Two proteins similar to β-amylases (enzymes usually associated with starch breakdown) possess a BZR1-type DNA binding domain and are nuclear localized. They bind a G box-containing motif and regulate the expression of genes, many of which also respond to brassinosteroids, to influence shoot growth. Similar proteins occur in other plants, implying functional conservation.
Abstract
Plants contain β-amylase–like proteins (BAMs; enzymes usually associated with starch breakdown) present in the nucleus rather than targeted to the chloroplast. They possess BRASSINAZOLE RESISTANT1 (BZR1)-type DNA binding domains—also found in transcription factors mediating brassinosteroid (BR) responses. The two Arabidopsis thaliana BZR1-BAM proteins (BAM7 and BAM8) bind a cis-regulatory element that both contains a G box and resembles a BR-responsive element. In protoplast transactivation assays, these BZR1-BAMs activate gene expression. Structural modeling suggests that the BAM domain’s glucan binding cleft is intact, but the recombinant proteins are at least 1000 times less active than chloroplastic β-amylases. Deregulation of BZR1-BAMs (the bam7bam8 double mutant and BAM8-overexpressing plants) causes altered leaf growth and development. Of the genes upregulated in plants overexpressing BAM8 and downregulated in bam7bam8 plants, many carry the cis-regulatory element in their promoters. Many genes that respond to BRs are inversely regulated by BZR1-BAMs. We propose a role for BZR1-BAMs in controlling plant growth and development through crosstalk with BR signaling. Furthermore, we speculate that BZR1-BAMs may transmit metabolic signals by binding a ligand in their BAM domain, although diurnal changes in the concentration of maltose, a candidate ligand produced by chloroplastic β-amylases, do not influence their transcription factor function.
INTRODUCTION
Growth is underpinned by carbohydrate metabolism (Smith and Stitt, 2007). Sugars serve as substrates for the biosynthesis of major cellular components and are derived from photosynthesis during the day. During the night, when photosynthesis is not possible, sugars are derived from the degradation of chloroplastic transitory starch. This is mediated primarily by the action of chloroplast-localized β-amylases, which generate maltose from starch for export to the cytosol (Chia et al., 2004; Niittylä et al., 2004; Fulton et al., 2008). Sugars also act as regulatory signals. High sugar levels trigger the repression of photosynthesis and the induction of carbohydrate storage, whereas low levels trigger a cessation of growth and the induction of catabolic processes (Koch, 1996; Bläsing et al., 2005; Rolland et al., 2006; Baena-González et al., 2007). During the diurnal cycle, fluctuations in endogenous sugar levels have been shown to be the major factor driving changes in the Arabidopsis thaliana transcriptome (Bläsing et al., 2005).
Sugar signaling in plants is not well understood, but there is evidence that several mechanisms exist (Rolland et al., 2006). For example, hexokinase1 (HXK1) has been shown to act as a Glc sensor in addition to metabolizing Glc (Moore et al., 2003; Cho et al., 2006). The Arabidopsis mutant deficient in HXK1 (glucose insensitive2 [gin2]) is insensitive to high levels of exogenous Glc, which repress postgerminative growth of the wild-type seedlings. The gin2 mutant exhibits reduced vegetative growth in high light conditions and has altered responses to auxin and cytokinin. Such crosstalk between sugar and hormonal signaling could provide a mechanism of growth control (Moore et al., 2003). Suc is also sensed, although the signal transduction mechanism has not been elucidated. Again, crosstalk between Suc signaling and hormone (abscisic acid) signaling has been demonstrated (Rook et al., 2001).
Vascular plants have multiple genes encoding β-amylases and β-amylase–like proteins (BAMs; Arabidopsis has nine). One of these BAM proteins was recently shown to be noncatalytic and to exert a regulatory role over starch degradation in the chloroplast (Fulton et al., 2008). Others are predicted to be extraplastidial and their functions are unknown. We noticed that two BAM proteins, BAM7 (At2g45880, also called BMY4) and BAM8 (At5g45300, also called BMY2), were unusual because in addition to a well-defined glucosyl-hydrolase domain, they possess an N-terminal domain with sequence similarity to transcription factors of the BRASSINAZOLE RESISTANT1 (BZR1) type.
BZR1 together with BRI1-EMS-SUPPRESSOR1 (BES1) and their homologs (BES1 HOMOLOGs [BEHs]) represent a plant-specific family of transcriptional regulators essential for mediating the transcriptional response to BRs. BRs are plant steroid hormones that are perceived at the cell surface and function in diverse developmental and growth processes (Clouse et al., 1996; Li and Chory, 1997; Kinoshita et al., 2005). Mutant plants lacking the ability to synthesize or perceive BRs are severely dwarfed and have altered developmental patterns (Chory et al., 1991; Clouse et al., 1996; Kauschmann et al., 1996; Vert and Chory, 2006). The targets of BEH transcription factors include genes controlling growth responses and genes controlling BR homeostasis (Wang et al., 2002; Yin et al., 2002; He et al., 2005; Kim et al., 2009). BR signaling interacts with other hormone signaling pathways. In particular, BRs and auxin act synergistically to trigger similar cellular responses (Goda et al., 2004; Nemhauser et al., 2004; Mouchel et al., 2006).
Here, we show that BAM7 and BAM8 localize to the nucleus, activate gene expression via a specific DNA target motif, and play a role in controlling plant growth and development. The two-domain structure of BAM7 and BAM8 proteins is widely conserved in plants, suggesting a fundamental role for this type of transcription factor, which could potentially communicate the status of metabolism to control growth and development.
RESULTS
BZR1-BAMs Are Nuclear Proteins
BAM7 and BAM8 both possess full-length glucosyl–hydrolase domains (belonging to family 14). Both proteins also have N-terminal extensions that are not present in the other members of the BAM family (Figure 1A). The extensions share sequence similarity to the transcriptional regulator BZR1 and its immediate homologs (BEH proteins). Blast searches did not reveal any other proteins in the Arabidopsis genome carrying a BZR1-like domain. We identified amino acids likely to be involved in DNA binding and putative bipartite nuclear localization sequences (NLS), rich in basic amino acids (i.e., Lys, His, and Arg; see Supplemental Figure 1 online). Genes encoding similar BZR1-BAMs are present in other higher plant genomes, including gymnosperm and angiosperm species (see Supplemental Figure 1 online). We used fluorescence microscopy to determine the subcellular localization of BZR1-BAMs with green fluorescent protein (GFP) or yellow fluorescent protein (YFP) fused to the C-terminal end, in stably transformed plants and in transfected protoplasts. In all cases, GFP/YFP fluorescence colocalized with the diamidino-2-phenylindole-stained nucleus (Figure 1B; see Supplemental Figure 2A online). The amino-terminus of each protein containing the BZR1 domain was sufficient to target YFP to the nucleus. We created mutated forms of the BAM8 protein in which we substituted the basic residues of the predicted bipartite NLS with glutamines. These mutated forms were fused to GFP and transiently expressed in tobacco leaves. Mutation of either of the two basic regions comprising the NLS was sufficient to exclude the BAM8-GFP from the nucleus (see Supplemental Figure 2B online). Antibodies raised against the recombinant BAM7 or BAM8 proteins were used to localize the native proteins in the wild-type plants. The anti-BAM7 antibodies recognized recombinant BAM7 in extracts of stably transformed plants overexpressing HA- or YFP-tagged versions of the protein, but did not detect BAM7 in extracts of the wild-type plants (Figure 2), possibly because the endogenous protein is too low in abundance. The anti-BAM8 antibodies identified a protein of the predicted molecular weight (77 kD) in crude homogenates of the wild-type leaves, but not in bam8 knockout mutants (Figure 2). This protein sedimented with the cell debris, and was enriched in preparations of nuclei, showing that endogenous BAM8 protein is present in the nucleus of the wild-type plants (Figure 1C). The stably overexpressed HA-tagged BAM7 protein (Figure 2B), was similarly enriched in preparations of nuclei (Figure 1D).
Figure 1.
BAM7 and BAM8 Are Nuclear Proteins Comprising a Putative DNA Binding Domain of the BZR1-Type and a Glycoside Hydrolase–Like Domain.
(A) Protein models of Arabidopsis BAM3, BAM7, BAM8, and BZR1. Red, BZR1 domain; yellow, glycoside–hydrolase family 14 domain; green, transit peptide (TP; Fulton et al. 2008) for chloroplast localization; cyan, putative bipartite nuclear localization signal. aa, amino acids.
(B) Transiently expressed BAM7-GFP and BAM8-GFP in Arabidopsis protoplasts localize to nucleus. Localization studies were performed using C-terminal GFP fusions with the complete BAM7 and BAM8 sequence. Chlorophyll autofluorescence (Chl) and 4′,6-diamidino-2-phenylindole (DAPI) staining serve as markers for chloroplasts and nuclei, respectively.
(C) Immunoblots of total homogenate, soluble, and nuclear-enriched fractions from the wild-type plants using an anti-BAM8 antibody. The majority of the endogenous BAM8 protein was detected in the nuclear fraction.Disproportionating enzyme2 (DPE2) and Histone3 (H3) are used as cytosolic and nuclear markers, respectively. Molecular weight markers are indicated in kilodaltons.
(D) Immunoblots of total homogenate, soluble, and nuclear-enriched fractions from HA-BAM7–overexpressing plants using an anti-HA antibody. HA-BAM7 protein was primarily detected in the nuclear fraction.
Figure 2.
Isolation of BZR1-BAM Mutants and Overexpression Lines.
(A) Gene models of BAM7 (At2g45880) and BAM8 (At5g45300) and the position of the T-DNA insertions and stop codons in the mutant alleles. The two bam7 knockout mutants each carry a mutation in the fifth exon resulting in premature stop codons (W291* in bam7-1, W305* in bam7-2). The bam8 mutants are T-DNA insertion lines disrupting the gene in the 1st and 8th exon, respectively (bam8-1, SALK_000892; bam8-2, GK-243B11).
(B) Protein gel blots probed with antibodies against BAM7 (α−BAM7) or BAM8 (α−BAM8) using total leaf homogenates. BAM7-antiserum detects the recombinant BAM7 protein (lanes 4 and 5) but no endogenous protein (lane 1). BAM8 antiserum detects an endogenous protein in the wild type (lane 1), which appears as a double band of a molecular weight slightly greater than 75 kD. Both bands are absent in the two bam8 knockout lines (lanes 6 and 7). 1, wild type; 2, bam7-1; 3, bam7-2; 4, BAM7-OX‐2 (HA-tag); 5, BAM7-OX-1 (YFP fusion); 6, bam8-1; 7, bam8-2; 8, BAM8-OX‐3 (HA-tag); 9, BAM8-OX-1 (YFP fusion). Arrows indicate the positions of molecular weight markers. Single and double asterisks indicate the position of the HA- and YFP-fusion proteins, respectively.
[See online article for color version of this figure.]
We modeled the BAM domain of BAM8, based on the crystal structure of the soybean β-amylase. Most of the amino acids lining the glucan binding pocket, including the two catalytic glutamic acid residues, are conserved (see Supplemental Figures 1 and 3A online). The modeled active sites can accommodate a glucan substrate such as maltotetraose, although the overall surface charge of the pockets is predicted to be less electronegative than of known active β-amylases. We determined whether the BAM7 and BAM8 proteins have β-amylase activity. Recombinant proteins were assayed either using amylopectin as a substrate (monitoring maltose release) or using the Betamyl assay kit (which contains the chlorogenic substrate p-nitrophenyl maltopentaoside). Both proteins had very low β-amylase activity in vitro with both assay methods (Figure 3). The specific activity was at least 1000 times lower than that of the recombinant chloroplastic enzymes BAM1 and BAM3. These localization and activity data show that BAM7 and BAM8 do not function in starch breakdown in the chloroplast but may have another role in the nucleus. We suggest that BAM domain may bind a ligand without necessarily hydrolyzing it.
Figure 3.
BZR1-BAMs Have Very Low Glucan Hydrolytic Activity In Vitro.
(A) Purified recombinant proteins used in activity assays. Protein concentration was estimated based on Coomassie staining. Asterisks indicate the protein band of interest immediately to the left. FL, full-length protein; CAT, putative glycoside hydrolase domains alone.
(B) β−Amylase activities of recombinant BAM7 and BAM8 proteins (as in A) were determined in vitro using the Betamyl assay. Recombinant BAM1 and BAM3 proteins served as positive controls. Values are the means ± sd from duplicate experiments. Note the logarithmic scale. ND, not detected.
(C) Maltose release by recombinant BAM7 and BAM8 proteins (as in A) after incubation with maltoheptaose (G7) or amylopectin substrate. Values are the means ± sd from duplicate experiments. Note the logarithmic scale.
[See online article for color version of this figure.]
The BBRE: A Novel cis-Regulatory Element
BZR1 domains from different proteins have been reported to bind to specific DNA sequences. To determine whether the BZR1 domain of BZR1-BAMs bind to a specific DNA sequence, a random binding site selection (RBSS) experiment was performed using the immobilized BZR1 domain of BAM7. Initially, the protein was incubated with random oligonucleotides. Protein-bound oligonucleotides were isolated, PCR-amplified, and used in subsequent RBSS rounds. After four rounds, electrophoretic mobility shift assays (EMSAs) showed that a visible fraction of oligonucleotides could be bound by the BZR1 domain of BAM7 or BAM8. The fraction of bound oligonucleotides increased in subsequent RBSS rounds (Figure 4A). After seven rounds, DNA sequencing revealed a highly enriched DNA motif (Figure 4B). Interestingly, this motif contained within it other well-known cis-regulatory elements, including the G box sequence (5′-CACGTG-3′), which is the target of BES1 and other transcriptional regulators such as basic helix-loop-helix proteins (Toledo-Ortiz et al., 2003; Yin et al., 2005, 2011; Yu et al., 2011) and the BR-responsive element (5′-CGTG[T/C]G-3′), which is the target of BZR1 (He et al., 2005). No sequence-specific enrichment was observed in a control RBSS experiment where the BZR1 domain was omitted. EMSA experiments showed that the core sequence of the motif (5′-CACGTGTG-3′) was sufficient for DNA binding of recombinant BZR1 domains and full-length BAM7 and BAM8 proteins (Figures 4A and 4C). This sequence was designated BBRE for BZR1-BAM-Responsive Element. Binding of the labeled BBRE-oligonucleotides could be reduced by addition of an excess of unlabeled oligonucleotides with the same sequence (Figure 4C). Mutation of the BBRE to 5′-CACTTGTG-3′ (mBBRE) abolished BZR1-BAM binding. This mutated sequence also failed to compete with the binding of the BBRE (Figure 4C).
Figure 4.
BAM7 and BAM8 Proteins Bind a Specific Cis-Regulatory Element and Activate Gene Expression.
(A) EMSA using DIG-labeled oligonucleotides obtained by sequential rounds of RBSS. Oligonucleotides were incubated with recombinant BZR1-domains of BAM7 and BAM8. Input and eluates from rounds 4 to 7, and the final round 7 eluate of the control experiment (C), are shown. Full-length BAM7 and BAM8 (FL) bind to the most enriched oligonucleotide (EO). Single and double asterisks mark positions of unbound and bound oligonucleotides, respectively.
(B) The conserved BBRE motif isolated by RBSS contains known binding sites for transcriptional regulators including the BR-responsive element (BRRE) and the palindromic G box. The percentage value given below the residues indicates the conservation at this location in the motif from an alignment of 1,072,225 Illumina sequence reads. The asterisk indicates the base that was mutated to a thymine in the mBBRE.
(C) BZR1-domains and full-length BZR1-BAM proteins bind the BBRE but a single base mutation in the mBBRE is able to abolish the binding. Comp., a 300-fold excess of unlabeled BBRE (c) reduces binding, but a 300-fold excess of unlabeled mBBRE (cm) does not. Only the location of the shifted oligonucleotides is shown. Note that lanes 5 to 7 were exposed for a shorter time than the rest of the EMSA. Input, dotblot of DIG-labeled oligonucleotides, shows equal labeling of the BBRE and the mBBRE (dilutions are indicated on the right).
(D) BAM8 activates reporter gene expression via the BBRE. Arabidopsis protoplasts were transfected with the Luc reporter under the control of the minimal CaMV 35S promoter (min35S), or with one (1xBBRE) or three copies of the BBRE sequence (3xBBRE) upstream of the min35S, or with three copies of mBBRE (3xmBBRE) upstream of the min35S. HA-tagged, full-length BAM7 or BAM8 served as effectors, and salmon sperm DNA was used as control DNA. Reporter activity is relative to the transfection control GUS and normalized to the average value obtained with the min35S using control DNA. Values are the mean LUC/GUS ratios of three replicate transformations performed in parallel (±sd).
To test if BAM7 and BAM8 can act as transcriptional regulators, we performed transactivation assays in Arabidopsis protoplasts cotransfected with three plasmids. We used a plasmid containing the β-glucuronidase (GUS) gene as a transfection control, a plasmid containing BAM7 or BAM8 as the effector genes, and a plasmid containing the luciferase (LUC) gene as a reporter. The LUC gene was placed downstream of the minimal CaMV 35S promoter (min35S) and the effect of introducing the BBRE or the mBBRE was tested. We observed a small induction of luciferase activity, even in the absence of an effector gene, when one or three copies of the BBRE were present compared with the min35S control experiment (Figure 4D). This may be due to endogenous transcription factors recognizing the BBRE. When protoplasts were cotransfected with a construct constitutively expressing the full-length BAM8 reporter gene, expression was strongly and consistently induced above control levels if the BBRE was present upstream of the min35S. Induction was further enhanced by multiple copies of the BBRE (Figure 4D). Induction was strictly dependent on the BBRE, as no induction of BAM8-mediated reporter gene expression was obtained with the mBBRE sequence. This shows that BAM8 can act as an activator of gene expression via the BBRE in gene promoter sequences. However, in protoplasts expressing the full-length BAM7 protein, induction of the reporter gene beyond control levels was either much weaker than for BAM8 (data not shown) or not observed (Figure 4D).
BZR1-BAMs Are Transcriptional Activators That Control Shoot Development
We analyzed the phenotypes of plants overexpressing BAM7 or BAM8 (either as HA- or YFP-tagged proteins). When grown on soil, independent transgenic lines overexpressing BAM7 (BAM7-OX) were indistinguishable from the wild type, whereas plants overexpressing BAM8 (BAM8-OX) showed altered growth and developmental phenotypes (see Supplemental Figure 4A online). Compared with the wild type, the BAM8-OX plants had reduced shoot fresh weight and developed smaller rosettes, which had short petioles and rounded, dark-green, hyponastic leaves (Figure 5; see Supplemental Figure 4A online). This phenotype was observed in independent transgenic lines. We produced and analyzed bam7- and bam8-null mutants. There were no consistent differences in the morphology of the bam7- and bam8-null mutants compared with the wild type, but the shoot fresh weight in bam7 mutants was decreased (see Supplemental Figure 4 online). The bam7bam8 double mutant also had a reduced fresh weight (Figure 5; see Supplemental Figure 4B online). Remarkably, the bam7bam8 double mutant displayed a leaf morphology phenotype that was in some ways the opposite of the BAM8-OX lines; the rosettes had long petioles and epinastic leaves compared with the wild type (Figure 5; see Supplemental Figure 4A online). We investigated whether deregulation of the BZR1-BAMs altered endogenous carbohydrate levels. Starch, Suc, Glc, Fru, and maltose levels in the knockout mutants and the BAM-OX plants were comparable to the wild type (see Supplemental Figure 5 online). These data show that BZR-BAMs are not directly involved in carbohydrate metabolism, but rather influence plant growth and development.
Figure 5.
Deregulation of BZR1-BAMs Impairs Plant Growth and Alters Leaf Development.
Mutant plants lacking BZR1-BAMs (bam7bam8, middle) or overexpressing BAM8 (BAM8-OX, right) were grown together with the wild type for 21 d on soil. For fresh weight determinations (FW, ±se, n = 28), whole rosettes were used. For relative petiole length (petiole/leaf, ±se, n ≥ 11) and leaf flattening (f index, ±se, n ≥ 11), mature leaf no. 5 was analyzed. *P < 0.005, **P < 0.0005. Scale bar = 5 mm.
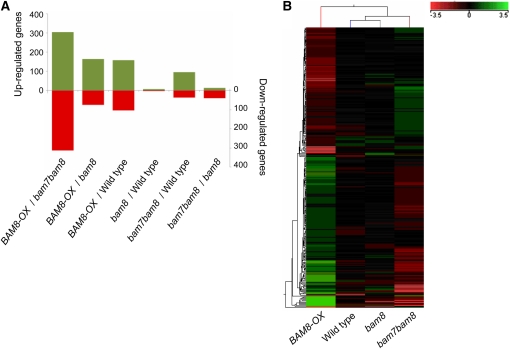
To understand how BAM7 and BAM8 control shoot development, we conducted microarray analyses using RNA extracted from the aerial parts of 14-d-old wild-type, bam8, bam7bam8, and BAM8-OX seedlings harvested 4 h into the dark period. We observed only minor differences between the transcriptional profiles of bam8 and the wild type, with only 11 genes significantly changed by more than a factor of 2. However, BAM8-OX and bam7bam8 both differed markedly from the wild type (with 263 and 132 genes changed, respectively; Figure 6A; see Supplemental Table 1 online). Interestingly, most of the changes in BAM8-OX were the inverse of those in bam7bam8, resulting in transcriptional profiles that were very different from each other (Figure 6B), with 618 genes differing significantly in expression (see Supplemental Table 1 online).
Figure 6.
Deregulation of BZR1-BAMs Causes Distinct Changes in Gene Expression.
(A) Pairwise comparison between genotypes. Bar height indicates the number of genes that change by at least twofold (t test, P < 0.1). Green, upregulated genes; red, downregulated genes.
(B) Heat map obtained from hierarchical clustering of 576 genes, the expression of which changes across genotypes (analysis of variance, P < 0.1; fold-change > 2). Fold-changes relative to the average expression across 12 arrays is indicated by color coding (given as log2 values).
We analyzed the 1000-bp promoter sequences of the 500 most up- or downregulated genes between genotypes. This revealed that the BBRE motif was significantly over-represented in the promoters of genes upregulated in BAM8-OX (where it was the most abundant eight-letter motif), and/or repressed in bam7bam8, compared with the wild type. By contrast, the BBRE was not enriched in the promoters of genes downregulated in BAM8-OX or upregulated in bam7bam8 (Table 1; see Supplemental Table 2 online). Over 80% of the BBREs were present within the 500 bp of the transcriptional initiation site, and we focused on these regions. We identified 312 genes represented on the ATH1 array that carry the BBRE in their 500-bp promoters. Of these BBRE genes, many more were upregulated in BAM8-OX compared with the wild type than would be expected by chance (Figure 7A; see Supplemental Table 1 online). This was especially true among genes that were highly upregulated. Similarly, more BBRE genes than expected were repressed in bam7bam8 relative to the wild type. Eighty percent of the BBRE genes that changed in BAM8-OX and bam7bam8 relative to the wild type showed inverse regulation (Figure 7B). Importantly, this shows that genes that are changed in BAM8-OX plants are also under the control of the endogenous BZR1-BAMs in the wild-type plants. Collectively, the microarray data, the RBSS, and the transactivation analyses in protoplasts provide compelling evidence that BZR1-BAMs function as transcriptional activators in vivo by binding to the BBRE in the promoters of target genes.
Table 1.
The Most Abundant Motifs Present within the 1000-Bp Promoters of Deregulated Genes
| Genotype | Upregulated | Downregulated |
| BAM8-OX | cacgtgtg (1.5E−14) | acggttaa (3.6E−02) |
| bam8 | cctgacga (7.9E−02) | gcacgtgt (3.0E−02) |
| bam7bam8 | taagccga (3.2E−01) | cacgtgtg (2.2E−11) |
The most abundant eight-letter motifs identified in the promoters of the 500 genes showing the greatest deregulation in BAM8-OX, bam8, and bam7bam8 relative to the wild type (for bam8 upregulated genes, n = 293). E values are given in parentheses (see also Supplemental Table 2 online).
Figure 7.
Expression of BBRE Genes and BL-Responsive Genes Is Altered in BAM8-OX and bam7bam8.
(A) The relative occurrence of significantly (P < 0.1) deregulated genes carrying a BBRE in their 500-bp promoters. Relative occurrence is defined as the ratio of observed changes to those expected by chance, given the total number of deregulated genes (see Methods). More BBRE genes than expected are highly upregulated in BAM8-OX (red), and very downregulated in bam7bam8 (blue), relative to other genotypes. The bar patterns indicate the fold-change categories, as indicated. Genotypes are as follows: B8OX, BAM8-OX; b7b8, bam7bam8; b8, bam8; WT, wild type. Pale bars indicate that, between the compared genotypes, fewer than 50 genes in total are deregulated to the extent indicated. The numbers of genes are given in Supplemental Table 1 online.
(B) Expression of BBRE genes in BAM8-OX and bam7bam8 relative to the wild type, showing inverse regulation. Blue circles indicate genes that are changed in either genotype (P < 0.1); red crosses indicate genes that are changed in both genotypes; gray circles indicate genes that are unchanged.
(C) Relative occurrence of BL-repressed genes among significantly deregulated genes, annotated as in (A).
(D) Relative occurrence of BL-induced genes among significantly deregulated genes, annotated as in (A). The numbers of genes for (C) and (D) are given in Supplemental Table 3 online.
As the BBRE motif has similarities to the G box and BR-responsive element motifs—known binding sites for BR-responsive transcription factors—we analyzed the expression of genes previously reported to be responsive to BL (brassinolide, a potent BR; Nemhauser et al., 2004). Genes repressed by BL showed greater changes in our microarrays than BL-induced genes. Of the BL-repressed genes, more than expected were upregulated in BAM8-OX relative to the wild type, while more than expected were repressed in bam7bam8 (Figures 7C and 7D; see Supplemental Table 3 online). BL-induced genes tended to be repressed in BAM8-OX (see Supplemental Figure 6 online). Thus, some genes are regulated by BZR1-BAMs and BRs, but usually in opposite ways. However, a few BL-repressed genes were also downregulated in BAM8-OX or induced in bam7bam8.
MAPMAN software (Thimm et al., 2004) revealed major transcriptional changes in BAM8-OX relative to the wild type in genes involved in cell wall metabolism (see Supplemental Figure 7A online). Several xyloglucan endotransglycosylases, xyloglucan endotransglucosylase/hydrolases, and expansin-like proteins were repressed in BAM8-OX. Xyloglucan endotransglycosylases and expansins are positively regulated by BR signaling. Certain BR-related growth phenotypes have been proposed to be caused by their deregulation (Xu et al., 1995; Kauschmann et al., 1996; Goda et al., 2002; Goda et al., 2004). The repression of these genes could explain some of the growth defects observed in BAM8-OX plants. Genes involved in flavonoid and phenylpropanoid metabolism were also repressed in BAM8-OX. Flavonoids have been proposed to control auxin fluxes and therefore may also affect plant growth (Ringli et al., 2008). Among transcription factors, several genes encoding AUX/IAA proteins were repressed in BAM8-OX seedlings, again supporting the inverse effect of BAM8-OX to BRs, which induce AUX/IAAs and act synergistically with auxins (Goda et al., 2002; Nakamura et al., 2003; Goda et al., 2004). Other transcription factors were also deregulated, including members of the WRKY, MYB, and the AP2/EREBP families, but did not show consistent patterns of repression or activation. MAPMAN revealed far fewer changes in specific pathways in the bam7bam8 double mutant (see Supplemental Figure 7B online). As BAM8 activates transcription, genes that are repressed in BAM8-OX are unlikely to be under its direct control, but rather controlled by downstream components (i.e., other transcriptional regulators). Among the genes upregulated in BAM8-OX and carrying a BBRE in their promoters (putative direct targets) are four genes encoding proteins with annotated functions as regulators of gene expression: SWN (At4g02020; Swinger), ANAC102 (At5g63790; Arabidopsis Nac Domain Containing Protein 102), ZAT10 (At1g27730; zinc finger [C2H2-type] family protein), and a WRKY transcription factor (At2g44745). These are candidates for regulatory factors that act downstream in the BZR1-BAM signaling pathway.
Chloroplast BAMs release maltose, which is exported to the cytosol for further metabolism (Niittylä et al., 2004; Fulton et al., 2008). Based on our in silico analysis (see Supplemental Figure 3 online), we reasoned that the BAM domain, despite having a very low hydrolytic activity, may still bind a ligand. We considered it possible that BZR1-BAMs could bind cytosolic maltose, thereby sensing changes in starch-derived sugar levels during the day-night cycle. Maltose levels are at their highest 4 h into the dark (Chia et al., 2004). Therefore, we repeated the microarray experiment with seedlings harvested 4 h into the light period, when sugars are derived from photosynthesis rather than starch degradation (Zeeman et al., 2007; Fulton et al., 2008) and maltose levels are very low. There were marked differences in gene expression patterns between the light- and dark-harvested samples, consistent with previous studies (Bläsing et al., 2005). However, we observed similar genotype-specific patterns as in the first microarray experiment: the BBRE was over-represented in the promoters of genes induced in BAM8-OX and repressed in bam7bam8, and BBRE genes changed more often and with greater amplitudes than other genes (see Supplemental Figure 8 and Supplemental Tables 2 and 4 online). Genes showing deregulation between genotypes at one time point tended to change in a similar way at the other time point (see Supplemental Figure 8 online). This suggests that BZR1-BAMs regulate transcription at both time points and that fluctuations in endogenous maltose levels did not influence their activity.
DISCUSSION
To our knowledge, the BZR1-BAMs represent a unique class of transcription factors in plants. BAM-like proteins are conserved across the plant kingdom and are present in some amoebal and bacterial genomes. BZR1-like proteins, however, are only found in plants. The presence of BZR1-BAMs in distantly related higher plants shows that the two-domain structure was established before the divergence of gymnosperm and angiosperm species. It is reasonable to suggest that their function in the control of gene expression has been conserved. Thus, BZR1-BAMs may also influence the growth and morphology of agronomically important crop species.
Our data provide compelling evidence that BZR1-BAMs regulate transcription through the BBRE cis-regulatory element. The BBRE was identified via an in vitro approach but is strongly over-represented in the promoters of genes with decreased expression when the BZR1-BAMs were mutated and increased expression when BAM8 was overexpressed. The similarities between the BBRE and target motifs in BR-responsive genes mean that BZR1-BAMs may compete with BZR1 and its homologs at the promoters of common target genes, thereby directly modulating BR responses to control growth and development. This could explain the over-representation of BL-responsive genes among BZR1-BAM-regulated genes and the growth phenotypes we observed. It is also possible that the signaling pathways interact further downstream rather than (or in addition to) having common targets (Figure 8). Public microarray data suggest that BAM7 and BAM8 are expressed at a low level throughout the plant, but are highest in the shoot apical meristems (http://www.bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi), consistent with a developmental role.
Figure 8.
A Model for the Integration of BZR1-BAM and BR Signaling.
BZR1-BAMs activate gene expression through binding to the BBRE in the promoters of their direct targets. BZR1-BAMs and BZR1/BEHs may compete for common targets. Signal integration may also occur downstream of the direct targets. We speculate that BZR1-BAMs may respond to sugars or other as-yet uncharacterized metabolic signals. Thereby, metabolic and developmental signals could be simultaneously monitored and translated into an appropriate transcriptional response to control leaf growth.
BAM8 activated reporter gene transcription in the protoplast transactivation assay and its overexpression caused significant growth and developmental abnormalities, presumably via changes in gene transcription. We did not see consistent transcriptional activation activity for BAM7 in the protoplast system and overexpression of BAM7 did not cause a comparable growth phenotype to overexpression of BAM8. Nevertheless, our analyses suggest that the two proteins have overlapping functions in vivo. The proteins have a high degree of sequence similarity (50% amino acid identity) and both recognize the BBRE. Furthermore, the loss of BAM8 alone caused little or no phenotypic or transcriptional changes, whereas the bam7bam8 double mutant had a distinctive transcript profile in which many genes were inversely regulated compared with the BAM8-OX plants. This suggests that the BAM7 protein was functioning as a transcriptional activator in the bam8 mutant background. The bam7 mutants, while similar in appearance to the wild-type plants, are significantly smaller. Therefore, we do not exclude a role for BAM7 in vivo. It is remarkable that, in some ways, the growth phenotype of bam7bam8 double mutant is the opposite to that of the BAM8-OX lines (long rather than short petioles, epinastic rather than hyponastic leaves). It is likely that these differences are the direct result of the inverse mis-expression of the direct and downstream target genes in bam7bam8 compared with BAM8-OX, as many of these genes have roles in the process of cell growth or its coordination.
The BZR1 domain in BZR1-BAMs may have other functions besides DNA binding. BES1 can interact with other transcription factors and has been proposed to adopt a tertiary structure similar to basic helix-loop-helix proteins, which bind to DNA as dimers (Yin et al., 2005; Li et al., 2009). Thus, BAM7 and BAM8 may also dimerize with themselves, with each other, or with other proteins. Interaction with BEH proteins may contribute to the altered BR responses and would be consistent with the similarity between the BBRE and the known BEH target motifs.
The low specific activity of the wild-type BZR1-BAM proteins against glucan substrates suggests that β-amylolysis is not their primary function in vivo. However, many of the amino acids lining the active site and involved in substrate binding, including the catalytic residues, are conserved with respect to active BAMs (Fulton et al., 2008). We propose that the function of the BAM domain is to bind a glycan or sugar ligand. Interaction with a ligand could influence the function of BZR1-BAMs either through modulating their affinity for the BBRE or by altering their ability to recruit other protein factors required to activate transcription. The most obvious candidate sugar ligand for BZR1-BAMs is maltose, the product of starch breakdown by BAMs. However, our microarray experiments with BZR1-BAM mutants at the two time points during the diurnal cycle when endogenous maltose levels differ most suggest that BZR1-BAM–mediated target gene expression was not dependent on maltose levels. Future work will identify whether a ligand binds to BZR1-BAMs, and if so, how it influences their activity.
Overall, we propose that the function of BZR1-BAMs is to provide a metabolic signal to influence the rate and/or pattern of plant growth (Figure 8). This discovery adds to the roles played by BAM proteins. In addition to the BZR1-BAMs described here and the catalytically active BAMs in the chloroplast (BAMs 1–3), a noncatalytic isoform (BAM4), also present in the chloroplasts, is believed to regulate starch degradation (Fulton et al., 2008). Given that there are numerous examples in plants of gene families encoding enzymes and enzyme-like proteins, we suggest that there may be many examples like BZR1-BAMs where metabolite signaling rather than catalysis is the protein’s primary function.
METHODS
Plant Material, Growth Conditions, and Growth Measurements
Arabidopsis thaliana plants (all ecotype Columbia) were grown as described (Fulton et al., 2008). For protein and transcript analyses, seedlings were grown for 14 d. For metabolite analyses, plants were grown for 22 to 24 d. Mutant seed stocks were obtained from the ABRC or from the European Arabidopsis Stock Centre: CS95972 (bam7-1), CS88564 (bam7-2), SALK_000892 (bam8-1), and GK-24B11 (bam8-2). Double mutants were obtained by crossing. Homozygosity of single and double mutants was confirmed by PCR-based genotyping. For T-DNA insertion mutations, a T-DNA specific primer was used in combination with a gene-specific primer. Primer pairs (all given 5′ to 3′) were ATGCATACTCTCAACAACACCATC and GGTTCACGTAGTGGGCCATCG for the bam8-1 allele, ATGCATACTCTCAACAACACCATC and TGAGATTCAATTGCAGCCTTAG for the wild-type allele, respectively; AAAGTCACGAGGATTCACTTTC and ATATTGACCATCATACTCATTGC for the bam8-2 allele, AAAGTCACGAGGATTCACTTTC and GGCCTTGAGGAGAGAATAAACA for the wild-type allele, respectively. bam7-1 and bam7-2 are ethyl methanesulfonate-mutagenized lines identified by the Arabidopsis Tilling Program (http://tilling.fhcrc.org/). A 109-bp PCR fragment produced with primers ATTGGCTGATCGAGATGGAC and GGGGGAATGACCCTCAACTA is cleaved by BccI into 8-, 45-, and 56-bp fragments in the wild type and into 8-, 22-, 34-, and 45-bp fragments in bam7-1. A 159-bp product produced with TCGCGGACCATCTGAAAAAGCTGCCTGGAACCAGT and ATTGGCTGATCGAGATGGAC is susceptible to XmnI digestion in bam7-2 but not in the wild type.
The BAM7 cDNA sequence was obtained from the RIKEN Resource Center (Tsukuba, Japan) and the BAM8 cDNA from the ABRC. These were cloned into pB7YWG2.0 (Karimi et al., 2002), pEZT-NL (D. Ehrhardt, Carnegie Institution of Washington), and pEarleyGate201 (Earley et al., 2006) by recombinant cloning. These constructs, driven by the 35S CaMV promoter, express BAM7 and BAM8 with GFP or YFP fused to their C termini (pEZT-NL and pB7YWG2.0, respectively) or with an HA-tag fused to their N termini (pEarleyGate201). Wild-type (BAM7 constructs) or bam8-1 plants (BAM8 constructs), transformed using Agrobacterium tumefaciens (Clough and Bent, 1998; Chung et al., 2000), expressed fluorescent- or HA-tagged proteins. If not stated differently, BAM8-OX refers to BAM8-OX-1 (YFP fusion).
Multiple Sequence Alignment
Protein sequences of BZR1-domain proteins and β-amylase–like proteins were trimmed manually to the conserved core sequences, aligned using the web-based tool Multalin (Corpet, 1988), and displayed using Jalview (Clamp et al., 2004).
Subcellular Localization Analysis
Protoplasts from stable transgenic Arabidopsis plants or from the wild type (for transient expression experiments with constructs encoding GFP- and YFP-tagged proteins, above) were prepared as described (Yoo et al., 2007). 4′,6-diamidino-2-phenylindole staining was used to visualize nuclei. For localization in Nicotiana benthamiana, BAM8 plasmids were transformed into A. tumefaciens LBA 4404 and infiltrated into leaves as described (Sparkes et al., 2006). Tissues were imaged using a Nikon TE 2000-E inverted microscope (Nikon) fitted with a Chroma ET-GFP filter set or a Zeiss Axioplan2 fluorescence microscope (Carl Zeiss, Germany).
Biochemical Analysis
Rabbit polyclonal antisera were raised against the full-length BAM7 and BAM8 proteins (Eurogentec, Seraing, Belgium). Nuclear proteins were enriched as described (Kinkema et al., 2000). SDS-PAGE and immunoblot analyses were performed using standard protocols (Chia et al., 2004).
Modeling of Arabidopsis BAM8 Protein Structure
MODELER (Eswar et al., 2006) was used to model the structure of the BAM8 β-amylase–like domain using the structure of soybean β-amylase Gm BMY1 as a template (1Q6C.pdb). The sequences of Gm BMY1 and At BAM8 β-amylase domains share 41% sequence identity. Five models were built and the structure with the lowest DOPE score was chosen for further analysis. No restraint violations were reported near the active site of the models. Least squared superpositions of the template and model structures, and figures, were made using PyMol (DeLano, 2002).
Production of Recombinant Proteins
Full-length BAM7 and BAM8 coding sequences and the sequences encoding the putative catalytic domains (amino acids 251–691 for BAM7 and 258–689 for BAM8) were cloned into pET29a(+) (Novagen, Merck KGaA, Darmstadt, Germany). Sequences encoding the BZR1 domains (amino acids 65–229 and 82–245 for BAM7 and BAM8, respectively) were introduced into pET21a(+) (Novagen). BAM1 and BAM3 lacking their predicted transit peptides (amino acids 1–41 and 1–85, respectively) were cloned into pET29a(+). pET21a(+) and pET29a(+) constructs were expressed in Rosetta(DE3)pLysS (Novagen) and BL21Codon-Plus (DE3)-RIL (Stratagene, Amsterdam, Netherlands), respectively. Soluble proteins were extracted and affinity-purified using the ProBond purification system (Invitrogen, Basel, Switzerland).
β-Amylase Activity Assays
β-Amylase activity of recombinant proteins [in 100 mM 3-(N-morpholino)-propanesulfonic acid, pH 7.2, 1 mM dithiothreitol (DTT), 1 mM EDTA, 10 mg/mL BSA, and 10% (v/v) ethanediol] was determined using the Betamyl assay kit according to the manufacturer’s instructions (Megazyme, Bray, UK). Alternatively, maltose release from glucan substrates by recombinant proteins was assayed. Purified recombinant proteins were incubated with 10 mg/mL amylopectin or 5 mg/mL G7 in 50 mM Na-acetate, pH 5.6, 5 mM EDTA, and 5 mM DTT at 30°C for 30 min. The reaction was stopped by boiling the samples for 10 min. Released maltose was quantified as described under “quantification of soluble sugars.”
Quantification of Starch and Soluble Sugars
Entire rosettes of nonflowering plants were harvested, weighed, and immediately frozen in liquid nitrogen. Extraction of starch and soluble sugars was done as described (Fulton et al., 2008). Quantification of starch from the insoluble fraction was done by the determination of the total amount of Glc released after complete digestion with α-amylase and amyloglucosidase, as described previously (Smith and Zeeman, 2006). Maltose, Suc, Fru, and Glc in the soluble fraction were determined using high-performance anion-exchange chromatography and detected with pulsed amperometric detection. Samples were prepared as described (Chia et al., 2004). Sugars were separated on a Dionex PA-20 column according to the following conditions: Eluent A, 100 mM NaOH; eluant B, 150 mM NaOH and 500 mM sodium acetate. The gradient was as follows: 0 to 15 min, 100% A; 15 to 26 min, a concave gradient to 20% A and 80% B (mono- and disaccharide elution); 26 to 32 min, kept at 20% A and 80% B (column wash step); 32 to 40 min, step to 100% A (column re-equilibration). Peaks were identified by coelution with known sugar standards. Peak areas were determined using Chromeleon software.
RBSS
Double stranded oligo-nucleotides containing a core sequence of 15 random nucleotides were synthesized: TGGAGAAGAGGAGAGTGGGCNNNNNNNNNNNNNNNCTCTTTTGCATTCTTCTTCGATTCCGGG. Recombinant BZR1-domains of BAM7 and BAM8 were immobilized on Ni-Sepharose Fast Flow (GE Healthcare, Glattbrugg, Switzerland). Three hundred nanograms of purified double-stranded random oligonucleotides were incubated with the protein charged resin, or uncharged resin as a control, in DNA binding buffer (20 mM Tris-HCl, pH 8.0, 50 mM KCl, 0.5 mM EDTA, 1 mM DTT, 20 μg/mL BSA, 2 μg/mL poly[(dI)-(dC)], and 10% glycerol) for 20 min at 22°C. The resin was washed seven times with 1 mL of DNA binding buffer to remove unbound DNA. After the last wash, 10 μL of H2O was added and the resin was then boiled for 10 min and centrifuged at 18,000g for 5 min. Three microliters of the supernatant was PCR-amplified using the primers TGGAGAAGAGGAGAGTGGGC and CCCGGAATCGAAGAAGAATGCAAAAGAG. PCR products were gel-purified using the MinElute Kit (QIAGEN, Basel, Switzerland). Up to 150 ng of eluted DNA was used for the following round of RBSS. After seven rounds, the gel eluates were subjected to automated sequencing using a Solexa Genome Analyzer (Fasteris, Geneva, Switzerland). A consensus sequence from 1,072,225 sequence reads was calculated using Bioprospector (Liu et al., 2001) and MEME (Bailey et al., 2006). Weblogo (Crooks et al., 2004) was used to display the consensus motif from an alignment of 3600 randomly chosen sequences. Multalin (Corpet, 1988) was used to calculate the alignment.
EMSA
The EMSA was performed using the digoxigenin Gel Shift Kit, 2nd generation (Roche, Rotkreuz, Switzerland) according to the manufacturer’s instructions. One hundred fifty nanograms of recombinant BZR1-domain or 500 ng recombinant full-length BAM7 or BAM8 protein and 30 fmol of digoxigenin-labeled oligonucleotides were used per assay.
Protoplast Transactivation Assay
A pUC18 vector carrying the LUC gene under the control of the CaMV minimal 35S promoter (min35S) was kindly provided by Dr. Bruno Müller (University of Zürich, Switzerland). One or three copies of a BBRE-containing sequence (5′-GTCTGTTTCTACACGTGTGATCGATTTATTTT-3′) or three copies of an mBBRE containing sequence (5′-GTCTGTTTCTACACTTGTGATCGATTTATTTT-3′) were introduced upstream of the min35S, yielding the reporter plasmids 1xBBRE, 3xBBRE, and 3xmBBRE. The Ubq10:GUS:nosT (Yoo et al., 2007) served as the transfection control plasmid. Effector plasmids were pEarleyGate201-BAM7 and pEarleyGate201-BAM8. In control experiments, the effector plasmid was replaced by sheared salmon testis DNA (Sigma-Aldrich). Effector, reporter, and transfection control plasmids were mixed at a ratio of 5:4:1. Three replicate protoplast transformations were performed for each effector-reporter pair. Protoplasts were incubated in WI solution supplemented with 15 mM Suc. After 36 h, protoplasts were sedimented at 100g for 2 min and analyzed as describe previously (Yoo et al., 2007). Triplicated fluorescence and luminescence measurements were performed using an Infinite® M1000 microtiter plate reader (Tecan Trading AG, Männedorf, Switzerland). Three replicate transformations were performed for each plasmid combination, and the entire experiment repeated at least three times.
Transcriptional Analysis
Two-week-old soil-grown Arabidopsis seedlings were harvested 4 h into the light period and 4 h into the dark period. Three experimental replicates were grown for the microarray analyses. Sample preparation and transcript analysis using ATH1 GeneChips was done as described (Stettler et al., 2009). The full data described in the results are given as Supplemental Data Sets 1 to 6 online. The expression of 17 genes that showed deregulation in our microarrays was quantified independently by quantitative RT-PCR. For quantitative PCR, Fast SYBR Green Master Mix (Applied Biosystems, Rotkreuz, Switzerland) was used on a 7500 Fast Real-Time PCR system (Applied Biosystems). PP2A (At1g13320) was used as a reference gene (see Supplemental Tables 5 and 6 online).
Bioinformatic Analysis
We identified genes that showed genotype-specific deregulation by analysis of variance. We performed pairwise comparisons between all genotypes using the two-group analysis application from the R-server (http://fgcz-bfabric.uzh.ch/b-fabric/), selecting all genes that showed detectable expression with a signal greater than 25 for at least one of the 12 arrays (arrays from each time point were analyzed separately). We then selected the genes showing a statistically significant change between two genotypes (t test, P < 0.1) and determined the fold-changes as a log2 ratio between two genotypes. Genespring software (Agilent Technologies, Inc.) was used to display hierarchical clustering.
For the promoter analysis, the 500 probes showing either the greatest increase analysis of variance or the greatest reduction in expression between two genotypes were selected and the 1000 bp and 500 bp upstream sequences of the corresponding genes were retrieved from the TAIR database. The most over-represented eight-letter motif was identified by the oligo-analysis tool from Regulatory Sequence Analysis Tools (http://rsat.ulb.ac.be/rsat/; van Helden et al., 1998) with Arabidopsis as the background model.
Based on the total number of expressed genes (“number present”), we calculated the percentage of genes that showed a statistically significant change between pairs of genotypes. We also calculated the percentages of genes showing an up- or downregulation of at least two-, four-, or eightfold (i.e., a logarithmic fold-change of at least 1-, 2-, and 3 or -1, -2, and -3, respectively). For a given list of genes (e.g., BL-induced genes, or genes with the BBRE in their promoters), we determined first the total “number present” (i.e., those that were expressed in our microarrays), and second, the number from each list that we observed as deregulated between genotypes, according to the fold-change categories above. For each gene list, we then calculated the number that would be expected by chance in each fold-change category (e.g., the number of genes carrying a BBRE in their 500-bp promoter region upregulated by twofold or more between BAM8-OX and bam7bam8) by multiplying the number of present genes in the list by the percentage of total present genes in that fold-change category (207 × 2.4% in this example, giving an expected value of approximately five genes; see Supplemental Table 1 online). This expected number (Exp) was then compared with the observed number present (37 in this example) to give a “Present/Exp” value (i.e., 7.45 times more genes were twofold or more upregulated in BAM8-OX relative to bam7bam8 than would be expected by chance). Thus, the higher the value, the greater the over-representation of the genes in the list for a given fold change category.
MAPMAN software (Thimm et al., 2004) was used to display changes in expression between two genotypes.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: BAM7, At2g45880; BAM8, At5g45300; BZR1, At1g75080; UGT85A2, At1g22360; NAC102, At5g63790; uncharacterized gene, At3g05500; HB4, At2g44910; MYB56, At5g17800; uncharacterized gene, At5g12110; cycp3.1, At2g45080; uncharacterized gene, At5g22580; BEH2, At4g36780; WES1, At4g27260; uncharacterized gene, At1g62660; uncharacterized gene, At5g62210; DFR, At5g42800; F3H, At3g51240; TT5, At3g55120; uncharacterized gene, At5g05270; PAL1, At2g37040; and PP2A, At1g13320.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. The Two-Domain Structure of BAM7 and BAM8 Is Conserved in Higher Plants.
Supplemental Figure 2. BAM7 and BAM8 Localize to the Nucleus.
Supplemental Figure 3. Molecular Modeling of the BAM Domain of the BAM8 Protein.
Supplemental Figure 4. Phenotypes of BAM7 and BAM8 Knockout and Overexpressing Lines.
Supplemental Figure 5. Starch and Soluble Sugar Levels in BZR1-BAM Null Mutants and Overexpression Lines.
Supplemental Figure 6. Expression of BL-Responsive Genes in the Dark Microarray Experiment.
Supplemental Figure 7. MAPMAN Display of Deregulated Genes Involved in Metabolism.
Supplemental Figure 8. Relative Expression in BAM8-OX versus bam7bam8 at 4 h into the Light and 4 h into the Dark Period.
Supplemental Table 1. Regulation of BBRE Genes upon Deregulation of BZR1-BAMs in the Dark.
Supplemental Table 2. Occurrence of the BBRE in Promoters of Deregulated Genes.
Supplemental Table 3. Specific Changes of BL-Responsive Genes upon Deregulation of BZR1-BAMs.
Supplemental Table 4. Regulation of BBRE Genes upon Deregulation of BZR1-BAMs in the Light.
Supplemental Table 5. Relative Expression of Selected Genes Determined by Microarray and Quantitative PCR.
Supplemental Table 6. Primers Used for Quantitative RT-PCR.
Supplementary Data Set 1. Analysis of Variance of Transcript Levels in 14-d-Old Seedlings of the Wild Type, bam8, bam7bam8, and BAM8-OX Harvested 4 h into the Dark Period, Determined by Affymetrix ATH1 Genechips.
Supplementary Data Set 2. Pairwise Comparisons in Transcript Levels in 14-d-Old Seedlings of the Wild Type, bam8, bam7bam8, and BAM8-OX Harvested 4 h into the Dark Period, Determined by Affymetrix ATH1 Genechips.
Supplementary Data Set 3. Genes with Altered Transcript Levels and Expression of BR-Responsive Genes in Seedlings of the Wild Type, bam8, bam7bam8, and BAM8-OX Harvested 4 h into the Dark Period.
Supplementary Data Set 4. Analysis of Variance of Transcript Levels in 14-d-Old Seedlings of the Wild Type, bam8, bam7bam8, and BAM8-OX Harvested 4 h into the Light Period, Determined by Affymetrix ATH1 Genechips.
Supplementary Data Set 5. Pairwise Comparisons in Transcript Levels in 14-d-Old Seedlings of the Wild Type, bam8, bam7bam8, and BAM8-OX Harvested 4 h into the Light Period, Determined by Affymetrix ATH1 Genechips.
Supplementary Data Set 6. Genes with Altered Transcript Levels and Expression of BR-Responsive Genes in Seedlings of the Wild Type, bam8, bam7bam8, and BAM8-OX Harvested 4 h into the Light Period.
Acknowledgments
We thank Catherine Aquino and Stefan Zoller for help with microarrays, which were performed at the Functional Genomics Centre Zurich, Bruno Müller for providing protoplast transactivation vectors, Christoph Ringli and Cris Kuhlemeier for valuable advice and helpful comments on the manuscript, Alex Bannigan for assistance with microscopy, Ernst Aichinger and Simona Eicke for technical help, Matthias Hirsch-Hoffmann for bioinformatic help, and Fasteris SA (Geneva, Switzerland) for advice in sequencing. This work was funded partly by the Swiss National Science Foundation (National Centre for Competence in Research–Plant Survival), by the SystemsX.ch initiative (Plant Growth in a Changing Environment), partly by ETH Zurich, and partly by Grant J-881 from the Thomas F. Jeffress and Kate Miller Jeffress Memorial Trust (to J.D.M.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- Baena-González E., Rolland F., Thevelein J.M., Sheen J. (2007). A central integrator of transcription networks in plant stress and energy signalling. Nature 448: 938–942 [DOI] [PubMed] [Google Scholar]
- Bailey T.L., Williams N., Misleh C., Li W.W. (2006). MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 34 (Web Server issue): W369-W373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bläsing O.E., Gibon Y., Günther M., Höhne M., Morcuende R., Osuna D., Thimm O., Usadel B., Scheible W.R., Stitt M. (2005). Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. Plant Cell 17: 3257–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia T., Thorneycroft D., Chapple A., Messerli G., Chen J., Zeeman S.C., Smith S.M., Smith A.M. (2004). A cytosolic glucosyltransferase is required for conversion of starch to sucrose in Arabidopsis leaves at night. Plant J. 37: 853–863 [DOI] [PubMed] [Google Scholar]
- Cho Y.H., Yoo S.D., Sheen J. (2006). Regulatory functions of nuclear hexokinase1 complex in glucose signaling. Cell 127: 579–589 [DOI] [PubMed] [Google Scholar]
- Chory J., Nagpal P., Peto C.A. (1991). Phenotypic and genetic analysis of det2, a new mutant that affects light-regulated seedling development in Arabidopsis. Plant Cell 3: 445–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung M.H., Chen M.K., Pan S.M. (2000). Floral spray transformation can efficiently generate Arabidopsis transgenic plants. Transgenic Res. 9: 471–476 [DOI] [PubMed] [Google Scholar]
- Clamp M., Cuff J., Searle S.M., Barton G.J. (2004). The Jalview Java alignment editor. Bioinformatics 20: 426–427 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Clouse S.D., Langford M., McMorris T.C. (1996). A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 111: 671–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet F. (1988). Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16: 10881–10890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. (2004). WebLogo: A sequence logo generator. Genome Res. 14: 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano W.L. (2002). The PyMOL Molecular Graphics System. (San Carlos, CA: DeLano Scientific; ) [Google Scholar]
- Earley K.W., Haag J.R., Pontes O., Opper K., Juehne T., Song K.M., Pikaard C.S. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Eswar N., Webb B., Marti-Renom M.A., Madhusudhan M.S., Eramian D., Shen M.-Y., Pieper U., Sali A. (2006). Comparative protein structure modeling using Modeller. Curr. Protoc. Bioinformatics Chapter 5. Unit 5: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton D.C., et al. (2008). Beta-AMYLASE4, a noncatalytic protein required for starch breakdown, acts upstream of three active beta-amylases in Arabidopsis chloroplasts. Plant Cell 20: 1040–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H., Sawa S., Asami T., Fujioka S., Shimada Y., Yoshida S. (2004). Comprehensive comparison of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis. Plant Physiol. 134: 1555–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H., Shimada Y., Asami T., Fujioka S., Yoshida S. (2002). Microarray analysis of brassinosteroid-regulated genes in Arabidopsis. Plant Physiol. 130: 1319–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J.X., Gendron J.M., Sun Y., Gampala S.S., Gendron N., Sun C.Q., Wang Z.Y. (2005). BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307: 1634–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kauschmann A., Jessop A., Koncz C., Szekeres M., Willmitzer L., Altmann T. (1996). Genetic evidence for an essential role of brassinosteroids in plant development. Plant J. 9: 701–713 [Google Scholar]
- Kim T.W., Guan S., Sun Y., Deng Z., Tang W., Shang J.X., Sun Y., Burlingame A.L., Wang Z.Y. (2009). Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat. Cell Biol. 11: 1254–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinkema M., Fan W., Dong X. (2000). Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell 12: 2339–2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T., Caño-Delgado A., Seto H., Hiranuma S., Fujioka S., Yoshida S., Chory J. (2005). Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature 433: 167–171 [DOI] [PubMed] [Google Scholar]
- Koch K.E. (1996). Carbohydrate-modulated gene expression in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47: 509–540 [DOI] [PubMed] [Google Scholar]
- Li J., Chory J. (1997). A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90: 929–938 [DOI] [PubMed] [Google Scholar]
- Li L., Yu X., Thompson A., Guo M., Yoshida S., Asami T., Chory J., Yin Y. (2009). Arabidopsis MYB30 is a direct target of BES1 and cooperates with BES1 to regulate brassinosteroid-induced gene expression. Plant J. 58: 275–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Brutlag D.L., Liu J.S. (2001). BioProspector: Discovering conserved DNA motifs in upstream regulatory regions of co-expressed genes. Pac. Symp. Biocomput. 2001: 127–138 [PubMed] [Google Scholar]
- Moore B., Zhou L., Rolland F., Hall Q., Cheng W.H., Liu Y.X., Hwang I., Jones T., Sheen J. (2003). Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 300: 332–336 [DOI] [PubMed] [Google Scholar]
- Mouchel C.F., Osmont K.S., Hardtke C.S. (2006). BRX mediates feedback between brassinosteroid levels and auxin signalling in root growth. Nature 443: 458–461 [DOI] [PubMed] [Google Scholar]
- Nakamura A., Higuchi K., Goda H., Fujiwara M.T., Sawa S., Koshiba T., Shimada Y., Yoshida S. (2003). Brassinolide induces IAA5, IAA19, and DR5, a synthetic auxin response element in Arabidopsis, implying a cross talk point of brassinosteroid and auxin signaling. Plant Physiol. 133: 1843–1853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser J.L., Mockler T.C., Chory J. (2004). Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol. 2: E258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niittylä T., Messerli G., Trevisan M., Chen J., Smith A.M., Zeeman S.C. (2004). A previously unknown maltose transporter essential for starch degradation in leaves. Science 303: 87–89 [DOI] [PubMed] [Google Scholar]
- Ringli C., Bigler L., Kuhn B.M., Leiber R.M., Diet A., Santelia D., Frey B., Pollmann S., Klein M. (2008). The modified flavonol glycosylation profile in the Arabidopsis rol1 mutants results in alterations in plant growth and cell shape formation. Plant Cell 20: 1470–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland F., Baena-Gonzalez E., Sheen J. (2006). Sugar sensing and signaling in plants: Conserved and novel mechanisms. Annu. Rev. Plant Biol. 57: 675–709 [DOI] [PubMed] [Google Scholar]
- Rook F., Corke F., Card R., Munz G., Smith C., Bevan M.W. (2001). Impaired sucrose-induction mutants reveal the modulation of sugar-induced starch biosynthetic gene expression by abscisic acid signalling. Plant J. 26: 421–433 [DOI] [PubMed] [Google Scholar]
- Smith A.M., Stitt M. (2007). Coordination of carbon supply and plant growth. Plant Cell Environ. 30: 1126–1149 [DOI] [PubMed] [Google Scholar]
- Smith A.M., Zeeman S.C. (2006). Quantification of starch in plant tissues. Nat. Protoc. 1: 1342–1345 [DOI] [PubMed] [Google Scholar]
- Sparkes I.A., Runions J., Kearns A., Hawes C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1: 2019–2025 [DOI] [PubMed] [Google Scholar]
- Stettler M., Eicke S., Mettler T., Messerli G., Hörtensteiner S., Zeeman S.C. (2009). Blocking the metabolism of starch breakdown products in Arabidopsis leaves triggers chloroplast degradation. Mol. Plant 2: 1233–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O., Bläsing O., Gibon Y., Nagel A., Meyer S., Krüger P., Selbig J., Müller L.A., Rhee S.Y., Stitt M. (2004). MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Toledo-Ortiz G., Huq E., Quail P.H. (2003). The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15: 1749–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Helden J., André B., Collado-Vides J. (1998). Extracting regulatory sites from the upstream region of yeast genes by computational analysis of oligonucleotide frequencies. J. Mol. Biol. 281: 827–842 [DOI] [PubMed] [Google Scholar]
- Vert G., Chory J. (2006). Downstream nuclear events in brassinosteroid signalling. Nature 441: 96–100 [DOI] [PubMed] [Google Scholar]
- Wang Z.Y., Nakano T., Gendron J., He J., Chen M., Vafeados D., Yang Y., Fujioka S., Yoshida S., Asami T., Chory J. (2002). Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2: 505–513 [DOI] [PubMed] [Google Scholar]
- Xu W., Purugganan M.M., Polisensky D.H., Antosiewicz D.M., Fry S.C., Braam J. (1995). Arabidopsis TCH4, regulated by hormones and the environment, encodes a xyloglucan endotransglycosylase. Plant Cell 7: 1555–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Vafeados D., Tao Y., Yoshida S., Asami T., Chory J. (2005). A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 120: 249–259 [DOI] [PubMed] [Google Scholar]
- Yin Y., Wang Z.Y., Mora-Garcia S., Li J., Yoshida S., Asami T., Chory J. (2002). BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109: 181–191 [DOI] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Yu X., Li L., Zola J., Aluru M., Ye H., Foudree A., Guo H., Anderson S., Aluru S., Liu P., Rodermel S., Yin Y. (2011). A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J. 65: 634–646 [DOI] [PubMed] [Google Scholar]
- Zeeman S.C., Smith S.M., Smith A.M. (2007). The diurnal metabolism of leaf starch. Biochem. J. 401: 13–28 [DOI] [PubMed] [Google Scholar]