This work reveals the significant roles of myo-inositol in Arabidopsis embryogenesis. Experiments show that depletion of myo-inositol by knocking out multiple MIPS genes, which encode d-myo-inositol-3-phosphate synthase, could affect the membrane trafficking via the phosphatidylinositol metabolism by leading to altered auxin distribution in developing embryos.
Abstract
In animal cells, myo-inositol is an important regulatory molecule in several physiological and biochemical processes, including signal transduction and membrane biogenesis. However, the fundamental biological functions of myo-inositol are still far from clear in plants. Here, we report the genetic characterization of three Arabidopsis thaliana genes encoding d-myo-inositol-3-phosphate synthase (MIPS), which catalyzes the rate-limiting step in de novo synthesis of myo-inositol. Each of the three MIPS genes rescued the yeast ino1 mutant, which is defective in yeast MIPS gene INO1, and they had different dynamic expression patterns during Arabidopsis embryo development. Although single mips mutants showed no obvious phenotypes, the mips1 mips2 double mutant and the mips1 mips2 mips3 triple mutant were embryo lethal, whereas the mips1 mips3 and mips1 mips2+/− double mutants had abnormal embryos. The mips phenotypes resembled those of auxin mutants. Indeed, the double and triple mips mutants displayed abnormal expression patterns of DR5:green fluorescent protein, an auxin-responsive fusion protein, and they had altered PIN1 subcellular localization. Also, membrane trafficking was affected in mips1 mips3. Interestingly, overexpression of PHOSPHATIDYLINOSITOL SYNTHASE2, which converts myo-inositol to membrane phosphatidylinositol (PtdIns), largely rescued the cotyledon and endomembrane defects in mips1 mips3. We conclude that myo-inositol serves as the main substrate for synthesizing PtdIns and phosphatidylinositides, which are essential for endomembrane structure and trafficking and thus for auxin-regulated embryogenesis.
INTRODUCTION
myo-inositol is the most important and abundant stereoisomer of the six-carbon cyclitol inositol. It exists in all eukaryotes and is also found in some prokaryotes (Eagle et al., 1957; Bachhawat and Mande, 2000). myo-inositol emerged early during evolution and evolved into diverse derivatives, such as phosphoinositides (PIs), inositol phosphates, glycosylphosphatidylinositol, and various inositol conjugates (Michell, 2008). Inositol and its derivatives are involved in various biochemical and physiological processes, including intracellular signal transduction (Irvine and Schell, 2001), membrane construction and trafficking (Cullen et al., 2001), membrane-related protein anchoring (Tiede et al., 1999; Peskan et al., 2000; Borner et al., 2005), and cell wall construction (Loewus 1973). They also act as protein cofactors (Macbeth et al., 2005; Tan et al., 2007) and play roles in auxin storage and trafficking in plant seeds (Hall and Bandurski, 1986).
In mammalian cells, myo-inositol and its derivatives play important roles in reproduction (Chiu et al., 2003; Papaleo et al., 2009), embryo development (Kane et al., 1992; Greene and Copp, 1997; Hynes et al., 2000), and neuron function (Hanley et al., 1988; Fisher et al., 2002). Disruption of inositol homeostasis is reported to be a factor in bipolar disorder (Shaltiel et al., 2004) and neurodegenerative diseases, including Alzheimer’s disease (Miller et al., 1993), Huntington’s disease, and Parkinson’s disease (Sarkar et al., 2005). Recently, inositol deficiency was shown to induce a neonatally lethal phenotype in mice (Buccafusca et al., 2008).
The cellular myo-inositol pool in mammalian cells is maintained by a combination of de novo biosynthesis, uptake from intracellular matrix, and recycling from PIs. d-MYO-INOSITOL-3-PHOSPHATE SYNTHASE (MIPS) has the most important role in de novo synthesis of inositol by catalyzing the rate-limiting step using glucose-6-phosphate as the substrate. MIPS is highly conserved among eukaryotes (Loewus and Murthy, 2000; Majumder et al., 2003). The first gene encoding MIPS to be identified was yeast INO1 (Culbertson et al., 1976; Donahue and Henry, 1981). Since then, many MIPS genes have been identified in various prokaryotic and eukaryotic cells. In plants, characterization of MIPSs from diverse plant species revealed their important roles in stress responses (Nelson et al., 1998; Ghosh Dastidar et al., 2006; Murphy et al., 2008) and in the regulation of phytate, the fully phosphorylated form of myo-inositol and the major phosphorus storage compound in seeds (Hegeman et al., 2001; Brinch-Pedersen et al., 2002; Stevenson-Paulik et al., 2005). There are three MIPS genes in Arabidopsis; the MIPS1 gene was the first to be functionally identified by complementation of the yeast ino1 mutant and to date is the best-characterized MIPS gene (Johnson and Sussex, 1995). Recently, a loss-of-function MIPS1 mutant, mips1, was reported to exhibit impaired pathogen resistance (Murphy et al., 2008), programmed cell death (PCD) in leaves (Meng et al., 2009; Donahue et al., 2010), and deformed cotyledon development (Chen and Xiong, 2010).
Once synthesized, myo-inositol acts as a substrate for the formation of various inositol-derived molecules, for instance, phosphatidylinositol (PtdIns) and indole-3-acetic acid (IAA)-myo-inositol conjugates. PtdIns contribute 21% of the phospholipids in nonphotosynthetic plant membranes (Harwood, 1980), and the various phosphorylated forms of PtdIns, known as PIs, have critical roles in cytoskeletal rearrangements, membrane trafficking, and organelle labeling (Simonsen et al., 2001; Cantley, 2002; Thole and Nielsen, 2008). In mammalian systems, Ins(1,4,5)P3, the hydrolyzed product of PtdIns(4,5)P2, plays important roles in intracellular signal transduction as a second messenger. To date, however, there is no direct proof that Ins(1,4,5)P3-mediated signaling exists in plants (Wheeler and Brownlee, 2008). The Arabidopsis thaliana PHOSPHATIDYLINOSITOL SYNTHASE1 (PIS1) gene (At1g68000) and PIS2 gene (At4g38570) are responsible for the net synthesis of PtdIns from myo-inositol and cytidine 5′-diphospho-1,2-diacyl-sn-glycerol (CDP-diacylglycerol) (Collin et al., 1999; Xue et al., 2000). As demonstrated in maize (Zea mays), myo-inositol can also form an IAA-myo-inositol conjugate (Normanly, 2010), the reaction of which involves two steps (Kesy and Bandurski, 1990). The first and rate-limiting step of this reaction, which is reversible, is specifically catalyzed by UDP-GLUCOSYL TRANSFERASE 84B1, encoded by UGT84B1 (At2g23260) in Arabidopsis (Jackson et al., 2001). For the second step in maize, IAA moiety was transacylated from IAA-glucose to myo-inositol with a large negative free energy change (Kesy and Bandurski, 1990). However, no corresponding gene responsible for the second step has yet been found in Arabidopsis. Therefore, it remains to be investigated whether IAA-myo-inositol conjugates are present in Arabidopsis.
Plant MIPS genes are highly expressed in developing seeds (Johnson and Wang, 1996; Yoshida et al., 1999; Hegeman et al., 2001; Abreu and Aragão, 2007; Mitsuhashi et al., 2008), suggesting that myo-inositol has an important role in seed/embryo development. In this study, we used genetic approaches to investigate the roles of three MIPS genes in Arabidopsis development, especially in embryo development. Of the three genes, MIPS1 showed the most comprehensive and long-lasting expression during seed development. For each of the MIPS genes, single null mutants showed no visible phenotypes, but the double mutants mips1 mips2+/− and mips1 mips3 displayed severe defects in embryogenesis, producing altered numbers of cotyledons (one to four) with deformed shapes. Furthermore, the homozygous mips1 mips2 and mips1 mips2 mips3 triple mutants were embryonic lethal, suggesting that de novo synthesis of myo-inositol is critical. Our data suggest that the developmental defects of mips mutants are caused by altered auxin transport and responses in the double and triple mutants. Interestingly, overexpression of PIS2 largely rescued the abnormal embryo phenotypes in the mips1 mips3 double mutant, restoring the shape and number of cotyledons, and also the endomembrane function, whereas overexpression of UGT84B1 enhanced the cotyledon phenotypes. The fact that PIS2 largely rescued the myo-inositol deficiency phenotype suggests that the defects in mips1 mips3 resulted from decreased levels of membrane PtdIns. We conclude that MIPS-mediated de novo synthesis of myo-inositol is essential for maintaining the normal function of endomembrane trafficking and for maintaining endomembrane structure, which is critical for the correct auxin transport and, thus, correct auxin localization during embryo pattern formation.
RESULTS
Arabidopsis MIPS Genes Rescued the Yeast Inositol Auxotrophy Mutant ino1
As a highly conserved enzyme, MIPS is found in all eukaryotes and a few prokaryotes. In the phylogenetic tree constructed with more than 100 eukaryotic MIPS protein sequences using PHYLIP (Felsenstein, 2005), MIPSs from plants form an independent subgroup (see Supplemental Figure 1 online), and multiple copies of the MIPS gene emerge only in land plant species. There are three putative MIPS homologous genes in the Arabidopsis genome. It appears that MIPS3 diverted from the ancestor of MIPS1 and MIPS2 at an early stage of evolution as the result of duplication and then a recent duplication event produced MIPS1 and MIPS2. We first investigated whether all three genes are functional. The yeast myo-inositol auxotrophy mutant ino1 cannot survive in the absence of myo-inositol because of a deletion in the INO1 gene (Nunez et al., 2008). We generated constructs containing each of the Arabidopsis MIPS genes driven by a constitutive GPD promoter and transformed them into the yeast mutant ino1, with empty vector–transformed ino1 as the negative control and yeast innate INO1-transformed ino1 as the positive control. The yeast INO1 gene rescued the ino1 mutant as expected (Figure 1). The three Arabidopsis MIPS genes also rescued the yeast ino1 mutant (Figure 1). This result showed that each of the three MIPS genes is a functional counterpart of yeast INO1 in Arabidopsis, furthermore showing that the inositol biosynthetic pathways are highly conserved between yeast and plants. Since the three Arabidopsis MIPSs shared ~90% identity at the amino acid level and were all able to rescue yeast ino1 mutant, the evolution of these MIPS genes possibly represents diversification of temporal and spatial regulation rather than changes in amino acid sequences or catalytic capacity.
Figure 1.
Complementation of the Yeast ino1 Mutant.
Expression of each of the three Arabidopsis MIPS complements the inositol auxotrophy phenotype of the yeast ino1 mutant. The yeast cells transformed with INO1 or empty vector were used as positive and negative controls, respectively. Plates were spotted with 10-fold serial dilutions and incubated at 30°C for 2 d.
The Three MIPS Genes Showed Different Expression Patterns and Levels during Embryogenesis
We first used quantitative RT-PCR to examine the transcript levels of each of the MIPS genes in different Arabidopsis tissues and organs. All of the MIPS genes were expressed in the tested tissues (Figure 2; see Supplemental Figure 2 online), consistent with a previous report (Donahue et al., 2010). The highest expression levels of MIPS1 were observed in siliques (Figure 2), which is consistent with results from microarray analyses (Zimmermann et al., 2004).
Figure 2.
Relative Expression Level of MIPS Genes in Different Arabidopsis Tissues.
MIPS1, MIPS1, and MIPS3 mRNA transcript levels in 8-DAG seedlings grown on half-strength Murashige and Skoog medium and in different organs of soil-grown Arabidopsis plants were quantified by real-time PCR. The expression levels for all genes were normalized to that of TUB2. Error bars were calculated as se for three independent experiments.
To analyze the expression patterns in more detail, we generated constructs with 2-kb promoter fragments of each of the MIPS genes driving the uidA reporter gene and introduced these constructs into Arabidopsis. The overall β-glucuronidase (GUS) staining patterns in siliques differed among the three MIPS genes (i.e., MIPS1 was specifically expressed in developing seeds, MIPS2 was highly expressed in the seedpods and seeds, and MIPS3 was expressed in the funiculi and vascular tissues of seedpods) (Figures 3A to 3C). MIPS1 expression was detectable throughout embryo development, from the early globular stage to the mature stage in both the embryo and the endosperm (Figures 3D to 3G), but not in the seed coat from the torpedo (Figure 3F) to the mature stages (Figure 3G). At the globular stage, the expression patterns of MIPS2 and MIPS3 were similar to that of MIPS1 (Figures 3H and 3L). However, unlike MIPS1, MIPS2 and MIPS3 were not expressed in embryos after the globular stage. We detected some MIPS2 signals in seed coats at the heart stage (Figure 3I) and in endosperm at the torpedo stage (Figure 3J). At the heart and torpedo stages, MIPS3 was expressed only in the funiculi (Figures 3M and 3N). MIPS2 and MIPS3 were not expressed in mature seeds (Figures 3K and 3O). The results suggest that the transcription of the three MIPS genes is coordinately regulated during seed development and that they might function differently in different tissues.
Figure 3.
Dynamic Expression of MIPS Genes in Seed Development.
(A) to (C) GUS staining pattern of siliques from MIPS1:GUS (A), MIPS2:GUS (B), and MIPS3:GUS (C) transgenic lines. These seeds contain early torpedo stage embryos. Bars = 1 mm.
(D) to (G) GUS staining of seeds from MIPS1:GUS transgenic lines in sequential developmental stages as shown at the bottom of the figure. Bars = 100 μm.
(H) to (K) GUS staining of seeds from pMIPS2:GUS transgenic lines in sequential developmental stages. Bars = 100 μm.
(L) to (O) GUS staining of seeds from pMIPS3:GUS transgenic lines in sequential developmental stages. Bars = 100 μm.
Single mips Mutants Showed No Obvious Phenotypes during Embryo and Leaf Development
To examine the biological roles of the three MIPS genes, we obtained the T-DNA insertion lines for MIPS1 (SALK_023626) and MIPS2 (SALK_031685) from the ABRC and identified a T-DNA insertion line of MIPS3 from the mutant collection generated in our own laboratory (Qin et al., 2005). The insertion sites of the three single mutants were all confirmed by sequencing (Figure 4A). RT-PCR analysis showed that no transcripts were detectable in the homozygous mips1, mips2, and mips3 mutants (Figure 4B). We observed no abnormal phenotypes, including the embryo development, for three homozygous single mutants under optimal growth conditions (120 μmol ·m−2 ·s−1, 16 h light/8 h dark) (Figure 4C). To further confirm that the single mutants of these three MIPS genes are normal, we analyzed more T-DNA insertion alleles of each MIPS gene (Figure 4A). No abnormal phenotypes were observed during embryogenesis under optimal growth conditions (Figure 4D). These results suggest that the three MIPS genes are probably functionally redundant.
Figure 4.
Expression and Phenotype Analysis of T-DNA Insertion Mutants for Each MIPS Gene.
(A) Gene structures of MIPS1, MIPS2, and MIPS3 with the location of T-DNA insertions. The left border of each insertion was confirmed by sequencing. Exons (dark-green boxes indicate coding regions; lighter boxes indicate 5′ [left] and 3′ [right] untranslated regions) and T-DNA insertion sites (purple arrows) are indicated. SALK lines written in black are those used in further analysis and crossing. SALK lines written in gray are used in phenotype confirmation. LB, T-DNA left border; RB, T-DNA right border. Gene organizations are depicted in proportion.
(B) Transcript levels of MIPS1, MIPS2, MIPS3 (left panel), and the internal control TUB2 (right panel) in wild-type (WT) and three mips T-DNA insertion mutants were determined using RT-PCR (32 cycles).
(C) The 20-DAG seedling (top row) and seed (bottom row) phenotypes of mips1 (left panels), mips2 (middle panels), and mips3 (right panels) single knockout mutants under the 16-h-light and 8-h-dark optimal growth condition. Bars = 1 cm.
(D) Normal cotyledon phenotype in different homozygous mips single mutants. T-DNA insertion lines for three MIPS genes were grown under the long-day light (120 μmol ·m−2 ·s−1) optimal condition. The T-DNA insertion sites and the genotypes were confirmed by PCR. Bars = 1mm.
Single mips Mutants Were Sensitive to High Light Intensity Stress
Although under the optimal growth condition no abnormal phenotype was observed for mips1 (SALK_023626) in this study and another previous study (Murphy et al., 2008), this mutant was recently reported to be abnormal either during embryogenesis (i.e., affected cotyledon development) (Chen and Xiong, 2010; Donahue et al., 2010) or during postembryogenesis (i.e., induced lesion-mimic phenotypes in its rosette leaves under long-day conditions) (Meng et al., 2009; Donahue et al., 2010). To help clarify the phenotype discrepancy of mips1 among different research groups, we grew the mips1 single mutant plants under long-day conditions (16 h light/8 h dark) with different light intensities and examined embryogenesis and leaf development. We found that, while plants grown under lower light intensity showed no obvious phenotypes, plants grown under high light intensity (i.e., 160 μmol ·m−2 ·s−1 and 220 μmol ·m−2 ·s−1) showed lesion-mimic phenotypes and that the higher light intensity applied the more severe lesion-mimic phenotypes developed (Figure 5A). To further investigate whether cotyledon development was affected, we examined cotyledon development in the seeds of mips1 plants grown under different light intensity conditions. The results showed that, while displaying normal embryo and postembryo development under the 60 μmol ·m−2 ·s−1 or 120 μmol ·m−2 ·s−1 light conditions, mips1 produced offspring with abnormal cotyledons under high light intensity conditions (i.e., 3.1% abnormal cotyledons under 160 μmol ·m−2 ·s−1 and 5.4% under 220 μmol ·m−2 ·s−1) (Figure 5B). The severity of mips1 phenotypes positively correlated to the light intensity (Figures 5A and 5B). These results suggest that loss of function of the major MIPS would lead to hypersensitivity of the plants to high light intensity stress. Together with the recent report that oxidative stress could greatly reduce the cellular myo-inositol level without affecting MIPS1 function (Chaouch and Noctor, 2010), it seems that the phenotype discrepancy is possibly due to the different growth conditions applied (e.g., light intensity) and that the observed phenotypes are possibly attributed to the coaction of light intensity and partial loss of MIPS function.
Figure 5.
The Phenotypes of mips1 under Different Light Intensity Conditions.
(A) Rosette leaf phenotypes of mips1 (SALK_023626) grown under different light intensity conditions in comparison with the wild type (WT). Yellow circles indicate lesion-mimic patches. Bars = 0.5 cm.
(B) Cotyledon phenotypes of the seeds from mips1 (SALK_023626) grown under different light intensity conditions. Each pie chart shows the percentage of the cotyledon phenotype proportion. Lime sectors represent the normal cotyledons and yellow sectors represent the abnormal cotyledons. Bars = 1 mm.
MIPS Mutant Combinations Produced Abnormal Cotyledons
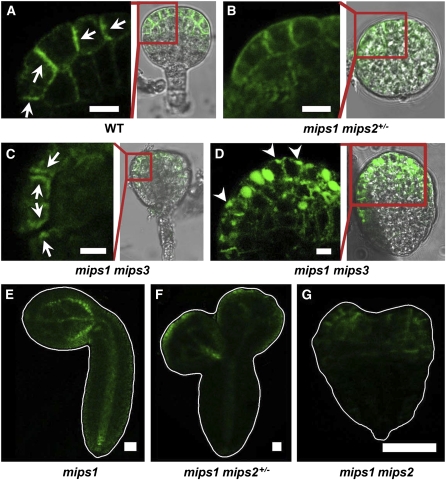
To elucidate the physiological function of MIPS genes, we generated higher orders of MIPS mutants. Compared with the wild type (Figure 6A), the mips1+/− mips2 mips3 triple mutant had no obvious phenotypes (see Supplemental Figure 3B online), possibly due to the higher expression level of MIPS1 in the mips2 mutant (Donahue et al., 2010), while mips1 mips2+/− displayed lesions in rosette and cauline leaves (see Supplemental Figure 3C online) under optimal growth condition (120 μmol ·m−2· s−1, 16 h light/8 h dark). The mips1 mips3 double mutant displays severe postembryonic phenotypes, including anthocyanidin accumulation in rosette leaves and much smaller adult plants compared with the wild type (see Supplemental Figure 3D online). In plants, anthocyanidin was reported to accumulate under stress conditions and to play important roles against UV irradiation and oxidative damage (Winkel-Shirley, 2002). The fact that the mips1 mips3 mutants showed anthocyanidin accumulation under optimal growth conditions suggests that the depletion of myo-inositol causes constitutive stress response in Arabidopsis. Trypan blue staining results indicated programmed cell death in mips1 mips2+/− leaf lesions (see Supplemental Figure 3G online), while leaves from the wild type and mips1 mips3 double mutant showed negative staining results (see Supplemental Figure 3H online).
Figure 6.
Phenotypic Analysis of mips Double Mutant Cotyledons.
(A) to (H) Cotyledon phenotypes of wild-type (WT; [A]), mips1 mips2+/− ([B] to [D]), mips1 mips3 ([E] to [G]), and the complementation line of mips1 mips3 with genomic MIPS1 gene driven by its own promoter (H). Bars = 1 mm.
(I) to (K) The vascular patterns of cotyledons of mips1 mips2+/− (I) and mips1 mips3 ([J] and [K]). Red asterisks in (I) indicate the paralleled veins in cotyledon petioles, and the asterisk in (J) indicates irregular vein cluster. The arrow in (I) indicates the open end of a vascular bundle and in (J) indicates the isolated vascular segment. Cotyledon fusion site of mips1 mips3 seedlings is indicated by arrow in (K). P, petiole. Bars = 500 μm.
(L) to (N) Pavement cell and stomatal guard cell morphology of the wild-type (L), mips1 mips2+/− (M), and mips1 mips3 (N) double mutant cotyledons. Extremely large guard cells (arrows) and small guard cells (red arrowheads) were found in mips double mutant cotyledons. Bars = 50 μm.
(O) Relative cell ploidy ratio of the wild-type and mips1 mips3 cotyledons. More than 10,000 nuclei were counted for each sample.
(P) myo-inositol content in mature seeds of wild-type and mips double mutants. Asterisks indicate P value <0.001. Error bars were calculated as se for three independent experiments.
It is interesting to notice that both mips1 mips2+/− (Figures 6B to 6D) and mips1 mips3 (Figures 6E to 6G) double mutants had cotyledons that were fused at the basal end. In addition, the number of cotyledons in mips1 mips3 was also altered, ranging from one to four, and approximately half of the two-cotyledon seedlings lacked bilateral symmetry (Table 1, Figures 6E to 6G). These cotyledon phenotypes of mips1 mips3 were complemented by reintroduction of a MIPS1 gene (Figure 6H). Symmetrical and asymmetrical cotyledons of both mips1 mips2+/− and mips1 mips3 showed distorted vascular systems, such as parallel veins (Figure 6I), clustered bundles of irregular veins (Figure 6J), and discontinuous or poorly axialized venation patterns (Figure 6K). Scanning electron microscopy analysis revealed that, unlike the wild type (Figure 6L), abnormal cotyledons of both mips1 mips2+/− and mips1 mips3 showed uneven cotyledon surfaces with unequal pavement cell and stomata cell sizes (Figures 6M and 6N). To investigate whether the abnormal size of the cotyledon epidermal cells was caused by endoreduplication (Melaragno et al., 1993), we conducted flow cytometry analysis on the cotyledon cells from mips1 mips3 seedlings at 10 d after germination (DAG). The results showed that both 2C and 32C cells were more abundant in the mips1 mips3 mutants than in the wild type (Figure 6O), suggesting that cell cycle regulation is affected in mips1 mips3 cotyledon cells.
Table 1.
Frequencies of Cotyledon Phenotypes from Seeds of a Single Plant of the Genotypes Listed
| Cotyledon Nos. and Frequency |
|||||||
| Two |
|||||||
| Parental Genotype | One | Symmetry | Asymmetry | Three | Four | Ungerminated | Total No. of Seedlings |
| mips1 mips3 (1) | 8 | 103 | 123 | 48 | 5 | 0 | 287 |
| mips1 mips3 (2) | 6 2.8% | 79 36.2% | 97 43.7% | 30 15.5% | 4 1.8% | 0 | 216 |
| +/− (1) | 7 | 17 | 23 | 9 | 1 | 9 | 66 |
| +/minus (2) | 14 11.7% | 40 31.7% | 30 29.4% | 12 1% | 0 0.6% | 18 15% | 114 |
The aborted seeds in mips1 mips2+/− were excluded before plating on the half-strength Murashige and Skoog medium and were not counted as ungerminated. All the seedlings that were counted as mips1 mips2+/− were genotyped. Numbers in parentheses represent the number of independent lines of each genotype.
We used gas chromatography–time-of-flight–mass spectrometry (MS) to determine whether the myo-inositol content is indeed reduced in the mature seeds of the double mutants, since MIPS genes were highly expressed in the developing seeds (Figure 3). The result showed that, while the myo-inositol level in wild-type seeds was 509 ± 11 μg/g tissue, the myo-inositol contents in mips1 mips3, mips1 mips2+/−, and mips2 tmips3 double mutants were 120 ± 6 μg/g tissue, 127 ± 12 μg/g tissue, and 413 ± 16 μg/g tissue, respectively (Figure 6P). These results indicate that the myo-inositol level was indeed reduced in the higher-order MIPS mutants and that MIPS1 is the most important member responsible for the myo-inositol synthesis in Arabidopsis seeds.
MIPS Genes Are Required for Embryo Development
Because the initiation of cotyledons begins at the late globular stage of embryo development (Chandler, 2008) and many mutants defective in embryo development also displayed abnormal cotyledons (Berleth and Jürgens, 1993; Bennet et al., 1995; Aida et al., 1997; Friml et al., 2003), we investigated embryo development in the double mutants. Whereas normal embryo development was observed in the wild type (Figures 7A to 7D), disorganized embryo basal shapes were observed in globular stage embryos of mips1 mip2+/− (Figure 7E) and tmips1 mips3 (Figure 7I). From the heart stage on, single and triple cotyledon primordia were visible in the double mutant misp1 mips2+/− (Figures 7F and 7G). The mips1 mips3 double mutant showed asymmetrical cotyledon primordia (Figure 7J) and abnormal bulges in the hypocotyls (Figure 7K). Both of these double mutants produced abnormal mature embryos (Figures 7H and 7L).
Figure 7.
Embryo Development of the Wild Type and mips Double and Triple Mutants.
(A) to (L) Nomarski images of cleared Arabidopsis seeds from siliques of the wild type (WT; [A] to [D]), mips1 mips2+/− ([E] to [H]), and mips1 mips3 ([I] to [L]) at the four developmental stages noted at the top of the figure. Arrows in (E) indicate the abnormal cell division orientation, and red arrowheads in (F) indicate three cotyledon primordia. The arrow pair in (K) indicates the abnormal bulges in hypocotyl. C, cotyledon primordium; H, hypocotyl; Er, embryonic root. Bars = 20 μm.
(M) to (O) Dissected siliques of the wild type (M), mips1 mips2+/− (N), and mips1+/− mips2 mips3 (O). Red asterisks indicate the flat white seeds that are distinguishable from the adjacent round green seeds.
(P) to (S) Nomarski images of cleared Arabidopsis seeds from mips1+/− mips2 mips3 siliques. In one silique, although most of the embryos reached the heart stage (Q), about a quarter of the embryos remained at the globular stage (P). When most of the embryos reached the torpedo stage (S), about a quarter of the embryos from the same silique remained at the globular stage (R). Bars = 20 μm.
We failed to obtain any mips1 mips2 homozygous double mutants. Normal green round seeds developed in the wild-type siliques (Figure 7M), whereas in the siliques of mips1 mips2+/−, we observed both green round seeds and white wrinkled seeds, with a ratio of ~3:1 (525:191, χ2 = 0.99; P > 0.05; Figure 7N), suggesting that the mips1 mips2 homozygous mutant is embryo lethal. Like mips1 mips2+/−, approximately one-quarter of the seeds in siliques of the mips1+/− mips2 mips3 triple mutant were white; the green-to-white ratio was 672:197 (χ2 = 2.39; P > 0.05; Figure 7O). These white embryos were at the late globular shape to heart shape stages. After the globular stage, the development of mips1 mips2 mips3 mutant embryos was mostly arrested (Figures 7P and 7R), while mips2 mips3 and mips1+/− mips2 mips3 embryos from the same silique developed beyond the globular stage (Figures 7Q and 7S).
In summary, if mips1 was heterozygous in the double mutant, then no embryo defective phenotype was observed. In the mips1 homozygous background, plants heterozygous for mips2 or, by contrast, homozygous for mip3, produced severe embryo defective phenotypes, whereas the homozygous mips1 mips2 double mutant was embryo lethal (Table 2). Our data demonstrate that, among the three MIPS genes, MIPS1 has the most important role during Arabidopsis embryo development.
Table 2.
The Phenotypes of mips Single, Double, and Triple Mutants under Optimal Growth Conditions
| Genotype | Embryo Phenotype | Postembryo Phenotype |
| mips1 | No | No |
| mips2 | No | No |
| mips3 | No | No |
| mips2 mips3 | No | No |
| mips1 mips3 | Deformed embryo | Dwarf and small; anthocyanidin accumulation |
| mips1+/− mips2 | No | 1/4 Bearing seeds aborted |
| mips1 mips2+/− | Deformed embryo; ungerminated seeds | Leaf PCD; 1/4 bearing seeds aborted |
| mips1 mips2 | Embryo lethal | NA |
| mips1+/− mips2 mips3 | No | 1/4 Bearing seeds aborted |
| mips1 mips2+/− mips3 | Deformed embryo | Fragile; small; anthocyanidin accumulation |
| mips1 mips2 mips3 | Embryo lethal | NA |
Seedling phenotypes of some double and triple mips mutants are shown in Supplemental Figure 3 online. NA, not applicable.
Defective Embryo Development in mips Mutants Is Associated with Impaired Distribution and Polar Transport of Auxin
Symmetrical outgrowth of cotyledon primordia depends on appropriate apical auxin maxima. To investigate whether auxin distribution was affected in mips mutants, we crossed the mips double and triple mutants with the auxin reporter DR5:green fluorescent protein (GFP) and examined the GFP signals in the embryos. In the embryo of wild-type Arabidopsis, auxin accumulated in the uppermost suspensor cells (hypophysis) at the globular stage (Figure 8A) and in cotyledon primordial tips and prevascular cells at the heart and torpedo stages in addition to the hypophysis (Figures 8B and 8C), consistent with previous reports (Tanaka et al., 2006; Chandler, 2008; Nawy et al., 2008). In the embryos from mips1 mips2+/− siliques, in addition to the signal from the uppermost hypophysis, we observed an extra signal from suspensor cells at the late globular stage (Figure 8D). At the heart stage, asymmetrical auxin maxima were observed in cotyledon primordia, although the embryo was morphologically symmetrical (Figure 8E). In most of the mips1 mips2+/− mature embryos, an apical expression pattern of DR5:GFP signal was observed (Figure 8F). In mips1 mips3 embryos, a very weak DR5:GFP signal was observed in the hypophysis (Figure 8G). Meanwhile, auxin distribution was severely altered (e.g., globular-shaped mips1 mips3 embryos at the heart or torpedo stages showed two auxin level peaks) (Figure 8H), and a bulged embryo proper showed dispersed GFP signals (Figure 8I). In all the embryos of mips1 mips2+/− and mips1 mips3, we detected GFP signals at the root pole (Figures 8F and 8I). Interestingly, we observed distorted distribution of the GFP signals in the mips1 mips2 mips3 embryos dissected from mature siliques of mips1+/− mips2 mips3. This result suggested that the cells are alive, even though the embryo development had ceased and that the distribution of auxin was greatly distorted (Figures 8J to 8L). The fact that the aberrant DR5:GFP signals were observed before the asymmetrical morphology emerged in the embryos suggests that auxin distortion is probably responsible for abnormal embryo development.
Figure 8.
Auxin Maxima Distribution in the Wild-Type, mips Double, and Triple Mutant Embryos.
(A) to (C) DR5:GFP distribution in the wild-type (WT) embryos at the globular stage (A), the late heart stage (B), and the mature stage (C).
(D) to (F) DR5:GFP distribution in embryos dissected from mips1 mips2+/− siliques at the globular stage (D), the late heart stage (E), and the mature stage (F). Arrow in (D) indicates the extra GFP signal in suspensor cells.
(G) to (I) DR5:GFP distribution in mips1 mips3 embryos at the globular stage (G), the late heart stage (H), and the torpedo stage (I). The dashed line frame in (G) indicates the hypophysis cells with decreased DR5:GFP signals.
(J) to (L) DR5:GFP distribution in embryos dissected from the flat white seeds of mips1+/− mips2 mips3 mature silique ([J] and [K]). Red arrowhead in (L) indicates the abnormal bulge in shoot apical meristem.
Bars = 25 μm in (A) to (L).
PIN1 Localization Was Altered in mips Mutant Embryos
The correct establishment and maintenance of auxin maxima is important for normal bilateral axis formation in Arabidopsis embryos. These processes rely on the polar localization of the auxin efflux carrier, PIN1. Incorrect localization of PIN1 can result from cytoskeleton disruption and changes in the sterol component of the plasma membrane (Kleine-Vehn and Friml, 2008). To investigate whether PIN1 localization was altered in mutant embryos, we crossed mips1 mips2+/− and mips1 mips3 double mutants with a PIN1:GFP marker line. At the globular stage, PIN1:GFP was expressed in the epidermal cells in mips1 mips2+/− and mips1 mips3 embryos, similar to the expression in the wild type. However, whereas PIN1 showed polar plasma membrane localization in the wild-type embryo (Figure 9A), localization of PIN1:GFP was less polar in the mips1 mips2+/− embryos (Figure 9B) and localized at two opposite membranes (Figure 9C) or outward membranes of epidermal cells (Figure 9D) in mips1 mips3 embryos. Within the same silique of mips1 mips2+/−, we found later-stage embryos of mips1, mips1 mips2+/−, and mips1 mips2 in which PIN1 localization differed (Figures 9E to 9G). These data suggest that PIN1 polar localization was altered in the mutant embryos at the globular stage. Because at this stage most of the double mutant embryos showed normal morphology with no apical DR5:GFP signals, it is likely that the altered PIN1 localization at the globular stage is responsible for distorted apical auxin distribution at later developmental stages.
Figure 9.
PIN1 Localization in the Wild-Type and mips Double Mutant Embryos.
(A) PIN1:GFP signals in a globular stage embryo of the wild type (WT). Arrows show the PIN1:GFP polar localization, which indicates the direction of auxin flux toward the differentiating cotyledon primordium.
(B) A globular shape embryo dissected from a silique of mips1 mips2+/− showing evenly distributed PIN1:GFP signals on plasma membrane.
(C) and (D) PIN1:GFP signals in globular-stage embryo of mips1 mips3. Arrows in (C) indicate the disorganized PIN1 localization on lateral plasma membranes. Arrowheads in (D) indicate PIN proteins facing outwards. In (A) to (D), the left panels show the fluorescence image of the area in the red rectangle in the right Nomarski image. Bars = 5 μm in (A) to (D).
(E) to (G) Three phenotypically distinct embryos dissected from the same silique of mips1 mips2+/− showing different PIN1:GFP signals. The predicted genotypes are amips1 (E), mips1 mips2+/− (F), and mips1 mips2 (G). Bars = 25 μm.
Embryo-Defective Phenotypes of mips Double Mutants Are Rescued by PIS Overexpression but Enhanced by UGT84B1 Overexpression
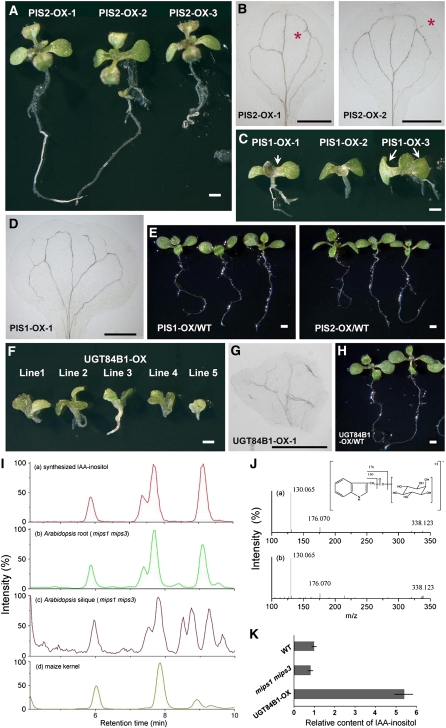
After synthesis, myo-inositol, together with CDP-diacylglycerol, can be converted to PtdIns in a reaction catalyzed by PIS1 and PIS2 (Collin et al., 1999). To clarify the primary defects caused by myo-inositol deficiency, we cloned PIS1 and PIS2, generated overexpression constructs driven by their native promoters plus four cauliflower mosaic virus 35S enhancers, and transformed these constructs into both the mips1 mips3 double mutant and the wild type. In the PIS2 overexpressor lines in the mips1 mips3 background, plants with two normal symmetric cotyledons accounted for >65% of the population, compared with 36% in mips1 mips3, indicating that PIS2 overexpression had largely rescued the cotyledon symmetry phenotypes in the mips1 mips3 double mutants (Figures 10A and 10B, Table 3). Moreover, PIS1 overexpression partially rescued the cotyledon phenotype of mips1 mips3 double mutants but not the short-root phenotype (Figures 10C and 10D, Table 3). By contrast, overexpression of PIS2 or PIS1 in wild-type plants did not affect the cotyledons (Figure 10E). Moreover, recent studies showed that the levels of PtdIns species were reduced in mips1 single mutants (Chen and Xiong, 2010; Donahue et al., 2010). These results suggest that the defects in mips mutants were caused by decreased levels of PtdIns and that myo-inositol synthesized by MIPS is essential for synthesis of PtdIns.
Figure 10.
Phenotypic Analysis and Biochemical Characterization of PIS2-, PIS1-, and UGT84B1-Overexpressor Lines in the mips1 mips3 Background.
(A) to (H) The 12-DAG PIS2-OX (A), PIS1-OX (C), and UGT84B1-OX (F) transgenic lines in the mips1 mips3 background. The T1 generation lines shown above were selected by growing on half-strength Murashige and Skoog agar plate containing 50 μg/mL hygromycin. PIS2-OX lines in (A) show regularly symmetry two cotyledons. Arrow (left) in (C) indicates the broader cotyledon petiole; arrow pair (right) in (C) indicates the two asymmetric cotyledons. Cotyledon venations of PIS2-OX (B), PIS1-OX (D), and UGT84B1-OX (G) transgenic lines. Red asterisks in (B) indicate the extra vascular structures. Wild-type Arabidopsis overexpressing PIS2 (PIS2-OX) or PIS1 (PIS1-OX) (E) or UGT84B1 (UGT84B1-OX) (H) are also shown. Bars = 1 mm.
(I) Liquid chromatography–MS chromatograms of IAA-myo-inositol quantification assay using synthesized IAA-myo-inositol (a), mips1 mips3 root (b), mips1 mips3 silique (c), and maize kernel (d). Four peaks in chromatogram (a) show four isomers of IAA-myo-inositol.
(J) MS2 spectra of the first isomer (retention time, 5.9 min) in synthesized IAA-myo-inositol (a) and wild-type Arabidopsis root (b). The fraction of m/z 130 and m/z 176, as well as the [M+H]+ m/z 338.123, confirmed the IAA-myo-inositol structure of the first isomer.
(K) Relative content of IAA-myo-inositol in Arabidopsis root tissue of different genotypes. Duplicate experiments were performed for each genotype. Error bars show se. WT, wild type.
Table 3.
Frequencies of Cotyledon Phenotypes from Seeds of Single Plants
| Cotyledon Nos. and Frequency |
||||||
| Two |
||||||
| Parental Genotype | One | Symmetry | Asymmetry | Three | Four | Total No. of Seedlings |
| PIS2-OX-1 | 2 2.1% | 83 87.4% | 7 7.4% | 3 3.2% | 0 | 95 |
| PIS2-OX-2 | 5 5.4% | 61 65.6% | 18 19.8% | 9 9.7% | 0 | 93 |
| PIS2-OX-3 | 1 1.1% | 68 71.6% | 19 20.0% | 7 7.4% | 0 | 95 |
| PIS1-OX-1 | 4 4.3% | 42 44.7% | 36 37.5% | 12 12.5% | 0 | 94 |
| PIS1-OX-2 | 2 2.3% | 55 61.8% | 22 24.7% | 10 11.2% | 0 | 89 |
| PIS1-OX-3 | 3 3.7% | 38 46.9% | 31 38.3% | 9 11.1% | 0 | 81 |
A single transgenic plant was used for statistical analysis of each transgenic line.
In maize, myo-inositol can also be conjugated with IAA to form myo-inositol–conjugated IAA molecules. We then generated UGT84B1 overexpression constructs driven by their native promoters plus four cauliflower mosaic virus 35S enhancers and transformed these constructs into both the mips1 mips3 double mutant and the wild-type Arabidopsis plants. In contrast with PIS overexpressor lines, most of the UGT84B1 overexpressor lines in the mips1 mips3 mutant background exhibited stronger cotyledon phenotypes, and some developed no roots (Figures 10F and 10G), whereas overexpression of UGT84B1 in wild-type plants did not affect the cotyledons (Figure 10H). Since there is no report of the existence of IAA-myo-inositol in Arabidopsis up to now, we first adopted HPLC-MS to clarify this question. The result showed that four IAA-myo-inositol isomers existed both in Arabidopsis roots and siliques (the data of mips1 mips3 are shown as an example), while two isomers existed in maize kernels, by comparing the chromatogram and spectra with those of chemically synthesized IAA-myo-inositol (Figures 10I and 10J). Then, we performed the relative quantification analysis on the roots from germinating seeds of the wild type, mips 1 mips3 double mutants, and UGT84B1 overexpressor lines using HPLC-MS. The result showed that overexpression of UGT84B1 in the mips1 mips3 background resulted in a significant increase in IAA-myo-inositol level, whereas the relative level of the IAA-myo-inositol content in mips1 mips3 double mutant was slightly lower than the wild type (Figure 10K). It is possible that the more severe cotyledon phenotypes in the UGT84B1 overexpressor lines in the mips1 mips3 background are attributed to the further reduction of the free myo-inositol in the mutant, which suggests the essential role of free myo-inositol in cotyledon development. Another possibility is that increase of the IAA-myo-inositol level also further reduced the level of free active IAA, the distribution and transport of which had already been severely distorted in the mutant (Figures 8 and 9), which further confirmed the role of auxin distortion as the major factor that causes the abnormal cotyledon phenotypes in the mips mutants.
Endomembrane Function in Embryo Cells Was Impaired in the mips1 mips3 Double Mutant
We first used the endomembrane trafficking inhibitor brefeldin A to analyze PIN1:GFP trafficking in both the wild type and mips1 mips3 double mutant. However, the PIN1 proteins abnormally aggregated in mips1 mips3 embryo cells even without brefeldin A treatment (see Supplemental Figure 4 online), suggesting that the PIN1 trafficking in the double mutant embryo cells was affected, possibly due to the impaired PIN1 exocytosis. To investigate whether the endomembrane function is affected, we then used the fluorescent styryl dyes (e.g., FM4-64 and FM1-43) to either mark the plasma membrane or monitor the clathrin-dependent endocytosis process (Bolte et al., 2004) in mips double mutant embryo cells at early stages. After treatment, the dye first stains the plasma membrane, then reaches the endosomal network through endocytic internalization, and finally labels the surface of vacuole (Ueda et al., 2001). We used the dye AM1-43, the fixable form of FM1-43 with similar emission and absorption spectra, for the assay. The results showed that, in the embryo cells at the globular stage, after 5-min treatment, AM1-43 dye was extensively endocytosed and was detectable in the vacuole membrane in the wild type (Figure 11A) but not in mips1 mips3 mutants (Figure 11D). Most of the globular embryo cells of mips1 mips3 remained in the plasma membrane (Figure 11D). In the embryo cells at the early heart stage, 5-min treatment produced dye-stained vesicles in the wild type (Figure 11B), while mutant cells had much less stained vesicles (Figures 11B and 11E). The fact that the dye uptake rate is slower in the mutant suggests that the endocytosis is affected in the mutant embryo cells. After 10-min treatment, the dye was endocytosed completely into the cells and trafficked to the vesicle membrane both in the wild type and the mutant (Figures 11C and 11F). Interestingly, some mips1 mips3 embryo cells showed a massive intake of the fluorescent dye (Figure 11F), implying that the integrity of the plasma membrane is also impaired in the mutant embryo cells (Schapire et al., 2008).
Figure 11.
Endomembrane of the Wild-Type and mips1 mips3 Embryo Cells.
(A) to (F) Arabidopsis wild-type (WT; [A] to [C]) and mips1 mips3 double mutant ([D] to [F]) embryo cells treated with the endocytosis marker AM1-43. More than 20 siliques were analyzed for each treatment. Bars = 10 μm.
(G) Endomembrane system of a wild-type embryo cell at the torpedo stage. Arrowhead indicates a double membrane bound vesicle; arrow indicates a budding vesicle. ER, endoplasmic reticulum. Bar = 200 nm.
(I) Endomembrane system of an mips1 mips3 embryo cell at the torpedo stage. MT, mitochondria. Bar = 200 nm.
(K) Endomembrane system of a torpedo embryo cell from PIS2-OX–transformed mips1 mips3 plant; arrow indicates a trafficking vesicle. Bar = 200 nm.
(H), (J), and (L) Magnification of rectangle regions in (G), (I), and (K), respectively. Arrows in (H) and (L) indicate the single membrane bound trafficking vesicles, which were not found in (J).
To investigate whether the endomembrane structure was affected in the mips1 mips3 double mutant, we carefully analyzed the endomembrane structures in torpedo stage embryos of the wild type and mips1 mips3 double mutant by transmission electron microscopy. Compared with those of the wild type (Figure 11G), the vesicle budding structures were much less prominent in mips1 mips3 (Figure 11I). Single-layer vesicles were commonly found in wild-type embryo cells (Figure 11H) but were rarely observed in mips1 mips3 (Figure 11J; see Supplemental Figure 5 online). These data, together with the AM1-43 staining results, suggest that both the endomembrane trafficking process and the endomembrane structure were damaged in mips1 mips3. This hypothesis is further supported by the complementation result that these vesicles were restored in the mips1 mips3 mutant embryos when PIS2 was overexpressed (Figures 11K and 11L; see Supplemental Figure 5 online). Since PIN1 polar localization depends on polar trafficking of recycled endosomes, which is initiated by endocytosis from the plasma membrane (Kleine-Vehn and Friml, 2008), the defective polar localization of PIN1 in the mips embryos could therefore be attributed to the defects in membrane dysfunction caused by the lack of myo-inositol.
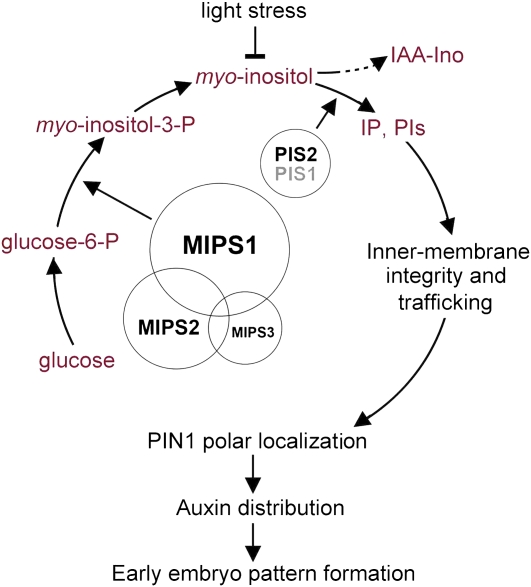
In conclusion, our study showed that MIPS-catalyzed de novo synthesis of myo-inositol is essential for auxin-mediated embryo development in Arabidopsis. The myo-inositol serves as the main substrate for synthesis of PtdIns, the membrane component essential for keeping normal function of endomembrane trafficking and maintenance of endomembrane structure, which is critical for the correct auxin transport and, thus, correct auxin localization during embryo pattern formation (Figure 12).
Figure 12.
A Proposed Model for the Biological Roles of the Three MIPS Genes in Arabidopsis Embryo Pattern Formation.
The myo-inositol produced by sequential reactions, in which MIPS enzymes catalyze the rate-limiting step, can be either conjugated to IAA or converted to membrane component PtdIns. Sufficient PtdIns is important for endomembrane structure and trafficking and, thus, for auxin transport and localization to regulate proper embryo development.
DISCUSSION
In this study, we demonstrated distinct dynamic expression patterns of three MIPS genes that are essential for normal Arabidopsis embryo development. Moreover, we provided evidence for the direct involvement of PtdIns, a derivative of myo-inositol, in auxin-regulated embryo pattern formation.
Although previous studies on other plant species revealed high levels of expression of MIPS genes during seed development (Johnson and Wang, 1996; Yoshida et al., 1999; Hegeman et al., 2001; Abreu and Aragão, 2007), the exact function of MIPS in this process remains elusive, possibly because of the multiple copies of MIPS genes in plants. For instance, in Arabidopsis, the detailed expression pattern of each of the MIPS proteins was difficult to distinguish in developing seeds because of the high amino acid sequence identity among the three MIPSs. In this study, we found that MIPS1 was expressed at all stages during embryo development. By contrast, expression of MIPS2 and MIPS3 was detected in the seed coat and funiculus of maternal tissues after the globular stage (Figure 3) but decreased to undetectable levels in the embryo and endosperm. The overlapping expression of the three MIPS genes at the globular stage, but not at later stages, suggests an essential role of myo-inositol synthesis at early stages of embryo development. As embryo development proceeds, it is likely that MIPS1 alone synthesizes sufficient myo-inositol for the later embryo stages. The fact that a complete knockout of MIPS1 showed no phenotype during the entire process of embryo development suggests that the amount of myo-inositol synthesized by both MIPS2 and MIPS3 at the globular stage is sufficient for normal embryo development. Expression of MIPS2 and MIPS3 in the endosperm and maternal tissues after the globular stage provides another route by which the myo-inositol synthesized in the endosperm and maternal tissues can be transported to the embryo. The double mutants mips1 mips2+/− and mips1 mips3 displayed impaired embryo symmetry after the late globular stage, whereas the embryo development of mips2 mips3 was normal. This result indicates that MIPS1 plays the most important role in synthesizing myo-inositol for embryo development, consistent with the high expression levels of MIPS1 at all stages of embryo development. It appears that MIPS2 has a more important role in embryo development than MIPS3 because MIPS2 had a broader expression pattern than that of MIPS3 (Figure 3), and the mips1 mips2 double mutant is embryo lethal. In addition to the production of phytate in seed phosphorus and cation storage (Brinch-Pedersen et al., 2002; Stevenson-Paulik et al., 2005), here, we used genetic approaches to demonstrate that the coordinated expression of the three MIPS genes is also essential for embryo pattern formation in Arabidopsis.
myo-inositol is transformed into various derivatives that have roles in numerous biological pathways. In mammalian systems, myo-inositol–derived molecules have two major roles: one as membrane components (i.e., PtdIns and PIs) and the other in signal transduction (i.e., inositol phosphates). However, the Ins(1,4,5)P3 intracellular signal transduction pathway appears to be absent from plants, since no Ins(1,4,5)P3 receptor has been found in land plants (Wheeler and Brownlee, 2008). On the other hand, myo-inositol can be conjugated with IAA to form myo-inositol–conjugated IAA molecules in Arabidopsis (Figure 10). The DR5:GFP (Figure 8) and PIN1:GFP (Figure 9) signals were severely altered in the embryos of mips1 mips2+/− and mips1 mips3, indicating that there is a connection between myo-inositol and auxin maxima/transport. Once produced, myo-inositol is transformed into different derivatives with roles in various biochemical processes and pathways. Therefore, the phenotypes caused by myo-inositol deficiency could result from many affected pathways. In this study, we used genetic and microscopy approaches to clarify which pathway(s) is most affected and to determine how myo-inositol production is associated with auxin during embryo development. Interestingly, we found that overexpression of PIS2, driven by its own promoter plus four 35S enhancers, largely rescued the cotyledon phenotype of mips1 mips3 (Figure 10). Recently it was reported that overexpression of PIS2 in Arabidopsis increased levels of PtdIns(4)P and PtdIns(4,5)P2, which are the major PI species in plasma membranes, the endoplasmic reticulum, the Golgi complex, and trafficking vesicles (Löfke et al., 2008). Therefore, these results suggest that, under myo-inositol insufficient conditions, there are limited amounts of the membrane components PtdIns and PIs, possibly PtdIns(4)P and PtdIns(4,5)P2. Defects in membrane function resulting from the limited amounts of membrane components might be the major reason for the embryo defects in mips1 mips3. Overexpression of PIS2 would transform the limited amount of myo-inositol into PtdIns, thereby maintaining the membrane function and consequently largely rescuing the cotyledon phenotype. As a major component of nonphotosynthetic membranes, PtdIns and their phosphorylated forms, PIs, are involved in intracellular vesicular transport and function as the organelle identity markers. Each distinct organelle membrane has its unique PI composition (Simonsen et al., 2001; Krauß and Haucke, 2007). Our conclusion that endomembrane function was affected in mips mutants is supported by the transmission electron microscopy observation that the structures of inner membrane systems were altered, and there were significantly fewer membrane-derived trafficking vesicles in cells of mips1 mips3, which was largely rescued by the overexpression of PIS2 (Figure 11). The lack of vesicle budding and trafficking could also explain the depolarization and mistransport of PIN:GFP in mips double mutant embryos.
Although Arabidopsis PIS1 and PIS2 share 87% identity at the amino acid level, overexpression of PIS1 only partially rescued the cotyledon phenotype of mips1 mips3, while overexpression of PIS2 more completely rescued it. One explanation could be that PIS2 is specifically expressed in developing seeds but PIS1 is not, according to the microarray data (Zimmermann et al., 2004). Another possibility is that PIS1 and PIS2 have different biochemical properties because overexpression of PIS1 did not affect PI content (Löfke et al., 2008).
Recently, the single knockout mutant mips1 was reported to display leaf PCD and seed phenotype (Meng et al., 2009; Donahue et al., 2010). However, we did not observe any differences between the wild type and the mips single mutant (SALK_023626) under our experimental conditions (120 μmol ·m−2 ·s−1, 16 h light/8 h dark). Our result is consistent with a previous report in which no developmental defects were observed in the mips1 single mutant (Murphy et al., 2008). However, we observed stable occurrence of PCD patches in the mips1 mips2+/− leaves under the same (120 μmol ·m−2 ·s−1, 16 h light/8 h dark) growth condition (see Supplemental Figure 3 online). Furthermore, our light intensity experiment results showed that both rosette leaf and embryo development of mips1 were sensitive to high light conditions, suggesting that the leaf PCD phenotype is attributed to the coaction of high light intensity stress and partial lack of the major MIPS activity. This hypothesis is further supported by a recent report that stress-induced reactive oxygen species would greatly reduce the cellular myo-inositol content without affecting the expression of MIPS1 gene. Notably, the embryo phenotypes of mips double and triple mutants in our study were inheritably stable and were less affected by environmental factors, making them good materials for further genetic and cellular analysis.
In summary, we demonstrate that myo-inositol is the main substrate for synthesis of PtdIns, which are essential to maintain endomembrane function during embryo development. De novo synthesis of myo-inositol is required for the correct transport and localization of auxin during embryo pattern formation in Arabidopsis. Our findings suggest that there is a link between the lipid components and the dynamic function of endomembrane system.
METHODS
Yeast Complementation Assay
The yeast ino1 strain SJY425 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 ino1Δ::HIS3MX6) was kindly provided by Susan A. Henry (Nunez et al., 2008). Arabidopsis thaliana MIPS1, MIPS2, and MIPS3 and yeast INO1 genes were cloned into the yeast expression vector pAG425GPD-ccdB (Saccharomyces cerevisiae Advanced Gateway Destination vector from Addgene) (Alberti et al., 2007), and recombinants were transformed into ino1 with leu2 as the selection marker. The yeast synthetic complete medium contained the following ingredients per 1 liter double-distilled water:20 g glucose, 5 g ammonium sulfate, 1 g potassium phosphate, 0.5 g magnesium sulfate, 0.1 g sodium chloride, 0.1 g calcium chloride, 0.5 mg boric acid, 0.04 mg cupric sulfate, 0.1 mg potassium iodide, 0.2 mg ferric chloride, 0.4 mg manganese sulfate, 0.2 mg sodium molybdate, 0.4 mg zinc sulfate, 2 μg biotin, 400 μg calcium pantothenate, 2 μg folic acid, 400 μg niacin, 200 μg p-aminobenzoic acid, 400 μg pyridoxine hydrochloride, 200 μg riboflavin, 400 μg thiamine, 20 mg adenine sulfate, 20 mg Arg, 20 mg His, 60 mg Leu, 230 mg Lys, 20 mg Met, 300 mg Thr, 20 mg Trp, and 40 mg uracil. In this experiment, Leu was omitted from the yeast synthetic complete medium. When transformed yeast cell suspensions reached 0.5 OD600/mL, they were serially diluted (1:10) and spotted onto plates either with or without 75 μM myo-inositol. Plates were incubated at 30°C for 2 d before analysis.
Expression Analysis
Total RNA was isolated from wild-type seedlings grown on solid half-strength Murashige and Skoog medium under continuous light conditions at 8 DAG and from tissues of 30-DAG wild-type plants grown under 16-h-light/8-h-dark conditions using TRIzol reagent (Invitrogen) followed by RNase-free DNase (TaKaRa) treatment. For each sample, 500 ng RNA was reverse transcribed into cDNA using Superscript II reverse transcriptase (Invitrogen). Two microliters of diluted cDNA samples (1:10) were mixed with 10 μL SYBR Green Real-Time PCR Master Mix (TOYOBO) and 100 nM gene-specific primers that were designed from sequences close to the 3′ end (see Supplemental Table 1 online), and then quantitatively analyzed by an Opticon continuous fluorescence detector (MJ Research) (Liu et al., 2008). Three biological replications were performed for each template and primer combination, and TUB2 expression level was used as the internal control. We used the cycle threshold (Ct) 2−ΔΔCt method (Livak and Schmittgen, 2001) to calculate the relative expression level of target genes according to the expression level of TUB2.
cDNA samples were synthesized from RNA extracted from mips1, mips2, mips3, and wild-type Arabidopsis rosette leaves using the methods described above. Gene-specific primers for the MIPS gene and the TUB2 gene (see Supplemental Table 1 online) were used for RT-PCR.
Plant Materials and Growth Conditions
The mips1 (SALK_023626) and mips2 (SALK_031689) Arabidopsis mutants were obtained from the ABRC, and the left borders of insertion T-DNA were confirmed by sequencing of amplified PCR products. The MIPS3 knockout mutant (mips3) with PPT resistance was identified from a pool of Arabidopsis (Columbia ecotype) T-DNA insertion lines (Qin et al., 2005), and the insertion site was detected by thermal asymmetric interlaced PCR and confirmed by DNA sequencing of the T-DNA inserts, segregation analysis, and RT-PCR. All the other T-DNA insertion lines used in this study were requested from the ABRC, and the homozygous lines and the T-DNA insertion sites were confirmed by PCR and sequencing. After sterilization and plating on solid half-strength Murashige and Skoog medium, seeds were stratified for 3 d at 4°C and then transferred to a continuously lit growth chamber at 22°C for germination. The seedlings were transplanted into potting soil 10 d later and were grown under long-day conditions (16 h light/8 h dark) with a light intensity of 120 μmol ·m−2 ·s−1 at 22°C and 60% relative humidity, unless stated otherwise. Double and triple mutants were obtained by crosses between the three single mutants mentioned above, and genotypes were determined by PCR (for primers, see Supplemental Table 1 online). For cotyledon phenotype analysis, we used seedlings at 8 DAG that had been grown under continuous light conditions. pPIN1:PIN1-GFP and pDR5rev:GFP lines (Friml et al., 2003) were crossed with the mips1+/− mips2 mips3 mutant.
Phylogenetic Analysis
A total of 101 full-length amino acid sequences of MIPS proteins from eukaryotes (see Supplemental Data Set 1 online) were obtained from the National Center for Biotechnology Information by databank searching using the BLAST program (Altschul et al., 1990). Sequence alignment was performed using the ClustalW program (Thompson et al., 1994). The resultant PHYLIP file (see Supplemental Data Set 1 online) was analyzed by phylogenetic software PHYLIP 3.68 (Felsenstein, 2005). Yeast INO1 sequence was used as the outgroup for making rooted phylogenetic tree. A total of 1000 bootstrapped sequence sets were produced using SEQBOOT, and the tree file was processed by PROTPARS and CONSENSE programs. Visualization of the tree was performed using TreeView program (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html).
Scanning Electron Microscopy
Cotyledons from 8-DAG mips1 mips2+/−, mips1 mips3, and wild-type seedlings were collected and fixed in FAA (50% ethanol, 6% glacial acetic acid, and 5% formaldehyde) for 4 h at 25°C, followed by serial ethanol dehydration and isoamyl acetate substitution. Cotyledons were critical-point dried in liquid CO2 and examined under a scanning electron microscope (Hitachi 4700) as described previously (Zhang et al., 2007).
Flow Cytometry
Cotyledons (1 to ~2 cm2 each line) of 8-DAG mips1 mips3 and wild-type seedlings that had been grown on solid half-strength Murashige and Skoog medium were chopped in 50 μL chopping buffer (Galbraith et al., 1983) for 30 s, and then the suspension was filtered through a 50-μm nylon mesh and completed to a final volume of 500 mL with chopping buffer. The mixture was incubated with 10 μL 10 μg/mL RNase at 37°C for 20 min before addition of 2 μL 5 mg/mL propidium iodide and incubation in the dark for 20 min at 4°C. The resuspended nuclei were analyzed using a FACSCalibur flow cytometer (Becton Dickinson) with laser excitation at 488 nm. The DNA contents of nuclei were calculated by Cellquest (Becton Dickinson).
Whole-Mount Clearing and Fluorescence Detection of Embryos
For whole-mount preparations, embryos at different stages were mounted in a chloral hydrate/glycerol/water (8:1:3) mixture and cleared for 5 to 30 min, according to specific embryo developmental stage, before microscopy analysis. The mutant embryos containing pPIN1:PIN1-GFP or pDR5rev:GFP were dissected from seeds at specific developmental stages and mounted in a 6% glucose solution. Observation of GFP signals was performed using a Leica TCS SPE confocal with DM4000B microscope (Leica). The excitation wavelength of samples was 488 nm and emission was detected between 520 and 580 nm. For AM1-43 staining, Arabidopsis embryos at a specific stage were carefully dissected from developing seeds and incubated on ice with 5 μM of AM1-43 (Biotium) prepared in a 6% glucose solution, washed with 6% glucose solution, and mounted in the same solution for visualization. Embryos were then analyzed under a Leica TCS SPE confocal microscope.
Gas Chromatography Analysis
Three biological replications were performed for each genotype. For each experiment, 20 mg seeds were frozen and ground in liquid nitrogen, and 1 mL 80% methanol was added with 100 mM of ribitol as an internal standard. The mixture was incubated in 60°C for 1 h and centrifuged at 10,000g and 4°C for 15 min. The soluble part was removed to a new tube, centrifuged again, and dried at 45°C under a clean nitrogen stream. Freshly prepared derivatization reagent [250 μL; 1:1 mixture of pyridine and N,O-bis(trimethylsilyl)trifluoroacetamide + 1% trimethylchlorosilane] was added to each sample and vortexed for 20 min, and the mixture was incubated in 80°C for 15 min. Each sample was added with 250 μL hexane before injection. Gas chromatography–mass spectrometry analysis was performed by Agilent 6890 GC/5975B MSD (Agilent Technologies). Samples were loading to an autosampler and injected with a split of 4.8 mL min−1. Separation was performed by helium gas stream with a controlled flow pressure of 8.81 p.s.i. and a linear velocity of 1 mL min−1. The injection port was set as described (Torabinejad et al., 2009). The mass spectrometer was equipped with a DB-225MS capillary column (30 m × 0.25 mm i.d.; J&W Scientific). Quantification of myo-inositol was calculated by the standard curve of myo-inositol and the recovery of internal standard ribitol. All the chemicals used in this experiment were purchased from Sigma-Aldrich.
Histochemical Staining
Supplemental Table 1 online shows primers used in vector construction for GUS assays. Promoters for MIPS1, MIPS2, and MIPS3 were amplified from Arabidopsis genomic DNA and cloned into vector pCAMBIA1391Xa (CAMBIA), pCAMBIA1391Xb (CAMBIA), and the gateway vector pBGWFS7 (VIB-Ghent University), respectively. Siliques were dissected from both sides and fixed in 80% acetone for 20 min. Samples were washed with 0.1 M sodium phosphate buffer, pH 7.0, and immersed in GUS staining solution (0.5 mg/mL 5-bromo-4-chloro-3-indolyl glucuronide, 10 mM Na2EDTA, 0.5 mM potassium ferricyanide/ferrocyanide, and 0.06% Triton X-100 in 0.1 M Na2HPO4, pH 7.0) (Guo et al., 2009). The staining solution was vacuum infiltrated into samples for 10 min, and samples were then incubated at 37°C for 4 h. Siliques were cleared in 70% ethanol before microscopy. Trypan blue staining of the wild type and mips double mutants was performed as described by Koch and Slusarenko (1990).
Transmission Electron Microscopy
Developing seeds at specific stages were collected and fixed in 2% glutaraldehyde and 2% formaldehyde in 0.1 M sodium phosphate buffer, pH 6.8, for 3 h at room temperature and then for 24 h at 4°C. Seeds were then washed three times with 0.1 M sodium phosphate buffer and postfixed in 1% OSO4 for 1 h, followed by dehydration in a graded acetone series. Seeds were embedded in a complete resin mixture (Spi-chem Spurr) and incubated in 70°C for 9 h. Samples were sectioned using a Leica EM UC6 ultramicrotome and stained with uranyl acetate and lead stain solutions. Sections were examined with a JEOL electron microscope.
HPLC-MS Analysis
The synthesis of IAA-myo-inositol was performed by reaction of imidazolated IAA and sodium myo-inositolate in DMSO as decribed before (Nowaki et al., 1978). The reaction mixture was stored at −20°C until use. The synthesized IAA-myo-inositol was diluted 1000-fold using 1% aqueous formic acid before HPLC-MS sample loading.
Roots of Arabidopsis germinating seeds were dissected and pooled. For a single experiment, 10 mg roots were grounded in liquid nitrogen and extracted with 500 μL prechilled methanol for 1 h (120 rpm, 4°C). After centrifugation, the residue was reextracted for 20 min. The supernatant was combined and passed through a 0.45-μm filter. The filtrate was evaporated in vacuo and resolved in 15 μL 1% aqueous formic acid. For Arabidopsis siliques and maize (Zea mays) kernels, 50 mg tissues were grounded in liquid nitrogen and treated following the same procedure described above. One injection of 10 μL was analyzed by Agilent HPLC 1200 coupled with a ZORBAX Eclipse Plus C18 column (150 mm × 2.1 mm; 5 μm; Agilent) that was used for the separation. The column thermostat was set at 30°C. The mobile phase was consisted of A, methanol, and B, 0.02% aqueous formic acid, at flow rate of 0.25 mL/min. The initial mobile phase was 20% A and 80% B. A linear gradient to 30% A and 70% B in 10 min was performed. The MS conditions were as follows: gas temperature, 350°C; gas flow, 12 L/min; nebulizer, 35 p.s.i.; capilliary voltage, 2000 V; fragmentor, 125 V. The IAA-myo-inositol contents were calculated by the peak area of extracted ion current of [M+H]+(338.123). Every isomer of IAA-myo-inositol from roots extract was confirmed by comparing MS2 spectra with those of synthesized IAA-myo-inositol.
Accession Numbers
Sequence data can be found in the GenBank/EMBL database under the following accession numbers: MPS1 (NM_120143; At4g39800), MIPS2 (NM_127790; At2g22240), MIPS3 (NM_121055; At5g10170), PIS1 (NM_105470; At1g68000), PIS2 (NM_120018; At4g38570), and UGT84B1 (NM_127890; At2g23260), respectively. Accession numbers used for the phylogenetic analysis in Supplemental Figure 1 online can be found in Supplemental Table 2 online. T-DNA insertion lines used for mutant analysis were as follows: mips1 (SALK_023626), mips2 (SALK_031685), and mips3 (N.070710 from the T-DNA insertion mutant collection generated in our laboratory). Other T-DNA insertion lines that were used for phenotype confirmation (Figure 4A) were SALK_023813, CS851587, SALK_108779C, SALK_120131, and SALK_068584.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Phylogenetic Tree of Eukaryotic MIPS Proteins.
Supplemental Figure 2. Expression Patterns of the Three MIPS Genes in Different Arabidopsis Tissues.
Supplemental Figure 3. Phenotypes of mips Double and Triple Mutants and Their Trypan Blue Staining Assays.
Supplemental Figure 4. PIN1 Localization in Globular Stage Embryos of Wild-Type and mips1 mips3 Double Mutant upon BFA Treatment.
Supplemental Figure 5. Numbers of Trafficking Vesicles in Wild-Type, mips1 mips3 Double Mutant, and PIS2-OX Embryo Cells.
Supplemental Table 1. Primers Used in This Study.
Supplemental Table 2. Accession Numbers for the Sequences Used in the Alignment in Supplemental Figure 1 and Supplemental Data Set 1.
Supplemental Data Set 1. Text File of the Sequences and Alignment Used for the Phylogenetic Analysis Shown in Supplemental Figure 1.
Acknowledgments
We thank Yunde Zhao (University of California at San Diego), Ruixi Li and Hongwei Guo (Peking University), and Xing-Wang Deng (Yale University) for helpful suggestions and valuable discussions. We also thank Zhiqiang Ma and Miss Wenyuan Wang (Peking University) for technical assistance. The work was supported by the National Natural Science Foundation of China (Grants 90717003 and 30625002 to L.-J.Q. and 90717002 to M.Z.) and the National Basic Research Program of China (Grant 2009CB941503) and partially supported by the 111 Project.
References
- Abreu E.F.M., Aragão F.J.L. (2007). Isolation and characterization of a myo-inositol-1-phosphate synthase gene from yellow passion fruit (Passiflora edulis f. flavicarpa) expressed during seed development and environmental stress. Ann. Bot. (Lond.) 99: 285–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida M., Ishida T., Fukaki H., Fujisawa H., Tasaka M. (1997). Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell 9: 841–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti S., Gitler A.D., Lindquist S. (2007). A suite of Gateway cloning vectors for high-throughput genetic analysis in Saccharomyces cerevisiae. Yeast 24: 913–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Bachhawat N., Mande S.C. (2000). Complex evolution of the inositol-1-phosphate synthase gene among archaea and eubacteria. Trends Genet. 16: 111–113 [DOI] [PubMed] [Google Scholar]
- Bennet S.R.M., Alvarez J., Bossinger G., Smyth D. (1995). Morphogenesis in pinoid mutants of Arabidopsis thaliana. Plant J. 8: 505–520 [Google Scholar]
- Berleth T., Jürgens G. (1993). The role of the monopteros gene in organising the basal body region of the Arabidopsis embryo. Development 118: 575–587 [Google Scholar]
- Bolte S., Talbot C., Boutte Y., Catrice O., Read N.D., Satiat-Jeunemaitre B. (2004). FM-dyes as experimental probes for dissecting vesicle trafficking in living plant cells. J. Microsc. 214: 159–173 [DOI] [PubMed] [Google Scholar]
- Borner G.H.H., Sherrier D.J., Weimar T., Michaelson L.V., Hawkins N.D., Macaskill A., Napier J.A., Beale M.H., Lilley K.S., Dupree P. (2005). Analysis of detergent-resistant membranes in Arabidopsis. Evidence for plasma membrane lipid rafts. Plant Physiol. 137: 104–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinch-Pedersen H., Sørensen L.D., Holm P.B. (2002). Engineering crop plants: Getting a handle on phosphate. Trends Plant Sci. 7: 118–125 [DOI] [PubMed] [Google Scholar]
- Buccafusca R., Venditti C.P., Kenyon L.C., Johanson R.A., Van Bockstaele E., Ren J., Pagliardini S., Minarcik J., Golden J.A., Coady M.J., Greer J.J., Berry G.T. (2008). Characterization of the null murine sodium/myo-inositol cotransporter 1 (Smit1 or Slc5a3) phenotype: myo-inositol rescue is independent of expression of its cognate mitochondrial ribosomal protein subunit 6 (Mrps6) gene and of phosphatidylinositol levels in neonatal brain. Mol. Genet. Metab. 95: 81–95 [DOI] [PubMed] [Google Scholar]
- Cantley L.C. (2002). The phosphoinositide 3-kinase pathway. Science 296: 1655–1657 [DOI] [PubMed] [Google Scholar]
- Chandler J.W. (2008). Cotyledon organogenesis. J. Exp. Bot. 59: 2917–2931 [DOI] [PubMed] [Google Scholar]
- Chaouch S., Noctor G. (2010). Myo-inositol abolishes salicylic acid-dependent cell death and pathogen defence responses triggered by peroxisomal hydrogen peroxide. New Phytol. 188: 711–718 [DOI] [PubMed] [Google Scholar]
- Chen H., Xiong L. (2010). myo-Inositol-1-phosphate synthase is required for polar auxin transport and organ development. J. Biol. Chem. 285: 24238–24247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu T.T.Y., Rogers M.S., Briton-Jones C., Haines C. (2003). Effects of myo-inositol on the in-vitro maturation and subsequent development of mouse oocytes. Hum. Reprod. 18: 408–416 [DOI] [PubMed] [Google Scholar]
- Collin S., Justin A.-M., Cantrel C., Arondel V., Kader J.-C. (1999). Identification of AtPIS, a phosphatidylinositol synthase from Arabidopsis. Eur. J. Biochem. 262: 652–658 [DOI] [PubMed] [Google Scholar]
- Culbertson M.R., Donahue T.F., Henry S.A. (1976). Control of inositol biosynthesis in Saccharomyces cerevisiae: Properties of a repressible enzyme system in extracts of wild-type (Ino+) cells. J. Bacteriol. 126: 232–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen P.J., Cozier G.E., Banting G., Mellor H. (2001). Modular phosphoinositide-binding domains–Their role in signalling and membrane trafficking. Curr. Biol. 11: 882–893 [DOI] [PubMed] [Google Scholar]
- Donahue J.L., Alford S.R., Torabinejad J., Kerwin R.E., Nourbakhsh A., Ray W.K., Hernick M., Huang X., Lyons B.M., Hein P.P., Gillaspy G.E. (2010). The Arabidopsis thaliana Myo-inositol 1-phosphate synthase1 gene is required for Myo-inositol synthesis and suppression of cell death. Plant Cell 22: 888–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue T.F., Henry S.A. (1981). myo-Inositol-1-phosphate synthase. Characteristics of the enzyme and identification of its structural gene in yeast. J. Biol. Chem. 256: 7077–7085 [PubMed] [Google Scholar]
- Eagle H., Oyama V.I., Levy M., Freeman A.E. (1957). Myo-Inositol as an essential growth factor for normal and malignant human cells in tissue culture. J. Biol. Chem. 226: 191–205 [PubMed] [Google Scholar]
- Felsenstein J. (2005). PHYLIP (Phylogeny Inference Package) Version 3.6. (Seattle, WA: University of Washington; ). [Google Scholar]
- Fisher S.K., Novak J.E., Agranoff B.W. (2002). Inositol and higher inositol phosphates in neural tissues: Homeostasis, metabolism and functional significance. J. Neurochem. 82: 736–754 [DOI] [PubMed] [Google Scholar]
- Friml J., Vieten A., Sauer M., Weijers D., Schwarz H., Hamann T., Offringa R., Jürgens G. (2003). Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426: 147–153 [DOI] [PubMed] [Google Scholar]
- Galbraith D.W., Harkins K.R., Maddox J.M., Ayres N.M., Sharma D.P., Firoozabady E. (1983). Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 220: 1049–1051 [DOI] [PubMed] [Google Scholar]
- Ghosh Dastidar K., Maitra S., Goswami L., Roy D., Das K.P., Majumder A.L. (2006). An insight into the molecular basis of salt tolerance of L-myo-inositol 1-P synthase (PcINO1) from Porteresia coarctata (Roxb.) Tateoka, a halophytic wild rice. Plant Physiol. 140: 1279–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene N.D.E., Copp A.J. (1997). Inositol prevents folate-resistant neural tube defects in the mouse. Nat. Med. 3: 60–66 [DOI] [PubMed] [Google Scholar]
- Guo Y., Qin G., Gu H., Qu L.-J. (2009). Dof5.6/HCA2, a Dof transcription factor gene, regulates interfascicular cambium formation and vascular tissue development in Arabidopsis. Plant Cell 21: 3518–3534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall P.J., Bandurski R.S. (1986). [3H]Indole-3-acetyl-myo-inositol hydrolysis by extracts of Zea mays L. vegetative tissue. Plant Physiol. 80: 374–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley M.R., Jackson T.R., Vallejo M., Patterson S.I., Thastrup O., Lightman S., Rogers J., Henderson G., Pini A. (1988). Neural function: Metabolism and actions of inositol metabolites in mammalian brain. Philos. Trans. R. Soc. Lond. B Biol. Sci. 320: 381–398 [DOI] [PubMed] [Google Scholar]
- Harwood J.L. (1980). Plant acyl lipids: Structure, distribution and analysis. The Biochemistry of Plants, Stumpf P.K., (New York: Academic Press; ), pp. 1–55 [Google Scholar]
- Hegeman C.E., Good L.L., Grabau E.A. (2001). Expression of D-myo-inositol-3-phosphate synthase in soybean. Implications for phytic acid biosynthesis. Plant Physiol. 125: 1941–1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes A.C., Sreenan J.M., Kane M.T. (2000). Uptake and incorporation of myo-inositol by bovine preimplantation embryos from two-cell to early blastocyst stages. Mol. Reprod. Dev. 55: 265–269 [DOI] [PubMed] [Google Scholar]
- Irvine R.F., Schell M.J. (2001). Back in the water: The return of the inositol phosphates. Nat. Rev. Mol. Cell Biol. 2: 327–338 [DOI] [PubMed] [Google Scholar]
- Jackson R.G., Lim E.-K., Li Y., Kowalczyk M., Sandberg G., Hoggett J., Ashford D.A., Bowles D.J. (2001). Identification and biochemical characterization of an Arabidopsis indole-3-acetic acid glucosyltransferase. J. Biol. Chem. 276: 4350–4356 [DOI] [PubMed] [Google Scholar]
- Johnson M.D., Sussex I.M. (1995). 1 L-myo-inositol 1-phosphate synthase from Arabidopsis thaliana. Plant Physiol. 107: 613–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M.D., Wang X. (1996). Differentially expressed forms of 1-L-myo-inositol-1-phosphate synthase (EC 5.5.1.4) in Phaseolus vulgaris. J. Biol. Chem. 271: 17215–17218 [DOI] [PubMed] [Google Scholar]
- Kane M.T., Norris M., Harrison R.A.P. (1992). Uptake and incorporation of inositol by preimplantation mouse embryos. J. Reprod. Fertil. 96: 617–625 [DOI] [PubMed] [Google Scholar]
- Kesy J.M., Bandurski R.S. (1990). Partial purification and characterization of indol-3-ylacetylglucose:myo-inositol indol-3-ylacetyltransferase (indoleacetic acid-inositol synthase). Plant Physiol. 94: 1598–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J., Friml J. (2008). Polar targeting and endocytic recycling in auxin-dependent plant development. Annu. Rev. Cell Dev. Biol. 24: 447–473 [DOI] [PubMed] [Google Scholar]
- Koch E., Slusarenko A. (1990). Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell 2: 437–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauß M., Haucke V. (2007). Phosphoinositide-metabolizing enzymes at the interface between membrane traffic and cell signalling. EMBO Rep. 8: 241–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., et al. (2008). Targeted degradation of the cyclin-dependent kinase inhibitor ICK4/KRP6 by RING-type E3 ligases is essential for mitotic cell cycle progression during Arabidopsis gametogenesis. Plant Cell 20: 1538–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Loewus F.A.ed (1973). Biogenesis of Plant Cell Wall Polysaccharides. (New York: Academic Press; ). [Google Scholar]
- Loewus F.A., Murthy P.P.N. (2000). myo-Inositol metabolism in plants. Plant Sci. 150: 1–19 [Google Scholar]
- Löfke C., Ischebeck T., König S., Freitag S., Heilmann I. (2008). Alternative metabolic fates of phosphatidylinositol produced by phosphatidylinositol synthase isoforms in Arabidopsis thaliana. Biochem. J. 413: 115–124 [DOI] [PubMed] [Google Scholar]
- Macbeth M.R., Schubert H.L., Vandemark A.P., Lingam A.T., Hill C.P., Bass B.L. (2005). Inositol hexakisphosphate is bound in the ADAR2 core and required for RNA editing. Science 309: 1534–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder A.L., Chatterjee A., Ghosh Dastidar K., Majee M. (2003). Diversification and evolution of L-myo-inositol 1-phosphate synthase. FEBS Lett. 553: 3–10 [DOI] [PubMed] [Google Scholar]
- Melaragno J.E., Mehrotra B., Coleman A.W. (1993). Relationship between endopolyploidy and cell size in epidermal tissue of Arabidopsis. Plant Cell 5: 1661–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng P.H., Raynaud C., Tcherkez G., Blanchet S., Massoud K., Domenichini S., Henry Y., Soubigou-Taconnat L., Lelarge-Trouverie C., Saindrenan P., Renou J.P., Bergounioux C. (2009). Crosstalks between myo-inositol metabolism, programmed cell death and basal immunity in Arabidopsis. PLoS ONE 4: e7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michell R.H. (2008). Inositol derivatives: Evolution and functions. Nat. Rev. Mol. Cell Biol. 9: 151–161 [DOI] [PubMed] [Google Scholar]
- Miller B.L., Moats R.A., Shonk T., Ernst T., Woolley S., Ross B.D. (1993). Alzheimer disease: Depiction of increased cerebral myo-inositol with proton MR spectroscopy. Radiology 187: 433–437 [DOI] [PubMed] [Google Scholar]
- Mitsuhashi N., Kondo M., Nakaune S., Ohnishi M., Hayashi M., Hara-Nishimura I., Richardson A., Fukaki H., Nishimura M., Mimura T. (2008). Localization of myo-inositol-1-phosphate synthase to the endosperm in developing seeds of Arabidopsis. J. Exp. Bot. 59: 3069–3076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy A.M., Otto B., Brearley C.A., Carr J.P., Hanke D.E. (2008). A role for inositol hexakisphosphate in the maintenance of basal resistance to plant pathogens. Plant J. 56: 638–652 [DOI] [PubMed] [Google Scholar]
- Nawy T., Lukowitz W., Bayer M. (2008). Talk global, act local-patterning the Arabidopsis embryo. Curr. Opin. Plant Biol. 11: 28–33 [DOI] [PubMed] [Google Scholar]
- Nelson D.E., Rammesmayer G., Bohnert H.J. (1998). Regulation of cell-specific inositol metabolism and transport in plant salinity tolerance. Plant Cell 10: 753–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normanly J. (2010). Approaching cellular and molecular resolution of auxin biosynthesis and metabolism. Cold Spring Harb. Perspect. Biol. 2: a001594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowaki J., Cohen J.D., Bandurski R.S. (1978). Synthesis of 14C-indole-3-acetic-myo-inositol. J. Labelled Compd. Rad. 15: 325–329 [Google Scholar]
- Nunez L.R., Jesch S.A., Gaspar M.L., Almaguer C., Villa-Garcia M., Ruiz-Noriega M., Patton-Vogt J., Henry S.A. (2008). Cell wall integrity MAPK pathway is essential for lipid homeostasis. J. Biol. Chem. 283: 34204–34217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaleo E., Unfer V., Baillargeon J.P., Chiu T.T. (2009). Contribution of myo-inositol to reproduction. Eur. J. Obstet. Gynecol. Reprod. Biol. 147: 120–123 [DOI] [PubMed] [Google Scholar]
- Peskan T., Westermann M., Oelmüller R. (2000). Identification of low-density Triton X-100-insoluble plasma membrane microdomains in higher plants. Eur. J. Biochem. 267: 6989–6995 [DOI] [PubMed] [Google Scholar]
- Qin G., Gu H., Zhao Y., Ma Z., Shi G., Yang Y., Pichersky E., Chen H., Liu M., Chen Z., Qu L.-J. (2005). An indole-3-acetic acid carboxyl methyltransferase regulates Arabidopsis leaf development. Plant Cell 17: 2693–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S., Floto R.A., Berger Z., Imarisio S., Cordenier A., Pasco M., Cook L.J., Rubinsztein D.C. (2005). Lithium induces autophagy by inhibiting inositol monophosphatase. J. Cell Biol. 170: 1101–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schapire A.L., Voigt B., Jasik J., Rosado A., Lopez-Cobollo R., Menzel D., Salinas J., Mancuso S., Valpuesta V., Baluska F., Botella M.A. (2008). Arabidopsis synaptotagmin 1 is required for the maintenance of plasma membrane integrity and cell viability. Plant Cell 20: 3374–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaltiel G., Shamir A., Shapiro J., Ding D., Dalton E., Bialer M., Harwood A.J., Belmaker R.H., Greenberg M.L., Agam G. (2004). Valproate decreases inositol biosynthesis. Biol. Psychiatry 56: 868–874 [DOI] [PubMed] [Google Scholar]
- Simonsen A., Wurmser A.E., Emr S.D., Stenmark H. (2001). The role of phosphoinositides in membrane transport. Curr. Opin. Cell Biol. 13: 485–492 [DOI] [PubMed] [Google Scholar]
- Stevenson-Paulik J., Bastidas R.J., Chiou S.T., Frye R.A., York J.D. (2005). Generation of phytate-free seeds in Arabidopsis through disruption of inositol polyphosphate kinases. Proc. Natl. Acad. Sci. USA 102: 12612–12617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X., Calderon-Villalobos L.I.A., Sharon M., Zheng C., Robinson C.V., Estelle M., Zheng N. (2007). Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446: 640–645 [DOI] [PubMed] [Google Scholar]
- Tanaka H., Dhonukshe P., Brewer P.B., Friml J. (2006). Spatiotemporal asymmetric auxin distribution: A means to coordinate plant development. Cell. Mol. Life Sci. 63: 2738–2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thole J.M., Nielsen E. (2008). Phosphoinositides in plants: Novel functions in membrane trafficking. Curr. Opin. Plant Biol. 11: 620–631 [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiede A., Bastisch I., Schubert J., Orlean P., Schmidt R.E. (1999). Biosynthesis of glycosylphosphatidylinositols in mammals and unicellular microbes. Biol. Chem. 380: 503–523 [DOI] [PubMed] [Google Scholar]
- Torabinejad J., Donahue J.L., Gunesekera B.N., Allen-Daniels M.J., Gillaspy G.E. (2009). VTC4 is a bifunctional enzyme that affects myoinositol and ascorbate biosynthesis in plants. Plant Physiol. 150: 951–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda T., Yamaguchi M., Uchimiya H., Nakano A. (2001). Ara6, a plant-unique novel type Rab GTPase, functions in the endocytic pathway of Arabidopsis thaliana. EMBO J. 20: 4730–4741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler G.L., Brownlee C. (2008). Ca2+ signalling in plants and green algae—Changing channels. Trends Plant Sci. 13: 506–514 [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley B. (2002). Biosynthesis of flavonoids and effects of stress. Curr. Opin. Plant Biol. 5: 218–223 [DOI] [PubMed] [Google Scholar]
- Xue H.-W., Hosaka K., Plesch G., Mueller-Roeber B. (2000). Cloning of Arabidopsis thaliana phosphatidylinositol synthase and functional expression in the yeast pis mutant. Plant Mol. Biol. 42: 757–764 [DOI] [PubMed] [Google Scholar]
- Yoshida K.T., Wada T., Koyama H., Mizobuchi-Fukuoka R., Naito S. (1999). Temporal and spatial patterns of accumulation of the transcript of Myo-inositol-1-phosphate synthase and phytin-containing particles during seed development in rice. Plant Physiol. 119: 65–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Chen Y., Wang Z.-Y., Chen Z., Gu H., Qu L.-J. (2007). Constitutive expression of CIR1 (RVE2) affects several circadian-regulated processes and seed germination in Arabidopsis. Plant J. 51: 512–525 [DOI] [PubMed] [Google Scholar]
- Zimmermann P., Hirsch-Hoffmann M., Hennig L., Gruissem W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]