This work reveals that anthocyanin biosynthesis is coupled to floral transition by miR156-targeted SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) genes. SPL negatively regulates anthocyanin accumulation by directly preventing expression of anthocyanin biosynthetic genes through destabilization of a MYB-bHLH-WD40 transcriptional activation complex.
Abstract
Flavonoids are synthesized through an important metabolic pathway that leads to the production of diverse secondary metabolites, including anthocyanins, flavonols, flavones, and proanthocyanidins. Anthocyanins and flavonols are derived from Phe and share common precursors, dihydroflavonols, which are substrates for both flavonol synthase and dihydroflavonol 4-reductase. In the stems of Arabidopsis thaliana, anthocyanins accumulate in an acropetal manner, with the highest level at the junction between rosette and stem. We show here that this accumulation pattern is under the regulation of miR156-targeted SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) genes, which are deeply conserved and known to have important roles in regulating phase change and flowering. Increased miR156 activity promotes accumulation of anthocyanins, whereas reduced miR156 activity results in high levels of flavonols. We further provide evidence that at least one of the miR156 targets, SPL9, negatively regulates anthocyanin accumulation by directly preventing expression of anthocyanin biosynthetic genes through destabilization of a MYB-bHLH-WD40 transcriptional activation complex. Our results reveal a direct link between the transition to flowering and secondary metabolism and provide a potential target for manipulation of anthocyanin and flavonol content in plants.
INTRODUCTION
Flavonoids are major plant phenylpropanoid metabolites found throughout the plant kingdom, including the model species Arabidopsis thaliana. These diphenylchroman compounds play vital roles in growth and development by providing plants with red, blue, and purple pigments that protect against UV radiation, attract pollinators and other beneficial organisms, or mediate plant–microbe interactions (Buer et al., 2010). In addition, many simple flavonoid compounds have antioxidant properties and can therefore potentially be used as dietary nutraceutics for human health (Winkel-Shirley, 2001; Liu et al., 2006). Flavonoid biosynthesis leads to a variety of structurally distinct subclasses, including the flavonols, flavones, isoflavones, anthocyanins, and proanthocyanidins. The most common flavonols in Arabidopsis are kaempferol glycosides, with quercetin glycoside levels increasing after exposure to UV radiation (Veit and Pauli, 1999). Another group of flavonoids are anthocyanins that accumulate in variable amounts in Arabidopsis leaves and stems, depending on light level and nutrition (Holton and Cornish, 1995). The glycosylated and acylated cyanidin derivatives constitute the major anthocyanins present in the leaves and stems of mature Arabidopsis plants (Bloor and Abrahams, 2002).
The flavonoid backbone is formed through the central flavonoid biosynthetic pathway, in which the key enzymes chalcone synthase (CHS), chalcone isomerase (CHI), and flavanone 3-hydroxylase (F3H) condense and convert the phenylpropanoid precursor p-coumaroyl-CoA, with three molecules of malonyl-CoA, to dihydrokaempferol (Figure 1A). Dihydrokaempferol can be further catalyzed into dihydroquercetin by flavonoid 3′-hydroxylase (F3′H). Dihydrokaempferol and dihydroquercetin, two major components of dihydroflavonols, serve as common precursors that can either be catalyzed by flavonol synthase (FLS) to form a variety of copigment flavonols and glycosidic derivatives or by dihydroflavonol reductase (DFR) in the first dedicated step for anthocyanin biosynthesis (Figure 1A) (Harborne and Williams, 2000; Winkel-Shirley, 2002). In Arabidopsis, there is a single gene for DFR, whereas FLS is encoded by a small family of six genes (Owens et al., 2008; Preuss et al., 2009).
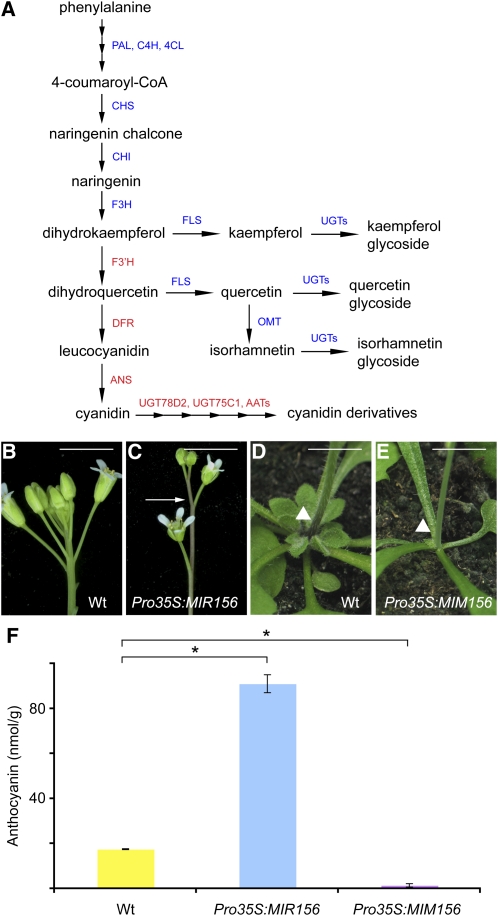
Figure 1.
Anthocyanin Accumulation.
(A) Schematic diagram of flavonoid biosynthetic pathways. Anthocyanin biosynthetic genes are labeled in red. 4CL, 4-coumarate:CoA ligase; AAT, anthocyanin acyltransferase; C4H, cinnamate-4-hydroxylase; OMT, O-methyltransferase; PAL, phenylalanine ammonia lyase; UGT, UDP-dependent glycosyltransferase.
(B) and (C) Inflorescences of wild-type (Wt, ecotype Col-0) and Pro35S:MIR156 plants. Arrow indicates the purple pigment in the stem of Pro35S:MIR156 plants. Bar = 1 cm.
(D) and (E) Stem-rosette junction (arrowheads). Compared with the wild type, Pro35S:MIM156 plants accumulated less purple pigment (arrowheads). Bar = 1 cm.
(F) Quantification of anthocyanin in stems. Errors bars indicate ±sd (n = 3). *Student’s test, P < 0.01.
The transcriptional control of flavonoid biosynthesis has been intensively studied (Broun, 2005), and genetic screens have identified several classes of transcriptional regulators. The first group includes four MYB proteins, PRODUCTION OF ANTHOCYANIN PIGMENTS1 (PAP1), PAP2, MYB113, and MYB114. Overexpression of any one of these results in an increase of anthocyanin accumulation (Borevitz et al., 2000; Gonzalez et al., 2008). The second group encodes three redundant bHLH factors, TRANSPARENT TESTA8 (TT8), GLABROUS3 (GL3), and ENHANCER OF GLABRA3 (EGL3), with their simultaneous inactivation causing anthocyanin deficiency (Zhang et al., 2003). MYB and bHLH proteins combine with the WD40 repeat–containing protein TRANSPARENT TESTA GLABRA1 (TTG1) to form a transcriptional complex that activates anthocyanin biosynthetic genes, including ANTHOCYANIDIN SYNTHASE (ANS), DFR, F3′H, LEUCOANTHOCYANIN DIOXYGENASE, UDP-GLUCOSYL TRANSFERASE 78D2 (UGT78D2), and UDP-GLUCOSYL TRANSFERASE 75C1 (UGT75C1) (Figure 1A) (Tohge et al., 2005; Gonzalez et al., 2008). Three other closely related MYB proteins, MYB11, MYB12, and MYB111, regulate early steps of the flavonoid pathway, including those catalyzed by the enzymes encoded by CHS, CHI, F3H, and FLS1 (Stracke et al., 2007). In addition, MYBL2, an R3-MYB–related protein, acts as a repressor by interfering with the formation of the MYB-bHLH-WD40 complex (Dubos et al., 2008; Matsui et al., 2008). Finally, three members of the LATERAL ORGAN BOUNDARY DOMAIN (LBD) gene family, LBD37, LBD38, and LBD39, have recently been identified as negative regulators of anthocyanin biosynthesis (Rubin et al., 2009).
Whereas the response of flavonoid biosynthetic genes to various biotic and abiotic stress factors has been extensively studied (Lea et al., 2007; Lillo et al., 2008), less is known about the developmental regulation of flavonol and anthocyanin accumulation in Arabidopsis. Recent studies have identified a microRNA (miRNA), miR156, as a pleiotropic regulator of plant development. miR156, the levels of which gradually decrease with age, targets a group of transcription factors called SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) (Wu and Poethig, 2006; Wang et al., 2009). SPLs in turn affect a broad range of developmental processes in Arabidopsis. For example, they promote vegetative phase transition by activating another miRNA, miR172, and induce flowering through MADS box genes and LEAFY (Wang et al., 2009; Wu et al., 2009; Yamaguchi et al., 2009). In addition, SPLs play critical roles in regulating embryonic development (Nodine and Bartel, 2010), cell size (Wang et al., 2008; Usami et al., 2009), leaf production rates (Wang et al., 2008), and the formation of trichomes on floral organs (Yu et al., 2010), as well as being required for fertility (Xing et al., 2011).
Here, we show that the transition from the formation of leaves to the formation of flowers on the Arabidopsis shoot goes hand in hand with a change in the ratio of flavonols to anthocyanins. Both are directly regulated by at least one miR156-targeted SPL factor, SPL9. We conclude that SPLs coordinate the transition to flowering with the biosynthesis of anthocyanin by disruption of the MYB-bHLH-WD40 transcription complex.
RESULTS
Inverse Relationship of Anthocyanin Content and SPL Levels
While carefully comparing the inflorescences of wild-type and transgenic plants that have reduced SPL activity because of miR156 overexpression (Pro35S:MIR156) (Schwab et al., 2005), we noticed hyperaccumulation of purple pigments. In wild-type inflorescences, internodes were green, whereas Pro35S:MIR156 plants had purplish internodes (Figures 1B and 1C). Purple pigment was found in wild-type plants at the junction of the stem and rosette leaves (Figure 1D). This was lacking in Pro35S:MIM156 plants (Figure 1E), which have increased SPL activity due to reduced miR156 action (Franco-Zorrilla et al., 2007).
In the wild type, another tissue with high amounts of purple pigment is the junction between the hypocotyl and cotyledons. Although Pro35S:MIR156 accumulated the same amount of purple pigment as the wild type, the hypocotyl-cotyledon junction in Pro35S:MIM156 seedlings was completely green (see Supplemental Figure 1 online).
We extracted pigment from the stems of wild-type and transgenic plants and measured absorption at 530 nm, which is indicative of anthocyanins. Compared with the wild type, anthocyanin content was about 4-fold higher in Pro35S:MIR156 stems and reduced to about one-tenth in Pro35S:MIM156 stems (Figure 1F), which is consistent with the visible color change.
Regulation of Anthocyanin Biosynthesis by the SPL9 Subfamily
The Arabidopsis genome contains 11 SPL genes that are targeted by miR156. These SPL genes can be classified into two major groups, represented by SPL3 and SPL9, respectively (Cardon et al., 1999). To learn which group of SPL genes regulates anthocyanin biosynthesis, we investigated SPL loss-of-function mutants. The spl3-1 mutant was identified in the Wassilewskija (Ws) background (Wu and Poethig, 2006). spl3-1 had the same amount of purple pigments in seedlings and inflorescences as wild-type Ws plants (see Supplemental Figure 2 online). In the SPL9 subfamily, SPL9 and SPL15 are particularly important (Schwarz et al., 2008; Wang et al., 2008). spl9-4 spl15-1 double mutants, however, did not accumulate increased levels of anthocyanin at the hypocotyl-cotyledon junction or in inflorescences (see Supplemental Figures 1 and 2 online), suggesting redundant regulation of anthocyanin biosynthesis by several miR156-targeted SPLs.
We then examined plants that overexpressed individual SPL genes. Overexpression of either SPL9 or SPL10, both from the SPL9 subfamily, but not of SPL3, decreased anthocyanin accumulation at the junction between stem and rosette (see Supplemental Figure 2 online). Together, these results suggested that upregulation of SPL9-type SPL genes was responsible for the phenotype of Pro35S:MIM156 plants and that the SPL3 subfamily plays only a minor role, if any, in regulating anthocyanin biosynthesis.
Accumulation of Flavonoids in Transgenic Plants
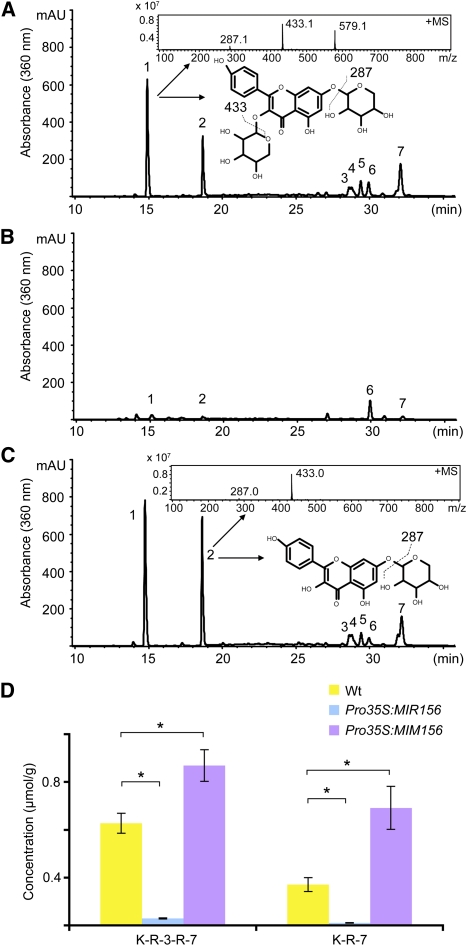
Because flavonols and anthocyanins are synthesized from common precursors, we also analyzed the flavonol content of plants with increased and decreased SPL activity. Soluble phenolic compounds were extracted from stems with methanol and subjected to liquid chromatography–mass spectrometry (LC-MS) analysis. Considering the array of flavonoid glycosidic derivatives including glucosides that have been reported before in Arabidopsis (Yonekura-Sakakibara et al., 2008), we treated phenolic extracts with β-glucosidase, to simplify LC-MS identification of compounds and to facilitate their quantification. After digestion, two kaempferol derivatives, kaempferol 3-O-rhamnopyranoside 7-O-rhamnopyranoside and kaempferol 7-O-rhamnopyranoside (K-7-R), were identified and characterized as the major flavonols in the wild type (Figure 2A). These results are consistent with a previous report of three major flavonol derivatives, kaempferol 3-O-β-[β-d-glucopyranosyl(1-6)d-glucopyranoside]-7-O-α-l-rhamnopyranoside, kaempferol 3-O-β-d-glucopyranoside-7-O-α-l-rhamnopyranoside, and kaempferol 3-O-α-l-rhamnopyranoside-7-O-α-l-rhamnopyranoside (Veit and Pauli, 1999). After β-glucosidase digestion, the first two conjugates can be converted to K-7-R.
Figure 2.
LC-MS Analyses of Flavonol Level.
(A) to (C) LC-MS profiles of soluble phenolic compounds from the stems of wild-type (A), Pro35S:MIR156 (B), and Pro35S:MIM156 plants (C). Insets show the mass spectrum and structures of the peaks 1 and 2, which correspond to kaempferol 3-O-rhamnopyranoside 7-O-rhamnopyranoside (K-3-R-7-R; shown in [A]) and K-7-R (shown in [C]), respectively. mAU, milliabsorbance unit.
(D) Quantification of flavonols in the stems of wild-type, Pro35S:MIR156, and Pro35S:MIM156 plants. Data represent means of three trials. Errors bars indicate ± sd (n = 3). *Student’s test, P < 0.01. Wt, wild type.
In Pro35S:MIR156 plants, the levels of both kaempferol derivatives were very low, less than one-tenth of the amount detected in the wild type (Figures 2B and 2D). By contrast, levels of the two kaempferol derivatives were increased by 1.6- to 2.9-fold in Pro35S:MIM156 plants (Figures 2C and 2D). Another type of flavonol, quercetin, and/or its derivatives were not detected in wild-type stems under our analytic conditions; in addition, only negligible levels of molecular and adduct ions at mass-to-charge ratios of 449 and 303, which are indicative of quercetin rhamnopyranoside, were found in the multistage MS analysis on phenolic extracts from both Pro35S:MIR156 and Pro35S:MIM156 stems. Taken together, these data reveal an antagonistic relationship between the accumulations of flavonols, primarily kaempferol derivatives, and anthocyanins in Arabidopsis stems. When SPL levels were reduced in Pro35S:MIR156 plants, the levels of anthocyanins rose, while those of flavonols fell. Opposite effects were seen with the increased SPL levels in Pro35S:MIM156 plants.
In addition to flavonols, several phenolic compounds of higher molecular weight were detected in Arabidopsis stem extracts (Figure 2A). Although the nature of these metabolites remains to be conclusively determined, four were decreased in Pro35S:MIR156 plants to about a quarter of wild-type levels (see Supplemental Figure 3 online). Similar to the flavonols, the levels of these putative phenolic derivatives were increased in Pro35S:MIM156 stems (Figure 2A; see Supplemental Figure 3 online).
Expression of Flavonoid Biosynthetic and Regulatory Genes
To understand the molecular basis of the changes in anthocyanin and flavonol levels, we first examined the expression of early biosynthetic genes encoding phenylalanine ammonia lyase1 (PAL1), CHS, and CHI (Figure 1A) by reverse transcription followed by quantitative PCR (qRT-PCR). Expression of PAL1 was slightly increased in both Pro35S:MIR156 and Pro35S:MIM156 plants. CHI and CHS showed the same expression pattern: transcript levels of both genes were higher in Pro35S:MIM156 than in the wild type but lower in Pro35S:MIR156 (Figure 3A). Of the six FLS-encoding genes, FLS1 to FLS6, FLS1 is the most important, with a small contribution from FLS3 (Owens et al., 2008). Compared with the wild type, neither FLS1 nor FLS3 expression levels were greatly changed in either Pro35S:MIM156 or Pro35S:MIR156 plants (Figure 3A).
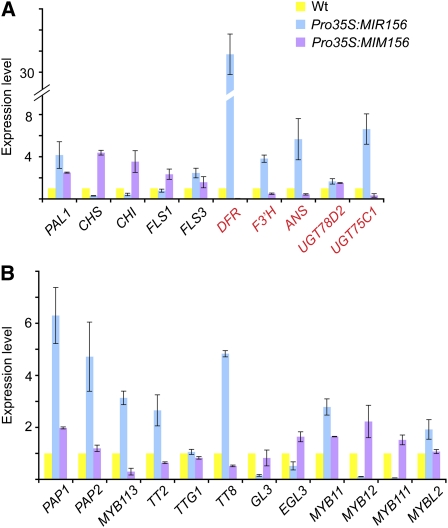
Figure 3.
Expression of Flavonoid Biosynthetic and Regulatory Genes.
(A) Expression of flavonoid biosynthetic genes. Anthocyanin biosynthetic genes are labeled in red. Expression level in the wild type (Wt) was set to 1. Errors bars indicate ± se (n = 3). Two biological replicates were analyzed, with similar results.
(B) Expression of flavonoid regulatory genes.
Two-centimeter-long stems above the rosette were harvested and subjected to qRT-PCR analyses. Errors bars indicate ± se (n = 3). Two biological replicates were analyzed, with similar results.
We then monitored the expression of anthocyanin biosynthetic genes, such as ANS, DFR, F3′H, UGT75C1, and UGT78D2 (Figure 3A). DFR transcripts were greatly increased, by over 30-fold, in Pro35S:MIR156 plants and correspondingly decreased in Pro35S:MIM156 plants. The expression of ANS, F3′H, and UGT75C1 showed the same trend as that of DFR, whereas the transcript level of UGT78D2 remained unchanged in both Pro35S:MIR156 and Pro35S:MIM156 plants. These observations suggest that miR156-regulated SPLs negatively regulate anthocyanin levels by repressing biosynthetic genes.
We further examined the expression of several regulatory genes in the flavonoid biosynthetic pathway, including PAP1, PAP2, MYB113, TT2, TT8, GL3, EGL3, MYB11, MYB12, MYB111, and MYBL2 (Figure 3B). The transcript levels of PAP1, PAP2, MYB113, TT2, and TT8 were not dramatically changed in Pro35S:MIM156 plants but moderately increased in Pro35S:MIR156 plants (Figure 3B). Interestingly, the expression of MYB12 and MYB111 was greatly decreased in the stems of Pro35S:MIR156 plants. These two MYBs are positive regulators of CHS and CHI (Stracke et al., 2007), and the low levels of MYB12 and MYB111 transcripts might explain why CHI and CHS levels were decreased in Pro35S:MIR156 plants as well. Compared with the wild type, there was no significant change of GL3, EGL3, and MYBL2 expression in the stems of either Pro35S:MIM156 or Pro35S:MIR156 plants.
Spatial Expression Pattern of DFR and SPLs
Since the expression of DFR is very sensitive to SPL activity, subsequent analyses focused on DFR. To compare the expression patterns of DFR and SPLs during the transition from leaves to flowers, the basal, middle, and apical regions of wild-type stems were collected (Figure 4A). Expression of DFR was lowest in the apical portion and highest in the basal section, whereas SPL9 and SPL15 showed an opposite pattern, suggesting a potential negative regulation of DFR by SPLs. The expression of two genes from the SPL3 subfamily, SPL3 and SPL4, did not notably change along the stem (Figure 4A), consistent with the findings that these genes have less of an impact on anthocyanin biosynthesis along the stem.
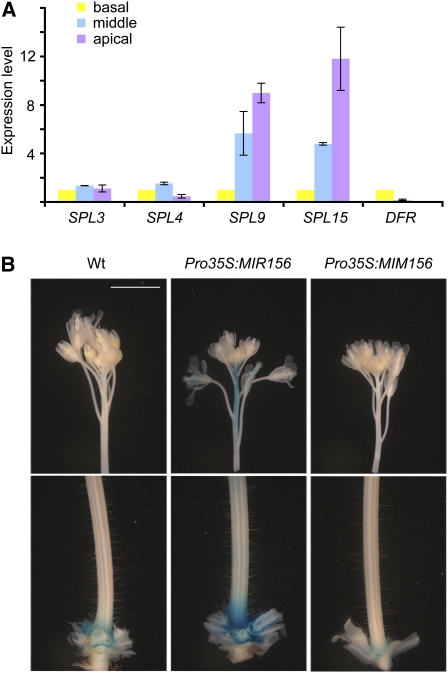
Figure 4.
Spatial Expression of SPLs and DFR.
(A) Transcript levels of SPLs and DFR. The apical, middle, and basal parts of stems were harvested and subjected to qRT-PCR analyses. Expression level in the basal part of stems was set to 1. Errors bars indicate ± se (n = 3). Two biological replicates were analyzed, with similar results.
(B) GUS reporter for DFR promoter activity. Twenty-day-old plants expressing GUS under the control of the DFR promoter were stained for 1 h. Bar = 1 cm. Wt, wild type.
To investigate further the regulation of DFR by SPLs, we generated a reporter line in which the β-glucuronidase (GUS) coding region was placed under control of a 2.5-kb DFR promoter fragment (ProDFR:GUS). After the main shoot had elongated, GUS activity was easily detected at the junction between the stem and rosette leaves, whereas GUS activity in the remainder of the stem, including the internodes between the flowers, was much lower (Figure 4B). GUS activity at the base of the stem was enhanced and reduced in Pro35S:MIR156 and Pro35S:MIM156 plants, respectively (Figure 4B). In addition, GUS activity was apparent in the internodes between the flowers of Pro35S:MIR156 plants. This pattern overall mimicked that seen for anthocyanin accumulation (Figures 1B to 1E).
Requirement of the MYB-bHLH-WD40 Complex for Effects of SPL Activity on Anthocyanin Accumulation
DFR expression is positively regulated by a complex that includes a MYB protein of the PAP1/PAP2/MYB113/MYB114 group, a bHLH protein of the TT8/GL3/EGL3 group, and the WD40 protein TTG1 (Gonzalez et al., 2008). To test whether the high level of DFR transcripts in Pro35S:MIR156 plants depended on the MYB-bHLH-WD40 complex, we overexpressed miR156 in the plants lacking PAP1, TT8, and TTG1 activity. Since a pap1 mutant in the Columbia-0 (Col-0) background was not available, we knocked down PAP1 using a miRNA-induced silencing method, which relies on an miRNA trigger and artificial transacting small RNAs (Felippes and Weigel, 2009). pap1-migs plants in Col-0 had a similar phenotype as a pap1 mutant described for the Nossen (No-0) accession (Teng et al., 2005), with much reduced anthocyanin levels at the junctions between hypocotyl and cotyledons, and rosette and stems. Both pap1-migs and ttg1-1 were sufficient to suppress anthocyanin accumulation in the inflorescences of Pro35S:MIR156 plants (see Supplemental Figure 4 online). TT8 is functionally redundant with GL3 and EGL3 (Zhang et al., 2003). Accordingly, tt8-1 mutation only reduced anthocyanin accumulation in Pro35S:MIR156 inflorescences. Taken together, these results indicate that the MYB-bHLH-WD40 complex is necessary for the upregulation of DFR in Pro35S:MIR156 stems.
Binding of SPL9 to the DFR Promoter
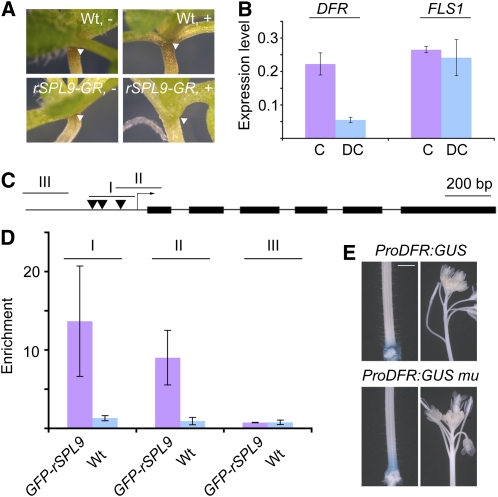
We then tested whether DFR was directly regulated by SPL9. To this end, we used transgenic plants in which expression of an in-frame fusion of a miRNA nontargetable version of SPL9 (rSPL9) to the rat glucocorticoid receptor (GR) is driven by SPL9 regulatory sequences (Wang et al., 2009). Treatment with the hormone ligand dexamethasone (DEX) of 1-week-old ProSPL9:rSPL9-GR seedlings visibly decreased anthocyanin accumulation at the base of the cotyledons (Figure 5A). Furthermore, after 6 h of treatment with DEX and the translation inhibitor cycloheximide (CHX), we observed a 60% decrease in DFR transcripts compared with mock-treated controls (Figure 5B). The expression of FLS1 was not affected.
Figure 5.
DFR Is a Direct Target of SPL9.
(A) Seven-day-old, long day–grown DEX- (+) or mock-treated (−) seedlings. The accumulation of anthocyanin was greatly reduced in DEX-treated ProSPL9:rSPL9-GR plants (arrows). Wt, wild type.
(B) Induced expression of DFR and FLS1 in ProSPL9:rSPL9-GR plants. Seven-day-old, long day–grown seedlings were treated with either DEX and CHX (DC) or CHX alone (C). Seedlings were harvested 6 h after treatment. Expression was normalized relative to that of β-TUBULIN-2. Errors bars indicate ± se (n = 3). Two biological replicates were analyzed, with similar results.
(C) Diagram of DFR genomic region. Solid lines, black boxes, and arrow indicate promoter/intron, exons, and transcription start site, respectively. Three regions were chosen for qRT-PCR analyses. Black triangles stand for GTAC boxes.
(D) ChIP analyses of 1-week-old wild-type and ProSPL9:GFP-rSPL9 seedlings. Crude chromatin extracts were immunopercipated with either anti-Myc or anti-GFP antibody. Purified ChIP and input DNAs were used for qRT-PCR analyses. Relative enrichment of each fragment was calculated by comparing the samples treated with anti-GFP or anti-Myc antibodies (2−(Ct@Myc–Ct@GFP)). Errors bars indicate ± se (n = 3). Two biological replicates were analyzed, with similar results.
(E) GUS staining of ProDFR:GUS and ProDFR:GUS mu (GTAC boxes mutated) plants. Tissue of 20-d-old plants was stained for 1 h. Bar = 0.5 cm.
To confirm that SPL9 directly interacts with cis-regulatory sequences at the DFR locus, we performed chromatin immunoprecipitation (ChIP) assays using a transgenic line that expresses a fusion of green fluorescent protein (GFP) to rSPL9 under its native promoter (Wang et al., 2009). A GTAC consensus sequence has been identified as the core binding motif of SPLs (Klein et al., 1996; Kropat et al., 2005; Liang et al., 2008). There are three GTAC boxes within the 1.5-kb promoter of DFR (Figure 5C), and we analyzed three regions, two (I and II) of which covered the three GTAC boxes. Chromatin extracted from 10-d-old seedlings was immunoprecipitated with either anti-GFP or anti-Myc antibodies, and the presence of DFR promoter sequences in the precipitate was monitored by qPCR. There was no apparent enrichment of any of the GTAC containing fragments in wild-type samples (Figure 5D). By contrast, regions I to II, but not region III, were readily amplified in ProSPL9:GFP-rSPL9 samples after pulldown with anti-GFP, but not with anti-Myc antibodies (Figure 5D), indicating that SPL9 is able to bind to the promoter region of DFR.
To assess whether SPL9 binding to GTAC boxes is required for normal promoter DFR activity, we generated a GUS reporter (ProDFR:GUS mu), in which all three GTAC boxes were mutated. If negative regulation of DFR is mediated by SPL9 binding to these GTAC boxes, one would expect that the ProDFR:GUS mu reporter will be ectopically active in inflorescences. However, there was no obvious difference between ProDFR:GUS and ProDFR:GUS mu plants (Figure 5E), suggesting that SPL9 is indirectly recruited to the DFR promoter.
SPL9 Interferes with MYB-bHLH-WD40 Complex Formation
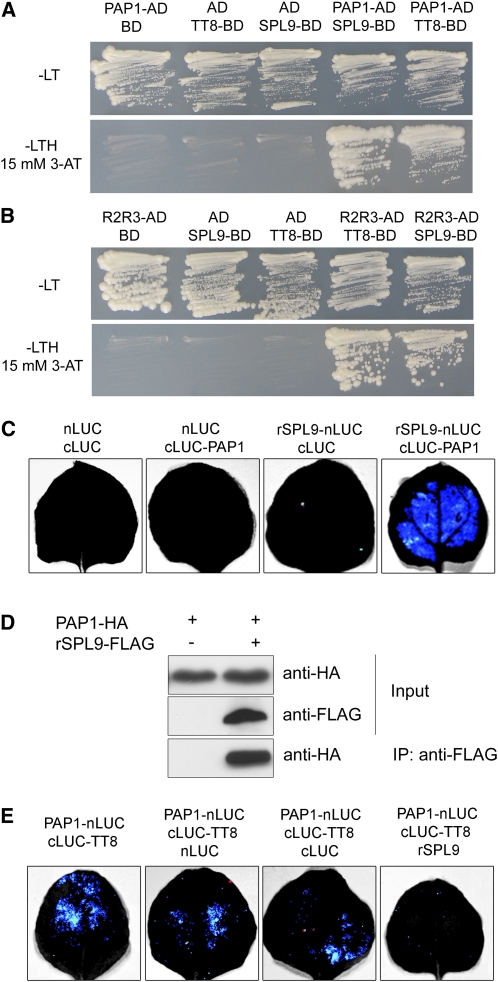
The SPLs belonging to the SPL9 group have been identified as transcriptional activators of several target genes involved in flowering and trichome development (Shikata et al., 2009; Wang et al., 2009; Wu et al., 2009; Yamaguchi et al., 2009; Yu et al., 2010). However, our expression analyses suggested that SPL9 exerted a repressive role in the case of DFR expression. In addition, SPL9 is likely to be recruited to the DFR promoter independently of the GATC boxes. One scenario is that SPL9 binds to the well-known MYB-bHLH-WD40 activator complex and interferes with its activity. To test this possibility, we first performed a yeast two-hybrid assay. Consistent with a previous report (Zimmermann et al., 2004), PAP1 could directly bind to TT8. A similar interaction was observed between SPL9 and PAP1, and its paralog, MYB113 (Figure 6A; see Supplemental Figure 5 online). In addition, both SPL9 and TT8 bound to the same domain, R2R3 domain, of PAP1 (Figure 6B) (Zimmermann et al., 2004).
Figure 6.
SPL9 Binds to PAP1.
(A) SPL9 and TT8 binding to PAP1 in yeast. TT8 and SPL9 were in-frame fused to the GAL4 binding domain (BD) in pGBKT7, whereas PAP1 was fused to the GAL4 activation domain (AD) in pGADT7. Transformed yeast cells were grown on SD –Leu Trp (–LT) (top). The direct interactions between TT8-BD, SPL9-BD, and PAP1-AD were assayed on a SD –Leu Trp His (–LTH) plate, supplemented with 15 mM 3-amino-1,2,4,-triazole (3-AT) (bottom). The pGADT7 (AD) and pGBKT7 (BD) were used as controls.
(B) Role of R2R3 domain of PAP1 in mediating binding of PAP1 to SPL9 in yeast. The R2R3 domain of PAP1 was fused to AD in pGADT7.
(C) BiFC assay. The leaves of N. benthamiana were infiltrated with agrobacteria as indicated. Blue luminescence indicates a direct protein–protein interaction between rSPL9 and PAP1. Constructs were combined at a 1:1 ratio.
(D) CoIP analysis. Proteins were transiently expressed in N. benthamiana. Protein was immunoprecipitated with anti-FLAG antibody, and the IP fraction was analyzed in a protein blot with anti-HA antibody. Input fraction was analyzed by immunoblotting using either anti-FLAG or anti-HA antibody.
(E) Competition of SPL9 and TT8 for PAP1 binding. Constructs were combined at a 1:1:4 ratio for PAP1-LUC-N: LUC-C-TT8: LUC-N, LUC-C, or rSPL9. Blue luminescence indicates a direct interaction between TT8 and PAP1.
To confirm our yeast two-hybrid results, we performed a bimolecular fluorescence complementation (BiFC) assay based on firefly luciferase (LUC) (Chen et al., 2008). We fused rSPL9 to the N-terminal half of LUC (rSPL9-nLUC) and PAP1 to the C-terminal half (cLUC-PAP1) and transiently introduced both fusion proteins into Nicotiana benthamiana by infiltration with Agrobacterium tumefaciens. LUC activity was barely detectable in leaves infiltrated with combinations of Pro35S:nLUC and Pro35S:cLUC, Pro35S:cLUC and Pro35S:rSPL9-nLUC, or Pro35S:nLUC and Pro35S:cLUC-PAP1. By contrast, leaves that coexpressed Pro35S:cLUC-PAP1 and Pro35S:nLUC-rSPL9 produced a strong LUC signal (Figure 6C).
Next, we performed a coimmunoprecipitation (CoIP) experiment. Both hemagglutinin (HA)-epitope–tagged PAP1 (PAP1-HA) and FLAG-tagged rSPL9 (rSPL9-FLAG) were transiently expressed in N. benthamiana. PAP1-HA proteins were readily detected in the sample immunoprecipitated with the anti-FLAG antibody but not in the mock control (Figure 6D), confirming that SPL9 can directly bind to PAP1 in vivo.
Finally, we used transient expression in N. benthamiana to test whether the SPL9-PAP1 interaction influences the integrity of the MYB-bHLH-WD40 complex. In agreement with the yeast two-hybrid results, LUC activity was readily detected in leaves that coexpressed Pro35S:PAP1-nLUC and Pro35S:cLUC-TT8 but not in leaves that coexpressed Pro35S:PAP1-nLUC and Pro35S:cLUC, or Pro35S:nLUC and Pro35S:cLUC-TT8 (Figure 6E). Moreover, LUC activity in leaves that coexpressed Pro35S:PAP1-nLUC and Pro35S:cLUC-TT8 was greatly suppressed when Pro35S:rSPL9, but not Pro35S:nLUC or Pro35S:cLUC, was coinfiltrated (Figure 6E), indicating that SPL9 is indeed able to compete with TT8 for binding to PAP1.
DISCUSSION
A Model for SPL Regulation of Metabolic Flux in the Flavonoid Pathway
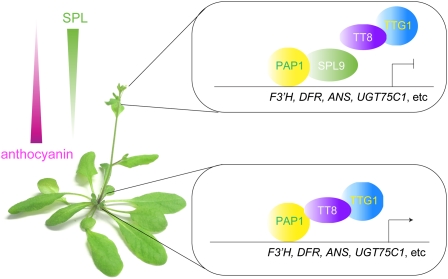
We propose a model for how metabolic flux through the branched flavonoid biosynthetic pathway is affected by miR156-regulated SPL transcription factors (Figure 7). In the places with high anthocyanin concentration, such as the junction between stems and rosette, high levels of miR156 reduce SPL activity and thereby permit elevated expression of F3′H, DFR, and other anthocyanin biosynthetic genes. As a consequence, dihydrokaempferol is directed into the anthocyanin branch. Expression of SPLs gradually increases along the growing stem because miR156 levels decline as the plant progresses during development (Wu and Poethig, 2006; Wang et al., 2009; Wu et al., 2009). As SPL level rises, the expression of anthocyanin biosynthetic genes is more and more repressed, resulting in the increased conversion of dihydrokaempferol to kaempferol by FLS and production of flavonols.
Figure 7.
A Model for Negative Regulation of Anthocyanin Biosynthesis by miR156-Targeted SPLs.
Anthocyanins accumulate in an acropetal manner, with the highest level at the junction between rosette and stem. This pattern is regulated by miR156-targeted SPL genes. High levels of SPLs in the inflorescences repress anthocyanin accumulation by directly preventing expression of anthocyanin biosynthetic genes, such as ANS, F3′H, DFR, and UGT75C1, through destabilization of the MYB-bHLH-WD40 transcriptional activation complex.
It has been reported that inactivation of DFR increases accumulation of quercetin (Pelletier et al., 1999), whereas fls1 mutants have elevated anthocyanin content (Owens et al., 2008; Stracke et al., 2009). Our results agree with these findings and indicate that metabolic flux in the flavonoid biosynthetic pathway is controlled by substrate competition between FLS and DFR. Given that FLS is encoded by multiple genes in the Arabidopsis genome and that each member has different catalytic activities and expression patterns (Owens et al., 2008; Preuss et al., 2009), it is likely that the FLS-to-DFR ratio varies among different tissue and organs. How this ratio is spatially and temporally controlled requires further examination.
MYB11, MYB12, and MYB111 encode three functionally redundant MYBs regulating the expression of several early flavonoid biosynthetic genes, including CHS, CHI, F3H, and FLS1 (Stracke et al., 2007). Interestingly, we found that the expression of MYB12 and MYB111 was high in Pro35S:MIR156 and low in Pro35S:MIM156 plants (Figure 3B). The impact of SPL on MYB12 and MYB111 is likely to be indirect, probably through a feedback loop. The high level of anthocyanin in Pro35S:MIR156 plants might lead to a decrease of dihydrokaempferol biosynthesis through the repression of early flavonoid biosynthetic genes by MYB12 and MYB111.
Coordination of Anthocyanin-to-Flavonol Ratio with the Floral Transition
Several lines of evidence indicate that miR156-targeted SPL genes control a suite of temporal changes during plant development. SPLs promote the transition from the juvenile to adult phase of vegetative development by activating expression of miR172 (Wu et al., 2009), and they induce flowering through a suite of MADS box genes (Wang et al., 2009; Yamaguchi et al., 2009). Recently, SPL9 has been shown to regulate trichome initiation in floral organs by activation of TRICHOMELESS1 (TCL1) (Yu et al., 2010). Regulated activity of SPLs is also essential for embryonic development (Nodine and Bartel, 2010), cell size (Wang et al., 2008; Usami et al., 2009), and fertility (Xing et al., 2011). We have uncovered an additional role of SPL proteins in influencing metabolic flux in the flavonoid biosynthetic pathway.
It is intriguing that there is a close connection between trichome formation and flavonoid biosynthesis in Arabidopsis. Previous studies along with results presented here reveal two molecular links between these two developmental processes. The first link comprises TTG1, a common WD40 factor that is required for anthocyanin biosynthesis, as a partner of PAP1/PAP2/MYB113 and TT8, and trichome development, in which it forms a complex with GL1 and GL3/EGL3. Loss-of-function mutants of TTG1 abolish both trichome initiation and anthocyanin accumulation (Walker et al., 1999). The miR156-targeted SPLs constitute a second link between these processes. Both trichome initiation on floral organs and anthocyanin accumulation are coupled to the onset of flowering through the SPLs. Why might the formation of trichomes and the flavonol-to-anthocyanin ratio both change upon the transition to flowering? One hypothesis is that the acropetal accumulation of anthocyanins and formation of trichomes on stems contribute to coordinated defenses against crawling herbivores. Anthocyanins provide visual cues that distract herbivores (Lev-Yadun and Gould, 2009), while trichomes endow plants with physical protection against herbivores (Juniper and Southwood, 1986).
miR156 is widely present in all land plants (Axtell and Bowman, 2008), and a recent study indicates that the expression pattern, as well as the role, of miR156 is conserved between annual and perennial plants (Wang et al., 2011). Overexpression of miR156 in poplar (Populus spp) trees causes similar phenotypes as in Arabidopsis and maize (Zea mays), such as decreased leaf size, increased leaf initiation rate, and reduced apical dominance. Whether miR156 has a similarly conserved role in anthocyanin accumulation in perennial plants awaits further investigation.
Regulation of a MYB-bHLH-WD40 Complex by SPLs
Our results also provide new insights into how SPL transcription factors regulate gene expression. Previous studies have shown that SPL9 activates expression of several downstream target genes, including APETALA1, FRUITFULL, MIR172b, and TCL1, by directly binding to their regulatory sequences (Wang et al., 2009; Wu et al., 2009; Yamaguchi et al., 2009; Yu et al., 2010). In support of SPL9 being a transcriptional activator is the observation that a close homolog, SPL10, has a transcriptional activation domain, as assayed in a transient expression system (Shikata et al., 2009). Our results indicate that SPL9 is a bifunctional transcription factor, similar to what has recently been shown for AP2-like proteins (Yant et al., 2010). Specifically, our data reveal that SPL9 suppresses DFR expression by interfering with the integrity of a MYB-bHLH-WD40 transcriptional-activation complex. This conclusion is supported by at least four lines of evidence. First, ChIP analysis has shown that SPL9 is recruited to the DFR promoter (Figure 5D). Second, the binding of SPL9 on the DFR promoter is indirect, as the putative SPL binding sites in the DFR promoter have no functional relevance (Figure 5E). Third, the expression of anthocyanin biosynthetic genes, such as DFR, ANS, F3′H, and UGT75C1, was accordingly changed in Pro35S:MIR156 and Pro35S:MIM156 plants (Figure 3A). Furthermore, SPL9 is able to compete with TT8 for their binding to PAP1 (Figures 6A and 6E).
Similar to PAP1 and paralogs, TT2 is another MYB protein that binds to bHLH factors. TT2 protein accumulates mainly in immature seeds and forms a ternary complex with TT8 and TTG1. The TT2-TT8-TTG1 complex controls proanthocyanin biosynthesis by activating the expression of BANYULS, which encodes a anthocyanidin reductase (Baudry et al., 2004). It is still unclear whether TT2 plays a similar role as PAP1/PAP2/MYB113 in regulation of anthocyanin biosynthesis. We have no evidence that SPL suppresses the function of TT2 at the protein level, as we have failed to detect a direct interaction between SPL9 and TT2 in yeast (data not shown). However, TT2 transcript levels were increased in Pro35S:MIR156 and decreased in Pro35S:MIM156 plants, raising the possibility that SPL9 regulates anthocyanin biosynthesis by modulating TT2 expression.
As mentioned before, another MYB-bHLH-WD40 complex, named GL1-GL3/EGL3-TTG1, controls trichome initiation. We tested the interaction between SPL9 and GL1 in a yeast two-hybrid assay. However, no direct interaction between them was observed (data not shown). Since TT2 couldn’t bind to SPL9 either, these results indicate a specific interaction between SPL9 and anthocyanin-related R2R3 MYB proteins.
The mechanism by which SPL9 regulates anthocyanin biosynthetic genes shares similarities with that of MYBL2. Both SPL and MYBL2 destabilize a MYB-bHLH-WD4 complex by competing with bHLHs for their binding to PAP1. Furthermore, in both myb2l and Pro35S:MIR156 plants, the transcript levels of PAP1 and TT8 are increased (Figure 3A). Since TT8 expression is controlled by a positive feedback loop (Baudry et al., 2006), these observations suggest that PAP1 is able to reinforce its own expression as well. It will be interesting to see whether this self-regulation of PAP1 requires the MYB-bHLH-WD40 complex.
METHODS
Plant Materials
Arabidopsis thaliana plants, ecotypes Col-0 and Ws, and Nicotiana benthamiana were grown at 23°C in long days (16 h light/8 h dark). spl3-1 (Wu and Poethig, 2006), spl9-4 (Wang et al., 2008), spl15-1 (Wang et al., 2008), spl9-4 spl15-1 (Wang et al., 2008), Pro35S:MIR156 (Wang et al., 2008), Pro35S:MIM156 (Wang et al., 2008), Pro35S:rSPL3 (Wang et al., 2009), Pro35S:rSPL9 (Wang et al., 2008), ProSPL9:rSPL9 (Wang et al., 2008), ProSPL10:rSPL10 (Wang et al., 2008), ProSPL9:rSPL9-GR (Wang et al., 2009), and ProSPL9:GFP-rSPL9 (Wang et al., 2009) have been described. tt8-1 (NASC ID: N111) and ttg1-1 (NASC ID: N89) mutants were obtained from European Arabidopsis Stock Centre (NASC).
For DEX treatment, 1-week-old wild-type and ProSPL9:rSPL9-GR seedlings were sprayed with 10 μM DEX plus 10 μM CHX, 10 μM DEX, or ethanol (mock) for 6 h.
Anthocyanin Measurements
Stems from 20-d-old plants were collected, ground into fine powder in liquid nitrogen, and extracted with 80% methanol containing 5% HCl overnight at 4°C. After centrifugation at 14,000g for 20 min, the extracts were removed to new tubes and the amounts of anthocyanins were quantified photometrically (DU 640 spectrophotometer; Beckman Instruments).
Soluble Phenolic Extraction and LC-MS Analyses
Stem samples were dried overnight in a 95°C oven, ground in liquid nitrogen, and extracted with 80% methanol. The extracts were dried under nitrogen gas and resuspended and digested with β-glucosidase (Sigma-Aldrich) overnight at 37°C in 100 mM acetate buffer, pH 5.0. The digested products were extracted with an equal amount of water-saturated ethyl acetate three times. The ethyl acetate extracts were dried under nitrogen gas and dissolved in methanol. The samples were resolved with a reverse-phase C18 column (XDB-18, 5 μm; Agilent) at a flow rate of 0.8 mL/min with a mixture of solvent A (0.2% acetic acid in water) and solvent B (0.2% acetic acid in acetonitrile). The gradient of the solvent B was started from 5% for 5 min, increased to 30% at 15 min, 80% at 20 min and 100% at 30 min, and then maintained at 100% for 5 min. The mass spectrum was collected with an ion trap mass spectrometer equipped with an APCI source in positive mode at 4 kV spray voltage, 60 p.s.i. nebulizer pressure, 5 L/min dry gas at 350°C, and 400°C vaporizer temperature.
Constructs and Plant Transformation
For yeast two-hybrid constructs, cDNAs of SPL9, TT2, GL1, TT8, MYB113, and PAP1 were cloned into pGBKT7 and pGADT7 (Clontech). For BiFC constructs, the Pro35S:nLUC (pUC19-nLUC; Chen et al., 2008) and Pro35S:cLUC (pUC19-cLUC; Chen et al., 2008) cassette were first cloned into pCAMBIA2300 (CAMBIA). The resulting constructs were named JW771 and JW772. The coding regions of TT8, PAP1, and SPL9 were PCR amplified and cloned into JW771 and JW772, respectively. The promoter of DFR was PCR amplified using Pfusion DNA polymerase (New England Biolabs) and fused to the GUS coding region. A mutated version of the DFR promoter was generated by fusion PCR. For CoIP constructs, the coding regions of PAP1 and rSPL9 were linked to either HA or FLAG epitope tags by PCR and cloned into pCAMBIA2300. For PAP1-MIGS construct, the coding region of PAP1 was cloned into 173_CH42 (Felippes and Weigel, 2009) by replacing the CH42 fragment. Detailed information on all the constructs and primers can be found in Supplemental Tables 1 and 2 online.
The constructs were introduced into Agrobacterium tumefaciens strain GV3101 (pMP90) by the freeze-thaw method. Arabidopsis plants were transformed using the flower dip method (Clough and Bent, 1998). Transgenic seedlings were selected with 50 μg/mL kanamycin on plates or 0.1% glufosinate (BASTA) on soil. At least 50 T1 seedlings were analyzed for each construct.
Expression Analyses
Total RNA was extracted from stems or seedlings with the Plant RNeasy Mini kit (Qiagen). One microgram of total RNA was DNase I treated and used for cDNA synthesis with oligo(dT) primers and Superscript reverse transcriptase (Invitrogen). Quantitative RT-PCR was performed with SYBR-Green PCR Mastermix (Invitrogen), and amplification was real-time monitored on an Opticon Continuous Fluorescence Detection System (MJR). β-TUBULIN-2 was amplified as internal control. Oligonucleotide primers are given in Supplemental Table 1 online.
GUS staining was performed as described (Wang et al., 2008).
Yeast Two-Hybrid Assay
Plasmids were transformed into yeast strain AH109 (Clontech) by the LiCl-PEG method. Transformants were selected on SD -Leu -Trp plates. Interactions were tested on SD -Leu -Trp -His plates supplemented with 15 mM 3-amino-1,2,4,-triazole. Three independent clones for each transformation were tested.
ChIP Analysis
ChIP was performed as described (Wang et al., 2009). Briefly, crude chromatin extracts were split into three parts. One part was saved as input control and the other two parts were treated with anti-Myc or anti-GFP antibodies (Santa Cruz Biotechnology). After several washes, ChIP-ped DNA samples were reverse cross-linked and then purified using a PCR purification kit (Qiagen). One microliter of DNA was used for real-time PCR analyses. Relative enrichment of each fragment was calculated by normalizing the value for pulldown by anti-Myc against the value pulldown by anti-GFP using the following equation: 2−(Ct@Myc–Ct@GFP).
BiFC Analysis
The BiFC assay was performed as described (Chen et al., 2008). A. tumefaciens was infiltrated into N. benthamiana leaves as described (de Felippes and Weigel, 2010), with bacteria resuspended in infiltration buffer at OD600 = 0.8. Pro35S:P19-HA was coinfiltrated to inhibit gene silencing (Papp et al., 2003). Luciferin (1 mM) was infiltrated before LUC activity was monitored after 3 d.
CoIP Analysis
N. benthamiana leaves were infiltrated with A. tumefaciens harboring Pro35S:PAP1-HA or Pro35S:rSPL9-FLAG constructs. Leaves were harvested after 3 d and frozen in liquid nitrogen. Proteins were extracted with CoIP buffer (50 mM Tris-HCl, 100 mM NaCl, 10% glycerol, 5 mM EDTA, 0.1% Triton X-100, 0.2% Nonidet P-40, 50 μM MG132, and complete protease inhibitor cocktail tablet [Roche Diagnostics], pH 7.5) and centrifuged twice at 18,000g for 15 min. Monoclonal Anti-FLAG agarose (20 μL; Sigma-Aldrich) was added to 100 μL protein extract and incubated for 2 h. Beads were washed three times with CoIP buffer. Bound proteins were released by adding 2× protein loading buffer and boiling for 2 min at 95°C. The fusion proteins were detected by immunoblotting using monoclonal anti-HA-HRP (Sigma-Aldrich) and monoclonal anti-FLAG M2 antibody (Sigma-Aldrich).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: SPL3 (At2g33810), SPL9 (At2g42200), DFR (At5g42800), FLS1 (At5g208640), and PAP1 (At1g56650).
Author Contributions
J.-W.W. designed the research. J.-W.W., F.F.F., and J.-Y.G. performed the experiments. J.-Y.G., C.-J.L., D.W., and J.-W.W. analyzed the data. C.-J.L., D.W., and J.-W.W. wrote the article.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Seedlings of Mutant and Transgenic Plants.
Supplemental Figure 2. Role of SPLs in Anthocyanin Accumulation.
Supplemental Figure 3. Quantification of Putative Phenolic Derivatives.
Supplemental Figure 4. Inflorescences of Pro35S:MIR156 in tt8-1, pap1-migs, and ttg1-1 Backgrounds.
Supplemental Figure 5. SPL9 Binding to MYB113 in Yeast.
Supplemental Table 1. Oligonucleotide Primer Sequences.
Supplemental Table 2. Constructs.
Acknowledgments
We thank the European Arabidopsis Stock Centre for seeds, James C. Carrington for the Pro35S:P19-HA construct, Jian-Min Zhou for BiFC constructs, and Eunyoung Chae and Yasushi Kobayashi for discussion. This work was supported by an EMBO Long-Term Fellowship to J.-W.W. (ALTF 274-2006), by a DFG-SFB 446 grant, by European Community FP6 IPs SIROCCO (Contract LSHG-CT-2006-037900) and AGRON-OMICS (Contract LSHG-CT-2006-037704) to D.W., by the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the U.S. Department of Energy through Grant DEAC0298CH10886, and by the Pilot Program for Plant Epigenetic Study from the Office of Biological and Environmental Research, Department of Energy, to C.-J.L.
References
- Axtell M.J., Bowman J.L. (2008). Evolution of plant microRNAs and their targets. Trends Plant Sci. 13: 343–349 [DOI] [PubMed] [Google Scholar]
- Baudry A., Caboche M., Lepiniec L. (2006). TT8 controls its own expression in a feedback regulation involving TTG1 and homologous MYB and bHLH factors, allowing a strong and cell-specific accumulation of flavonoids in Arabidopsis thaliana. Plant J. 46: 768–779 [DOI] [PubMed] [Google Scholar]
- Baudry A., Heim M.A., Dubreucq B., Caboche M., Weisshaar B., Lepiniec L. (2004). TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 39: 366–380 [DOI] [PubMed] [Google Scholar]
- Bloor S.J., Abrahams S. (2002). The structure of the major anthocyanin in Arabidopsis thaliana. Phytochemistry 59: 343–346 [DOI] [PubMed] [Google Scholar]
- Borevitz J.O., Xia Y., Blount J., Dixon R.A., Lamb C. (2000). Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12: 2383–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broun P. (2005). Transcriptional control of flavonoid biosynthesis: A complex network of conserved regulators involved in multiple aspects of differentiation in Arabidopsis. Curr. Opin. Plant Biol. 8: 272–279 [DOI] [PubMed] [Google Scholar]
- Buer C.S., Imin N., Djordjevic M.A. (2010). Flavonoids: New roles for old molecules. J. Integr. Plant Biol. 52: 98–111 [DOI] [PubMed] [Google Scholar]
- Cardon G., Höhmann S., Klein J., Nettesheim K., Saedler H., Huijser P. (1999). Molecular characterisation of the Arabidopsis SBP-box genes. Gene 237: 91–104 [DOI] [PubMed] [Google Scholar]
- Chen H., Zou Y., Shang Y., Lin H., Wang Y., Cai R., Tang X., Zhou J.-M. (2008). Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 146: 368–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- de Felippes F.F., Weigel D. (2010). Transient assays for the analysis of miRNA processing and function. Methods Mol. Biol. 592: 255–264 [DOI] [PubMed] [Google Scholar]
- Dubos C., Le Gourrierec J., Baudry A., Huep G., Lanet E., Debeaujon I., Routaboul J.-M., Alboresi A., Weisshaar B., Lepiniec L. (2008). MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J. 55: 940–953 [DOI] [PubMed] [Google Scholar]
- Felippes F.F., Weigel D. (2009). Triggering the formation of tasiRNAs in Arabidopsis thaliana: The role of microRNA miR173. EMBO Rep. 10: 264–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla J.M., Valli A., Todesco M., Mateos I., Puga M.I., Rubio-Somoza I., Leyva A., Weigel D., García J.A., Paz-Ares J. (2007). Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 39: 1033–1037 [DOI] [PubMed] [Google Scholar]
- Gonzalez A., Zhao M., Leavitt J.M., Lloyd A.M. (2008). Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 53: 814–827 [DOI] [PubMed] [Google Scholar]
- Harborne J.B., Williams C.A. (2000). Advances in flavonoid research since 1992. Phytochemistry 55: 481–504 [DOI] [PubMed] [Google Scholar]
- Holton T.A., Cornish E.C. (1995). Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 7: 1071–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juniper B., Southwood R. (1986). Insects and the Plant Surface. (London, Baltimore: Edward Arnold; ). [Google Scholar]
- Klein J., Saedler H., Huijser P. (1996). A new family of DNA binding proteins includes putative transcriptional regulators of the Antirrhinum majus floral meristem identity gene SQUAMOSA. Mol. Gen. Genet. 250: 7–16 [DOI] [PubMed] [Google Scholar]
- Kropat J., Tottey S., Birkenbihl R.P., Depège N., Huijser P., Merchant S. (2005). A regulator of nutritional copper signaling in Chlamydomonas is an SBP domain protein that recognizes the GTAC core of copper response element. Proc. Natl. Acad. Sci. USA 102: 18730–18735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea U.S., Slimestad R., Smedvig P., Lillo C. (2007). Nitrogen deficiency enhances expression of specific MYB and bHLH transcription factors and accumulation of end products in the flavonoid pathway. Planta 225: 1245–1253 [DOI] [PubMed] [Google Scholar]
- Lev-Yadun S., Gould K.S. (2009). Role of anthocyanins in plant defence. In Anthocyanins: Biosynthesis, Functions, and Applications, Gould K., Davies K., Winefield C., eds (New York: Springer: ), pp. 21–48 [Google Scholar]
- Liang X., Nazarenus T.J., Stone J.M. (2008). Identification of a consensus DNA-binding site for the Arabidopsis thaliana SBP domain transcription factor, AtSPL14, and binding kinetics by surface plasmon resonance. Biochemistry 47: 3645–3653 [DOI] [PubMed] [Google Scholar]
- Lillo C., Lea U.S., Ruoff P. (2008). Nutrient depletion as a key factor for manipulating gene expression and product formation in different branches of the flavonoid pathway. Plant Cell Environ. 31: 587–601 [DOI] [PubMed] [Google Scholar]
- Liu C.-J., Deavours B.E., Richard S.B., Ferrer J.-L., Blount J.W., Huhman D., Dixon R.A., Noel J.P. (2006). Structural basis for dual functionality of isoflavonoid O-methyltransferases in the evolution of plant defense responses. Plant Cell 18: 3656–3669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui K., Umemura Y., Ohme-Takagi M. (2008). AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J. 55: 954–967 [DOI] [PubMed] [Google Scholar]
- Nodine M.D., Bartel D.P. (2010). MicroRNAs prevent precocious gene expression and enable pattern formation during plant embryogenesis. Genes Dev. 24: 2678–2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens D.K., Alerding A.B., Crosby K.C., Bandara A.B., Westwood J.H., Winkel B.S.J. (2008). Functional analysis of a predicted flavonol synthase gene family in Arabidopsis. Plant Physiol. 147: 1046–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp I., Mette M.F., Aufsatz W., Daxinger L., Schauer S.E., Ray A., van der Winden J., Matzke M., Matzke A.J. (2003). Evidence for nuclear processing of plant micro RNA and short interfering RNA precursors. Plant Physiol. 132: 1382–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier M.K., Burbulis I.E., Winkel-Shirley B. (1999). Disruption of specific flavonoid genes enhances the accumulation of flavonoid enzymes and end-products in Arabidopsis seedlings. Plant Mol. Biol. 40: 45–54 [DOI] [PubMed] [Google Scholar]
- Preuss A., Stracke R., Weisshaar B., Hillebrecht A., Matern U., Martens S. (2009). Arabidopsis thaliana expresses a second functional flavonol synthase. FEBS Lett. 583: 1981–1986 [DOI] [PubMed] [Google Scholar]
- Rubin G., Tohge T., Matsuda F., Saito K., Scheible W.-R. (2009). Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell 21: 3567–3584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R., Palatnik J.F., Riester M., Schommer C., Schmid M., Weigel D. (2005). Specific effects of microRNAs on the plant transcriptome. Dev. Cell 8: 517–527 [DOI] [PubMed] [Google Scholar]
- Schwarz S., Grande A.V., Bujdoso N., Saedler H., Huijser P. (2008). The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol. Biol. 67: 183–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikata M., Koyama T., Mitsuda N., Ohme-Takagi M. (2009). Arabidopsis SBP-box genes SPL10, SPL11 and SPL2 control morphological change in association with shoot maturation in the reproductive phase. Plant Cell Physiol. 50: 2133–2145 [DOI] [PubMed] [Google Scholar]
- Stracke R., De Vos R.C., Bartelniewoehner L., Ishihara H., Sagasser M., Martens S., Weisshaar B. (2009). Metabolomic and genetic analyses of flavonol synthesis in Arabidopsis thaliana support the in vivo involvement of leucoanthocyanidin dioxygenase. Planta 229: 427–445 [DOI] [PubMed] [Google Scholar]
- Stracke R., Ishihara H., Huep G., Barsch A., Mehrtens F., Niehaus K., Weisshaar B. (2007). Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 50: 660–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng S., Keurentjes J., Bentsink L., Koornneef M., Smeekens S. (2005). Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiol. 139: 1840–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohge T., et al. (2005). Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J. 42: 218–235 [DOI] [PubMed] [Google Scholar]
- Usami T., Horiguchi G., Yano S., Tsukaya H. (2009). The more and smaller cells mutants of Arabidopsis thaliana identify novel roles for SQUAMOSA PROMOTER BINDING PROTEIN-LIKE genes in the control of heteroblasty. Development 136: 955–964 [DOI] [PubMed] [Google Scholar]
- Veit M., Pauli G.F. (1999). Major flavonoids from Arabidopsis thaliana leaves. J. Nat. Prod. 62: 1301–1303 [DOI] [PubMed] [Google Scholar]
- Walker A.R., Davison P.A., Bolognesi-Winfield A.C., James C.M., Srinivasan N., Blundell T.L., Esch J.J., Marks M.D., Gray J.C. (1999). The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11: 1337–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.-W., Czech B., Weigel D. (2009). miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138: 738–749 [DOI] [PubMed] [Google Scholar]
- Wang J.-W., Park M.Y., Wang L.-J., Koo Y., Chen X.-Y., Weigel D., Poethig R.S. (2011). miRNA control of vegetative phase transition in trees. PLoS Genet. 7: e1002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.-W., Schwab R., Czech B., Mica E., Weigel D. (2008). Dual effects of miR156-targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell 20: 1231–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B. (2001). It takes a garden. How work on diverse plant species has contributed to an understanding of flavonoid metabolism. Plant Physiol. 127: 1399–1404 [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B. (2002). Biosynthesis of flavonoids and effects of stress. Curr. Opin. Plant Biol. 5: 218–223 [DOI] [PubMed] [Google Scholar]
- Wu G., Park M.Y., Conway S.R., Wang J.-W., Weigel D., Poethig R.S. (2009). The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138: 750–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Poethig R.S. (2006). Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133: 3539–3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing S., Salinas M., Höhmann S., Berndtgen R., Huijser P. (2011). miR156-targeted and nontargeted SBP-box transcription factors act in concert to secure male fertility in Arabidopsis. Plant Cell 22: 3935–3950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi A., Wu M.F., Yang L., Wu G., Poethig R.S., Wagner D. (2009). The microRNA-regulated SBP-Box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL, and APETALA1. Dev. Cell 17: 268–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yant L., Mathieu J., Dinh T.T., Ott F., Lanz C., Wollmann H., Chen X., Schmid M. (2010). Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2. Plant Cell 22: 2156–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K., Tohge T., Matsuda F., Nakabayashi R., Takayama H., Niida R., Watanabe-Takahashi A., Inoue E., Saito K. (2008). Comprehensive flavonol profiling and transcriptome coexpression analysis leading to decoding gene-metabolite correlations in Arabidopsis. Plant Cell 20: 2160–2176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu N., Cai W.-J., Wang S., Shan C.-M., Wang L.-J., Chen X.-Y. (2010). Temporal control of trichome distribution by microRNA156-targeted SPL genes in Arabidopsis thaliana. Plant Cell 22: 2322–2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Gonzalez A., Zhao M., Payne C.T., Lloyd A. (2003). A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130: 4859–4869 [DOI] [PubMed] [Google Scholar]
- Zimmermann I.M., Heim M.A., Weisshaar B., Uhrig J.F. (2004). Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J. 40: 22–34 [DOI] [PubMed] [Google Scholar]