This study uses in vivo targeting, subcellular fractionation, and bioinformatics approaches to investigate the signal sequences that determine whether signal-anchored proteins are targeted to the endoplasmic reticulum or to endosymbiotic organelles.
Abstract
Proteins localized to various cellular and subcellular membranes play pivotal roles in numerous cellular activities. Accordingly, in eukaryotic cells, the biogenesis of organellar proteins is an essential process requiring their correct localization among various cellular and subcellular membranes. Localization of these proteins is determined by either cotranslational or posttranslational mechanisms, depending on the final destination. However, it is not fully understood how the targeting specificity of membrane proteins is determined in plant cells. Here, we investigate the mechanism by which signal-anchored (SA) proteins are differentially targeted to the endoplasmic reticulum (ER) or endosymbiotic organelles using in vivo targeting, subcellular fractionation, and bioinformatics approaches. For targeting SA proteins to endosymbiotic organelles, the C-terminal positively charged region (CPR) flanking the transmembrane domain (TMD) is necessary but not sufficient. The hydrophobicity of the TMD in CPR-containing proteins also plays a critical role in determining targeting specificity; TMDs with a hydrophobicity value >0.4 on the Wimley and White scale are targeted primarily to the ER, whereas TMDs with lower values are targeted to endosymbiotic organelles. Based on these data, we propose that the CPR and the hydrophobicity of the TMD play a critical role in determining the targeting specificity between the ER and endosymbiotic organelles.
INTRODUCTION
Newly synthesized organellar proteins are distributed to their destinations by two different means: direct targeting from the cytosol to organelles and vesicle trafficking between organelles (Walter and Johnson, 1994). Direct targeting is used for proteins destined to the endoplasmic reticulum (ER), plastids, mitochondria, nucleus, and peroxisomes, and vesicle trafficking is employed for proteins destined to various endomembrane compartments as well as for secretory proteins after targeting to the ER. Additionally, class II peroxisomal membrane proteins are targeted to peroxisomes indirectly via the ER after cotranslational ER targeting (Platta and Erdmann, 2007).
Organellar proteins that are transported as cargo proteins need specific tags that act as targeting or sorting signals. Such targeting signals include the hydrophobic leader sequence of ER proteins and the transit peptide of chloroplast proteins (Rapoport, 1991; Bruce, 2000). In addition, numerous sorting signals have been identified from proteins destined to various endomembrane compartments (Rodriguez-Boulan and Müsch, 2005; Hwang, 2008; Braulke and Bonifacino, 2009). The targeting and sorting signals of organellar proteins display various characteristics depending on the target compartments; these characteristics serve as the basis for the development of a variety of algorithms to predict the localization of organellar proteins (Petsalaki et al., 2006; Acencio and Lemke, 2009; Assfalg et al., 2009). Proteins targeted to the ER contain a signal peptide consisting of 7 to 20 highly hydrophobic amino acid residues. However, the exact amino acid sequence varies greatly depending on individual proteins (Gierasch, 1989; Nielsen et al., 1997). In luminal proteins, the signal peptide is located at the N terminus and removed after translocation into the ER. By contrast, in membrane proteins, the signal peptide can be placed at various positions within a molecule and also functions as a transmembrane domain (TMD) to anchor the protein to the ER membrane. The hydrophobic signal peptide of both ER luminal and membrane proteins is recognized by the signal recognition particle (SRP) during translation and targeted to the ER by interaction between the SRP and the SRP receptor (Egea et al., 2005; Halic and Beckmann, 2005). However, tail-anchored membrane proteins are also transported to the ER by additional pathways involving the SRP, heat shock protein 40 kD–heat shock 70 kD protein 8, or arsenite-stimulated ATPase 1/TMD recognition complex 40 kD ATPase subunit–mediated posttranslational targeting mechanisms (Stefanovic and Hegde, 2007; Rabu et al., 2009).
Chloroplast and mitochondrial proteins also are targeted directly from the cytosol. These two organelles are thought to have evolved from endosymbiotic bacteria, and the majority of their constituent proteins are imported posttranslationally from the cytosol (Bruce, 2000; Neupert and Herrmann, 2007; Agne and Kessler, 2009; Balsera et al., 2009). Multiple pathways exist for targeting proteins to these two organelles (Bolender et al., 2008; Jarvis, 2008; Dhanoa et al., 2010). For interior proteins of these organelles, an N-terminal signal peptide, called the transit peptide and the presequence for plastid and mitochondrial proteins, respectively, is sufficient to direct targeting from the cytosol. The exact nature of sequence information in these signal sequences is not fully understood. These signal peptides have a highly divergent sequence that usually contains 50 to 70 amino acid residues. An amphiphatic α-helix in the presequence and small sequence motifs embedded in the transit peptide are critical for protein import into mitochondria and chloroplasts, respectively (Klaus et al., 1996; Lee et al., 2008). Both the transit peptide and the presequence are removed from the mature portion of organellar proteins after translocation into these organelles (Bruce, 2000; Neupert and Herrmann, 2007).
In addition, a large number of membrane proteins are found at the outer envelope membrane (OEM) of chloroplasts and mitochondria and are targeted by multiple mechanisms (Lee et al., 2001; Walther and Rapaport, 2009; Dhanoa et al., 2010). OEM proteins do not contain any cleavable signal sequence but they do have a hydrophobic TMD to anchor them to the OEM. OEM proteins of chloroplasts or mitochondria have an intrinsic complication in their targeting; these OEM proteins also have a hydrophobic TMD that can be recognized by the SRP; thus, there is potential for mistargeting them to the ER. For this reason, these endosymbiotic organellar membrane proteins must have a mechanism to evade SRP-mediated cotranslational targeting to the ER. Recently, a C-terminal positively charged region (CPR) flanking the TMD has been identified as an SRP-evading signal in a few chloroplast OEM proteins (Lee et al., 2001, 2004). Similarly, mitochondrial proteins in animal cells also contain the CPR flanking to the TMD and the CPR is important in mitochondrial targeting (Kanaji et al., 2000; Waizenegger et al., 2003; Walther and Rapaport, 2009). However, positively charged amino acids are also found around the TMD, and they play a critical role in determining the topology of membrane proteins (von Heijne, 1992). Thus, it is not clear how the CPR plays a role in determining chloroplast or mitochondrial targeting. Previous studies have shown that moderate hydrophobicity of the TMD and the presence of the CPR are important for targeting tail-anchored and signal-anchored (SA) proteins to endosymbiotic organelles (Kanaji et al., 2000; Lee et al., 2001; Hwang et al., 2004; Maggio et al., 2007).
In this study, we investigated the signal sequence that distinguishes between targeting SA proteins to the ER and targeting them to endosymbiotic organelles. To simplify analysis of the SA protein targeting to these organelles, we focused on the targeting specificity conferred by the N-terminal TMD and CPR of CPR-containing SA proteins. Here, we present evidence that the targeting specificity of CPR-containing SA proteins between the ER and endosymbiotic organelles is determined by a combination of the CPR and the hydrophobicity of the TMD.
RESULTS
The CPR Is Necessary, but Not Sufficient, to Determine Targeting Specificity of SA Proteins between the ER and Endosymbiotic Organelles
To gain insight into the mechanism by which SA proteins are specifically delivered to chloroplasts and mitochondria, we compared sequences of OEM proteins from Arabidopsis thaliana chloroplasts or mitochondria (see Supplemental Figure 1 online). A common feature of these sequences is positively charged amino acid residues, Lys and Arg, in the C-terminal flanking region of the TMD. In previous studies, the CPR of the TMD has been shown to play a critical role in the targeting of SA proteins to the chloroplast OEM (Lee et al., 2001, 2004). In addition, a mitochondrial protein mtOM64 from Arabidopsis also contained the CPR (see Supplemental Figure 1 online), similar to chloroplast OEM proteins in plant cells and mitochondrial OEM proteins found in animal cells (Lee et al., 2001, 2004; Chew et al., 2004). Therefore, we hypothesized that the CPR plays a critical role in the targeting of SA proteins to both chloroplast and mitochondrial OEMs. However, the amino acid sequences of the CPRs vary greatly depending on the individual proteins. Based on chloroplast and mitochondrial OEM proteins (see Supplemental Figure 1 online), we defined the CPR as a region that contains at least three positively charged amino acid residues, consisting of Lys and/or Arg residues, within eight amino acid residues of the C-terminal end of a TMD.
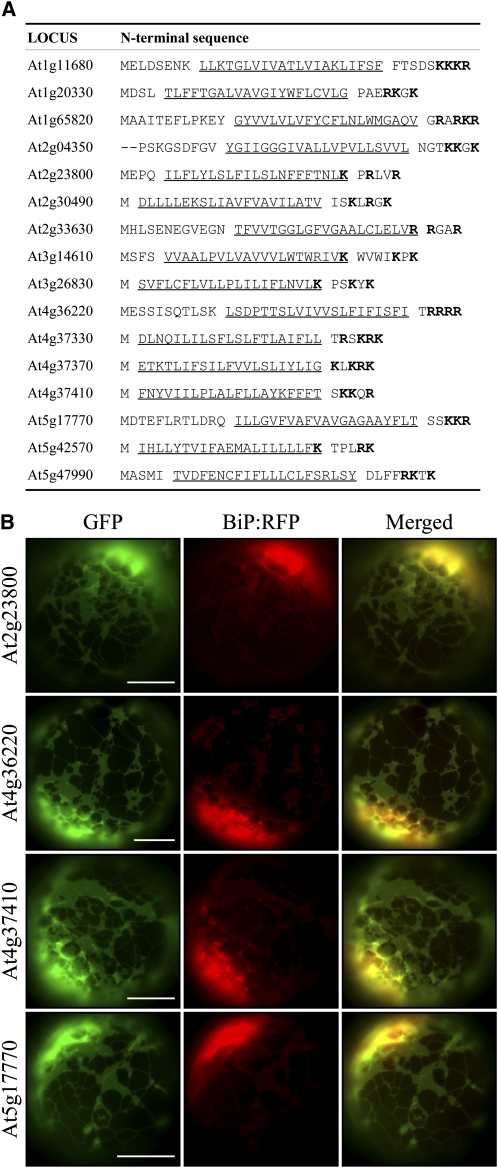
Next, we examined whether the CPR is sufficient for targeting a SA protein to the endosymbiotic organelles. Recently, a large number of membrane proteins have been identified from various organelles by proteomics (Millar et al., 2001; Rolland et al., 2003; Baginsky and Gruissem, 2004; Ephritikhine et al., 2004; Heazlewood et al., 2004). Close examination of these proteins revealed that a large number of ER-targeted membrane proteins also contain the CPR flanking their TMDs (Figure 1A). This observation raised the question of how the targeting specificity of CPR-containing SA proteins between the ER and endosymbiotic organelles is determined. SA proteins without the CPR are likely targeted to the ER. Consistent with this idea, two putative SA proteins without the CPR were targeted to the ER when expressed as green fluorescent protein (GFP) fusion proteins (see Supplemental Figure 2 online). In membrane proteins, positive charges around the TMD play a role in determining the topology of membrane proteins, as summarized by the positive-inside rule (von Heijne, 1992). Thus, these results raise the possibility that the CPR is necessary, but insufficient alone, for specific targeting of SA proteins to endosymbiotic organelles in plant cells. Additional sequence information appears to be needed. We sought to confirm this by in vivo targeting experiments with ER proteins At2g23800, At4g36220, At4g37410, and At5g17770, which each have a CPR next to the TMD (Figure 1A), and generated GFP fusion constructs (At2g23800:GFP, At4g36220:GFP, At4g37410:GFP, and At5g17770:GFP) using N-terminal segments containing the TMD and CPR of these proteins. The GFP fusion constructs were introduced into Arabidopsis protoplasts, and their localizations were examined via a fluorescence microscopy. Indeed, these GFP fusion proteins each produced an ER localization pattern (Figure 1B). This indicated that the presence of the CPR is not sufficient to determine targeting specificity to endosymbiotic organelles and that additional factors are required.
Figure 1.
Certain CPR-Containing Proteins Also Are Targeted to the ER.
(A) A list of putative ER-targeted SA proteins. The sequences of putative ER-targeted SA proteins that contain the CPR were obtained from proteomics data (SUBA database) based on the sorting criteria (see Methods). The TMDs are underlined, and Lys and Arg residues in the CPR are highlighted in bold.
(B) In vivo targeting. At2g23800, At4g36220, At4g37410, and At5g17770 were randomly selected from the list in (A), and the N-terminal region (40 to 50 amino acid residues) of these proteins, containing the TMD and CPR, was fused N-terminally to GFP. The resulting constructs were introduced into Arabidopsis protoplasts together with BiP:RFP, and localization of fusion proteins was examined under a fluorescence microscope. To simplify the labels, GFP was omitted from the construct names throughout. Bars = 20 μm.
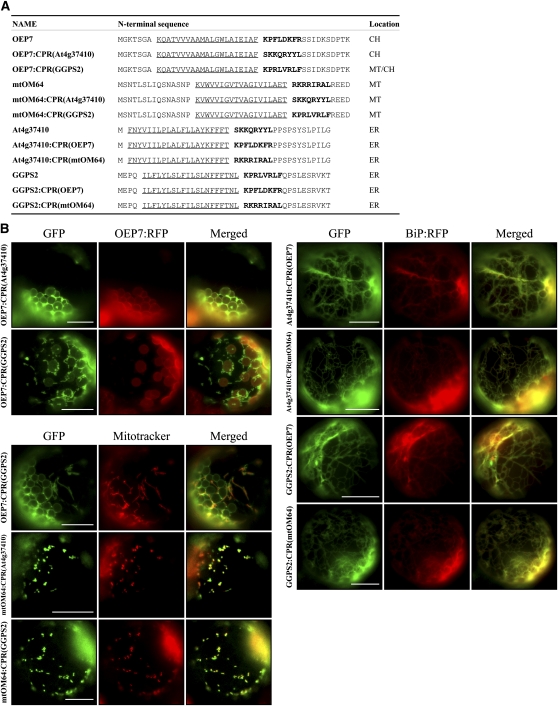
However, despite the apparent similarity between the CPRs of ER and endosymbiotic organellar proteins, the CPR of ER proteins does not play a role in SRP evasion as observed for the CPR of mitochondrial proteins (Kanaji et al., 2000). Thus, another possibility is that CPRs differ functionally from each other. To distinguish between the two possibilities, we examined the targeting of mutant ER (At4g37410) and geranylgeranyl pyrophosphate synthase 2 (GGPS2) and endosymbiotic proteins (OEP7 and mtOM64) with swapped CPR domains (Figure 2A). The chloroplast protein mutants OEP7:CPR(At4g37410) and OEP7:CPR(GGPS2) with the CPRs of ER proteins At4g37410 and GGPS2, respectively, were targeted to chloroplasts with a minor portion being targeted to mitochondria. Similarly, the mitochondrial protein mutants mtOM64:CPR(At4g37410) and mtOM64:CPR(GGPS2) with the CPRs of the ER proteins At4g37410 and GGPS2, respectively, were targeted to mitochondria. Furthermore, four ER protein mutants, At4g37410:CPR(OEP7), At4g37410:CPR(mtOM64), GGPS2:CPR(OEP7), and GGPS2:CPR(mtOM64), that had the CPR of either OEP7 or mtOM64 were properly targeted to the ER (Figure 2B). These results support the idea that the CPR is necessary but not sufficient to determine the targeting specificity between the ER and endosymbiotic organelles and that additional factors are required.
Figure 2.
Localization of CPR Swapping Mutants in Protoplasts.
(A) Sequences of CPR swapping mutants. CPRs were swapped between ER and chloroplast/mitochondria C-SA proteins and the resulting constructs were fused to GFP. Localizations listed in the table are from results shown in (B). Underlined and bold sequences are TMD and CPR, respectively.
(B) Targeting of CPR swapping mutants. The CPR swapping mutants in (A) were introduced into protoplasts together with OEP7:RFP or BiP:RFP, and their localization was examined under a fluorescent microscope. Mitotracker was used to stain mitochondria. To simplify the labels, GFP was omitted from the construct names throughout. Bars = 20 μm.
Hydrophobicity of the TMD Is Critical in Determining the Targeting Specificity of CPR-Containing SA Proteins between the ER and Endosymbiotic Organelles
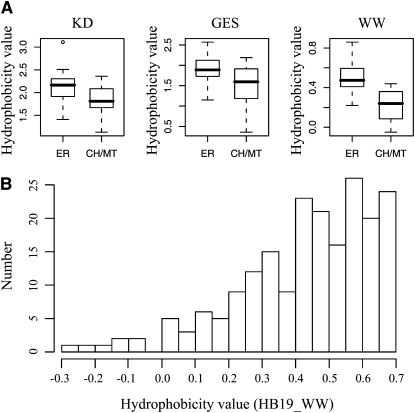
The results shown in Figure 1 and Supplemental Figure 1 online strongly suggest that an additional factor is necessary for targeting specificity of CPR-containing SA (C-SA) proteins. One possibility is that the hydrophobicity of the TMD influences the targeting specificity of C-SA proteins between the ER and endosymbiotic organelles. In animal cells, mitochondrial proteins tend to have a TMD with moderate hydrophobicity (Kanaji et al., 2000; Waizenegger et al., 2003). Accordingly, we examined the hydrophobicity of TMDs of ER, chloroplast, and mitochondrial proteins. We focused only on proteins that had a TMD within the 40 N-terminal amino acid residues and excluded ones with a signal anchor positioned after larger N-terminal domains. The SUBA database (Heazlewood et al., 2005), which provides proteomics data for protein localizations, was searched for candidate proteins, and 46 ER membrane proteins and 54 endosymbiotic organellar membrane proteins were found (see Supplemental Table 1 online). Then, the hydrophobicity of the N-terminal TMDs between these two groups was compared using the Kyte and Doolittle (KD) hydrophobicity scale (Kyte and Doolittle, 1982) (see Supplemental Table 1 online). The hydrophobicity values of the TMD of the ER and endosymbiotic proteins range from 1.35 to 3.22 and 1.04 to 2.79 with average hydrophobicity values of 2.18 and 1.76, respectively. The majority (72%) of ER SA proteins had a hydrophobicity value >2.0 on the KD hydrophobicity scale, whereas 80% of endosymbiotic organellar SA proteins had a hydrophobicity value <2.0 on the KD hydrophobicity scale (see Supplemental Table 1 and Supplemental Figure 3 online). However, the hydrophobicity values of ER-targeted SA proteins significantly overlapped with those of endosymbiotic SA proteins. Many different methods for determining hydrophobicity have been proposed (Engelman et al., 1986; Wimley and White, 1996). Therefore, we explored whether other methods provide better differentiation of hydrophobicity of the TMD between ER and endosymbiotic organellar SA proteins. We reexamined the hydrophobicity of the ER and endosymbiotic organellar SA proteins listed in Supplemental Table 1 online using Wimley and White (WW) and Engelman (GES) hydrophobicity scales (Engelman et al., 1986; Wimley and White, 1996). With the WW hydrophobicity scale, the majority (78%) of the ER-targeted SA proteins had a hydrophobicity value >0.4, whereas the majority (83%) of the endosymbiotic organellar SA proteins had a hydrophobicity value <0.4 (see Supplemental Figure 3 online), indicating that although there was still a certain degree of overlap between ER and endosymbiotic organellar targeted SA proteins, the WW hydrophobicity scale provided better differentiation between ER and endosymbiotic SA proteins. By contrast, the GES hydrophobicity scale produced poor differentiation in this regard. This result suggested that TMDs with a hydrophobicity value >0.4 on the WW hydrophobicity scale are targeted to the ER, whereas TMDs with lower values are targeted to endosymbiotic organelles.
Given the results shown in Figure 1 and Supplemental Figure 3 online, we next sought to confirm the localization of C-SA proteins listed in Supplemental Table 1 online by in vivo targeting experiments. We selected several proteins (At1G26710, At1g66770, and At5g42590) containing highly hydrophobic TMDs from the list in Supplemental Table 1 online. The N-terminal region containing their TMD and CPR was fused to GFP, and the resulting At1G26710:GFP, At1g66770:GFP, and At5g42590:GFP constructs were transformed into protoplasts from Arabidopsis leaf tissues (Jin et al., 2001; Kim et al., 2001). Localization of these fusion proteins was examined by fluorescence microscopy. At1G26710, At1g66770, and At5g42590, which were listed as chloroplast proteins, produced a cytosolic pattern or an ER pattern when their N-terminal regions were fused to GFP and expressed in protoplasts (see Supplemental Figure 4 online). To test if this difference in localization was caused by the N-terminal fragment containing the TMD and CPR, the full-length proteins were fused to GFP and their localizations were examined in protoplasts. The full-length fusion proteins (At1g26710-FL:GFP, At1g66770-FL:GFP, and At5g42590-FL:GFP) were also localized in the cytosol or targeted to the ER (see Supplemental Figure 4 online), strongly suggesting that At1g26710, At1g66770, and At5g42590 are cytosolic or ER proteins.
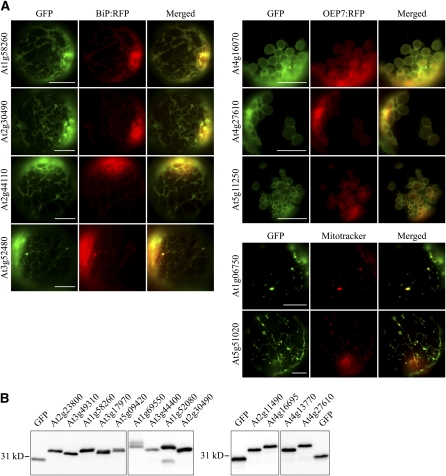
To define the determinants of C-SA protein targeting to the ER or endosymbiotic organelles, we decided to include only proteins whose localizations had been confirmed by in vivo targeting experiments. However, there were only a limited number of plant C-SA proteins whose localizations had been confirmed. Thus, reliable sequence information for differential targeting of C-SA proteins between ER and endosymbiotic organelles could not be easily obtained. Accordingly, we performed a genome-wide analysis of Arabidopsis C-SA proteins, combining computational and experimental approaches. Through a computational pipeline including TMD predictions using Conpred II (Arai et al., 2004), the exclusion of signal sequence–containing ER luminal proteins using SPOCTOPUS (Viklund et al., 2008), and the detection of CPRs based on a custom script, we obtained 217 putative membrane proteins that have an N-terminal TMD sequence with a CPR. From the putative membrane proteins identified, we selected 50 proteins whose localizations were not confirmed by in vivo targeting experiments; we then determined their localizations by in vivo targeting experiments in protoplasts using GFP fusion constructs generated with the N-terminal region containing the TMD and CPR. The GFP fusion constructs were cotransformed with OEP7:RFP (for red fluorescent protein), which is a fusion protein consisting of OEP7 and RFP that is targeted to the chloroplast OEM (Lee et al., 2001). The localizations of these fusion proteins were examined under a fluorescence microscope. The localization of reporter proteins to mitochondria was confirmed by colocalization with Mitotracker. Initially, we examined the targeting specificity of the N-terminal TMD and CPR of proteins At1g58260, At2g30490, At2g44110, and At3g52480 whose TMDs have WW hydrophobicity values of 0.47, 0.22, 0.41, and 0.6, respectively. The GFP fusion proteins At1g58260:GFP, At2g30490:GFP, At2g44110:GFP, and At3g52480:GFP (with the N-terminal TMDs and CPRs of the indicated proteins) were targeted to the ER (Figure 3A). In addition, we examined the targeting specificity of the N-terminal TMD and CPR of proteins At5g11250, At4g27610, and At4g16070 that had TMDs with WW hydrophobicity values ranging from 0.21 to 0.39. These GFP fusion proteins each produced a ring pattern surrounding the chloroplasts and overlapped closely with OEP7:RFP. Fusion proteins generated from At5g51020 and At1g06750, in which the TMD also displayed WW hydrophobicity values below 0.2, produced punctate staining patterns that also stained with Mitotracker. To confirm the expression of these fusion proteins, protein extracts from the protoplasts were subjected to immunoblot analysis using an anti-GFP antibody (Figure 3B; see Supplemental Figure 5 online). Except for At2g30490:GFP, Figure 3A showed that all GFP proteins with a hydrophobicity value >0.4 on the WW hydrophobicity scale were targeted to the ER, otherwise targeting was to endosymbiotic organelles. To expand our findings, we examined the localizations of additional GFP fusion constructs with the N-terminal TMDs and CPRs of 41 other C-SA proteins in in vivo targeting experiments. In addition, for some representative proteins, we examined the localizations of full-length GFP fusion proteins to determine whether the N-terminal TMD and CPR are sufficient to determine targeting specificity (see Supplemental Figure 6 online). Tables 1 and 2 summarize the in vivo targeting results for all proteins examined in this study together with seven SA proteins that had been confirmed previously in other studies (Okada et al., 2000; Lee et al., 2001, 2004; Schnurr et al., 2002; Oikawa et al., 2003; Asano et al., 2004; Chew et al., 2004).
Figure 3.
In Vivo Localization of GFP Fusion Constructs in Protoplasts.
(A) Localization of GFP fusion proteins. The N-terminal 40 to 50 amino acid residues of the indicated proteins were fused to the N terminus of GFP. The resulting constructs were transformed into protoplasts together with the ER marker BiP:RFP or the chloroplast marker OEP7:RFP, and localization of these fusion proteins was examined. In addition, protoplasts were stained with Mitotracker, a marker for mitochondria. To simplify the labels, GFP was omitted from the construct names. Bars = 20 μm.
(B) Immunoblot analysis of fusion proteins in protoplasts. Total protein extracts from transformed protoplasts were analyzed by immunoblotting using anti-GFP antibody. GFP alone was included as a control.
Table 1.
Hydrophobicity Analysis of ER-Targeted SA Proteins Confirmed by in Vivo Targeting
| Locus | HB19_KD | HB19_GES | HB19_WW |
| At1g11000 | 2.30 | 1.97 | 0.41 |
| At1g11680 | 2.26 | 1.84 | 0.44 |
| At1g53610 | 3.11 | 2.36 | 0.41 |
| At1g54370 | 2.41 | 2.12 | 0.44 |
| At1g58260 | 1.83 | 1.73 | 0.47 |
| At1g69550 | 2.49 | 2.57 | 0.67 |
| At1g73340 | 2.23 | 2.28 | 0.65 |
| At1g76090 | 1.95 | 1.89 | 0.43 |
| At2g23800 | 1.85 | 1.81 | 0.86 |
| At2g30490 | 2.36 | 1.27 | 0.22 |
| At2g30770 | 2.16 | 1.81 | 0.56 |
| At2g40890 | 1.86 | 1.47 | 0.30 |
| At2g44110 | 2.17 | 2.01 | 0.41 |
| At3g04220 | 1.92 | 2.25 | 0.41 |
| At3g44400 | 2.27 | 1.89 | 0.49 |
| At3g52480 | 2.29 | 2.16 | 0.60 |
| At4g00360 | 1.41 | 1.41 | 0.48 |
| At4g09100 | 1.91 | 1.89 | 0.58 |
| At4g13770 | 2.46 | 1.47 | 0.40 |
| At4g22690 | 2.16 | 2.06 | 0.44 |
| At4g31500 | 2.51 | 1.95 | 0.43 |
| At4g36220 | 2.13 | 2.22 | 0.59 |
| At4g37340 | 2.16 | 2.11 | 0.69 |
| At4g37410 | 2.04 | 1.73 | 0.69 |
| At5g17770 | 2.31 | 2.13 | 0.29 |
| At5g44620 | 1.72 | 1.82 | 0.59 |
| At5g45340 | 2.27 | 1.70 | 0.55 |
| At5g47990 | 1.86 | 1.15 | 0.61 |
Hydrophobicity analysis was performed for the TMD of ER-targeted SA proteins. Localization of 50 putative SA proteins was determined by in vivo targeting experiments. We included as positive controls seven SA proteins that had been previously confirmed by in vivo targeting. Among 57 proteins, 28 proteins localized to the ER. The three different hydrophobicity scales (KD, WW, and GES) were used in the analysis.
Table 2.
Hydrophobicity Analysis of Chloroplast and Mitochondrial SA Proteins Confirmed by in Vivo Targeting
| Locus | HB19_KD | HB19_GES | HB19_WW | Localization |
| At1g06750 | 1.78 | 1.58 | 0.19 | MT |
| At1g34470 | 1.80 | 1.54 | 0.23 | CH/MT |
| At1g52080 | 2.19 | 2.00 | 0.42 | CH/MT |
| At1g53000 | 1.57 | 1.57 | 0.03 | MT |
| At1g68220 | 2.36 | 2.14 | 0.44 | CH/ER |
| At1g74550 | 1.50 | 0.94 | 0.35 | CH/ER |
| At1g77590 | 1.79 | 1.65 | 0.34 | CH/ER |
| At3g17970 | 1.91 | 1.83 | 0.37 | CH |
| At3g25690 | 1.81 | 0.95 | 0.01 | CH |
| At3g49310 | 2.14 | 1.61 | 0.25 | CH/ER |
| At3g50460 | 1.66 | 1.25 | 0.06 | CH/ER |
| At3g50470 | 1.52 | 1.20 | −0.05 | CH/CY |
| At3g52420 | 2.06 | 1.69 | 0.13 | CH |
| At3g54510 | 2.11 | 1.83 | 0.44 | MT/CH/ER |
| At4g16070 | 2.12 | 2.04 | 0.21 | CH |
| At4g16695 | 1.99 | 2.16 | 0.31 | MT/ER |
| At4g27610 | 1.13 | 0.95 | 0.26 | CH |
| At5g09420 | 1.87 | 1.17 | 0.11 | MT |
| At5g11250 | 1.81 | 2.19 | 0.39 | CH |
| At5g51020 | 1.68 | 0.36 | 0.02 | MT |
Hydrophobicity analysis was performed for the TMD of chloroplast or mitochondrial SA proteins. Continued to proteins listed in Table 1, 20 proteins were targeted to chloroplasts or mitochondria. The remaining nine proteins were not membrane proteins; three and six were ER-luminal and cytosolic proteins, respectively. The three different hydrophobicity scales (KD, WW, and GES) were used in the analysis. CH, MT, and CY indicate chloroplast, mitochondria, and cytosol, respectively.
Using this new data set, we analyzed again the hydrophobicity of TMDs using the KD, WW, and GES hydrophobicity scales to see if there was any distinction in the hydrophobicity of TMDs between ER and endosymbiotic organellar C-SA proteins. Again, the WW hydrophobicity scale produced much better differentiation in TMD hydrophobicity values between ER and endosymbiotic organellar C-SA proteins (Figure 4A). Among the C-SA proteins we analyzed, 89% of ER C-SA proteins had a WW hydrophobicity value above 0.4, whereas 85% of endosymbiotic organellar C-SA proteins had a WW hydrophobicity value below 0.4, raising the possibility that the hydrophobicity value of 0.4 could be an important criterion for distinguishing the localizations of ER and endosymbiotic C-SA proteins. In mammals and yeast, mitochondria-targeted SA proteins have lower hydrophobicity values than do ER-targeted proteins (see Supplemental Table 2 online). To test this idea further, we analyzed 217 C-SA proteins (see Supplemental Table 3 online) screened from the database using the WW hydrophobicity scale. Figure 4B shows the distribution pattern of C-SA proteins according to hydrophobicity value. Proteins that have hydrophobicity values between 0.35 and 0.4 were underrepresented, thus separating total C-SA proteins into two groups delimited by the hydrophobicity value of 0.4. The group of proteins that have hydrophobicity values above 0.4 may be targeted to the ER, and the group of proteins that have hydrophobicity values below 0.4 may be targeted to endosymbiotic organelles. This result supports the idea that the hydrophobicity value of 0.4 may be an important criterion for distinguishing the localization of C-SA proteins. However, the hydrophobicity of the TMD did not differ between chloroplast and mitochondrial SA proteins. The average hydrophobicity values of ER and endosymbiotic organellar C-SA proteins were 0.50 and 0.23, respectively. In this new data set, 89% of ER-targeted C-SA proteins had TMD hydrophobicity values >0.4 on the WW hydrophobicity scale, and 85% of endosymbiotic organellar C-SA proteins had values <0.4. The lack of complete differentiation between groups suggests that factors in addition to hydrophobicity are involved in determining targeting specificity to the ER or endosymbiotic organelles.
Figure 4.
Distribution of C-SA Proteins According to the Hydrophobicity of their TMD.
(A) Hydrophobicity between ER and chloroplast/mitochondrial SA proteins. Protein sequences of C-SA proteins that had been confirmed by in vivo targeting experiments (Figure 3; see Supplemental Figures 5 and 6 online) were analyzed for hydrophobicity of the TMD using three different hydrophobicity scales (KD, WW, and GES) as described in Methods. Hydrophobicity of the TMD was compared between ER and chloroplast/mitochondrial (CH/MT) membrane proteins through box graphs (Wilcoxon’s rank-sum test, P value of KD and WW < 0.001; P value of GES < 0.05).
(B) Distribution of C-SA proteins according to their hydrophobicity values. A total of 217 C-SA proteins from Supplemental Table 3 online were analyzed for their hydrophobicity values using the WW hydrophobicity scale. The distribution of SA proteins is presented using a 0.05 interval for the hydrophobicity value.
Hydrophobicity Changes in the TMD of C-SA Proteins Switch Targeting Specificities between the ER and Endosymbiotic Organelles
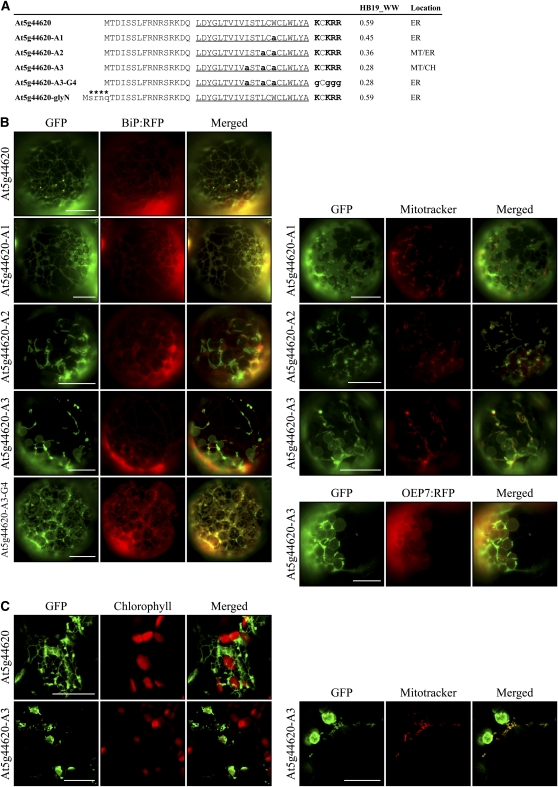
To confirm experimentally that the hydrophobicity of the TMD is critical in determining target specificity, we selected At5g44620, an ER-targeted C-SA protein, and gradually lowered the hydrophobicity of the TMD by substituting one, two, or three Ala residues for Trp alone, Trp and Leu, or Trp, Leu, and Ile in the TMD, respectively (Figure 5A). The TMDs of At5g44620-A1, At5g44620-A2, and At5g44620-A3 had WW hydrophobicity values of 0.45, 0.36, and 0.28, respectively, which were lower than the 0.59 for the wild-type TMD. N-terminal fragments containing the TMD and CPR of wild-type At5g44620 and the three substitution mutants were fused to GFP to generate At5g44620:GFP, At5g44620-A1:GFP, At5g44620-A2:GFP, and At5g44620-A3:GFP, respectively. These constructs were introduced into protoplasts together with BiP:RFP, a chimeric ER marker protein in which BiP and RFP are fused (Lee et al., 2002b), or OEP7:RFP by polyethylene glycol (PEG)-mediated transformation or into intact leaf tissues by agroinfiltration. Subsequently these transiently expressed GFP proteins were examined via fluorescence microscopy ( F igures 5B and 5C). At5g44620:GFP produced an ER pattern in both protoplasts and intact cells in infiltrated leaf tissues. In addition, At5g44620-A1:GFP that had a Trp-to-Ala substitution still produced the ER pattern. However, the other two mutants, At5g44620-A2:GFP and At5g44620-A3:GFP, produced new GFP patterns; At5g44620-A2:GFP produced the ER pattern together with a mitochondrial pattern, and A5g44620-A3:GFP produced the mitochondrial pattern together with a chloroplast pattern. The mitochondrial localization of the GFP fusion proteins generated from At5g44620-A2 and At5g44620-A3 was confirmed by colocalization with Mitotracker staining (Figures 5B and 5C). To test whether the CPRs of these mutants played a role in their targeting to mitochondria, we examined the localization of a new mutant, At5g44620-A3-G4:GFP, generated by substituting the CPR from At5g44620-A3:GFP with Gly residues. It was targeted to the ER instead of mitochondria/chloroplasts (Figure 5B). These results indicate that a decrease in the hydrophobicity of the TMD in ER-targeted C-SA proteins is sufficient for the targeting of these proteins to endosymbiotic organelles instead of the ER. The results are consistent with the hypothesis that the hydrophobicity of the TMD plays a critical role in determining the targeting specificity of C-SA proteins to the ER or endosymbiotic organelles.
Figure 5.
Decreasing the Hydrophobicity of the TMD of ER-Targeted At5g44620 Shifts Targeting Specificity to Chloroplasts and Mitochondria.
(A) The N-terminal sequences of At5g44620 and its mutant constructs. To reduce hydrophobicity, Leu, Ile, and Trp in the TMD were substituted with Ala, as indicated by bold and lowercase letters. In addition, the CPR was removed by Gly substitution. Hydrophobicity was determined using the WW scale (HB19_WW). The TMDs are underlined, and Lys and Arg residues in the CPR are highlighted in bold. An N-glycosylation site indicated by asterisks was added to the N terminus of At5g44620.
(B) and (C) In vivo localization.
(B) Protoplasts were transformed with the indicated constructs, and localization was examined. In addition, transformed protoplasts were stained with Mitotracker.
(C) Leaves were infiltrated with Agrobacterium tumefaciens carrying a GFP fusion construct of At5g44620 or At5g44620-A3. The agroinfiltrated leaves were examined through a confocal laser scanning microscope. To simplify the labels, GFP was omitted from the construct names. Bars = 20 μm.
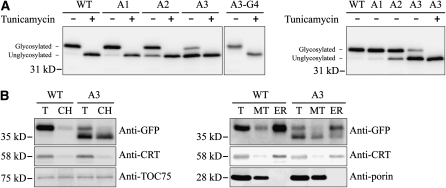
Next, we confirmed the image analysis results at the biochemical level using immnuoblot analysis. To demonstrate more clearly the change in targeting specificity between the ER and endosymbiotic organelles, we used the N-glycosylation that occurs at the ER and thus can be used as evidence for the ER localization. We generated At5g44620:GFP(glyNC), At5g44620-A1:GFP(glyNC), At5g44620-A2:GFP(glyNC), At5g44620-A3:GFP(glyNC), and At5g44620-A3-G4:GFP(glyNC) by adding two glycosylation sites to their corresponding constructs, one at the N terminus of At5g44620 and the other at the C terminus of the GFP, respectively. The modifications did not change the localization patterns (see Supplemental Figure 7 online). Protein extracts from the transformed protoplasts that had been incubated with or without tunicamycin, an inhibitor of N-glycosylation, were analyzed by immunoblotting using anti-GFP antibody. At5g44620:GFP(glyNC) from tunicamycin-treated protoplasts migrated faster than that from untreated protoplasts (Figure 6A), indicating that At5g44620:GFP(glyNC) is N-glycosylated and thus targeted to the ER. Comparison of the N-glycosylation of wild-type and mutant forms of At5g44620:GFP(glyNC) showed that the wild type, At5g44620-A1:GFP(glyNC), and At5g44620-A3-G4:GFP(glyNC) were fully N-glycosylated. By contrast, 31 and 76% of At5g44620-A2:GFP(glyNC) and At5g44620-A3:GFP(glyNC) proteins were detected as the unglycosylated form, respectively, confirming the data obtained with image analysis (Figure 6A). To obtain independent evidence for the localization of these GFP reporter proteins, we analyzed subcellular fractions containing the ER, chloroplasts, or mitochondria by immunoblot analysis using antibodies directed against GFP and organelle markers. Anti-TOC75, anti-calreticulin, and anti-porin antibodies were used for chloroplasts, ER, and mitochondria, respectively. Consistent with the results of the image and glycosylation pattern analyses, the unglycosylated form of At5g44620-A3:GFP(glyNC) was detected primarily in the chloroplast fraction, although a small portion was also detected in the mitochondrial fraction. On the other hand, glycosylated forms of At5g44620:GFP(glyNC) and At5g44620-A3:GFP(glyNC) were detected primarily in the ER fraction, with barely any detection of these proteins in the chloroplast and mitochondrial fractions (Figure 6B).
Figure 6.
Immunoblot Analyses of Localization and Subcellular Fractionation.
(A) Immunoblot analysis of localization. The indicated constructs were transformed into protoplasts and incubated in the presence (+) or absence (−) of tunicamycin (10 μg/mL) for 14 h after transformation. Protein extracts were prepared from the transformed protoplasts and analyzed by immunoblotting using anti-GFP antibody. Glycosylated, glycosylated form; Unglycosylated, unglycosylated form. glyNC indicates the two glycosylation sites at the N and C termini of the construct. WT, At5g44620:GFP(glyNC); A1, At5g44620-A1:GFP(glyNC); A2, At5g44620-A2:GFP(glyNC); A3, At5g44620-A3:GFP(glyNC); A3-G4, At5g44620-A3-G4:GFP(glyNC).
(B) Subcellular fractions containing chloroplasts, mitochondria, and ER were obtained from protoplasts transformed with a GFP fusion construct of At5g44620:GFP(glyNC) or At5g44620-A3:GFP(glyNC). The isolated fractions were analyzed by immunoblotting using the indicated antibodies. T, total extracts; CH, protein extracts of purified chloroplasts; MT, protein extracts of purified mitochondria; ER, protein extracts of purified ER. Anti-TOC75, anti-CRT (calreticulin), and anti-porin antibodies were used as markers for chloroplast, ER, and mitochondria, respectively. WT, At5g44620:GFP(glyNC); A3, At5g44620-A3:GFP(glyNC).
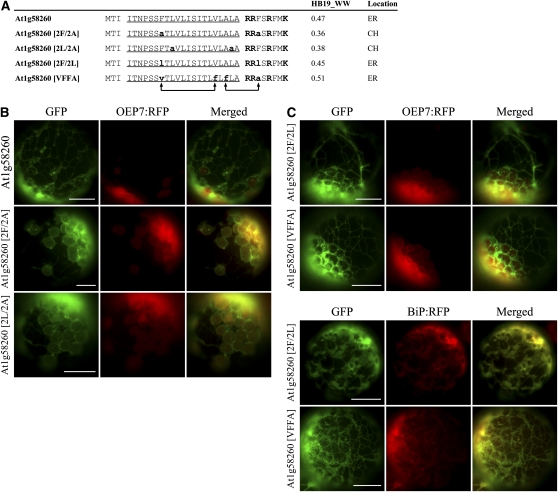
To get a clue whether this is a general phenomenon in targeting specificity determination, we selected another ER-targeted C-SA protein, At1g58260, and examined whether its localization switches from the ER to endosymbiotic organelles depending on the hydrophobicity of TMD. The hydrophobicity was lowered by substituting two Ala for two Phe residues or two Ala for two Leu residues; subsequently, the N-terminal TMD and CPR of the wild type and mutants was fused to GFP to give At1g58260:GFP, At1g58260[2F/2A]:GFP and At1g58260[2L/2A]:GFP (Figure 7A). The wild-type At1g58260:GFP produced an ER pattern. However, the mutants At1g58260[2F/2A]:GFP and At1g58260[2L/2A]:GFP produced a pattern indicating that the major portion of these proteins is targeted to chloroplasts with a minor portion to the ER (Figure 7B). As a control, we generated two additional TMD mutants; At1g58260[2F/2L] had a substitution of two Leu residues for Phe residues, and At1g58260[VFFA] had a shuffling of two Phe residues in the TMD and CPR (Figure 7A). The substitution of Phe to Leu caused a minor decrease in hydrophobicity from 0.47 to 0.45 on the WW hydrophobicity scale. The substitution mutants were fused to GFP and introduced into protoplasts. As expected, both At1g58260[2F/2L]:GFP and At1g58260[VFFA]:GFP produced an ER pattern as observed with the wild-type sequence (Figure 7C), confirming that the hydrophobicity of TMD, rather than the position or presence of Phe residues, is a critical determinant of targeting specificity. Similar results were obtained with At2g44110. When Ala was substituted for Leu or Ile, hydrophobicity was reduced to 0.32 or 0.33 from 0.41 on the WW hydrophobicity scale, respectively, and targeting was changed from the ER to chloroplasts (see Supplemental Figure 8 online).
Figure 7.
Lowering the Hydrophobicity of the TMD of ER-Targeted At1g58260 Shifts Targeting Specificity to Chloroplasts.
(A) N-terminal sequences of At1g58260 and its mutant constructs. Phe residues in the TMD and CPR were either substituted with Ala (indicated by bold and lowercase letters) (2F/2A) or swapped (indicated with arrows) (VFFA). In addition, Phe residues were substituted with Leu (indicated by bold and lowercase letters) (2F/2L). In the case of 2L/2A, two Leu residues were substituted with two Ala residues. Hydrophobicity was determined using the WW hydrophobicity scale (HB19_WW). The TMDs are underlined, and Lys and Arg residues in the CPR are highlighted in bold.
(B) and (C) In vivo targeting of GFP fusion proteins. GFP fusion constructs were introduced into protoplasts together with OEP7:RFP or BiP:RFP, and localization was examined under a fluorescence microscope. Bar = 20 μm.
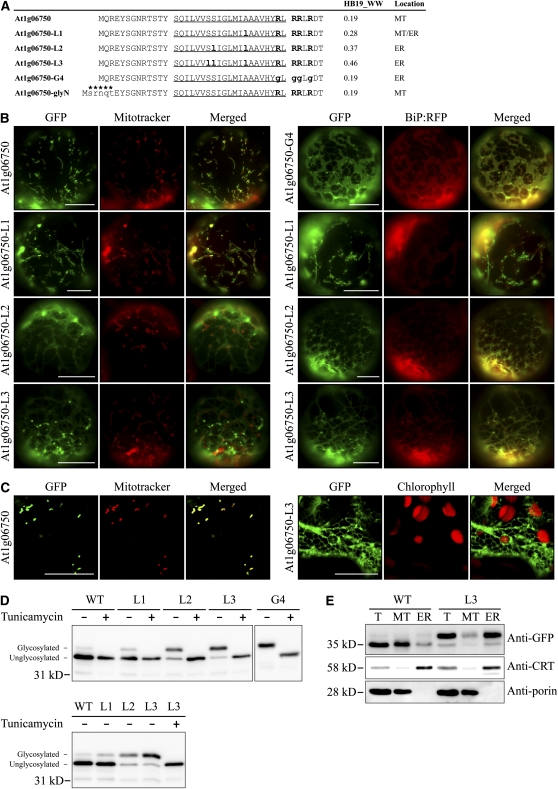
Next, we tested whether a mitochondrial membrane protein can be rerouted to the ER by increasing the hydrophobicity of TMD. We selected At1g06750, a mitochondrial C-SA protein. The N-terminal region of At1g06750 that contained the TMD and the CPR was fused to GFP, and the resulting construct, At1g06750:GFP, was introduced into protoplasts by PEG-mediated transformation or into leaf tissues by agroinfiltration. At1g06750:GFP produced a punctate staining pattern that closely overlapped with that of Mitotracker, indicating that At1g06750:GFP is targeted to mitochondria. Next, we generated three mutants, At1g06750-L1, At1g06750-L2, and At1g06750-L3, which had in the TMD substitutions of A27L, S21L/A27L, and S20L/S21L/A27L that resulted in hydrophobicity values of 0.28, 0.37, and 0.46, respectively, on the WW hydrophobicity scale (Figure 8A). The resulting mutants were introduced into protoplasts as GFP fusion constructs, and localization was examined under a fluorescence microscope (Figure 8B). Transformed protoplasts were also stained with Mitotracker to locate mitochondria. At1g06750-L1:GFP still produced the punctate staining pattern that closely overlapped with Mitotracker. However, At1g06750-L2:GFP and At1g06750-L3:GFP produced an ER pattern that did not overlap with Mitotracker. In addition, At1g06750-L3:GFP produced a punctate staining pattern that was not stained with Mitotracker, raising the possibility that At1g06750-L3:GFP is targeted to the Golgi apparatus or other endomembrane compartments by vesicle trafficking. To confirm that the ER pattern represented the ER, the three mutants of At1g06750:GFP were introduced into protoplasts together with BiP:RFP, and their localization was examined. Figure 8B shows that both At1g06750-L2:GFP and At1g06750-L3:GFP closely overlapped with BiP:RFP, confirming that At1g06750-L2:GFP and At1g06750-L3:GFP localize to the ER. In addition, At1g06750-L3:GFP also showed an ER pattern in intact cells of infiltrated leaf tissues (Figure 8C), confirming the ER localization of At1g06750-L3:GFP. To test the importance of the CPR in the mitochondrial targeting of these proteins, we generated a new mutant, At1g06750-G4:GFP, that had a Gly substitution in its CPR. Instead of being targeted to mitochondria, it was targeted to the ER, confirming that the CPR is necessary for mitochondrial targeting (Figure 8B).
Figure 8.
Increasing the Hydrophobicity of the TMD of Mitochondria Targeted At1g06750 Shifts Targeting Specificity to the ER.
(A) N-terminal sequences of At1g06750 and its mutant constructs. The Ala and Ser in the TMD were substituted with Leu, as indicated by bold and lowercase letters. In addition, the CPR was removed by a Gly substitution. The hydrophobicity of TMD was determined using the WW scale (HB19_WW). The TMDs are underlined, and Lys and Arg residues in the CPR are highlighted in bold. An N-glycosylation site, SRNQT, indicated by asterisks was added to the N terminus of At1g06750.
(B) and (C) In vivo localization.
(B) Protoplasts were transformed with the indicated constructs, and localization was examined. In addition, transformed protoplasts were stained with Mitotracker.
(C) Leaves were infiltrated with Agrobacterium carrying a GFP fusion construct of At1g06750 or At1g06750-L3. The agroinfiltrated leaves were examined through a confocal laser scanning microscope. Bars = 20 μm.
(D) Immunoblot analysis of localization. The indicated constructs were transformed into protoplasts and incubated in the presence (+) or absence (−) of tunicamycin (10 μg/mL) for 14 h after transformation. Protein extracts were prepared from the transformed protoplasts and analyzed by immunoblotting using anti-GFP antibody. Glycosylated, glycosylated form; Unglycosylated, unglycosylated form. glyNC indicates the two glycosylation sites at the N and C termini of the construct. WT, At1g06750:GFP(glyNC); L1, At1g06750-L1:GFP(glyNC); L2, At1g06750-L2:GFP(glyNC); L3, At1g06750-L3:GFP(glyNC); G4, At1g06750-G4:GFP(glyC).
(E) Subcellular fractionation. Mitochondria and ER fractions were obtained from protoplasts transformed with a GFP fusion construct of At1g06750:GFP(glyNC) or At1g06750-L3:GFP(glyNC). The isolated fractions were analyzed by immunoblotting using the indicated antibodies. T, total extracts; MT, protein extracts of purified mitochondria; ER, protein extracts of purified ER; anti-CRT (calreticulin) and anti-porin antibodies were used as markers for ER and mitochondria, respectively. WT, At1g06750:GFP(glyNC); L3, At1g06750-L3:GFP(glyNC).
To confirm this further at the biochemical level, we performed immunoblot analysis using proteins from transformed protoplasts. Again, two N-glycosylation sites were introduced into the GFP fusion constructs, one at the N terminus of At1g06750 (Figure 8A) and the other at the C terminus of GFP. The modifications did not change the localization patterns (see Supplemental Figure 7 online). Protoplasts were transformed with these modified constructs and incubated in the presence or absence of tunicamycin. Protein extracts from the transformed protoplasts were analyzed by immunoblotting using anti-GFP antibody (Figure 8D). The upper and lower bands indicated N-glycosylated and unglycosylated reporter proteins that were targeted to the ER and endosymbiotic organelles, respectively. Consistent with image analysis, the amount of the upper band gradually increased with increasing hydrophobicity. To obtain supporting data for the localization of reporter proteins, we performed immunoblot analysis of subcellular fractions using antibodies directed against GFP and organelle markers. Consistent with the localization data of image and glycosylation pattern analyses, At1g06750:GFP(glyNC) and At1g06750-L3:GFP(glyNC) proteins were detected primarily in the mitochondria and the ER fraction, respectively (Figure 8E).
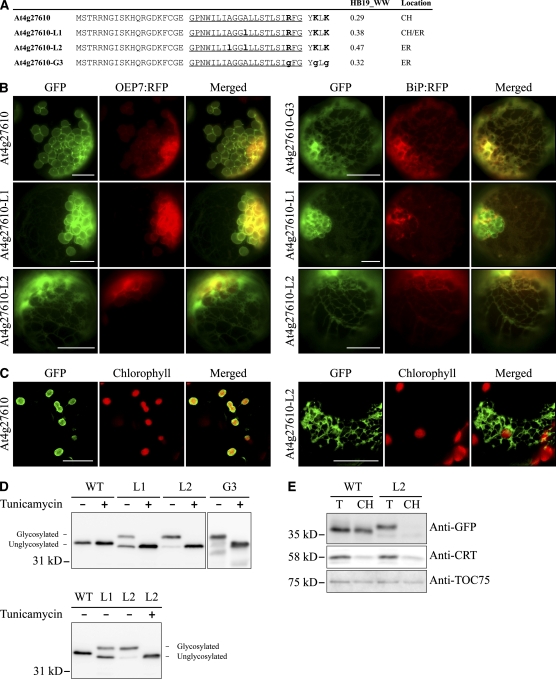
Next, we tested whether a chloroplast C-SA protein can also be targeted to the ER by increasing the hydrophobicity of the TMD. At4g27610, having a TMD with a hydrophobicity value of 0.29 on the WW hydrophobicity scale, was targeted to chloroplasts when expressed as a GFP fusion protein. The TMD hydrophobicity of At4g27610 was increased by substituting one or two Leu residues for one or two Ala residues, respectively. In addition, to test the importance of the CPR in chloroplast targeting, At4g27610-G3 was generated by substituting its CPR with three Gly residues (Figure 9A). GFP fusion protein constructs generated using the N-terminal TMD and CPR of the wild type and mutants of At4g27610 were introduced into protoplasts together with OEP7:RFP or BiP:RFP, and localization of these proteins was examined (Figure 9B). At4g27610-L1:GFP with only one Leu substitution for Ala and with a TMD hydrophobicity value of 0.38 on the WW hydrophobicity scale produced both chloroplast and ER patterns. In addition, At4g27610-L2:GFP with a substitution of two Leu for Ala residues and with a TMD hydrophobicity value of 0.47 on the WW hydrophobicity scale primarily produced the ER pattern, again confirming that the hydrophobicity is critical for targeting specificity of SA proteins. By contrast, At4g27610-G3:GFP produced the ER pattern, confirming that the CPR is also necessary for chloroplast targeting of the wild-type protein. Consistent with the results obtained with protoplasts, At4g27610:GFP and At4g27610-L2:GFP showed chloroplast and ER localization patterns, respectively, in the intact cells of agroinfiltrated leaf tissues (Figure 9C).
Figure 9.
Increasing the Hydrophobicity of the TMD of a Chloroplast-Targeted Protein Shifts Targeting Specificity to the ER.
(A) N-terminal sequences of At4g27610 and its mutant constructs. Ala residues in the TMD were substituted with Leu, as indicated by bold and lowercase letters. In addition, the CPR was removed by a Gly substitution. Hydrophobicity was determined using the WW scale (HB19_WW). The TMDs are underlined, and Lys and Arg residues in the CPR are highlighted in bold. An N-glycosylation site was added to the C terminus of GFP.
(B) and (C) In vivo localization.
(B) Protoplasts were transformed with the indicated constructs, and localization was examined.
(C) Leaves were infiltrated with Agrobacterium carrying a GFP fusion construct of At4g27610 or At4g27610-L2. The agroinfiltrated leaves were examined through a confocal laser scanning microscope. Bars = 20 μm.
(D) Immunoblot analysis of localization. The indicated constructs were transformed into protoplasts and incubated in the presence (+) or absence (−) of tunicamycin (10 μg/mL) for 14 h after transformation. Protein extracts were prepared from the transformed protoplasts and analyzed by immunoblotting using anti-GFP antibody. Glycosylated, glycosylated form; Unglycosylated, unglycosylated form. glyC indicates the N-glycosylation at the C terminus of GFP. WT, At4g27610:GFP(glyC); L1, At4g27610-L1:GFP(glyC); L2, At4g27610-L2:GFP(glyC); G3, At4g27610-G3:GFP(glyC).
(E) Subcellular fractionation. Chloroplast fractions were obtained from protoplasts transformed with a GFP fusion construct of At4g27610:GFP(glyC) or At4g27610-L2:GFP(glyC). The isolated fractions were analyzed by immunoblotting using the indicated antibodies. T, total extracts; CH, protein extracts of purified chloroplasts; anti-TOC75 and anti-CRT (calreticulin) antibodies were used as markers for chloroplasts and ER, respectively. WT, At4g27610:GFP(glyC); L2, At4g27610-L2:GFP(glyC).
To confirm the results obtained from image analysis at the biochemical level, we fused the N-terminal regions of the wild type and mutants of At4g27610 to GFP(glyC) that had an N-glycosylation site at the C terminus of GFP, and the resulting constructs were introduced into protoplasts. The transformed protoplasts were incubated in the presence or absence of tunicamycin, and protein extracts from the transformed protoplasts were analyzed by immunoblotting using anti-GFP antibody. As expected, the chloroplast-targeted wild-type protein did not produce the glycosylated form (Figure 9D). However, the amount of the glycosylated form increased gradually with increasing hydrophobicity, confirming that increased hydrophobicity causes ER targeting. In addition, At4g27610-G3:GFP(glyC) with a substitution of Gly residues for its CPR resulted in N-glycosylation, confirming that the CPR is important for evading ER targeting. Next, to obtain supporting data for the localization of reporter proteins, we analyzed subcellular fractions containing chloroplasts by immunoblot analysis using antibodies directed against GFP and organelle markers. Unglycosylated At4g27610:GFP(glyC) and At4g27610-L2:GFP(glyC) were detected in the chloroplast fraction. By contrast, the N-glycosylated form of At4g27610-L2:GFP(glyC) was not detected in the chloroplast fraction (Figure 9E), confirming the localization data from the image and glycosylation pattern analyses.
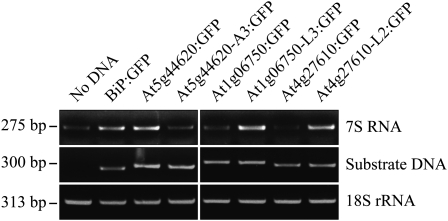
ER Targeting Correlates with SRP Binding to TMDs of High Hydrophobicity
Next, we examined whether the SRP is involved in the ER targeting of chloroplast and mitochondrial C-SA protein mutants that had increased hydrophobicity values in their TMDs. The SRP is known to recognize the hydrophobic TMD for the targeting of membrane proteins (Halic and Beckmann, 2005; Rapoport, 2007). To test SRP binding to TMDs of C-SA proteins, we used an in vitro transcription-translation coupled system. GFP fusion constructs At5g44620:GFP, At5g44620-A3:GFP, At1g06750:GFP, At1g06750-L3:GFP, At4g27610:GFP, and At4g27610-L2:GFP were truncated so that they would produce polypeptides of N-terminal 89 to 95 amino acid residues that contain only the TMD, CPR, and the first 40 amino acid residues of GFP. The truncated constructs were translated in rabbit reticulocyte lysates. Since the truncated constructs had no termination codon, ribosomes could not be released from the nascent polypeptide but instead remained stalled at the end of polypeptide. Ribosome/mRNA/nascent polypeptide complexes (RNCs) were precipitated by ultracentrifugation. The presence of the SRP in the precipitates was determined by RT-PCR using 7S RNA-specific primers. Since 7S RNA is one of the major SRP components (Halic and Beckmann, 2005), it can be conveniently used to examine for the presence of the SRP. Samples without any DNA or with BiP:GFP encoding an ER luminal protein were included as negative and positive controls, respectively. 7S RNA was only detected weakly in the sample without DNA, which likely resulted from the weak binding affinity of the SRP for ribosomes (Figure 10). By contrast, BiP:GFP produced a much stronger signal, confirming the enhanced interaction between the SRP and BiP:GFP. A hydrophobic leader sequence in the nascent polypeptide increases the SRP binding affinity for the ribosome, and the SRP also binds to the hydrophobic leader sequence, resulting in enhanced binding of the SRP to the RNC (Berndt et al., 2009). Next, we compared the 7S RNA intensity in the precipitates of samples containing ER- and endosymbiotic-targeted versions of GFP fusion proteins. ER-targeted GFP fusion proteins, At5g44620:GFP, At1g06750-L3:GFP, and At4g27610-L2:GFP, showed stronger 7S RNA signals than did their chloroplast/mitochondria-targeted counterparts, At5g44620-A3:GFP, At1g06750:GFP, and At4g27610:GFP, respectively (Figure 10). These results indicate that increasing the TMD hydrophobicity value >0.4 in mitochondrial and chloroplast C-SAs causes SRP binding and the ER targeting of GFP fusion proteins.
Figure 10.
ER Targeting of SA Proteins Correlates with SRP Binding to the TMD.
GFP fusion protein constructs At5g44620:GFP, At5g44620-A3:GFP, At1g06750:GFP, At1g06750-L3:GFP, At4g27610:GFP, At4g27610-L2:GFP, and BiP:GFP were amplified by PCR. The PCR products were used for in vitro transcription and translation in the rabbit reticulocyte lysate system. After transcription/translation reactions, RNCs were isolated by ultracentrifugation and RNA was purified. RT-PCR was performed to detect the presence of SRP 7S RNA and GFP fusion proteins. 18S rRNA was included in the analysis as an internal control for RT-PCR.
DISCUSSION
To define the exact sequence information that differentiates the targeting of C-SA proteins between the ER and endosymbiotic organelles, we performed massive in vivo targeting and subcellular fractionation experiments. From these data, we concluded that the targeting of a C-SA protein to endosymbiotic organelles requires two conditions, the CPR and a TMD with a hydrophobicity value <0.4 on the WW hydrophobicity scale. However, in this study, we focused on the targeting specificity conferred by a segment containing the TMD and its short flanking region when expressed in the form of an SA GFP fusion protein. Thus, despite the fact that we confirmed targeting in some cases with full-length proteins, we still cannot rule out the possibility that the targeting specificity conferred by the TMD and its short flanking region is different from that of the full-length protein from which the TMD and its short flanking region were derived. Previously, the importance of the CPR in targeting C-SA membrane proteins to chloroplasts has been demonstrated with two chloroplast proteins, OEP7 and TOC64 (Lee et al., 2001, 2004). In addition, four Arg residues and one Lys residue are found at the C-terminal flanking region of the TMD of the mitochondrial protein mtOM64, which is consistent with the finding that positively charged residues are important in protein targeting to mitochondria in animal cells (Kanaji et al., 2000; Waizenegger et al., 2003; Chew et al., 2004). Consistent with this observation, chloroplast or mitochondrial protein mutants lacking a CPR were targeted to the ER. This raises the possibility that the CPR is a general requirement for targeting C-SA proteins to endosymbiotic organelles in all eukaryotic cells. Thus, one possible explanation is that the presence of the CPR right next to the TMD prevents binding of the SRP to the hydrophobic TMD of nascent C-SA proteins during translation, thereby preventing SRP-mediated ER targeting of chloroplast or mitochondrial C-SA proteins, as suggested by previous studies (Kanaji et al., 2000; Lee et al., 2001). In addition, the CPR may also play an additional role in targeting C-SA proteins to chloroplast or mitochondrial OEMs (Bae et al., 2008). Consistent with this hypothesis, AKR2A, which plays a critical role in targeting C-SA proteins to the chloroplast OEM, binds to a TMD in vitro only when the TMD has a CPR (Bae et al., 2008). However, a protein factor that recognizes the TMD and CPR in SA mitochondrial proteins has not been identified.
Many ER-targeted SA proteins also have a TMD that is flanked by a CPR (Nelson and Strobel, 1988; Kanaji et al., 2000). In this study, we demonstrated that the hydrophobicity of the TMD plays a critical role in determining the targeting specificity of C-SA proteins to the ER and endosymbiotic organelles. Of the three different hydrophobicity scales we tested, the WW hydrophobicity scale was superior in differentiating the hydrophobicity of the TMD between ER and endosymbiotic organellar C-SA proteins. Previous reports demonstrated that the WW scale predicts TMD helices more accurately than did other hydrophobicity scales by considering the free energy of dehydrating peptide bonds that affect the stability of TMD helices (Jayasinghe et al., 2001; White and von Heijne, 2008). Thus, the WW hydrophobicity scale may provide a better indication of the hydrophobicity of TMD helices. The TMDs of endosymbiotic organellar C-SA proteins had an average WW hydrophobicity value of 0.23, which was lower than the average WW hydrophobicity value of 0.50 for ER C-SA proteins. This finding is consistent with the results for animal cells showing that the TMDs of mitochondrial OEM proteins have moderate hydrophobicity, whereas the TMDs of ER proteins have higher hydrophobicity (Kanaji et al., 2000; Waizenegger et al., 2003; Walther and Rapaport, 2009). However, in animal cells, the concept of moderate hydrophobicity is ambiguous, and different hydrophobicity scales can produce different hydrophobicity values for the same TMD. Therefore, it is difficult to predict the localization of SA proteins based on the hydrophobicity value. In this study, we demonstrated that TMDs with a hydrophobicity value >0.4 on the WW hydrophobicity scale are targeted primarily to the ER, whereas TMDs with lower values are targeted to endosymbiotic organelles. With this result, it could be possible to predict localization of C-SA proteins.
However, it is not clear how hydrophobicity affects targeting. The interaction between SRP54 and the signal sequence is mediated by hydrophobic interactions (Halic and Beckmann, 2005; Janda et al., 2010). One possibility is that the difference in the hydrophobicity influences the strength of interaction between the targeting signal and cytosolic targeting factors, such as the SRP and AKR2 of ER and chloroplast proteins, respectively. When the TMD has high hydrophobicity, the interaction between the SRP and the TMD is strong enough to override any effects of the CPR (Kanaji et al., 2000). By contrast, when the hydrophobicity value of the TMD is <0.4 on the WW hydrophobicity scale, the interaction between the TMD and the SRP is too weak to overcome the inhibitory effect of the CPR on the interaction between the SRP and the TMD.
Exceptions to these general effects of hydrophobicity were evident. Approximately 11% of ER C-SA proteins and 15% of endosymbiotic organellar C-SA proteins had TMDs with hydrophobicity values that did not follow the expected tendencies. These exceptions are not clearly understood. The length, structural features, conserved amino acids, and amino acid composition of TMDs, as well as the net charge of CPRs, has been suggested to affect the localization of tail-anchored and SA proteins (Kanaji et al., 2000; Lee et al., 2001; Borgese et al., 2003; Hwang et al., 2004; Maggio et al., 2007). In addition, other targeting factors may be involved in the determination of targeting specificity. Thus, other factors still have to be considered before the localization of a particular SA protein can be predicted accurately.
Here, we focus on how C-SA proteins are differentially targeted between the ER and endosymbiotic organelles. Another important question we did not address is how C-SA membrane proteins that escape SRP-mediated ER targeting are specifically targeted to chloroplast or mitochondrial OEMs. In animal cells and yeast that have only mitochondria, this question is not relevant. However, this question is important in plant cells. The N-terminal domain we used in in vivo targeting experiments is sufficient for specific targeting to either chloroplasts or mitochondria, suggesting that the N-terminal domain contains an additional signal for specific targeting to these organelles. However, the additional signal for specific targeting to chloroplasts or mitochondria is unknown.
The SRP binds to ribosomes weakly, and its binding affinity is enhanced when the hydrophobic leader sequence of ER luminal proteins or the TMD in membrane proteins emerges from the ribosome (Flanagan et al., 2003; Berndt et al., 2009). This raises the possibility that all SA proteins are recognized and sorted by the SRP during translation for targeting to the ER. However, in the cell, many SA proteins are targeted to endosymbiotic organelles and even some ER proteins are transported posttranslationally (Rapoport, 2007). Thus, the mechanism for targeting SA proteins to endosymbiotic organelles should function in such a way that avoiding SRP-mediated targeting is a prerequisite for targeting to endosymbiotic organelles. This finding may have an important implication for eukaryotic cell evolution; the targeting signals of various organellar proteins may contain clues about the cellular environment of the host cell when new organelles were established during evolution. The SRP-mediated protein targeting mechanism in bacteria must be one of the earliest protein targeting mechanisms (Driessen and Nouwen, 2008). Thus, during evolution, when endosymbiotic bacteria were converted to endosymbiotic organelles, the SRP-mediated protein targeting mechanism presumed to operate in the host cell must have been avoided in order to target SA proteins to newly establishing chloroplasts or mitochondria. Indeed, the presence of a CPR in SA proteins targeted to chloroplasts and mitochondria is consistent with this reasoning.
METHODS
Growth of Plants
Arabidopsis thaliana (Colombia ecotype) was cultivated on Gamborg B5 plates (Duchefa; G0210.0050) under conditions of 40% relative humidity, 22°C, and a 16-h-light/8-h-dark cycle in a growth chamber. Leaf tissues of 2- to 3-week-old plants were used to isolate protoplasts.
Plasmid DNA Construction and PCR-Based Mutagenesis
DNA fragments encoding a putative signal anchor region of membrane proteins (listed in Tables 1 and 2) were amplified from Arabidopsis genomic DNA or cDNA by PCR using specific primers. All sequences of the primers are described in Supplemental Table 4 online. The PCR products were digested with restriction enzymes and ligated into a pUC-GFP vector, containing the cauliflower mosaic virus 35S promoter, GFP, and Nos terminator (Jin et al., 2001) in such a way that the N-terminal signal anchor region was fused in frame to the N terminus of GFP. For in vitro transcription/translation-coupled reactions, GFP fusion protein constructs, At5g44620:GFP, At5g44620-A3:GFP, At1g06750:GFP, At1g06750-L3:GFP, At4g27610:GFP, At4g27610-L2:GFP, and BiP:GFP, were digested with XhoI/EcoRI restriction enzymes and ligated into pBluescript II SK(+). For generating site-specific amino acid substitution mutants, each pair of complementary upper and lower primers, of which the central regions were mutated according to the replaced amino acid residues, were designed. In the first round of PCR, upper and lower PCR products were obtained with each primer set of the upper/Nos and the lower/35S Pro, respectively (Nos primer for the Nos terminator and 35S Pro for the 5′ end region of the 35S promoter in the pUC-GFP vector). In the second round of PCR, the upper/lower PCR products were used as templates, and mutated PCR products were amplified using the 35S Se/Nos primer set (35S se for the 3′ end of the 35S promoter in the pUC-GFP vector).
To induce N-glycosylation of SA proteins in the ER lumen, a consensus N-glycosylation sequence was introduced at the N-terminal region of each SA protein or at the C-terminal region of the GFP. The Asn of each N-glycosylation sequence was flanked by a single Lys or Arg residue to be more accessible to oligosaccharidyl transferase (Chang et al., 1994). To generate the pUC-GFP-glyC vector that had an N-glycosylation site at the C terminus, the GFP DNA fragment was amplified using the primer set of 35S Se/GFP-N-gly-R. The PCR product was digested with XbaI/NotI restriction enzymes and ligated into the pUC vector. The N-terminal N-glycosylation site was introduced by PCR using primers At5g44620-N-gly-F/Nos for At5g44620 and its mutant constructs and At1g06750-N-gly-F/Nos for At1g06750 and its mutants. The PCR products were digested with XhoI/BamHI and ligated into the pUC-GFP-glyC vector. The nucleotide sequence of all the PCR products was confirmed by sequencing.
Prediction of Putative SA Proteins and Calculation of Hydrophobicity
To select C-SA membrane proteins rather than luminal proteins, a two-step approach was devised using Conpred II and SPOCTOPUS algorithms. In the first step, through Conpred II, transmembrane domain profiles of protein sequences were obtained, and among them membrane proteins that have a transmembrane domain within the N-terminal 40 amino acid residues were selected. In the second step, putative membrane proteins were reanalyzed with SPOCTOPUS to remove ER signal sequence–containing proteins, which are often confused with SA proteins by transmembrane domain predictors. The TMD was determined by algorithm SPOCTOPUS. To obtain SA proteins listed in Figure 1A and Supplemental Table 1 online, we selected them from among proteins whose localizations were inferred by GFP or tandem mass spectrometry assays from the SUBA database using a two-step approach with and without the CPR criterion, respectively. In the case of Supplemental Table 3 online, we used protein sequences downloaded from the Swiss-Prot/Trembl database for the C-SA protein screening.
To measure the hydrophobicity of the TMD of each protein, the mean hydrophobicity of 19 amino acid residues was calculated in order from the N terminus and the highest value was taken to represent the hydrophobicity of each TMD. Three different hydrophobicity scales (KD, WW, and GES) were used to calculate hydrophobicity (Kyte and Doolittle, 1982; Engelman et al., 1986; Wimley and White, 1996).
Transient Expression in Protoplasts and Treatment of Tunicamycin
All DNA plasmids were purified with Qiagen plasmid purification kits. Arabidopsis protoplasts were prepared from leaf tissues, and purified DNA plasmids were introduced into protoplasts by PEG-mediated transformation following previous studies (Jin et al., 2001; Lee et al., 2002a). After a 14- to 16-h incubation at 22°C in a growth chamber, transformed protoplasts were observed with a microscope or sampled for immunoblot analysis. For the Mitotracker staining, transformed protoplasts were incubated in W5 medium (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, 5 mM glucose, and 1.5 mM MES, pH 5.6) containing 100 nM Mitotracker for 10 min at room temperature in the dark. Stained protoplasts were washed two times with fresh W5 medium and observed with a microscope after a 3-h incubation at 22°C in the dark. To examine the glycosylation pattern of GFP fusion proteins, transformed protoplasts were incubated with or without tunicamycin (10 μg/mL) for 14 h in the dark.
Immunoblotting and Subcellular Fractionation
The preparation of protein extracts from transformed protoplasts followed previous methods (Kim et al., 2001). Protoplasts were collected in an E-tube, and incubation solution was removed by quick centrifugation. After discarding the incubation solution, protoplasts were lysed by vortexing in denaturation buffer (2.5% SDS and 2% β-mercaptoethanol) and sampling buffer (250 mM Tris-Cl, pH 6.8, 0.5 M DTT, 10% SDS, 0.05% Bromophenol blue, and 50% glycerol). The lysed sample was boiled for 7 min and centrifuged at 10,000g and 4°C to remove cell debris. Proteins were separated by SDS-PAGE and transferred to polyvinylidene fluoride membranes using protein electrophoresis and blotting apparatuses (Hoefer). Protein blots were developed with an enhanced chemiluminescence kit (Amersham Pharmacia Biotech) and visualized using the LAS3000 image capture system (FUJIFILM). Anti-GFP (Clontech), anti-CRT, anti-porin (Calbiochem), and anti-TOC75 antibodies were used, as described previously (Lee et al., 2006; Song et al., 2006).
Subcellular fractions containing chloroplasts, mitochondria, or the ER were obtained from protoplasts by modified fractionation protocols (Hawes and Satiat-Jeunemaitre, 2001). Transformed protoplasts were resuspended in 3 mL HMS buffer (330 mM sorbitol, 50 mM HEPES-KOH, pH 7.6, and 3 mM MgCl2) and filtered gently three times through two layers of 11-μm nylon net filters (Millipore) for homogenization. To obtain subcellular fractions containing chloroplasts, the homogenate was incubated on ice for 30 min after the addition of 10 mM EDTA. After the incubation, the homogenate was loaded onto 5 mL of 40% Percoll (Sigma-Aldrich) in HMS buffer and centrifuged at 7500g at 4°C for 90 min (SW41 swing rotor; Beckman Coulter). Intact chloroplasts formed a loose pellet at the base of the tube. The upper fraction was removed carefully with a pipette, and the remaining solution with a volume <0.5 mL containing the loose pellet was collected and diluted with 1 mL HMS buffer in an eppendorf tube and centrifuged at 7500g at 4°C for 20 min. After centrifugation, the loose pellet at the base of the tube was collected with a 1-mL pipette and resuspended gently in 1 mL HMS buffer. The chloroplasts were then pelleted at the bottom of the tube by centrifugation at 7500g at 4°C for 15 min. To obtain fractions containing mitochondria or the ER, the homogenate was incubated on ice for 30 min in 3 mL HMS buffer supplemented with 20 mM EDTA and 10 mM EGTA and centrifuged at 900g at 4°C for 6 min to remove chloroplasts. The supernatant was then loaded onto a two-step sucrose gradient (from bottom to top: 2 mL of 60% and 4 mL of 36% sucrose in 50 mM HEPES-KOH, pH 7.6, 0.1 mM MgCl2, and 3 mM EDTA) and centrifuged at 40,000g at 4°C for 90 min (SW41 swing rotor). ER and mitochondrial fractions were collected at the top of the 36% sucrose layer and at the 36/60% sucrose interface, respectively. Each fraction was diluted in 9 mL HMS buffer, and organelles were pelleted by centrifugation at 40,000g at 4°C for 40 min (SW41 swing rotor).
Agroinfiltration and Microscopy
DNA constructs selected for agroinfiltration were digested with XbaI/EcoRI restriction enzymes and ligated into the pBIB binary vector (Becker, 1990), and the resulting constructs were transformed into Agrobacterium tumefaciens. Agrobacteria were cultured overnight at 28°C in 5 mL Luria-Bertani medium containing 50 μg/mL rifampicin and 50 μg/mL kanamycin. The overnight culture (0.5 mL) was inoculated into 5 mL of fresh Luria-Bertani medium containing 50 μg/mL kanamycin and grown to OD 1 to 2 at 600 nm. The bacteria were harvested by centrifugation at 3000g and resuspended in induction medium (50 mM MES, pH 5.6, 0.5% glucose, 2 mM NaH2PO4·2H2O, 200 μM acetosyringone, and 1× AB salts [20× AB salts: 20 g/L NH4Cl, 6 g/L MgSO4·7H2O, 3 g/L KCl, 0.2 g/L CaCl2·2H2O, and 50 mg/L FeSO4·7H2O]) to OD 0.2 at 600 nm and then incubated at 28°C for 6 h (Yang et al., 2000). After incubation, the culture was diluted with induction medium to 0.2 OD and injected into leaves using a 1-mL syringe without a needle as described previously (Wroblewski et al., 2005). Agroinfiltrated plants were kept in the dark for 1 d, and infected leaves were observed through a confocal laser scanning microscope after 3 d. To stain mitochondria with Mitotracker, agroinfiltrated leaf tissues were incubated in B5 medium containing 500 nM Mitotracker for 5 min at room temperature and washed three times with fresh B5 medium (Ueda et al., 2006).
The GFP, RFP, and Mitotracker images of protoplasts were obtained with a Zeiss Axioplan fluorescence microscope and a cooled CCD camera (Zeiss). The filter settings were XF116 (exciter, 474AF20; dichroic, 500DRLP; emitter, 510AF23), XF33/E (exciter, 535DF35; dichroic, 570DRLP; emitter, 605DF50), and XF137 (exciter, 540AF30; dichroic, 570DRLP; emitter, 585ALP) (Omega) for GFP, RFP, and autofluorescence of chlorophyll, respectively.
Agroinfiltrated leaves were observed through a confocal laser scanning microscope (Carl Zeiss LSM 510 META system). The excitation wavelengths/emission filters were 488 nm (argon-ion laser)/505 to 530 band-pass for GFP, 543 nm (HeNe laser)/560 long-pass for chlorophyll autofluorescence, and 543 nm/560 to 615 band-pass for Mitotracker. Images are presented in pseudocolor.
In Vitro Transcription and Translation Couple Reaction and RT/PCR for SRP Detection
Truncated GFP fusion constructs were generated by PCR using GFP fusion protein constructs At5g44620:GFP, At5g44620-A3:GFP, At1g06750:GFP, At1g06750-L3:GFP, At4g27610:GFP, At4g27610-L2:GFP, and BiP:GFP in pBluescript II SK(+) as templates. PCR primers used were 5′-CCGCCGCGCTTAATGCGCCG-3′ (200-bp upstream region from the T7 promoter) and 5′-GTAGGTGGCATCGCCCTC-3′ (corresponding to the N-terminal 40 amino acid residues of GFP). PCR product (300 ng), 1 μL of the T7 TNT PCR enhancer, and 1 μL of 1 mM Met were added to 40 μL of TNT T7 master mix containing rabbit reticulocyte lysates, amino acids, T7 RNA polymerase, and reaction buffer (TNT Quick Coupled Transcription/Translation System; Promega). Final reaction volume was adjusted to 50 μL with nuclease-free water. The reaction mixture was gently mixed by pipetting and incubated at 30°C for 70 min according to the manufacturer’s protocol.
After the in vitro translation reaction, samples were used to purify RNCs. The reaction mixtures (50 μL) were layered onto 500 μL isolation buffer [25% sucrose, 50 mM HEPES-KOH, pH 7.5, 550 mM KOAc, pH 7.5, 5 mM Mg(OAc)2] and centrifuged at 100,000g at 4°C for 50 min (TLA 120.2 rotor; Beckman Coulter). The pellet was resuspended gently in 25 μL isolation buffer without sucrose. Total RNA was isolated from the pellet using Trizol reagent (Invitrogen) and used for RT-PCR. PCR was performed using primers for 7S RNA, 18S rRNA, and GFP fusion proteins (see Supplemental Table 4 online) under the following conditions: 94°C for 30 s, 52°C for 30 s, and 72°C for 20 s with 20 cycles.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: OEP7, At3g52420; mtOM64, At5g09420; BiP, At5g28540; GGPS2, At2g23800; At5g20520; At3g48890; At1g26710; At1g66770; and At5g42590. Other genes are listed in Tables 1 and 2.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Chloroplast and Mitochondrial Targeted SA Proteins from Arabidopsis Contain the CPR.
Supplemental Figure 2. Localization of Putative Membrane Proteins without CPRs.
Supplemental Figure 3. SA Proteins of the ER Tend to Have TMDs with Higher Hydrophobicity Than Those of Endosymbiotic Organelles.
Supplemental Figure 4. Reexamination of Localization of Certain Membrane Proteins from the Proteomics Data.
Supplemental Figure 5. Immunoblot Analysis of GFP Fusion Proteins.
Supplemental Figure 6. In Vivo Targeting of Putative C-SA Proteins in Protoplasts.
Supplemental Figure 7. The N- and C-Terminal N-Glycosylation Sites Do Not Affect the Localization.
Supplemental Figure 8. Lowering the Hydrophobicity of the TMD of ER-Targeted At3g44110 Shifts Targeting Specificity to Chloroplasts and Mitochondria.
Supplemental Table 1. The Hydrophobicity Values of ER and Endosymbiotic Organellar SA Proteins Obtained from Proteomics Data.
Supplemental Table 2. Mitochondria-Targeted SA Proteins of Mammals and Yeast.
Supplemental Table 3. 217 Putative Signal-Anchored Proteins Containing a CPR.
Supplemental Table 4. Sequences of Primers Used in This Study.
Acknowledgments
We thank Hsou-min Li (Academi of Sinica, Nankang, Taiwan) for the anti-TOC75 antibody. This work was supported in parts by the World Class University program (R31-2008-000-10105-0), the National Research Foundation of Korea (Grant 20110000025), and the Advenced Biomass R&D Center (20100029720) from the Ministry of Education, Science, and Technology (Korea).
References
- Acencio M.L., Lemke N. (2009). Towards the prediction of essential genes by integration of network topology, cellular localization and biological process information. BMC Bioinformatics 10: 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agne B., Kessler F. (2009). Protein transport in organelles: The Toc complex way of preprotein import. FEBS J. 276: 1156–1165 [DOI] [PubMed] [Google Scholar]
- Arai M., Mitsuke H., Ikeda M., Xia J.X., Kikuchi T., Satake M., Shimizu T. (2004). ConPred II: a consensus prediction method for obtaining transmembrane topology models with high reliability. Nucleic Acids Res. 32(Web Server issue): W390–W393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano T., Yoshioka Y., Kurei S., Sakamoto W., Machida Y.; Sodmergen (2004). A mutation of the CRUMPLED LEAF gene that encodes a protein localized in the outer envelope membrane of plastids affects the pattern of cell division, cell differentiation, and plastid division in Arabidopsis. Plant J. 38: 448–459 [DOI] [PubMed] [Google Scholar]
- Assfalg J., Gong J., Kriegel H.P., Pryakhin A., Wei T., Zimek A. (2009). Supervised ensembles of prediction methods for subcellular localization. J. Bioinform. Comput. Biol. 7: 269–285 [DOI] [PubMed] [Google Scholar]
- Bae W., Lee Y.J., Kim D.H., Lee J., Kim S., Sohn E.J., Hwang I. (2008). AKR2A-mediated import of chloroplast outer membrane proteins is essential for chloroplast biogenesis. Nat. Cell Biol. 10: 220–227 [DOI] [PubMed] [Google Scholar]
- Baginsky S., Gruissem W. (2004). Chloroplast proteomics: potentials and challenges. J. Exp. Bot. 55: 1213–1220 [DOI] [PubMed] [Google Scholar]
- Balsera M., Soll J., Bölter B. (2009). Protein import machineries in endosymbiotic organelles. Cell. Mol. Life Sci. 66: 1903–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker D. (1990). Binary vectors which allow the exchange of plant selectable markers and reporter genes. Nucleic Acids Res. 18: 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndt U., Oellerer S., Zhang Y., Johnson A.E., Rospert S. (2009). A signal-anchor sequence stimulates signal recognition particle binding to ribosomes from inside the exit tunnel. Proc. Natl. Acad. Sci. USA 106: 1398–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolender N., Sickmann A., Wagner R., Meisinger C., Pfanner N. (2008). Multiple pathways for sorting mitochondrial precursor proteins. EMBO Rep. 9: 42–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgese N., Colombo S., Pedrazzini E. (2003). The tale of tail-anchored proteins: Coming from the cytosol and looking for a membrane. J. Cell Biol. 161: 1013–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braulke T., Bonifacino J.S. (2009). Sorting of lysosomal proteins. Biochim. Biophys. Acta 1793: 605–614 [DOI] [PubMed] [Google Scholar]
- Bruce B.D. (2000). Chloroplast transit peptides: Structure, function and evolution. Trends Cell Biol. 10: 440–447 [DOI] [PubMed] [Google Scholar]
- Chang X.B., Hou Y.X., Jensen T.J., Riordan J.R. (1994). Mapping of cystic fibrosis transmembrane conductance regulator membrane topology by glycosylation site insertion. J. Biol. Chem. 269: 18572–18575 [PubMed] [Google Scholar]
- Chew O., Lister R., Qbadou S., Heazlewood J.L., Soll J., Schleiff E., Millar A.H., Whelan J. (2004). A plant outer mitochondrial membrane protein with high amino acid sequence identity to a chloroplast protein import receptor. FEBS Lett. 557: 109–114 [DOI] [PubMed] [Google Scholar]
- Dhanoa P.K., Richardson L.G., Smith M.D., Gidda S.K., Henderson M.P., Andrews D.W., Mullen R.T. (2010). Distinct pathways mediate the sorting of tail-anchored proteins to the plastid outer envelope. PLoS ONE 5: e10098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driessen A.J., Nouwen N. (2008). Protein translocation across the bacterial cytoplasmic membrane. Annu. Rev. Biochem. 77: 643–667 [DOI] [PubMed] [Google Scholar]
- Egea P.F., Stroud R.M., Walter P. (2005). Targeting proteins to membranes: Structure of the signal recognition particle. Curr. Opin. Struct. Biol. 15: 213–220 [DOI] [PubMed] [Google Scholar]
- Engelman D.M., Steitz T.A., Goldman A. (1986). Identifying nonpolar transbilayer helices in amino acid sequences of membrane proteins. Annu. Rev. Biophys. Biophys. Chem. 15: 321–353 [DOI] [PubMed] [Google Scholar]
- Ephritikhine G., Ferro M., Rolland N. (2004). Plant membrane proteomics. Plant Physiol. Biochem. 42: 943–962 [DOI] [PubMed] [Google Scholar]
- Flanagan J.J., Chen J.C., Miao Y., Shao Y., Lin J., Bock P.E., Johnson A.E. (2003). Signal recognition particle binds to ribosome-bound signal sequences with fluorescence-detected subnanomolar affinity that does not diminish as the nascent chain lengthens. J. Biol. Chem. 278: 18628–18637 [DOI] [PubMed] [Google Scholar]
- Gierasch L.M. (1989). Signal sequences. Biochemistry 28: 923–930 [DOI] [PubMed] [Google Scholar]
- Halic M., Beckmann R. (2005). The signal recognition particle and its interactions during protein targeting. Curr. Opin. Struct. Biol. 15: 116–125 [DOI] [PubMed] [Google Scholar]
- Hawes C.R., Satiat-Jeunemaitre B. (2001). Plant Cell Biology: A Practical Approach. (Oxford; New York: Oxford University Press; ). [Google Scholar]
- Heazlewood J.L., Tonti-Filippini J., Verboom R.E., Millar A.H. (2005). Combining experimental and predicted datasets for determination of the subcellular location of proteins in Arabidopsis. Plant Physiol. 139: 598–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heazlewood J.L., Tonti-Filippini J.S., Gout A.M., Day D.A., Whelan J., Millar A.H. (2004). Experimental analysis of the Arabidopsis mitochondrial proteome highlights signaling and regulatory components, provides assessment of targeting prediction programs, and indicates plant-specific mitochondrial proteins. Plant Cell 16: 241–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I. (2008). Sorting and anterograde trafficking at the Golgi apparatus. Plant Physiol. 148: 673–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang Y.T., Pelitire S.M., Henderson M.P., Andrews D.W., Dyer J.M., Mullen R.T. (2004). Novel targeting signals mediate the sorting of different isoforms of the tail-anchored membrane protein cytochrome b5 to either endoplasmic reticulum or mitochondria. Plant Cell 16: 3002–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda C.Y., Li J., Oubridge C., Hernández H., Robinson C.V., Nagai K. (2010). Recognition of a signal peptide by the signal recognition particle. Nature 465: 507–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis P. (2008). Targeting of nucleus-encoded proteins to chloroplasts in plants. New Phytol. 179: 257–285 [DOI] [PubMed] [Google Scholar]
- Jayasinghe S., Hristova K., White S.H. (2001). Energetics, stability, and prediction of transmembrane helices. J. Mol. Biol. 312: 927–934 [DOI] [PubMed] [Google Scholar]
- Jin J.B., Kim Y.A., Kim S.J., Lee S.H., Kim D.H., Cheong G.W., Hwang I. (2001). A new dynamin-like protein, ADL6, is involved in trafficking from the trans-Golgi network to the central vacuole in Arabidopsis. Plant Cell 13: 1511–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaji S., Iwahashi J., Kida Y., Sakaguchi M., Mihara K. (2000). Characterization of the signal that directs Tom20 to the mitochondrial outer membrane. J. Cell Biol. 151: 277–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.H., Eu Y.J., Yoo C.M., Kim Y.W., Pih K.T., Jin J.B., Kim S.J., Stenmark H., Hwang I. (2001). Trafficking of phosphatidylinositol 3-phosphate from the trans-Golgi network to the lumen of the central vacuole in plant cells. Plant Cell 13: 287–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaus C., Guiard B., Neupert W., Brunner M. (1996). Determinants in the presequence of cytochrome b2 for import into mitochondria and for proteolytic processing. Eur. J. Biochem. 236: 856–861 [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R.F. (1982). A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157: 105–132 [DOI] [PubMed] [Google Scholar]
- Lee D.W., Kim J.K., Lee S., Choi S., Kim S., Hwang I. (2008). Arabidopsis nuclear-encoded plastid transit peptides contain multiple sequence subgroups with distinctive chloroplast-targeting sequence motifs. Plant Cell 20: 1603–1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.W., Lee S., Lee G.J., Lee K.H., Kim S., Cheong G.W., Hwang I. (2006). Functional characterization of sequence motifs in the transit peptide of Arabidopsis small subunit of rubisco. Plant Physiol 140: 466–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.H., Kim D.H., Lee S.W., Kim Z.H., Hwang I. (2002a). In vivo import experiments in protoplasts reveal the importance of the overall context but not specific amino acid residues of the transit peptide during import into chloroplasts. Mol. Cells 14: 388–397 [PubMed] [Google Scholar]
- Lee M.H., Min M.K., Lee Y.J., Jin J.B., Shin D.H., Kim D.H., Lee K.H., Hwang I. (2002b). ADP-ribosylation factor 1 of Arabidopsis plays a critical role in intracellular trafficking and maintenance of endoplasmic reticulum morphology in Arabidopsis. Plant Physiol. 129: 1507–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.J., Kim D.H., Kim Y.W., Hwang I. (2001). Identification of a signal that distinguishes between the chloroplast outer envelope membrane and the endomembrane system in vivo. Plant Cell 13: 2175–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.J., Sohn E.J., Lee K.H., Lee D.W., Hwang I. (2004). The transmembrane domain of AtToc64 and its C-terminal lysine-rich flanking region are targeting signals to the chloroplast outer envelope membrane [correction]. Mol. Cells 17: 281–291 [PubMed] [Google Scholar]
- Maggio C., Barbante A., Ferro F., Frigerio L., Pedrazzini E. (2007). Intracellular sorting of the tail-anchored protein cytochrome b5 in plants: A comparative study using different isoforms from rabbit and Arabidopsis. J. Exp. Bot. 58: 1365–1379 [DOI] [PubMed] [Google Scholar]
- Millar A.H., Sweetlove L.J., Giegé P., Leaver C.J. (2001). Analysis of the Arabidopsis mitochondrial proteome. Plant Physiol. 127: 1711–1727 [PMC free article] [PubMed] [Google Scholar]
- Nelson D.R., Strobel H.W. (1988). On the membrane topology of vertebrate cytochrome P-450 proteins. J. Biol. Chem. 263: 6038–6050 [PubMed] [Google Scholar]
- Neupert W., Herrmann J.M. (2007). Translocation of proteins into mitochondria. Annu. Rev. Biochem. 76: 723–749 [DOI] [PubMed] [Google Scholar]
- Nielsen H., Engelbrecht J., Brunak S., von Heijne G. (1997). Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10: 1–6 [DOI] [PubMed] [Google Scholar]
- Oikawa K., Kasahara M., Kiyosue T., Kagawa T., Suetsugu N., Takahashi F., Kanegae T., Niwa Y., Kadota A., Wada M. (2003). Chloroplast unusual positioning1 is essential for proper chloroplast positioning. Plant Cell 15: 2805–2815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K., Saito T., Nakagawa T., Kawamukai M., Kamiya Y. (2000). Five geranylgeranyl diphosphate synthases expressed in different organs are localized into three subcellular compartments in Arabidopsis. Plant Physiol. 122: 1045–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petsalaki E.I., Bagos P.G., Litou Z.I., Hamodrakas S.J. (2006). PredSL: A tool for the N-terminal sequence-based prediction of protein subcellular localization. Genomics Proteomics Bioinformatics 4: 48–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platta H.W., Erdmann R. (2007). Peroxisomal dynamics. Trends Cell Biol. 17: 474–484 [DOI] [PubMed] [Google Scholar]
- Rabu C., Schmid V., Schwappach B., High S. (2009). Biogenesis of tail-anchored proteins: The beginning for the end? J. Cell Sci. 122: 3605–3612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapoport T.A. (1991). Protein transport across the endoplasmic reticulum membrane: Facts, models, mysteries. FASEB J. 5: 2792–2798 [DOI] [PubMed] [Google Scholar]
- Rapoport T.A. (2007). Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature 450: 663–669 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Boulan E., Müsch A. (2005). Protein sorting in the Golgi complex: Shifting paradigms. Biochim. Biophys. Acta 1744: 455–464 [DOI] [PubMed] [Google Scholar]
- Rolland N., Ferro M., Seigneurin-Berny D., Garin J., Douce R., Joyard J. (2003). Proteomics of chloroplast envelope membranes. Photosynth. Res. 78: 205–230 [DOI] [PubMed] [Google Scholar]
- Schnurr J.A., Shockey J.M., de Boer G.J., Browse J.A. (2002). Fatty acid export from the chloroplast. Molecular characterization of a major plastidial acyl-coenzyme A synthetase from Arabidopsis. Plant Physiol. 129: 1700–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Lee M.H., Lee G.J., Yoo C.M., Hwang I. (2006). Arabidopsis EPSIN1 plays an important role in vacuolar trafficking of soluble cargo proteins in plant cells via interactions with clathrin, AP-1, VTI11, and VSR1. Plant Cell 18: 2258–2274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovic S., Hegde R.S. (2007). Identification of a targeting factor for posttranslational membrane protein insertion into the ER. Cell 128: 1147–1159 [DOI] [PubMed] [Google Scholar]
- Ueda M., Arimura S., Yamamoto M.P., Takaiwa F., Tsutsumi N., Kadowaki K. (2006). Promoter shuffling at a nuclear gene for mitochondrial RPL27. Involvement of interchromosome and subsequent intrachromosome recombinations. Plant Physiol. 141: 702–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viklund H., Bernsel A., Skwark M., Elofsson A. (2008). SPOCTOPUS: A combined predictor of signal peptides and membrane protein topology. Bioinformatics 24: 2928–2929 [DOI] [PubMed] [Google Scholar]
- von Heijne G. (1992). Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J. Mol. Biol. 225: 487–494 [DOI] [PubMed] [Google Scholar]
- Waizenegger T., Stan T., Neupert W., Rapaport D. (2003). Signal-anchor domains of proteins of the outer membrane of mitochondria: Structural and functional characteristics. J. Biol. Chem. 278: 42064–42071 [DOI] [PubMed] [Google Scholar]
- Walter P., Johnson A.E. (1994). Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu. Rev. Cell Biol. 10: 87–119 [DOI] [PubMed] [Google Scholar]
- Walther D.M., Rapaport D. (2009). Biogenesis of mitochondrial outer membrane proteins. Biochim. Biophys. Acta 1793: 42–51 [DOI] [PubMed] [Google Scholar]
- White S.H., von Heijne G. (2008). How translocons select transmembrane helices. Annu. Rev. Biophys. 37: 23–42 [DOI] [PubMed] [Google Scholar]
- Wimley W.C., White S.H. (1996). Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat. Struct. Biol. 3: 842–848 [DOI] [PubMed] [Google Scholar]
- Wroblewski T., Tomczak A., Michelmore R. (2005). Optimization of Agrobacterium-mediated transient assays of gene expression in lettuce, tomato and Arabidopsis. Plant Biotechnol. J. 3: 259–273 [DOI] [PubMed] [Google Scholar]
- Yang Y., Li R., Qi M. (2000). In vivo analysis of plant promoters and transcription factors by agroinfiltration of tobacco leaves. Plant J. 22: 543–551 [DOI] [PubMed] [Google Scholar]