This study examines the role of the PHYTOCHROME INTERACTING FACTOR3 homolog SPATULA (SPT) in the control of the developing seedling and shows that SPT is a potent regulator of cotyledon size, acting in parallel to DELLAs. As DELLAs negatively regulate SPT abundance, the light regulation of DELLAs drives the DELLA-SPT counterbalance, enforcing growth restraint across a range of ambient light conditions that are prevalent in nature.
Abstract
The period following seedling emergence is a particularly vulnerable stage in the plant life cycle. In Arabidopsis thaliana, the phytochrome-interacting factor (PIF) subgroup of basic-helix-loop-helix transcription factors has a pivotal role in regulating growth during this early phase, integrating environmental and hormonal signals. We previously showed that SPATULA (SPT), a PIF homolog, regulates seed dormancy. In this article, we establish that unlike PIFs, which mainly promote hypocotyl elongation, SPT is a potent regulator of cotyledon expansion. Here, SPT acts in an analogous manner to the gibberellin-dependent DELLAs, REPRESSOR OF GA1-3 and GIBBERELLIC ACID INSENSITIVE, which restrain cotyledon expansion alongside SPT. However, although DELLAs are not required for SPT action, we demonstrate that SPT is subject to negative regulation by DELLAs. Cross-regulation of SPT by DELLAs ensures that SPT protein levels are limited when DELLAs are abundant but rise following DELLA depletion. This regulation provides a means to prevent excessive growth suppression that would result from the dual activity of SPT and DELLAs, yet maintain growth restraint under DELLA-depleted conditions. We present evidence that SPT and DELLAs regulate common gene targets and illustrate that the balance of SPT and DELLA action depends on light quality signals in the natural environment.
INTRODUCTION
To complete a successful life cycle, growth of newly germinated seedlings needs to be optimized in accordance with the immediate surroundings and prevailing season. To a large extent, this is achieved by channeling information from the environment to the growth-regulating hormonal pathways. During the early stages of the plant life cycle, it is essential that photosynthetic competence is achieved to sustain photoautotrophic growth. For this reason, light has a major influence on seedling development, shaping plant architecture and plant organ growth, leading to a fine-tuning of the plant’s organ growth.
A subfamily of basic-helix-loop-helix (bHLH) transcription factors, which includes the phytochrome-interacting factors (PIFs), are central integrators of environmental and hormone signals during seedling development and represent a principal mechanism through which seedling growth is controlled (Toledo-Ortiz et al., 2003; Duek and Fankhauser, 2005). When seedlings are grown in the dark, they follow a skotomorphogenic program of development, where rapid elongation of the hypocotyl and folding of the underdeveloped cotyledons allow for fast growth while the plant is seeking a light source. At the molecular level, the dark developmental program is driven by PIFs because mutants null for multiple pifs lose their ability to grow rapidly in darkness and instead adopt a photomorphogenic-like program of development (Leivar et al., 2008, 2009; Shin et al., 2009).
Several PIFs have been shown to be targeted for degradation by the phytochrome photoreceptors. Upon photoactivation, the pigment-bearing phytochromes are activated and rapidly migrate into the nucleus, where they interact with PIFs through their AP domains (Ni et al., 1998, 1999; Kircher et al., 1999, 2002; Martínez-García et al., 2000; Huq and Quail, 2002; Huq et al., 2003; Khanna et al., 2004; Oh et al., 2004). This physical association results in PIF phosphorylation and degradation by the proteasome machinery (Bauer et al., 2004; Shen et al., 2005, 2008; Al-Sady et al., 2006; Oh et al., 2006; Lorrain et al., 2008). Nozue et al. (2007) demonstrated that PIF4 and PIF5 have a pivotal role in integrating light and clock signaling to drive daily growth rhythms. In seedlings grown under diurnal conditions, PIF4 and PIF5 transcript levels and protein accumulate during the dark period, promoting growth at the end of the night. Light-triggered destabilization of PIF4 and PIF5 at dawn results in growth cessation during the early morning (Nozue et al., 2007). The strong coupling between PIFs and phytochrome also provides a mechanism to trigger elongation growth under vegetation shade conditions where active phytochrome B (phyB) Pfr levels are depleted (Lorrain et al., 2009).
Interestingly, recent reports have shown that PIFs impose reciprocal control on phyB because a reduction in PIF levels has been shown to promote phyB accumulation over time (Monte et al., 2007; Al-Sady et al., 2008; Leivar et al., 2008). Under continuous red light irradiation, pif knockout mutants present enhanced de-etiolation phenotypes, characterized by shorter hypocotyls, and in some conditions, larger cotyledons (Ni et al., 1998; Huq and Quail, 2002; Leivar et al., 2008). Thus, in constant light, PIFs appear to act at least partly by modulating phyB levels to promote hypocotyl cell expansion and, to a lesser extent, repress cotyledon growth.
During early seedling development, the plant hormones are important internal regulators of growth and cell expansion. Gibberellins (GAs), the tetracyclic diterpenoid plant hormones, are known to promote growth by destabilizing growth suppressors in the DELLA subfamily of GRAS transcriptional regulators (Silverstone et al., 1997, 1998, 2001; Pysh et al., 1999; Dill and Sun, 2001; Harberd, 2003; Feng et al., 2008; Achard and Genschik, 2009). Here, GA binds to the GIBBERELLIN INSENSITIVE DWARF1 (GID1) receptors to form GID1–GA complexes that then bind to the DELLA growth repressors (Dill et al., 2001; Griffiths et al., 2006; Nakajima et al., 2006; Willige et al., 2007; Ariizumi et al., 2008; Murase et al., 2008; Shimada et al., 2008). This interaction enhances DELLA recognition by the F box SLY1/GID2 subunit of the E3 ligase SCFSLY1/GID2 complex, which promotes its subsequent degradation by the 26S proteasome (Dill et al., 2004).
In a broader context, DELLAs are well known for their prominent role in the green revolution spearheaded by Norman Borlaug that allowed food production to keep pace with worldwide population growth (Salamini, 2003; Swaminathan, 2009). Dominant DELLA gene mutations, selected by traditional breeding, led to the production of improved, higher yield dwarf crop varieties (Sun and Gubler, 2004; Fukao and Bailey-Serres, 2008; Gao et al., 2008). Therefore, in our recent history, DELLAs have played an important role in food security.
Recent studies have suggested that the regulation of growth by PIFs and by DELLAs is integrated. DELLAs have been shown to directly interact with PIF3 and PIF4 in vivo, inhibiting their transcriptional activity (de Lucas et al., 2008; Feng et al., 2008). These findings may be linked with the observation that phyB promotes the gradual accumulation of DELLAs in hypocotyl tissue following exposure to light (Achard et al., 2007). Here, phyB modulation of GA metabolic gene expression is proposed to suppress GA production and boost DELLA levels (Achard et al., 2007; Alabadí and Blázquez, 2009). DELLAs are able to inhibit PIF3 and PIF4 activity (de Lucas et al., 2008; Feng et al., 2008); this may represent an additional route for phyB to modulate PIF activity and therefore seedling growth.
The focus of studies on the seedling hypocotyl means that we currently have large deficiencies in our understanding of how cotyledon growth is regulated. Because seed leaves are the major photosynthesizing organs in the immature seedling, their growth control is vital to the success of the young plant. To dissect the molecular control of this process, we focused our activity on SPATULA (SPT), a gene in the PIF class of bHLH transcription factors that we had shown previously to regulate cotyledon size under red light conditions (Penfield et al., 2005). Although SPT shares a high similarity with PIF3 and PIF4 (Leivar and Quail, 2011), the full-length SPT protein is unable to bind phyB and lacks any active phytochrome binding motif (Khanna et al., 2004). Several members of the PIF family of transcription factors, including PIF3-LIKE 1 and LONG HYPOCOTYL IN FAR-RED, share this inability to bind to phytochrome (Khanna et al., 2004). Nevertheless, they participate in phytochrome signaling and are able do dimerize with other members of the PIF family (Toledo-Ortiz et al., 2003; Leivar and Quail, 2011). SPT belongs to this category of PIF-like transcription factors. Previous work demonstrated that SPT regulates seed germination in response to cold and light signaling, in part by manipulating GA3ox expression in the seed (Oh et al., 2004, 2006, 2007; Penfield et al., 2005). In the present study, we show that SPT is functionally distinct at the seedling stage, where it operates over a broad temperature range and independently of light to regulate cotyledon expansion. In seedlings, SPT restrains growth, in a manner similar to the unrelated DELLAs. Our study also demonstrates that the levels of SPT and DELLAs are tightly coordinated to prevent the detrimental effects on growth that would result from either an excess or a deficiency in these potent growth regulators.
RESULTS
SPT Is a Major Regulator of Cotyledon Expansion
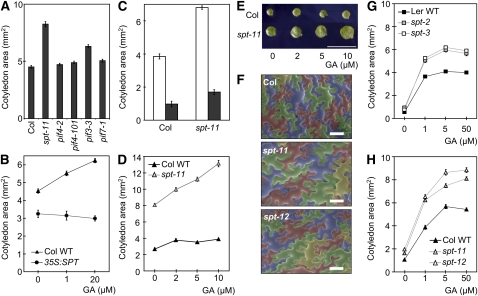
We have shown previously that spt mutants have a significantly expanded cotyledon phenotype under red light conditions (Penfield et al., 2005). We observed similarly enlarged cotyledons in an extended range of spt mutant alleles: spt-11 and spt-12, null alleles in the Columbia (Col) background (Ichihashi et al., 2010), as well as the previously described Landsberg erecta (Ler) alleles: spt-1, a weak allele; spt-2, carrying a point mutation in the bHLH domain; and spt-3, a strong knockdown allele (Heisler et al., 2001). Cotyledon expansion is severely impeded in 35S:SPT seedlings that have a 30-fold increase in SPT transcript levels (Figures 1A and 1B; see Supplemental Figures 1A and 1B online). Contrasting with this, pif3-3 has a relatively subtle impact, whereas pif4-101 and pif7-1 have no detectable impact on cotyledon size under conditions where we observe strong, short hypocotyl phenotypes for all these mutant alleles (Figure 1A; see Supplemental Figure 1C online). These data indicate that SPT has a more prominent role in suppressing cotyledon expansion than the gene paralogs PIF3, PIF4, and PIF7, which mainly regulate hypocotyl growth. We also noted that the spt phenotype was observed at 15 to 25°C, suggesting that in seedlings, SPT operates over a broad ambient temperature range to regulate this response (see Supplemental Figure 1D online).
Figure 1.
spt Mutants Display Altered Cotyledon Expansion in Response to GA.
(A) Cotyledon area of 12-d-old Col (wild-type), spt-11, pif4-2, pif4-101, pif3-3, and pif7-1 seedlings grown under red light at 20°C. Bars indicate se.
(B) Cotyledon area of 12-d-old Col (wild-type [WT]) and 35S:SPT seedlings in control conditions and in the presence of increasing concentrations of GA3, grown under red light, 20°C.
(C) Cotyledon area of 7-d-old Col (wild-type) and spt-11 seedlings grown under red light at 20°C with (black column) or without (white column) 0.2 μM PAC. Bars = se.
(D) Cotyledon area of 7-d-old Col (wild-type) and spt-11 seedlings grown under red light at 20°C in the presence of increasing concentrations of GA3. Bars = se.
(E) Cotyledon phenotype of the seedlings measured in (D). Bar = 10 mm.
(F) SEM false-color images of cotyledon pavement cells in fully expanded 11-d-old Col, spt-11, and spt-12 seedlings grown under red light on media containing 50 μM GA3 (without PAC). Bar = 100 μm.
(G and H) Cotyledon area of 8-d-old Ler, spt-2, and spt-3 (G) and Col, spt-11, and spt-12 (H) seedlings grown under red light at 20°C on 0.2 μM PAC-supplemented medium in the presence of increasing concentrations of GA3. Bars = se.
spt Mutant Alleles Enhance Sensitivity to GA
Our earlier work demonstrated that, during germination, SPT regulates GA biosynthesis genes. To investigate whether the spt-11 cotyledon phenotype was a consequence of enhanced GA levels, we first tested whether the spt phenotype could be eliminated by applying paclobutrazol (PAC), a GA biosynthesis inhibitor. PAC treatment strongly suppressed the large spt-11 cotyledon phenotype, suggesting that it may result from altered GA levels or GA responsiveness (Figure 1C). To distinguish between these possibilities, we assessed the impact of elevated or reduced SPT levels on gibberellic acid (GA3) mediated cotyledon expansion. If SPT simply altered GA levels, we would expect the GA saturation threshold in the already enlarged spt-11 to be lower than in the wild type and the 35S:SPT small cotyledon phenotype to be rescued with GA. However, spt-11 cotyledons continued to respond to GA3 at concentrations that were saturating for the wild type (Figures 1D and 1E). Conversely, cotyledon size was severely reduced in 35S:SPT seedlings that were insensitive to GA3 application (Figure 1B). These data suggest that SPT is unlikely to control cotyledon expansion by moderating GA levels. Rather, SPT appears to impose growth restraint even following GA-mediated destruction of the DELLA growth repressors. Therefore, SPT appears to counter the impact of GA on cotyledons because SPT depletion leads to an excessive expansion following GA application. Because GA levels have been shown to control ABA signaling (Zentella et al., 2007; Piskurewicz et al., 2008, 2009), it is also possible that the spt phenotype is influenced by ABA.
Using scanning electron microscopy (SEM), we were able to show that GA-treated spt-11 and spt-12 mutants had a higher proportion of large pavement cells when compared with the wild type (Figure 1F; see Supplemental Figure 2 online). Because GA is known to regulate cell size, this observation is consistent with our whole-organ data, which suggests that SPT antagonizes GA action in cotyledons (Achard et al., 2009; Ubeda-Tomás et al., 2009). Interestingly, in contrast to the Col lines, Ler wild-type cotyledons were completely unresponsive to GA3, whereas spt-2 and spt-3 were only moderately responsive to GA3, suggesting that the response to GA may be saturated in the Ler accession (see Supplemental Figure 3 online). This appeared to be the case, as when we reduced endogenous GA levels by applying PAC, we observed robust GA-induced cotyledon expansion in Ler and Col wild-type seedlings, whereas spt-2, spt-3 (Ler), and spt-11, spt-12 (Col) alleles exhibited altered GA sensitivity when compared with their respective wild types (Figures 1G and 1H; see Supplemental Figure 3 online). Therefore, we used PAC for our follow-up analysis using the Ler accession.
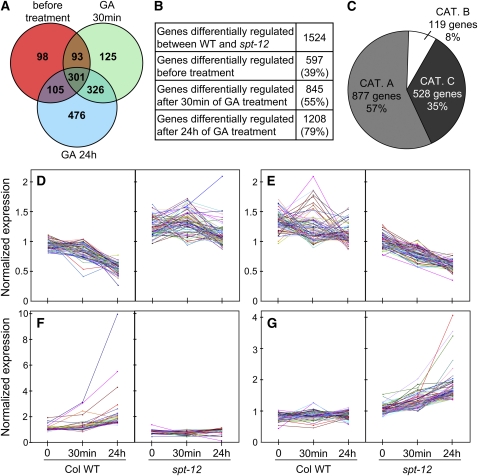
SPT Regulates Gene Subsets in a GA-Dependent and -Independent Manner
Given the strong GA-related spt cotyledon phenotype, we wanted to examine the spt mutant transcriptome to establish whether SPT activity was restricted to GA signaling and/or whether we could find evidence for altered GA responsiveness. Here, we performed triplicate microarray experiments in wild-type Col and spt-12 mutant seedlings in (GA−) controls and then 30 min and 24 h following GA3 application. First, we noted that the number of genes misexpressed ≥1.5-fold in spt-12 versus wild type rose from 597 to 845 and again to 1208, 30 min and 24 h postGA3 application. Therefore, exposure to GA3 for 24 h doubled the number of genes with altered expression in spt-12, strongly implicating SPT as a suppressor of GA-mediated expression in wild-type seedlings (Figures 2A and 2B; see Supplemental Data Set 1 online). Of the 1524 genes that registered a ≥1.5-fold change in transcript levels in spt-12 versus wild type across our experiment, 57% fell into category A: genes that were unaffected by GA (Figure 2C; see Supplemental Data Set 2 online). The substantial size of this category indicates that SPT can operate independently of GA. Eight percent of the total fell into category B: GA-controlled genes with comparable regulation in spt-12 and wild type (see Supplemental Data Set 3 online). This suggests that a small proportion of SPT-controlled genes are independently regulated by GA. The remaining 35%, category C, exhibited altered GA regulation in spt-12 versus wild type, representing GA-controlled genes that were influenced by SPT presence (see Supplemental Data Set 4 online). Within this gene set, four prominent coregulated groups emerged (Figures 2D to 2G; see Supplemental Data Set 5 online). Group 1 contains genes that were downregulated by GA in the wild type and invariant or upregulated in spt-12. Group 2 genes were unaffected by GA in the wild type and downregulated in response to GA in spt-12. Group 3 comprises genes that were GA upregulated in the wild type but repressed and GA unresponsive in spt-12, whereas group 4 genes have the opposite response: GA unresponsive in wild type and upregulated in spt-12. Thus, SPT can enhance or repress GA-mediated expression of specific gene subsets.
Figure 2.
Microarray Analysis of SPT-Regulated Genes.
(A) Venn diagram showing the distribution of the SPT-regulated genes, determined from microarray analysis of 4-d-old red light-grown Col and spt-12 untreated controls (0) and 30 min or 24 h following treatment with 50 mM GA3. SPT targets are defined as genes presenting at least a 1.5-fold change in mean expression between triplicate wild-type and spt-12 samples. The number of genes in each category is shown.
(B) Table summarizing the increasing number of genes scored as SPT regulated with GA3 treatment. WT, wild type.
(C) Pie chart representing the distribution of the SPT-regulated genes in terms of their GA regulation. Category A shows genes that are not regulated by GA3 treatment in our arrays. Category B shows genes that exhibit similar GA-dependent regulation in the wild type and spt-12. Category C shows genes that exhibit different GA regulation in the wild type and spt-12.
(D) to (G) Expression profiles of category C gene subsets that show synergistic regulation by SPT and GA.
(D) Genes that are downregulated by GA in the wild type, upregulated in spt-12 mutant, and not regulated or upregulated by GA in spt-12.
(E) Genes that are not GA regulated in the wild type but are downregulated by GA in spt-12.
(F) Genes upregulated by GA in the wild type but repressed and GA unresponsive in spt-12.
(G) Genes that are repressed and GA unresponsive in the wild type but GA responsive in the spt-12 mutant.
RGA and GAI Restrain Cotyledon Expansion
The altered responsiveness of spt mutants and 35S:SPT seedlings to GA suggested to us that SPT may compensate for GA-mediated DELLA depletion in wild-type seedlings. Therefore, we wanted to establish the principal DELLAs controlling cotyledon growth under our conditions. To do this, we analyzed mutants depleted in one, two, or multiple DELLAs in a ga1-3 background, which severely restricts GA biosynthesis (Figure 3A; Sun and Kamiya, 1994). The rga-t2 mutation relieved ga1-3-imposed repression, restoring cotyledon expansion to near wild-type levels. While gai-t6 was ineffective on its own, loss of both REPRESSOR OF GIBBERELLIC ACID INSENSITIVE3 (RGA) and GIBBERELLIC ACID INSENSITIVE (GAI) led to grossly expanded cotyledons, revealing a redundant role for RGA with GAI in the control of this response. Because ga1-3 gai-t6 rga-t2 cotyledons were almost as large as the ga1-3 della5 (rga-t2 gai-t6 rgl1-1 rgl2-1 rgl3-4) mutant, this identified RGA and GAI as the primary DELLAs regulating this process. These DELLA family members share the highest sequence homology, which may account for this functional overlap (Lee et al., 2002).
Figure 3.
RGA and GAI Regulate GA-Mediated Cotyledon Expansion, Cotyledon-Located RGA Is Depleted in the Light, and PIFs Are Modest Regulators of Cotyledon Expansion.
(A) Cotyledon area of 11-d-old Ler (wild-type [WT]), ga1-3, ga1-3 gai-t6, ga1-3 rga-t2, ga1-3 gait-6 rga-t2, and ga1-3 della5 (rga-t2 gai-t6 rgl1-1 rgl2-1 rgl3-4) seedlings grown under red light. Bars = se.
(B) and (C) GFP-RGA detection by confocal microscopy in 4-d-old seedlings expressing pRGA:GFP-RGA. Seedlings were grown ±0.2 μM PAC and exposed to 4 d of dark or 3 d of dark followed by 24 h of red light.
(D) Cotyledon area of 12-d-old seedlings (genotypes as indicated) grown under red light at 20°C and exposed to zero or increasing concentrations of GA3 (white, untreated; gray, 1 μM GA3; black, 20 μM GA3). Bars = se.
phyB Depletes DELLA Levels in Seedling Cotyledons
Previous studies using seedling hypocotyl tissue have shown that phyB promotes green fluorescent protein (GFP)-RGA accumulation in red light, while levels fall following a period of darkness (Achard et al., 2007). These data suggest that hypocotyl-located GFP-RGA operates during the daytime to suppress hypocotyl growth. The inhibitory effect of red light on hypocotyl elongation is well known; however, the opposite response is induced in seedling cotyledons, which expand following exposure to light (Franklin and Quail, 2010). Likewise, when kept in darkness, hypocotyl and cotyledon tissues undergo opposing growth responses, suggesting that DELLA proteins may be subject to differential regulation in these distinct tissues. Confocal imaging showed that in dark-grown seedlings, as expected, nuclear GFP-RGA was largely absent from hypocotyl cells, and as noted previously, nuclear GFP-RGA was detected with increasing frequency through the apical hook region (Figure 3B; see Supplemental Figure 4 online; Vriezen et al., 2004). However, we also detected high levels of GFP-RGA throughout the cotyledon (Figure 3B; Supplemental Figure 4 online). Exposure to red light led to a significant depletion in the pool of cotyledon-located GFP-RGA, whereas PAC application reversed this trend, boosting seedling GFP-RGA levels (Figure 3C). We also observed higher incidents of nuclear GFP-RGA in cotyledons of phyB-9 null and wild-type seedlings exposed to low red light supplemented with far-red (R + FR), which reduces active phyB Pfr levels (see Supplemental Figure 5 online). Therefore, it appears that phyB activity simultaneously boosts RGA levels in the hypocotyl to restrict growth and reduces RGA levels in the cotyledons, allowing their expansion. Because PAC treatment can overcome the ability of red light to suppress RGA accumulation, this suggests that phyB may moderate RGA levels by controlling GA levels in seedling cotyledons. This interpretation is consistent with a previous report that demonstrates that phyB controls hypocotyl-located RGA abundance, at least in part, by regulating the levels of bioactive GA (Achard et al., 2007). A similar observation has been detected in seeds, where red light treatment leads to a destabilization of RGA and GAI, in a process involving de novo GA biosynthesis (Oh et al., 2007). Interestingly, in contrast to DELLAs that are light regulated, SPT levels are not altered by shift from darkness to red light and remain relatively constant for several hours in the light (see Supplemental Figure 6 online).
DELLAs and PIFs Have Distinct Roles in the Cotyledon and Hypocotyl
In the regulation of seedling hypocotyl growth, DELLAs are thought to bind to PIFs through the bHLH DNA-recognition domain and block their transcriptional activity (de Lucas et al., 2008; Feng et al., 2008). Because the bHLH domain is highly conserved among the PIF subfamily (see Supplemental Figure 7 online), in vitro interactions between PIF family members and DELLAs are readily detected (Gallego-Bartolomé et al., 2010). We have also observed binding among RGA, GAI, and SPT in a yeast two-hybrid assay (see Supplemental Figure 8 online). However, the mode of action proposed for the hypocotyl relies on PIF and DELLA antagonism, which is incongruous with the comparable behavior of SPT, PIF3, and DELLAs in the cotyledon response (Figure 1; Ni et al., 1998; Huq and Quail, 2002; Leivar et al., 2008). To establish whether other PIFs operated in a qualitatively similar manner to SPT and PIF3 in our conditions, we examined the combined effects of spt-11 with pif3-3, pif4-101,or pif7-1 ± GA (Figure 3D). Here, the aim was to identify genetic interactions between pif and spt alleles or reveal redundant roles in the absence of the dominant growth suppressor SPT. Our data show that in the absence of GA, pif3-3 acts additively with spt-11 to control cotyledon size, whereas the impact of pif4-101 or pif7-1 is only observed in a spt-11 background. Therefore, like SPT and DELLAs, PIFs also suppress cotyledon expansion but have more minor roles, with PIF4 and PIF7 acting redundantly with SPT. This provides confirmation that in contrast to the opposing roles of PIFs and DELLAs in hypocotyls, when regulating cotyledon expansion, SPT, PIFs, and DELLAs have analogous roles. Interestingly, the GA responsiveness of the double mutants was unchanged when compared with the spt-11 monogenic mutant, indicating that SPT is the principal antagonist of GA-mediated cotyledon expansion (Figure 3D).
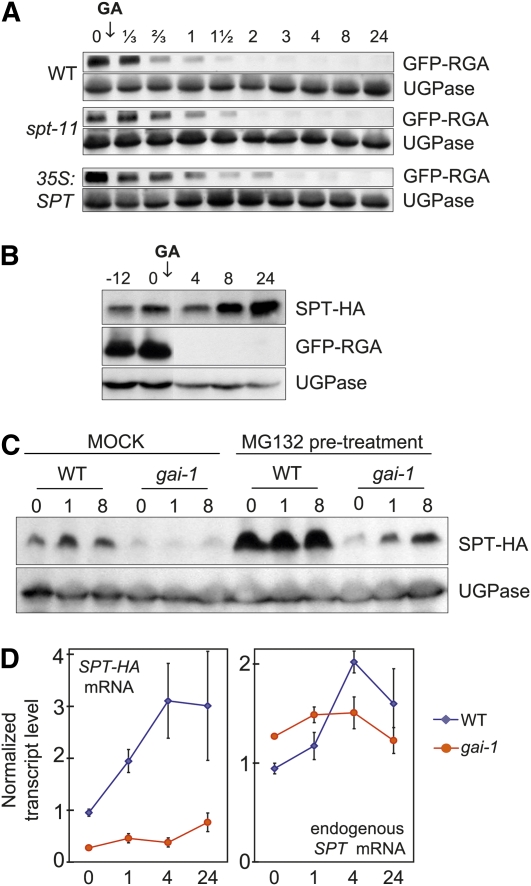
SPT Does Not Enhance DELLA Destruction
The hypersensitivity of spt mutants to GA provided the possibility that SPT operates by reducing GA-induced DELLA destruction. To test this, GFP-RGA levels were determined by immunoblot analysis in the spt-11 null, wild-type, and 35S:SPT seedlings (Figure 4A; see Supplemental Figure 9 online). Consistent with previous reports, we observed a gradual dose-dependent reduction in DELLA levels following GA3 application (see Supplemental Figure 9 online; Silverstone et al., 2001). RGA destruction kinetics were unaffected in the spt-11 mutant and only marginally delayed in 35S:SPT seedlings, indicating that the spt mutant phenotype does not result from enhanced DELLA proteolysis (Figure 4A). This finding is compatible with our microarray analysis because we found no evidence for SPT regulation of DELLAs and GA biosynthesis genes (see Supplemental Data Set 1 online).
Figure 4.
DELLAs Negatively Regulate SPT Protein Levels.
(A) GFP-RGA accumulation in 6-d-old red light-grown seedlings detected by immunoblot. Seedlings were harvested at the times shown (in hours), post-50 μM GA3 application. UGPase detection was used as a loading control. WT, wild type.
(B) SPT-HA and GFP-RGA accumulation detected by immunoblot before and following a treatment with GA3 (50 μM) of 3-d-old red light-grown seedlings expressing both recombinant proteins. Seedlings were grown on 0.2 μM PAC-containing medium and exposed to a period of 12 h of darkness before GA treatment. UGPase detection was used as a loading control.
(C) SPT-HA accumulation in 5-d-old red light-grown seedlings expressing 35S:SPT-HA in a wild-type or a gai-1 genetic background, detected by immunoblot. Harvest times (in hours) post-10 μM GA3 application, with or without MG132 pretreatment, are shown. UGPase detection was used as a loading control.
(D) SPT-HA and endogenous SPT mRNA levels in 5-d-old red light-grown seedlings expressing 35S:SPT-HA in a wild-type or a gai-1 genetic background, detected by real-time PCR. Values are given relative to the housekeeping gene ACT7. Harvest times (in hours) post-10 μM GA3 application are shown. Bars = se from three biological replicates.
[See online article for color version of this figure.]
DELLAs Suppress SPT Protein Accumulation
The altered sensitivity of spt mutants to GA treatment suggested to us that SPT abundance or activity could be regulated by DELLAs. Therefore, we tested whether SPT abundance was altered or remained the same when DELLA levels were manipulated, using 35S:SPT-HEMAGGLUTININ (HA) seedlings. To achieve a wide range of DELLA levels, we first applied PAC to boost DELLA abundance and then treated seedlings with GA3 to deplete DELLA levels (Figure 4B). As expected, GA3 application led to a reduction in RGA-GFP levels but a gradual rise in SPT-HA abundance. This inverse relationship between RGA-GFP and SPT-HA levels suggested that DELLAs, possibly RGA and/or GAI, may influence SPT protein accumulation. We next utilized gai-1, a semidominant allele that contains a 51-bp in-frame deletion in the DELLA region of GAI, producing a stable, constitutively active, and GA-insensitive form of GAI (Dill et al., 2001). Our immunoblot analysis established that gai-1 strongly represses SPT-HA protein accumulation in a GA-independent manner (Figure 4C). Application of the proteasome inhibitor MG132 enhanced SPT-HA levels in wild-type seedlings, indicating that SPT is subject to proteasome-mediated proteolysis. However, the impact of gai-1 was still evident in MG132-treated seedlings, where SPT-HA levels rose gradually over time. Because MG132 application led to a slow accumulation of SPT-HA, we reasoned that gai-1 is unlikely to regulate SPT through a proteasome-dependent mechanism.
Because the gai-1-dependent alteration in SPT protein levels was evident even though transcription was driven by the 35S promoter, this suggested that GAI may exert control on SPT posttranscriptionally. To explore this possibility, we examined transcript levels by quantitative PCR (qPCR) of both the SPT-HA transgene and the endogenous SPT mRNA in 35S:SPT-HA seedlings. Here, we detected a moderate but persistent GA-mediated rise in the SPT-HA transgene mRNA that was absent in gai-1 seedlings (Figure 4D). Qualitative observations were also made for the endogenous SPT transcript (Figure 4D). This implicates GAI in posttranscriptional regulation of SPT mRNA abundance. Because the impact of gai-1 on SPT-HA transgene transcript and protein abundance was comparable, we concluded that GAI may regulate SPT mainly by limiting transcript accumulation. Thus, GAI and probably other DELLAs appear to repress SPT transcript accumulation, whereas GA-activated DELLA depletion relieves this repression, leading to a rise in SPT transcript levels.
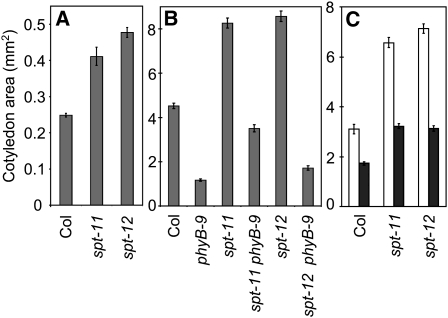
phyB-9 Attenuates the spt Cotyledon Phenotype
To establish whether we could obtain further evidence for DELLA cross-regulation of SPT, we exploited our finding that cotyledon-located DELLAs are phyB modulated. Because DELLA levels are elevated by phyB depletion, we would expect phyB-9 to reduce the impact of spt alleles. First, we established that the enlarged cotyledon phenotype was evident in spt-11 and spt-12 etiolated seedlings, suggesting that SPT operated independently of light and therefore phyB (Figure 5A). However, when grown under red light, phyB-9 dramatically reduced the effectiveness of spt-11 and spt-12 in promoting cotyledon expansion (Figure 5B). These results illustrate that the efficacy of spt alleles is influenced by phyB, a potent regulator of DELLA levels (see Supplemental Figure 5 online).
Figure 5.
spt Phenotype Severity Is Influenced by Light Quality.
(A) Cotyledon area of 12-d-old Col (wild-type), spt-11, and spt-12 seedlings grown at 20°C in the dark. Bars = se.
(B) Cotyledon area of 12-d-old Col (wild-type), phyB-9, spt-11, spt-12, phyB-9 spt-11, and phyB-9 spt-12 seedlings grown under red light at 20°C. Bars = se.
(C) Cotyledon area of 12-d-old Col (wild-type), spt-11, and spt-12 seedlings grown at 20°C under red light (white columns) or red light supplemented with far-red light (R:FR < 0.02; black columns). Bars indicate se.
Light Quality Determines the Dynamic Balance of SPT and DELLA
The proposed DELLA cross-regulatory control of SPT, together with our findings that cotyledon-located DELLAs are phyB regulated, suggested to us that the SPT:DELLA equilibrium may be determined by the prevailing light conditions. To examine this, we compared spt-11 and spt-12 seedlings grown under R + FR, which depletes active phyB Pfr and raises nuclear DELLA levels (see Supplemental Figure 5C online). The provision of supplementary FR led to a significant cotyledon size reduction in wild-type seedlings and a marked reduction of the impact of the spt-11 and spt-12 mutations on cotyledon expansion (Figure 5C). The increased potency of the spt alleles under light conditions that deplete DELLA levels support our proposal that SPT is subject to negative regulation by DELLAs. Because R + FR mimics vegetative shade conditions, this indicates that the dynamic balance of SPT and DELLA action would be determined by light quality signals in the natural environment. Our data infer that DELLAs are more important for maintaining growth suppression under vegetative shade, whereas SPT action dominates in nonshaded environments.
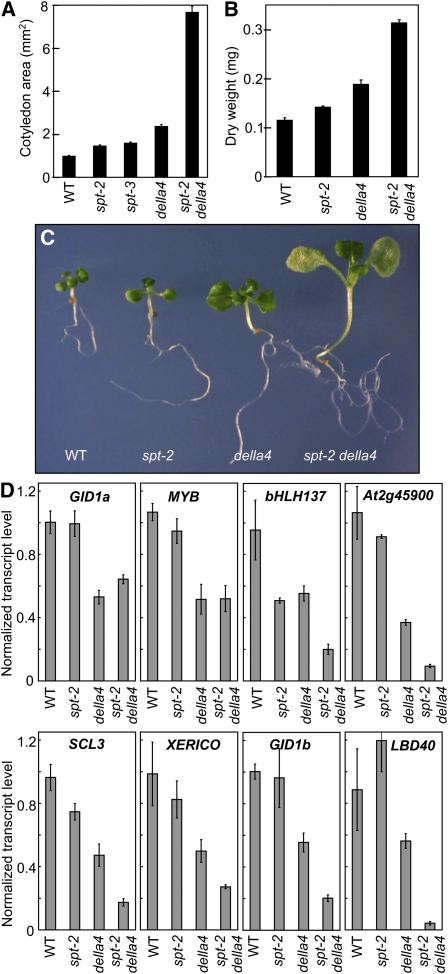
The spt-2 della4 Mutant Seedlings Have Unrestrained Growth
To establish whether we could obtain genetic support for SPT operating to counter the effects of DELLA loss, spt-2 was introgressed into the della4 (gai-t6 rga-t2 rgl1-1 rgl2-1) mutant background. To enhance the DELLA-mediated cotyledon response in Ler seedlings, we grew them on PAC (Figure 1G; see Supplemental Figure 3 online). Under these conditions, the spt-2, spt-3, and della4 mutants exhibit modest cotyledon expansion phenotypes when compared with the wild type (Figure 6A). However, loss of SPT and DELLA activity is strongly synergistic, giving rise to significantly larger seedlings with increased biomass (Figures 6A to 6C). The retention of substantial growth inhibition in SPT-deficient and DELLA-deficient mutants indicates that DELLAs and SPT can operate independently. SPT and DELLA functional redundancy is not only observed in the cotyledon but also in the hypocotyl, which is very elongated in spt-2 della4 compared with spt-2 and della4 (Figure 6C).
Figure 6.
SPT and DELLAs Act in Parallel to Regulate a Common Gene Subset.
(A) Cotyledon area of 12-d-old Ler (wild-type [WT]), spt-2, spt-3, della4, and spt-2 della4 (rga-t2 gai-t6 rgl1-1 rgl2-1) seedlings grown on 0.2 μM PAC under red light. Dry weight (B) and whole-seedling phenotype (C) of the corresponding genotypes. Bars = se.
(D) GID1a, MYB, BHLH137, AT2G45900, SCL3, XERICO, GID1a, and LBD40 transcript levels determined by quantitative real-time PCR in 5-d-old seedlings grown under the same conditions as (A). Values are given relative to the housekeeping gene UBQ10. Bars = se from three biological replicates.
[See online article for color version of this figure.]
SPT and DELLA Activity Converges at Common Target Genes
Our physiological analysis illustrates that SPT and DELLAs have a high degree of functional overlap (Figures 1A to 1E, 3A, and 6A to 6C). Furthermore, of the 43% of GA-regulated genes identified in our microarray data, 528 of 647 (82%) exhibited altered GA-dependent expression in spt-12 (Figure 2C, category C). This suggested to us that SPT and DELLAs may regulate a common gene subset. Therefore, it was of interest when we established that there was a significant enrichment for SPT-regulated genes within subsets identified as DELLA-regulated genes in published array data (Cao et al., 2006; Zentella et al., 2007; Achard et al., 2008; Hou et al., 2008). Although our study indicates that SPT regulates 6.2% of the Arabidopsis thaliana ATH1 genome array, we observed between 11.6 and 18.3% of SPT-regulated genes among the published DELLA-regulated gene sets (see Supplemental Figure 10A and Supplemental Data Sets 6 to 11 online). Because the plant material and conditions used in the comparative studies differed from our own, we anticipate that the observed values for DELLA and SPT coregulated genes may be underestimated. Likewise, there was a twofold enrichment for DELLA-regulated genes among our SPT-regulated gene set (see Supplemental Figure 10B and Supplemental Data Set 11 online). Of the genes that were coregulated by SPT and DELLA, 29% were subject to opposite regulation, whereas 64% were regulated in the same direction by SPT and DELLAs (see Supplemental Figure 11 online).
Recently, a number of gene promoters have been identified as putative RGA targets in chromatin immunoprecipitation (ChIP) assays (Zentella et al., 2007). Among these are SCARECROW-LIKE3 (SCL3); XERICO, a RING-H2 zinc finger gene; MYB, a putative MYB transcription factor; BHLH137, a putative bHLH transcription factor; AT2G45900 (unknown function); LOB DOMAIN-CONTAINING PROTEIN40 (LBD40); and GA INSENSITIVE DWARF1 (GID1a) and GID1b GA receptors. To test the possibility that SPT and DELLAs regulate a common set of genes, we tested the impact of spt-2 and della4 on these known DELLA targets. Consistent with previous findings, our qPCR analysis demonstrated that transcript levels of these genes are severely depleted in the della4 mutant that lacks RGA, GAI, RGL1, and RGL2 (Figure 6D; Zentella et al., 2007). Our data also illustrate that GID1a and MYB transcript levels are unaffected by the presence of the spt-2 mutation, suggesting that under our conditions, GID1a and MYB are regulated by DELLAs but not SPT. Because our microarray data indicate that SPT can also regulate genes independently of GA, this strengthens the argument that SPT and DELLAs can act in discrete pathways (Figure 2C; category A). In contrast to GID1a and MYB, bHLH137 is strongly regulated by SPT as well as DELLAs. The remaining genes, SCL3, XERICO, At2g45900, LBD40, and GID1b are unaffected or moderately suppressed by spt-2 alone, but this response is strongly augmented by della4. This spt-2 and della4 synergism, which is also observed for physiological outputs, is compatible with our proposed mode of action that invokes DELLA-regulated SPT compensation (Figure 6). The data also indicate that although SPT and DELLAs belong to completely unrelated gene families, they have a degree of functional overlap in terms of gene targets and physiological outputs.
The genetic redundancy, coupled with DELLA cross-regulation of SPT (Figures 4D and 4C), suggests that the SPT and DELLA growth suppressors operate in a molecular circuit that can switch between SPT and DELLA dominance (Figure 7). This simple control balances SPT and DELLA activity to maintain growth restraint and to prevent extreme behavior that would arise from an excess or a depletion of these potent growth regulators.
Figure 7.
The SPT-DELLA Compensatory Genetic Circuit Maintains Growth Restraint.
SPT and DELLAs are potent growth suppressors that regulate distinct and common gene subsets. Although SPT and DELLAs can operate independently, SPT (the responder) is subject to negative regulation by DELLAs (the inducers). This cross-regulation generates a compensatory action where SPT responds reciprocally to changes in the levels of DELLAs. Therefore, the molecular circuit provides growth restraint that is adjusted to DELLA levels. Compensatory regulation is more frequently observed among paralogous genes; however, this is a nice example where the unrelated genes SPT and DELLAs fulfill this role. DELLA abundance is also regulated by light quality; therefore, the balance between DELLA and SPT circuit arms is determined by the ambient light conditions in the natural environment.
[See online article for color version of this figure.]
DISCUSSION
Previously, we presented evidence that SPT regulates seed dormancy and the transcription of GA biosynthesis genes (Penfield et al., 2005). In this article, we demonstrate that SPT-regulated signaling is manifestly different at the seedling stage. During seedling development, SPT is a potent repressor of cotyledon expansion, where it acts alongside, but in cooperation, with DELLAs. We show that SPT is subject to cross-regulation by DELLAs and that the resultant compensatory behavior is important for maintaining growth repression over a range of light conditions that are prevalent in nature.
SPT, RGA, and GAI Are the Principal Repressors of Cotyledon Growth
bHLH transcription factors have important roles in the early life of the plant, integrating environmental information about the daily photoperiod, light quality, and ambient temperature (Josse et al., 2008). Through seedling development, PIFs drive hypocotyl elongation, which occurs at the expense of cotyledon expansion, whereas PIF depletion restricts hypocotyl growth (Ni et al., 1998; Huq and Quail, 2002; Khanna et al., 2006; Leivar et al., 2008). We have shown that in contrast to the PIFs, SPT has a relatively minor role in regulating hypocotyl elongation. Instead, SPT is a robust suppressor of cotyledon expansion, whereas PIFs have only limited or redundant regulatory roles (Figure 1A). Another key contributor to this response is the DELLA growth repressor RGA, which operates with its redundant counterpart GAI (Figure 3A). We have recently demonstrated that in the adult plant, SPT has a more prominent role in regulating growth at cooler ambient temperatures (Sidaway-Lee et al., 2010). Therefore, it is interesting that at the seedling stage, SPT appears to act over a wide temperature range (see Supplemental Figure 1D online).
PIFs and DELLAs Are Functionally Distinct in the Hypocotyl and Cotyledon
In the seedling hypocotyl, PIFs promote growth, whereas DELLAs suppress growth. This antagonism is central to a proposed mechanism of action where DELLA proteins restrain hypocotyl growth by inactivating PIFs (de Lucas et al., 2008; Feng et al., 2008). Here, it has been established that DELLAs bind the highly conserved DNA recognition domain of PIF3 and PIF4 to block their transcriptional activity (de Lucas et al., 2008; Feng et al., 2008). Because DELLAs appear to bind several PIFs, it has been intimated that this class of bHLHs could be common targets for DELLAs (Gallego-Bartolomé et al., 2010). We have established that SPT, which also possesses the conserved DELLA binding domain, interacts with DELLAs in a yeast two-hybrid assay (see Supplemental Figure 8 online). However, in contrast to the hypocotyl, where PIFs and DELLAs act in opposition, we and others have shown that SPT, PIFs, and DELLAs have analogous growth-suppressing roles in the cotyledon (Figures 1 and 3; Ni et al., 1998; Huq and Quail, 2002; Leivar et al., 2008). These observations infer that SPT, PIFs, and DELLAs regulate cotyledon expansion via a process that is unequivocally distinct from that described for hypocotyl elongation.
SPT Is a GA Antagonist
Our previous finding that SPT regulates GA biosynthesis enzymes during germination suggested that the spt cotyledon phenotype may simply result from elevated GA levels. However, we found no evidence of altered GA biosynthesis gene expression in spt-12 seedlings by microarray analysis (see Supplemental Data Set 1 online), although it is possible that the use of whole seedlings could have obscured the identification of cotyledon-specific GA metabolism genes. We found that 57% (category A) of SPT-regulated genes were unaffected by GA application, which suggested that SPT can regulate gene expression independently of GA signaling (Figure 2C). The remaining 42% (categories B and C) were scored as GA responsive, indicating that although SPT may not specifically target GA biosynthesis genes, it does regulate GA response genes. We also noted that 82% (category C) of the GA-regulated subset exhibited an altered GA response in spt-12 compared with the wild type. These results concur with our physiological data where we also observed dramatic alterations in GA responsiveness of cotyledon expansion in spt mutants (Figure 1, C–G). Therefore, SPT appears to counter the impact of GA on gene expression and cotyledon expansion. Because SPT loss results in markedly expanded cotyledons following GA application, this signifies that SPT is required to prevent the excessive growth that would otherwise result from DELLA depletion in wild-type seedlings.
SPT Can Operate Independently of DELLAs
GA is known to promote growth by triggering the proteolytic degradation of the growth-suppressing DELLA proteins (Silverstone et al., 1997, 1998, 2001; Pysh et al., 1999; Dill et al., 2001; Harberd, 2003; Achard and Genschik, 2009). GA binds to the GID1 nuclear receptors to form GID1–GA complexes that interact with DELLA proteins, resulting in their polyubiquitination by SCFGID2/SLY1 and subsequent degradation by the 26S proteasome (Dill et al., 2001, 2004; Griffiths et al., 2006; Nakajima et al., 2006; Willige et al., 2007; Ariizumi et al., 2008; Murase et al., 2008; Shimada et al., 2008). Because spt alleles exhibited altered responsiveness to GA, we wanted to test whether this could result from elevated GA-mediated DELLA destruction (Figure 1, D–H). Our immunoblot analysis indicated that this was unlikely to be the case because GA destruction kinetics for RGA-GFP were comparable in wild type and spt-11 (Figure 4A). Moreover, we also noted that the impact of the spt alleles is observed in both controls without GA and at GA concentrations that severely deplete DELLA levels, suggesting that DELLAs are not required for the spt response (Figures 1G and 1H). Therefore, it appears that SPT suppresses cotyledon expansion through a process that does necessitate DELLA presence.
SPT and DELLAs Operate as a Compensatory Growth Suppressor Module
As we had observed modified GA-mediated gene expression and cotyledon growth responses in spt mutants, this suggested to us that SPT may be subject to regulation by DELLAs (Figures 1 and 2). Reduction of DELLA levels by GA-induced proteolysis led to a rise in SPT protein levels (Figure 4B). Furthermore, in gai-1 seedlings that express a mutant GAI protein that is constitutively active and GA resistant, SPT protein levels were strongly suppressed (Figure 4C). The impact of the gai-1 mutation on SPT protein levels was still evident in MG132-treated seedlings, inferring that gai-1 was unlikely to control SPT destruction via a mechanism that depended on proteasome activity. However, we did observe a GA-dependent rise in the 35S-driven SPT transgene transcript, a response that was negated by the presence of the GA-insensitive gai-1. The rise in transcript levels, which is detectable 1 h post-GA application, precedes the observed rise in protein levels (between 4 and 8 h), inferring that this control is unlikely to result from SPT protein feedback (Figure 4D). Rather, GAI and possibly other DELLAs may influence SPT protein levels by regulating transcript stability. This proposition could be tested further by establishing the impact of gai and rga loss of function alleles on SPT protein abundance, although such a role for GAI and RGA has been reported for the regulation of anther development and short-day flowering time (Achard et al., 2004). Here, GAI and RGA were shown to modulate levels of miRNA159, which mediates mRNA cleavage of GAMYB targets, although the precise operational mechanism is unknown. SPT transcript does not appear to contain the conserved miRNA159 target sequence, so it is unlikely to be regulated by miRNA159. However, it would be interesting to establish whether SPT is targeted by an analogous or related process.
The negative regulation of SPT by DELLAs ensures that SPT protein production is coupled to DELLA abundance. This molecular counterbalance has features in common with compensatory genetic circuits that have been described in numerous biological networks (Baggs et al., 2009; Kafri et al., 2009). However, these circuits frequently comprise functionally redundant duplicates that maintain a target response through cross-regulation. Here, we have shown that like DELLAs, SPT is a potent growth suppressor, and although these genes are unrelated, they maintain growth restraint through a counterbalance mechanism (Figure 7).
phyB Promotes Nuclear RGA Accumulation in Cotyledons
Previously, phyB has been reported to promote nuclear GFP-RGA accumulation in seedling hypocotyls cells following exposure to red light (Achard et al., 2007). In contrast, we found that in cotyledons, GFP-RGA accumulated to high levels in dark-grown seedlings, whereas exposure to red light led to GFP-RGA depletion (Figures 3B and 3C). We also noted that cotyledon-located GFP-RGA levels were elevated in phyB-9-null seedlings and when phyB was inactivated by the provision of supplementary FR (see Supplemental Figure 5 online). Therefore, DELLAs are subject to differential phyB-mediated control that is dependent upon their tissue location within the seedling. Contrasting with this, SPT levels remain fairly constant in seedlings exposed to a prolonged period of red light (see Supplemental Figure 6 online). Because we also noted that the spt mutant phenotype was evident in etiolated as well as light-grown seedlings, we concluded that unlike DELLAs, SPT was not obviously light regulated (Figures 3B, 3C, 5A; see Supplemental Figure 5 online).
The External Light Environment Shifts the SPT:DELLA Balance
The coupling of DELLAs levels to phyB suggested to us that the balance of SPT and DELLA action could be dependent upon the external light environment. If this is the case, we would anticipate that phyB depletion, which elevates cotyledon DELLA levels, should reduce the severity of the spt mutation (see Supplemental Figure 5 online). Compliant with this prediction, the phyB-9 allele greatly attenuates the spt-11 and spt-12 cotyledon phenotype (Figure 5B). In the natural environment, vegetative shade can greatly deplete the proportion of active phyB (Pfr). Under R + FR, which simulates vegetation shade, the spt-11 and spt-12 alleles were less effective than under red light conditions, which are more indicative of open habitats (Figure 5C). Therefore, the balance between SPT- and DELLA-dominated control of cotyledon expansion is dictated by ambient light conditions. DELLAs predominate in light conditions indicative of vegetation-dense habitats, whereas SPT action prevails in light that typifies unshaded, open habitats. Tethering SPT levels to DELLAs guarantees growth suppression across a spectral range of light conditions that are prevalent in nature.
SPT and DELLAs Converge at Common Genes
Our genetic and molecular data indicate that SPT is a potent growth repressor that has a specific role in controlling growth when DELLA levels are low (Figure 7). Strong support for this proposal is provided by our finding that growth restraint is sustained in spt-2 and della4, but not in the spt-2 della4 mutant seedlings that have significantly larger cotyledons, longer hypocotyls, and increased biomass (Figure 6, A–C). These data demonstrate that, although SPT and DELLAs belong to completely unrelated gene families, they have a high degree of functional overlap. Moreover, the dramatic impact that the combined loss of SPT and the DELLAs GAI, RGA, RGL1, and RGL2 (in della4) have on seedling size provides an appreciation for the importance of this counterbalance circuit in maintaining growth suppression.
Our microarray data illustrated that although SPT clearly regulates genes independently of GA, 43% of misregulated genes in spt-12 versus wild type were GA regulated. Interestingly, for 82% of genes in this group (category C), the GA response was dependent on SPT presence, suggesting that SPT and DELLA signaling may converge at a common gene subset (Figure 2C). In support of this notion, in our SPT-regulated gene subset, we observed a strong enrichment of DELLA-regulated genes identified in other microarray studies (see Supplemental Figure 10 and Supplemental Data Sets 6 to11 online). To test this proposition, we tested the impact of spt-2 and della4 alone or combined on transcript levels of GID1a, SCL3, XERICO, At2g45900, LBD40 and GID1b, and BHLH137, genes previously identified by ChIP as targets for DELLA regulation (Figure 6D; Zentella et al., 2007). In our assays, GID1a and MYB mRNA levels were suppressed by della4 but not by spt-2, suggesting that these genes are regulated by DELLAs and not by SPT in our conditions. BHLH137 emerged as a common target for SPT and DELLAs, as BHLH137 transcript levels were reduced in spt-2 and della4, whereas SCL3, XERICO, At2g45900, LBD40, and GID1b exhibited synergistic regulation by spt-2 and della4. Collectively, our qPCR analysis reinforces the notion that SPT and DELLAs can act independently of each other because known DELLA targets are unaffected by SPT loss. It illustrates that SPT and DELLAs also regulate a subset of common genes. Furthermore, the synergistic control of dual targets by spt-2 and della4 is compatible with the expected behavior of SPT and DELLAs in a compensatory genetic circuit.
This study highlights SPT as a principal regulator of cotyledon growth, which contrasts with the behavior of PIF gene paralogs that mainly promote hypocotyl elongation. SPT has a high degree of functional overlap with the unrelated DELLAs proteins because signals from these potent growth repressors converge at common gene targets (Figure 7). Importantly, SPT levels are negatively regulated by DELLA proteins, compelling SPT to act as a molecular counterbalance to DELLAs. This control circuit prevents excessive growth restriction when DELLAs are abundant, yet maintains restraint on growth when DELLA levels are low. We have shown that in seedling cotyledons, DELLAs are depleted by phyB action following exposure to light, whereas SPT levels remain constant. The light regulation of DELLAs drives the DELLA-SPT counterbalance, which enforces growth restraint across a range of ambient light conditions that are prevalent in nature. The importance of maintaining growth suppression throughout seedling development is exemplified by the oversized spt-2 della4 seedlings, where growth is unrestrained.
METHODS
Plant Materials and Growth Conditions
The wild-type Arabidopsis thaliana ecotypes used in this study are Landsberg erecta and Columbia-0. The spt-2, spt-3 (Heisler et al., 2001), 35S:SPT, 35S:SPT-HA (Penfield et al., 2005), spt-11, spt-12 (Ichihashi et al., 2010), pif4-101 (Lorrain et al., 2008), pif3-3 (Monte et al., 2004), pif4-2, pif7-1 (Leivar et al., 2008), ga1-3 (Sun et al., 1992), pRGA:GFP-RGA (Silverstone et al., 1998), gai-1 (Dill et al., 2001), and della multiple (Cao et al., 2005; Koini et al., 2009) mutants have been described previously. spt-11 pRGA:GFP-RGA; 35S:SPT pRGA:GFP-RGA, 35S:SPT-HA pRGA:GFP-RGA, gai-1 35S:SPT-HA, spt2 gai-t6 rga-t2 rgl1-1 rgl2-1, spt-11 pif3-3, spt-11 pif4-101, and spt-11 pif7-1 were obtained by cross-pollination and were all verified by PCR (and sequenced in the case of the spt-2 mutation).
Seeds were surface sterilized and sown on Gilroy-agar media (no Suc; supplemented with phytohormones where indicated; Wymer et al., 1997). The seeds were sown in an evenly spaced manner to prevent shading during seedling growth. Where possible, and for the majority of experiments, all genotypes were sown on the same plate to remove plate to plate variation. When multiple genotypes were used, mutant alleles were always sown on the same plate at their isogenic wild type. Seeds were stratified in darkness for 3 d at 4°C and were then light pulsed for 6 h under white light at 20°C to promote even germination. Seedlings in all experiments were subsequently grown at 20°C under 40 μmol m−2 s−1 fluence rate red light provided by light-emitting diodes with a 660-nm emission peak, unless otherwise stated. All plates in an individual experiment were exposed to identical light conditions (fluence rate and spectral range). Where ga1-3 mutation was present, seeds were pretreated with 50 μM GA3 for 3 d at 4°C before being thoroughly cleaned, sterilized, and sown.
Physiological Measurements
For hypocotyl length and cotyledon area measurements, seedlings were flattened on their agar plates to reveal the full extent of their hypocotyl and cotyledon phenotype, and images were taken using a digital camera. Cotyledon area and hypocotyl length were measured using the ImageJ software (http://rsbweb.nih.gov/ij/), and values ± se were obtained for n ≥ 25. For dry weight measurements, entire seedlings were harvested from agar plates, blotted, pooled per 20, and dried in aluminum foil at 93°C for 3 d. Values ± se were obtained from five pools of 20 seedlings per genotype.
SEM
Whole cotyledons were mounted flat, adaxial surface uppermost, on a modified Gatan cryospecimen carrier using colloidal graphite (Agar Scientific G303) as a cryoadhesive. Specimens were cryofixed by plunging into liquid nitrogen at about −210°C. The specimen carrier was transferred under low vacuum to the specimen stage of a Gatan Alto 2500 cryopreparation unit at about −185°C. The specimens were sputter coated with 6 to 8 nm of 60:40 gold:palladium alloy (Testbourne Ltd.) in an atmosphere of argon gas (Messer UK Ltd.) and transferred under high vacuum to the cryospecimen stage of a Hitachi 4700 II cold field-emission scanning electron microscope (Hitachi High Technologies) at <−170°C. Specimens were examined at nominal image magnifications up to ×50,000 using a beam accelerating voltage of 5.0 kV and filament current of 10 μA and working distances between 4 and 12 mm. Digital images were captured at a resolution of 2560 × 1920 pixels using the signal from the upper (semi–in-lens) secondary electron detector.
Individual cells were artificially colored using Adobe Photoshop CS3, and cell area and perimeter were measured using the ImageJ software. The nonparametric Kolmogorov-Smirnov test was performed on the populations of values obtained to determine whether cell area and cell perimeter were increased in the GA-treated spt alleles.
Fluorescence Microscopy
Whole seedlings were mounted in water between two cover slips and observed using a Nikon eclipse TE 2000-U confocal microscope with a 60× objective. Specimens were excited with a 480 nm laser beam, and images were visualized between 510 and 540 nm (GFP emission) and above 600 nm (chlorophyll fluorescence). Images were overlaid using Adobe Photoshop CS3.
GA Treatments and Immunoblots
A minimum of 50 seedlings were grown on small plates of Gilroy-agar media (with 0.2 μM PAC where indicated; this concentration allowed seed germination without exogenous treatment, indicating that some GA biosynthesis remained possible). At time point 0, 4 mL of GA3 (at the concentration indicated) was added to each plate and was allowed to entirely cover the surface of the seedlings. When harvested, seedlings were dry-blotted and then snap-frozen in liquid nitrogen.
Where indicated, 4 mL of MG132 (100 μM) or 4 mL of water (MOCK) was added to the plates 2 h prior to the GA3 treatment.
Total proteins were extracted as described (Duek et al., 2004). Twenty microliters of each sample was run on 10% SDS-PAGE, followed by a wet transfer to a polyvinylidene fluoride membrane. The HA tag was detected by probing the membrane with a rat anti-HA antibody (3F10; Roche) at a dilution of 1:1000 followed by a horseradish peroxidase (HRP)-conjugated sheep anti-rat (Abcam) at a dilution of 1:5000. The GFP tag was detected by probing the membrane with a sheep anti-GFP antibody (gift from Kevin Hardwick) at a dilution of 1:1000 followed by a HRP-conjugated donkey anti-sheep antibody (AbD SEROTEC) at a dilution of 1:5000. Loading was checked by directly reprobing membranes using a goat anti-UDP-Glc pyrophosphorylase (anti-UGPase) antibody (AGRISERA) at a dilution of 1:1000 followed by a HRP-conjugated sheep anti-rat (Bio-Rad) at a dilution of 1:5000. Signals were detected using the Amersham ECL kit as per instructed by the manufacturer.
Transcript Analysis
For real-time qPCR analysis, whole seedlings were grown as indicated in the figure legends, and triplicate samples were harvested in RNA Later (Qiagen). RNA was extracted using the plant RNeasy extraction kit (Qiagen). Two micrograms of RNA was reverse-transcribed using the ReverseAid FirstStrand cDNA synthesis kit (Fermentas), and the real-time PCR was performed on the cDNA using a Rotor Gene 300 (Corbett Research). Gene expression was quantified as a relation to the expression of the housekeeping genes UBQ10 and ACT7. The primers used are listed in Supplemental Table 1 (online).
For microarray analysis, Col and spt-12 seedlings were grown on Gilroy-agar plates for 4 d at 20°C under red light (40 μmol·m−2 s−1) and treated as described previously with 50 μM GA3. Triplicate samples were harvested individually at time points 0 min, 30 min, and 24 h. RNA was extracted as described previously, and over 1 μg of RNA from each sample was sent to the NASC Affy Gene ChIP service, who performed an ATH1 Genome Array. The full data set is available to download from the NASCarrays database (http://affymetrix.Arabidopsis.info/; reference: NASCARRAYS-505). Background correction, normalization of all the arrays together (18 arrays), and gene expression analysis of the array data were performed using the GeneChip-robust multiarray analysis routine (Wu et al., 2004) in GeneSpring, version 7.2 (Silicon Genetics). SPT targets were defined as genes presenting at least a 1.5-fold change in mean expression between triplicate wild-type and spt-12 samples (±GA treatment). A cutoff P value of 0.05 in a one-way analysis of variance test corrected by a Benjamini and Hochberg false discovery rate was applied. Nonnuclear encoded and absent genes (raw expression < 50 in all six conditions) were excluded from the analysis.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: SPT, AT4G36930; PIF3, AT1G09530; PIF4, AT2G43010; PIF7, AT5G61270; GA1, AT4G02780; GAI, AT1G14920; RGA, AT2G01570; RGL1, AT1G66350; RGL2, AT3G03450; RGL3, AT5G17490; PHYB, AT2G18790; SCL3, AT1G50420; XERICO, AT2G04240; MYB, AT3G11280; BHLH137, AT5G50915; LBD40, AT1G67100; GID1A, AT3G05120; GID1B, AT3G63010; and AT2G45900.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Characterization of spt Alleles, pif Alleles, and 35S:SPT Seedlings.
Supplemental Figure 2. spt Mutants Exposed to GA Have Larger Cotyledon Pavement Cells Than Wild Type.
Supplemental Figure 3. The Ler and Col Accessions Respond to GA Differently.
Supplemental Figure 4. Cotyledon-Located GFP-RGA Accumulates in the Dark.
Supplemental Figure 5. RGA Accumulation in the Cotyledon Is Controlled by phyB.
Supplemental Figure 6. SPT Protein Levels Are Stable in Red Light.
Supplemental Figure 7. SPT, PIF3, and PIF4 Share a Highly Conserved bHLH Domain.
Supplemental Figure 8. SPT Interacts with RGA and GAI in a Yeast Two-Hybrid Assay.
Supplemental Figure 9. Dose-Dependent GA-Mediated GFP-RGA Degradation.
Supplemental Figure 10. SPT and DELLAs Regulate a Common Gene Subset.
Supplemental Figure 11. SPT and DELLAs Mostly Regulate Genes in a Similar Direction.
Supplemental Table 1. List of the Primers Used in the qPCR Experiments.
Supplemental Data Set 1. List of the 1524 Genes Presenting at Least a 1.5-fold Change in Mean Expression between Triplicate Wild-Type and spt-12 Samples.
Supplemental Data Set 2. Category A (877 Genes): Genes Regulated by SPT and Not Regulated by GA3 Treatment in Our Arrays.
Supplemental Data Set 3. Category B (119 Genes): Genes Regulated by SPT That Exhibit Similar GA-Dependent Regulation in Wild Type and spt-12.
Supplemental Data Set 4. Category C (528 Genes): Genes Regulated by SPT That Exhibit Different GA Regulation in Wild Type and spt-12.
Supplemental Data Set 5. Expression Profiles of Category C Gene Subsets That Show Synergistic Regulation by SPT and GA.
Supplemental Data Set 6. Comparison of Our Transcriptomics Data with Microarrays from Achard et al. (2008).
Supplemental Data Set 7. Comparison of Our Transcriptomics Data with Arrays from Cao et al. (2006; Seed Data Set).
Supplemental Data Set 8. Comparison of Our Transcriptomics Data with Arrays from Cao et al. (2006; Flower Bud Data Set).
Supplemental Data Set 9. Comparison of Our Transcriptomics Data with Arrays from Hou et al. (2008).
Supplemental Data Set 10. Comparison of Our Transcriptomics Data with Arrays from Zentella et al. (2007).
Supplemental Data Set 11. List of the 319 Genes Differentially Regulated by Both SPT and One or More DELLAs and Comparison of Their Regulation in the Different Data Sets Analyzed in This Article.
Acknowledgments
We thank Liz Alvey and Nick Harberd for providing the della multiple mutants; Pablo Leivar and Peter Quail for the pif3-3, pif4-2, and pif7-1 mutants; Kevin Hardwick for the anti-GFP antibodies; and members of the Halliday laboratory for discussions and comments. This work was supported by UK Biotechnology and Biological Sciences Research Council Grants BBE0003631 to K.J.H. and BBE0005411 to I.A.G., by a Scottish Universities Life Sciences Alliance research grant to F.N., and by funds from the Spanish Ministerio de Ciencia e Innovación and the Fondo Europeo de Desarrollo Regiona (BIO2008-00169) to J.F.M.-G.
References
- Achard P., Genschik P. (2009). Releasing the brakes of plant growth: How GAs shutdown DELLA proteins. J. Exp. Bot. 60: 1085–1092 [DOI] [PubMed] [Google Scholar]
- Achard P., Gusti A., Cheminant S., Alioua M., Dhondt S., Coppens F., Beemster G.T.S., Genschik P. (2009). Gibberellin signaling controls cell proliferation rate in Arabidopsis. Curr. Biol. 19: 1188–1193 [DOI] [PubMed] [Google Scholar]
- Achard P., Herr A., Baulcombe D.C., Harberd N.P. (2004). Modulation of floral development by a gibberellin-regulated microRNA. Development 131: 3357–3365 [DOI] [PubMed] [Google Scholar]
- Achard P., Liao L., Jiang C., Desnos T., Bartlett J., Fu X., Harberd N.P. (2007). DELLAs contribute to plant photomorphogenesis. Plant Physiol. 143: 1163–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achard P., Renou J.P., Berthomé R., Harberd N.P., Genschik P. (2008). Plant DELLAs restrain growth and promote survival of adversity by reducing the levels of reactive oxygen species. Curr. Biol. 18: 656–660 [DOI] [PubMed] [Google Scholar]
- Alabadí D., Blázquez M.A. (2009). Molecular interactions between light and hormone signaling to control plant growth. Plant Mol. Biol. 69: 409–417 [DOI] [PubMed] [Google Scholar]
- Al-Sady B., Kikis E.A., Monte E., Quail P.H. (2008). Mechanistic duality of transcription factor function in phytochrome signaling. Proc. Natl. Acad. Sci. USA 105: 2232–2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Sady B., Ni W., Kircher S., Schäfer E., Quail P.H. (2006). Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol. Cell 23: 439–446 [DOI] [PubMed] [Google Scholar]
- Ariizumi T., Murase K., Sun T.P., Steber C.M. (2008). Proteolysis-independent downregulation of DELLA repression in Arabidopsis by the gibberellin receptor GIBBERELLIN INSENSITIVE DWARF1. Plant Cell 20: 2447–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggs J.E., Price T.S., DiTacchio L., Panda S., Fitzgerald G.A., Hogenesch J.B. (2009). Network features of the mammalian circadian clock. PLoS Biol. 7: e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer D., Viczián A., Kircher S., Nobis T., Nitschke R., Kunkel T., Panigrahi K.C., Adám E., Fejes E., Schäfer E., Nagy F. (2004). Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell 16: 1433–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D., Cheng H., Wu W., Soo H.M., Peng J. (2006). Gibberellin mobilizes distinct DELLA-dependent transcriptomes to regulate seed germination and floral development in Arabidopsis. Plant Physiol. 142: 509–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D., Hussain A., Cheng H., Peng J. (2005). Loss of function of four DELLA genes leads to light- and gibberellin-independent seed germination in Arabidopsis. Planta 223: 105–113 [DOI] [PubMed] [Google Scholar]
- de Lucas M., Davière J.M., Rodríguez-Falcón M., Pontin M., Iglesias-Pedraz J.M., Lorrain S., Fankhauser C., Blázquez M.A., Titarenko E., Prat S. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484 [DOI] [PubMed] [Google Scholar]
- Dill A., Jung H.S., Sun T.P. (2001). The DELLA motif is essential for gibberellin-induced degradation of RGA. Proc. Natl. Acad. Sci. USA 98: 14162–14167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill A., Sun T. (2001). Synergistic derepression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics 159: 777–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill A., Thomas S.G., Hu J., Steber C.M., Sun T.P. (2004). The Arabidopsis F box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 16: 1392–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duek P.D., Fankhauser C. (2005). bHLH class transcription factors take centre stage in phytochrome signalling. Trends Plant Sci. 10: 51–54 [DOI] [PubMed] [Google Scholar]
- Duek P.D., Elmer M.V., van Oosten V.R., Fankhauser C. (2004). The degradation of HFR1, a putative bHLH class transcription factor involved in light signaling, is regulated by phosphorylation and requires COP1. Curr. Biol. 14: 2296–2301 [DOI] [PubMed] [Google Scholar]
- Feng S., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin K.A., Quail P.H. (2010). Phytochrome functions in Arabidopsis development. J. Exp. Bot. 61: 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao T., Bailey-Serres J. (2008). Submergence tolerance conferred by Sub1A is mediated by SLR1 and SLRL1 restriction of gibberellin responses in rice. Proc. Natl. Acad. Sci. USA 105: 16814–16819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Bartolomé J., Minguet E.G., Marín J.A., Prat S., Blázquez M.A., Alabadí D. (2010). Transcriptional diversification and functional conservation between DELLA proteins in Arabidopsis. Mol. Biol. Evol. 27: 1247–1256 [DOI] [PubMed] [Google Scholar]
- Gao X.H., Huang X.Z., Xiao S.L., Fu X.D. (2008). Evolutionarily conserved DELLA-mediated gibberellin signaling in plants. J. Integr. Plant Biol. 50: 825–834 [DOI] [PubMed] [Google Scholar]
- Griffiths J., Murase K., Rieu I., Zentella R., Zhang Z.L., Powers S.J., Gong F., Phillips A.L., Hedden P., Sun T.P., Thomas S.G. (2006). Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 18: 3399–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harberd N.P. (2003). Botany. Relieving DELLA restraint. Science 299: 1853–1854 [DOI] [PubMed] [Google Scholar]
- Heisler M.G., Atkinson A., Bylstra Y.H., Walsh R., Smyth D.R. (2001). SPATULA, a gene that controls development of carpel margin tissues in Arabidopsis, encodes a bHLH protein. Development 128: 1089–1098 [DOI] [PubMed] [Google Scholar]
- Hou X., Hu W.W., Shen L., Lee L.Y., Tao Z., Han J.H., Yu H. (2008). Global identification of DELLA target genes during Arabidopsis flower development. Plant Physiol. 147: 1126–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E., Al-Sady B., Quail P.H. (2003). Nuclear translocation of the photoreceptor phytochrome B is necessary for its biological function in seedling photomorphogenesis. Plant J. 35: 660–664 [DOI] [PubMed] [Google Scholar]
- Huq E., Quail P.H. (2002). PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J. 21: 2441–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichihashi Y., Horiguchi G., Gleissberg S., Tsukaya H. (2010). The bHLH transcription factor SPATULA controls final leaf size in Arabidopsis thaliana. Plant Cell Physiol. 51: 252–261 [DOI] [PubMed] [Google Scholar]
- Josse E.M., Foreman J., Halliday K.J. (2008). Paths through the phytochrome network. Plant Cell Environ. 31: 667–678 [DOI] [PubMed] [Google Scholar]
- Kafri R., Springer M., Pilpel Y. (2009). Genetic redundancy: New tricks for old genes. Cell 136: 389–392 [DOI] [PubMed] [Google Scholar]
- Khanna R., Huq E., Kikis E.A., Al-Sady B., Lanzatella C., Quail P.H. (2004). A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix-loop-helix transcription factors. Plant Cell 16: 3033–3044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R., Shen Y., Toledo-Ortiz G., Kikis E.A., Johannesson H., Hwang Y.S., Quail P.H. (2006). Functional profiling reveals that only a small number of phytochrome-regulated early-response genes in Arabidopsis are necessary for optimal deetiolation. Plant Cell 18: 2157–2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher S., Gil P., Kozma-Bognár L., Fejes E., Speth V., Husselstein-Muller T., Bauer D., Adám E., Schäfer E., Nagy F. (2002). Nucleocytoplasmic partitioning of the plant photoreceptors phytochrome A, B, C, D, and E is regulated differentially by light and exhibits a diurnal rhythm. Plant Cell 14: 1541–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher S., Kozma-Bognar L., Kim L., Adam E., Harter K., Schafer E., Nagy F. (1999). Light quality-dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant Cell 11: 1445–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koini M.A., Alvey L., Allen T., Tilley C.A., Harberd N.P., Whitelam G.C., Franklin K.A. (2009). High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr. Biol. 19: 408–413 [DOI] [PubMed] [Google Scholar]
- Lee S., Cheng H., King K.E., Wang W., He Y., Hussain A., Lo J., Harberd N.P., Peng J. (2002). Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev. 16: 646–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Monte E., Al-Sady B., Carle C., Storer A., Alonso J.M., Ecker J.R., Quail P.H. (2008). The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell 20: 337–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Quail P.H. (2011). PIFs: Pivotal components in a cellular signaling hub. Trends Plant Sci. 16: 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Tepperman J.M., Monte E., Calderon R.H., Liu T.L., Quail P.H. (2009). Definition of early transcriptional circuitry involved in light-induced reversal of PIF-imposed repression of photomorphogenesis in young Arabidopsis seedlings. Plant Cell 21: 3535–3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain S., Allen T., Duek P.D., Whitelam G.C., Fankhauser C. (2008). Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 53: 312–323 [DOI] [PubMed] [Google Scholar]
- Lorrain S., Trevisan M., Pradervand S., Fankhauser C. (2009). Phytochrome interacting factors 4 and 5 redundantly limit seedling de-etiolation in continuous far-red light. Plant J. 60: 449–461 [DOI] [PubMed] [Google Scholar]
- Martínez-García J.F., Huq E., Quail P.H. (2000). Direct targeting of light signals to a promoter element-bound transcription factor. Science 288: 859–863 [DOI] [PubMed] [Google Scholar]
- Monte E., Al-Sady B., Leivar P., Quail P.H. (2007). Out of the dark: How the PIFs are unmasking a dual temporal mechanism of phytochrome signalling. J. Exp. Bot. 58: 3125–3133 [DOI] [PubMed] [Google Scholar]
- Monte E., Tepperman J.M., Al-Sady B., Kaczorowski K.A., Alonso J.M., Ecker J.R., Li X., Zhang Y., Quail P.H. (2004). The phytochrome-interacting transcription factor, PIF3, acts early, selectively, and positively in light-induced chloroplast development. Proc. Natl. Acad. Sci. USA 101: 16091–16098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murase K., Hirano Y., Sun T.P., Hakoshima T. (2008). Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 456: 459–463 [DOI] [PubMed] [Google Scholar]
- Nakajima M., et al. (2006). Identification and characterization of Arabidopsis gibberellin receptors. Plant J. 46: 880–889 [DOI] [PubMed] [Google Scholar]
- Ni M., Tepperman J.M., Quail P.H. (1998). PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95: 657–667 [DOI] [PubMed] [Google Scholar]
- Ni M., Tepperman J.M., Quail P.H. (1999). Binding of phytochrome B to its nuclear signalling partner PIF3 is reversibly induced by light. Nature 400: 781–784 [DOI] [PubMed] [Google Scholar]
- Nozue K., Covington M.F., Duek P.D., Lorrain S., Fankhauser C., Harmer S.L., Maloof J.N. (2007). Rhythmic growth explained by coincidence between internal and external cues. Nature 448: 358–361 [DOI] [PubMed] [Google Scholar]
- Oh E., Kim J., Park E., Kim J.I., Kang C., Choi G. (2004). PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell 16: 3045–3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Yamaguchi S., Hu J., Yusuke J., Jung B., Paik I., Lee H.S., Sun T.P., Kamiya Y., Choi G. (2007). PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell 19: 1192–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Yamaguchi S., Kamiya Y., Bae G., Chung W.I., Choi G. (2006). Light activates the degradation of PIL5 protein to promote seed germination through gibberellin in Arabidopsis. Plant J. 47: 124–139 [DOI] [PubMed] [Google Scholar]
- Penfield S., Josse E.M., Kannangara R., Gilday A.D., Halliday K.J., Graham I.A. (2005). Cold and light control seed germination through the bHLH transcription factor SPATULA. Curr. Biol. 15: 1998–2006 [DOI] [PubMed] [Google Scholar]
- Piskurewicz U., Jikumaru Y., Kinoshita N., Nambara E., Kamiya Y., Lopez-Molina L. (2008). The gibberellic acid signaling repressor RGL2 inhibits Arabidopsis seed germination by stimulating abscisic acid synthesis and ABI5 activity. Plant Cell 20: 2729–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskurewicz U., Turecková V., Lacombe E., Lopez-Molina L. (2009). Far-red light inhibits germination through DELLA-dependent stimulation of ABA synthesis and ABI3 activity. EMBO J. 28: 2259–2271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pysh L.D., Wysocka-Diller J.W., Camilleri C., Bouchez D., Benfey P.N. (1999). The GRAS gene family in Arabidopsis: Sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J. 18: 111–119 [DOI] [PubMed] [Google Scholar]
- Salamini F. (2003). Plant biology. Hormones and the green revolution. Science 302: 71–72 [DOI] [PubMed] [Google Scholar]
- Shen H., Moon J., Huq E. (2005). PIF1 is regulated by light-mediated degradation through the ubiquitin-26S proteasome pathway to optimize photomorphogenesis of seedlings in Arabidopsis. Plant J. 44: 1023–1035 [DOI] [PubMed] [Google Scholar]
- Shen H., Zhu L., Castillon A., Majee M., Downie B., Huq E. (2008). Light-induced phosphorylation and degradation of the negative regulator PHYTOCHROME-INTERACTING FACTOR1 from Arabidopsis depend upon its direct physical interactions with photoactivated phytochromes. Plant Cell 20: 1586–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada A., Ueguchi-Tanaka M., Nakatsu T., Nakajima M., Naoe Y., Ohmiya H., Kato H., Matsuoka M. (2008). Structural basis for gibberellin recognition by its receptor GID1. Nature 456: 520–523 [DOI] [PubMed] [Google Scholar]
- Shin J., Kim K., Kang H., Zulfugarov I.S., Bae G., Lee C.H., Lee D., Choi G. (2009). Phytochromes promote seedling light responses by inhibiting four negatively acting phytochrome-interacting factors. Proc. Natl. Acad. Sci. USA 106: 7660–7665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidaway-Lee K., Josse E.M., Brown A., Gan Y., Halliday K.J., Graham I.A., Penfield S. (2010). SPATULA links daytime temperature and plant growth rate. Curr. Biol. 20: 1493–1497 [DOI] [PubMed] [Google Scholar]
- Silverstone A.L., Ciampaglio C.N., Sun T. (1998). The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 10: 155–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone A.L., Jung H.S., Dill A., Kawaide H., Kamiya Y., Sun T.P. (2001). Repressing a repressor: Gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell 13: 1555–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone A.L., Mak P.Y., Martínez E.C., Sun T.P. (1997). The new RGA locus encodes a negative regulator of gibberellin response in Arabidopsis thaliana. Genetics 146: 1087–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T., Goodman H.M., Ausubel F.M. (1992). Cloning the Arabidopsis GA1 locus by genomic subtraction. Plant Cell 4: 119–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T.P., Gubler F. (2004). Molecular mechanism of gibberellin signaling in plants. Annu. Rev. Plant Biol. 55: 197–223 [DOI] [PubMed] [Google Scholar]
- Sun T.P., Kamiya Y. (1994). The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthetase A of gibberellin biosynthesis. Plant Cell 6: 1509–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan M.S. (2009). Obituary: Norman E. Borlaug (1914–2009). Nature 461: 894. [DOI] [PubMed] [Google Scholar]
- Toledo-Ortiz G., Huq E., Quail P.H. (2003). The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15: 1749–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubeda-Tomás S., Federici F., Casimiro I., Beemster G.T.S., Bhalerao R., Swarup R., Doerner P., Haseloff J., Bennett M.J. (2009). Gibberellin signaling in the endodermis controls Arabidopsis root meristem size. Curr. Biol. 19: 1194–1199 [DOI] [PubMed] [Google Scholar]
- Vriezen W.H., Achard P., Harberd N.P., Van Der Straeten D. (2004). Ethylene-mediated enhancement of apical hook formation in etiolated Arabidopsis thaliana seedlings is gibberellin dependent. Plant J. 37: 505–516 [DOI] [PubMed] [Google Scholar]
- Willige B.C., Ghosh S., Nill C., Zourelidou M., Dohmann E.M., Maier A., Schwechheimer C. (2007). The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell 19: 1209–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Irizarry R.A., Gentleman R., Murillo F.M., Spencer F. (2004). A model based background adjustment for oligonucleotide expression arrays. Department of Biostatistics Working Papers, Working Paper 1. http://www.bepress.com/jhubiostat/paper1
- Wymer C.L., Bibikova T.N., Gilroy S. (1997). Cytoplasmic free calcium distributions during the development of root hairs of Arabidopsis thaliana. Plant J. 12: 427–439 [DOI] [PubMed] [Google Scholar]
- Zentella R., Zhang Z.L., Park M., Thomas S.G., Endo A., Murase K., Fleet C.M., Jikumaru Y., Nambara E., Kamiya Y., Sun T.P. (2007). Global analysis of della direct targets in early gibberellin signaling in Arabidopsis. Plant Cell 19: 3037–3057 [DOI] [PMC free article] [PubMed] [Google Scholar]