Survey sequence and array hybridization data from flow-sorted barley chromosomes were integrated using a comparative genomics model to define an ordered gene map of the barley genome that contains approximately two-thirds of its estimated 32000 genes. The resulting high-resolution framework facilitated a genome-wide structural analysis of the barley genome and a detailed comparative analysis with wheat.
Abstract
We used a novel approach that incorporated chromosome sorting, next-generation sequencing, array hybridization, and systematic exploitation of conserved synteny with model grasses to assign ~86% of the estimated ~32,000 barley (Hordeum vulgare) genes to individual chromosome arms. Using a series of bioinformatically constructed genome zippers that integrate gene indices of rice (Oryza sativa), sorghum (Sorghum bicolor), and Brachypodium distachyon in a conserved synteny model, we were able to assemble 21,766 barley genes in a putative linear order. We show that the barley (H) genome displays a mosaic of structural similarity to hexaploid bread wheat (Triticum aestivum) A, B, and D subgenomes and that orthologous genes in different grasses exhibit signatures of positive selection in different lineages. We present an ordered, information-rich scaffold of the barley genome that provides a valuable and robust framework for the development of novel strategies in cereal breeding.
INTRODUCTION
Access to a genome sequence is now considered pivotal for unraveling key questions in crop plant biology and interrogating the molecular mechanisms that underpin trait formation. A genome sequence is central to the development of true genomics-informed breeding strategies and for unlocking the full potential of natural genetic variation for future crop improvement. Unfortunately for several key crops, deciphering a complete genome sequence to date has been precluded by the size and/or complexity of their genomes. Given the combined challenges of food security and climate change, it is vital that this situation is resolved and resources are developed that, even if not meeting an optimal gold standard, in the interim provide a high value and high utility surrogate.
Despite their importance in global agriculture, the Triticeae species wheat (Triticum aestivum; 2n=6x=42) and barley (Hordeum vulgare; 2n=2x=14), ranked 1 and 5 in world food production (FAOSTAT, 2007; http://faostat.fao.org/), are two such crops where genome size and complexity (17 Gbp for wheat [Bennett and Smith, 1976] and 5.1 Gbp for barley [Doležel et al., 1998]) so far preclude the development of such a gold standard reference genome sequence. Genomic data both from sequenced BAC clones and the application of next-generation sequencing (NGS) methodologies are available at a limited scale (Steuernagel et al., 2009; Wicker et al., 2009; http://www.cerealsdb.uk.net/) but lack the context required for broad and general utility. Given a close evolutionary relationship (divergence 13 million years ago [MYA]; Gaut, 2002) that has resulted in extensive conservation of synteny (Moore et al., 1995; Devos, 2005), it is generally accepted that elucidating a genome sequence for barley, a genetically tractable diploid inbreeder, would serve both its own genetics and breeding communities well while providing a faithful proxy for the genomically taxing 17 Gbp hexaploid bread wheat genome. This proposition is supported by agronomic traits such as flowering time and vernalization response being shared with wheat and the causal genes located at conserved genomic regions (Fu et al., 2005; Turner et al., 2005; Yan et al., 2006; Beales et al., 2007). Even race-specific disease resistance, a paradigm for species-specific genetic control in plants, shares conserved genetic elements in barley and wheat. Recently, a functional allele of the barley gene Mla, which confers resistance to the powdery mildew fungus (Zhou et al., 2001), was isolated from Triticum monococcum (Jordan et al., 2010). Indeed, an increasing body of information supports the notion of treating the Triticeae as a single genetic system.
Barley is itself an important crop. In addition to being the raw material for the brewing and distilling industry, barley is an important component of animal feed, can contribute health benefits in the human diet, and is agroecologically important, being planted worldwide on >57 million hectares (FAOSTAT, 2010; http://www.fao.org/faostat), often as an integral component of crop rotation management. Historically, it also has been an important model for classical genetics where its diploid genome has facilitated genetic analysis, a position that extended into the genomics era where early EST sequences provided resources for microarray design that in turn established routine functional genomics (Close et al., 2004; Druka et al., 2006). Subsequently, the same sequences were exploited to generate high-density gene maps using innovative marker technology (Stein et al., 2007; Potokina et al., 2008; Close et al., 2009; Sato et al., 2009a), and these opened the way for in-depth comparative analyses with other grass genomes (Bolot et al., 2009; Thiel et al., 2009; Abrouk et al., 2010; Murat et al., 2010). More recently, detailed information about barley genome composition has been accumulated using NGS technologies (Wicker et al., 2006, 2008, 2009). Despite the significance of each of these advances, the difficulties associated with fully unraveling the complex and repeat-rich 5.1-Gbp barley genome remain a significant challenge.
Recently, we demonstrated the potential of a cost-efficient and integrated cytogenetics, molecular genetics, and bioinformatics approach for generating a specific gene index for an entire barley chromosome. From a Roche 454 data set of 1.3-fold coverage generated from flow-sorted barley chromosome 1H, sequence signatures of >5000 genes were extracted and integrated with data from the rice (Oryza sativa) and sorghum (Sorghum bicolor) genomes to deliver a comprehensive virtual linear gene order model (Mayer et al., 2009). Here, we extended this approach by incorporating full-length cDNA (fl-cDNA) and DNA hybridization microarray data and applied it to the whole barley genome. This has allowed us to develop the first blueprint of a diploid Triticeae genome: a genome-wide putative linear gene index of barley embedded in a comparative grass genome organization model. The model is founded in an assembled series of genome zippers, a bioinformatics framework that exploits the extensive conservation of synteny observed between fully sequenced grass genomes.
RESULTS
Gene Content of Barley
We purified separately an entire barley chromosome (1H) and 12 chromosome arms (2HS to 7HL) by flow cytometry, amplified the DNA by multiple displacement amplification (MDA), and then shotgun sequenced the resulting preparations to 1.04- to 2.00-fold coverage using Roche 454 technology (Table 1; see Supplemental Table 1 online). At this depth of sequencing, base pair coverage for the individual samples was estimated to range between 64.7 and 86.5% according to Lander-Waterman genome assembly statistics (Lander and Waterman, 1988). We tested this estimate by comparing the individual sequence collections against a genetic map comprised of 2785 nonredundant gene-based single nucleotide polymporphism markers (Close et al., 2009). The observed gene (marker) discovery rate (i.e., the sensitivity) from individual chromosome arms ranged from 81.0 to 98.0% (average sensitivity of 85.9%; see Supplemental Data Set 1 online) exceeding the estimated values.
Table 1.
Sequence and Coverage Statistics of Individual Barley Chromosomes and Chromosome Arms
| Chromosome/Chromosome Arm | Size (Mbp) | Sequences (Mbp) | Sequences of High Quality (Mbp) | Reached Coverage (X-Fold) | Reached Coverage of High-Quality Sequences (X-Fold) | Expected Lander Waterman | Expected Lander Waterman of High-Quality Sequences | Observed Marker Detection Rate (Sensitivity) of High-Quality Sequences |
| 1H Morex | 622 | 798 | 675 | 1.28 | 1.09 | 72.00% | 66.38% | 95.18 |
| 1H Betzes | 622 | 813 | 569 | 1.31 | 0.91 | 73.01% | 59.74% | 88.55 |
| 1H (MoBe) | 622 | 1,611 | 1,244 | 2.60 | 2.00 | 92.57% | 86.46% | 98.19 |
| 2HS | 362 | 528 | 377 | 1.46 | 1.04 | 76.78% | 64.65% | 82.35 |
| 2HL | 428 | 924 | 670 | 2.16 | 1.57 | 88.47% | 79.20% | 86.24 |
| 3HS | 336 | 657 | 470 | 1.96 | 1.40 | 85.91% | 75.34% | 80.58 |
| 3HL | 419 | 1,155 | 744 | 2.76 | 1.78 | 93.67% | 83.14% | 85.95 |
| 4HS | 336 | 653 | 452 | 1.94 | 1.35 | 85.63% | 74.08% | 80.55 |
| 4HL | 393 | 911 | 605 | 2.32 | 1.54 | 90.17% | 78.56% | 83.01 |
| 5HS | 301 | 760 | 546 | 2.52 | 1.81 | 91.95% | 83.63% | 90.29 |
| 5HL | 459 | 949 | 651 | 2.07 | 1.42 | 87.38% | 75.83% | 83.03 |
| 6HS | 332 | 830 | 570 | 2.50 | 1.72 | 91.79% | 82.09% | 86.29 |
| 6HL | 357 | 981 | 587 | 2.75 | 1.64 | 93.61% | 80.60% | 86.38 |
| 7HS | 382 | 640 | 505 | 1.67 | 1.32 | 81.17% | 73.29% | 80.97 |
| 7HL | 373 | 636 | 468 | 1.70 | 1.25 | 81.73% | 71.35% | 84.89 |
| (Σ) 5,100 | (Σ) 11,235 | (Σ) 7,889 | (∅) 2.20 | (∅) 1.55 | (∅) 88.91% | (∅) 78.77% | (∅) 86.16 |
Basic statistics for chromosome (arm)-based shotgun sequencing of the barley genome. The table lists individual chromosome (arm) sizes, sequence data generated, coverage reached, the theoretical coverage as defined by the Lander Waterman equation, and the marker detection rate for the individual chromosome (arms). The accession used for sequencing was barley cultivar Betzes. For chromosome 1H, data previously generated in the barley cultivar Morex (Mayer et al., 2009) were combined with data generated in the cv Betzes. Statistics are given for the individual cultivars as well as the combined data set. Summary values given are from the combined Morex/Betzes data rather than the individual data sets.
We then assessed the purity of the chromosome/chromosome arm fractions by counting the proportion of false positive and true negative matches in the data set (i.e., the specificity). Specificities ranged from 88 to 98% (average 96.8%; see Supplemental Data Set 1 online). Applying a confusion matrix, the probability for correct classification reached between 0.89 and 0.97 (average 0.96) for individual chromosome arms (see Supplemental Table 2 online). These findings are consistent with a purity of enrichment estimated by fluorescent in situ hybridization analysis of the individual sorted chromosomal fractions (see Supplemental Table 3 online). Overall the data indicated >95% confidence that genes detected in a chromosome arm sequence data set originated from the assigned source.
To both validate and extend the 454 sequencing-based observations, we generated a complementary chromosome arm gene content data set by hybridizing individual preparations (in three replications) to barley long-oligonucleotide microarrays. In total, we were able to assign 16,804 genes on the array to individual chromosome arms at high confidence (see Supplemental Figure 1 online). Using the previously defined criteria, the genes assigned by array hybridization revealed an average specificity of 99%.
Given the high purity of the flow sorted chromosome samples, we attempted to determine a minimum set of genes for the barley genome. Both 454 sequence and array hybridization–based data sets were compared against complete model grass genomes using BLASTX (similarity ≥ 75% and ≥ 30 amino acids). From the 454 data, 17,290, 18,340, and 19,289 genes were detected from rice, sorghum, and Brachypodium distachyon, respectively, resulting in a cumulative set of 21,240 nonredundant homologous genes (Table 2). Sequence comparison of the 16,804 array-based unigenes assigned to barley chromosome arms identified an overlapping set of 11,708 genes that were also detected in the 454 sequence data. In total, 10,865 (93%) provided the same chromosomal assignment, consistent with chromosome purity estimates. Of these, 5096 genes were exclusively detected by microarray hybridization leading to an additional 3357, 3438, and 3908 homologous genes identified in rice, sorghum, and Brachypodium, respectively (totaling 4046 nonredundant genes) (Table 2). Thus, a cumulative set of 25,286 genes was detected by comparing 454 sequence and array-based data against all three model genomes (Table 2).
Table 2.
Estimated Gene Content of Barley
| Nonredundant Genes |
Nonredundant Genes (Cumulative) | |||
| Data Sets | Brachypodium | Rice | Sorghum | |
| Chr. arm 454 data | 19,289 | 17,290 | 18,340 | 21,240 |
| Chr. arm–specific array probes (16,804) | 12,382 (74%) | 10,617 (63%) | 10,915 (65%) | 12,755 (76%) |
| Chr. arm–specific array probes not overlapping with 454 data set (5,196) | 3,908 (75%) | 3,357 (65%) | 3,438 (66%) | 4,046 (78%) |
| Genes detected from 454 data and array hybridization | 23,197 | 20,647 | 21,778 | 25,286 |
| Nonredundant fl-cDNA (23,588) | 17,622 (75%) | 15,340 (65%) | 15,419 (65%) | 18,204 (77%) |
| Barley genes detected from 454, array hybridization, and fl-cDNA data | 29,163 | 28,895 | 29,947 | 30,670 |
| Estimated number of homologs considering complete genome 454 data | 22,429 (85%) | 20,104 (71%) | 21,325 (77%) | 24,698 |
| Number of matching nonredundant fl-cDNA against reference genomes (out of 23,588) | 17,622 (75%) | 15,340 (65%) | 15,419 (65%) | 18,204 (77%) |
| Estimated total (24,698/77 × 100) | 32,075 | |||
BLASTX comparisons against the reference genomes of Brachypodium, rice, and sorghum were undertaken using a stringent filter criterion of ≥75% sequence similarity spanning ≥30 amino acids. Sequence-tagged genes of barley deduced from similarity comparisons of Roche 454, array-based, and flcDNA data sets against reference genomes.
To determine how many barley genes can be detected in the three model genomes by stringent homology searches, we used a set of 23,588 nonredundant barley fl-cDNAs. These can be considered as an unbiased reference that represent randomly selected complete coding sequence of genes. In total, 5384 fl-cDNA’s remained without a corresponding match (similarity ≥ 75%, length ≥ 30 amino acids). Thus, some 23% of all barley genes lack sufficient sequence similarity to any gene of the three model grass genomes (Table 2). This is consistent with the value found for the hybridization-based results indicating that the array-based unigene set is a representative collection. Taking the 25,286 nonredundant barley genes detected from 454 and array-based data together with 5384 fl-cDNA that do not match homologs in the three model genomes gives an overall set of 30,670 sequence-supported barley genes.
Based on the experimental sensitivity of 86% for the 454 sequence data, the maximum cumulative overlap of nonredundant homologous genes between barley and the three model genomes would increase from 21,240 to 24,698 genes (Table 2). Since only 77% of the barley genes have a homolog in any of the three model genomes of rice, Brachypodium, or sorghum at the stringency applied, an overall content of ~32,000 (24,698/77 × 100) genes can be postulated for the entire barley genome (Table 2). This is in the range of the gene counts provided for the annotated Brachypodium, rice, and sorghum genomes (International Rice Genome Sequencing Project, 2005; Paterson et al., 2009; The International Brachypodium Initiative, 2010). In summary, we estimate that as many as 96% (30,670/32,000) of the barley gene repertoire is represented by either 454 sequence data, array-based unigenes, or fl-cDNAs used in this study.
A First Draft of the Linear Gene Order in the Barley Genome
To establish a hypothetical order for the genes assigned to chromosome arms, we constructed a multilayered scaffold based on conserved synteny for all barley chromosomes (see Supplemental Figure 2 online). We first identified syntenic regions for each chromosome arm in each of the three model grass genomes by sequence comparison of (repeat-masked) 454 sequences and hybridization probes. Figures 1 and 2 show the comparisons with Brachypodium and rice, respectively, and the sorghum comparison is presented in Supplemental Figure 3 online. The respective conserved syntenic regions were selected, and only genes that exhibited a corresponding match from barley 454 sequences and/or hybridization probes were used for integration into the barley scaffold. The mapped and ordered barley gene-based marker map comprising 2785 markers (Close et al., 2009) formed the integration scaffold for the detected orthologous genes and formed a genome-wide framework of sequence-based homology bridges upon which we interlaced all of the intervening genes present in the model genome sequences. Finally, we compiled (i.e., zipped up) the complementary sets of information to form a combined and ordered gene content model for seven barley pseudochromosomes. We call these genome zippers (see Supplemental Data Sets 2 to 8 online). They contain all of the genes in each of the three model species organized on a barley genetic framework associated with the corresponding barley genomic sequence tags, barley ESTs, and barley full-length cDNAs.
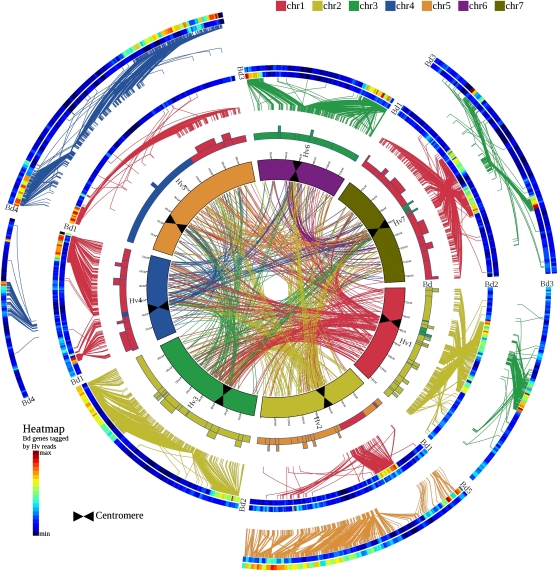
Figure 1.
High-Resolution Comparative Analysis between Barley and B. distachyon.
High-density comparative analysis of the linear gene order of the barley genome zippers versus the sequenced model grass genome of Brachypodium. The figure includes four sets of concentric circles: the inner circle represents the seven chromosomes of barley scaled according to the barley genetic map (bars at 10-cM intervals). Each barley chromosome is assigned a color according to the sequence on the color key, starting with chr1 through chr7. The positions of the barley centromeres are indicated by black bars. Moving outwards, the second circle illustrates a schematic model of the seven barley chromosomes, but this time color-coded according to blocks of conserved synteny with the model genome. The color coding is again based on the sequence on the color key, but this time is based on the model genome linkage groups, starting with chr1 through chr5 for Brachypodium. Boxes extending from these colored bars indicate regions involved in larger-scale structural changes (e.g., inversions). The outer partially complete circles of heat map colored bars represent pseudomolecules of the model genome linkage groups arranged according to conserved synteny with barley 1H-7H. When pairs of adjacent heat map bars are shown, they illustrate where the homologs of a short (inner heat map bar) or a long (outer heat map bar) barley chromosome arm data set is allocated to the respective model genome pseudochromosome. The heat maps illustrate the density of genes hit by the 454 shotgun reads from the relevant barley chromosome arm. Conserved syntenic regions are highlighted by yellow-red–colored regions. Putative orthologs between barley and the model genomes are connected with lines (colored according to model genome chromosomes) between the second and third circles. Colored lines in the center represent putative paralogous relationships between barley chromosomes on the basis of fl-cDNA supported genes included in the genome zipper models of the seven barley chromosomes.
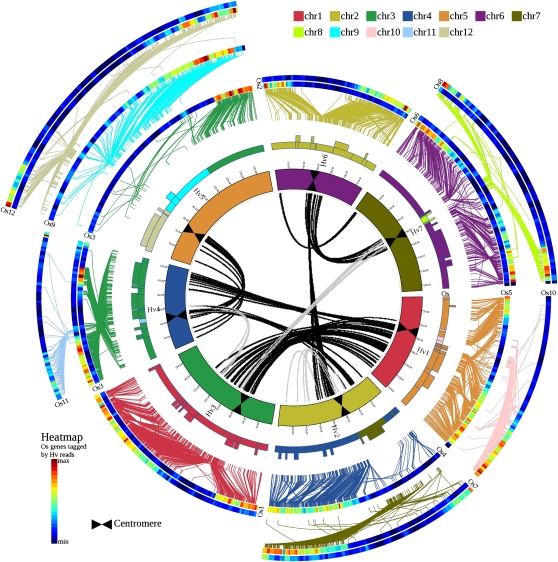
Figure 2.
High-Resolution Comparative Analysis between Barley and Rice.
High-density comparative analysis of the linear gene order of the barley genome zippers versus the sequenced model grass genome of rice. Details are as provided in the Figure 1 legend. Putative orthologs between barley and the rice genomes are connected with lines (colored according to model genome chromosomes) between second, third, and fourth circles. In the center, nine major segmental duplications of the barley genome are visualized as statistically significant groups of paralogous genes. Each line represents a duplicated gene (paralogous gene pair). Black lines indicate ancestral duplications shared with the model grass genomes, and gray lines highlight barley-specific duplications.
By this procedure, between 2261 and 3616 genes were tentatively positioned along each of the individual barley chromosomes, representing a cumulative set of 21,766 genes across the entire barley genome (Table 3, Figures 1 and 2; see Supplemental Figure 3 and Supplemental Data Sets 2 to 8 online). An additional set of 5815 genes could not be integrated into the genome zippers based on conserved synteny models but were associated with individual chromosomes/chromosome arms. Overall, we were able to tentatively position 27,581 barley genes, or 86% of the estimated 32,000 gene repertoire of the barley genome, into chromosomal regions.
Table 3.
Genome Zipper Statistics: Genes, ESTs, and 454 Reads Associated with the Genome Zipper
| Data Sets | 1H MoBe | 1H Morex | 1H Betzes | 2H | 3H | 4H | 5H | 6H | 7H | All |
| Number of markers | 332 | 332 | 332 | 468 | 445 | 314 | 492 | 337 | 397 | 2,785 |
| Number of markers with associated gene from reference genome(s) | 210 | 196 | 191 | 286 | 295 | 217 | 299 | 198 | 214 | 1,719 |
| Number of matched array hybridization probes | 732 | n.d. | n.d. | 2,044 | 1,502 | 1,242 | 1,935 | 1,407 | 2,003 | 10,865 |
| Number of matched fl-cDNAs | 1,676 | 1,287 | 1,247 | 1,619 | 1,628 | 1,255 | 1,474 | 1,058 | 1,395 | 10,105 |
| Number of nonredundant sequence reads | 51,972 | 28,485 | 17,716 | 29,250 | 30,576 | 21,402 | 25,262 | 19,536 | 22,420 | 200,418 |
| Number of nonredundant ESTs | 3,543 | 2,631 | 2,354 | 3,678 | 3,392 | 2,605 | 3,354 | 2,387 | 3,120 | 22,079 |
| Number of Brachypodium genes | 2,141 | 1,888 | 1,875 | 2,379 | 2,363 | 1,876 | 2,159 | 1,588 | 1,915 | 14,421 |
| Number of rice genes | 1,845 | 1,541 | 1,321 | 2,073 | 2,016 | 1,614 | 1,576 | 1,348 | 1,621 | 12,093 |
| Number of sorghum genes | 1,833 | 1,669 | 1,432 | 1,946 | 2,039 | 1,284 | 1,695 | 1,369 | 1,721 | 11,887 |
| Number of nonredundant anchored gene loci in Genome Zipper | 3,331 | 2,456 | 2,261 | 3,616 | 3,394 | 2,709 | 3,208 | 2,304 | 3,204 | 21,766 |
The table gives an overview of the data associated with and anchored along the chromosomal zippers. The number of markers is allocated to individual chromosomes. Data for the sequence collections of the individual cultivars used for 1H (Betzes and Morex) are listed separately as well as a combined data set (MoBe). n.d., not determined.
Positioning of Barley Centromeres
The genetic centromere of barley chromosomes is characterized by large clusters of genes/markers whose order cannot be genetically resolved due to insufficient recombination in relatively small mapping populations (n = 100 to 200). The analysis of DNA samples from individual arms of barley chromosomes 2H to 7H enabled us to deduce the transition from proximal (short) to distal (long) chromosome arms (i.e., the centromere position; see Supplemental Data Sets 2 to 8 online; genome zippers). For barley 1H, only entire chromosomes could be sorted. However, arm-specific information could be deduced based on available sorted chromosome arm shotgun sequence data of the highly collinear homoeologous chromosome 1A of wheat (T. Wicker, K.F.X. Mayer, and N. Stein, unpublished results). For all chromosomes, a single position (1H = 50 centimorgans [cM], 2H = 59.21 cM, 3H = 55.57cM, 4H = 48.72 cM, 5H = 51.3 cM, 6H = 55.36 cM, and 7H = 78.22 cM) was identified that contained genes allocated by 454 sequence reads to either the short or the long arm DNA data sets. Hence, we defined this to be the genetic position of the respective centromeres and ordered the genes here according to conserved synteny with the genomic models. Among 21,766 genes anchored to the genome zipper, 3125 (14%) genes were allocated to these genetic centromeres. Based on the 454 sequence- and array-based gene assignment to chromosome arms, we could distribute all but nine of these 3125 genes to specific arms of chromosomes 1H to 7H.
A Mosaic of Collinearity Is Observed between Barley and Model Grass Genomes
Shotgun sequencing and array hybridization provided chromosome arm gene content that was translated into tentative linear gene orders using conserved synteny-based genome zippers. This order provided an opportunity to step back and reappraise the overall extent of collinearity between barley and each of the three model grass genomes independently. Overall, 47, 20, and 33% of the loci anchored along the genome zippers were supported by conserved synteny in one, two, or all three model genomes, respectively. When barley gene order was compared with individual model genomes, we found that the number of conserved syntenic loci was similar in comparison with rice and sorghum (12,093 and 11,887, respectively) but was considerably higher with Brachypodium (14,422) reflecting a closer phylogenetic relationship. Overall, 20% of the loci anchored along the genome zippers were supported only by their order in the Brachypodium genome, while 14.5 and 13% were exclusively supported by either rice or sorghum, respectively.
To reach the highest stringency and to reduce the risk of paralogous gene comparisons between species, we restricted all further steps of comparative genome analysis to genes incorporated in the genome zipper that had barley fl-cDNA support. Blocks of conserved synteny were apparent between barley and the model genomes, and these were consistent with previous observations among the different clades of grasses (Bolot et al., 2009) (Figures 1 to 3). Since the gene order in barley was guided by a dense genetic map, we first assigned and then systematically compared the order and orientation of intervals among pairs or groups of genes to the model genomes. We identified numerous local inversions that appear to have either occurred specifically in barley, in one of the model genomes, or are shared between two genomes (Figure 3). For example, all inversions detected on the corresponding model genome segments of barley chromosome 3HL appear to be barley specific, since the order is conserved in all of the three model grass genomes. We then investigated patterns of ancestral whole-genome duplication in the barley genome. While this has been reported previously (Salse et al., 2009b; Thiel et al., 2009), the considerably increased gene coverage, particularly those with fl-cDNA support, along the genome zippers allowed us to recalculate paralogous relationships within the barley genome. This revealed a complex pattern of putatively duplicated genome segments (center of Figure 1). Using the alignment parameters and statistical tests defined by Salse et al. (2009a, 2009b), we identified nine major duplications (212 paralogous pairs) that cover 48% of the barley genome (center of Figure 2). Six of these corresponded to previously described ancestral segmental duplications shared between grass genomes. Three were considered barley specific. We thus substantiated in this analysis the previously reported paralogous gene content and duplicated block boundaries of such ancestral shared duplications in the Triticeae (Salse et al., 2008; Thiel et al., 2009).
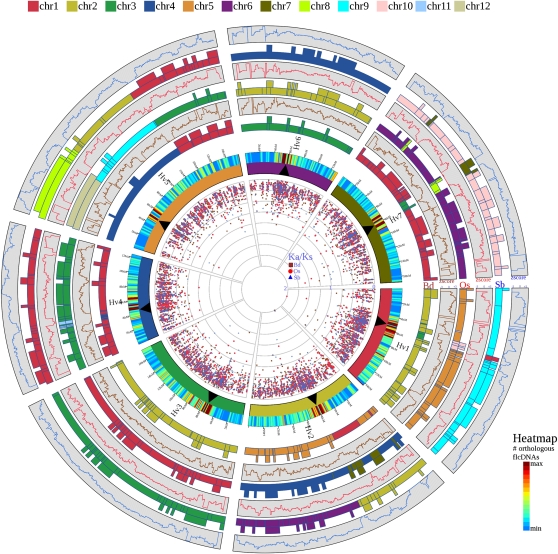
Figure 3.
Barley-Centered Four-Genome Comparative View of Grass Genome Collinearity.
The seven barley chromosomes (Hv1 to Hv7) are depicted by the inner circle of colored bars exactly as in Figure 1. The heat map attached to each chromosome indicates the density of barley fl-cDNAs anchored and positioned along the chromosomes according to the genome zipper models. Gene density is colored according to the heat map scale. Moving outwards, the bars represent a schematic diagram of the barley chromosomes colored according to conserved synteny with the genomes of Brachypodium (Bd), rice (Os), and sorghum (Sb), respectively. In each case, the chromosome numbers and segments are colored according to the chromosome color code (i.e., chr1 through chr5 for Bd, chr1 through chr12 for Os, and chr1 through chr10 for Sb). As in Figure 1, boxes extending from the colored bars indicate structural changes (e.g., inversions) between the gene order in barley and the respective model genome. To the outside of each model genome chromosome, box graphs show the z-score derived from a sliding window analysis of the frequency of fl-cDNAs present at a conserved syntenic position with their corresponding orthologs in Bd, Os, and Sb, respectively (see Methods for a full description of the analysis). A z-score >0 indicates higher than the average conservation of synteny, and a z-score <0 highlights decreased syntenic conservation. The data points in the center of the diagram depict the Ka/Ks ratios between barley full-length genes and their orthologs in Bd, Os, and Sb. Values against Bd are plotted as dark red rectangles, against Os in red circles, and against Sb in blue triangles.
There Is No Single Best Genomic Model for Barley
The principle uses of genomic models (certainly for wheat and barley) have been as predictors of regional candidate genes in positional cloning projects or for the development of gene-based markers that are tightly linked to a gene of interest. While these have been valid approaches, they frequently fail due to regional breakdown in the conservation of synteny. Given our newly available genomic information, we estimated the predictive value of individual model grass genomes for barley. We first associated the fl-cDNA supported linearly ordered barley genes with their orthologous counterparts in Brachypodium, rice, and sorghum. For this analysis, between 1247 and 1676 fl-cDNAs for each barley chromosome (average density of 9.3 fl-cDNAs per cM; 10,105 fl-cDNA/1090 cM) were tested. The extent of conserved synteny is not continuous for each barley genome segment/model genome species comparison. Therefore, a z-score within a sliding window (3-cM window, 0.1-cM shift) was calculated for comparison between each model species and barley to identify regions where conserved synteny was above or below average (z > 0 and z < 0, respectively) (Figure 3). Pronounced differences were observed along each chromosome, pinpointing regions where the degree of conserved synteny with individual model genomes was greater than with others. These differences highlighted the advantage of adopting an integrative approach that used three model genomes in parallel to overcome limitations imposed by species-specific regional differences. It enabled us to anchor and order loci even in regions where one or two of the model genomes may have contained structural rearrangements, gene loss, or translocations.
Fast-Evolving Genes
All full-length coding sequences (fl-cDNAs) that were ordered and positioned in the genome zippers at conserved syntenic positions (10,105) were then used to calculate the ratio of nonsynonymous (Ka) to synonymous substitutions (Ks) against their orthologs in the respective model genomes. We calculated the Ka/Ks ratios for all compared genes. The Ka/Ks ratio measures the strength of selection acting on a protein sequence under the assumption that synonymous substitutions evolve neutrally. A ratio <1 indicates purifying selection, and a ratio of >1 positive selection. The average Ka/Ks ratio of fl-cDNAs analyzed against Brachypodium (8160 genes), rice (7009 genes), and sorghum (6871 genes) is 0.21, 0.23, and 0.23, respectively, which indicates that the vast majority evolve under strong purifying selection. We chose a Ka/Ks ratio >0.8 as a cutoff to identify rapidly evolving genes that includes genes with few evolutionary constraints or positively selected genes. In total, 105 barley genes exhibited Ka/Ks values >0.8 in comparison to one (82 genes), two (15 genes), or all three (eight genes) model species, respectively (Figure 3; see Supplemental Figure 4 and Supplemental Data Set 9 online). These are assigned a wide range of putative molecular functions, including transcription factors and hormone responsive genes. Based on Ka/Ks ratios alone, these are candidates for conferring barley or Triticeae-specific phenotypic characteristics.
Rearrangements in Wheat A, B, and D Subgenomes
Within the Triticeae, the Hordeum (including barley) and the Triticum (including wheat) lineages split ~11 to 13 MYA (Gaut, 2002; Huang et al., 2002a), with the Triticum subgenomes radiating ~2.5 to 4.5 MYA. The tetraploid genome of Triticum turgidum (genome composition AABB) formed ~0.4 to 0.5 MYA, with a subsequent hybridization with Aegilops tauschii (DD) (~8000 years ago) forming the modern genome of allohexaploid bread wheat (genome composition AABBDD [Huang et al., 2002b]). Using the genome zipper derived fl-cDNA gene indices assembled into pseudochromosomes, we tested the widely held view that barley (HH) contains an archetypal Triticeae genome by comparing it to the previously constructed high-density physical marker map of wheat (Qi et al., 2004) (Figure 4; see Supplemental Figure 5 online). As expected, most of the chromosome arms exhibit well-conserved synteny with previously reported chromosomal translocations involving wheat 4A, 5A, and 7B accurately identified (Figure 4A; see Supplemental Figure 5 online). The availability of the barley genome zipper model allowed us also to estimate the gene content of the chromosomal fragments involved in such rearrangements (Figure 4B). Patterns of pericentric inversions could be deduced that confirmed previous observations involving wheat 2B, 3B, 4A, and 5A (Qi et al., 2006). The density of the compared data sets revealed regions that appear to be present in barley but lack counterparts in any of the homeologous wheat chromosomes (e.g., 1AS, 1AL, 2AL, and 2DL, all long arms of homeologous group 5 chromosomes; see Supplemental Figure 5 online); hence, blocks of barley genes cannot be assigned blocks of orthologs in the wheat bin map. Whether these regions have (1) been lost before the radiation of the wheat subgenomes, (2) have been integrated into barley independently, or (3) are simply not represented in the wheat EST bin map will only be resolved on the basis of more comprehensive data sets (e.g., by comparison to 454 sequence data of sorted wheat chromosomes). In addition, many small regions appeared to be absent in only one wheat subgenome, suggesting segmental loss possibly during or after major polyploidization events. Overall, at a structural level, no wheat subgenome was more similar to barley than any other and in terms of overall structural similarity and integrity, no conclusive evidence for more rapid structural evolution of any wheat subgenome was found. We conclude that most structural variation between A, B, and D genomes acts at a regional, maybe functional, level.
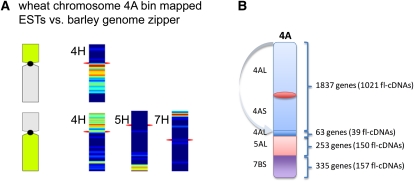
Figure 4.
Structure of Wheat Chromosome 4A in Relation to the Barley Genome Zipper.
Wheat subgenome specific markers of chromosome 4A have been compared against the genome zipper chromosome model of barley (for a genome-wide overview, see Supplemental Figure 5 online). Orthologous regions are depicted and visualized by a heatmap.
(A) Wheat EST markers allocated to 4AS cross-match to barley genes on 4HL and markers allocated to 4AS, a small region on 4AL, 5AL, and 7BS cross-match to 4HL. Thus, a reciprocal translocation involving chromosomes 4A and 5A and a translocation from 7BS to 4AL was detected. Compared with barley 4H, wheat chromosome 4A contains a pericentromeric inversion.
(B) The barley genome zipper model allows the size of the affected regions to be estimated and the minimal number of genes located in these rearranged regions of the wheat chromosomes to be predicted.
DISCUSSION
A complete reference genome sequence remains an aspiration for the barley research community, primarily due to technical and economic constraints resulting from the size and inherent complexity of its 5.1-Gbp genome. As a step toward that goal, we report here a high resolution sequence-based gene map containing an estimated 86% of the genes in the barley genome. We present the genome as a set of seven genome zippers that embrace the well-established conservation of synteny shown to exist among grass genomes. We propose that these genome zippers provide a high utility surrogate for both the barley genome itself and for closely related Triticeae cereals and are a high-resolution infrastructure upon which structural genomic information, such as physical maps, can be superimposed (Schulte et al., 2009).
The data used to derive the genome zippers were generated from low-pass 454 shotgun sequencing of individual flow-sorted barley chromosome/chromosome arm preparations and hybridization of equivalent subgenomic DNA preparations against a barley long oligonucleotide (gene) array. Both data sets are independent, exhibit high sensitivity and specificity, and show excellent concordance (>95%). Combining a recently developed 2785 gene-based genetic marker map (Close et al., 2009) with synteny information from model grass genomes provided the framework that enabled us to produce a highly structured and ordered sequence-based map comprising of 21,766 ordered barley genes. We consider that this ordering of genes along the chromosomes has reached a density and precision that can only be exceeded by a complete barley genome sequence.
This high-resolution view of the barley genome illuminates issues that have been faced in cereal genetics and breeding for many years. For example, we observed that 3125 genes fall into regions of the genome classified as genetic centromeres. These are regions where gene order cannot be established by meiotic mapping and where even crude assignment of genes to either proximal or distal chromosome arms has previously proved impossible. We were not only able to assign all but nine of these 3125 genes to the proximal or distal arms but also to propose a linear order. This allowed us to undertake genome scale analyses that included a fine-detail reappraisal of conservation of synteny with sequenced grass genomes, including an assessment of regional variation in the degree of conservation, an exploration of large-scale ancestral duplications, rearrangements, and more recent and local duplications. We present these for immediate exploitation by the Triticeae genetics and genomics community for both fundamental (i.e., physical map anchoring) or applied (i.e., candidate gene identification) purposes.
The clustering of genes toward genetic centromeres of barley has been well documented (Stein et al., 2007). In this study, one-third of all genes (6788 genes) in the genome zippers are located within 10-cM intervals that encompass each genetic centromere (6.4% of the entire barley genetic map). In wheat, sequencing megabase-sized BAC contigs selected from distributed regions of the chromosome 3B physical map revealed the presence of genes throughout the physical length of the chromosome, with a twofold higher concentration toward the telomeres (Choulet et al., 2010). Since regions with low recombination frequency per physical unit (hence, the regions around genetic centromeres) may extend in barley over as much as half a barley chromosome (Künzel et al., 2000), it can be expected that gene distribution in barley will follow a similar pattern as observed for wheat chromosome 3B. Unfortunately, this will place severe constraints on positional gene isolation for as many as one-third of barley genes. While the genome zippers will still provide a rich source of information for gene-based marker development and candidate gene identification in these regions, it is likely that innovative genetic strategies, such as deletion mapping or genome-wide association studies in highly diverse (e.g., wild) populations that have had orders of magnitude more opportunity for recombination, may be required (Waugh et al., 2009).
Due to their close evolutionary relationship, we investigated the degree of structural conservation between barley and wheat in more detail. As reported previously by comparing transcript map data to sequenced model genomes (Bolot et al., 2009), at a global level, a high degree of similarity was confirmed between the two species. Wheat chromosome 4A represents a notable exception, being a highly rearranged chromosome involving a large-scale inversion and two interchromosomal translocations (Mickelson-Young et al., 1995; Nelson et al., 1995; Miftahudin et al., 2004). The novelty of comparing the genome zipper model of barley to the wheat EST deletion bin map is that a better estimate of the genes involved can be made than by comparison to more distantly related models. Thus, several centromeric inversions that have been reported for the wheat genome (Qi et al., 2006) could also be deduced from our high-density comparison. These rearrangements appear to be wheat specific, not occurring at this frequency in the diploid barley genome. An apparent pericentromeric inversion shared by all wheat group one chromosomes likely indicates that the inversion occurred in barley in the period between the separation of the barley lineage and the radiation of wheat (i.e., some 11 to 4.5 to 2.5 MYA). Confirming this will require further experimentation. Based on the resolution of the bin-mapped wheat EST markers, many small regions appear to be missing from the individual wheat subgenomes. In contrast with all previous comparative analyses in the Triticeae, the genome zippers allow both the genetic size and the conserved (syntenic) gene content of the affected regions to be determined.
On a structural basis, none of the individual wheat A, B, or D subgenomes was more closely or distantly related to the H genome with numerous variations apparent in only one or two wheat subgenomes. This implies a highly complex, mosaic type, structural evolution of the A, B, and D subgenomes after radiation and the two subsequent polyploidization events that lead to the genomic composition of modern wheat (AABBDD). Such an outcome may have been predicted as a consequence of profound changes in genome structure and function induced by genomic shock in the early generations following the development of the allopolyploid (Chen, 2007). Indeed, in newly formed synthetic wheats, the reproducible elimination of specific sequences accounting for up to ~14% of the genomic DNA has been demonstrated and proposed to provide a physical mechanism for genetic diploidization in new allopolyploids (Feldman et al., 1997; Ozkan et al., 2001; Shaked et al., 2001). While local rearrangements, expansions, and single gene loss is beyond the currently available resolution, once a more complete genome sequence is available, the evolutionary dynamics between the H genome and the A, B, and D genomes of wheat can be expected to give important insights into genomic evolution and the structural and functional consequences of allopolyploidization.
We estimate that the barley genome contains in the order of 32,000 genes. Our estimate was based on (1) a stringent comparison of a comprehensive set of barley fl-cDNAs against sequenced model grass genomes and (2) the number of genes detected in 454 sequence and array-based data obtained from sorted barley chromosomes that matched a model genome homolog. Comparisons against model genomes detected 21,240 nonredundant genes. Given a sensitivity of 0.86, this would scale to 24,700 barley genes with a sequence homolog for the complete genome. Analysis of a set of 23,588 nonredundant barley fl-cDNAs revealed that using our stringent criteria 23% lack a sequence homologous counterpart in the model genomes. Taking this observation into account, we expect ~32,000 genes to be present in the barley genome. This number is remarkably consistent with gene number estimates for diploid grass model genomes (International Rice Genome Sequencing Project, 2005; Paterson et al., 2009; The International Brachypodium Initiative, 2010).
An estimate of 50,000 genes was given for a diploid wheat genome on the basis of megabase-sized BAC contig sequencing of chromosome 3B and short-read (Illumina/Solexa) survey sequencing of sorted 3B chromosomes (Choulet et al., 2010). Since the approaches used and the underlying sequence data differ, our analysis is not directly comparable to that of wheat 3B. For example, analysis of closely related expanded gene families, such as locally duplicated genes or translocated duplicated genes, cannot be appropriately addressed in shotgun sequences. Thus, paralogous gene families might in part have been interpreted as single genes, and consequently our gene number estimate may represent a lower limit.
The barley fl-cDNAs at conserved positions in all four genomes in the genome zipper allowed us to conduct a global survey for fast-evolving genes in barley by comparison to one, two, or all three sequenced model grass genomes and identified 105 genes with significant Ka/Ks values. We identified only eight barley genes that exhibited Ka/Ks ratios >0.8 in comparison to all three model grass genomes. Three genes were of unknown function and the remaining five genes can all be assigned to developmental roles based on their annotation. Two are transcription factors: one (NIASHv2057H16; see Supplemental Data Set 9 online) exhibiting strong similarity to a homeobox transcription factor Oshox24 (Agalou et al., 2008), which in rice shows differential expression in roots and panicle tissues at maturation. One was a rapid alkalinization factor, a class of genes shown to be involved in root and maybe also pollen development in different plant species (Germain et al., 2005; Wu et al., 2007; Zhang et al., 2010). Two genes encode homologs of pectin-methylesterase inhibitors (PMEIs). PMEIs inhibit the enzyme pectin-methylesterase, which is required for demethoxylation of methylated pectins, a necessary step before degradation by pectin-depolymerizing enzymes. pectin-methylesterases are ubiquitious enzymes in plants and their fine-tuned regulation (i.e., by PMEI) may be crucial during steps of development that require cell wall modifications (for review, see Jolie et al., 2010). It is tempting to speculate about the possible role of these five genes in specific developmental processes in barley. However, the significance of our observations as well as other possible mechanisms leading to evolution of species- and clade-specific traits like diversification of gene expression regulation (reviewed in Rosin and Kramer, 2009) will require future experimental testing.
Linear gene order information as provided by the barley genome zippers will be vital for the generation of a complete genome reference for barley. The development of a high information content fingerprint BAC-based physical map of the barley genome is well advanced (Schulte et al., 2009), and this effort will likely profit from the presented data sets for anchoring the physical map to a genetic/syntenic framework. Referring to the model character of barley for other Triticeae genomes, such a detailed barley framework will play a pivotal role in the assembly of data that could be generated for other Triticeae species. An obvious primary target is of course wheat (Kubaláková et al., 2002) and survey sequencing of chromosomes for the construction of a genome-wide collection of wheat genome zippers has already been initiated (IWGSC; http://www.wheatgenome.org/Projects). The approach is equally attractive for rye (Secale cereale; Kubaláková et al., 2003). More generally, the approach may be adopted as an economic and technical paradigm for other unsequenced orphan crop genomes where individual chromosomes, chromosome arms, or translocations can be separated by flow sorting techniques. These include legumes such as chickpea (Cicer arietinum; Vláčilová et al., 2002), garden pea (Pisum sativum; Neumann et al., 2002), and field bean (Phaseolus vulgaris; Doležel and Lucretti, 1995) where the feasibility of chromosome flow sorting has previously been demonstrated.
The genome zipper–based linear gene order model of two-thirds of all barley genes will open a path toward contextualized genome-wide diversity analysis in barley. Currently available NGS technology allows for whole-genome shotgun sequencing and de novo assembly to draft sequence quality even of complex mammalian genomes (Li et al., 2010). With the currently available technology, a similar attempt in barley could lead to assembled gene sequence information and thus provide a genomic reference for genes of the genome zipper. Using this information as reference for resequencing, polymorphism surveys will become a realistic endeavor for the majority of the barley gene space. In combination with the appropriate plant material, such as the well-characterized mutant collections available in barley (Druka et al., 2010), we may soon be able to clone the genes that are responsible for many phenotypic traits by direct resequencing, similar to approaches successfully applied in Arabidopsis thaliana (Schneeberger et al., 2009).
METHODS
Purification and Amplification of Chromosomal DNA
Intact mitotic chromosomes/arms were isolated by flow cytometric sorting from barley Hordeum vulgare cultivar Morex and cv Betzes (1H) and wheat (Triticum aestivum)-barley telosome addition lines (2HS-7HL arms originating from cv Betzes). The purity in the sorted fractions was determined by fluorescence in situ hybridization essentially as described previously (Suchánková et al., 2006). The DNA of sorted chromosomes was purified and amplified by MDA as described previously (Šimková et al., 2008).
Roche 454 Sequencing
DNA amplified from sorted chromosomes was used for 454 shotgun sequencing. Five micrograms of individual chromosome arm MDA DNAs were used to prepare the 454 sequencing libraries using the GS Titanium General Library preparation kit following the manufacturer’s instructions (Roche Diagnostics). The 454 sequencing libraries were processed using the GS FLX Titanium LV emPCR (Lib-L) and GS FLX Titanium Sequencing (XLR70) kits (Roche Diagnostics) according to the manufacturer's instructions. Sequencing details are summarized in Table 1 and Supplemental Table 1 online.
Microarray Construction and Analysis
A custom microarray SCRI_Hv35_44k_v1 (Agilent design 020599) representing 42,302 barley sequences was generated. Barley sequences for this design were selected from a total of 50,938 unigenes from HarvEST assembly 35 (http://www.harvest-web.org/) representing ~450,000 ESTs. Selection criteria were based upon the ability to define orientation derived from (1) homology to members of the nonredundant protein database (NCBI nr), (2) homology to ESTs known to originate from directional cDNA libraries, and (3) presence of a significant poly(A) tract. The microarray was designed with one 60mer probe per selected unigene in 4 × 44k format using default parameters in the Web-based Agilent eArray software (https://earray.chem.agilent.com/earray/) and includes recommended QC control probes. Full details of array design, probe sequences, and unigene accession numbers can be found at ArrayExpress (http://www.ebi.ac.uk/microarray-as/ae/; accession number A-MEXP-1728). Due to the redundancy in the EST-based unigene data set used as a basis for array design, the microarray comprised an estimated 25 to 32,000 nonredundant barley genes (Michael Bayer, personal communication; each gene was represented on average by ~1.3 to 1.7 probes per genes).
Fluorescent Labeling of Chromosome DNA and Hybridization to Barley Microarrays
Amplified chromosomal DNA was labeled using a modified Bioprime DNA labeling system (Invitrogen). For each sample, 2 μg amplified genomic DNA in 21 μL was added to 20 μL Random Primer Reaction Buffer and denatured at 95°C for 5 min prior to cooling on ice. To this, 5 μL modified 10× deoxynucleotide triphosphate mix (1.2 mM each of dATP, dGTP, and dTTP, 0.6 mM dCTP, 10 mM Tris, pH 8.0, and 1 mM EDTA), 3 μL of either Cy3 or Cy5 dCTP (1 mM), and 1 μL Klenow enzyme was added and incubated for 16 h at 37°C. Labeled samples for each array were combined and unincorporated dyes removed using the MinElute PCR purification kit (Qiagen) as recommended, eluting twice with 1× 10 μL sterile water. Specific activities of incorporated dyes (nmol/μg DNA) were estimated using spectrophotometry.
The design of the microarray experiment is detailed in ArrayExpress (accession number E-TABM-1063) and ensured that independent replicate samples of each amplified chromosome arm were labeled once with each of two fluorescent dyes, Cy3 and Cy5, to minimize dye bias. Microarray hybridization and washing were conducted according to the manufacturer's protocols as for gene expression arrays (Agilent Two-Color Microarray-Based Gene Expression Analysis, version 5.5). For each array, 20 μL purified labeled samples were added to 5 μL 10× blocking aent and heat denatured at 98°C for 3 min then cooled to room temperature. GE Hybridization Buffer HI-RPM (25 μL) was added and mixed prior to hybridization at 65°C for 17 h at 10 rpm. Array slides were dismantled in Agilent Wash 1 buffer and washed in Wash 1 buffer for 1 min, then Agilent Wash 2 buffer for 1 min, and centrifuged dry. Hybridized slides were scanned using an Agilent G2505B scanner at resolution of 5 μm at 532 nm (Cy3) and 633 nm (Cy5) wavelengths with extended dynamic range (laser settings at 100 and 10%).
Microarray Data Extraction and Analysis
Microarray images were imported into Agilent Feature Extraction (FE v.10.5.1.1) software and aligned with the appropriate array grid template file (020599_D_F_20080612). Intensity data and QC metrics were extracted using a suitable FE protocol (GE2-v5_95_Feb07), and data from each array were normalized in FE using the LOWESS (locally weighted polynomial regression) algorithm to minimize differences in dye incorporation efficiency (Yang et al., 2002). Entire normalized data sets for both channels of each array were loaded into GeneSpring (v.7.3.1) software for further analysis. Data were subjected to additional normalization whereby values were set to a minimum of 5.0, data from each array were scaled to the 50th percentile of all measurements on the array, and the signal from each probe was subsequently normalized to the median of its values. Unreliable data with consistently low probe intensity levels (raw values <100) in all replicate samples were discarded. Statistical filtering of data for each experiment was performed using analysis of variance with Benjamini and Hochberg (Benjamini and Hochberg, 1995) false discovery rate for multiple testing correction (P value <0.005). Heat maps were generated from filtered probe/gene lists using an average linkage clustering algorithm based upon Pearson correlation using default parameters in GeneSpring. Clustered probes enriched for each chromosome arm were selected manually from the gene tree.
General Sequence Analysis
Repeat Masking of 454 Sequence Data
To determine genic regions covered by 454 sequencing data, the content of repetitive DNA per sequence read was masked after being identified using Vmatch (http://www.vmatch.de) against the MIPS-REdat Poaceae v8.2 repeat library (contains known grass transposons from the Triticeae Repeat Database, http://wheat.pw.usda.gov/ITMI/Repeats, as well as de novo detected LTR retrotransposon sequences from several grass species, specifically, maize [Zea mays],12434; sorghum [Sorghum bicolor], 7500; rice [Oryza sativa], 1928; Brachypodium distachyon, 466; wheat, 356; and barley, 86 sequences) by applying the following parameters: 60% identity cutoff, 30-bp minimal length, seed length 14, exdrop 5, and e-value 0.001.
Identification of Genetic Markers in the 1H-7H Data Sets
The repeat-masked sequence collections from all seven barley chromosomes were compared (BLASTN) against 2785 nonredundant (of total 2943) EST-based markers (Close et al., 2009; http://harvest.ucr.edu) under optimized parameters (-r 1 -q -1 -W 9 -G 1 -E 2: -r reward for a nucleotide match, default = 1; -q penalty for a nucleotide mismatch, default = -3; -W word size, default; -G cost to open a gap, default = -1; -E cost to extend a gap, default = -1). Only BLAST matches exceeding an identity threshold of 98% and an alignment length of 50 bp were considered.
A Nonredundant Set of Barley fl-cDNA
In this study, a set of 5006 (Sato et al., 2009b) and a set of 23,623 barley full-length cDNAs (Matsumoto et al., 2011) was used for sequence comparison. All redundant cDNA sequences were removed and a database of 23,588 nonredundant fl-cDNAs was generated for further steps of analysis using CD-HIT-EST (http://www.bioinformatics.org/cd-hit/) applying the following parameter settings: -c 0.98 and -n 8 (-c sequence identity threshold, default 0.9; -n word length, default 5).
Overall Gene Content in the Combined Chromosome-Specific Barley Sequence Data Set
To estimate the number of barley genes that have been captured in the barley sequence collection generated by Roche 454 sequencing, BLASTX (Altschul et al., 1990) comparisons were performed with the repeat-filtered 454 sequence reads, the microarray probe sets, and the nonredundant fl-cDNAs against Brachypodium, rice, and sorghum proteins (Brachypodium genome annotation v1.2 [ftp://ftpmips.helmholtz-muenchen.de/plants/brachypodium/v1.2]; rice RAP-DB genome build 4 [http://rapdb.dna.arc.go.jp]; sorghum genome annotation v1.4 [http://genome.jgi-psf.org/Sorbi1/Sorbi1.download.ftp.html]; Paterson et al., 2009). The number of tagged genes and the number of gene matching reads and fl-cDNAs were counted after filtering according to the following criteria: (1) the best hit display with a similarity >75% and (2) an alignment length ≥30 amino acids. To increase specificity, microarray probes (length of 60 nucleotides) were associated with their respective cognate EST. These were used for subsequent integration using the parameters above.
Association of Barley fl-cDNA and EST to Individual Barley Chromosomes (Arms)
The putative chromosomal origin of barley cDNA and EST collections (HarvEST barley v1.73, assembly 35; http://harvest.ucr.edu/) was determined by BLASTN comparison against the repeat masked shotgun sequence reads from all seven barley chromosomes. Only the best hits with an identity of >98% and a minimal alignment length of 50 bp were considered. Each cDNA or EST was assigned to a particular chromosome (arm) if at least 80% of associated shotgun sequence reads were assigned to the same chromosome.
Assessment of Linear Gene Order in Barley (Genome Zipper)
Conserved synteny between three model grass genomes was used as a template to develop a linear gene order model (genome zipper) of the genes assigned to individual barley chromosomes by the analysis steps described above. The workflow toward a so-called genome zipper of a given barley chromosome was designed to structure and order barley genes identified either by 454 shotgun sequencing of or microarray hybridization to sorted chromosomal DNA on the basis of collinearity to model grass genomes. As a first step, the repeat masked shotgun sequences and array probes associated with each individual chromosome/chromosome arm were compared (BLASTX) against the three reference genomes Brachypodium, sorghum, and rice. Genes from syntenic regions, as defined by the density of homology matches, from the three genomes were selected and compared with the dense gene-based marker map of barley, which served as a scaffold to anchor collinear segments from model genomes. This step was performed for the three model grass genomes and results are interlaced based on joint marker associations as well as best bidirectional hit (bbh) classification. Sequence-tagged genes are anchored to the marker scaffold and additional tagged genes without barley marker association were ordered following the concept of conserved synteny and closest evolutionary distance. Finally the integrated syntenic scaffolds were associated with fl-cDNAs, array probes, ESTs, and shotgun reads that exhibited matches to the syntenic genes and the barley EST-based marker. Genome zipper–based tentative gene order, including associated information, is provided in Supplemental Data Sets 2 to 8 online.
Analysis of Conserved Synteny
The degree of conserved synteny against each of the model grass genomes rice, sorghum, and Brachypodium was calculated using a sliding window approach. For each genetic position (3-cM window, window shift 0.1 cM), the number of syntenic genes (classified as syn+) divided by the sum of all genes (syntenic and nonsyntenic, syn+ and syn-) was calculated (=conserved synteny). Genome-wide local differences were analyzed by calculating the z-score to indicate regions with above average and below average conservation (z > 0 and z < 0, respectively).
Calculation of Synonymous and Nonsynonymous (Ka/Ks) Substitution Rates
Sequence divergence as well as speciation event dating analysis based on the rate of nonsynonymous (Ka) versus synonymous (Ks) substitutions was calculated using the YY00 program within the PAML suite (phylogenetic analysis by maximum likelihood) (Nei and Gojobori, 1986; Yang, 2007). Only high-quality alignments and depending on the number of detectable orthologs 2, 3, or 4 sequences were used.
Analysis of Traces of Genome Duplications in Barley
Analysis was performed using the procedure and definitions defined previously (Salse et al., 2009a, 2009b) as well as by a best BLAST hit (bbh) strategy. Sequence divergence and speciation event dating analysis based on the rate of nonsynonymous (Ka) versus synonymous (Ks) substitutions was calculated and an average substitution rate (r) of 6.5 × 10−9 substitutions per synonymous site per year (Gaut et al., 1996; SanMiguel et al., 1998). The time (T) since gene insertion has been estimated using the formula T = Ks/r.
Analysis of Synteny between Barley and Homoeologous Wheat Chromosomes
Barley fl-cDNAs integrated in the barley genome zipper were concatenated following the order assigned in the genome zipper (with spacer sequences between individual genes) to result in approximated chromosome scaffolds. These scaffolds were compared against the high-density physical wheat transcript map (deletion bin map; Qi et al., 2004) using BLASTN (identity ≥85%, match length ≥100 nucleotides). Matching and nonmatching genes were depicted independently for the A, B, and D derived markers in a heat map following the assigned gene order from the barley genome zippers.
Data Availability and Accession Numbers
The nonredundant set of 23,588 fl-cDNAs was generated from a set of 5006 fl-cDNAs (Sato et al., 2009b; accession numbers AK248134 to AK253139) and a set of 23,623 fl-cDNAs (Matsumoto et al., 2011; accession numbers AK353559 to AK377172). All 454 sequence information in this study generated from flow-sorted chromosomes was submitted to the European Bioinformatics Institute sequence read archive under accession number ERP000445. A database for sequence homology search (BLAST) is provided at http://webblast.ipk-gatersleben.de/barley/. All data contained in the genome zipper models can be downloaded as Excel spread sheets from http://mips.helmholtz-muenchen.de/plant/triticeae/genomes/index.jsp.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Hierarchical Clustering of Microarray Hybridization to Sorted Chromosomal DNA of Barley.
Supplemental Figure 2. Flow Chart for the Genome Zipper Analysis Pipeline.
Supplemental Figure 3. Conservation of Synteny between Barley and Sorghum.
Supplemental Figure 4. Number of Genes with Ka/Ks Values of >0.8 between Barley and Brachypodium, Rice, and Sorghum.
Supplemental Figure 5. Global Analysis of Barley/Wheat Conserved Synteny on the Basis of the Genome Zipper Model.
Supplemental Table 1. Sequencing Statistics for Individual Chromosomes and Chromosome Arm.
Supplemental Table 2. Accuracy (the Proportion of True Results) of Sequence Read Distribution to Mapped Barley Markers.
Supplemental Table 3. Summary of Flow-Sorted Chromosome Fractions and Their Purities as Determined by FISH.
Supplemental Data Set 1. 454 Sequence Read Distribution to Barley EST-Based Markers.
Supplemental Data Sets 2 to 8. Genome Zipper of Barley Chromosomes 1H to 7H, Respectively.
Supplemental Data Set 9. Genes with Evidence for Positive Selection as Based on Ka/Ks Signatures.
Acknowledgments
We thank Jarmila Číhalíková, Romana Šperková, and Zdenka Dubská for assistance with chromosome sorting and DNA amplification as well as Susanne König and Jana Schellwat for Roche 454 sequencing of sorted chromosomes. We are grateful to the Cereal Research Institute Kroměříž for the seeds of barley cultivar Betzes and the National Bioresource Project-Wheat (Japan) for seeds of wheat-barley telosome addition lines. We thank David Marshall, Karl Schmid, and three anonymous reviewers for constructive and helpful comments on the manuscript. We kindly acknowledge Bjoern Usadel, Birgit Kersten Diego Riano-Pachon, and Doreen Pahlke from GABI-PD (Genome Analysis in the Biological system plant–Primary Database) for support with submission of sequence data sets to European Bioinformatics Institute. This work was financially supported by the following grants: 0314000 GABI Barlex from the Bundesministerium für Bildung und Forschung to N.S., K.F.X.M., M.P., and U.S.; FP7-212019 TriticeaeGenome from the European Union commission to N.S., R.W., K.F.X.M., and J.D.; and the Ministry of Education, Youth, and Sports of the Czech Republic and the European Regional Development Fund (Operational Programme Research and Development for Innovations No. CZ.1.05/2.1.00/01.0007). J.D., R.W., K.F.X.M., and N.S. collaborated in the frame of European Cooperation in Science and Technology action FA0604 Tritigen.
References
- Abrouk M., Murat F., Pont C., Messing J., Jackson S., Faraut T., Tannier E., Plomion C., Cooke R., Feuillet C., Salse J. (2010). Palaeogenomics of plants: Synteny-based modelling of extinct ancestors. Trends Plant Sci. 15: 479–487 [DOI] [PubMed] [Google Scholar]
- Agalou A., et al. (2008). A genome-wide survey of HD-Zip genes in rice and analysis of drought-responsive family members. Plant Mol. Biol. 66: 87–103 [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Beales J., Turner A., Griffiths S., Snape J.W., Laurie D.A. (2007). A pseudo-response regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum L.). Theor. Appl. Genet. 115: 721–733 [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57: 289–300 [Google Scholar]
- Bennett M.D., Smith J.B. (1976). Nuclear DNA amounts in angiosperms. Philos. Trans. R. Soc. Lond. B Biol. Sci. 274: 227–274 [DOI] [PubMed] [Google Scholar]
- Bolot S., Abrouk M., Masood-Quraishi U., Stein N., Messing J., Feuillet C., Salse J. (2009). The ‘inner circle’ of the cereal genomes. Curr. Opin. Plant Biol. 12: 119–125 [DOI] [PubMed] [Google Scholar]
- Chen Z.J. (2007). Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu. Rev. Plant Biol. 58: 377–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choulet F., et al. (2010). Megabase level sequencing reveals contrasted organization and evolution patterns of the wheat gene and transposable element spaces. Plant Cell 22: 1686–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Close T.J., et al. (2009). Development and implementation of high-throughput SNP genotyping in barley. BMC Genomics 10: 582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Close T.J., Wanamaker S.I., Caldo R.A., Turner S.M., Ashlock D.A., Dickerson J.A., Wing R.A., Muehlbauer G.J., Kleinhofs A., Wise R.P. (2004). A new resource for cereal genomics: 22K barley GeneChip comes of age. Plant Physiol. 134: 960–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos K.M. (2005). Updating the ‘crop circle’. Curr. Opin. Plant Biol. 8: 155–162 [DOI] [PubMed] [Google Scholar]
- Doležel J., Greilhuber J., Lucretti S., Meister A., Lysák M.A., Nardi L., Obermayer R. (1998). Plant genome size estimation by flow cytometry: Inter-laboratory comparison. Ann. Bot. (Lond.) 82: 17–26 [Google Scholar]
- Doležel J., Lucretti S. (1995). High-resolution flow karyotyping and chromosome sorting in Vicia faba lines with standard and reconstructed karyotypes. Theor. Appl. Genet. 90: 797–802 [DOI] [PubMed] [Google Scholar]
- Druka A., Franckowiak J., Lundqvist U., Bonar N., Alexander J., Houston K., Radovic S., Shahinnia F., Vendramin V., Morgante M., Stein N., Waugh R. (2010). Genetic dissection of barley morphology and development. Plant Physiol. 155: 617–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druka A., et al. (2006). An atlas of gene expression from seed to seed through barley development. Funct. Integr. Genomics 6: 202–211 [DOI] [PubMed] [Google Scholar]
- Feldman M., Liu B., Segal G., Abbo S., Levy A.A., Vega J.M. (1997). Rapid elimination of low-copy DNA sequences in polyploid wheat: A possible mechanism for differentiation of homoeologous chromosomes. Genetics 147: 1381–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu D., Szűcs P., Yan L., Helguera M., Skinner J.S., von Zitzewitz J., Hayes P.M., Dubcovsky J. (2005). Large deletions within the first intron in VRN-1 are associated with spring growth habit in barley and wheat. Mol. Genet. Genomics 273: 54–65 [DOI] [PubMed] [Google Scholar]
- Gaut B.S. (2002). Evolutionary dynamics of grass genomes. New Phytol. 154: 15–28 [Google Scholar]
- Gaut B.S., Morton B.R., McCaig B.C., Clegg M.T. (1996). Substitution rate comparisons between grasses and palms: Synonymous rate differences at the nuclear gene Adh parallel rate differences at the plastid gene rbcL. Proc. Natl. Acad. Sci. USA 93: 10274–10279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain H., Chevalier É., Caron S., Matton D.P. (2005). Characterization of five RALF-like genes from Solanum chacoense provides support for a developmental role in plants. Planta 220: 447–454 [DOI] [PubMed] [Google Scholar]
- Huang S., Sirikhachornkit A., Faris J.D., Su X., Gill B.S., Haselkorn R., Gornicki P. (2002a). Phylogenetic analysis of the acetyl-CoA carboxylase and 3-phosphoglycerate kinase loci in wheat and other grasses. Plant Mol. Biol. 48: 805–820 [DOI] [PubMed] [Google Scholar]
- Huang S., Sirikhachornkit A., Su X., Faris J., Gill B., Haselkorn R., Gornicki P. (2002b). Genes encoding plastid acetyl-CoA carboxylase and 3-phosphoglycerate kinase of the Triticum/Aegilops complex and the evolutionary history of polyploid wheat. Proc. Natl. Acad. Sci. USA 99: 8133–8138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Brachypodium Initiative (2010). Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463: 763–768 [DOI] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project (2005). The map-based sequence of the rice genome. Nature 436: 793–800 [DOI] [PubMed] [Google Scholar]
- Jolie R.P., Duvetter T., Van Loey A.M., Hendrickx M.E. (2010). Pectin methylesterase and its proteinaceous inhibitor: A review. Carbohydr. Res. 345: 2583–2595 [DOI] [PubMed] [Google Scholar]
- Jordan T., Seeholzer S., Schwizer S., Keller B. (2010). The wheat Mla homolog TmMla1 exhibits an evolutionary conserved function against powdery mildew in both wheat and barley. Plant J. 65: 610–621 [DOI] [PubMed] [Google Scholar]
- Kubaláková M., Vrána J., Cíhalíková J., Simková H., Doležel J. (2002). Flow karyotyping and chromosome sorting in bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 104: 1362–1372 [DOI] [PubMed] [Google Scholar]
- Kubaláková M., Valárik M., Barto J., Vrána J., Cíhalíková J., Molnár-Láng M., Doležel J. (2003). Analysis and sorting of rye (Secale cereale L.) chromosomes using flow cytometry. Genome 46: 893–905 [DOI] [PubMed] [Google Scholar]
- Künzel G., Korzun L., Meister A. (2000). Cytologically integrated physical restriction fragment length polymorphism maps for the barley genome based on translocation breakpoints. Genetics 154: 397–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E.S., Waterman M.S. (1988). Genomic mapping by fingerprinting random clones: A mathematical analysis. Genomics 2: 231–239 [DOI] [PubMed] [Google Scholar]
- Li R., et al. (2010). The sequence and de novo assembly of the giant panda genome. Nature 463: 311–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto T., et al. (2011). Comprehensive sequence analysis of 24,783 barley full-length cDNAs derived from twelve clone libraries. Plant Physiol. 156: 20–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer K.F.X., et al. (2009). Gene content and virtual gene order of barley chromosome 1H. Plant Physiol. 151: 496–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mickelson-Young L., Endo T.R., Gill B.S. (1995). A cytogenetic ladder-map of the wheat homoeologous group-4 chromosomes. Theor. Appl. Genet. 90: 1007–1011 [DOI] [PubMed] [Google Scholar]
- Miftahudin R.K., et al. (2004). Analysis of expressed sequence tag loci on wheat chromosome group 4. Genetics 168: 651–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore G., Devos K.M., Wang Z., Gale M.D. (1995). Cereal genome evolution. Grasses, line up and form a circle. Curr. Biol. 5: 737–739 [DOI] [PubMed] [Google Scholar]
- Murat F., Xu J.-H., Tannier E., Abrouk M., Guilhot N., Pont C., Messing J., Salse J. (2010). Ancestral grass karyotype reconstruction unravels new mechanisms of genome shuffling as a source of plant evolution. Genome Res. 20: 1545–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Gojobori T. (1986). Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3: 418–426 [DOI] [PubMed] [Google Scholar]
- Nelson J.C., Sorrells M.E., Van Deynze A.E., Lu Y.H., Atkinson M., Bernard M., Leroy P., Faris J.D., Anderson J.A. (1995). Molecular mapping of wheat: Major genes and rearrangements in homoeologous groups 4, 5, and 7. Genetics 141: 721–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann P., Pozárková D., Vrána J., Doležel J., Macas J. (2002). Chromosome sorting and PCR-based physical mapping in pea (Pisum sativum L.). Chromosome Res. 10: 63–71 [DOI] [PubMed] [Google Scholar]
- Ozkan H., Levy A.A., Feldman M. (2001). Allopolyploidy-induced rapid genome evolution in the wheat (Aegilops-Triticum) group. Plant Cell 13: 1735–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson A.H., et al. (2009). The Sorghum bicolor genome and the diversification of grasses. Nature 457: 551–556 [DOI] [PubMed] [Google Scholar]
- Potokina E., Druka A., Luo Z., Wise R., Waugh R., Kearsey M. (2008). Gene expression quantitative trait locus analysis of 16 000 barley genes reveals a complex pattern of genome-wide transcriptional regulation. Plant J. 53: 90–101 [DOI] [PubMed] [Google Scholar]
- Qi L., Friebe B., Gill B.S. (2006). Complex genome rearrangements reveal evolutionary dynamics of pericentromeric regions in the Triticeae. Genome 49: 1628–1639 [DOI] [PubMed] [Google Scholar]
- Qi L.L., et al. (2004). A chromosome bin map of 16,000 expressed sequence tag loci and distribution of genes among the three genomes of polyploid wheat. Genetics 168: 701–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosin F.M., Kramer E.M. (2009). Old dogs, new tricks: Regulatory evolution in conserved genetic modules leads to novel morphologies in plants. Dev. Biol. 332: 25–35 [DOI] [PubMed] [Google Scholar]
- Salse J., Abrouk M., Bolot S., Guilhot N., Courcelle E., Faraut T., Waugh R., Close T.J., Messing J., Feuillet C. (2009b). Reconstruction of monocotelydoneous proto-chromosomes reveals faster evolution in plants than in animals. Proc. Natl. Acad. Sci. USA 106: 14908–14913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salse J., Abrouk M., Murat F., Quraishi U.M., Feuillet C. (2009a). Improved criteria and comparative genomics tool provide new insights into grass paleogenomics. Brief. Bioinform. 10: 619–630 [DOI] [PubMed] [Google Scholar]
- Salse J., Bolot S., Throude M., Jouffe V., Piegu B., Quraishi U.M., Calcagno T., Cooke R., Delseny M., Feuillet C. (2008). Identification and characterization of shared duplications between rice and wheat provide new insight into grass genome evolution. Plant Cell 20: 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- SanMiguel P., Gaut B.S., Tikhonov A., Nakajima Y., Bennetzen J.L. (1998). The paleontology of intergene retrotransposons of maize. Nat. Genet. 20: 43–45 [DOI] [PubMed] [Google Scholar]
- Sato K., Nankaku N., Takeda K. (2009a). A high-density transcript linkage map of barley derived from a single population. Heredity 103: 110–117 [DOI] [PubMed] [Google Scholar]
- Sato K., Shin-I T., Seki M., Shinozaki K., Yoshida H., Takeda K., Yamazaki Y., Conte M., Kohara Y. (2009b). Development of 5006 full-length cDNAs in barley: A tool for accessing cereal genomics resources. DNA Res. 16: 81–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeberger K., Ossowski S., Lanz C., Juul T., Petersen A.H., Nielsen K.L., Jørgensen J.-E., Weigel D., Andersen S.U. (2009). SHOREmap: Simultaneous mapping and mutation identification by deep sequencing. Nat. Methods 6: 550–551 [DOI] [PubMed] [Google Scholar]
- Schulte D., Close T.J., Graner A., Langridge P., Matsumoto T., Muehlbauer G., Sato K., Schulman A.H., Waugh R., Wise R.P., Stein N. (2009). The international barley sequencing consortium—At the threshold of efficient access to the barley genome. Plant Physiol. 149: 142–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaked H., Kashkush K., Ozkan H., Feldman M., Levy A.A. (2001). Sequence elimination and cytosine methylation are rapid and reproducible responses of the genome to wide hybridization and allopolyploidy in wheat. Plant Cell 13: 1749–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šimková H., Svensson J.T., Condamine P., Hribová E., Suchánková P., Bhat P.R., Bartoš J., Safár J., Close T.J., Doležel J. (2008). Coupling amplified DNA from flow-sorted chromosomes to high-density SNP mapping in barley. BMC Genomics 9: 294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein N., Prasad M., Scholz U., Thiel T., Zhang H., Wolf M., Kota R., Varshney R.K., Perovic D., Grosse I., Graner A. (2007). A 1,000-loci transcript map of the barley genome: New anchoring points for integrative grass genomics. Theor. Appl. Genet. 114: 823–839 [DOI] [PubMed] [Google Scholar]
- Steuernagel B., et al. (2009). De novo 454 sequencing of barcoded BAC pools for comprehensive gene survey and genome analysis in the complex genome of barley. BMC Genomics 10: 547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchánková P., Kubaláková M., Kovárová P., Bartoš J., Cíhalíková J., Molnár-Láng M., Endo T.R., Doležel J. (2006). Dissection of the nuclear genome of barley by chromosome flow sorting. Theor. Appl. Genet. 113: 651–659 [DOI] [PubMed] [Google Scholar]
- Thiel T., Graner A., Waugh R., Grosse I., Close T.J., Stein N. (2009). Evidence and evolutionary analysis of ancient whole-genome duplication in barley predating the divergence from rice. BMC Evol. Biol. 9: 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner A., Beales J., Faure S., Dunford R.P., Laurie D.A. (2005). The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science 310: 1031–1034 [DOI] [PubMed] [Google Scholar]
- Vláčilová K., Ohri D., Vrána J., Cíhalíková J., Kubaláková M., Kahl G., Doležel J. (2002). Development of flow cytogenetics and physical genome mapping in chickpea (Cicer arietinum L.). Chromosome Res. 10: 695–706 [DOI] [PubMed] [Google Scholar]
- Waugh R., Jannink J.-L., Muehlbauer G.J., Ramsay L. (2009). The emergence of whole genome association scans in barley. Curr. Opin. Plant Biol. 12: 218–222 [DOI] [PubMed] [Google Scholar]
- Wicker T., Narechania A., Sabot F., Stein J., Vu G.T.H., Graner A., Ware D., Stein N. (2008). Low-pass shotgun sequencing of the barley genome facilitates rapid identification of genes, conserved non-coding sequences and novel repeats. BMC Genomics 9: 518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T., Schlagenhauf E., Graner A., Close T.J., Keller B., Stein N. (2006). 454 sequencing put to the test using the complex genome of barley. BMC Genomics 7: 275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T., Taudien S., Houben A., Keller B., Graner A., Platzer M., Stein N. (2009). A whole-genome snapshot of 454 sequences exposes the composition of the barley genome and provides evidence for parallel evolution of genome size in wheat and barley. Plant J. 59: 712–722 [DOI] [PubMed] [Google Scholar]
- Wu J., Kurten E.L., Monshausen G., Hummel G.M., Gilroy S., Baldwin I.T. (2007). NaRALF, a peptide signal essential for the regulation of root hair tip apoplastic pH in Nicotiana attenuata, is required for root hair development and plant growth in native soils. Plant J. 52: 877–890 [DOI] [PubMed] [Google Scholar]
- Yan L., Fu D., Li C., Blechl A., Tranquilli G., Bonafede M., Sanchez A., Valarik M., Yasuda S., Dubcovsky J. (2006). The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc. Natl. Acad. Sci. USA 103: 19581–19586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Dudoit S., Luu P., Lin D., Peng V., Ngai J., Speed T. (2002). Normalization for cDNA microarray data: A robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. (2007). PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24: 1586–1591 [DOI] [PubMed] [Google Scholar]
- Zhang G.Y., Wu J., Wang X.W. (2010). Cloning and expression analysis of a pollen preferential rapid alkalinization factor gene, BoRALF1, from broccoli flowers. Mol. Biol. Rep. 37: 3273–3281 [DOI] [PubMed] [Google Scholar]
- Zhou F.S., Kurth J.C., Wei F.S., Elliott C., Valè G., Yahiaoui N., Keller B., Somerville S., Wise R., Schulze-Lefert P. (2001). Cell-autonomous expression of barley Mla1 confers race-specific resistance to the powdery mildew fungus via a Rar1-independent signaling pathway. Plant Cell 13: 337–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The nonredundant set of 23,588 fl-cDNAs was generated from a set of 5006 fl-cDNAs (Sato et al., 2009b; accession numbers AK248134 to AK253139) and a set of 23,623 fl-cDNAs (Matsumoto et al., 2011; accession numbers AK353559 to AK377172). All 454 sequence information in this study generated from flow-sorted chromosomes was submitted to the European Bioinformatics Institute sequence read archive under accession number ERP000445. A database for sequence homology search (BLAST) is provided at http://webblast.ipk-gatersleben.de/barley/. All data contained in the genome zipper models can be downloaded as Excel spread sheets from http://mips.helmholtz-muenchen.de/plant/triticeae/genomes/index.jsp.