Abstract
Brassinosteroid (BR) signal transduction research has progressed rapidly from the initial discovery of the BR receptor to a complete definition of the basic molecular components required to relay the BR signal from perception by receptor kinases at the cell surface to activation of a small family of transcription factors that regulate the expression of more than a thousand genes in a BR-dependent manner. These mechanistic advances have helped answer the intriguing question of how a single molecule, such as a hormone, can have dramatic pleiotropic effects on a broad range of diverse developmental pathways and have shed light on how BRs interact with other plant hormones and environmental cues to shape the growth of the whole plant. This review summarizes the current state of BR signal transduction research and then examines recent articles uncovering gene regulatory networks through which BR influences both vegetative and reproductive development.
INTRODUCTION
Brassinosteroids (BRs) are growth-promoting steroid hormones that occur widely across the plant kingdom, regulating multiple aspects of physiological responses essential to both vegetative and reproductive development. After their discovery in the 1970s, an array of experiments over the next decade suggested that these newly discovered plant compounds with structural similarity to animal steroid hormones promoted cell expansion and division, regulated senescence, male fertility, pollen development, and fruit ripening, and modulated the plant’s response to numerous environmental signals (Mandava, 1988). The biochemical definition of the BR biosynthetic pathway (Fujioka and Yokota, 2003; Bishop, 2007) and the discovery of BR-deficient and -insensitive mutants in Arabidopsis thaliana and several crop plants in the 1990s provided convincing evidence that BRs were as essential for normal plant development as their better-known hormone counterparts, including auxins, cytokinins, and gibberellins (GAs) (Clouse and Sasse, 1998; Altmann, 1999; Bishop, 2003).
BRs are perceived at the cell surface by BRASSINOSTEROID INSENSITIVE1 (BRI1), a member of the large family of leucine-rich repeat receptor-like kinases (LRR RLKs) found in plants (Shiu et al., 2004; Belkhadir and Chory, 2006). Several independent genetic screens have revealed over two dozen mutant alleles of bri1 in Arabidopsis, most of which exhibit the extreme dwarfism, altered leaf morphology, abnormal vascular development, delayed flowering and senescence, and reduced male fertility characteristic of severe BR-deficient mutants (Clouse et al., 1996; Kauschmann et al., 1996; Li and Chory, 1997; Noguchi et al., 1999; Friedrichsen et al., 2000). BR-insensitive mutants have also been identified and BRI1 orthologs cloned in numerous crop species, including tomato (Solanum lycopersicum), rice (Oryza sativa), barley (Hordeum vulgare), cotton (Gossypium hirsutum), and pea (Pisum sativum) (Koka et al., 2000; Yamamuro et al., 2000; Montoya et al., 2002; Chono et al., 2003; Nomura et al., 2003; Sun et al., 2004; Holton et al., 2007). Mutational analysis in Arabidopsis and in several crop species has shown conclusively that the BRI1 receptor is required for normal BR perception and plant growth.
BRI1, like other plant LRR RLKs, has an organization of functional domains similar to mammalian receptor tyrosine kinases (RTKs) and transforming growth factor-β (TGF-β) receptor kinases (Pawson, 2004; Hubbard and Miller, 2007), including an extracellular domain involved in ligand binding and receptor oligomerization, a single-pass transmembrane sequence, and a cytoplasmic kinase domain (CD) consisting of a catalytic kinase domain (KD) and flanking regulatory sequences: the juxtamembrane region (JM) and the C-terminal (CT) domain (Vert et al., 2005). BR initiates a cascade of cellular events by binding directly to a novel steroid binding motif in the extracellular domain of BRI1, which leads to phosphorylation and activation of the BRI1-CD and transduction of the signal via a phosphor-relay through an intracellular kinase to the nucleus, where several novel BR-responsive transcription factors alter the expression of genes promoting cell elongation, division, and differentiation (Li and Nam, 2002; He et al., 2005; Kinoshita et al., 2005; Yin et al., 2005; Vert and Chory, 2006).
The discovery of additional LRR RLKs that interact with BRI1 in vitro and in vivo suggested that receptor kinase oligomerization plays an important role in BR signal transduction (Li et al., 2002; Nam and Li, 2002), similar to the mechanism of action of many animal receptor kinases that signal through ligand-mediated homo- or heterooligomerization of the receptor followed by phosphorylation and activation of the intracellular kinase domain. Kinase activation results in recognition and phosphorylation of downstream components of the signal transduction pathway, leading ultimately to alterations in gene expression. To thoroughly characterize BRI1 function, it was essential to understand the role of ligand-dependent receptor oligomerization and kinase domain phosphorylation, including identification of specific phosphorylation sites and their functional significance in protein–protein interactions and modification of protein function. Identification of BRI1-KD substrates that propagate the specific signal downstream, resulting in regulated expression of specific gene networks, was also required for a complete picture of BR signaling.
ADVANCES IN BR SIGNAL TRANSDUCTION
Early Events: Phosphorylation and Oligomerization of the BRI1 Receptor Kinase
BRI1 immunoprecipitation from BR-treated Arabidopsis seedlings followed by liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis identified at least 11 sites of in vivo phosphorylation in the JM, KD, and CT domains of BRI1 and also revealed that phosphorylation on many of these residues was BR dependent (Wang et al., 2005b). Functional characterization of each identified site by biochemical and genetic analyses showed that the highly conserved kinase activation loop residues, Ser-1044 and Thr-1049, were critical for kinase function in vitro and proper BR signaling in vivo (Figure 1), while multiple JM and CT residues were required for optimal substrate phosphorylation by the BRI1 KD (Wang et al., 2005b). Similar approaches in conjunction with phospho-specific antibodies demonstrated that BRI1 was a dual-function kinase capable of phosphorylation on both Ser/Thr and Tyr residues and that Tyr phosphorylation was critical to certain aspects of BR signal transduction in planta (Oh et al., 2009).
Figure 1.
Functional Analysis of BRI1 Phosphorylation Sites.
The bri1-5 receptor kinase mutant is a weak allele with semidwarf phenotype, altered leaf structure, and shortened petioles. Expression of wild-type BRI1-Flag in bri1-5 rescues the mutant phenotype, while expression of a mutant construct in which the critical Thr-1049 kinase domain activation loop phosphorylation site is substituted with Ala leads to a dominant-negative effect with an extreme dwarf phenotype similar to bri1 null mutants. (Adapted from Figure 8 of Wang et al. [2005b] with permission.)
BRI1 can exist in plant membranes as a ligand-independent homodimer that is stabilized and activated by BR binding (Russinova et al., 2004; Wang et al., 2005a; Hink et al., 2008). However, full expression of BR signaling requires heterooligomerization of BRI1 with members of the SOMATIC EMBROYGENESIS RECEPTOR KINASE (SERK) subfamily of LRR RLKs (Hecht et al., 2001). BRI1-ASSOCIATED RECEPTOR KINASE1 (BAK1), also known as SERK3, interacts both in vitro and in vivo with BRI1 (Li et al., 2002; Nam and Li, 2002; Russinova et al., 2004), and this association is promoted by BR, as is in vivo phosphorylation of both BRI1 and BAK1 (Wang et al., 2005b, 2008). SERK4, alternatively named BAK1-LIKE (BKK1), also interacts with BRI1 in vivo in a BR-dependent manner (He et al., 2007). Moreover, BAK1 interacts with other LRR RLKs, including FLS2, and promotes their function in plant defense responses (Heese et al., 2007; Chinchilla et al., 2009; Bar et al., 2010; Postel et al., 2010; Schulze et al., 2010). Thus, BAK1 functions in independent pathways by enhancing the signaling output of distinct LRR RLK partners that bind different ligands (Chinchilla et al., 2009). Furthermore, SERK1, known to be involved in embryogenesis, also heterodimerizes with BRI1 and enhances BR signaling (Karlova et al., 2006), suggesting that SERKs in general are coreceptors that regulate multiple independent pathways by association with different LRR RLKs (Li, 2010a).
To elucidate the detailed mechanisms of phosphorylation and oligomerization of BRI1 and BAK1 in response to BR, different combinations of kinase-inactive and wild-type tagged versions of Arabidopsis BRI1 and BAK1 were expressed in the same transgenic plant, showing that an active BRI1 kinase, but not BAK1 kinase, was required for BR-dependent association of the pair. Moreover, when BAK1-green fluorescent protein (GFP) was expressed in the bri1-1 null mutant background, phosphorylation levels were dramatically reduced in BAK1-GFP. A range of in vitro kinase assays also showed that BAK1 stimulates BRI1 activity and that both BRI1 and BAK1 can transphosphorylate each other on specific residues (Wang et al., 2008). As with BRI1, LC-MS/MS analysis identified multiple in vivo and in vitro phopshorylation sites for BAK1, and functional characterization of each site revealed that the kinase activation loop residue Thr-455 appears essential for BAK1 function, just as the corresponding BRI1 residue Thr-1049 was also required for kinase function and BR signaling in planta (Wang et al., 2005b). Thus, this highly conserved residue may represent a fundamental site for kinase activation in LRR RLKs. These studies, combining LC-MS/MS analysis, functional characterization in mutant backgrounds, and in vitro biochemical studies allowed development of a novel sequential transphosphorylation model of BRI1/BAK1 interaction (Figure 2) that shows plant receptor kinases share some of the properties of both RTKs and TGF-β receptor kinases in mammals while retaining unique plant-specific features (Wang et al., 2008). Like BRI1, BAK1 also autophosphorylates on Tyr residues, and mutational analysis demonstrated that Tyr phosphorylation regulates a subset of BAK1 functions in vivo (Oh et al., 2010).
Figure 2.
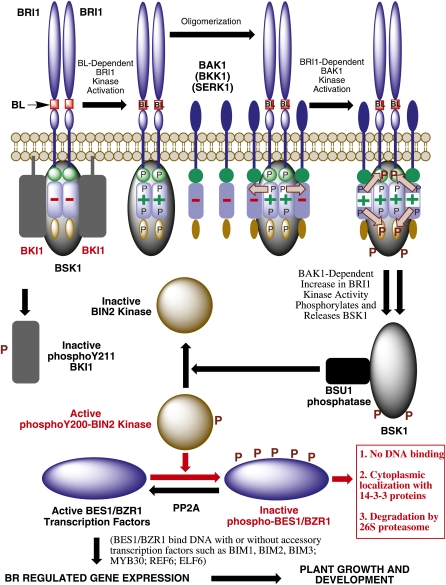
Current Model of BR Signal Transduction.
The active BR, brassinolide (BL), binds to the extracellular domain of the BRI1 receptor kinase, promoting a basal BRI1 kinase activity that phosphorylates the negative regulator BKI1 on Y211, releasing it from the membrane and allowing BRI1 to associate with BAK1 or its homologs BKK1 and SERK1. BRI1 and BAK1 transphosphorylate each other on specific residues, which enhances the signaling capacity of BRI1, leading to phosphorylation of BSK1 and its release from the receptor complex. Activated BSK1 associates with and activates BSU1, which dephosphorylates the BIN2 kinase on Tyr-200, inactivating it. The unphosphorylated forms of BES1 and BZR1 transcription factors then accumulate (with aid of PP2A) and bind to the promoters of BR-regulated target genes, eliciting a specific physiological response such as cell elongation. Positive regulators of the pathway are shown in black, with negative regulators in red. For simplicity, the nuclear or cytoplasmic localization of BSU1, BIN2, BES1, and BZR1 are not shown in this model but are discussed in more detail in the text.
Downstream Events: Inhibitors, Kinases, and Phosphatases
One of the principal effects of BR signal transduction is the inactivation of the BRASSINOSTEROID INSENSITIVE2 (BIN2) kinase, identified by genetic and biochemical approaches as a downstream negative regulator of BR signaling (Li and Nam, 2002). BIN2 has sequence similarity to mammalian GLYCOGEN SYNTHASE KINASE3 (GSK3) and in Arabidopsis is a member of a small family of 10 related genes, several of which have been implicated in BR signaling (De Rybel et al., 2009; Yan et al., 2009; Rozhon et al., 2010). Inactivation of BIN2 leads to activation of two closely related transcription factors near the terminal end of BR signaling, BRASSINAZOLE-RESISTANT1 (BZR1) (Wang et al., 2002; He et al., 2005) and BRI1-EMS SUPPRESSOR1 (BES1) (Yin et al., 2002; Yin et al., 2005), also known as BZR2 (Wang et al., 2002). Until recently, a major gap in our understanding of BR signal transduction lay in the events that followed BRI1/BAK1 heterooligomerization and phosphorylation and preceded inactivation of BIN2. Work in several laboratories (reviewed in Kim and Wang, 2010; Li, 2010b) has greatly clarified the downstream portions of BR signaling in Arabidopsis and connected BRI1/BAK1 activation with downstream events.
A yeast two-hybrid screen for BRI1 interactors identified BRI1 KINASE INHIBITOR1 (BKI1) as a negative regulator of BR signaling. It is membrane associated in the absence of BR and binds to BRI1, presumably inactivating its function by preventing association with BAK1 (Wang and Chory, 2006). BR treatment causes dissociation of BKI1 from the membrane, which releases repression of the BR signaling pathway. BRI1 and BKI1 interact directly in vitro and in vivo, and BKI1 is phosphorylated by recombinant BRI1-CD in vitro. Recent work has fine-tuned the BRI1/BKI1 interaction and the mechanism of negative regulation of BR signaling by BKI1 (Jaillais et al., 2011). This work demonstrates that BKI1 binds BRI1 through a C-terminal 20-residue conserved segment and that a peptide encompassing this binding domain inhibits the association of BRI1 with BAK1. Moreover, a Lys-Arg–rich domain within BKI1 targets the protein to the plasma membrane, and phosphorylation of Tyr-211 within this motif releases BKI1 from the membrane in a BR- and BRI1-dependent manner. Recombinant BRI1-CD phosphorylates wild-type BKI1 on Tyr residues, but a Y211F BKI1 mutant is not Tyr phosphorylated by BRI1-CD, suggesting that Tyr-211 is the major site of Tyr phosphorylation. Consistent with the in vitro data, overexpression of a BKI1-Y211F mutant construct in transgenic Arabidopsis results in severely dwarfed plants, and the mutant BKI1-Y211F protein is constitutively membrane localized, even after BR treatment. Taken together, these data strongly suggest that upon BR perception, BRI1 phosphorylates BKI1 on Tyr-211, causing it to dissociate from the membrane, thus allowing BRI1 to associate with BAK1 and initiate BR signaling. This further suggests that in addition to the previously reported autophosphorylation of BRI1 on Tyr residues (Oh et al., 2009), BRI1 may also phosphorylate downstream substrates on Tyr, providing yet another similarity in mechanisms with animal RTKs (Lemmon and Schlessinger, 2010; Lim and Pawson, 2010).
A proteomic screen of BR-regulated proteins identified several members of the receptor-like cytoplasmic kinase subfamily RLCK-XII, named BR-SIGNALING KINASES (BSKs), as direct substrates of BRI1 and positive regulators of BR signaling (Tang et al., 2008b). BSKs, while not having a predicted transmembrane sequence, show putative N-myrisylation sites that could direct their membrane localization, and BSK1 and BSK3 have been show to interact directly with BRI1 in vivo in the absence of ligand. BRI1 phosphorylates BSK1, most likely on Ser-230, causing its activation and release from the receptor complex (Tang et al., 2008b). Phosphorylated BSK1 then interacts with BRI SUPPRESSOR1 (BSU1) phosphatase (Mora-García et al., 2004), promoting its interaction with the negative regulator BIN2 (Kim et al., 2009). The precise mechanism by which BSK1 activates BSU1 is not clear, but substantial genetic and biochemical evidence suggests that BSK1 binding to BSU1 promotes BSU1-mediated dephosphorylation of phosphoTyr 200 in BIN2, thus inactivating this critical negative regulator of BR signaling (Kim et al., 2009). This work closes the gap between the highly studied events of BR perception at the cell surface to the inactivation of BIN2, the key inhibitory protein preventing expression of BR-regulated genes through the BZR1 and BES1 transcription factors.
Further Downstream Events: Regulation of the BZR1 and BES1 Transcription Factors
While it is clear that BIN2 negatively regulates BES1 and BZR1 function by phosphorylation of specific residues in these essential transcription factors, the mechanism underlying this inhibition has been somewhat controversial. It was originally proposed that BIN2-mediated phosphorylation of BES1 and BZR1 led to degradation of these proteins, while BR-promoted inactivation of BIN2 resulted in altered subcellular localization of the transcription factors with accumulation of the unphosphorylated forms of BES1 and BZR1 in the nucleus, resulting in regulation of a host of BR-responsive genes (Wang et al., 2002; Yin et al., 2002). Subsequently, it was proposed that BES1 localization was instead constitutively nuclear and that rather than promoting protein degradation, BIN2-mediated phosphorylation of BES1 inhibited DNA binding and dimerization with other transcription factors (Vert and Chory, 2006). However, independent analysis of BES1 and BZR1 function and subcellular localization by several groups convincingly demonstrated that a BR-dependent nucleocytoplasmic shuttle was involved in the regulation of both transcription factors and that 14-3-3 proteins bound directly to the phosphorylated BZR1 and BES1 to enhance their nuclear export and/or cytoplasmic retention, thus providing yet another mechanism by which BIN2 phosphorylation negatively affects the function of these transcription factors (Bai et al., 2007; Gampala et al., 2007; Ryu et al., 2007, 2010a, 2010b). It may be that all three mechanisms contribute to BES1 and BZR1 regulation depending on tissue type and developmental stage and the expression levels of BIN2 in the specific tissue.
Several recent studies have shed further light on the detailed mechanisms of the BSU1/BIN2 interaction and the phosphorylation of BES1 and BZR1 by BIN2. It appears from these studies that BIN2 phosphorylates BES1 and BZR1 in the nucleus on specific Ser and Thr residues that promote 14-3-3 binding and export of the transcription factors to the cytoplasm (Ryu et al., 2010a), while BSU1 is activated by BR in the cytoplasm and inactivates BIN2 in the nucleus (Ryu et al., 2010b). However, an alternative model in which BIN2 phosphorylates BES1 and BZR1 primarily in the cytoplasm to enhance their cytoplasmic retention by binding 14-3-3 proteins has also been proposed (Bai et al., 2007; Gampala et al., 2007; Kim et al., 2009). BIN2 has sequence similarity to animal GSK3, which plays negative regulatory roles in several signaling pathways by phosphorylating and inactivating specific substrates (Frame and Cohen, 2001). Phosphorylation by GSK3 often requires a priming phosphorylation of the substrate by a second kinase or a scaffold protein that simultaneously binds GSK3 and its substrate (Harwood, 2001). It appears that neither of these mechanisms is required for BIN2 phosphorylation of its substrate BZR1. Instead, a 12–amino acid BIN2 docking motif adjacent to the C terminus of BZR1 allows interaction with BIN2 and subsequent phosphorylation on specific BZR1 residues. Deletion of the docking motif prevents BIN2-BZR1 interaction and in vivo phosphorylation of BZR1 and leads to the nuclear accumulation of BZR1-GFP in dark-grown hypocotyls. Thus, plant and animal GSK3s show distinct mechanisms of substrate binding. Interestingly, fusing the BIN2 docking motif to the protein Armadillo allowed BIN2 to phosphorylate this known substrate of the Drosophila GSK3 kinase Shaggy (Peng et al., 2010).
While nearly all of the major steps of BR signaling were identified by the end of 2009, the mechanism by which phosphorylated, cytoplasmic BZR1 and BES1 were dephosphorylated and transported into the nucleus in their active forms remained an unanswered question. It was originally proposed that BSU1 phosphatase performed this function (Mora-García et al., 2004), but subsequent work revealed that BSU1 did not dephosphorylate BZR1 in vitro and instead interacted directly with BIN2 both in vitro and in vivo to modify its function in BR signaling (Kim et al., 2009). Using a combination of affinity purification followed by LC-MS/MS analysis, yeast two-hybrid assays, and a variety of genetic and biochemical approaches, it has now been demonstrated that cytoplasmic protein phosphatase 2A (PP2A) is responsible for dephosphorylating BZR1 and BZR2/BES1, thus increasing the active form of these transcription factors and promoting BR signaling (Tang et al., 2011). The PP2A holoenzyme is composed of a scaffolding subunit A, a regulatory subunit B, and a catalytic subunit C (Janssens et al., 2008). A range of biochemical and genetic analyses demonstrated that members of the PP2A B’ subunit family bound directly to the Pro-, Glu-, Ser-, and Thr-rich (PEST) domain of BZR1, a putative regulatory sequence containing the original bzr1-1D mutation (Tang et al., 2011). The PEST domain is essential for PP2A binding in vivo and for BL-dependent dephoshphorylation of BZR1 sites phosphorylated by BIN2, including Ser-173, the critical 14-3-3 binding residue. Moreover, PP2A mutants showed reduced BR signaling and in some cases a typical BR dwarf phenotype. Interestingly, in vivo constructs expressing BZR1 with the PEST domain deleted accumulate the phosphorylated form of BZR1 and show reduced growth and dark-green leaves, typical of a BR mutant, suggesting that phosphorylated BZR1 actually has an inhibitory effect on BR signaling. Taken together, the data of Tang et al. (2011) demonstrate that PP2A is a positive regulator of BR signaling that acts by dephosphorylating BZR1, allowing the unphosphorylated and active form of BZR1 (and BZR2/BES1) to accumulate and initiate BR-regulated gene expression. Whether or not the PP2A/BZR1 interaction is regulated by BR, as is the BSU1/BIN2 interaction, or is constitutive, remains to be determined.
DEFINING BR-REGULATED TRANSCRIPTIONAL NETWORKS
The research summarized above has focused on defining each step of BR signaling and examining specific mechanisms of protein interaction and phosphorylation that lead to the activation of the closely related BZR1 and BES1 transcription factors. Several studies have now expanded our understanding of the detailed mechanism by which these BR-activated transcription factors subsequently regulate BR-responsive gene sets. Mutant analysis combined with microarray studies revealed that BZR1 and BES1 regulate the expression of hundreds of genes in a BR-dependent manner and directly bind to the promoters of many of these genes at defined target sequences (He et al., 2005; Yin et al., 2005). Moreover, the specificity and strength of promoter binding and transcriptional activation can be modulated by association with other transcription factors, including those in the basic helix-loop-helix (bHLH), MYB, Interact-With-Spt6, and jumonji N/C domain families of nuclear proteins (Yin et al., 2005; Yu et al., 2008; Li et al., 2009, 2010b).
bHLH Transcription Factors Modulate BR Signaling and Growth Responses
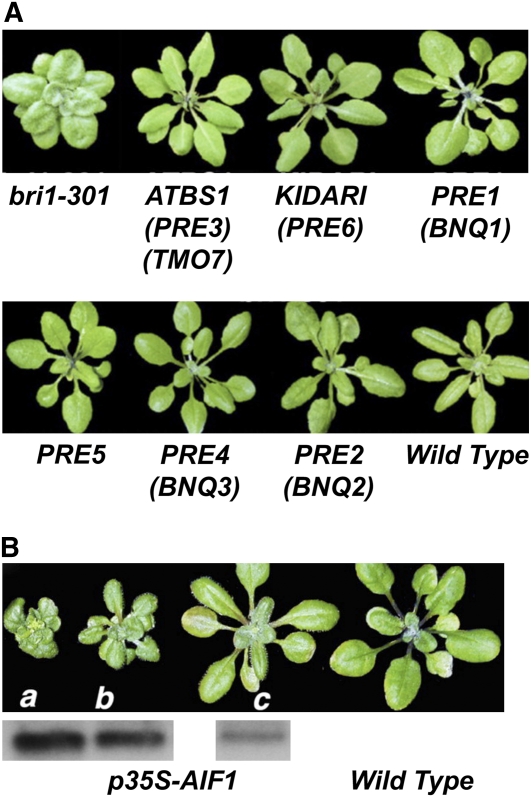
The Arabidopsis genome encodes over 160 bHLH proteins defined by a 15– to 17–amino acid region containing several basic residues potentially involved in DNA binding and the HLH region responsible for homo- or heterodimerization (Toledo-Ortiz et al., 2003). Dimerization with a classical bHLH protein enhances DNA binding to the E-box (CANNTG) promoter element in target genes, while dimerization with an atypical, non-DNA binding bHLH inhibits DNA binding of the partner as well. An activation tagging genetic screen for suppressors of the dwarf phenotype of the weak bri1-301 allele (Wang et al., 2009), revealed a 93–amino acid atypical bHLH protein, which was named ACTIVATION-TAGGED BRI1-SUPPRESSOR1 (ATBS1). ATBS1 is a positive regulator of BR signaling that rescues multiple aspects of bri1-301, including cell expansion, leaf shape, and flowering time (Figure 3). Since ATBS1 is an atypical, non-DNA binding bHLH, it was predicted that its positive effect on BR signaling resulted from heterodimerizing and sequestering other bHLH proteins that were negative regulators of BR signaling. Indeed, a yeast two-hybrid screen identified four related atypical bHLH proteins, named ATBS1 INTERACTING FACTOR1 (AIF1) to AIF4, all of which interact with ATBS1 (Wang et al., 2009). Overexpression of AIF1 abolishes the suppression of bri1-301 by ATBS1 and results in a strong bri1 dwarf phenotype, indicating that AIF1 is a negative regulator of BR signaling. The exact mechanism of this negative regulation is under investigation, but it is antagonized by ATBS1.
Figure 3.
ATypical bHLH Transcription Factor in BR-Regulated Growth.
(A) All six members of the PRE family of atypical bHLH transcription factors rescue the dwarf phenotype, altered leaf shape, and shortened petioles of the weak bri1-301 mutant allele when overexpressed, indicating a positive role in BR signaling.
(B) Overexpression of the AIF1 bHLH transcription factor in wild-type Arabidopsis results in an extreme dwarf phenotype, suggesting that AIF1 is a negative regulator of BR signaling. The extent of the phenotype is correlated with the level of transgene expression. (Adapted with permission from Figures 4 and 7 of Wang et al. [2009].)
ATBS1 itself is a member of a small family of atypical bHLH proteins containing six proteins that were originally identified as positive regulators of GA signaling, named PACLOBUTRAZOL RESISTANT1 (PRE1) to PRE6, whose overexpression resulted in enhanced seed germination, elongated hypocotyls, and early flowering, all indicative of activated GA signaling (Lee et al., 2006). Interestingly, besides ATBS1 (which is equivalent to PRE3), overexpression of the remaining five PRE family members also rescued the bri1-301 dwarf phenotype (Figure 3), suggesting that the entire PRE family is redundantly involved in both BR and GA signal transduction (Wang et al., 2009). A connection with auxin signaling was also suggested in an independent study in which ATBS1/PRE3 was identified as a direct target of the auxin-dependent transcription factor MONOPTEROS (MP), required for embryonic root initiation and specification, and named TARGET OF MP7 in that study (Schlereth et al., 2010). Finally, a connection with light signaling was also suggested by independent studies in which PRE6 was identified as a repressor of photomorphogenesis (named KIDARI in that study), which interacted with the bHLH transcription factor LONG HYPOCOYTL IN FAR RED1 (HFR1), inhibiting its activity (Hyun and Lee, 2006); and the identification of PRE1, PRE2, and PRE4 as BANQUO1 (BNQ1) to BNQ3, which also interact with HFR1 to regulate photomorphogenesis (Mara et al., 2010). Thus, this small family of atypical bHLH proteins appears to play essential roles in modulating plant growth and morphogenesis by participating in multiple hormone and light signaling pathways. The somewhat complex role of PRE family proteins in regulating multiple signaling pathways has also been discussed in some detail in a recent review (Li, 2010b).
Rice lamina joint cells are highly responsive to BR, and the inclination of the lamina joint has been used as a sensitive BR bioassay for cell elongation (Wada et al., 1984). A dominant mutant, ili1-D, with increased lamina joint inclination and hypersensitivity to BR, was identified by screening a collection of rice T-DNA insertion lines (Zhang et al., 2009). The mutant phenotype was caused by overexpression of an atypical bHLH rice protein that is an ortholog of Arabidopsis PRE1. A yeast two-hybrid screen in rice identified IBH1 (ILI1 Binding bHLH Protein1), a classical bHLH protein that interacts with ILI1 both in vitro and in vivo. The Arabidopsis ortholog of IBH1 also interacts with PRE1 in planta. Overexpression of ILI1 or PRE1 resulted in increased cell elongation and BR sensitivity, while overexpression of IBH1 caused decreased cell elongation and reduced BR sensitivity. Moreover, PRE1 is expressed at high levels in young growing tissue, while IBH1 is more highly expressed in mature organs that have ceased growth. This suggests that PRE1/ILI1 antagonize IBH1 by heterodimerization and interference of IBH1’s function as a negative regulator of BR-responsive genes required for cell elongation. Significantly, both PRE1 and IBH1 are direct targets of BZR1, with PRE1 RNA levels being enhanced by BZR1 promoter binding and IBH1 RNA levels being repressed, as would be expected from the positive and negative roles, respectively, of PRE1 and IBH1 in BR signaling (Zhang et al., 2009). Thus, these two BR-regulated antagonistic factors provide a growth regulating mechanism that enhances cell elongation in immature tissue that is highly responsive to BR while arresting growth in mature tissue. Furthermore, another rice atypical bHLH termed BRASSINOSTEROID UPREGULATED1, which has sequence similarity to ILI1 and Arabidopsis PRE6 and PRE1, has also been shown to positively regulate BR signaling and enhance bending of the lamina joint (Tanaka et al., 2009), suggesting that multiple bHLH proteins regulate BR signaling in both monocots and dicots.
ChIP-chip Analyses and the Identification of BZR1 and BES1 Direct Targets
It is now clear that a large part of BR signaling results from the dephosphorylation and activation of two transcription factors, BES1 and BZR1, which then regulate the expression of sets of genes leading to observed BR physiological responses. Tissue- and developmental-specific regulation of target gene sets in different physiological responses might be explained by heterodimerization of BES1 and BZR1 with different factors that enhanced or repressed binding to specific promoters of genes involved in that particular response. Two recent independent studies employing chromatin immunoprecipitation microarray (ChIP-chip) experiments have greatly expanded our understanding of the range of genes that are direct targets of BES1 and BZR1 and have uncovered extensive gene regulatory networks that modulate BR-promoted growth in Arabidopsis (Sun et al., 2010; Yu et al., 2011).
A total of 953 BR-regulated genes were shown by ChIP-chip analysis to be direct targets of BZR1, with 450 being activated, 462 repressed, and 41 affected in complex ways by BZR1 binding to their promoters (Sun et al., 2010). A similar analysis identified 250 direct targets of BES1 that were also regulated by BR treatment, with 165 being activated and 85 repressed by BES1 binding (Yu et al., 2011). It was previously thought that BZR1 was primarily a repressor of transcription that bound to the BR response element (BRRE) sequence CGTG(T/C)G (He et al., 2005), while BES1 was primarily a transcriptional activator binding to the E-box element (CANNTG) either as a homodimer or heterodimer with other transcription factors (Yin et al., 2005). The global analyses of Sun et al. (2010) and Yu et al. (2011) clearly showed that both BZR1 and BES1 can function as either activators or repressors, depending on the specific target gene promoter, and DNA binding experiments in both studies demonstrated that BZR1 and BES1 can recognize both the BRRE and E-Box motifs. Moreover, comparison of the 953 BZR1 targets with the 250 BES1 targets shows an overlap of 120 genes, indicating that BZR1 and BES1 can regulate the same individual gene by binding to its promoter. Whether BZR1 and BES1 bind to the same promoter at the same time or whether binding is regulated temporally and spatially by the presence of accessory factors has yet to be determined.
Examination of the demonstrated or predicted function of the BES1 and BZR1 target genes confirms many previously proposed physiological roles for BR signaling and also reveals novel interactions with other signaling pathways. Numerous transcription factors of various classes are direct targets of BES1 and BZR1, suggesting that BR signaling is amplified and expanded by a transcriptional regulatory network of primary and secondary target genes (Sun et al., 2010; Yu et al., 2011). BZR1 binds to promoters of many genes involved in hormone biosynthesis, transport, and signal transduction, including five BR biosynthesis genes as well as the BRI1 receptor to provide negative feedback regulation, and BIN2, BSU1, and BZR1 itself to positively regulate downstream BR signaling. The interaction of BR and auxin signaling is well known, and the global ChIP-chip studies revealed that many auxin-regulated transcription factors are BZR1 targets in addition to the PIN genes involved in auxin transport. Moreover, several genes involved in the biosynthesis or signaling of GA, abscisic acid, ethylene, cytokinin, and jasmonate are direct targets of BZR1 and BES1, suggesting that besides BR-specific target genes, BR signaling affects the activity of most other plant hormones in a variety of ways that may help integrate signals leading to normal growth and development (Sun et al., 2010; Yu et al., 2011). Conversely, other hormones may affect BR responses. A recent study in Arabidopsis showed that auxin enhanced BR biosynthesis, in part by inhibiting BZR1 feedback repression of the DWARF4 gene encoding a steroid hydroxylase that is a rate-limiting step in BR biosynthesis (Chung et al., 2011). BR also promotes expression of a bHLH protein, TCP1, that binds to and activates the DWARF4 promoter (Guo et al., 2010).
NOVEL MECHANISMS OF BR-REGULATED GROWTH
Vegetative Growth
Promotion of cell expansion through BR-regulated expression of genes involved in cell wall modifications, ion and water transport, and cytoskeleton rearrangements has been well documented in individual genetic studies and global microarray analyses (Clouse and Sasse, 1998; Vert et al., 2005; Kim and Wang, 2010). Many of these previously characterized genes have now also been demonstrated to be direct targets of the BZR1 and BES1 transcription factors (Sun et al., 2010; Yu et al., 2011). Additionally, novel genes involved in BR-regulated cell expansion have recently been uncovered. DEVELOPMENTALLY REGULATED PLASMAMEMBRANE POLYPEPTIDE (DREPP) was identified as a BR-regulated protein in proteomic studies (Tang et al., 2008a), and the DREPP gene has now been confirmed to be a BR-induced direct target of BZR1 (Sun et al., 2010). Overexpression of DREPP enhances cell elongation in a BR-deficient mutant, indicating DREPP is a positive regulator of BR response. A gene closely related to DREPP encodes a microtubule-associated protein functioning in directional cell growth (Wang et al., 2007), suggesting that DREPP might be involved in BR-mediated microtubule reorganization, a process long thought to be promoted by BR during cell elongation (Mayumi and Shibaoka, 1995). A putative zinc finger transcription factor, At4g39070, is repressed by direct binding of BZR1 to its promoter and appears to negatively regulate BR signaling since repression of At4g39070 enhances hypocotyl elongation while its overexpression inhibits elongation (Sun et al., 2010).
In addition to cell expansion, the role of BRs in cell division and differentiation has also been documented, but the mechanisms involved in cell proliferation and organ initiation are not as clear as those in BR-promoted elongation. Two recent studies have shown that BRs participate not only in cell elongation but also regulate progression of the cell cycle and meristem size in the developing Arabidopsis root. BR mutants have reduced root meristem size and altered expression of specific cell cycle markers, which can be rescued by overexpression of cyclin D3 (González-García et al., 2011), a gene previously shown to be BR regulated in Arabidopsis cell suspension cultures (Hu et al., 2000). Furthermore, BR signaling through BRI1 in the epidermis, and not the endodermis, quiescent center or stele, is sufficient to control root meristem size through a mobile element that is likely different from BES1 or BZR1 (Hacham et al., 2011). Thus, like auxin and cytokinin, BRs are not merely involved in simple cell elongation but also modulate timing of cell division and differentiation in a complex process such as organ formation.
BR, Light, and Photomorphogenesis
The interplay of light and hormones in regulating seedling growth is a widely studied topic, but the precise mechanisms of how environmental signals such as light impact BR function in regulating plant growth and development are not well understood. There is still a great deal to be learned about molecular mechanisms of deetiolation and the developmental switch from skotomorphogenesis to photomorphogenesis and how light and BR signals are integrated in this process. The deetiolated phenotype of many Arabidopsis-deficient and insensitive mutants suggests a connection between light and BR action and a negative role for BR in photomorphogenesis, although some have questioned this role because BR levels remained the same in the dark and the light in tomato and pea (Symons and Reid, 2003; Symons et al., 2008). However, the recent ChIP-chip and microarray analyses in Arabidopsis have pointed to BR regulation of light signaling as a plausible mechanism of interaction (Sun et al., 2010; Yu et al., 2011).
RNA transcript profiling showed that more than 750 genes were regulated in the same way by red light or by loss of BR signaling in the bri1-116 mutant, suggesting that red light and BR antagonistically affect seedling photomorphogenesis by regulating a common set of target genes in opposite ways (Sun et al., 2010). Moreover, HY5, the basic leucine zipper transcription factor that functions as a positive regulator of photomorphogenesis, binds to the promoters of 1170 genes that are also BZR1 direct targets. These genes are generally regulated in opposite ways by BZR1 and HY5, again supporting an antagonistic interaction between BR and light by targeting common downstream genes in the two pathways (Sun et al., 2010). BR can also directly regulate important light signaling components. A recent study demonstrated that BZR1 represses expression of the GATA2 transcription factor, a positive regulator of photomorphogenesis, by directly binding to its promoter more strongly in the dark than in the light and that GATA2 induces many light-activated but BR-repressed genes while downregulating light-repressed but BR-activated genes (Luo et al., 2010). Additional connections between light and BR pathways in regulating morphogenesis were recently provided by the observation that the GOLDEN2-LIKE (GLK) transcription factors, required for chloroplast development (Waters et al., 2009), are direct targets of the BES1 transcription factor (Yu et al., 2011). Microscopy analysis suggests that BR inhibits chloroplast development through BES1 repression of GLK1 and GLK2 transcription.
Reproductive Development
The transition from vegetative to reproductive growth in plants is a major developmental switch regulated by a set of complex, integrated signal transduction pathways that respond to photoperiod, GA, and cold treatment (Li et al., 2010a). It has been recognized for more than a decade that BR-deficient and insensitive mutants in Arabidopsis exhibit delayed flowering, suggesting an additional role for BRs in regulating the timing of floral initiation (Chory et al., 1991; Li and Chory, 1997; Azpiroz et al., 1998). This idea was strengthened by the discovery that BRI1-dependent BR signaling appears to promote flowering in wild-type Arabidopsis by repressing the expression of FLOWERING LOCUS C (FLC), which itself quantitatively represses flowering (Domagalska et al., 2007).
A possible mechanism for this BR-mediated inhibition of flowering was further suggested by the demonstration that RELATIVE OF EARLY FLOWERING6 (REF6), a Jumonji N/C domain-containing transcriptional regulator that functions in chromatin modification, directly interacts with BES1 (Yu et al., 2008). REF6 is a repressor of FLC, and ref6 mutants accumulate FLC transcripts, leading to late flowering. Therefore, it is conceivable that BR regulates FLC expression through a BES1/REF6 dimer, but this requires experimental confirmation (Clouse, 2008). Recent experiments employing BR- and GA-deficient double mutants and Arabidopsis plants overexpressing key BR and GA biosynthetic enzymes also suggest an interaction between BR and GA in regulating time of flowering with the role of BRs being dependent on the presence and concentration of GAs (Domagalska et al., 2010).
Another well-known defect in BR biosynthetic or signaling mutants is male infertility, but the underlying mechanisms of how BR regulates male fertility are unknown. A recent study combining microscopy analysis with transcript profiling showed that many of the well-studied genes that regulate anther and pollen development have altered expression in BR mutants, leading to observable defects in anther and pollen morphology. Moreover, the same study showed by ChIP analysis that BES1 directly binds to the promoters of many of these genes, indicating there is a direct BR response on the expression of genes essential for anther and pollen development (Ye et al., 2010). A novel role for BRs in gynoecium and ovule development has also been uncovered recently through genetic screens that identified a BR biosynthetic enzyme as a suppressor of mutations in the Arabidopsis SEUSS gene, encoding a transcriptional adaptor protein involved in floral organ identity and the development of the carpel margin meristem (Nole-Wilson et al., 2010).
CONCLUSION
The large body of work reviewed above clearly shows that BRs regulate multiple aspects of plant growth and development through a signal transduction pathway whose major components are now firmly established. Furthermore, the precise mechanisms of how steroid perception at the cell surface leads to significant alterations in gene expression and consequent changes in specific developmental programs has become more clearly defined in the past few years. It appears that the pleiotropic effects of BR action on plant physiology may be due to the activation of sets of transcription factors that are direct targets of the BR-activated transcriptional regulators, BZR1 and BES1, and that in turn regulate subsets of genes in a specific response, such as cell elongation, flowering, senescence, leaf morphogenesis, etc. While the volume and quality of recent BR signaling research has been impressive, much remains to be accomplished if the understanding of molecular mechanisms in BR action is to reach the level of detail available for many of the well-studied mammalian receptor kinases.
The island domain in the extracellular region of the BRI1 receptor kinase clearly is involved in BR binding (Kinoshita et al., 2005), but solving a three-dimensional crystal structure for the BRI1 extracellular domain is required to understand whether the steroid ligand binds to a monomer or homodimer of BRI1 and how ligand binding leads to initial BRI1 kinase activation, which is required for BKI1 release and association with the BAK1 coreceptor. It would also precisely define the unique steroid binding motif in plants, which is distinctly different from animal steroid hormone receptors. The crystal structure of the human Toll-Like Receptor 3 ectodomain, which has some sequence similarity to the BRI1 ectodomain, has been solved (Choe et al., 2005), suggesting it is technically feasible to determine the BRI1 ectodomain structure.
Multiple in vivo phosphorylation sites have been identified in BRI1 and BAK1, and it is possible that differential phosphorylation in these receptor kinases may lead to alternative downstream signaling and consequent activation of different gene sets involved in specific physiological responses (Wang et al., 2005b, 2008). For example, a null mutant of bri1 shows delayed flowering, but a site-directed Y831F BRI1 mutant, which retains overall kinase function but with altered phosphorylation on Tyr residues, actually flowers earlier than the wild type (Oh et al., 2009). Differential phosphorylation of BAK1 may also determine whether this coreceptor binds to BRI1 or FLS2 in response to specific stimuli (Wang et al., 2008). In vivo phosphorylation sites for BKI1, BSK1, BIN2, BES1, and BZR1 have also been identified (Tang et al., 2008a, 2008b; Kim et al., 2009; Jaillais et al., 2011). Continued functional analysis of individual phosphorylation sites in BRI1 and downstream signaling components, including use of LC-MS/MS techniques that quantitatively monitoring individual phosphorylation states in response to BR (Clouse et al., 2008), may help fine-tune BR signaling. Finally, while BSK1 has been shown to be phosphorylated by BRI1, the mechanism by which this phoshporylation allows interaction with and activation of BSU1 has not been described.
The identification of hundreds of direct targets of BES1 and BZR1 (Sun et al., 2010; Yu et al., 2011) sets the stage for genetic and biochemical analysis of those targets with previously undefined functions, which may further reveal previously unknown physiological processes affected by BR signaling. Moreover, the demonstration that BES1 and BZR1 can bind to the same gene promoter in many cases suggests that detailed studies to determine whether this binding is coordinated or independent may be informative. While the majority of the BR signal appears to flow through BES1 and BZR1, it may be that additional, undiscovered transcription factors are involved in some specific BR responses, and continued searches for BRI1, BAK1, and BIN2 interacting proteins may be productive. The finding that BIN2 phosphorylates AIF1 in vitro is intriguing and suggests BIN2 has additional targets besides BES1 and BZR1 (Wang et al., 2009).
The recent rate of publication on BR signaling suggests that 2011 will again be a productive year in understanding how these steroid hormones regulate multiple aspects of plant growth and development. Moreover, as an outcome of intensive work on BR signal transduction, BRI1 is becoming the best understood plant receptor kinase with respect to mechanism of action and may serve as a model to approach functional studies in other members of this large family of membrane proteins mediating many environmental responses and internal signals in plant development.
Acknowledgments
Work in the author’s laboratory is supported by the National Science Foundation (MCB-1021363 and MCB-0742411) and the North Carolina Agricultural Research Service. I thank Jianming Li for providing images of ATBS1 and AIF1.
References
- Altmann T. (1999). Molecular physiology of brassinosteroids revealed by the analysis of mutants. Planta 208: 1–11 [DOI] [PubMed] [Google Scholar]
- Azpiroz R., Wu Y., LoCascio J.C., Feldmann K.A. (1998). An Arabidopsis brassinosteroid-dependent mutant is blocked in cell elongation. Plant Cell 10: 219–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai M.Y., Zhang L.Y., Gampala S.S., Zhu S.W., Song W.Y., Chong K., Wang Z.Y. (2007). Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc. Natl. Acad. Sci. USA 104: 13839–13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar M., Sharfman M., Ron M., Avni A. (2010). BAK1 is required for the attenuation of ethylene-inducing xylanase (Eix)-induced defense responses by the decoy receptor LeEix1. Plant J. 63: 791–800 [DOI] [PubMed] [Google Scholar]
- Belkhadir Y., Chory J. (2006). Brassinosteroid signaling: A paradigm for steroid hormone signaling from the cell surface. Science 314: 1410–1411 [DOI] [PubMed] [Google Scholar]
- Bishop G.J. (2003). Brassinosteroid mutants of crops. J. Plant Growth Regul. 22: 325–335 [DOI] [PubMed] [Google Scholar]
- Bishop G.J. (2007). Refining the plant steroid hormone biosynthesis pathway. Trends Plant Sci. 12: 377–380 [DOI] [PubMed] [Google Scholar]
- Chinchilla D., Shan L., He P., de Vries S., Kemmerling B. (2009). One for all: The receptor-associated kinase BAK1. Trends Plant Sci. 14: 535–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe J., Kelker M.S., Wilson I.A. (2005). Crystal structure of human toll-like receptor 3 (TLR3) ectodomain. Science 309: 581–585 [DOI] [PubMed] [Google Scholar]
- Chono M., Honda I., Zeniya H., Yoneyama K., Saisho D., Takeda K., Takatsuto S., Hoshino T., Watanabe Y. (2003). A semidwarf phenotype of barley uzu results from a nucleotide substitution in the gene encoding a putative brassinosteroid receptor. Plant Physiol. 133: 1209–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory J., Nagpal P., Peto C.A. (1991). Phenotypic and genetic analysis of det2, a new mutant that affects light-regulated seedling development in Arabidopsis. Plant Cell 3: 445–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung Y., et al. (2011). Auxin stimulates DWARF4 expression and brassinosteroid biosynthesis in Arabidopsis. Plant J. 66: 564–578 [DOI] [PubMed] [Google Scholar]
- Clouse S.D. (2008). The molecular intersection of brassinosteroid-regulated growth and flowering in Arabidopsis. Proc. Natl. Acad. Sci. USA 105: 7345–7346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse S.D., Goshe M.B., Huber S.C., Li J. (2008). Functional analysis and phosphorylation site mapping of leucine-rich repeat receptor-like kinases. In Plant Proteomics: Technologies, Strategies and Applications, Agrawal G.K., Rakwal R., eds (Hoboken, NJ: John Wiley & Sons; ), pp. 469–484 [Google Scholar]
- Clouse S.D., Langford M., McMorris T.C. (1996). A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 111: 671–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse S.D., Sasse J.M. (1998). BRASSINOSTEROIDS: Essential regulators of plant growth and development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49: 427–451 [DOI] [PubMed] [Google Scholar]
- De Rybel B., et al. (2009). Chemical inhibition of a subset of Arabidopsis thaliana GSK3-like kinases activates brassinosteroid signaling. Chem. Biol. 16: 594–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domagalska M.A., Sarnowska E., Nagy F., Davis S.J. (2010). Genetic analyses of interactions among gibberellin, abscisic acid, and brassinosteroids in the control of flowering time in Arabidopsis thaliana. PLoS ONE 5: e14012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domagalska M.A., Schomburg F.M., Amasino R.M., Vierstra R.D., Nagy F., Davis S.J. (2007). Attenuation of brassinosteroid signaling enhances FLC expression and delays flowering. Development 134: 2841–2850 [DOI] [PubMed] [Google Scholar]
- Frame S., Cohen P. (2001). GSK3 takes centre stage more than 20 years after its discovery. Biochem. J. 359: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrichsen D.M., Joazeiro C.A., Li J., Hunter T., Chory J. (2000). Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol. 123: 1247–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka S., Yokota T. (2003). Biosynthesis and metabolism of brassinosteroids. Annu. Rev. Plant Biol. 54: 137–164 [DOI] [PubMed] [Google Scholar]
- Gampala S.S., et al. (2007). An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev. Cell 13: 177–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-García M.P., Vilarrasa-Blasi J., Zhiponova M., Divol F., Mora-García S., Russinova E., Caño-Delgado A.I. (2011). Brassinosteroids control meristem size by promoting cell cycle progression in Arabidopsis roots. Development 138: 849–859 [DOI] [PubMed] [Google Scholar]
- Guo Z., Fujioka S., Blancaflor E.B., Miao S., Gou X., Li J. (2010). TCP1 modulates brassinosteroid biosynthesis by regulating the expression of the key biosynthetic gene DWARF4 in Arabidopsis thaliana. Plant Cell 22: 1161–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacham Y., Holland N., Butterfield C., Ubeda-Tomas S., Bennett M.J., Chory J., Savaldi-Goldstein S. (2011). Brassinosteroid perception in the epidermis controls root meristem size. Development 138: 839–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood A.J. (2001). Regulation of GSK-3: A cellular multiprocessor. Cell 105: 821–824 [DOI] [PubMed] [Google Scholar]
- He J.X., Gendron J.M., Sun Y., Gampala S.S., Gendron N., Sun C.Q., Wang Z.Y. (2005). BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307: 1634–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He K., Gou X., Yuan T., Lin H., Asami T., Yoshida S., Russell S.D., Li J. (2007). BAK1 and BKK1 regulate brassinosteroid-dependent growth and brassinosteroid-independent cell-death pathways. Curr. Biol. 17: 1109–1115 [DOI] [PubMed] [Google Scholar]
- Hecht V., Vielle-Calzada J.P., Hartog M.V., Schmidt E.D., Boutilier K., Grossniklaus U., de Vries S.C. (2001). The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASE 1 gene is expressed in developing ovules and embryos and enhances embryogenic competence in culture. Plant Physiol. 127: 803–816 [PMC free article] [PubMed] [Google Scholar]
- Heese A., Hann D.R., Gimenez-Ibanez S., Jones A.M., He K., Li J., Schroeder J.I., Peck S.C., Rathjen J.P. (2007). The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl. Acad. Sci. USA 104: 12217–12222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hink M.A., Shah K., Russinova E., de Vries S.C., Visser A.J. (2008). Fluorescence fluctuation analysis of Arabidopsis thaliana somatic embryogenesis receptor-like kinase and brassinosteroid insensitive 1 receptor oligomerization. Biophys. J. 94: 1052–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holton N., Caño-Delgado A., Harrison K., Montoya T., Chory J., Bishop G.J. (2007). Tomato BRASSINOSTEROID INSENSITIVE1 is required for systemin-induced root elongation in Solanum pimpinellifolium but is not essential for wound signaling. Plant Cell 19: 1709–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Bao F., Li J. (2000). Promotive effect of brassinosteroids on cell division involves a distinct CycD3-induction pathway in Arabidopsis. Plant J. 24: 693–701 [DOI] [PubMed] [Google Scholar]
- Hubbard S.R., Miller W.T. (2007). Receptor tyrosine kinases: Mechanisms of activation and signaling. Curr. Opin. Cell Biol. 19: 117–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun Y., Lee I. (2006). KIDARI, encoding a non-DNA Binding bHLH protein, represses light signal transduction in Arabidopsis thaliana. Plant Mol. Biol. 61: 283–296 [DOI] [PubMed] [Google Scholar]
- Jaillais Y., Hothorn M., Belkhadir Y., Dabi T., Nimchuk Z.L., Meyerowitz E.M., Chory J. (2011). Tyrosine phosphorylation controls brassinosteroid receptor activation by triggering membrane release of its kinase inhibitor. Genes Dev. 25: 232–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens V., Longin S., Goris J. (2008). PP2A holoenzyme assembly: in cauda venenum (the sting is in the tail). Trends Biochem. Sci. 33: 113–121 [DOI] [PubMed] [Google Scholar]
- Karlova R., Boeren S., Russinova E., Aker J., Vervoort J., de Vries S. (2006). The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1 protein complex includes BRASSINOSTEROID-INSENSITIVE1. Plant Cell 18: 626–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauschmann A., Jessop A., Koncz C., Szekeres M., Willmitzer L., Altmann T. (1996). Genetic evidence for an essential role of brassinosteroids in plant development. Plant J. 9: 701–713 [Google Scholar]
- Kim T.W., Guan S., Sun Y., Deng Z., Tang W., Shang J.X., Sun Y., Burlingame A.L., Wang Z.Y. (2009). Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat. Cell Biol. 11: 1254–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.W., Wang Z.Y. (2010). Brassinosteroid signal transduction from receptor kinases to transcription factors. Annu. Rev. Plant Biol. 61: 681–704 [DOI] [PubMed] [Google Scholar]
- Kinoshita T., Caño-Delgado A., Seto H., Hiranuma S., Fujioka S., Yoshida S., Chory J. (2005). Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature 433: 167–171 [DOI] [PubMed] [Google Scholar]
- Koka C.V., Cerny R.E., Gardner R.G., Noguchi T., Fujioka S., Takatsuto S., Yoshida S., Clouse S.D. (2000). A putative role for the tomato genes DUMPY and CURL-3 in brassinosteroid biosynthesis and response. Plant Physiol. 122: 85–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Lee S., Yang K.Y., Kim Y.M., Park S.Y., Kim S.Y., Soh M.S. (2006). Overexpression of PRE1 and its homologous genes activates Gibberellin-dependent responses in Arabidopsis thaliana. Plant Cell Physiol. 47: 591–600 [DOI] [PubMed] [Google Scholar]
- Lemmon M.A., Schlessinger J. (2010). Cell signaling by receptor tyrosine kinases. Cell 141: 1117–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. (2010a). Multi-tasking of somatic embryogenesis receptor-like protein kinases. Curr. Opin. Plant Biol. 13: 509–514 [DOI] [PubMed] [Google Scholar]
- Li J. (2010b). Regulation of the nuclear activities of brassinosteroid signaling. Curr. Opin. Plant Biol. 13: 540–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Chory J. (1997). A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90: 929–938 [DOI] [PubMed] [Google Scholar]
- Li J., Li Y., Chen S., An L. (2010a). Involvement of brassinosteroid signals in the floral-induction network of Arabidopsis. J. Exp. Bot. 61: 4221–4230 [DOI] [PubMed] [Google Scholar]
- Li J., Nam K.H. (2002). Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 295: 1299–1301 [DOI] [PubMed] [Google Scholar]
- Li J., Wen J., Lease K.A., Doke J.T., Tax F.E., Walker J.C. (2002). BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110: 213–222 [DOI] [PubMed] [Google Scholar]
- Li L., Ye H., Guo H., Yin Y. (2010b). Arabidopsis IWS1 interacts with transcription factor BES1 and is involved in plant steroid hormone brassinosteroid regulated gene expression. Proc. Natl. Acad. Sci. USA 107: 3918–3923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Yu X., Thompson A., Guo M., Yoshida S., Asami T., Chory J., Yin Y. (2009). Arabidopsis MYB30 is a direct target of BES1 and cooperates with BES1 to regulate brassinosteroid-induced gene expression. Plant J. 58: 275–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim W.A., Pawson T. (2010). Phosphotyrosine signaling: Evolving a new cellular communication system. Cell 142: 661–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X.M., et al. (2010). Integration of light- and brassinosteroid-signaling pathways by a GATA transcription factor in Arabidopsis. Dev. Cell 19: 872–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandava N.B. (1988). Plant growth-promoting brassinosteroids. Annu. Rev. Plant Physiol. Plant Mol. Biol. 39: 23–52 [Google Scholar]
- Mara C.D., Huang T., Irish V.F. (2010). The Arabidopsis floral homeotic proteins APETALA3 and PISTILLATA negatively regulate the BANQUO genes implicated in light signaling. Plant Cell 22: 690–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayumi K., Shibaoka H. (1995). A possible double role for brassinolide in the re-orientation of cortical microtubules in the epidermal cells of Azuki bean epicotyls. Plant Cell Physiol. 36: 173–181 [Google Scholar]
- Montoya T., Nomura T., Farrar K., Kaneta T., Yokota T., Bishop G.J. (2002). Cloning the tomato curl3 gene highlights the putative dual role of the leucine-rich repeat receptor kinase tBRI1/SR160 in plant steroid hormone and peptide hormone signaling. Plant Cell 14: 3163–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora-García S., Vert G., Yin Y., Caño-Delgado A., Cheong H., Chory J. (2004). Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev. 18: 448–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam K.H., Li J. (2002). BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110: 203–212 [DOI] [PubMed] [Google Scholar]
- Noguchi T., Fujioka S., Choe S., Takatsuto S., Yoshida S., Yuan H., Feldmann K.A., Tax F.E. (1999). Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiol. 121: 743–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nole-Wilson S., Rueschhoff E.E., Bhatti H., Franks R.G. (2010). Synergistic disruptions in seuss cyp85A2 double mutants reveal a role for brassinolide synthesis during gynoecium and ovule development. BMC Plant Biol. 10: 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T., Bishop G.J., Kaneta T., Reid J.B., Chory J., Yokota T. (2003). The LKA gene is a BRASSINOSTEROID INSENSITIVE 1 homolog of pea. Plant J. 36: 291–300 [DOI] [PubMed] [Google Scholar]
- Oh M.H., Wang X., Kota U., Goshe M.B., Clouse S.D., Huber S.C. (2009). Tyrosine phosphorylation of the BRI1 receptor kinase emerges as a component of brassinosteroid signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 658–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh M.H., Wang X., Wu X., Zhao Y., Clouse S.D., Huber S.C. (2010). Autophosphorylation of Tyr-610 in the receptor kinase BAK1 plays a role in brassinosteroid signaling and basal defense gene expression. Proc. Natl. Acad. Sci. USA 107: 17827–17832 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Pawson T. (2004). Specificity in signal transduction: From phosphotyrosine-SH2 domain interactions to complex cellular systems. Cell 116: 191–203 [DOI] [PubMed] [Google Scholar]
- Peng P., Zhao J., Zhu Y., Asami T., Li J. (2010). A direct docking mechanism for a plant GSK3-like kinase to phosphorylate its substrates. J. Biol. Chem. 285: 24646–24653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postel S., Küfner I., Beuter C., Mazzotta S., Schwedt A., Borlotti A., Halter T., Kemmerling B., Nürnberger T. (2010). The multifunctional leucine-rich repeat receptor kinase BAK1 is implicated in Arabidopsis development and immunity. Eur. J. Cell Biol. 89: 169–174 [DOI] [PubMed] [Google Scholar]
- Rozhon W., Mayerhofer J., Petutschnig E., Fujioka S., Jonak C. (2010). ASKtheta, a group-III Arabidopsis GSK3, functions in the brassinosteroid signalling pathway. Plant J. 62: 215–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russinova E., Borst J.W., Kwaaitaal M., Caño-Delgado A., Yin Y., Chory J., de Vries S.C. (2004). Heterodimerization and endocytosis of Arabidopsis brassinosteroid receptors BRI1 and AtSERK3 (BAK1). Plant Cell 16: 3216–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H., Cho H., Kim K., Hwang I. (2010a). Phosphorylation dependent nucleocytoplasmic shuttling of BES1 is a key regulatory event in brassinosteroid signaling. Mol. Cells 29: 283–290 [DOI] [PubMed] [Google Scholar]
- Ryu H., Kim K., Cho H., Hwang I. (2010b). Predominant actions of cytosolic BSU1 and nuclear BIN2 regulate subcellular localization of BES1 in brassinosteroid signaling. Mol. Cells 29: 291–296 [DOI] [PubMed] [Google Scholar]
- Ryu H., Kim K., Cho H., Park J., Choe S., Hwang I. (2007). Nucleocytoplasmic shuttling of BZR1 mediated by phosphorylation is essential in Arabidopsis brassinosteroid signaling. Plant Cell 19: 2749–2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlereth A., Möller B., Liu W., Kientz M., Flipse J., Rademacher E.H., Schmid M., Jürgens G., Weijers D. (2010). MONOPTEROS controls embryonic root initiation by regulating a mobile transcription factor. Nature 464: 913–916 [DOI] [PubMed] [Google Scholar]
- Schulze B., Mentzel T., Jehle A.K., Mueller K., Beeler S., Boller T., Felix G., Chinchilla D. (2010). Rapid heteromerization and phosphorylation of ligand-activated plant transmembrane receptors and their associated kinase BAK1. J. Biol. Chem. 285: 9444–9451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu S.H., Karlowski W.M., Pan R., Tzeng Y.H., Mayer K.F., Li W.H. (2004). Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 16: 1220–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., et al. (2010). Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev. Cell 19: 765–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Fokar M., Asami T., Yoshida S., Allen R.D. (2004). Characterization of the brassinosteroid insensitive 1 genes of cotton. Plant Mol. Biol. 54: 221–232 [DOI] [PubMed] [Google Scholar]
- Symons G.M., Reid J.B. (2003). Hormone levels and response during de-etiolation in pea. Planta 216: 422–431 [DOI] [PubMed] [Google Scholar]
- Symons G.M., Smith J.J., Nomura T., Davies N.W., Yokota T., Reid J.B. (2008). The hormonal regulation of de-etiolation. Planta 227: 1115–1125 [DOI] [PubMed] [Google Scholar]
- Tanaka A., et al. (2009). BRASSINOSTEROID UPREGULATED1, encoding a helix-loop-helix protein, is a novel gene involved in brassinosteroid signaling and controls bending of the lamina joint in rice. Plant Physiol. 151: 669–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Deng Z., Oses-Prieto J.A., Suzuki N., Zhu S., Zhang X., Burlingame A.L., Wang Z.Y. (2008a). Proteomics studies of brassinosteroid signal transduction using prefractionation and two-dimensional DIGE. Mol. Cell. Proteomics 7: 728–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Kim T.W., Oses-Prieto J.A., Sun Y., Deng Z., Zhu S., Wang R., Burlingame A.L., Wang Z.Y. (2008b). BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 321: 557–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., et al. (2011). PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat. Cell Biol. 13: 124–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G., Huq E., Quail P.H. (2003). The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15: 1749–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G., Chory J. (2006). Downstream nuclear events in brassinosteroid signalling. Nature 441: 96–100 [DOI] [PubMed] [Google Scholar]
- Vert G., Nemhauser J.L., Geldner N., Hong F., Chory J. (2005). Molecular mechanisms of steroid hormone signaling in plants. Annu. Rev. Cell Dev. Biol. 21: 177–201 [DOI] [PubMed] [Google Scholar]
- Wada K., Marumo S., Abe H., Morishita T., Nakamura K., Uchiyama M., Mori K. (1984). A rice lamina inclination test-amicro-quantitative bioassay for brassinosteroids. Agric. Biol. Chem. 48: 719–726 [Google Scholar]
- Wang H., Zhu Y., Fujioka S., Asami T., Li J., Li J. (2009). Regulation of Arabidopsis brassinosteroid signaling by atypical basic helix-loop-helix proteins. Plant Cell 21: 3781–3791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Chory J. (2006). Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science 313: 1118–1122 [DOI] [PubMed] [Google Scholar]
- Wang X., Goshe M.B., Soderblom E.J., Phinney B.S., Kuchar J.A., Li J., Asami T., Yoshida S., Huber S.C., Clouse S.D. (2005b). Identification and functional analysis of in vivo phosphorylation sites of the Arabidopsis BRASSINOSTEROID-INSENSITIVE1 receptor kinase. Plant Cell 17: 1685–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Kota U., He K., Blackburn K., Li J., Goshe M.B., Huber S.C., Clouse S.D. (2008). Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev. Cell 15: 220–235 [DOI] [PubMed] [Google Scholar]
- Wang X., Li X., Meisenhelder J., Hunter T., Yoshida S., Asami T., Chory J. (2005a). Autoregulation and homodimerization are involved in the activation of the plant steroid receptor BRI1. Dev. Cell 8: 855–865 [DOI] [PubMed] [Google Scholar]
- Wang X., Zhu L., Liu B., Wang C., Jin L., Zhao Q., Yuan M. (2007). Arabidopsis MICROTUBULE-ASSOCIATED PROTEIN18 functions in directional cell growth by destabilizing cortical microtubules. Plant Cell 19: 877–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.Y., Nakano T., Gendron J., He J., Chen M., Vafeados D., Yang Y., Fujioka S., Yoshida S., Asami T., Chory J. (2002). Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2: 505–513 [DOI] [PubMed] [Google Scholar]
- Waters M.T., Wang P., Korkaric M., Capper R.G., Saunders N.J., Langdale J.A. (2009). GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell 21: 1109–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamuro C., Ihara Y., Wu X., Noguchi T., Fujioka S., Takatsuto S., Ashikari M., Kitano H., Matsuoka M. (2000). Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 12: 1591–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z., Zhao J., Peng P., Chihara R.K., Li J. (2009). BIN2 functions redundantly with other Arabidopsis GSK3-like kinases to regulate brassinosteroid signaling. Plant Physiol. 150: 710–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q., Zhu W., Li L., Zhang S., Yin Y., Ma H., Wang X. (2010). Brassinosteroids control male fertility by regulating the expression of key genes involved in Arabidopsis anther and pollen development. Proc. Natl. Acad. Sci. USA 107: 6100–6105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Vafeados D., Tao Y., Yoshida S., Asami T., Chory J. (2005). A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 120: 249–259 [DOI] [PubMed] [Google Scholar]
- Yin Y., Wang Z.Y., Mora-Garcia S., Li J., Yoshida S., Asami T., Chory J. (2002). BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109: 181–191 [DOI] [PubMed] [Google Scholar]
- Yu X., Li L., Li L., Guo M., Chory J., Yin Y. (2008). Modulation of brassinosteroid-regulated gene expression by Jumonji domain-containing proteins ELF6 and REF6 in Arabidopsis. Proc. Natl. Acad. Sci. USA 105: 7618–7623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Li L., Zola J., Aluru M., Ye H., Foudree A., Guo H., Anderson S., Aluru S., Liu P., Rodermel S., Yin Y. (2011). A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J. 65: 634–646 [DOI] [PubMed] [Google Scholar]
- Zhang L.Y., et al. (2009). Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell 21: 3767–3780 [DOI] [PMC free article] [PubMed] [Google Scholar]