceptualized as a single species whose growth rate was determined by the concentration of a single substrate, according to Monod kinetics. Some of the processes integrated in this model included bulk flow into and out of the reactor, transport of solutes into the biofilm by Fickian diffusion, substrate consumption by the microorganism, microbial growth, transport of cells within the biofilm by advective displacement, detachment of biomass from the surface of the film, and killing of microorganisms in the presence of an antibiotic. Macroscopic material balances around the entire reactor vessel were coupled to one-dimensional differential material balances that described processes occurring within the biofilm at the microscale.
In the base case model, two cell states were simulated: live and dead (Fig. 1A). Both cell types occupied the same volume, but only live cells consumed substrate and were capable of growth. Live cells could be converted to dead cells upon exposure to the antibiotic. The rate of killing was taken to be directly proportional to the concentration of live cells, the concentration of antibiotic, and the local specific growth rate of the live cells. In a special version of the model, a third cell state was introduced (Fig. 1B). This state, termed “damaged,” was a hypothetical intermediate between live and dead. Damaged cells could still consume substrate, but they did not grow and were not capable of recovering to the live cell state. In actuality, cells might occupy a spectrum of states from viable and fully active to inactive and dead. The use of a single intermediate state is a convenient simplification. The damaged cell state is supported by some experimental evidence (8, 13).
FIG. 1.
Two conceptual models of antibiotic action against bacteria. In the first model (A), there are only two cell states: live and dead. Live cells are converted to dead cells upon exposure to antibiotic. The rate of this transformation is characterized by the coefficient k1 and is proportional to the growth rate of the cell. In the second conceptual model (B), three cell states are proposed and are labeled live, damaged, and dead, respectively. Live cells are converted to damaged cells upon exposure to antibiotic. The rate of this transformation is characterized by the coefficient k1 and is also proportional to the growth rate of the cell. Damaged cells are converted to dead cells at a fixed rate, independent of additional exposure to antibiotic; the rate of this process is characterized by the coefficient k2. The second step is assumed to be independent of growth rate. Live cells and damaged cells are capable of using substrate, while dead cells exhibit no metabolic activity.
Base case parameter values are summarized in Table 1. For this study, the dilution rate was set to a very large value (417 h−1, which is 1,000 times the maximum specific growth rate of 0.42 h−1). The effect of this was that the bulk fluid concentrations were essentially equal to the influent settings. All of the gradients that occurred in the system were inside the biofilm or in the concentration boundary layer immediately adjacent to the biofilm.
TABLE 1.
Parameter values for biofilm modeling
| Parameter | Symbol | Units | Base case value | Range |
|---|---|---|---|---|
| Maximum specific growth rate | μmax | h−1 | 0.417 | |
| Yield coefficient | Yxs | 0.8 | ||
| Monod coefficient | Ks | mg liter−1 | 0.1 | |
| Cell volume fraction | ɛc | 0.2 | ||
| Cell intrinsic density | ρx | mg liter−1 | 3.0 × 105 | |
| Steady-state biofilm thickness | Lf | μm | 300 | 10-500 |
| Liquid boundary layer thickness | LL | μm | 10 | 0.1-300 |
| Substrate influent concentration | Cs | mg liter−1 | 8 | 0.5-80 |
| Antibiotic influent concentration | Cb | mg liter−1 | 1 | |
| Antibiotic dose duration | tb | hr | 0-36 | |
| Substrate diffusion coefficient | Ds | m2 h−1 | 9.67 × 10−6 | |
| Antibiotic diffusion coefficient | Db | m2 h−1 | 1.80 × 10−6 | |
| Biofilm/bulk diffusivity ratio | τ | 0.45 | ||
| Live cell death rate coefficient | k1 | 5 | ||
| Reactor liquid volume | V | m3 | 0.001 | |
| Biofilm surface area | A | m2 | 0.01 | |
| Volumetric flow rate | Q | m3 h−1 | 0.417 |
RESULTS AND DISCUSSION
Once a biofilm is thick enough, it invariably experiences gradients in the concentrations of metabolic substrates. This phenomenon is illustrated by the results in Fig. 2A, which show the concentration profiles of the limiting substrate as predicted in our simulations. These profiles indicate that the biofilm becomes substrate limited if it is thicker than approximately 50 μm. The microscale variation in the availability of this substrate results in gradients in the local growth rate (Fig. 2B). These predictions are consistent, at least in qualitative terms, with experimental measurements of the concentration gradients in key substrates, for example, dissolved oxygen (3, 9). They are also consistent with the few experimental measurements of the spatial patterns of growth and metabolic activity within biofilms (10, 15, 17, 19). These experimental measurements indicate zones of metabolic and protein synthetic activity of approximately 10 to 50 μm.
FIG. 2.
Predicted substrate concentration (A) and growth rate (B) profiles within biofilms of various thicknesses. Biofilms thicker than approximately 50 μm are predicted to experience nonuniform spatial distributions of substrate concentration and microbial growth. The normalized distance is the distance from the substratum divided by the biofilm thickness.
The predicted time course of biofilm killing is shown in Fig. 3. Figure 3 illustrates two main points. First, biofilm bacteria are killed more slowly than growing free-floating cells. Second, the rate of killing in the biofilm decreases as the biofilm thickness increases. It is natural to wonder whether the second result reflects inadequate penetration of the antibiotic into the biofilm. However, the antibiotic is predicted to permeate throughout the biofilm within 20 min after dosing, even for the thickest biofilm. The antibiotic concentration in the biofilm remains at the applied concentration throughout the treatment period. Slow antibiotic penetration is therefore not an important factor in protection of the biofilm in these simulations.
FIG. 3.
Biofilm killing (solid lines) versus time as a function of initial biofilm thickness. The dashed line indicates the predicted killing of planktonic cells. The protection is predicted to be greater the thicker the biofilm is.
A bit more insight into the time course of killing can be gleaned by examining the profiles of live and dead cell concentrations within the biofilm (Fig. 4). Bacteria are killed first near the biofilm-bulk fluid interface. With time, the killing front progresses inward toward the substratum. The movement of the killing front (defined as the location in the biofilm where the live and dead cell concentrations are equal) is charted in Fig. 5. These results indicate that the killing front is predicted to move at a velocity ranging from about 20 to 50 μm h−1. The rate at which the killing front advances appears to be determined by the rate at which the substrate penetrates into the biofilm. It is important that even though dead bacteria are unable to consume substrate in this version of the model, their physical presence imposes a resistance to the transport of substrate into the depths of the biofilm. In other words, dead cells shield their more deeply embedded and living neighbors from substrate, and this shielding retards the advance of the killing effects of the antibiotic.
FIG. 4.
Temporal evolution of live (solid line) and dead (dotted line) cell spatial distributions within an antibiotic-treated biofilm. Prior to antibiotic treatment, all of the cells were live cells. Each solid and dashed line pair represents a different stage in the time course of antibiotic treatment, as labeled. This simulation was conducted for a biofilm that was initially 300 μm thick.
FIG. 5.
Movement of killing front into a biofilm during antibiotic treatment. The position of the killing front, defined as the location where the fractions of live and dead cells are equal, is measured as the distance from the substratum.
If substrate delivery controls the rate of killing, then increasing the substrate concentration in the system should improve the action of the antibiotic against the biofilm. The results of the simulations of this experiment are reported in Fig. 6. The time required to achieve a certain extent of killing (a 6-log reduction) is predicted to be a strong and nonlinear function of the substrate concentration in the bulk fluid. Increasing the substrate concentration decreases the time required to kill the biofilm, and decreasing the substrate concentration significantly prolongs the time required to kill the biofilm. For example, the killing time (i.e., the time required to achieve a 6-log reduction) is predicted to be 13 h in the presence of the base case substrate concentration of 8 mg/liter, but it lengthens to 66 h (2.7 days) when the substrate concentration bathing the biofilm is reduced to 1 mg/liter. In light of this prediction, it would be interesting to see experimental measurements of biofilm susceptibility as a function of the nutrient concentration.
FIG. 6.
Time required for a 6-log reduction in biofilm viable cell numbers as a function of bulk fluid substrate concentration (solid line). The dotted line indicates the base case oxygen concentration of 8 mg/liter, and the dashed line indicates the minimal killing time attained at very high oxygen concentrations. The killing time is strongly dependent on the substrate concentration, especially for low substrate concentrations. This simulation was conducted for a biofilm that was initially 300 μm thick.
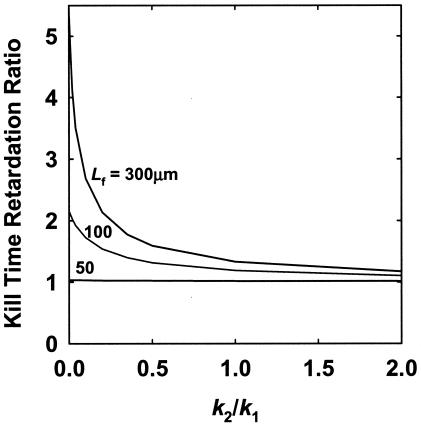
All of the preceding results are based on a model in which antibiotic-treated bacteria are assumed to lose all ability to consume substrate coincident with their loss of viability. This is probably not realistic. Bacteria that have been damaged by an antibiotic may continue to respire and even make new proteins for hours after they have lost the ability to form a colony on a plate. To account for this possibility, we developed a version of our model that allowed for three possible cell states, as shown in Fig. 1B. An intermediate cell state, which we have termed “damaged,” represents cells that continue to consume substrate, even though they would not score as viable in a conventional experimental measurement. When this intermediate cell state is included in the model, antibiotic killing of biofilm cells is retarded (Fig. 7). The degree of retardation ranges from 1 to more than 5 for a 300-μm-thick biofilm, depending on the relative rates of the transformation of live cells to damaged cells and damaged cells to dead cells. These relative rates are captured in the ratio k2/k1. Large values of k2/k1 indicate that damaged cells die more rapidly than they are formed, and therefore, damaged cells will be few and are expected to have little impact on the survival of the biofilm. Small values of k2/k1 indicate that damaged cells die slowly and that they will occupy a significant fraction of the biofilm and can affect the time course of killing. The rates that we have used are purely hypothetical, but these simulations illustrate that the existence of a nonviable, yet respiring cell state will add to the protection in the biofilm state. This intermediate cell state would not be expected to protect bacteria in the planktonic state because free-floating cells are all exposed to the same substrate concentration. The model predicts that damaged cells are found throughout the biofilm but that their numbers are greatest near the killing front.
FIG. 7.
Retardation of time required to achieve a 6-log reduction in biofilm viable cell numbers as a function of the ratio of damaged cell and live cell death rates (k2/k1). The kill time retardation ratio is the killing time computed for a biofilm with damaged cells to that for one in which no damaged cells are present.
Another process that would act to retard biofilm killing is maintenance utilization of substrate. Even bacteria that are not growing, if they consume substrate for maintenance purposes, would act to keep more deeply embedded cells in a state of nutrient deprivation.
Implicit in all of the preceding simulations is the assumption that the slowly moving fluid adjacent to the biofilm imposes minimal resistance to delivery of the substrate to the biofilm surface. More precisely, an external mass transfer film thickness of 10 μm was assumed. This might be an acceptable approximation for a biofilm system in which there is vigorous mixing or rapid flow resulting in turbulent conditions. Under laminar flow conditions, the external mass transfer resistance to substrate transport is likely to be larger. This has been simulated by increasing the external liquid film layer thickness in the model from 10 to 300 μm. This range of external film thicknesses is consistent with limited experimental measurements of mass transfer coefficients (6, 7). The result is significant retardation of biofilm killing (Fig. 8). Slower killing results because the substrate flux to the biofilm is reduced. This set of simulations shows that external mass transfer resistance exacerbates nutrient limitation and further protects biofilm bacteria from antibiotic killing.
FIG. 8.
Effect of external mass transfer resistance on biofilm killing. External mass transfer resistance is determined by the thickness of the external fluid film, LL. The base case external fluid film thickness used in all other simulations was 10 μm.
The combined effects of nutrient limitation, utilization of substrate by damaged, nonviable cells, and external mass transfer resistance have the potential to account for substantially reduced susceptibility in the biofilm state. For example, a 300-μm-thick biofilm that is bounded by a 300-μm-thick external fluid film and in which the value of the k1 rate coefficient is three times the value of k2 would be highly protected. These conditions lead to the prediction that the biofilm would be killed (6-log reduction) only after 52 h of continuous treatment. For comparison, planktonic cells would be killed to the same extent after only 2.7 h of antibiotic treatment.
These simulations collectively demonstrate that nutrient limitation and slow growth do constitute a plausible protective mechanism in biofilms when the antibiotic depends on substrate availability or growth for its killing action. These results do not prove that this is a sufficient explanation for antibiotic tolerance in biofilms. Our simulations suggest that nutrient-limited growth can retard killing in biofilms but cannot explain indefinite protection. Nutrient limitation and slow growth probably contribute to reduced biofilm susceptibility and operate in concert with other protective mechanisms to achieve the full degree of recalcitrance that is observed in vitro and in vivo.
We wish to emphasize that the variations in growth rate within the biofilm manifested in this model were predicted by calculating the simultaneous interaction of reaction and diffusion from first principles. This model serves as a basis for the design of experiments to test the effects of the substrate concentration on biofilm susceptibility and to investigate the possibility of substrate utilization by antibiotic-damaged cells.
Acknowledgments
This work was supported by an award from the W. M. Keck Foundation and by NIH award GM67245-01 to P.S.S.
Peg Dirckx drew Fig. 1.
REFERENCES
- 1.Brown, M. R. W., D. G. Allison, and P. Gilbert. 1988. Resistance of bacterial biofilms to antibiotics: a growth-rate related effect? J. Antimicrob. Chemother. 22:777-783. [DOI] [PubMed] [Google Scholar]
- 2.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 3.de Beer, D., P. Stoodley, F. Roe, and Z. Lewandowski. 1994. Effects of biofilm structure on oxygen distribution and mass transport. Biotechnol. Bioeng. 43:1131-1138. [DOI] [PubMed] [Google Scholar]
- 4.Donlan, R. M., and J. W. Costerton. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15:167-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Evans, D. J., D. G. Allison, M. R. W. Brown, and P. Gilbert. 1991. Susceptibility of Pseudomonas aeruginosa and Escherichia coli biofilms towards ciprofloxacin: effect of specific growth rate. J. Antimicrob. Chemother. 27:177-184. [DOI] [PubMed] [Google Scholar]
- 6.Horn, H., and D. C. Hempel. 1995. Mass transfer coefficients for an autotrophic and a heterotrophic biofilm system. Water Sci. Technol. 32:199-204. [Google Scholar]
- 7.Livingston, A. G., J. P. Arcangeli, A. T. Boam, S. F. Zhang, M. Marangon, and L. M. F. dos Santos. 1998. Extractive membrane bioreactors for detoxification of chemical industry wastes: process development. J. Membr. Sci. 151:29-44. [Google Scholar]
- 8.Mason, D. J., E. G. M. Power, H. Talsania, I. Phillips, and V. A. Gant. 1995. Antibacterial action of ciprofloxacin. Antimicrob. Agents Chemother. 39:2752-2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramsing, N. B., M. Kuhl, and B. B. Jorgensen. 1993. Distribution of sulfate-reducing bacteria, O2, and H2S in photosynthetic biofilms determined by oligonucleotide probes and microelectrodes. Appl. Environ. Microbiol. 59:3840-3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sternberg, C., B. B. Christensen, T. Johansen, A. T. Nielsen, J. B. Andersen, M. Givskov, and S. Molin. 1999. Distribution of bacterial growth activity in flow-chamber biofilms. Appl. Environ. Microbiol. 65:4108-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stewart, P. S. 1994. Biofilm accumulation model that predicts antibiotic resistance of Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 38:1052-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stewart, P. S., and J. W. Costerton. 2001. Antibiotic resistance of bacteria in biofilms. Lancet 358:135-138. [DOI] [PubMed] [Google Scholar]
- 13.Stewart, P. S., T. Griebe, R. Srinivasan, C.-I. Chen, F. P. Yu, D. de Beer, and G. A. McFeters. 1994. Comparison of respiratory activity and culturability during monochloramine disinfection of binary population biofilms. Appl. Environ. Microbiol. 60:1690-1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stewart, P. S., M. A. Hamilton, B. R. Goldstein, and B. T. Schneider. 1996. Modeling biocide action against biofilms. Biotechnol. Bioeng. 49:445-455. [DOI] [PubMed] [Google Scholar]
- 15.Walters, M. C., F. Roe, A. Bugnicourt, M. J. Franklin, and P. S. Stewart. 2003. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to the tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob. Agents Chemother. 47:317-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wanner, O., and P. Reichert. 1996. Mathematical modeling of mixed-culture biofilms. Biotechnol. Bioeng. 49:172-184. [DOI] [PubMed] [Google Scholar]
- 17.Wentland, E. J., P. S. Stewart, C.-T. Huang, and G. A. McFeters. 1996. Spatial variations in growth rate within Klebsiella pneumoniae colonies and biofilm. Biotechnol. Prog. 12:316-321. [DOI] [PubMed] [Google Scholar]
- 18.Xu, K. D., G. A. McFeters, and P. S. Stewart. 2000. Biofilm resistance to antimicrobial agents. Microbiology 146:547-549. [DOI] [PubMed] [Google Scholar]
- 19.Xu, K. D., P. S. Stewart, F. Xia, C.-T. Huang, and G. A. McFeters. 1998. Spatial physiological heterogeneity in Pseudomonas aeruginosa biofilm is determined by oxygen availability. Appl. Environ. Microbiol. 64:4035-4039. [DOI] [PMC free article] [PubMed] [Google Scholar]