Chloroplast NAD(P)H dehydrogenase (NDH) has been thought to function as an NAD(P)H:plastoquinone oxidoreductase in cyclic electron transport around photosystem I, although its electron donor binding site was unclear. This study indicates that CRR31, a novel NDH subunit, serves in the high-affinity binding of ferredoxin to NDH and proposes that chloroplast NDH is ferredoxin:plastoquinone oxidoreductase.
Abstract
Some subunits of chloroplast NAD(P)H dehydrogenase (NDH) are related to those of the respiratory complex I, and NDH mediates photosystem I (PSI) cyclic electron flow. Despite extensive surveys, the electron donor and its binding subunits have not been identified. Here, we identified three novel components required for NDH activity. CRRJ and CRRL are J- and J-like proteins, respectively, and are components of NDH subcomplex A. CRR31 is an Src homology 3 domain-like fold protein, and its C-terminal region may form a tertiary structure similar to that of PsaE, a ferredoxin (Fd) binding subunit of PSI, although the sequences are not conserved between CRR31 and PsaE. Although CRR31 can accumulate in thylakoids independently of NDH, its accumulation requires CRRJ, and CRRL accumulation depends on CRRJ and NDH. CRR31 was essential for the efficient operation of Fd-dependent plastoquinone reduction in vitro. The phenotype of crr31 pgr5 suggested that CRR31 is required for NDH activity in vivo. We propose that NDH functions as a PGR5-PGRL1 complex-independent Fd:plastoquinone oxidoreductase in chloroplasts and rename it the NADH dehydrogenase-like complex.
INTRODUCTION
The light reaction of photosynthesis involves electron transport in the thylakoid membranes of chloroplasts. Electrons excised from water in photosystem II (PSII) are ultimately accumulated in NADPH. Protons are translocated from the stroma to the lumen across thylakoid membranes, in steps coupled to electron transport through the cytochrome (Cyt) b6f complex, and the resulting ΔpH is used in ATP synthesis. In contrast with this linear electron transport, photosystem I (PSI) cyclic electron transport recycles electrons from ferredoxin (Fd) or NAD(P)H to plastoquinone (PQ), which mediates the electron transport between PSII and Cyt b6f (Shikanai, 2007a). Consequently, ΔpH is formed without accumulation of NADPH.
Since the discovery of PSI cyclic electron transport (Arnon et al., 1954), its physiological significance has been unclear. Arabidopsis thaliana mutants specifically defective in PSI cyclic electron transport were identified, and the mutant phenotypes suggested that the electron transport functioned in both photosynthesis and photoprotection. In Arabidopsis, PSI cyclic electron transport is mediated by two pathways that depend on the PROTON GRADIENT REGULATION5 (PGR5)-PGR5-LIKE1 (PGRL1) complex and NAD(P)H dehydrogenase (NDH) (Munekage et al., 2004; Shikanai, 2007a; DalCorso et al., 2008). In C3 plants, the main pathway depends on the PGR5-PGRL1 complex, whereas NDH functions in stress resistance (Endo et al., 1999; Munekage et al., 2004; Wang et al., 2006).
Eleven plastid genes, ndhA-ndhK, encoding subunits of chloroplast NDH, were identified based on their similarity to genes encoding subunits of mitochondrial NADH dehydrogenase (Complex I) (Matsubayashi et al., 1987). However, chloroplast NDH is more similar to cyanobacterial NDH-1, which is believed to be an origin of chloroplast NDH (Friedrich et al., 1995; Shikanai, 2007b). By extensive surveys using proteomic, genetic, and bioinfomatic approaches, over 15 nucleus-encoded subunits were identified (Shikanai, 2007b; Majeran et al., 2008; Peng et al., 2009; Suorsa et al., 2009; Takabayashi et al., 2009). Chloroplast NDH consists of four parts, A, B, membrane, and lumen subcomplexes, and it further associates with PSI to form the NDH-PSI supercomplex (see Supplemental Figure 1 online; Peng et al., 2008, 2009). Subunits included in A and membrane subcomplexes are also conserved in the cyanobacterial NDH-1 complex, but the B and lumen subcomplexes are specific to chloroplasts (Peng et al., 2009). On the other hand, the cyanobacterial NDH-1MS complex contains the specific subunits (CupA, CupB, and CupS) that are involved in the CO2-concentrating mechanism (Battchikova and Aro, 2007).
Recently, the three-dimensional structure of L-shaped complex I from the thermophilic bacterium Thermus thermophilus was reported (Sazanov and Hinchliffe, 2006; Efremov et al., 2010). The peripheral arm contains the NADH binding site; flavin mononucleotide (FMN), which primarily accepts electrons; and eight or nine iron-sulfur clusters (Sazanov and Hinchliffe, 2006; Efremov et al., 2010). Nqo1-Nqo6, Nqo9, and Nqo15 form the arm, whereas Nqo1-Nqo3 form the dehydrogenase domain for accepting two electrons from NADH (see Supplemental Figure 1A online). Electrons are transferred to menaquinone at its binding site in a membrane-embedded subunit through iron-sulfur clusters in the peripheral arm.
NAD(P)H-oxidizing subunits corresponding to Nqo1-Nqo3 have not been found in chloroplasts or cyanobacteria even after the recent progress in understanding their structure. The identities of electron donors to NDH are also still debated (Friedrich et al., 1995; Friedrich and Weiss, 1997; Shikanai, 2007b). Identification of novel peripheral subunits of NDH is necessary for the clarification of the mechanism of electron input to NDH from PSI. Recently, we identified novel NDH subunits in NDH-PSI by shotgun proteomics and reverse genetics (Peng et al., 2009). Here, we focus on three proteins detected in wild-type NDH-PSI but not in its intermediate complex-lacking subcomplex A in the ndhl mutant, as candidate peripheral subunits of NDH (Peng et al., 2009; see Supplemental Data Set 1 online). CRR31 contributes to the formation of an Fd binding site of NDH and probably interacts with NDH via J-protein, CRRJ, and J-like protein, CRRL, which are novel subunits of NDH subcomplex A. A complex structure of NDH-PSI is summarized in Supplemental Figure 1B online; this includes the information reported in this article. We propose that chloroplast NDH accepts electrons from Fd and functions as PGR5-PGRL1 complex-independent Fd:PQ oxidoreductase in Arabidopsis.
RESULTS
The crr31, crrj, and crrl Mutants Are Specifically Impaired in Chloroplast NDH Activity
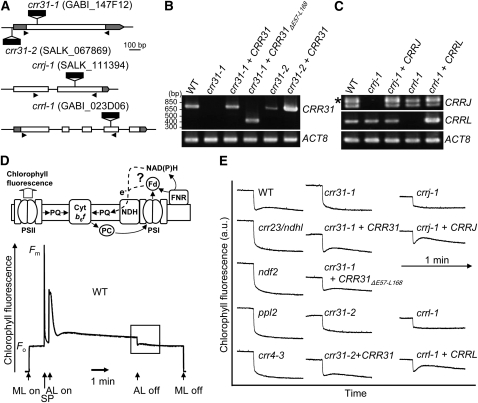
In the absence of subcomplex A, other parts of NDH-PSI accumulate stably and are detected as band II in blue-native (BN) gels, whereas full-size NDH-PSI is detected as band I (see Supplemental Figure 1 online; Peng et al., 2009). Because the putative subunits involved in electron donor binding are likely to associate with subcomplex A, we focused on unknown proteins At4g23890, At4g09350, and At5g21430, which were detected in band I but were absent from band II in our mass analysis (Peng et al., 2009; see Supplemental Data Set 1 online). They are shown to be components essential for NDH activity in this work; thus, we refer to them as CRR31, CRRJ, and CRRL. CRR31 does not contain any introns; T-DNA was inserted into the coding region (crr31-1) and 5′ untranslated region (crr31-2) (Figure 1A). RT-PCR showed leaky accumulation of transcript in crr31-2, whereas crr31-1 is a knockout allele (Figure 1B). CRRJ and CRRL consist of two and five exons, respectively, and T-DNA was inserted into the second exon of CRRJ (crrj-1) and the fourth exon of CRRL (crrl-1) (Figure 1A). RT-PCR did not detect any transcript in either mutant (Figure 1C).
Figure 1.
Identification of Novel NDH-Deficient Mutants.
(A) Structure of CRR31, CRRJ, and CRRL. Positions of T-DNA insertions are indicated. Open and gray boxes indicate exons and untranslated regions, respectively. Positions of primers used for RT-PCR in (B) and (C) are indicated by arrowheads.
(B) RT-PCR analysis of CRR31 mRNA in wild-type (WT), crr31, and transgenic lines. ACT8 was amplified as a control.
(C) RT-PCR analysis of CRRJ and CRRL mRNA in wild-type, crrj-1, crrl-1, and transgenic lines. The asterisk indicates the position of cDNA from prematured CRRJ RNA.
(D) Analysis of NDH activity by monitoring the transient increase in chlorophyll fluorescence after turning off AL. The schematic model indicates the electron transport from an unidentified electron donor, Fd or NAD(P)H, to the PQ pool via NDH. A typical trace of chlorophyll fluorescence in the wild type is shown. Leaves were exposed to AL (150 μmol photons m−2 s−1) for 5 min, and the subsequent transient increase in chlorophyll fluorescence (boxed area) was monitored in the dark. Fm, maximum chlorophyll fluorescence; Fo, minimal chlorophyll fluorescence; ML, measuring light; SP, saturating light pulse of white light.
(E) Monitoring of in vivo NDH activity by chlorophyll fluorescence. The boxed area in (D) is magnified in (E). a.u., arbitrary units. The fluorescence levels were normalized by the Fm levels.
Chloroplast NDH mediates electron flow from stromal reductants to PQ (Figure 1D). After actinic light (AL) illumination, NDH still donates electrons to PQ in the dark, and this PQ reduction can be monitored as a transient increase in chlorophyll fluorescence (Burrows et al., 1998; Shikanai et al., 1998) (Figure 1D). The crr23 mutant is defective in NdhL, a subunit of subcomplex A (Shimizu et al., 2008), and this postillumination increase in chlorophyll fluorescence was arrested in this mutant (Figure 1E). In crr31-1, crr31-2, crrj-1, and crrl-1, the transient increase in chlorophyll fluorescence was absent, as in crr23, indicating that CRR31, CRRJ, and CRRL are required for NDH activity.
The wild-type genomic sequences of CRR31, CRRJ, and CRRL were introduced into the corresponding mutants (Figures 1B and 1C). All the transformations fully complemented the postillumination increase in chlorophyll fluorescence, confirming that each T-DNA insertion resulted in the absence of NDH activity (Figure 1E). We also analyzed several chlorophyll fluorescence parameters that reflect even subtle alterations of photosynthetic electron transport (see Supplemental Figure 2 online). The results are consistent with the phenotypes of other crr mutants specifically defective in NDH (Shimizu et al., 2008; Peng et al., 2009), reflecting its minor contribution to photosynthesis in growth chamber conditions.
CRR31 Is a Peripheral Thylakoid Membrane Protein That Accumulates Independently of the Other Parts of NDH
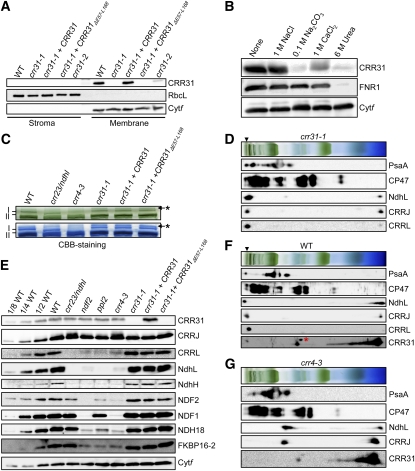
Localization of CRR31 was analyzed in protein blots with antibodies raised against mature CRR31 (Figure 2A). Chloroplasts were isolated from the wild type, crr31 alleles, and crr31-1 complemented by the introduction of the wild-type genomic CRR31 (crr31-1+CRR31) and further fractionated into the stroma and the membrane fraction containing thylakoids and envelopes. In the wild type, the antibody detected a protein of ~27 kD in the membrane fraction. This signal was absent in crr31-1 but was detected in crr31-1+CRR31. Consistent with the low-level accumulation of transcript (Figure 1B), a trace amount of CRR31 was detected in crr31-2. To clarify whether CRR31 is a peripheral or integral thylakoid protein, chloroplast membranes isolated from wild-type plants were incubated in 1 M NaCl, 0.1 M Na2CO3, 1 M CaCl2, and 6 M urea to release membrane-associated proteins (Figure 2B). CRR31 was significantly released from thylakoids under alkaline pH (0.1 M Na2CO3), saline (1 M CaCl2), and denaturing (6 M urea) conditions. During the treatments, an integral protein, Cytf, was retained in the membranes, while a stroma-side peripheral protein, Fd-NADP+ reductase 1 (FNR1), was released only in the presence of 6 M urea. These results indicate that CRR31 is a peripheral thylakoid protein.
Figure 2.
Characterization of crr31.
(A) Immunodetection of CRR31. Chloroplasts were isolated from the wild type (WT), crr31 alleles, and transgenic lines as indicated. Stroma and membrane protein extract corresponding to 2 μg chlorophyll were loaded. Ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit (RbcL) and cytochrome f (Cytf) were detected as a loading control of stroma and thylakoid proteins, respectively.
(B) Immunodetection of CRR31 in chloroplast membranes washed with salt or alkaline solutions. Chloroplast membranes isolated from the wild type were incubated with 1 M NaCl, 0.1 M Na2CO3, 1 M CaCl2, or 6 M urea for 30 min on ice and then the membranes were collected by ultracentrifugation. Membrane proteins corresponding to 2 μg chlorophyll were loaded. FNR1 and Cytf were detected as controls of peripheral membrane protein and integral membrane protein, respectively.
(C) Analysis of the NDH-PSI supercomplex by BN-PAGE. Magnification of the top part of the BN-PAGE gel in Supplemental Figure 3 is shown. The BN-PAGE gel (top panel) was stained with Coomassie Brilliant Blue (CBB) (bottom panel). The positions of bands I and II are indicated based on Peng et al. (2009). Asterisks indicate bands probably corresponding to aggregated proteins.
(D) Immunodetection of CRRJ, CRRL, and NDH-PSI in 2D-BN-SDS-PAGE. Protein complexes isolated from crr31-1were separated by BN-PAGE and further subjected to 12.5% SDS-PAGE. The position of band I is indicated by an arrowhead.
(E) Immunodetection of CRR31, CRRJ, CRRL, and NDH subunits in various genotypes as indicated across the top. Membrane protein extract corresponding to 2 μg chlorophyll was loaded onto each lane, as well as a dilution series of wild-type proteins. Antibodies used are indicated on the right. Cytf was detected as a loading control.
(F) and (G) Immunodetection of CRR31, CRRJ, CRRL, and NDH-PSI in 2D-BN-SDS-PAGE. Protein complexes isolated from the wild type (F) and crr4-3 (G) were separated as in (E). The position of band I is indicated by an arrowhead. A red asterisk indicates a nonspecific signal.
To analyze the localization of CRR31 in thylakoid membranes, high molecular weight protein complexes were solubilized in 1% dodecyl-maltoside and separated in BN gels (Figure 2C; see Supplemental Figure 3 online). As previously reported (Peng et al., 2009), a high molecular weight green band (band I) was absent in crr4-3, where NDH-PSI was destabilized in the absence of a membrane subunit, NdhD (Kotera et al., 2005). In crr23, which is defective in NdhL, band I was replaced by band II, corresponding to NDH-PSI lacking subcomplex A. However, band I was detected in crr31-1, whereas band II was not, suggesting that CRR31 is not required for stabilizing NDH-PSI (Figures 2C and 2D).
To analyze the impact of the crr31 defect on the accumulation of each NDH subunit, protein blots were probed with antibodies against NDH subunits NdhH and NdhL (subcomplex A); NDF1, NDF2, and NDH18 (subcomplex B); and FKBP16-2 (lumen subcomplex) (see Supplemental Figure 1B online). As previously reported (Peng et al., 2009), subunit levels were lower in the mutant backgrounds, except for the levels of NDF1, NDH18, and FKBP16-2 in crr23 (Figure 2E). By contrast, levels of any subunits were not affected in crr31-1. This observation is consistent with the results of the BN gels (Figure 2C), indicating that CRR31 is not required for the accumulation of the other parts of NDH-PSI. Furthermore, accumulation of CRR31 was not affected in the mutants defective in NDH subunits crr23, ndf2, ppl2, and crr4-3 (Figure 2E).
Although CRR31 and NDH-PSI accumulate independently of each other, they may interact in vivo because CRR31 was discovered in band I in a BN gel. To test this possibility, the BN gel was further separated by two-dimensional (2D) SDS-PAGE (Figure 2F). As previously reported (Peng et al., 2009), band I included both PsaA and NdhL. Consistent with the fact that CRR31 was discovered in band I (see Supplemental Data Set 1 online), a trace level of CRR31 was detected in band I. However, the majority of CRR31 was discovered in a putative free form, and a small amount of CRR31 was also detected in a 300-kD complex (Figure 2F). The 300-kD CRR31 complex does not contain NdhL or PsaA, and it accumulated in crr4-3 (Figure 2G). Due to its fragile nature, we could not conclude that CRR31 associates with NDH-PSI simply based on these biochemical analyses.
CRR31 Is a Novel Src Homology 3 Domain-Like Fold Protein Conserved in Phototrophs
CRR31 encodes a protein consisting of 250 amino acids, and the first 48 amino acids are predicted to be a plastid-targeting signal (http://www.cbs.dtu.dk/services/TargetP/) (Figure 3A). Proteins homologous to CRR31 are found in other terrestrial plants and also in cyanobacteria (see Supplemental Figure 4 online) but not in nonphototrophs. The green alga Chlamydomonas reinhardtii, which does not have chloroplast NDH, does not have the CRR31 homolog (http://genome.jgi-psf.org/Chlre4/Chlre4.home.html). Consistent with the analysis of salt-washed thylakoids (Figure 2B) suggesting that CRR31 is a peripheral thylakoid protein, CRR31 does not have any transmembrane domains. The C-terminal region of CRR31 is highly conserved among plants, as well as among cyanobacteria, but CRR31 proteins of terrestrial plants have a long N-terminal extension, which includes a tandem repeat sequence (Figure 3A; see Supplemental Figure 4 online). Eukaryotic phototroph-specific N-terminal extension is also observed in PsaE and PsaD, which are stroma-side peripheral subunits of PSI (Varotto et al., 2000; Ihnatowicz et al., 2004). To assess the function of this N-terminal extension, the CRR31ΔE57-L168 gene encoding the transit peptide fused to the C-terminal domain of CRR31 was introduced into crr31-1 (Figure 1B). The transformation complemented the postillumination increase in chlorophyll fluorescence, although the extent was slightly lower than that in the wild type (Figure 1E, crr31-1+CRR31ΔE57-L168). This result suggests that the N-terminal extension of CRR31 is not essential for NDH activity, but it might be required for the maximum activity or the stability of CRR31, which we could not evaluate in vivo due to the nature of the antibody (Figure 2A).
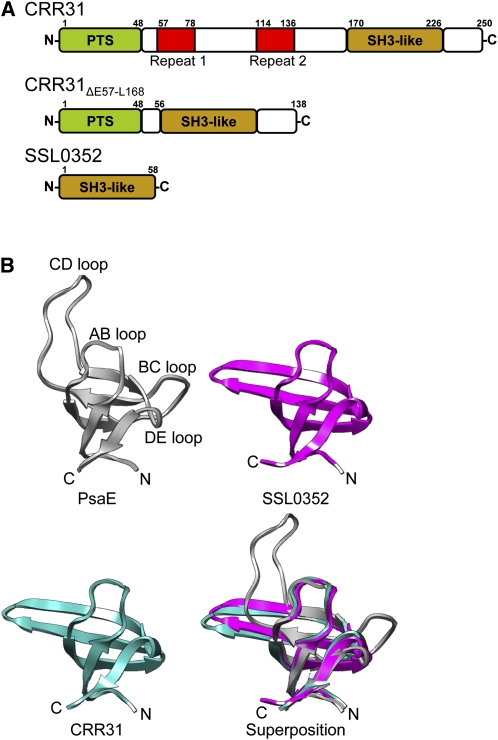
Figure 3.
Structures of CRR31, SSL0352, and PsaE.
(A) Schematic structures of CRR31, CRR31ΔE57-168, and Synechocystis SSL0352. Plastid targeting signal (PTS), repeat sequences 1 and 2, and SH3 domain-like fold (SH3-like) are indicated by green, red, and brown boxes, respectively.
(B) Comparison of the tertiary structures of the SH3 domain-like fold in PsaE, SSL0352, and CRR31. The tertiary structures of Synechococcus PsaE (PDB ID: 1PSAE) (dark gray), Synechocystis SSL0352 (PDB ID: 3C4S) (magenta), and CRR31 (cyan) were prepared by UCSF Chimera. The tertiary structure of the C-terminal region (L168-E223) of CRR31 was predicted by SwissModel using SSL0352 as a template.
SSL0352 of Synechocystis sp PCC 6803 is homologous to the C-terminal region of Arabidopsis CRR31, and its tertiary structure was resolved by x-ray crystallography (PDB ID: 3C4S). SSL0352 is composed of five β-sheets and forms an Src homology 3 (SH3) domain-like structure (Figure 3B), which has been reported to be involved in protein–protein interactions (Kishan and Agrawal, 2005). Originally, the SH3 domain was identified as a conserved sequence of src (sarcoma) protein Tyr kinases, which are encoded in viral oncogenes (Mayer et al., 1988; Musacchio et al., 1992). Furthermore, the overall structure of SSL0352 was predicted to be similar to that of PsaE, a stroma-side peripheral subunit of PSI, by the VAST program. PsaE forms the Fd-docking site of PSI with PsaC and PsaD. The SH3 domain-like structure in PsaE is involved in interactions with Fd and PsaC, which contain Fe-S clusters (Sétif et al., 2002; Amunts and Nelson, 2009). CRR31 is an oxygenic phototroph-specific SH3 domain-like fold protein required for NDH activity and may form the Fd-docking site for NDH.
J-Protein CRRJ and J-Like Protein CRRL Are Thylakoid Proteins Essential for NDH Activity
CRRJ and CRRL encode proteins composed of 249 and 218 amino acids, respectively (Figure 4A; see Supplemental Figure 5 online). The first 45 and 53 residues, respectively, were predicted by TargetP to be plastid-targeting signals. TMHMM and SOSUI programs predicted one transmembrane domain at the C-terminal regions of both proteins. Furthermore, SMART (http://smart.embl-heidelberg.de/) and Pfam 24.0 (http://pfam.sanger.ac.uk/) analyses predicted that CRRJ and CRRL have J-domain or J-domain-like structure in the middles of their mature forms. No homologs of CRRJ and CRRL were identified in cyanobacteria or in Chlamydomonas by BLAST.
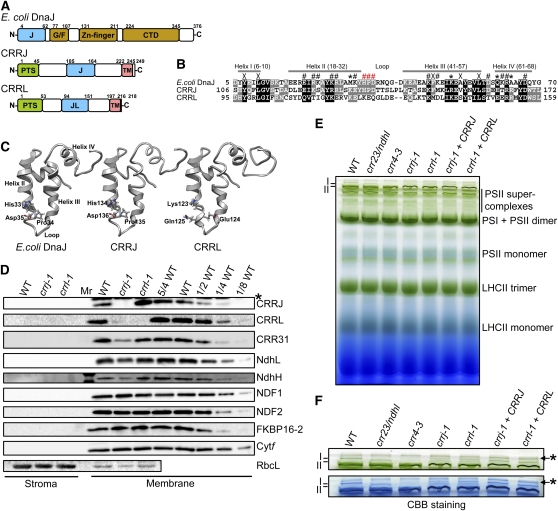
Figure 4.
Characterization of crrj-1 and crrl-1.
(A) Schematic structures of CRRJ, CRRL, and E. coli DnaJ. Boxes indicate the positions of plastid targeting signal (PTS), J- and J-like domains (J and JL), transmembrane domains (TM), Gly/Phe-rich motif (G/F), zinc-finger, and DnaJ C-terminal domains (CTD).
(B) Alignment of amino acid sequences of J- and J-like domains in E. coli DnaJ, CRRJ, and CRRL. The positions of helices I to IV in DnaJ are indicated by gray bars above sequences. Symbols above sequences indicate residues believed to be important in E. coli DnaJ for maintaining J-domain structure (X), for binding to Hsp70 chaperones (#), and for the specificity of this interaction (*) (Hennessy et al., 2000). The HPD motif is indicated by red #. The sequences were aligned by ClustalW2.
(C) Homology modeling of the tertiary structures of the J-domain in CRRJ and J-like domain in CRRL. The homology modeling was done by SwissModel using the J-domain of E. coli DnaJ (PDB ID: 1bq0) as a template. The models were constructed based on the amino acid sequences from Ser-103 to Gln-180 in CRRJ and from Thr-92 to Ile-176 in CRRL.
(D) Immunodetection of CRRJ, CRRL, CRR31, and NDH subunits. Chloroplasts were isolated from various genotypes as indicated across the top and further fractionated to stroma and membrane fractions. Protein extracts corresponding to 2 μg chlorophyll were loaded onto each lane as well as a dilution series in wild-type (WT) protein. Antibodies used are indicated on the right. RbcL (stroma) and Cytf (thylakoids) were detected as loading controls. An asterisk indicates nonspecific signal.
(E) Analysis of thylakoid protein complexes isolated by BN-PAGE. Chloroplasts were isolated from various genotypes as indicated across the top. Thylakoid protein extract corresponding to 10 μg chlorophyll was loaded onto each lane. The positions of bands I and II are indicated.
(F) Magnification of the top part of the BN-PAGE gel in (D). BN-PAGE gel (top panel) was stained with Coomassie Brilliant Blue (bottom panel). The positions of bands I and II are indicated. Asterisks indicate bands probably corresponding to aggregated proteins.
J-proteins are members of molecular chaperone DnaJ/heat shock protein 40 (Hsp40) and are ubiquitously conserved in organisms. J-protein assists protein folding, disassembly, and translocation across membranes by collaborating with DnaK/Hsp70 (Walsh et al., 2004). J-protein stimulates ATPase activity of DnaK/Hsp70 and stabilizes the interaction between Hsp70 and its substrates. J-domains are critical for the interaction with the ATPase domain of DnaK/Hsp70 (Walsh et al., 2004). The J-protein family is classified into four types on the basis of their domain structure and the presence or absence of an HPD (His, Pro, and Asp) motif in their J-domains (Walsh et al., 2004; Rajan and D’Silva, 2009). In CRRJ, the J-domain is highly conserved, including the HPD motif. By contrast, the conservation is low in the corresponding region of CRRL (J-like domain), and the HPD motif is absent (Figure 4B). Neither protein has a G/F region, zinc-finger, or C-terminal domain (Figure 4A); thus, CRRJ and CRRL are classified as type III and IV J-proteins, respectively. By homology modeling based on the tertiary structure of Escherichia coli DnaJ (Pellecchia et al., 1996), the J-domain of CRRJ consists of four α-helices, and the HPD motif was predicted to be located between helices II and III, as in other J-proteins (Figure 4C).
Specific antibodies raised against mature CRRJ and CRRL detected proteins whose molecular masses were similar to those of predicted mature forms of CRRJ (23.4 kD) and CRRL (18.7 kD). Consistent with the fact that CRRJ and CRRL each contain a transmembrane domain, they are localized to the membrane fraction of chloroplasts (Figure 4D). The crrl-1 mutant does not accumulate CRRL, and the introduction of the wild-type genomic CRRL complemented the protein accumulation (see Supplemental Figure 6A online), as observed in the complementation in NDH activity (Figure 1E). Although the CRRJ level was higher in crrl-1 than in the wild type (Figure 4D), the increased CRRJ did not complement the CRRL function because crrl-1 lacked NDH activity (Figure 1E). By contrast, neither CRRJ nor CRRL accumulated in crrj-1 (Figure 4D). Introduction of the wild-type genomic CRRJ into crrj-1 restored the levels of both CRRJ and CRRL (see Supplemental Figure 6A online). We conclude that CRRJ is required for the accumulation of CRRL, but CRRJ can accumulate independently of CRRL.
The J-domain is highly conserved in CRRJ. To assess the function of the J-domain, a mutation was introduced into the HPD motif of CRRJ, and this CRRJH134Q was introduced into crrj-1 (see Supplemental Figure 6 online). The mutation did not affect the accumulation of CRRJ, but the level of CRRL was not complemented by the transformation (see Supplemental Figure 6A online). NDH activity was not complemented by the transformation either (see Supplemental Figure 6B online), indicating that the HPD motif is essential for CRRJ function.
CRRJ Is Required for the Accumulation of Both the Subcomplex A of NDH and CRR31
Like CRR31, CRRJ and CRRL were discovered in band I (NDH-PSI) in the BN gel (see Supplemental Data Set 1 online). To confirm their localization, protein blots of 2D-BN-SDS-PAGE were probed with antibodies. As was the case for NdhL, both CRRJ and CRRL were specifically detected in the position of band I, suggesting that both proteins are components of NDH-PSI (Figure 2F).
Subsequently, we analyzed the accumulation of CRRJ and CRRL in the mutants lacking NDH. Although accumulation of CRRJ was not affected in any mutant background, CRRL was destabilized in the absence of NDH (Figure 2E), suggesting that CRRL interacts with NDH in thylakoid membranes.
If CRRL and CRRJ are subunits of NDH, their absence may destabilize NDH-PSI. To test this possibility, the levels of NDH subunits were analyzed in crrj-1 and crrl-1. crrl-1 accumulates the wild-type level of NDH subunits, indicating that CRRL is not required for stabilizing NDH (Figure 4D). By contrast, the levels of NdhH and NdhL were reduced to 25 to 50% of the wild-type levels in crrj-1 (Figure 4D). In crrj-1 transformed with the mutant CRRJH134Q, the levels of NdhH and NdhL were similarly affected (see Supplemental Figure 6 online). The levels of NDF1 and NDF2 (subcomplex B) and FKBP16-2 (lumen subcomplex) were not affected (Figure 4D); these results suggest that CRRJ is required for the accumulation of subcomplex A of NDH. Although CRRL is not required for stabilizing NDH subunits (Figure 4D), both bands I and II were detected in a BN gel in the crrl-1 and crrj-1 mutants (Figures 4E and 4F). Subcomplex A may dissociate from NDH-PSI in a BN gel in crrl-1, but CRRL is also likely to stabilize subcomplex A in vivo.
In summary, the accumulation of CRRL depends on CRRJ (Figure 4D) and also on NDH (Figure 2E). Although CRRJ accumulates independently of CRRL and NDH (Figures 2D and 4D), it is required for the accumulation of subcomplex A of NDH (Figure 4D). Because CRRJ is likely to be a functional J-protein, we do not eliminate the possibility that CRRJ plays a role in the folding of subcomplex A subunits. However, CRRJ comigrated exclusively with NDH-PSI in the BN gel, implying that CRRJ is associated with subcomplex A of NDH-PSI. It is also highly probable that CRRL is in this complex. We propose that CRRJ and CRRL are novel subunits of subcomplex A of NDH.
CRR31 accumulates independently of NDH-PSI, and it was detected mainly in a free form or in the putative 300-kD complex, although a trace level of CRR31 was detected in band I (Figure 2F). This contrasts with the fact that CRRJ and CRRL were detected exclusively in band I (Figure 2F). However, the CRR31 level was decreased to ~50% of the wild-type level in crrj-1, although the accumulation was not affected in crrl-1 (Figure 4D; see Supplemental Figure 6 online). The level of reduction is comparable to that of subcomplex A subunits NdhH and NdhL (Figure 4D), suggesting that the accumulation of CRR31 partially depends on CRRJ. We do not eliminate the possibility that CRRJ is required for the folding of CRR31, but a trace level of CRR31 was detected in band I, implying that CRR31 interacts with NDH-PSI via CRRJ. In the absence of the membrane subcomplex in crr4-3, CRRJ comigrated with NdhL in the BN gel, possibly in a putative partially stable subsupercomplex that was reported previously (Peng et al., 2009; Figure 2G). All the results support the hypothesis that CRRJ interacts with subcomplex A.
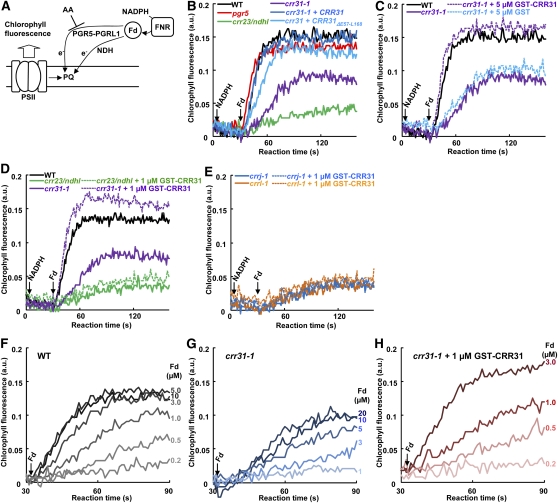
CRR31 Is Required for the Efficient Operation of Fd-Dependent PQ Reduction in Vitro
In PSI cyclic electron transport, electrons are recycled to PQ via two pathways that depend on the PGR5-PGRL1 complex and NDH. The activity can be monitored as Fd-dependent PQ reduction in the ruptured chloroplast system (Munekage et al., 2004), where NADPH is needed to reduce Fd via the reverse reaction of FNR (Miyake and Asada, 1994; Figure 5A). To monitor NDH-dependent PQ reduction specifically, antimycin A (AA) was added to inhibit PGR5-PGRL1–dependent PQ reduction in all of the assays except for pgr5. The addition of NADPH did not reduce PQ (Figure 5B); this is also true for NADH (Okegawa et al., 2008), implying that NDH does not accept electrons from NAD(P)H. In pgr5, PQ reduction activity was similar to that in the wild type in the presence of AA. In crr23, PQ reduction activity was almost completely inhibited. Although PQ reduction activity was higher in crr31-1 than in crr23, it was significantly lower than that in the wild type (Figure 5B). Since the Fd-dependent PQ reduction rate was slow in this system, the final PQ reduction level was lower in crr31-1 than in the wild type due to the competition with unknown PQ oxidizing reactions. Transformation of crr31-1 with wild-type genomic CRR31 fully complemented PQ reduction activity, whereas the PQ reduction activity was slightly lower in crr31-1 transformed with CRR31ΔE57-L168 (Figure 5B). This result is consistent with the observation that the postillumination increase in chlorophyll fluorescence was slightly lower in crr31-1 complemented by CRR31ΔE57-L168 (Figure 1E). We conclude that NDH activity monitored in ruptured chloroplasts was greatly but not completely inhibited in crr31-1.
Figure 5.
In Vitro Reconstitution of Active NDH in crr31-1 Ruptured Chloroplasts.
(A) Schematic model of Fd-dependent PQ reduction in ruptured chloroplasts. AA inhibits PGR5-PGRL1 complex-dependent PQ reduction.
(B) to (H) Fd-dependent PQ reduction activities were measured in the presence of 5 μM AA. a.u., arbitrary units.
(B) Fd-dependent PQ reduction activity in ruptured chloroplasts isolated from various genotypes as indicated. Increase in chlorophyll fluorescence level (Fo′) was monitored after consecutive addition of 0.25 mM NADPH and 5 μM Fd under weak illumination (1 μmol photons m−2 s−1) in osmotically ruptured chloroplasts (20 μg chlorophyll mL−1). The increase in fluorescence reflects NDH-dependent PQ reduction in the presence of AA. WT, wild type.
(C) Restoration of Fd-dependent PQ reduction activity in ruptured chloroplasts isolated from crr31-1 by the addition of GST-CRR31. Ruptured chloroplasts from crr31-1 were incubated with 5 μM GST or GST-CRR31 for 5 min prior to the consecutive addition of NADPH and Fd.
(D) and (E) Restoration of Fd-dependent PQ reduction activity with GST-CRR31 depends on NDH. Ruptured chloroplasts from crr31-1 and crr23/ndhl (D) and from crrj-1 and crrl-1 (E) were preincubated with 1 μM GST-CRR31 for 5 min, and the reaction was started as in (B).
(F) to (H) Fd concentration dependence of PQ reduction in ruptured chloroplasts isolated from the wild type (F), crr31-1 (G), and crr31-1 in the presence of 1 μM GST-CRR31 (H). Fd concentrations added to the reaction are indicated to the right of each curve.
Because NDH-PSI, which includes CRRJ and CRRL, is stable in crr31 (Figures 2D and 2F), it may be possible to reconstruct the active supercomplex by adding CRR31 to ruptured chloroplasts isolated from crr31. To test this possibility, recombinant CRR31 was expressed as a fusion protein with glutathione S-transferase (GST) in E. coli and was affinity purified. In the presence of AA, the addition of GST-CRR31 into the ruptured chloroplasts of crr31-1 restored PQ reduction activity to the wild-type level (Figure 5C). The addition of GST alone did not affect the activity, indicating that the complementation depended on CRR31. These results suggest that all the components required for NDH-dependent PQ reduction, except for CRR31, are present in crr31-1, and the addition of CRR31 can complement NDH activity.
To confirm that the addition of CRR31 activates PQ reduction activity via NDH, we added GST-CRR31 to ruptured chloroplasts isolated from crr23, which does not accumulate subcomplex A. The addition of CRR31 did not increase PQ reduction activity, indicating that CRR31 activates PQ reduction activity via NDH (Figure 5D).
We also added GST-CRR31 to ruptured chloroplasts isolated from crrj-1 and crrl-1 (Figure 5E). Accumulation of NDH-PSI was partially affected in crrj-1, whereas crrl-1 accumulated the wild-type level of other NDH subunits (Figure 2E). Despite the presence of NDH-PSI in crrj-1 and crrl-1, the addition of GST-CRR31 did not activate PQ reduction activity as in crr23 (Figure 5E). This result indicates that both CRRJ and CRRL are required for NDH activity in ruptured chloroplasts, consistent with the in vivo result (Figure 1E). These results suggest that CRRJ and CRRL are essential components of NDH activity, but CRR31 is an activator of the activity.
The predicted tertiary structure of CRR31 is similar to that of PsaE, a PSI subunit forming the Fd binding site (Sétif et al., 2002). Our assay system using ruptured chloroplasts implies that Fd, rather than NAD(P)H, is an electron donor to NDH (Munekage et al., 2004). Based on of these results, we hypothesize that CRR31 forms the Fd binding site in NDH-PSI and increases the affinity of NDH for Fd. To test this possibility, the effect of Fd concentration on PQ reduction activity was analyzed. In the wild type, 5 μM Fd was enough to saturate the reduction activity of PQ (Figure 5F). In crr31-1, however, even in the presence of 20 μM Fd, PQ reduction activity was comparable to that in the wild type with 1 μM Fd (Figure 5G). The addition of 1 μM GST-CRR31 to the ruptured chloroplasts of crr31-1 complemented PQ reduction activity in the presence of 3 μM Fd even more than did 20 μM Fd (Figure 5H). Taking all of these results together, we conclude that CRR31 is a factor required for the efficient operation of Fd-dependent PQ reduction in vitro and also that Fd is an electron donor to chloroplast NDH.
CRR31 Is Required for NDH Function in Vivo
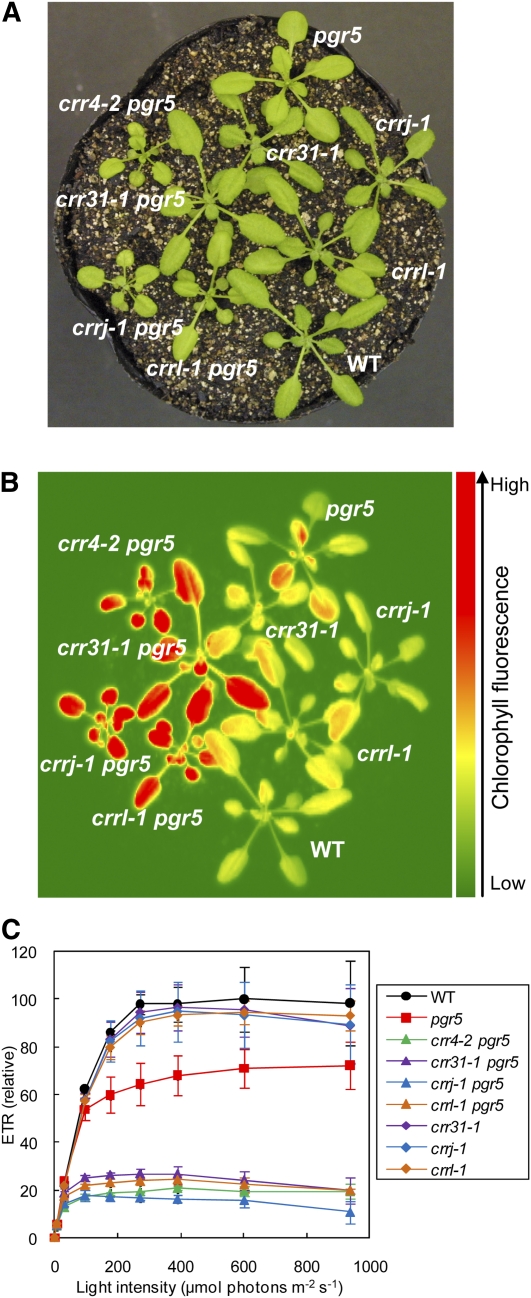
CRR31 is not essential for NDH activity, but its efficient operation required higher concentrations of Fd in the absence of CRR31 (Figure 5). To analyze the contribution of CRR31 to NDH activity in vivo, we created the double mutants crr31-1 pgr5, crrj-1 pgr5, and crrl-1 pgr5 (Figure 6). Because NDH is the machinery for stress resistance, the crr mutants specifically defective in NDH do not show particular phenotypes except for the minor alteration in chlorophyll fluorescence under growth chamber conditions (Shimizu et al., 2008). However, NDH is indispensable in pgr5, which is defective in the main pathway of PSI cyclic electron transport (Munekage et al., 2004), and the double mutant crr4-2 pgr5 showed severe reduction in growth and also showed high chlorophyll fluorescence (Figures 6A and 6B). Consistent with these phenotypes, electron transport is also impaired in crr4-2 pgr5 (Figure 6C). As in other single crr mutants, crr31-1, crrj-1, and crrl-1 did not exhibit any phenotype (Figure 6; see Supplemental Figure 7 online). Consistent with the fact that CRRJ and CRRL are required for NDH activity, the double mutants crrj-1 pgr5 and crrl-1 pgr5 showed definite phenotypes, as did crr4-2 pgr5 (Figure 6; see Supplemental Figure 7 online). Although the growth of crr31-1 pgr5 was comparable to that of the wild type, this double mutant showed high chlorophyll fluorescence (Figures 6A and 6B). Electron transport was also affected in crr31-1 pgr5, although the phenotype was milder than in the other double mutants (Figure 6C). These results suggest that CRR31 function is required for NDH activity in vivo.
Figure 6.
Characterization of crr31-1 pgr5, crrj-1 pgr5, and crrl-1 pgr5 Double Mutants.
(A) Growth phenotype of the double mutants. Seedlings were cultured at 50 μmol photons m−2 s−1 for 3 weeks after germination. WT, wild type.
(B) High chlorophyll fluorescence phenotype of crr31-1 pgr5, crrj-1 pgr5, and crrl-1 pgr5. Dark-adapted wild-type and mutant seedlings were illuminated at 100 μmol photons m−2 s−1 for 1 min, and then a chlorophyll fluorescence image was captured by a charge-coupled device camera.
(C) Light intensity dependence of relative electron transport rate (ETR) in wild-type and mutant leaves. ETR is presented as values relative to the maximum ETR in the wild type (100%). Means ± sd (n = 3).
DISCUSSION
Three novel components (CRR31, CRRJ, and CRRL) are required for NDH activity. Consistent with their specific function in NDH activity, their putative orthologs are highly expressed in maize (Zea mays) bundle sheath cells, which accumulate high levels of NDH (Friso et al., 2010). Based on the following experimental evidence, we conclude that CRRJ and CRRL are NDH subunits included in subcomplex A. (1) CRRJ and CRRL are detected in band I but not in band II when NDH-PSI was analyzed on a BN gel (see Supplemental Data Set 1 online). (2) CRRJ comigrates with NdhL as a putative partially stable complex in crr4-3 (Figure 2G). (3) The accumulation of CRRL depends on NDH (Figure 2E). (4) CRRJ is required for the accumulation of CRRL and NDH (Figure 4D). (5) The NDH-PSI level is lower in crrl-1 than in the wild type (Figure 4F). However, we do not eliminate the possibility that CRRJ assists the protein folding of subcomplex A subunits or CRR31 by interacting with DnaK/Hsp70 via its J-domain. The Arabidopsis genome encodes two copies of plastid-targeting Hsp70, and knockout of either gene did not result in defective NDH activity. The J-domain was also suggested to mediate heterodimer formation between J- and J-like proteins (Frazier et al., 2004; Kozany et al., 2004). In a protein translocator complex present in the inner membranes of yeast mitochondria, Tim16/Pam16 (J-like protein) interacts with Tim14/Pam18 (J-protein) to inhibit the activation of Hsp70 ATPase by Tim14/Pam18 (Li et al., 2004; D’Silva et al., 2005). CRRL is unstable in the absence of CRRJ (Figure 4), suggesting an interaction between CRRJ and CRRL. The CRRJ level was higher in crrl-1 than in the wild type (Figure 4), implying that the CRRJ/CRRL heterodimer may be replaced by a CRRJ homodimer. This idea is supported by the observation that CRRJH134Q carrying a mutation in the J-domain also reduced the level of CRRL (see Supplemental Figure 6 online). However, CRRJ cannot complement the function of CRRL for NDH activity (Figure 1E), and subcomplex A was only partially stabilized because both bands I and II were detected in crrl-1 and crrj-1+CRRJH134Q (Figures 4E and 4F; see Supplemental Figure 6C online). We propose a model in which CRRJ and CRRL form a heterodimer to interact further with subcomplex A of NDH (see Supplemental Figure 1B online).
This work provides solid evidence for the debate on the electron donor to chloroplast NDH (Friedrich et al., 1995; Friedrich and Weiss, 1997; Shikanai, 2007b). Our results strongly suggest that NDH accepts electrons from Fd rather than NAD(P)H. To avoid confusion, we propose to keep using the name NDH but to make it stand for the NADH dehydrogenase-like complex instead of for NAD(P)H dehydrogenase. Our conclusion is based on the following results. (1) In ruptured chloroplasts, NDH-dependent PQ reduction is not induced by NADH (Okegawa et al., 2008) or NADPH but by Fd (Figure 5). (2) The addition of GST-CRR31 restores the Fd concentration dependency of NDH activity in crr31 ruptured chloroplasts (Figures 5F to 5H). This in vitro reconstitution of PQ reduction activity requires NDH (Figure 5D). (3) The reduction rate of PQ by NDH was saturated at 5 μM Fd (Figure 5F). It could be assumed that the affinity of NDH for Fd is roughly comparable to that of FNR (Km for Fd = 2.8 μM) (Yonekura-Sakakibara et al., 2000), although our assay depending on chlorophyll fluorescence may not linearly reflect the PQ reduction. Consistent with these results, the phenotype of crr31 pgr5 (Figure 6) suggests the in vivo contribution of CRR31 in increasing the affinity of NDH for Fd.
From homology to Synechocystis SSL0352, the tertiary structure of the C-terminal region of Arabidopsis CRR31 was predicted to have an SH3 domain-like fold, and the backbone structure was highly similar to that of PsaE (Figure 3B). PsaE is an Fd binding subunit of PSI, and the lack of PsaE increases the Kd of PSI for Fd by a factor of two (Barth et al., 1998). Highly conserved positive charges in the C-terminal region of CRR31 might be involved in the interaction with negatively charged Fd, as in PsaE (Barth et al., 2000; see Supplemental Figure 4 online). In contrast with CRRJ and CRRL, CRR31 is mainly present as free protein and also in the putative 300-kD complex in the BN gel (Figure 2F). Furthermore, CRR31 is stable in the absence of NDH, and NDH accumulates in the absence of CRR31 (Figure 2E). From these results, we do not eliminate the possibility that CRR31 or the putative 300-kD CRR31 complex weakly interacts with NDH in thylakoid membranes. In any case, the accumulation of CRR31 partially depends on CRRJ (Figure 4), and a trace level of CRR31 was detected in band I (Figure 2F). Although further biochemical evidence is needed to conclude its localization and binding stability with NDH in vivo, we illustrate CRR31 on CRRJ to form the Fd binding site of NDH (see Supplemental Figure 1B online). However, the site of Fd oxidation and the route of electrons to the electron carriers present in subcomplex A are still unclear. Since information is still lacking on the active part of Fd oxidation, we do not completely eliminate the possibility that other electron donors exist. Future work will focus on the most fragile part of NDH, which is possibly included in the putative 300-kD CRR31 complex (see Supplemental Figure 1B online).
Why did plants alter the electron donor for NDH from NAD(P)H to Fd? In mitochondria, the major site of reactive oxygen generation is FMN in the NADH-oxidizing subunits (Hirst et al., 2008). If chloroplast NDH is the machinery that counteracts oxidative stress in chloroplasts, the presence of FMN to oxidize NAD(P)H would have been counterproductive, as FMN is the site of reactive oxygen generation.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana (Columbia) was grown in soil in a growth chamber (50 μmol photons m–2 s–1, 16 h photoperiod, 23°C) for 3 to 4 weeks. The SIGnAL T-DNA express database (http://signal.salk.edu/cgi-bin/tdnaexpress) was used to find T-DNA insertion mutants for crr31, crrj-1, and crrl-1. The T-DNA insertion lines SALK_067869 (crr31-2) and SALK_111394 (crrj-1) were provided by the Salk Institute Genomic Analysis Laboratory. GABI_147F12 (crr31-1) and GABI_023D06 (crrl-1) were provided by GABI-Kat (http://www.gabi-kat.de).
In Vivo Chlorophyll Fluorescence Analysis
Three to four plants of each genotype were analyzed, and average values and standard deviations were calculated. Chlorophyll fluorescence was measured using a MINI-pulse-amplitude modulation portable chlorophyll fluorometer (MINI-PAM; Walz). Minimal fluorescence at open PSII centers in the dark-adapted state (Fo) was excited by a weak measuring light (650 nm) at a PFD of 0.05 to 0.1 μmol photons m−2 s−1. A saturating pulse of white light (800 ms, 8000 μmol photons m−2 s−1) was applied to determine the maximal fluorescence at closed PSII centers in the dark-adapted state (Fm) and during AL illumination (Fm′). The steady state fluorescence level (Fs) was recorded during AL illumination (5 to 1000 μmol photons m−2 s−1). Maximum quantum yield of PSII was calculated as Fv/Fm. NPQ was calculated as (Fm − Fm′)/Fm′. The quantum yield of PSII (ΦPSII) was calculated as (Fm – Fs)/Fm′ (Genty et al., 1989). ETR was calculated as ΦPSII × light intensity (μmol photons m−2 s−1). qL, the fraction of open PSII center, was calculated as {ΦPSII/(1 − ΦPSII)}×[(1 − Fv/Fm)/(Fv/Fm)]×(NPQ + 1) (Miyake et al., 2009). The transient increase in chlorophyll fluorescence after turning off AL was monitored as described in the legend for Figure 1 (Shikanai et al., 1998).
RT-PCR Analysis
Total RNA was prepared from rosette leaves using an RNeasy plant mini kit (Qiagen). Contaminating DNA was digested with DNase I. Total RNA (2 μg) was reverse transcribed with random hexamers in a PrimeScript first-strand cDNA synthesis kit (TaKaRa Bio) in a total volume of 20 μL. After 10-fold dilution of the reaction mixture, a 1-μL aliquot containing cDNA was used in a subsequent PCR with TaKaRa Ex Taq DNA polymerase (TaKaRa Bio). The PCRs were performed in a final volume of 50 μL containing 1.25 units of DNA polymerase and 10 pmol of each primer. PCR primers used for amplification of CRR31, CRRJ, CRRL, and ACT8 are listed in Supplemental Table 1 online. cDNA was amplified so as to include at least one intron to distinguish cDNA from genomic DNA. PCRs consisted of 30-s denaturation at 94°C, 20-s annealing at 60°C, and 1-min extension at 72°C. RT-PCR products were separated on a 1.2% agarose gel and detected by ethidium bromide staining. The number of cycles was optimized so that the abundance of products could be compared within the linear phase of amplification.
Vector Construction and Plant Transformation
For complementation, the wild-type genomic sequences containing CRR31, CRRJ, and CRRL were amplified by PCR with primers listed in Supplemental Table 1 online and cloned into pDONR/Zeo (Invitrogen) by BP clonase reaction (Invitrogen) and then transferred to the binary vector pGWB-NB1 by LR clonase reaction (Invitrogen). The resultant vectors were transformed into Agrobacterium tumefeciens C58 by electroporation, and the bacteria were used to transform each Arabidopsis mutant by the floral dip method (Martinez-Trujillo et al., 2004). CRR31ΔE57-L168 and CRRJH134Q were constructed using a Quikchange II site-directed mutagenesis kit (Stratagene) with primers listed in Supplemental Table 1 online.
Production of Polyclonal Antisera against CRR31, CRRJ, and CRRL
The cDNAs encoding the mature form of CRR31 (amino acids 49 to 250) and the soluble parts of CRRJ (amino acids 45 to 221) and CRRL (amino acids 54 to 196) were amplified by PCR with primers listed in Supplemental Table 1 online and cloned into pCR4Blunt-TOPO by a Zero Blunt TOPO PCR cloning kit (Invitrogen). To express recombinant proteins as His-tagged fusion proteins, cDNAs encoding CRR31 and CRRJ were digested with NdeI and HindIII and cloned into pET22b (Novagen), and cDNA encoding CRRL was digested with NdeI and BamHI and cloned into pET16b (Novagen). Escherichia coli Rosetta (DE3) pLysS cells (Novagen) were transformed with the resultant plasmids. Expression of the recombinant proteins in E. coli was induced by 1 mM isopropyl β-d-thiogalactopyranoside for 8 h at 37°C, and the cells were collected by centrifugation. The collected cells were disrupted by sonication for 10 min, with one pause of 5 min, on ice in 20 mL of binding buffer (20 mM potassium phosphate buffer, pH 7.4, that contained 40 mM immidazole, 500 mM NaCl, 4 M urea, and Complete EDTA-free protease inhibitor cocktail [Roche]). The lysate was centrifuged at 1800g for 10 min, and then the supernatant was centrifuged at 48,000g for 1 h at 4°C. The resulting supernatant was filtered through a 0.4-μm filter and loaded onto a 1-mL HisTrap FF crude column (GE Healthcare) equilibrated with binding buffer. After the column was washed with 20 bed volumes of binding buffer, bound His-tagged fusion proteins were eluted from the column with 5 bed volumes of elution buffer (20 mM potassium phosphate buffer, pH 7.4, containing 500 mM immidazole, 500 mM NaCl, and 4 M urea). The purity of the fusion proteins was examined by SDS-PAGE, and the proteins were used as antigens.
Purification of GST-CRR31 Fusion Protein
A cDNA encoding the mature form CRR31 was amplified by PCR using primers 5′-AAATTTGCAATTGCCATGGGTAAATTCAATCTATGGG-3′ and 5′-AAATTTAAGCTTTTATGGTGCTGCCTCTTCC-3′. The amplified DNA was digested with MunI and HindIII and then cloned into pET41b (Novagen) to express CRR31 as a fusion protein with GST at the N terminus in E. coli. The resulting plasmid, pET41b-CRR31, was introduced into the E. coli Rosetta (DE3) pLysS strain. The transformed E. coli cells were grown in 250 mL Terrific Broth (1.2% tryptone, 2.4% yeast extract, 0.4% glycerol, 72 mM K2HPO4, and 17 mM KH2PO4) supplemented with 100 μg mL−1 kanamycin and 60 μg mL−1 chloramphenicol at 37°C for 7 h in the presence of 1.5 mM isopropyl β-d-thiogalactopyranoside to induce the expression of GST-CRR31 fusion protein. The E. coli cells were disrupted in 20 mL of PBS (pH 7.3, 10 mM Na2HPO4, 1.8 mM KH2PO4, 140 mM NaCl, and 2.7 mM KCl) containing Complete protease inhibitor (Roche) by sonication. The lysate was centrifuged at 1800g for 10 min and then the supernatant was centrifuged at 48,000g for 1 h at 4°C. The resulting supernatant was filtrated with a 0.4-μm filter and loaded onto a 1-mL GSTTrap FF column (GE Healthcare) equilibrated with PBS buffer. After the column was washed with 20 bed volumes of PBS buffer, GST-CRR31 fusion protein was eluted from the column with 5 bed volumes of elution buffer (50 mM Tris-HCl, pH 8.0, containing 10 mM reduced glutathione). The eluate containing the fusion protein was applied to PD-10 (GE Healthcare) equilibrated with PBS buffer containing 20% (v/v) glycerol to remove glutathione. GST protein was also purified by the same procedure from E. coli cells harboring pET41b. The concentration of protein was determined as reported by Bradford (1976) using BSA as the standard. The purified GST-CRR31 fusion protein and GST were frozen in liquid nitrogen and stored at −80°C until used.
Isolation of Intact Chloroplasts and Chloroplast Membranes
Intact chloroplasts were purified from leaves of 3- to 4-week-old plants as previously described (Munekage et al., 2002). The purified chloroplasts were suspended in 20 mM HEPES-KOH, pH 7.6, containing 5 mM MgCl2 and 2.5 mM EDTA. The insoluble fraction containing thylakoids and envelopes was separated from the stroma fraction by centrifugation for 5 min at 15,000g. The concentration of chlorophyll was determined as described previously (Porra et al., 1989). The salt washes of thylakoids were done as described previously with minor modifications (Peng et al., 2006). Isolated chloroplast membranes were resuspended to a final concentration of 100 μg chlorophyll mL−1 in 10 mM HEPES-KOH, pH 8.0, containing 10 mM MgCl2, 330 mM sorbitol, and 1 mM phenylmethylsulfonyl fluoride supplemented with 1 M NaCl, 100 mM Na2CO3, 1 M CaCl2, or 6 M urea, respectively. The membrane fraction without supplements was used as a control. These suspensions were incubated on ice for 30 min with gentle mixing. After the treatment, the membranes were centrifuged at 100,000g for 2 h at 4°C, washed with 20 mM HEPES-KOH, pH 7.6, containing 5 mM MgCl2 and 2.5 mM EDTA, and then solubilized with SDS-PAGE sample buffer. The solubilized membranes were used for SDS-PAGE and immunoblot analyses.
SDS-PAGE and Immunoblot Analyses
Proteins separated by 12.5% (w/v) SDS-PAGE were electrotransferred onto polyvinylidene fluoride membranes. The antibodies were added, and the protein-antibody complexes were labeled using an ECL Plus Western blotting detection kit (GE Healthcare). The chemiluminescence was detected with a lumino-image analyzer LAS3000 (FUJIFILM) and analyzed by Multi Gauge Version 3.0 software (FUJIFILM).
BN-PAGE and 2D-PAGE Analysis
BN and 2D-PAGE were performed as described before (Shimizu et al., 2008). Solubilized thylakoid membranes with 1% (v/v) dodecyl-maltoside corresponding to 10 μg chlorophyll were subjected to 5 to 12% BN-PAGE. The images of BN-PAGE gels were captured with an image scanner, and gels were stained with Bio-Safe Coomassie stain (Bio-Rad). For 2D-PAGE analysis, lanes excised from BN gels were applied to 12.5% SDS-PAGE.
In Vitro Assay of Fd-Dependent PQ Reduction
In vitro assay of Fd-dependent PQ reduction was performed as described before with minor modifications (Okegawa et al., 2008). Intact chloroplasts (20 μg of chlorophyll mL−1) were osmotically ruptured in 50 mM HEPES-NaOH, pH 8.0, for each assay. NADPH (0.25 mM) and the indicated concentrations of spinach (Spinacia oleracea) Fd (Sigma-Aldrich) were added, and an increase in apparent minimum chlorophyll fluorescence (Fo′) was recorded using a Mini-Pam (Walz). Fluorescence levels were normalized by Fm levels. To measure NDH-dependent PQ reduction, 5 μM AA (Sigma-Aldrich) was added to the assay to inhibit PGR5-PGRL1–depenedent PQ reduction activity as previously reported (Okegawa et al., 2008). For reconstitution of the active NDH complex, ruptured chloroplasts from mature leaves of crr31-1 were preincubated with the indicated concentrations of purified GST-CRR31 for 5 min in the dark before measuring chlorophyll fluorescence in the presence of 5 μM AA.
Modeling of the Tertiary Structure of Proteins
The tertiary structures of Synechocystis sp PCC 6803 SSL0352 (PDB ID: 3C4S) and Synechococcus sp PCC 7002 PsaE (PDB ID: 1PSE) were obtained from the Research Collaboratory for Structural Bioinformatics PDB (http://www.pdb.org/pdb/home/home.do). The amino acid sequences of SSL0352 and CRR31 were aligned using ClustalW2 (http://clustalw.ddbj.nig.ac.jp/top-j.html), and the alignment was used as query for homology modeling of the tertiary structure of the C-terminal region of CRR31 by SwissModel using alignment mode (http://swissmodel.expasy.org/). The similarity of the tertiary structures of SSL0352 and PsaE was analyzed by VAST (http://www.ncbi.nlm.nih.gov/Structure/VAST/vast.shtml). UCSF Chimera and MatchMaker (http://www.cgl.ucsf.edu/chimera/) were used for the visualization and superposition of tertiary structures of proteins. The models of tertiary structures of J- and J-like domains of CRRJ and CRRL, respectively, were constructed using the structure of the J-domain of E. coli DnaJ (PDB ID: 1bq0) as a template.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At (Arabidopsis thaliana) CRR31 (AT4G23890), Vv (Vitis vinifera) CRR31 (XP_002269878), Pt (Populus trichocarpa) CRR31_1 (XP_002297973.1), PtCRR31_2 (XP_002304561.1), Os (Oryza sativa) CRR31 (NP_001059119.1, Os07g0196200), Sb (Sorghum bicolor) CRR31 (XP_002445975.1), Zm (Zea mays) CRR31 (NP_001143622.1), Pp (Physcomitrella patens subsp patens) CRR31 (XP_001770269.1), T. BP-1 (NP_681425.1, TLR0636, Thermosynechococcus elongatus BP-1), N. 7120 (NP_484698.1, ASR0654, Nostoc sp PCC 7120), S. 6803 (NP_442353.1, SSL0352, Synechocystis sp PCC 6803), At CRRJ (AT4G09350), Vv CRRJ (XP_002272533.1), Pt CRRJ (XP_002327350.1), Os CRRJ (NP_001067508.1), Sb CRRJ (XP_002449374.1), Zm CRRJ (NP_001148857.1), Pp CRRJ (XP_001768838.1), At CRRL (AT5G21430), Vv CRRL (XP_002281227.1), Pt CRRL (XP_002309471.1), Rc (Ricinus communis) CRRL (XP_002516125.1), Os CRRL (NP_001066812.2), Sb CRRL (XP_002443210.1), Zm CRRL (NP_001148857.1), Pp CRRL_1 (XP_001767272.1), and Pp CRRL_2 (XP_001754379.1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Schematic Models of Bacterial Complex I and Chloroplast NDH-PSI.
Supplemental Figure 2. Chlorophyll Fluorescence Parameters of Wild-Type, crr31-1, crr31-2, crrj-1, and crrl-1 Leaves.
Supplemental Figure 3. Analysis of Thylakoid Protein Complexes in crr31-1.
Supplemental Figure 4. A Multiple Alignment of Amino Acid Sequences of CRR31 Homologs from Plants and Cyanobacteria.
Supplemental Figure 5. Multiple Alignments of Amino Acid Sequences of CRRJ and CRRL Homologs from Plants.
Supplemental Figure 6. The HPD Motif Is Essential for CRRJ Function.
Supplemental Figure 7. Chlorophyll Fluorescence Parameters of Wild-Type, crr31-1 pgr5, crrj-1 pgr5, and crrl-1 pgr5 Leaves.
Supplemental Table 1. Primer List and Sequences.
Supplemental Data Set 1. Proteins Identified in Band I but Not in Band II by LC-MS/MS Analysis.
Acknowledgments
We thank Asako Tahara (Kyoto University) for her skilled technical support and Tsuyoshi Endo (Kyoto University), Amane Makino (Tohoku University), and Toshiharu Hase (Osaka University) for their gifts of antibodies. We also thank T. Nakagawa (Shimane University) for providing the pGWB-NB1 vector. This work was supported by grants 17GS0316, 22114509, and 22247005 from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, a grant from the Ministry of Agriculture, Forestry, and Fisheries of Japan (Genomics for Agricultural Innovation; GPN0008), and a grant (10010050006) from Japan Science and Technology Agency.
References
- Amunts A., Nelson N. (2009). Plant photosystem I design in the light of evolution. Structure 17: 637–650 [DOI] [PubMed] [Google Scholar]
- Arnon D.I., Allen M.B., Whatley F.R. (1954). Photosynthesis by isolated chloroplasts. Nature 174: 394–396 [DOI] [PubMed] [Google Scholar]
- Barth P., Guillouard I., Sétif P., Lagoutte B. (2000). Essential role of a single arginine of photosystem I in stabilizing the electron transfer complex with ferredoxin. J. Biol. Chem. 275: 7030–7036 [DOI] [PubMed] [Google Scholar]
- Barth P., Lagoutte B., Sétif P. (1998). Ferredoxin reduction by photosystem I from Synechocystis sp. PCC 6803: Toward an understanding of the respective roles of subunits PsaD and PsaE in ferredoxin binding. Biochemistry 37: 16233–16241 [DOI] [PubMed] [Google Scholar]
- Battchikova N., Aro E.-M. (2007). Cyanobacterial NDH-1 complexes: Multiplicity in function and subunit composition. Physiol. Plant. 131: 22–32 [DOI] [PubMed] [Google Scholar]
- Bradford M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Burrows P.A., Sazanov L.A., Svab Z., Maliga P., Nixon P.J. (1998). Identification of a functional respiratory complex in chloroplasts through analysis of tobacco mutants containing disrupted plastid ndh genes. EMBO J. 17: 868–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DalCorso G., Pesaresi P., Masiero S., Aseeva E., Schünemann D., Finazzi G., Joliot P., Barbato R., Leister D. (2008). A complex containing PGRL1 and PGR5 is involved in the switch between linear and cyclic electron flow in Arabidopsis. Cell 132: 273–285 [DOI] [PubMed] [Google Scholar]
- D’Silva P.R., Schilke B., Walter W., Craig E.A. (2005). Role of Pam16’s degenerate J domain in protein import across the mitochondrial inner membrane. Proc. Natl. Acad. Sci. USA 102: 12419–12424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo T., Shikanai T., Takabayashi A., Asada K., Sato F. (1999). The role of chloroplastic NAD(P)H dehydrogenase in photoprotection. FEBS Lett. 457: 5–8 [DOI] [PubMed] [Google Scholar]
- Efremov R.G., Baradaran R., Sazanov L.A. (2010). The architecture of respiratory complex I. Nature 465: 441–445 [DOI] [PubMed] [Google Scholar]
- Frazier A.E., et al. (2004). Pam16 has an essential role in the mitochondrial protein import motor. Nat. Struct. Mol. Biol. 11: 226–233 [DOI] [PubMed] [Google Scholar]
- Friedrich T., Steinmüller K., Weiss H. (1995). The proton-pumping respiratory complex I of bacteria and mitochondria and its homologue in chloroplasts. FEBS Lett. 367: 107–111 [DOI] [PubMed] [Google Scholar]
- Friedrich T., Weiss H. (1997). Modular evolution of the respiratory NADH:ubiquinone oxidoreductase and the origin of its modules. J. Theor. Biol. 187: 529–540 [DOI] [PubMed] [Google Scholar]
- Friso G., Majeran W., Huang M., Sun Q., van Wijk K.J. (2010). Reconstruction of metabolic pathways, protein expression, and homeostasis machineries across maize bundle sheath and mesophyll chloroplasts: Large-scale quantitative proteomics using the first maize genome assembly. Plant Physiol. 152: 1219–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genty B., Briantais J.M., Baker N.R. (1989). The relationship between quantum yield of photosynthetic electron transport and quenching of chlorophyll fluorescence. Biochim. Biophys. Acta 990: 87–92 [Google Scholar]
- Hennessy F., Cheetham M.E., Dirr H.W., Blatch G.L. (2000). Analysis of the levels of conservation of the J domain among the various types of DnaJ-like proteins. Cell Stress Chaperones 5: 347–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J., King M.S., Pryde K.R. (2008). The production of reactive oxygen species by complex I. Biochem. Soc. Trans. 36: 976–980 [DOI] [PubMed] [Google Scholar]
- Ihnatowicz A., Pesaresi P., Varotto C., Richly E., Schneider A., Jahns P., Salamini F., Leister D. (2004). Mutants for photosystem I subunit D of Arabidopsis thaliana: Effects on photosynthesis, photosystem I stability and expression of nuclear genes for chloroplast functions. Plant J. 37: 839–852 [DOI] [PubMed] [Google Scholar]
- Kishan K.V.R., Agrawal V. (2005). SH3-like fold proteins are structurally conserved and functionally divergent. Curr. Protein Pept. Sci. 6: 143–150 [DOI] [PubMed] [Google Scholar]
- Kotera E., Tasaka M., Shikanai T. (2005). A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature 433: 326–330 [DOI] [PubMed] [Google Scholar]
- Kozany C., Mokranjac D., Sichting M., Neupert W., Hell K. (2004). The J domain-related cochaperone Tim16 is a constituent of the mitochondrial TIM23 preprotein translocase. Nat. Struct. Mol. Biol. 11: 234–241 [DOI] [PubMed] [Google Scholar]
- Li Y., Dudek J., Guiard B., Pfanner N., Rehling P., Voos W. (2004). The presequence translocase-associated protein import motor of mitochondria. Pam16 functions in an antagonistic manner to Pam18. J. Biol. Chem. 279: 38047–38054 [DOI] [PubMed] [Google Scholar]
- Majeran W., Zybailov B., Ytterberg A.J., Dunsmore J., Sun Q., van Wijk K.J. (2008). Consequences of C4 differentiation for chloroplast membrane proteomes in maize mesophyll and bundle sheath cells. Mol. Cell. Proteomics 7: 1609–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Trujillo M., Limones-Briones V., Cabrera-Ponce J.L., Herrera-Estrella L. (2004). Improving transformation efficiency of Arabidopsis thaliana by modifying the floral dip method. Plant Mol. Biol. Rep. 22: 63–70 [Google Scholar]
- Matsubayashi T., Wakasugi T., Shinozaki K., Yamaguchi-Shinozaki K., Zaita N., Hidaka T., Meng B.Y., Ohto C., Tanaka M., Kato A., Maruyama T., Sugiura M. (1987). Six chloroplast genes (ndhA-F) homologous to human mitochondrial genes encoding components of the respiratory chain NADH dehydrogenase are actively expressed: Determination of the splice sites in ndhA and ndhB pre-mRNAs. Mol. Gen. Genet. 210: 385–393 [DOI] [PubMed] [Google Scholar]
- Mayer B.J., Hamaguchi M., Hanafusa H. (1988). A novel viral oncogene with structural similarity to phospholipase C. Nature 332: 272–275 [DOI] [PubMed] [Google Scholar]
- Miyake C., Amako K., Shiraishi N., Sugimoto T. (2009). Acclimation of tobacco leaves to high light intensity drives the plastoquinone oxidation system—Relationship among the fraction of open PSII centers, non-photochemical quenching of Chl fluorescence and the maximum quantum yield of PSII in the dark. Plant Cell Physiol. 50: 730–743 [DOI] [PubMed] [Google Scholar]
- Miyake C., Asada K. (1994). Ferredoxin-dependent photoreduction of the monodehydroascorbate radical in spinach thylakoids. Plant Cell Physiol. 35: 539–549 [Google Scholar]
- Munekage Y., Hashimoto M., Miyake C., Tomizawa K., Endo T., Tasaka M., Shikanai T. (2004). Cyclic electron flow around photosystem I is essential for photosynthesis. Nature 429: 579–582 [DOI] [PubMed] [Google Scholar]
- Munekage Y., Hojo M., Meurer J., Endo T., Tasaka M., Shikanai T. (2002). PGR5 is involved in cyclic electron flow around photosystem I and is essential for photoprotection in Arabidopsis. Cell 110: 361–371 [DOI] [PubMed] [Google Scholar]
- Musacchio A., Gibson T., Lehto V.P., Saraste M. (1992). SH3—An abundant protein domain in search of a function. FEBS Lett. 307: 55–61 [DOI] [PubMed] [Google Scholar]
- Okegawa Y., Kagawa Y., Kobayashi Y., Shikanai T. (2008). Characterization of factors affecting the activity of photosystem I cyclic electron transport in chloroplasts. Plant Cell Physiol. 49: 825–834 [DOI] [PubMed] [Google Scholar]
- Pellecchia M., Szyperski T., Wall D., Georgopoulos C., Wüthrich K. (1996). NMR structure of the J-domain and the Gly/Phe-rich region of the Escherichia coli DnaJ chaperone. J. Mol. Biol. 260: 236–250 [DOI] [PubMed] [Google Scholar]
- Peng L., Fukao Y., Fujiwara M., Takami T., Shikanai T. (2009). Efficient operation of NAD(P)H dehydrogenase requires supercomplex formation with photosystem I via minor LHCI in Arabidopsis. Plant Cell 21: 3623–3640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L., Ma J., Chi W., Guo J., Zhu S., Lu Q., Lu C., Zhang L. (2006). LOW PSII ACCUMULATION1 is involved in efficient assembly of photosystem II in Arabidopsis thaliana. Plant Cell 18: 955–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L., Shimizu H., Shikanai T. (2008). The chloroplast NAD(P)H dehydrogenase complex interacts with photosystem I in Arabidopsis. J. Biol. Chem. 283: 34873–34879 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Porra R.J., Thompson W.A., Kriedemann P.E. (1989). Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophylls standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 975: 384–394 [Google Scholar]
- Rajan V.B.V., D’Silva P. (2009). Arabidopsis thaliana J-class heat shock proteins: Cellular stress sensors. Funct. Integr. Genomics 9: 433–446 [DOI] [PubMed] [Google Scholar]
- Sazanov L.A., Hinchliffe P. (2006). Structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus. Science 311: 1430–1436 [DOI] [PubMed] [Google Scholar]
- Sétif P., Fischer N., Lagoutte B., Bottin H., Rochaix J.-D. (2002). The ferredoxin docking site of photosystem I. Biochim. Biophys. Acta 1555: 204–209 [DOI] [PubMed] [Google Scholar]
- Shikanai T. (2007a). Cyclic electron transport around photosystem I: Genetic approaches. Annu. Rev. Plant Biol. 58: 199–217 [DOI] [PubMed] [Google Scholar]
- Shikanai T. (2007b). The NAD(P)H dehydrogenase complex in photosynthetic organisms: Subunit composition and physiological function. Funct. Plant Sci. Biotechnol. 1: 129–137 [Google Scholar]
- Shikanai T., Endo T., Hashimoto T., Yamada Y., Asada K., Yokota A. (1998). Directed disruption of the tobacco ndhB gene impairs cyclic electron flow around photosystem I. Proc. Natl. Acad. Sci. USA 95: 9705–9709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu H., Peng L., Myouga F., Motohashi R., Shinozaki K., Shikanai T. (2008). CRR23/NdhL is a subunit of the chloroplast NAD(P)H dehydrogenase complex in Arabidopsis. Plant Cell Physiol. 49: 835–842 [DOI] [PubMed] [Google Scholar]
- Suorsa M., Sirpiö S., Aro E.-M. (2009). Towards characterization of the chloroplast NAD(P)H dehydrogenase complex. Mol. Plant 2: 1127–1140 [DOI] [PubMed] [Google Scholar]
- Takabayashi A., Ishikawa N., Obayashi T., Ishida S., Obokata J., Endo T., Sato F. (2009). Three novel subunits of Arabidopsis chloroplastic NAD(P)H dehydrogenase identified by bioinformatic and reverse genetic approaches. Plant J. 57: 207–219 [DOI] [PubMed] [Google Scholar]
- Varotto C., Pesaresi P., Meurer J., Oelmüller R., Steiner-Lange S., Salamini F., Leister D. (2000). Disruption of the Arabidopsis photosystem I gene psaE1 affects photosynthesis and impairs growth. Plant J. 22: 115–124 [DOI] [PubMed] [Google Scholar]
- Walsh P., Bursać D., Law Y.C., Cyr D., Lithgow T. (2004). The J-protein family: Modulating protein assembly, disassembly and translocation. EMBO Rep. 5: 567–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Duan W., Takabayashi A., Endo T., Shikanai T., Ye J.Y., Mi H. (2006). Chloroplastic NAD(P)H dehydrogenase in tobacco leaves functions in alleviation of oxidative damage caused by temperature stress. Plant Physiol. 141: 465–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K., Onda Y., Ashikari T., Tanaka Y., Kusumi T., Hase T. (2000). Analysis of reductant supply systems for ferredoxin-dependent sulfite reductase in photosynthetic and nonphotosynthetic organs of maize. Plant Physiol. 122: 887–894 [DOI] [PMC free article] [PubMed] [Google Scholar]