RNA-seq assessment of the transcriptome of autotrophic and heterotrophic Chlamydomonas as a function of copper nutrition reveals changes in redox metabolism regulated by CRR1, an SBP domain transcription factor. The changes in RNA abundance impact the abundance of specific plastid-localized proteins and the level of saturation of plastid galactolipids.
Abstract
In this work, we query the Chlamydomonas reinhardtii copper regulon at a whole-genome level. Our RNA-Seq data simulation and analysis pipeline validated a 2-fold cutoff and 10 RPKM (reads per kilobase of mappable length per million mapped reads) (~1 mRNA per cell) to reveal 63 CRR1 targets plus another 86 copper-responsive genes. Proteomic and immunoblot analyses captured 25% of the corresponding proteins, whose abundance was also dependent on copper nutrition, validating transcriptional regulation as a major control mechanism for copper signaling in Chlamydomonas. The impact of copper deficiency on the expression of several O2-dependent enzymes included steps in lipid modification pathways. Quantitative lipid profiles indicated increased polyunsaturation of fatty acids on thylakoid membrane digalactosyldiglycerides, indicating a global impact of copper deficiency on the photosynthetic apparatus. Discovery of a putative plastid copper chaperone and a membrane protease in the thylakoid suggest a mechanism for blocking copper utilization in the chloroplast. We also found an example of copper sparing in the N assimilation pathway: the replacement of copper amine oxidase by a flavin-dependent backup enzyme. Forty percent of the targets are previously uncharacterized proteins, indicating considerable potential for new discovery in the biology of copper.
INTRODUCTION
Copper is an essential nutrient for all forms of aerobic life because of its role as a redox cofactor, especially in reactions involving oxygen chemistry, such as electron transfer during respiration and photosynthesis, neutralization of superoxide, and oxygenation of organic molecules. Copper deficiency occurs in organisms because of genetic disease, as in Menkes syndrome in humans, reduced content in the food supply, as in North Ronaldsay sheep in the Orkney islands or grazing sheep and cattle in sections of Australia, or reduced bioavailability, as in micro-aerobic or anaerobic aquatic environments where cyanobacteria and algae might bloom (Danks, 1988; Schaefer and Gitlin, 1999; Haywood et al., 2001; Mercer, 2001; Pierre et al., 2002). As is the case for any nutrient deficiency, a first line of defense is increased expression and function of copper assimilation pathways (Labbé and Thiele, 1999). Nevertheless, when nutritional supply is limited, organisms are unable to synthesize the full complement of copper-containing enzymes.
Chlamydomonas reinhardtii is an excellent reference organism for understanding metabolic acclimation to copper deficiency because, in addition to its well-characterized genetics and genomics, it grows in a simple salts medium from which it is easy to reduce the copper content (Quinn and Merchant, 1998; Merchant et al., 2006). In addition, although the genome encodes dozens of copper proteins, only three, plastocyanin, ferroxidase, and cytochrome oxydase, appear to be abundant in terms of contributing to the copper quota (Merchant et al., 2006). Plastocyanin is located in the chloroplast and functions as an electron carrier in photosynthesis, a ferroxidase on the plasma membrane functions in high affinity iron uptake, and cytochrome oxidase in the mitochondrion functions in respiration (Hanikenne et al., 2009).
When copper is limiting, plastocyanin can be replaced with cytochrome c6, a heme-containing cytochrome encoded by the CYC6 gene (Merchant and Bogorad, 1987a). The accumulation of plastocyanin versus cytochrome c6 is reciprocal and depends on the copper nutrition status (Merchant and Bogorad, 1986a). Upregulation of CYC6 occurs by transcriptional activation through copper-responsive elements (CuREs) in the 5′ flanking sequence. These elements serve as binding sites for the SBP domain of the copper-sensing transcription factor CRR1 (Quinn and Merchant, 1995; Kropat et al., 2005; Sommer et al., 2010). CRR1 is also required for downregulation of plastocyanin through regulated proteolysis and for upregulation of other subsequently discovered genes, including CPX1 and CRD1, which encode enzymes in the tetrapyrrole pathway, and CTRs encoding assimilatory transporters (Li et al., 1996; Moseley et al., 2000; Eriksson et al., 2004; Page et al., 2009). In each case, there are CuREs associated with the upregulated gene. Therefore, CRR1 acts as a global regulator of the nutritional copper regulon in Chlamydomonas.
For a more complete picture of nutritional copper homeostasis in Chlamydomonas, we used methods of high sensitivity, including digital gene expression tag profiling (DGE-TAG) and RNA-Seq to identify the Chlamydomonas copper regulon and quantitative proteomics by LC-MSE (liquid chromatography–mass spectrometry with elevated collision energy) to test whether the changes in RNA abundance are reflected in the abundance of the corresponding proteins. Comparison of wild-type and crr1 mutant strains identifies CRR1-dependent gene targets, and computational analysis of the promoters of these genes suggests that they are direct targets. Genes encoding redox enzymes and chloroplast proteins are overrepresented among the CRR1 targets: these include a previously undescribed flavin amine oxidase that is likely to substitute for a copper amine oxidase and many desaturases involved in lipid metabolism. Lipid profiles reveal a specific, CRR1-dependent modification of thylakoid membrane galactolipids resulting in increased desaturation that is likely to impact the function of the photosynthetic apparatus. We also identify factors responsible for downregulation of plastocyanin in copper deficiency: a putative chloroplast copper chaperone and a thylakoid membrane protease.
RESULTS
Transcriptome Analysis Reveals That Dozens of Genes Are Differentially Expressed in −Cu Relative to +Cu Cells
NlaIII Tags
In the DGE method (’t Hoen et al., 2008), oligo(dT) primed cDNAs are amplified, digested with a 4-base recognition site restriction enzyme, and processed to recover and sequence 21 nucleotides into the 3′ end of the digested product. Since each cDNA is sequenced only once, the 17-nucleotide sequence tags are recovered in proportion to the abundance of the cDNA, which in turn represents the abundance of the mRNA. The sequence tags are mapped to the genome and assigned to annotated genes, and the number of reads correlates with transcript abundance.
In the first iteration of the experiment, we used the DGE-TAG protocol from Illumina to compare the relative abundance of mRNAs from copper-replete (+Cu) versus copper-deficient (–Cu) wild-type cells (see Supplemental Data Set 1 online). To distinguish which changes are dependent on the copper response regulator, CRR1, we compared –Cu wild type versus crr1-2 cells. The NlaIII tags sequences were aligned to the version 3 genome, and mRNA abundance from each locus was estimated after application of normalizations (see Methods, Supplemental Methods Sections 1 and 2, and Supplemental Table 2 online). Although the protocol favors recovery of the most 3′ NlaIII site in a cDNA, this is not absolute, and additional more 5′ sites or alternative polyadenylation sites are recovered but at lower frequency (Figure 1A; see Supplemental Figure 1 online).
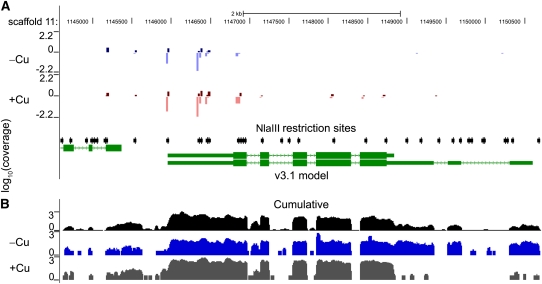
Figure 1.
Analysis of DGE-TAG and RNA-Seq Data at the Chlamydomonas CTH1 Locus.
This figure shows the UCSC browser view (http://genomes.mcdb.ucla.edu/CreCopper/) for the Chlamydomonas CTH1 locus. The browser view is centered at scaffold 11 at the base positions 114,500 to 1,150,500.
(A) UCSC browser view of DGE-TAG coverage for the CTH1 locus. JGI gene models are shown in green. The position of NlaIII sites in the genome are indicated with black arrows, and the positions to which 17-bp DGE-TAGs map are indicated with a vertical line where the height of the line indicates the intensity of the signal (on a log scale) and the color indicates the condition analyzed (blue for copper deficient and red for copper replete). The DGE-TAG method occasionally has unique hits that map to the antisense strand instead of the sense strand. Darker versus lighter coloration is used to show the DNA strand to which the tags align; darker coloration is used for the strand that goes 5′ to 3′ (from left to right), whereas the lighter color refers to the complementary strand. For example, the CTH1 locus is oriented 3′ to 5′ from left to right, and the majority of the tags are therefore lighter colored since they map on the bottom strand. DGE-TAG tracks shown in (A) represent the average tag intensity of replicate cultures for wild-type copper-replete (+Cu) and copper-deficient (–Cu) cultures.
(B) UCSC browser view of RNA-Seq coverage for the CTH1 locus. The track shown in black represents the normalized intensity, plotted on a log scale, of all RNA-Seq reads from 28 lanes mapped on to the genome. The tracks shown in gray and blue represent a single experiment: +Cu represents the RNA-Seq coverage for the locus from a wild-type copper-replete culture, while –Cu represents the coverage from a copper-deficient culture.
When we compared the abundance of tags from experimental replicates (+Cu compared with +Cu or –Cu compared with –Cu), we noted a very high correlation for moderately to highly expressed genes (>10 tags per million), but for lower abundance transcripts, the reproducibility was limited (see Supplemental Figure 2 online). Therefore, we used a 5-fold difference cutoff in tag frequency and a minimum of 10 counts per million in at least one condition to identify 32 differentially expressed genes with a false discovery rate (FDR) of <0.05 based on posterior probability of differential expression (see Supplemental Data Set 1 and Supplemental Methods Section 2 online). We recovered known CRR1 targets like CYC6, CPX1, CRD1, CTR2, and CTR3 (Merchant and Bogorad, 1987b; Hill and Merchant, 1995; Moseley et al., 2000; Page et al., 2009) along with new targets, such as CGL78, and two members of the GOX family that encode putative copper enzymes (glyoxal/galactose oxidase).
Comparison of the transcriptome of copper-deficient crr1-2 to wild-type cells indicated that greater than half (21 out 32) of the genes were likely CRR1 targets. Nevertheless, we noted that we did not recover all known CRR1 targets (e.g., CTR1 and CTH1; Moseley et al., 2002; Page et al., 2009); furthermore, many tags could not be assigned to a locus because they mapped outside the gene models. This occurs because many Chlamydomonas gene models in the FM3.1 set of the version 3 assembly (especially those without substantial EST support at the time of the analysis) do not include untranslated regions (UTRs). Chlamydomonas UTRs tend to be very long (35 to 40% of the total length of the transcript). Given that the DGE protocol recovers the most 3′ NlaIII tag, the probability that an NlaIII tag maps to a region outside a gene model that lacks a UTR is high. This is particularly true for some highly regulated genes for which there was no EST input into the generation of the FM3.1 gene models. Therefore, we applied the RNA-Seq method for better coverage of the transcriptome.
Whole Transcriptome Analysis: RNA-Seq
In the RNA-Seq method (Mortazavi et al., 2008; Nagalakshmi et al., 2008; Shendure, 2008), polyadenylated RNAs are fragmented, primed with random hexamers, and sequenced to obtain reads corresponding to the entire mRNA. Because the cDNAs are not cloned, sequences are recovered in proportion to their abundance in the population (subject to amplification bias). Because of the high correlation between replicate samples and the greater confidence of the pooled data (see Supplemental Figure 2 online), the RNA samples from independent experiments were pooled and reanalyzed by transcript resequencing protocols from Illumina. Approximately 500 Mb of 33- to 50-nucleotide sequences were obtained from each sample and aligned to the version 3 and version 4 genome assemblies with SOAP (Li et al., 2008) using an iterative approach (details in Supplemental Methods Sections 1 and 3 online). For the first round of alignment to the version 3 assembly, ~84% of the reads align with 0, 1, or 2 mismatches. In a second round of alignment, the remaining 33- to 50-nucleotide sequences are recursively trimmed (down to 21 to 25 nucleotides) until they align with no mismatches. This increases the fraction of aligned reads up to 89 to 95% in various experiments because it captures sequences that might span exon-exon junctions or sequences that might have mismatches at both ends [due to sequencing errors, poly(A) tails, or adaptors]. For version 4, we aligned 91% (average for all experiments) of the reads compared with 92% for the version 3 assembly (see Supplemental Data Set 2 online). For both assemblies, 90% of the aligned reads map uniquely to the genome; the remaining 10% map to two or more sites on the genome owing to the presence of repeat regions or because the length of the sequence is insufficient to distinguish between members of gene families. The frequency of recovery of sequences from each locus is dependent on the abundance of the mRNA in the starting material and after application of various corrections and normalization for transcript length is presented as RPKM (reads per kilobase of mappable transcript length per million mapped reads) (see Supplemental Methods Section 3.6 online; Figure 1B).
The transcript coverage across the genome is visualized on a local installation of the UCSC browser (http://genomes.mcdb.ucla.edu/CreCopper/). The normalized coverage graphs for each experiment provide an indication of transcript abundance, and if there are multiple transcript forms at a locus, the prevalence of one isoform versus another. In the example shown in Figure 1B, it is evident that the abundance of CTH1 transcripts is greater in +Cu cells versus –Cu cells and, furthermore, that the transcript form in –Cu cells is extended at the 5′ end relative to the shorter form in +Cu cells (cf. gray versus blue coverage graphs in Figure 1B), which is entirely in agreement with previous genetic analysis of this locus (Moseley et al., 2002).
When we compared relative RNA abundance in the +Cu versus –Cu transcriptome as determined by whole transcriptome analysis, we identified 149 genes in the FM3.1 set of gene models whose transcript abundances were changed by >2-fold (log2 ≥ 1) in –Cu relative to +Cu (FDR < 0.05, RPKM ≥ 10) (see comparison B in Figure 2A). Most of these involved increases in –Cu cells (128 of 149) (see Supplemental Data Set 3 online). To distinguish which of the changes in RNA abundance represent a programmed response to copper sensing versus compensatory or secondary responses resulting from loss of cuproprotein activity in copper deficiency, we analyzed the transcriptome of copper-deficient crr1 cells. The comparison of copper-deficient crr1-2 cells to copper-deficient wild-type (crr1:CRR1) cells, representing a previously characterized complemented strain (Kropat et al., 2005; Sommer et al., 2010) (Figure 2A), showed an impact on hundreds of transcripts. The overlap between the 821 genes in the comparison of copper-deficient crr1 to the copper-deficient rescued strain and the set of 149 genes regulated in the complemented strain by nutritional copper deficiency consisted of only 63 genes (Figure 2A, overlap between comparisons B and C). We conclude that the 63 genes represent the programmed response, while the changes in the expression of most other genes in the crr1 mutant versus the complemented strain likely result from the differences in growth rate and impaired metabolism resulting from loss of CRR1 function (Figures 2A and 2B). The crr1 mutant grows very poorly in copper-deficient medium (Eriksson et al., 2004; Kropat et al., 2005), and it is likely that most of the differences in RNA abundance are secondary, perhaps related to stress from loss of function of important cuproenzymes like cytochrome oxidase. Because of the genetic variability between so-called laboratory wild-type strains (Kropat et al., 2011; Siaut et al., 2011), the comparison between the mutant versus the rescued strain is more appropriate, especially for genes that display a low level of expression.
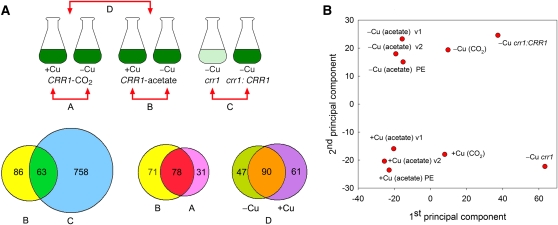
Figure 2.
Copper Deficiency Targets under Photoheterotrophic and Photoautotrophic Conditions.
(A) RNA-Seq identifies copper deficiency and CRR1 targets. RNA was extracted from batch cultures of Chlamydomonas cells. Genotypes, growth conditions, and comparisons A, B, C, and D are illustrated (top part). The crr1 and crr1:CRR1 cells were grown in the presence of acetate for comparison C. Note that the uncomplemented crr1 mutant grows very poorly in copper-deficient medium (pale green color). Cells were collected at a density of 3 × 106 cell mL−1. The Venn diagrams (bottom part) identify the number of differentially accumulating transcripts at ≥2-fold change (FDR < 0.05) in each pairwise comparison and the overlap of different sets. For comparison D, the number of differentially accumulating transcripts was computed at ≥5-fold change (FDR < 0.05). Transcripts with RPKM values <10 were not used in any of the analyses. The 63 putative CRR1 targets were used to retrieve sequences upstream of the transcription start site of those genes to identify CuREs (see text).
(B) Copper deficiency targets group into three categories. Principal component analysis of RNA-Seq data group the experiments into three different categories according to growth condition or genetic background (first component) and two different groups according to copper metabolism (second component). The type of library (single-end v1, v2, and paired end [PE]) is shown to be unimportant.
Since copper enzymes function in both respiration and photosynthesis, we compared the impact of copper nutrition on cells grown photoautotrophically versus photoheterotrophically (Figure 2A; see Supplemental Data Set 4 online). The total number of transcripts impacted by copper deficiency is similar to the number impacted in photoheterotrophic conditions. Nevertheless, there are some transcripts where copper deficiency has a greater impact in phototrophic cells and a few where a change in abundance is unique to the phototrophic growth situation (Figure 2A; 31 genes that were found in comparison A but not in comparison B). Among the 31 transcripts in these categories are those involved in iron assimilation and homeostasis, such as FEA2, FER1, FOX1, FTR1, and MSD3 (Allen et al., 2007a). This might be related to the increased iron content of photoautotrophic cells and the role of copper in iron assimilation (La Fontaine et al., 2002; Terauchi et al., 2010). When the data from all experiments is subjected to principal component analysis, the first component groups data by growth medium and genotype at the CRR1 locus (Figure 2B), and the second component groups by copper nutrition state. Both components combined explain 75% of the variance of the data. The analysis also shows that the method of sequencing (NlaIII tags, single- or paired-end whole transcriptome) is irrelevant.
Real-Time PCR versus DGE-TAG or mRNA-Seq
To assess the reliability of estimating fold changes in mRNA abundance by RNA-Seq, we compared fold changes with those estimated from real-time PCR experiments. Three experimental replicates were used in the real-time PCR experiments; two samples were identical to those analyzed by sequence-based methods and one sample was independent. For the DGE-TAG method, we observed a correlation of 0.88 with RT-PCR (see Supplemental Figure 3 online). We noted, in particular, that the correlation was poor where incomplete gene models precluded assignment of all tags to the locus, which means that the abundance of the corresponding transcript is underestimated. By contrast, the correlation between fold changes in the same two experiments measured by mRNA-Seq versus real-time PCR is 0.95 (Figure 3A). For the CYC6 gene, which is ~214-fold regulated, the data point falls close to the regression line, indicating the large dynamic range of the technique. When a third independent experiment is included for the PCR analysis, the r2 is 0.97, which gives an indication of the between-experiment reproducibility.
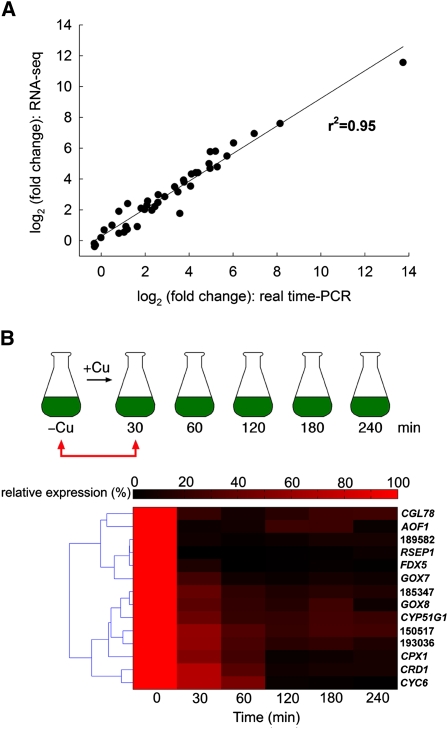
Figure 3.
Validation of mRNA Abundance Changes.
(A) Comparison of fold difference in RNA abundance between copper-replete versus copper-deficient cells estimated from RNA-Seq versus real-time PCR experiments. Each data point represents the change in abundance of one mRNA (average of two independent experiments). For real-time PCR, each sample was analyzed separately in technical triplicates. For RNA-Seq, the same two samples were pooled prior to preparation of libraries. The correlation coefficient was calculated from the regression line derived with a 95% confidence interval.
(B) The abundance of mRNAs of select CRR1 targets upon exposure of copper-deficient cells to copper. Top part: Copper-deficient cells, sampled at time 0, were exposed to 100 nM CuEDTA at a density of 3 × 106 cells mL−1 and sampled 30, 60, 120, 180, and 240 min later for preparation of total RNA as illustrated. Bottom part: The fold change in mRNA abundance at each time point was estimated by real-time PCR relative to the t = 0 RNA sample (100% expression) and is normalized to the endogenous reference gene CBLP, whose abundance does not change in this situation. The data presented are averages of three independent experiments.
We also tested the pattern of expression of a subset (13 out of 63) of CRR1 targets in a different experimental design (Figure 3B). Here, –Cu cells were supplemented with copper and sampled at various time points chosen on the basis of a previous time-course analysis of the decay of the CYC6 transcript in +Cu cells (0, 30, 60, 120,180, and 240 min) (Hill et al., 1991). Each of the tested targets shows a pattern of decay comparable to that of sentinel genes, CYC6, CPX1, and CRD1, regardless of the abundance of the transcript or the magnitude of regulation. The set of genes identified by comparison of exponential batch cultures therefore likely represents an authentic and specific response to nutritional copper deficiency.
Redox Proteins Are Overrepresented among CRR1 Targets
Use of Coverage Graphs for Manual Annotation of the Genome
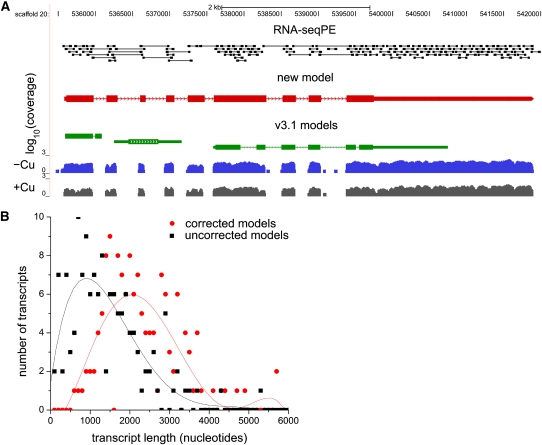
For estimating the transcript abundance, the sequence reads were mapped to the version 3 genome assembly, and reads that aligned to the FM3.1 gene catalog were used (see Methods and Supplemental Methods online). It is evident from examination of the coverage graphs for the 180 genes corresponding to differently accumulating transcripts (comparisons A and B in Figure 2A; see Supplemental Data Sets 3 and 4 online) that many gene models in the FM3.1 catalog are underpredicted (pathways.mcdb.ucla.edu provides a tool to convert FM3.1 protein IDs to FM4 protein IDs). Therefore, we used transcript coverage to extend the models, and used placement of paired-end reads to establish exon connectivity. For instance, the RNA-Seq data suggest that the gene model to the left of the CTH1 locus should be extended at its 3′ end (Figure 1B). Figure 4A shows a situation where paired-end alignments and coverage graphs suggested replacement of three adjacent gene models by a single larger model. In summary, ~71% (or 128) of the gene models were improved, resulting on average in an increase of ~1 kb in the length of gene models (Figure 4B).
Figure 4.
Transcript Length Distribution after Manual Gene Model Correction.
(A) UCSC browser view for a differentially expressed locus. Paired-end reads used to establish exon connectivity are shown as black rectangles joined by a line. The RNA-Seq coverage, on a log scale, from –Cu versus +Cu cells of one experiment is shown in blue or gray, respectively. The original JGI gene models, from FM3.1 of the Chlamydomonas genome, used to compute fold changes shown in Supplemental Data Set 3 online are shown in green. RNA-Seq data were used to inform the manual construction of a single gene model, shown in red.
(B) Distribution of transcript lengths. Models for 149 genes whose transcripts responded to copper deficiency were manually investigated and improved using both single- and paired-end RNA-seq reads. The availability of millions of new ESTs from 454 sequencing (http://genomes.mcdb.ucla.edu/Cre454/) allowed further improvements. The distribution of transcript lengths is shown for the uncorrected gene models in the FM3.1 frozen catalog (black squares) versus the gene models after data-dependent correction (red circles). A polynomial best-fit line is shown for both data sets.
The sequence reads were also aligned to the version 4 genome assembly, and fold changes were computed for the FM4 set of gene models as well as for a new set, Au10.2, that incorporated transcript coverage from >2 million 454 ESTs (released in 2011; http://genomes.mcdb.ucla.edu/Cre454/). A comparison of the alignment statistics is shown in Supplemental Data Set 2 online. There is not a large difference in the quality of the two assemblies (relative to each other), but given that the most recent set of gene models covers a larger fraction of the genome, there are more reads aligned to the Au10.2 transcripts relative to the FM3.1 set of transcripts. Nevertheless, a similar number of differentially expressed genes was identified regardless of the set of gene models used for the analysis (see Supplemental Data Set 5 online). The Au10.2 confirmed that incorporation of deep transcriptome coverage led to revisions of ~50% of the previous sets (FM4 or Au5) of gene models (http://genomes.mcdb.ucla.edu/CreCopper/ and http://www.phytozome.net/chlamy).
Functional Classification of Differentially Expressed Genes
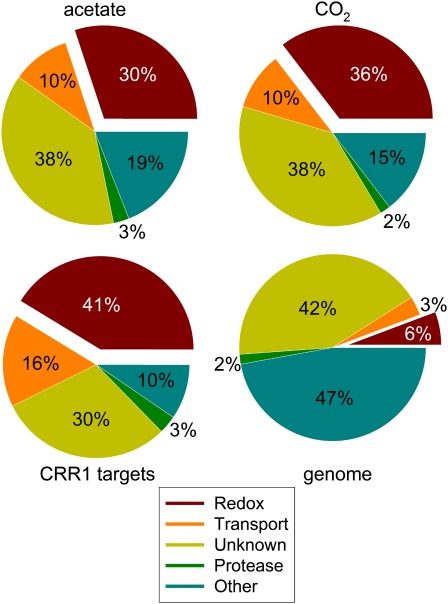
Each of the differentially expressed genes (see Supplemental Data Sets 3 and 4 online) was assessed for function by examining annotations (both user community and automated) on the protein page at the Chlamydomonas genome displayed on the Joint Genome Initiative (JGI) browser (http://genome.jgi-psf.org/Chlre3/Chlre3.home.html and http://genomeportal.jgi-psf.org/Chlre4/Chlre4.home.html) and by analysis of the predicted protein sequence for occurrence of domains that suggest activity or function (Figure 5). The HMMTOP algorithm (Tusnády and Simon, 2001) was used to distinguish whether the gene product might be membrane bound or soluble. Most are predicted to be soluble (75%). The distribution of functional categories is approximately the same regardless of whether we consider experiments in heterotrophic conditions or phototrophic conditions or whether we consider all copper-responsive genes or only the CRR1 targets.
Figure 5.
Predicted Protein Domain Function for Copper Deficiency and CRR1 Targets.
The deduced protein sequences of the transcript set of 149 and 109 copper deficiency targets from Supplemental Data Set 3 online (acetate, photoheterotrophic; top left) and Supplemental Data Set 4 online (CO2, photoautotrophic; top right), respectively, were analyzed at the Pfam site (E-value ≤1 × 10−4). The identified protein domains were grouped according to their function into five categories: redox (red), transport (orange), protease (green), unknown (gold), and other (blue). Unknowns refer to protein sequences for which a domain could not be identified or if the domain was identified as having an unknown function (DUF). Domains that could not be classified as redox, transport, or protease were grouped into the “other” category. The distribution of protein domain functions in the subset of CRR1 targets (63 genes, bottom left) identified under photoheterotrophic conditions (see Figure 2A) is also shown. Chlamydomonas gene model set FM3.1 was used to estimate the genome-wide distribution of protein functions in the same categories. For the analysis of the complete genome (bottom right), the protein sequences with a predicted domain function with a bit score ≤10 were labeled unknown.
Not surprisingly, there are differentially regulated transporters (Table 1). Among these are genes encoding the previously characterized members of the CTR family, all of which are CRR1 targets (Page et al., 2009), but also IRT2 encoding a ZIP family transporter that was previously identified as an iron nutrition–responsive gene (Allen et al., 2007a). This transporter could function either to allow iron uptake in a situation where ferroxidase function might be impaired (because of copper deficiency; La Fontaine et al., 2002; Chen et al., 2008), or it might serve as a secondary route for copper or zinc assimilation or distribution. Several genes involved in iron homeostasis are differently regulated in CO2-grown versus acetate-grown cells. These do not appear to be direct targets of CRR1. The pattern of expression in heterotrophic versus phototrophic conditions is not the same for each gene, indicating that there are multiple signals (copper, iron nutrition but also carbon source) contributing to the expression of this important pathway.
Table 1.
Copper Deficiency Impacts Various Metabolic Processes
| Process | Gene | Defline | CRR1 ─Cua | CRR1 +Cua | Fold Change | crr1:CRR1 ─Cua | crr1 ─Cua | Fold Change | CRR1 Target | Domain |
| Anaerobic metabolism | FDX5 | Apoferrodoxin | 2742 | 14 | 196 | 1605 | 1 | 1791 | Y | Redox |
| HYDEF | Fe hydrogenase assembly protein | 32 | 2.3 | 14 | 28 | 15 | 1.9 | N | Redox | |
| HYDG | Hydrogenase assembly factor | 79 | 11 | 7.2 | 136 | 127 | N | Redox | ||
| HCP3 | Hybrid cluster protein | 11 | 2.1 | 5.5 | 29 | 23 | 1.3 | N | Redox | |
| PFR1 | Pyruvate-ferredoxin oxidoreductase | 77 | 14 | 5.6 | 41 | 31 | 1.3 | N | Redox | |
| HYD1 | Fe hydrogenase | 184 | 44 | 4.2 | 676 | 146 | 4.6 | N | Redox | |
| 166317 | Prolyl 4-hydroxylase, α-subunit | 104 | 3.2 | 32 | 184 | 16 | 11 | Y | Redox | |
| 114525 | Prolyl 4-hydroxylase, α-subunit | 295 | 38 | 7.8 | 198 | 33 | 6.0 | Y | Redox | |
| 143800 | Prolyl 4-hydroxylase | 57 | 1.6 | 36 | 39 | 1.3 | 30 | Y | Redox | |
| HCP2 | Hybrid-cluster protein | 11 | 5.0 | 2.2 | 38 | 240 | 0.16 | N | Redox | |
| PAT1b | Phosphate acetyltransferase | 30 | 16 | 1.9 | 0.08 | 1.1 | N | |||
| Oxygen-dependent enzymes | 96954 | Prolyl 4-hydroxylase | 118 | 1.4 | 85 | 68 | 5.6 | 12 | Y | Redox |
| 143800 | Prolyl 4-hydroxylase | 57 | 1.6 | 36 | 39 | 1.3 | 30 | Y | Redox | |
| 166317 | Prolyl 4-hydroxylase, α-subunit | 104 | 3.2 | 32 | 184 | 16 | 11 | Y | Redox | |
| 114525 | Prolyl 4-hydroxylase, α-subunit | 295 | 38 | 7.8 | 198 | 33 | 6.0 | Y | Redox | |
| 193036 | Putative glyoxal or galactose oxidase | 528 | 10 | 54 | 430 | 15 | 30 | Y | Redox | |
| GOX17 | Glyoxal or galactose oxidase | 354 | 7.3 | 48 | 294 | 10 | 29 | Y | Redox | |
| GOX7 | Glyoxal or galactose oxidase | 37 | 1.3 | 28 | 26 | 1.6 | 17 | Y | Redox | |
| GOX8c | Glyoxal or galactose oxidase | 8.6 | 0.37 | 23 | 17 | 1.0 | 16 | Y | ||
| GOX3 | Glyoxal or galactose oxidase | 20 | 9.0 | 2.2 | 16 | 7.7 | 2.0 | Y | Redox | |
| HPD1 | 4-Hydroxyphenylpyruvate dioxygenase | 114 | 2.5 | 45 | 32 | 14 | 2.3 | Y | Redox | |
| CPX1 | Coproporphyrinogen III oxidase | 3483 | 232 | 15 | 1109 | 166 | 6.7 | Y | Redox | |
| CRD1 | Copper response defect 1 protein | 2047 | 176 | 12 | 1168 | 89 | 13 | Y | Redox | |
| CAO | Chlorophyll a oxygenase | 254 | 99 | 2.6 | 177 | 107 | 1.7 | N | Redox | |
| 113197 | Prolyl 4-hydroxylase, α-subunit | 18 | 7.8 | 2.3 | 6.3 | 6.5 | N | Redox | ||
| CDO1 | Cysteine dioxygenase | 14 | 6.2 | 2.2 | 9.5 | 12 | N | Redox | ||
| Lipid metabolism | CYP51G1 | Sterol 14 desaturase | 249 | 50 | 5.0 | 84 | 31 | 2.8 | Y | Redox |
| CYP745A1 | Cytochrome P450 | 120 | 24 | 4.9 | 57 | 18 | 3.3 | Y | Redox | |
| 59933 | SC5D, C-5 sterol desaturase | 189 | 41 | 4.6 | 77 | 30 | 2.6 | Y | Redox | |
| FAB2 | Plastid acyl-ACP desaturase | 866 | 187 | 4.6 | 592 | 121 | 4.9 | Y | Redox | |
| CYP710B1 | Cytochrome P450, C-22 desaturase | 177 | 52 | 3.4 | 175 | 46 | 3.8 | Y | Redox | |
| FAD5A | MGDG-specific palmitate Δ-7 desaturase | 285 | 97 | 2.9 | 142 | 40 | 3.5 | Y | Redox | |
| DES6 | ω-6-FAD, chloroplast isoform | 4029 | 1758 | 2.3 | 1538 | 2315 | 0.66 | N | Redox | |
| 135825 | Fatty acid desaturase, Δ-12 | 57 | 22 | 2.6 | 24 | 17 | 1.4 | N | Redox | |
| 40101 | Acyl-CoA dehydrogenase | 80 | 33 | 2.4 | 25 | 14 | 1.9 | N | Redox | |
| 177689 | CDP-alcohol phosphatidyltransferase | 29 | 10 | 3.0 | 11 | 9.4 | N | Other | ||
| CYP747A1 | Cytochrome P450 | 15 | 6.3 | 2.4 | 13 | 11 | N | Redox | ||
| SMT1 | Sterol-C24-methyltransferase | 474 | 232 | 2.0 | 264 | 146 | 1.8 | N | Other | |
| Copper assimilation | CTR1 | CTR-type copper ion transporter | 60 | 18 | 3.4 | 59 | 5.6 | 10 | Y | Transport |
| CTR2 | CTR-type copper ion transporter | 171 | 8.6 | 20 | 101 | 0.86 | 118 | Y | Transport | |
| CTR3 | CTR-type copper ion transporter | 209 | 19 | 11 | 269 | 8 | 34 | Y | Transport | |
| IRT2 | ZIP family transporter | 20 | 0.94 | 21 | 53 | 7.0 | 7.7 | Y | Transport | |
| Fe assimilation | FEA1 | Fe-assimilating protein | 448 | 226 | 2.0 | 3010 | 1760 | 1.7 | N | Other |
| Copper backup enzyme | AOF1 | Amine oxidase-like protein | 67 | 17 | 3.9 | 11 | 5.0 | 2.2 | Y | Redox |
| Chloroplastmetabolism | CYC6 | Cytochrome c6 | 2677 | 0.81 | 3315 | 1589 | 1.1 | 1427 | Y | Redox |
| FDX5 | Apoferrodoxin | 2742 | 14 | 196 | 1605 | 0.90 | 1791 | Y | Redox | |
| CGL78 | Conserved green lineage | 1514 | 194 | 7.8 | 699 | 78 | 9.0 | Y | Unknown | |
| FDX6 | Apoferrodoxin | 69 | 155 | 2.3 | 227 | 80 | 2.8 | Y | Redox |
The genes involved in anaerobic metabolism were identified previously (Mus et al., 2007). Genes without functional annotation in the FM3.1 set of Chlamydomonas gene models are indicated with a protein ID. To determine if the expression of genes is CRR1 dependent, the mRNA transcript abundances in crr1 and crr1:CRR1 cells were considered. A fold difference ≥ 2.0 with an FDR < 0.05 was considered significant, and the corresponding gene was considered a CRR1 target. The predicted domain function was determined via Pfam (E-value ≤1 × 10−4).
Transcript abundance, RPKM.
Transcript with fold change <2; not included in Supplemental Data Set 3 online.
Transcript abundance is <10; not included in Supplemental Data Set 3 online; verified fold change via RT-PCR.
Besides replacement of plastocyanin by cytochrome c6 (Merchant and Bogorad, 1987a), the analysis suggests that there are other copper sparing mechanisms operating in copper-deficient Chlamydomonas (i.e., mechanisms that allow allocation of copper to processes that are absolutely dependent on copper). We note upregulation of AOF1, which appears to encode an amine oxidase with a flavin as the redox cofactor; AOF1 might serve as a backup for one of the several copper-containing amine oxidases encoded in the Chlamydomonas genome (see Supplemental Data Set 3 online). AOF1 is also upregulated under N deficiency, suggesting that it may function to liberate ammonia from primary amines. Another example of metal sparing is offered by 142028, which encodes a protein with a metal binding domain (see Supplemental Data Set 3 online); its downregulation may reduce the copper quota and hence represent a metal sparing situation. Surprisingly, several members of the galactose/glyoxal oxidase family (GOX7, GOX17, GOX3, and 193036), which encode copper-containing enzymes, are upregulated as a result of copper deficiency. These proteins are ubiquitous in eukaryotes, but their physiological functions are not known. This is not likely to be a secondary effect (e.g., by feedback resulting from loss of oxidase function in copper deficiency) because the change in gene expression is CRR1 dependent.
When the differently regulated genes are classified according to function (Figure 5), the largest group corresponds to proteins that catalyze redox reactions. This is true for both CRR1-dependent and -independent responses. Given that only a small fraction of the genome encodes redox proteins, the enrichment among the copper deficiency response genes is significant (P < 10−15). We note also that certain pathways are well represented, such as the tetrapyrrole pathway (Figure 6; see Supplemental Data Set 6 online) in which three of six redox steps are impacted, namely, CPX1, CRD1, and CAO. While CPX1 and CRD1 are CRR1 targets, CAO is not. Lipid metabolism is also well represented with genes encoding primarily plastid proteins (but also some known extraplastidic ones). We identified genes affecting glycerolipid/membrane lipid metabolism (FAB2, FAD5A, and DES6), carotenoid biosynthesis (CYP745A1), potential sterol desaturases/hydroxylases (59933, CYP710B1, and CYP51G1), and Vitamin E or plastoquinone biosynthesis (HPD1), all of which are CRR1 targets (Table 1).
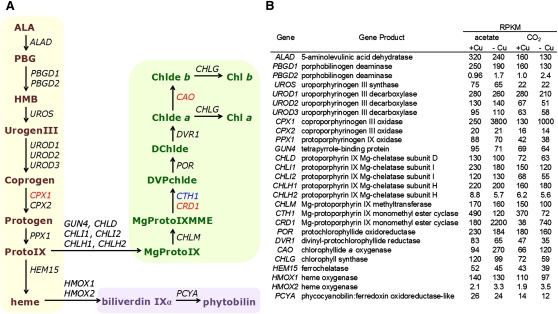
Figure 6.
Abundance of mRNAs at Each Step in the Tetrapyrrole Biosynthesis Pathway.
(A) The transcripts(s) encoding enzyme(s) at each step of the pathway are shown (Lohr et al., 2005; Tanaka and Tanaka, 2007). The heme branch is highlighted in beige, the chlorophyll branch in green, and the linear tetrapyrrole branch in blue. Transcripts whose abundance is impacted by copper nutrition are indicated in red (increased in –Cu) or blue (increased in +Cu).
(B) Relative mRNA abundances, in RPKM, are shown for copper-replete (+Cu) and copper-deficient (–Cu) photoheterotrophic (acetate) and photoautotrophic (CO2) cultures sampled at 3 × 106 cells mL−1. Three light-independent isoform proteins encoded by the chloroplast genes chlB, chlL, and chlN are not listed because RNA-Seq reads were not mapped to the chloroplast genome.
Among the redox proteins, we noted hydrogenase (HYD1), hydrogenase assembly factors (HYDEF and HYDG), and other genes whose expression is increased in anaerobic cells, like PFR1-encoding pyruvate ferredoxin oxidoreductase, and HCP3 encoding a hybrid cluster protein (Table 1; see Supplemental Figure 4 online; Ghirardi et al., 2007; Grossman et al., 2007; Mus et al., 2007). Given that the cells are well aerated during the experiment, especially the phototrophically grown ones, which are O2 evolving, it is unlikely that the O2-sensitive gene products would be functional. Therefore, we suspect that these responses, which mimic the anaerobic response (Mus et al., 2007), are a secondary consequence of copper deficiency. Indeed, none of these genes appears to be a target of CRR1 because the response is noted even in the crr1 mutant (see Supplemental Data Set 3 online). There is no difference in O2 evolution capacity in +Cu versus –Cu wild-type cells; therefore, it is unlikely that the copper-deficient cells are selectively anaerobic.
Changes in RNA Are Recapitulated by Analysis of the Corresponding Proteins and Have an Impact on Metabolic Physiology
Quantitative Analysis of the Soluble Proteome
Since most of the differentially accumulating RNAs encoded soluble proteins, we sought to analyze the soluble proteome of Chlamydomonas by quantitative proteomics to assess whether changes in the transcriptome resulted in corresponding changes in protein abundance. Total soluble proteins were fractionated by gel electrophoresis (see Supplemental Figure 5 online), and each fraction was analyzed after trypsin digestion by liquid chromatography–tandem mass spectrometry (LC-MS/MS) to identify peptides, which were searched against the predicted proteins encoded by a set of gene models (for both the version 3 and version 4 assemblies of the Chlamydomonas genome [http://genome.jgi-psf.org/Chlre3/Chlre3.home.html and http://genome.jgi-psf.org/Chlre4/Chlre4.home.html]).
To ensure that proteins were not lacking from our analysis due to missing or incorrect gene models, we searched the data using three independent gene models sets with slightly different gene repertoires: (1) FM3.1 corresponding to JGI models in the version 3 assembly (ftp://ftp.jgi-psf.org/pub/JGI_data/Chlamy/v3.0/proteins.Chlre3.fasta.gz), (2) FM4 corresponding to JGI models in the version 4 assembly (http://genomeportal.jgi-psf.org/Chlre4/download/Chlre4_best_proteins.fasta.gz), and (3) Au10.2 corresponding to Augustus models in the version 4 assembly (Stanke et al., 2008; http://www.phytozome.net/chlamy). The FM3.1 database was corrected by replacement of a subset of revised gene models including the set of copper proteins (see Supplemental Data Set 7 online), metal transporters (see Supplemental Data Set 8 online), and those corresponding to the differentially expressed transcripts.
Large-scale quantification of proteins was accomplished using a label-free data independent MS-scanning approach (Silva et al., 2006a, 2006b). The data-independent acquisition method uses alternating MS scans of low and elevated collision energy, providing a nearly 100% duty cycle for precursor ion and product ion measurement (see Methods). Compared with conventional data-dependent acquisition methods, a substantial increase in the number of total proteins identified and the sequence coverage of proteins is realized with the data-independent scanning, especially for quadrupole time-of-flight analyzers used in this study (Blackburn et al., 2010). The label-free quantification scheme is based on the relationship between the MS signal response and protein concentration; the average MS signal response for the three most intense tryptic peptides per mole of protein is constant within a coefficient of variation of approximately ±10% (Silva et al., 2006a, 2006b; Geromanos et al., 2009). Absolute quantification requires that a known quantity of a standard intact protein be spiked into the protein mixture of interest prior to digestion (with trypsin), or alternatively, a known quantity of predigested protein can be spiked into the mixture after it has been digested. The quantification method calculates the average MS signal response for the three most intense tryptic peptides for each protein in the mixture, including those from the internal standard. Although the dynamic range of proteomics data does not compare favorably with transcriptome data, this label-free proteomics approach compares well with other label-free and labeling schemes with three to four orders of dynamic range demonstrated (Silva et al., 2006a, 2006b; Bantscheff et al., 2007). Additionally, the absolute sensitivity of our MS experiments was determined to be in the low femtomole range. From three separate experiments, more than 1000 proteins were identified and quantified. To reduce the number of false positives, we required that a protein be identified in at least two out of three independent samples (resulting from triplicate cultures) for each condition.
Replicate proteome analysis of samples from replicate cultures and from different growth conditions showed good reproducibility (see Supplemental Figure 6 online). The distribution of data observed was similar to that found for the transcriptomics analysis, where a high correlation was observed for abundant proteins and a lower correlation was found for the low abundant proteins. The coefficient of variation between biological replicates is calculated to be ~0.3, which also demonstrates good biological reproducibility.
For 18 out of 27 proteins, we noted little or no impact in terms of which set of gene models was used. However, we identified three additional proteins (516174, 392175, and 516987) by including revised gene models in the database relative to a parallel search on an unmodified database (see Supplemental Data Set 9 online), highlighting the value of gene model correction based on RNA-Seq data. In four other cases, differences between gene model sets resulted in a difference with respect to protein abundance (AOF1, GOX17, UOX, and FEA2). The direction of regulation (accumulation or reduction) of protein amounts in −Cu cells relative to +Cu cells was not impacted by these changes in abundance. Lastly, two changes in the protein list were noted in only one out of the three database searches. The CUTA1 gene product was identified only from the FM4 set of models. Its identification relies on one peptide that is reassigned in FM3.1 searches and a second peptide that does not exist in the Augustus 10.2 gene models. The MMP2 gene product was not identified in the Au 10.2 set of proteins. This is attributed to two key peptides required for its assignment being reassigned to a different protein corresponding to Cre17.g718468.t1.1, which is missing from the FM3.1 and FM4 sets (see Supplemental Data Set 9 online).
We identified 18 proteins corresponding to differently accumulating transcripts (see Supplemental Data Set 9 online), which constitutes ~14% of the differentially accumulating transcript data set (see Supplemental Data Set 3 online). This fraction is comparable to the fraction of the proteome (~1000 unique proteins) captured by the technique. For 17 of the 18 proteins, the change in protein abundance mimics the change in RNA abundance (see Supplemental Data Set 9 online). The one exception is FEA1, but the corresponding gene is not under the control of CRR1. Given that FEA1 is an extracellular protein, it may not have been recovered quantitatively during the preparation of cell extracts. The increase in acyl-ACP desaturase and hydrogenase was validated by immunoblots (see Supplemental Figure 4 online).
We also examined the proteome for proteins that are known to be involved in metal trafficking or are known to be copper binding proteins (see Supplemental Data Sets 7 and 8 online). We found seven of these to have altered abundance under copper-deficient relative to copper-replete growth with no significant change in the amount of the corresponding mRNA. Among these is plastocyanin, a well-known example of a cuproprotein whose abundance is not controlled at the transcript level. The copper-dependent accumulation of plastocyanin is determined by regulated degradation of the apoprotein (Li and Merchant, 1995).
Membrane Proteins
In addition, in previous studies, we documented CRR1-dependent changes in the abundance of membrane proteins CTR1, CTR2, and CRD1 by immunoblot analyses, which increases the fraction of targets that is validated at the proteome level (Moseley et al., 2000; Page et al., 2009).
Chloroplast Metabolism
Besides the plastid-localized proteins discussed already, we identified several putative plastid proteins among the CRR1 targets: a novel but highly conserved protein CGL78, ferredoxins FDX5, FDX6, and 144679 with a polyferredoxin domain, and a thylakoid membrane protease RSEP1 (Table 1) (Riekhof et al., 2005; Merchant et al., 2007; Mus et al., 2007; Terauchi et al., 2009). CGL78 is a highly expressed transcript, especially in –Cu cells where its mRNA can constitute as much as 0.1% of the total mRNA pool (Table 1), and also in green organs in Arabidopsis thaliana (Schmid et al., 2005). Its function is not known, but it is restricted to photosynthetic organisms and, thus far, absolutely conserved in all organisms with a green plastid (Merchant et al., 2007). Sequence analysis indicates an N-terminal plastid-targeting transit sequence, and the protein has been localized to the chloroplast in proteomic studies; therefore, it is likely to have a function in a major chloroplast metabolic pathway (see Supplemental Figure 7 online). CGL78 is encoded in nearly all cyanobacterial genomes except for six Prochlorococcus species, all of which are from low light environments. Therefore, we hypothesize that CGL78 might have a function in the metabolism of chlorophyll binding proteins, perhaps in a process related to high light acclimation.
Impact on Fatty Acid Desaturation
Transcriptome profiling revealed increased abundance of RNAs encoding various desaturases (Table 1); for FAB2, this was verified also by increased abundance of the corresponding protein (see Supplemental Figure 4 and Supplemental Data Set 9 online). What is the significance of this with respect to membrane lipids? One interpretation is that the changes in desaturase activity are intended to compensate for a desaturation defect caused by nutritional copper deficiency. In this situation, the desaturation level of membrane lipids would be unchanged in wild-type copper-replete versus -deficient cells, but the copper-deficient crr1 mutant might have reduced levels of desaturation. Another interpretation is that there is a programmed increase in the level of desaturation in copper-deficient cells. In this case, we expect that membrane lipid desaturation will be unchanged in crr1 but increased in wild-type copper-deficient cells.
To distinguish between these possibilities and also to identify the lipid class that is impacted by copper deficiency, we compared the fatty acid composition of lipids extracted from copper-replete and copper-deficient cells (Figure 7A). There is a shift toward more unsaturated fatty acids in copper-deficient wild-type cells relative to the copper-replete situation. For instance, 16:2 and 16:3 species are increased with a concomitant decrease in 16:0 and 16:1; we also noted a decrease in the 18:1Δ9 species. The changes are significant based on statistical analysis (P < 0.05, two-sample unpaired t test) and the fact that they are not observed in the crr1 strain (see Supplemental Figure 8A online).
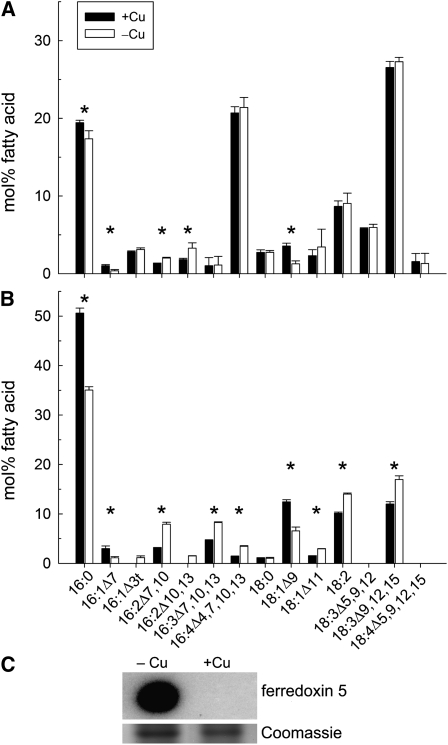
Figure 7.
Changes in Fatty Acid Composition and Ferrodoxin5 Abundance Parallel Changes in mRNA Abundance.
(A) Total fatty acids profile of whole CRR1 cells.
(B) The fatty acid composition of DGDG.
For (A) and (B), lipids were isolated from +Cu and −Cu CRR1 cells. Lipids were separated by thin layer chromatography and quantified by GC. The bars show lipid composition in mol % and indicate the means (±sd) of six independent experiments. Fatty acid types are shown as number of carbons:number and positions (Δ) of double bonds. Values marked with an asterisk are significantly different from each other (P < 0.05, nonpaired two-sample t test).
(C) For the immunodetection of ferredoxin5, proteins were separated on a nondenaturing polyacrylamide gel (15%) and transferred to PVDF for immunoblot analysis with anti-Fdx5. Protein loading was normalized by Coomassie blue staining.
To distinguish whether the observed changes are specific for a subset of lipids, we purified individual lipids prior to the analysis of the fatty acid composition. The most notable changes were observed for digalactosyldiacylglyceride (DGDG), which is normally enriched in saturated and monounsaturated fatty acids, and where nine changes meet statistical criteria for significance (Figure 7B). The 16:2, 16:3, 16:4, 18:2, and 18:3 species are enriched at the expense of the 16:0, 16:1, and 18:1Δ9 precursors. The 18:1Δ11 species is enriched as well. These changes are not observed in the crr1 mutant (see Supplemental Figure 8C online). In the monogalactosyldiacylglyceride (MGDG) profile from the wild type (see Supplemental Figure 9A online) but not from the crr1 mutant (see Supplemental Figure 8B online), we observed an increase in the 16:2 species. MGDG is typically highly enriched in 16:4 and 18:3 fatty acids, and major changes could therefore not be expected. However, the observed increase in polyunsaturated 16-carbon fatty acids indicates that under these conditions a higher proportion of DGDG is synthesized from MGDG. By contrast, for the endoplasmic reticulum–synthesized lipid diacylglyceryl-N,N,N-trimethylhomoserine, the only notable change occurs in 18:1Δ11, which is increased in the wild type but not in crr1 (see Supplemental Figures 8D and 9B online). Therefore, we conclude that copper deficiency impacts the composition of chloroplast membrane lipids by increasing the level of desaturation through increased expression of specific plastid-acting desaturases (FAB2, FAD5A, and DES6) (see Supplemental Figure 4 online).
Algal Homologs
The replacement of plastocyanin by cytochrome c6 has been documented in many algae (Sandmann et al., 1983; Quinn and Merchant, 1999), but the extent of metabolic acclimation to copper deficiency has not been investigated. Volvox carteri, which is most closely related to Chlamydomonas, has orthologs of 47 CRR1-dependent and 43 CRR1-independent differentially expressed genes, although surprisingly not CYC6, indicating that plastocyanin might be essential as it is in land plants (see Supplemental Data Set 10 online) (Pilon et al., 2009). Nevertheless, Volvox encodes two nearly identical isoforms of CRR1, one of which has short (~50 amino acid) N- and C-terminal extensions relative to Chlamydomonas CRR1, suggesting that the nutritional copper signaling pathway might be more complex. This might be necessitated by the occurrence of two different types of cells in Volvox each with its own function. Both forms have the C-terminal Cys-rich region, which has been implicated in zinc homeostasis in Chlamydomonas (Sommer et al., 2010).
CuREs
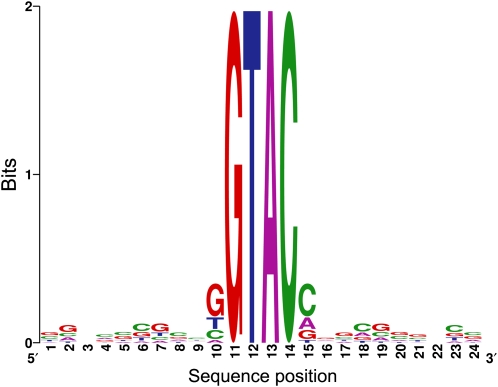
To assess whether the CRR1-dependent target genes might be direct targets of the DNA binding protein, we attempted to identify associated cis-acting sequence motifs in the 5′ flanking DNA. First, we adjusted gene models manually (see Supplemental Methods Section 4 online) based on coverage graphs and 454 ESTs to reflect the most 5′ end of the corresponding transcripts. Comparison of the sequences upstream of the CRR1 targets relative to the upstream regions of all gene models in the genome showed an enrichment of GTAC sequences, frequency of 1 in 156 versus 1 in 392. Since the SBP domain recognizes the GTAC core of the CuRE, it is likely that the set of 63 genes represents direct targets of CRR1 (Figure 8; see Supplemental Figure 10 online). In previous work, we tested each of the GTACs associated with the CYC6, CPX1, CRD1, and CTR1 genes and noted that only a subset was functional in activating transcription in copper-deficient cells (Quinn et al., 1999; Allen et al., 2008; Page et al., 2009). It is possible that the increased occurrence of GTACs (with perhaps weak affinity for the SBP domain) in the promoter regions might serve as a mechanism to concentrate the transcriptional activator in the vicinity of the promoter (see Supplemental Methods Section 4 online for details).
Figure 8.
Promoter Analysis of CRR1 Targets.
Motif-enrichment analysis of proximal promoters (~250 bp upstream of the transcription start site) of CRR1 targets shows overrepresentation of GTAC-core motifs (see Supplemental Methods online for details). The logo displays the sequence conservation 10 bp upstream and downstream of GATC sites.
[See online article for color version of this figure.]
DISCUSSION
Transcript Abundance
The power of RNA-Seq methodology for estimating transcript abundances lies in (1) the depth of coverage, which enables us to penetrate nearly every expressed gene; (2) the dynamic range of counting from 1 RPKM to >10,000 RPKM, which allows accurate regulation estimates as observed for the CYC6 gene, which is 214-fold upregulated in copper deficiency (Figure 3A); and (3) the specificity of the signal so that individual members of gene families can be readily distinguished. For example, even with short read sequences (33 to 50 nucleotides in this work), the TUA1 and TUA2 and TUB1 and TUB2 genes, which are 79% identical at the nucleotide level and hence difficult to discriminate by hybridization, are 60 and 54%, respectively, mappable in this work. When we incorporate digital tag sequencing into the experiment, we recover information about the orientation of the transcript (5′ to 3′) on the genome because the DGE protocol is strand specific. A comparison of technical replicates (individual lanes in a flow cell) or library types (single read versus paired end) and experimental replicates resulted in a very high degree of reproducibility. While we did not include standards for the determination of absolute transcript abundance, in previous work we estimated a copy number of 1 to 4 × 102 for the CYC6 transcript in copper-deficient cells (Hill and Merchant, 1992). On this basis, we can calculate that 6 to 24 RPKM is approximately equivalent to 1 mRNA molecule per cell. Given that the median transcript abundance is 5.6, this means that half of the genes have transcripts with copy numbers of only 1 per cell in a typical laboratory batch culture. The analysis pipeline simulation validated the significance of 2-fold changes in mRNA abundance for transcripts at or greater than the median (see Supplemental Figures 11 to 13 online).
Whole transcriptome analysis provides a picture of the relative contribution of individual metabolic pathways/processes in a cell. About 200 genes are very highly expressed in photoheterotrophically grown log-phase Chlamydomonas cells with mRNA abundances equivalent to >1000 RPKM. Among these are those encoding ribosomal proteins, a subset of the light-harvesting proteins, and components of the light reactions (see Supplemental Data Set 11 online). Interestingly, there are transcripts among the highly abundant set that have not been functionally characterized, such as 100620, 115491, and 135856 (encoding putative acetate transporters) and 184151 (a protein containing a Gly-rich RNA binding domain). In the case of the proteins of the photosynthetic apparatus, transcripts from 28 genes contribute 5% of the nucleus-encoded mRNA pool (see Supplemental Data Set 12 online). The promoters of the corresponding genes may be useful for ectopic/heterologous gene expression.
Tetrapyrrole Pathway
When we monitor individual pathways (Figure 6; see Supplemental Data Set 6 online), we can focus on each individual step in the pathway and the contribution of individual members of gene families. For instance, in the tetrapyrrole pathway, transcript abundance is in the range of 1 to 3 × 102 RPKM at each step. For some enzymes, one isoform contributes more to the mRNA pool than others (PBGD1, CPX1, and HMOX1), while for other enzymes, multiple isoforms seem to be equivalently expressed (UROD). This type of information is useful for the design and evaluation of reverse-genetic studies of individual members of a gene family. The impact of copper deficiency is evident at three of the oxygen-dependent steps in the tetrapyrrole pathway, those encoded by CPX1, CTH1/CRD1, and CAO. The physiological significance of this has not been determined, but loss of function of the –Cu isoform of the cyclase in the crd1 mutant results in a chlorotic phenotype in either copper-deficient cells or in O2-deficient cells, indicating the importance of this isoform in chlorophyll synthesis in these particular metabolic situations (Moseley et al., 2000; Quinn et al., 2002). The upregulation of CGL78 might be related to the changes in expression of tetrapyrrole pathway components.
Copper Proteins and Metal Transporters
The impact of copper nutrition on the abundance of transcripts encoding copper proteins and candidate metal transporters can be directly measured in our data. We searched the Chlamydomonas genome for known and potential copper binding proteins (see Supplemental Data Set 7 online). With some notable exceptions (PCY1, FOX1, and COX2A/B), most transcripts for copper binding proteins have low (1 RPKM) to median (5 RPKM) abundance, and none responds to copper deficiency. It is possible that copper-protein abundance is regulated, in most cases, at the level of protein accumulation as already documented for plastocyanin, ferroxidase, and cytochrome oxidase (Merchant and Bogorad, 1986b; La Fontaine et al., 2002) and as noted also in the proteomic analysis (see Supplemental Data Set 9 online). This is in contrast with the situation in Arabidopsis where reduced copper-protein abundance in copper-deficient cells occurs by miRNA-mediated downregulation of mRNAs encoding copper proteins (Abdel-Ghany and Pilon, 2008; Yamasaki et al., 2009). Based on transcript abundance, we note that three copper proteins, plastocyanin, ferroxidase, and cytochrome oxidase, constitute the bulk (~95%) of the copper-protein encoding mRNAs in the Chlamydomonas cell and, hence, the major contributors to the nutritional requirement for copper.
An inventory of the metal transporters in the genome indicated that most (10 of 10 copper transporters, 9 of 13 zinc transporters, 11 of 13 iron transporters, and 4 of 6 manganese transporters) are well expressed with abundances at or above the median (see Supplemental Data Set 8 online). Of the candidate copper transporters, only members of the CTR family (of which CTR1 and CTR2 are documented assimilatory transporters) are responsive to copper nutrition. IRT2 is also upregulated, dependent on CRR1, which suggests that it might facilitate iron uptake when ferroxidase function is compromised.
Transcriptome versus Proteome
Transcriptome analysis only allows us to measure the potential for modifying metabolic pathways and physiology, while protein levels provide more direct correlations with metabolic processes. Nevertheless, a large-scale proteome study suggested that for ~70% of the genes in Saccharomyces cerevisiae, the protein concentrations correlate with the RNA concentrations, indicating the importance of regulation through mRNA abundance (de Sousa Abreu et al., 2009). For copper-responsive gene expression involving CRR1, we are able to document a corresponding change in protein abundance for those mRNAs that are impacted by copper nutrition (Figure 7C; see Supplemental Data Set 9 online). Of the 63 CRR1 targets (overlap of comparisons B and C in Figure 2A), we could identify 12 targets in our subproteomic data, and we tested the abundance of another four by immunoblotting. In all cases, the increase or decrease in copper deficiency recapitulated the change in the increase or decrease of the corresponding RNA. Furthermore, in the case of fatty acid metabolism, we are able to document a physiological consequence of the change in gene expression. Specifically, the CRR1-dependent increase in FAB2 expression correlates with increased desaturation of chloroplast membrane lipids in copper-deficient cells (Figures 7A and 7B; see Supplemental Figure 9 online). We suspect that the immediate product of the FAB2 reaction does not accumulate because it is used as a substrate for additional desaturations to yield the dienoic and trienoic acids that are increased in copper-deficient cells. Previously, we showed that Chlamydomonas chloroplasts contain multiple ferredoxins, and we suggested that each has preferred substrates (Terauchi et al., 2009). On the basis of coexpression with nitrate assimilation genes, we suggested that FDX2 might be a substrate for nitrite reductase. In this work, we note coexpression of FDX5 with FAB2, and we therefore suggest that FDX5, whose abundance is increased in –Cu cells (Figure 7C; see Supplemental Figure 4 online), might be optimized for the acyl-ACP desaturase reaction (Jacobs et al., 2009; Terauchi et al., 2009).
Transcript Discovery and Coverage-Based Genome Annotation
For this and other Chlamydomonas projects, sequencing of the transcripts provides exon coverage information, which can be used to improve genome annotation and to distinguish alternate transcript forms (Figures 1 and 4). In this work, we adjusted a majority of the models for genes of interest. Most of the adjustments affected the length of the UTRs, which are quite long for Chlamydomonas (~35% of the transcript length), but a few impacted exon structure in the protein-coding regions (e.g., Figure 4A). Overall, for manually curated gene models, we increased transcript length by ~1 kb (Figure 4B). Although the DGE-TAG method is disadvantageous for genomes where the gene models do not have full-length UTRs (FM3.1 set for Chlamydomonas), it does provide strand specific transcription estimates and is useful for resolving overlapping transcripts.
The greater depth of sequence coverage in the transcript resequencing protocol (~12-fold average coverage per base of the exons) allowed us to identify more genes compared with the DGE-TAG method (see Supplemental Data Set 1 online) and nearly 50 times more than were found using microarrays in a previous study (Jamers et al., 2006). At the depth of coverage in this set of experiments, we can reliably quantitate mRNAs with as few as 1 copy per cell. When we used simulated data to compare our expression estimates with in silico libraries at much higher sequencing depth, we found that expression levels >7 to 10 RPKM are stable when sequencing depth is varied (see Discussion and Supplemental Methods Section 3.7 online). This validates the decision to use 10 RPKM as the cutoff in the definition of the nutritional copper transcriptome.
Biology of Copper Nutrition
Backups and Copper Sparing
Most of the significant changes in RNA abundance involve increases rather than decreases in gene expression in –Cu cells; about half of these are dependent on CRR1, indicating that they are primary responses to poor copper nutrition, and their pattern of expression is coordinate with that of sentinel genes like CYC6, CPX1, and CRD1 (e.g., Figure 3B). Given the role of copper as a cofactor involved in oxygen chemistry and redox reactions, it is appropriate that many of the CRR1 targets encode candidate redox enzymes. Since the repertoire of copper enzymes in Chlamydomonas is larger than anticipated in the pregenomic era, we expect that some of these targets might be new examples of backups, analogous to cytochrome c6 replacement of plastocyanin. AOF1 is certainly an obvious candidate for replacing one or more of the Chlamydomonas copper amine oxidases.
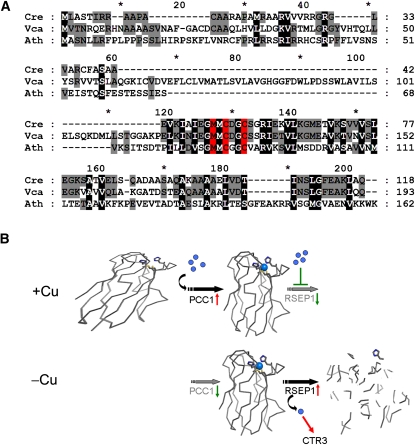
The copper-independent backups are likely to be important for copper sparing, which allows allocation of copper to processes that are absolutely dependent on copper. In parallel to upregulation of the backup proteins, the sparing phenomenon demands that the abundance of certain copper proteins be reduced. In the case of plastocyanin in Chlamydomonas, this occurs by regulated degradation, and this is likely the case for ferroxidase as well where the abundance of the protein (but not of the RNA) is lower in copper-deficient cells (Li and Merchant, 1995; La Fontaine et al., 2002; see Supplemental Data Set 9 online). Therefore, proteases that might be responsible for downregulation of copper-protein abundance are of interest. We discovered that one previously uncharacterized gene, now named RSEP1, encodes a protein related to a bacterial membrane metalloprotease (RseP in Escherichia coli). RSEP1 is tightly regulated (>20-fold) by copper nutrition and dependent on CRR1 (see Supplemental Data Set 3 online); in a time-course experiment, its mRNA is quickly reduced to basal levels within 30 min of copper supplementation (Figure 3B). Sequence analysis is consistent with thylakoid membrane localization. A related protein in Arabidopsis (albeit not an ortholog) is also organelle located (Bölter et al., 2006). RSEP1 is therefore a prime candidate for degrading plastocyanin in copper deficiency.
Another mechanism for changing copper allocation might be through the modification of copper distribution processes, which would occur through changes in the activity or expression of distributory copper transporters and copper chaperones. In this context, we note that the gene 142634, one of a few downregulated CRR1 targets, encodes a protein with a typical copper MxCxxC motif present in the copper chaperones and distributive copper transporters (Figure 9A) (Shikanai et al., 2003; Abdel-Ghany et al., 2005). Its downregulation might function to change intracellular copper flux, perhaps away from plastocyanin to make more copper available for cytochrome oxidase, an important copper enzyme in most aerobic cells (Figure 9B). Sequence analysis of the protein, which we have named PCC1 for plastid copper chaperone, is not incompatible with plastid localization. The protein is conserved also in Volvox (Figure 9A). By contrast, the mRNA encoding COX17 (an assembly factor required for copper incorporation into cytochrome oxidase; Glerum et al., 1996) is ~2-fold increased in copper deficiency (see Supplemental Data Set 4 online).
Figure 9.
Copper Status Determines the Fate of Plastocyanin.
(A) Sequence alignment of the PCC1 proteins from Chlamydomonas and V. carteri to the N-terminal domain of Arabidopsis PAA2 (GenBank AY297817). The putative copper binding motif (MxCxxC) is highlighted in red. Residues identical in all sequences are indicated by a black background, and conserved residues are indicated by a gray background.
(B) Our proposed pathway for changing the steady state abundance of plastocyanin by changing the expression of the hypothetical copper delivery protein PCC1 versus the putative protease RSEP1 as a function of copper nutrition (+Cu versus –Cu). Under copper-replete conditions (top), the expression of RSEP1 is repressed resulting in the accumulation of plastocyanin. Under copper-deficient conditions (bottom), the expression of PCC1 is repressed and RSEP1 actively degrades plastocyanin, thus liberating copper (blue spheres).
These two changes may serve as a mechanism for driving copper toward cytochrome oxidase over other copper proteins (Banci et al., 2010). However, we note that the change in COX17 RNA abundance is not CRR1 dependent and could be a response to other factors that control the synthesis of respiratory chain components. Indeed, a similar CRR1-independent increase in mRNA encoding COX15 (an assembly factor related to heme a synthesis) supports this idea (see Supplemental Data Set 3 online). An impact on the respiratory chain is also evident from CRR1-independent upregulation of AOX2 encoding an isoform of alternative oxidase. The increase in AOX2 mRNA is not recapitulated at the level of protein abundance nor by measurement of cyanide-insensitive respiration. It is possible that the AOX2 promoter simply responds to compromised cytochrome oxidase function in copper-deficient cells but posttranscriptional regulation mechanisms determine the abundance/activity of the enzyme.
The copper-containing glyoxal/galactose oxidases are ubiquitous eukaryotic enzymes, but their function is not known; therefore, we cannot evaluate the significance of their CRR1-dependent increase. Nevertheless, the fact that only a subset of GOX genes is upregulated might reflect a mechanism for increasing copper allocation to this subset. For instance, increased apoprotein production might provide a thermodynamic sink for copper.
Prolyl Hydroxylases
The Chlamydomonas genome encodes many prolyl hydroxylases, some are likely involved in the biosynthesis of cell wall proteins, but others may be involved in signaling or in modification of free Pro during amino acid metabolism (Schofield and Ratcliffe, 2004; Keskiaho et al., 2007). Five out of 17 genes appear to be responsive to copper nutrition, and four of these are CRR1 targets; 96954 and 166317 are physically linked with strong sequence identity (87% at the amino acid level except for divergent N termini) in both Chlamydomonas and Volvox (Table 1). Since we noticed a difference in the texture of copper-replete versus -deficient Chlamydomonas cells in culture, we speculate that some of the regulated prolyl-hydroxylases are responsible for changes in the structure of the extracellular matrix.
Oxygen Sensing
We also observed an overlap between some of the genes that are reported to be upregulated in a microarray study of anaerobic metabolism and genes that are upregulated in copper deficiency, including HYD1, HYDEF, HYDG, HCP2, HCP3, and PFR1 (Mus et al., 2007). However, none of these is a CRR1 target because the increase in transcript abundance is evident in the crr1 mutant as well (see Supplemental Data Set 3 and Supplemental Figure 4 online). One explanation for the upregulation of the anaerobic metabolic pathway is that O2 sensing might be compromised in copper deficiency because of a role for copper in the O2 sensor, an idea not inconsistent with the chemical properties of copper. Transition metals are well known for their compatibility for redox and O2 chemistry, and most of the well-characterized O2 sensors use redox-active or O2 binding prosthetic groups like heme or FeS centers (Beinert and Kiley, 1999; Rodgers, 1999; Swem et al., 2005; Aono, 2008).
Lipids
We showed that there is a highly specific modification of one set of lipids in copper-deficient Chlamydomonas cells, namely, the galactolipids in the thylakoid membrane (Figures 7A and 7B). This change is part of the copper deficiency program, since it is dependent on CRR1. Previous studies in both Chlamydomonas and Arabidopsis have documented the importance of fatty acid desaturation on galactolipids for structural integrity and function of the photosystem complexes and light harvesting in the photosystem II reaction center (Hugly et al., 1989; Sato et al., 1995, 1996; Vijayan and Browse, 2002). The change suggests that the physical properties of the thylakoid membrane from copper-deficient cells might be changed, perhaps to accommodate the use of cytochrome c6 as a mobile carrier between the b6f complex and photosystem I in place of plastocyanin. There is a single interaction site for c-type cytochromes with their substrates (electron donors and acceptors), whereas for plastocyanin there are two interaction sites, the hydrophobic north end and the negatively charged patch (Redinbo et al., 1994; Kerfeld et al., 1995). The demand for diffusion and mobility for each protein might be distinct.
METHODS
Strains and Cultures
Chlamydomonas reinhardtii strain CC-1021, the mutant crr1-2, and rescued crr1:CRR1 strains are available at the Chlamydomonas culture collection (Duke University). The rescued strain is more genetically similar to crr1 because it differs only at three loci: CRR1 and the site(s) of integration of the complementing gene and the pARG7.8 plasmid. The rescued strain has been well characterized along with several other independent transformants in two previous studies (Kropat et al., 2005; Sommer et al., 2010) as being wild-type for CRR1 function and nutritional copper homeostasis. Its properties are representative of those of other transformants and hence are not dependent on the sites of integration of the complementing CRR1 gene. We noted some strain-to-strain variation in the whole transcriptome of individual wild-type strains (Kropat et al., 2011), and a recent study documented considerable variation in various wild-type strain responses to nitrogen deprivation (Siaut et al., 2011).
Wild-type cultures were grown in Tris-acetate-phosphate (TAP) or minimal medium (referred to as TP) at 24°C with shaking (180 rpm) and under continuous light (~90 μmol m−2 s−1) (Harris, 1989). The mutant crr1-2 and rescued crr1:CRR1 strains were grown in TAP at 24°C with shaking (180 rpm) and under continuous light (~90 μmol m−2 s−1). Copper-deficient medium was prepared as described previously (Quinn and Merchant, 1998) and amended or not with the indicated amounts of CuEDTA. Inoculum cultures were maintained under copper-deficient conditions. TP was prepared by increasing the final concentration of phosphate (6.3 mM KH2PO4 and 5 mM KOH) 5-fold, Tris base 2.3-fold and omitting the acetate. The minimal medium cultures were bubbled constantly with air. Cells were collected at a density of 3 × 106 cell mL−1 (in exponential phase).
Nucleic Acid Analysis
Total RNA was prepared as described previously (Quinn and Merchant, 1998). RNA quality was assessed by an Agilent 2100 Bioanalyzer (electrophoresis and chromatographic separation on a microfluidics-based platform) and by hybridization as described previously (Quinn and Merchant, 1998). The probe used for detection, CBLP (also called RACK1), is a 915-bp EcoRI fragment from the cDNA cloned in pcf8-13 (Schloss, 1990).
DGE-NlaIII Tags and RNA-Seq
Total RNA was submitted to Illumina for Digital Gene Expression (21-nucleotide NlaIII-TAGs) or RNA-Seq (33- to 50-nucleotide reads). Over the course of the project, Illumina’s protocol was optimized from a beta test version (referred to as v1 in the data sets) to the present version (v2). In some experiments, the cDNA libraries were prepared for paired-end sequencing (35- to 50-nucleotide reads from each end, referred to as RNA-SeqPE in the data sets). The depth of sequencing is specified for each experiment in Supplemental Data Set 2 online. Sequence alignment, definition of mappable sets, disambiguation of multiple hits, quantitation of DGE-TAGs and RNA-Seq, imputation, normalization, and validation of the pipeline by simulation are described in the Supplemental Methods and Supplemental Figures 11 to 19 online.
Quantitative Real-Time PCR
The method for cDNA synthesis and real-time PCR was reported previously (Allen et al., 2007a). Primers are listed in Supplemental Table 1 online. Data are presented as the fold change in mRNA abundance relative to the copper-replete RNA sample and are normalized to the endogenous reference gene CBLP using the cycle threshold (CT) 2−ΔΔCT method. All experiments were performed in experimental replicates, and each sample was analyzed in technical triplicate. Real-time PCR procedures and analysis follow the MIQE guidelines (Bustin et al., 2009).
Metal Measurements
Chlamydomonas cells were collected as described previously (Allen et al., 2007b) with the exception that nitric acid was used to a final concentration of 2.4%. The metal content was analyzed on an Agilent 7500c quadrupole inductively coupled plasma-MS.
Ferredoxin5 Immunodetection
Proteins were separated on a nondenaturing polyacrylamide gel (15% monomer) and transferred in a semidry blotter onto a polyvinylidene difluoride membrane (0.45 mm; Millipore) for 40 min under constant current (4 W) in transfer buffer (25 mM Tris, 192 mM glycine, 0.0004% SDS [w/v], and 20% [v/v] methanol). The membrane was blocked overnight with 1% dried milk in PBS containing 0.3% (w/v) Tween 20 and 0.01% (w/v) sodium azide and incubated in primary antiserum; this solution was used as the diluent for both primary and secondary antibodies and for washing. The secondary antibody, used at 1:25,000, was goat anti-rabbit conjugated to horseradish peroxidase (Pierce Biotechnology). Bound antibody was detected using the Supersignal West Pico chemiluminescent substrate (Pierce Biotechnology).
Fatty Acid Analysis
Chlamydomonas strain 2137 and the mutant crr1-2 grown in TAP medium in the presence or absence of copper (6 × 10−6 M CuEDTA) were collected at a cell density of 3 × 106 cell mL−1. Lipids were extracted from lyophilized cell pellets using 1.5 mL chloroform:methanol (2:1 [v/v]). After centrifugation, the supernatant was transferred to a new tube, and the remaining pellet was reextracted with 1 mL chloroform:methanol as before. Extracts were pooled and phase separation induced with 0.9% KCl to yield a final ratio of chloroform:methanol:0.9% KCl (8:4:3 [v/v/v]) (Folch et al., 1957). After centrifugation, the upper water phase was discarded and the remaining lipid phase washed with 0.2 volumes of 0.9% KCl and phases were separated by centrifugation. The lower chloroform phase was transferred to a glass vial, evaporated under a nitrogen gas stream, and stored in airtight glass vials at −20°C until further use. Fatty acid content and composition of the extracts was determined by a gas chromatograph with flame ionization detector (GC-FID) analysis of fatty acid methyl esters as described previously (Rossak et al., 1997). The GC was equipped with a capillary DB-23 column (Agilent) using an initial temperature of 140°C, which was increased by 25°C/min to 160°C, followed by 8°C/min to 250°C and held at that temperature for 4 min. For the analysis of membrane lipids, an equivalent of 100 μg of fatty acid of the lipid extracts were developed on Si60 glass backed plates (EMD Chemicals) to a height of 7 cm with chloroform:methanol:acetic acid:water (75:13:9:3 [v/v/v/v]). After brief visualization in iodine vapor, the respective bands were scraped off the plate and transferred to a glass vial for the transesterification reaction and GC-FID analysis as described above.
Quantitative Proteome Analysis
Chlamydomonas (strain 2137) was grown in TAP medium in the presence or absence of 6 μM CuEDTA in triplicate cultures. Algal cells were collected at a density of ~6 × 106 cells mL−1 by centrifugation at 3440g for 5 min at 4°C. Cells were washed with 10 mM sodium phosphate, pH 7, collected by centrifugation, and resuspended in 10 mM sodium phosphate, pH 7.0, to a chlorophyll content of 1 mg mL−1 as measured according to Porra et al. (1989). Two freeze-thaw cycles at −80°C were performed to break open the cells. The insoluble material was removed after centrifugation at 16,000g for 10 min at 4°C followed by centrifugation at 253,000g for 20 min at 4°C. Proteins (30 μg of protein per lane) were separated by one-dimensional gel electrophoresis on 4 to 12% NuPage Bis-Tris gels (Invitrogen) and visualized by staining with Coomassie Brilliant Blue (Bio-Rad). Each gel lane was completely divided into bands of ~3 mm width. Each excised band was subjected to in-gel trypsin digestion (sequencing grade modified trypsin; Promega). Peptides were extracted into 50/50 water/acetonitrile containing 2.5% formic acid, lyophilized and resuspended into a 1% formic acid solution containing 25 fmol μL−1 of BSA trypsin digest and quantification standard (Waters). The peptides were analyzed using an HPLC system (nanoAcquity UltraPerformance UPLC) coupled to a Waters Xevo QToF (quadrupole time-of-flight) mass spectrometer. Peptides were eluted from the HPLC to the electrospray ionization mass spectrometer using a 5-μm Symmetry C18 180 μm × 20-mm reversed-phase trap column in line with a 1.7-μm BEH130, 75 μm × 100-mm reversed-phase C18 analytical column (Waters). The gradient used was 60 min 3% acetonitrile/0.1% formic acid to 40% acetonitrile/0.1% formic acid at a flow rate of 0.3 μL min−1. [Glu1]-Fibrinopeptide B was used as a mass calibration standard (100 fmol/μL) that was infused via a separate electrospray ionization sprayer, and the standard peptide was measured every minute of the chromatogram.
The LC mass spectrometer was operated in the MSE data-independent acquisition mode (Silva et al., 2006a, 2006b; Geromanos et al., 2009) in which no ion transmission window is applied with the first mass analyzer prior to collisionally activated dissociation. Alternating the energy applied to the collision cell between low energy (6 eV) and elevated energy (ramped from 15 to 40 eV) on a scan-to-scan basis (m/z 50 to 2000) provides high resolution (full width at half height resolution of >10,000) of precursor and associated product ion mass spectra from every ion above the limit of detection of the instrument. A nearly 100% duty cycle of data acquisition is achieved in this manner. Correlation of product ions to precursor ions is achieved using reconstructed retention time (i.e., alignment of LC retention times of the precursors and products) and chromatographic peak shapes. At least two injections from each gel band were analyzed by LC-MS.
Protein Lynx Global Server (PLGS version 2.4) was used to determine protein identification and quantification. Quantification of protein levels was achieved by the addition of an internal protein standard (BSA trypsin digest standard) to which the data set is normalized. The PLGS quantification package uses the concept that the average MS signal response for the three most intense tryptic peptides per mole of protein is a constant (Silva et al., 2006a, 2006b).
To account for differences between various gene model sets and genome assemblies, the MS data were searched against three different sets of gene models. For each of the two assemblies of the genome, there are multiple gene model predictions and what is referred to as a catalog track where the best model is chosen at each locus. The gene models catalog for the version 3 assembly is called FM3.1 or JGI3.1. For the version 4 assembly, there is the JGI catalog called FM4 as well as a new set of models called Au10.2. We searched against (1) a JGI v4 protein database (http://genomeportal.jgi-psf.org/Chlre4/download/Chlre4_best_proteins.fasta.gz); (2) a data set from FM3.1 (ftp://ftp.jgi-psf.org/pub/JGI_data/ Chlamy/v3.0/proteins.Chlre3.fasta.gz), which was modified by replacement of updated gene models from Supplemental Data Sets 3, 7, and 8 online in this work; and (3) the Augustus 10.2 protein database (http://www.phytozome.net/chlamy). All databases were further supplemented with sequences for keratin, trypsin, BSA, and chloroplast and mitochondrial proteins from the National Center for Biotechnology Information. The MS data files were searched using the PLGS algorithm that includes precursor and product ion tolerance, minimum number of peptide matches (1), minimum number of product ions per peptide (2), minimum number of product ions per protein (2), maximum number of missed tryptic cleavage sites (2), and maximum false positive rate (4%) (Li et al., 2009). All peptide matches obtained under the 4% maximum false positive rate were taken as correct assignments. Searches were limited to trypsin proteolysis fragments and peptide precursors, and product ion mass tolerances were set to 20 and 40 ppm, respectively. Carbamidomethylation of Cys residues was set as a fixed modification, and Met oxidation, Asn and Gln deamidation, and N-terminal acetylation were set as variable modifications. A protein was considered identified if it was detected using any of these databases. Also, in cases where related proteins had shared peptides, ions were manually assigned and quantification was determined using the three most abundant shared peptides.
Accession Numbers
All sequencing data have been deposited at the Gene Expression Omnibus database http://www.ncbi.nlm.nih.gov/geo/ with the accession number GSE25124. Other accession numbers from this study are as follows: Chlamydomonas CTH1 locus (AF337037 and AF426027); protein sequences used for alignment were Chlamydomonas PCC1 (protein ID 142634, http://genome.jgi-psf.org/Chlre3/Chlre3.home.html), Volvox carteri PCC1 (protein ID 90457; http://genome.jgi-psf.org/Volca1/Volca1.home.html), Arabidopsis PAA2 (AY297817), Chlamydomonas CGL78 (protein ID 162021; http://genome.jgi-psf.org/Chlre3/Chlre3.home.html) and its homologs from Arabidopsis thaliana (AT5G58250), Oryza sativa (Os03g21370), Physcomitrella patens ssp patens (PHYPADRAFT_121744), and Thermosynechococcus elongatus BP-1 (tll1499). The accession number for CBLP is X53574.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Graphical Alignment Results of DGE Data.
Supplemental Figure 2. Reproducibility and Differential Expression Analysis for DGE Data.
Supplemental Figure 3. Comparison of Fold Difference in RNA Abundance between Copper-Replete versus Copper-Deficient Cells Estimated from DGE-TAG versus Real-Time PCR Experiments.
Supplemental Figure 4. Change in Protein Abundance for Hydrogenase and Acyl-ACP Desaturase Parallels mRNA Abundance.
Supplemental Figure 5. Representative Lanes from Gel Electrophoresis.
Supplemental Figure 6. Reproducibility of Proteomics Data.
Supplemental Figure 7. Amino Acid Alignment of Chlamydomonas reinhardtii CGL78 to Homologs from Arabidopsis thaliana, Oryza sativa, Physcomitrella patens ssp patens, and Thermosynechococcus elongatus BP-1.
Supplemental Figure 8. Fatty Acid Content of crr1 Cells under Copper-Replete and -Deficient Conditions.
Supplemental Figure 9. Fatty Acid Composition of MGDG and DGTS for CRR1 Cells under Copper-Replete and -Deficient Conditions.
Supplemental Figure 10. Promoter Analysis of CRR1 Targets.
Supplemental Figure 11. Metaplot of RNA-Seq Coverage for Different Types of Libraries.
Supplemental Figure 12. Alignment of DGE Tags.
Supplemental Figure 13. Single-End RNA-Seq Simulated Library.
Supplemental Figure 14. Chlamydomonas Transcriptome Mappability for Different Read Lengths.
Supplemental Figure 15. Percentage of Trimming Hits as a Function of Average Exon Length for a Simulated Library of 35mers.
Supplemental Figure 16. Histogram of Fold Changes for a Simulated Library of 35mers.
Supplemental Figure 17. Allocation of Ambiguous Hits.
Supplemental Figure 18. Definition of Mappable Sets.
Supplemental Figure 19. Additional Corrections for the Estimation of the Transcript Relative Abundance.
Supplemental Table 1. Primers Used for Real-Time PCR.
Supplemental Table 2. Alignment Statistics of DGE Libraries.
Supplemental Data Set 1. Copper Deficiency–Responsive Genes Identified via DGE-TAGs.
Supplemental Data Set 2. Summary of RNA-Seq Statistics and Expression Estimates after Allocation of Ambiguous Hits.
Supplemental Data Set 3. RNA-Seq Identifies Copper Deficiency and CRR1 Targets under Photoheterotrophic Conditions.
Supplemental Data Set 4. RNA-Seq Identifies Copper Deficiency Targets in Photoautotrophically Grown Cells.
Supplemental Data Set 5. Identification of Copper Deficiency Targets via Alignment of Sequences to FM4 and Augustus 10.2.
Supplemental Data Set 6. Transcript Abundances for Genes Encoding Tetrapyrrole Biosynthesis Enzymes.
Supplemental Data Set 7. Transcript Abundance for Genes Encoding Copper-Containing Proteins and Copper Chaperones.
Supplemental Data Set 8. Transcript Abundance for Genes Encoding Metal Transporters.
Supplemental Data Set 9. Summary of Protein Abundance from MSE Analysis.
Supplemental Data Set 10. Orthologs of Chlamydomonas Copper Deficiency–Responsive Genes in V. carteri.
Supplemental Data Set 11. Most Abundant Transcripts in Chlamydomonas.
Supplemental Data Set 12. Transcript Abundance for Nucleus-Encoded Genes for Chlamydomonas Proteins of the Photosynthetic Apparatus.
Acknowledgments
We thank Thomas Happe for the ferredoxin5 antibody, John Shanklin for the acyl-ACP desaturase antibody, Peter Nixon for the hydrogenase antibody, Joan Valentine for use of the Agilent 7500ce, Deborah R. Francoleon for advice on proteomics, and Philo Juang for programming advice. This work was supported by the National Institutes of Health (GM42143), the UCLA-Department of Energy Institute of Genomics and Proteomics (DE-FC03-02ER63421), and U.S. Air Force Office of Scientific Research Grant FA9550-08-0165 (C.B. and A.V.). M.C. acknowledges support from Ruth L. Kirschstein National Research Service Award (F32 GM086006) and D.C. from the Spanish Foundation of Science and Technology (FECYT) and a California Nanosystems Institute (UCLA) grant. S.I.H. and S.J.K. acknowledge support from the Ruth L. Kirschstein National Research Service Award (GM07185).
References
- Abdel-Ghany S.E., Müller-Moulé P., Niyogi K.K., Pilon M., Shikanai T. (2005). Two P-type ATPases are required for copper delivery in Arabidopsis thaliana chloroplasts. Plant Cell 17: 1233–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdel-Ghany S.E., Pilon M. (2008). MicroRNA-mediated systemic down-regulation of copper protein expression in response to low copper availability in Arabidopsis. J. Biol. Chem. 283: 15932–15945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen M.D., del Campo J.A., Kropat J., Merchant S.S. (2007a). FEA1, FEA2, and FRE1, encoding two homologous secreted proteins and a candidate ferrireductase, are expressed coordinately with FOX1 and FTR1 in iron-deficient Chlamydomonas reinhardtii. Eukaryot. Cell 6: 1841–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen M.D., Kropat J., Merchant S.S. (2008). Regulation and localization of isoforms of the aerobic oxidative cyclase in Chlamydomonas reinhardtii. Photochem. Photobiol. 84: 1336–1342 [DOI] [PubMed] [Google Scholar]
- Allen M.D., Kropat J., Tottey S., Del Campo J.A., Merchant S.S. (2007b). Manganese deficiency in Chlamydomonas results in loss of photosystem II and MnSOD function, sensitivity to peroxides, and secondary phosphorus and iron deficiency. Plant Physiol. 143: 263–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aono S. (2008). Metal-containing sensor proteins sensing diatomic gas molecules. Dalton Trans. 28: 3137–3146 [DOI] [PubMed] [Google Scholar]
- Banci L., Bertini I., Ciofi-Baffoni S., Kozyreva T., Zovo K., Palumaa P. (2010). Affinity gradients drive copper to cellular destinations. Nature 465: 645–648 [DOI] [PubMed] [Google Scholar]
- Bantscheff M., Schirle M., Sweetman G., Rick J., Kuster B. (2007). Quantitative mass spectrometry in proteomics: A critical review. Anal. Bioanal. Chem. 389: 1017–1031 [DOI] [PubMed] [Google Scholar]
- Beinert H., Kiley P.J. (1999). Fe-S proteins in sensing and regulatory functions. Curr. Opin. Chem. Biol. 3: 152–157 [DOI] [PubMed] [Google Scholar]
- Blackburn K., Mbeunkui F., Mitra S.K., Mentzel T., Goshe M.B. (2010). Improving protein and proteome coverage through data-independent multiplexed peptide fragmentation. J. Proteome Res. 9: 3621–3637 [DOI] [PubMed] [Google Scholar]
- Bölter B., Nada A., Fulgosi H., Soll J. (2006). A chloroplastic inner envelope membrane protease is essential for plant development. FEBS Lett. 580: 789–794 [DOI] [PubMed] [Google Scholar]
- Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L., Vandesompele J., Wittwer C.T. (2009). The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55: 611–622 [DOI] [PubMed] [Google Scholar]
- Chen J.C., Hsieh S.I., Kropat J., Merchant S.S. (2008). A ferroxidase encoded by FOX1 contributes to iron assimilation under conditions of poor iron nutrition in Chlamydomonas. Eukaryot. Cell 7: 541–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danks D.M. (1988). Copper deficiency in humans. Annu. Rev. Nutr. 8: 235–257 [DOI] [PubMed] [Google Scholar]
- de Sousa Abreu R., Penalva L.O., Marcotte E.M., Vogel C. (2009). Global signatures of protein and mRNA expression levels. Mol. Biosyst. 5: 1512–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson M., Moseley J.L., Tottey S., Del Campo J.A., Quinn J.M., Kim Y., Merchant S. (2004). Genetic dissection of nutritional copper signaling in Chlamydomonas distinguishes regulatory and target genes. Genetics 168: 795–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folch J., Lees M., Sloane Stanley G.H. (1957). A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226: 497–509 [PubMed] [Google Scholar]
- Geromanos S.J., Vissers J.P., Silva J.C., Dorschel C.A., Li G.Z., Gorenstein M.V., Bateman R.H., Langridge J.I. (2009). The detection, correlation, and comparison of peptide precursor and product ions from data independent LC-MS with data dependant LC-MS/MS. Proteomics 9: 1683–1695 [DOI] [PubMed] [Google Scholar]
- Ghirardi M.L., Posewitz M.C., Maness P.C., Dubini A., Yu J., Seibert M. (2007). Hydrogenases and hydrogen photoproduction in oxygenic photosynthetic organisms. Annu. Rev. Plant Biol. 58: 71–91 [DOI] [PubMed] [Google Scholar]
- Glerum D.M., Shtanko A., Tzagoloff A. (1996). Characterization of COX17, a yeast gene involved in copper metabolism and assembly of cytochrome oxidase. J. Biol. Chem. 271: 14504–14509 [DOI] [PubMed] [Google Scholar]
- Grossman A.R., Croft M., Gladyshev V.N., Merchant S.S., Posewitz M.C., Prochnik S., Spalding M.H. (2007). Novel metabolism in Chlamydomonas through the lens of genomics. Curr. Opin. Plant Biol. 10: 190–198 [DOI] [PubMed] [Google Scholar]
- Hanikenne M., Merchant S.S., Hamel P.P. (2009). Transition metal nutrition: A balance between deficiency and toxicity. In The Chlamydomonas Sourcebook, D. Stern, ed (San Diego, CA: Academic Press), pp. 333–400. [Google Scholar]
- Harris E.H. (1989). The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use. (Academic Press, San Diego, CA: ). [DOI] [PubMed] [Google Scholar]
- Haywood S., Müller T., Müller W., Heinz-Erian P., Tanner M.S., Ross G. (2001). Copper-associated liver disease in North Ronaldsay sheep: A possible animal model for non-Wilsonian hepatic copper toxicosis of infancy and childhood. J. Pathol. 195: 264–269 [DOI] [PubMed] [Google Scholar]
- Hill K.L., Li H.H., Singer J., Merchant S. (1991). Isolation and structural characterization of the Chlamydomonas reinhardtii gene for cytochrome c6. Analysis of the kinetics and metal specificity of its copper-responsive expression. J. Biol. Chem. 266: 15060–15067 [PubMed] [Google Scholar]
- Hill K.L., Merchant S. (1992). In vivo competition between plastocyanin and a copper-dependent regulator of the Chlamydomonas reinhardtii cytochrome c6 gene. Plant Physiol. 100: 319–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill K.L., Merchant S. (1995). Coordinate expression of coproporphyrinogen oxidase and cytochrome c6 in the green alga Chlamydomonas reinhardtii in response to changes in copper availability. EMBO J. 14: 857–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugly S., Kunst L., Browse J., Somerville C. (1989). Enhanced thermal tolerance of photosynthesis and altered chloroplast ultrastructure in a mutant of Arabidopsis deficient in lipid desaturation. Plant Physiol. 90: 1134–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs J., Pudollek S., Hemschemeier A., Happe T. (2009). A novel, anaerobically induced ferredoxin in Chlamydomonas reinhardtii. FEBS Lett. 583: 325–329 [DOI] [PubMed] [Google Scholar]
- Jamers A., Van der Ven K., Moens L., Robbens J., Potters G., Guisez Y., Blust R., De Coen W. (2006). Effect of copper exposure on gene expression profiles in Chlamydomonas reinhardtii based on microarray analysis. Aquat. Toxicol. 80: 249–260 [DOI] [PubMed] [Google Scholar]
- Kerfeld C.A., Anwar H.P., Interrante R., Merchant S., Yeates T.O. (1995). The structure of chloroplast cytochrome c6 at 1.9 A resolution: Evidence for functional oligomerization. J. Mol. Biol. 250: 627–647 [DOI] [PubMed] [Google Scholar]
- Keskiaho K., Hieta R., Sormunen R., Myllyharju J. (2007). Chlamydomonas reinhardtii has multiple prolyl 4-hydroxylases, one of which is essential for proper cell wall assembly. Plant Cell 19: 256–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropat J., Hong-Hermesdorf A., Casero D., Ent P., Castruita M., Pellegrini M., Merchant S.S., Malasarn D. (2011). A revised mineral nutrient supplement increases biomass and growth rate in Chlamydomonas reinhardtii. Plant J., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropat J., Tottey S., Birkenbihl R.P., Depège N., Huijser P., Merchant S. (2005). A regulator of nutritional copper signaling in Chlamydomonas is an SBP domain protein that recognizes the GTAC core of copper response element. Proc. Natl. Acad. Sci. USA 102: 18730–18735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbé S., Thiele D.J. (1999). Pipes and wiring: The regulation of copper uptake and distribution in yeast. Trends Microbiol. 7: 500–505 [DOI] [PubMed] [Google Scholar]
- La Fontaine S., Quinn J.M., Nakamoto S.S., Page M.D., Göhre V., Moseley J.L., Kropat J., Merchant S. (2002). Copper-dependent iron assimilation pathway in the model photosynthetic eukaryote Chlamydomonas reinhardtii. Eukaryot. Cell 1: 736–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G.Z., Vissers J.P., Silva J.C., Golick D., Gorenstein M.V., Geromanos S.J. (2009). Database searching and accounting of multiplexed precursor and product ion spectra from the data independent analysis of simple and complex peptide mixtures. Proteomics 9: 1696–1719 [DOI] [PubMed] [Google Scholar]
- Li H.H., Merchant S. (1995). Degradation of plastocyanin in copper-deficient Chlamydomonas reinhardtii. Evidence for a protease-susceptible conformation of the apoprotein and regulated proteolysis. J. Biol. Chem. 270: 23504–23510 [DOI] [PubMed] [Google Scholar]
- Li H.H., Quinn J., Culler D., Girard-Bascou J., Merchant S. (1996). Molecular genetic analysis of plastocyanin biosynthesis in Chlamydomonas reinhardtii. J. Biol. Chem. 271: 31283–31289 [DOI] [PubMed] [Google Scholar]
- Li R., Li Y., Kristiansen K., Wang J. (2008). SOAP: Short oligonucleotide alignment program. Bioinformatics 24: 713–714 [DOI] [PubMed] [Google Scholar]
- Lohr M., Im C.S., Grossman A.R. (2005). Genome-based examination of chlorophyll and carotenoid biosynthesis in Chlamydomonas reinhardtii. Plant Physiol. 138: 490–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer J.F.B. (2001). The molecular basis of copper-transport diseases. Trends Mol. Med. 7: 64–69 [DOI] [PubMed] [Google Scholar]
- Merchant S., Bogorad L. (1986a). Regulation by copper of the expression of plastocyanin and cytochrome c552 in Chlamydomonas reinhardi. Mol. Cell. Biol. 6: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant S., Bogorad L. (1986b). Rapid degradation of apoplastocyanin in Cu(II)-deficient cells of Chlamydomonas reinhardtii. J. Biol. Chem. 261: 15850–15853 [PubMed] [Google Scholar]
- Merchant S., Bogorad L. (1987a). Metal ion regulated gene expression: use of a plastocyanin-less mutant of Chlamydomonas reinhardtii to study the Cu(II)-dependent expression of cytochrome c-552. EMBO J. 6: 2531–2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant S., Bogorad L. (1987b). The Cu(II)-repressible plastidic cytochrome c. Cloning and sequence of a complementary DNA for the pre-apoprotein. J. Biol. Chem. 262: 9062–9067 [PubMed] [Google Scholar]
- Merchant S.S., Allen M.D., Kropat J., Moseley J.L., Long J.C., Tottey S., Terauchi A.M. (2006). Between a rock and a hard place: Trace element nutrition in Chlamydomonas. Biochim. Biophys. Acta 1763: 578–594 [DOI] [PubMed] [Google Scholar]
- Merchant S.S., et al. (2007). The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Moseley J., Quinn J., Eriksson M., Merchant S. (2000). The Crd1 gene encodes a putative di-iron enzyme required for photosystem I accumulation in copper deficiency and hypoxia in Chlamydomonas reinhardtii. EMBO J. 19: 2139–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley J.L., Page M.D., Alder N.P., Eriksson M., Quinn J., Soto F., Theg S.M., Hippler M., Merchant S. (2002). Reciprocal expression of two candidate di-iron enzymes affecting photosystem I and light-harvesting complex accumulation. Plant Cell 14: 673–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mus F., Dubini A., Seibert M., Posewitz M.C., Grossman A.R. (2007). Anaerobic acclimation in Chlamydomonas reinhardtii: Anoxic gene expression, hydrogenase induction, and metabolic pathways. J. Biol. Chem. 282: 25475–25486 [DOI] [PubMed] [Google Scholar]
- Nagalakshmi U., Wang Z., Waern K., Shou C., Raha D., Gerstein M., Snyder M. (2008). The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320: 1344–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page M.D., Kropat J., Hamel P.P., Merchant S.S. (2009). Two Chlamydomonas CTR copper transporters with a novel cys-met motif are localized to the plasma membrane and function in copper assimilation. Plant Cell 21: 928–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre J.L., Fontecave M., Crichton R.R. (2002). Chemistry for an essential biological process: the reduction of ferric iron. Biometals 15: 341–346 [DOI] [PubMed] [Google Scholar]
- Pilon M., Cohu C.M., Ravet K., Abdel-Ghany S.E., Gaymard F. (2009). Essential transition metal homeostasis in plants. Curr. Opin. Plant Biol. 12: 347–357 [DOI] [PubMed] [Google Scholar]
- Porra R.J., Thompson W.A., Kriedemann P.E. (1989). Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophyll a and chlorophyll b extracted with 4 different solvents - verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 975: 384–394 [Google Scholar]
- Quinn J.M., Eriksson M., Moseley J.L., Merchant S. (2002). Oxygen deficiency responsive gene expression in Chlamydomonas reinhardtii through a copper-sensing signal transduction pathway. Plant Physiol. 128: 463–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn J.M., Merchant S. (1995). Two copper-responsive elements associated with the Chlamydomonas Cyc6 gene function as targets for transcriptional activators. Plant Cell 7: 623–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn J.M., Merchant S. (1998). Copper-responsive gene expression during adaptation to copper deficiency. Methods Enzymol. 297: 263–279 [DOI] [PubMed] [Google Scholar]
- Quinn J.M., Merchant S. (1999). Adaptation of Scenedesmus obliquus (Chlorophyceae) to copper-deficiency: Transcriptional regulation of Pcy1 but not Cpx1. J. Phycol. 35: 1253–1263 [Google Scholar]
- Quinn J.M., Nakamoto S.S., Merchant S. (1999). Induction of coproporphyrinogen oxidase in Chlamydomonas chloroplasts occurs via transcriptional regulation of Cpx1 mediated by copper response elements and increased translation from a copper deficiency-specific form of the transcript. J. Biol. Chem. 274: 14444–14454 [DOI] [PubMed] [Google Scholar]
- Redinbo M.R., Yeates T.O., Merchant S. (1994). Plastocyanin: Structural and functional analysis. J. Bioenerg. Biomembr. 26: 49–66 [DOI] [PubMed] [Google Scholar]
- Riekhof W.R., Sears B.B., Benning C. (2005). Annotation of genes involved in glycerolipid biosynthesis in Chlamydomonas reinhardtii: Discovery of the betaine lipid synthase BTA1Cr. Eukaryot. Cell 4: 242–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers K.R. (1999). Heme-based sensors in biological systems. Curr. Opin. Chem. Biol. 3: 158–167 [DOI] [PubMed] [Google Scholar]
- Rossak M., Schäfer A., Xu N., Gage D.A., Benning C. (1997). Accumulation of sulfoquinovosyl-1-O-dihydroxyacetone in a sulfolipid-deficient mutant of Rhodobacter sphaeroides inactivated in sqdC. Arch. Biochem. Biophys. 340: 219–230 [DOI] [PubMed] [Google Scholar]
- Sandmann G., Reck H., Kessler E., Boger P. (1983). Distribution of plastocyanin and soluble plastidic cytochrome c in various classes of algae. Arch. Microbiol. 134: 23–27 [Google Scholar]
- Sato N., Sonoike K., Tsuzuki M., Kawaguchi A. (1996). Photosynthetic characteristics of a mutant of Chlamydomonas reinhardtii impaired in fatty acid desaturation in chloroplasts. Biochim. Biophys. 1274: 112–118 [Google Scholar]
- Sato N., Tsuzuki M., Matsuda Y., Ehara T., Osafune T., Kawaguchi A. (1995). Isolation and characterization of mutants affected in lipid metabolism of Chlamydomonas reinhardtii. Eur. J. Biochem. 230: 987–993 [DOI] [PubMed] [Google Scholar]
- Schaefer M., Gitlin J.D. (1999). Genetic disorders of membrane transport. IV. Wilson’s disease and Menkes disease. Am. J. Physiol. 276: G311–G314 [DOI] [PubMed] [Google Scholar]
- Schloss J.A. (1990). A Chlamydomonas gene encodes a G protein beta subunit-like polypeptide. Mol. Gen. Genet. 221: 443–452 [DOI] [PubMed] [Google Scholar]
- Schmid M., Davison T.S., Henz S.R., Pape U.J., Demar M., Vingron M., Schölkopf B., Weigel D., Lohmann J.U. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Schofield C.J., Ratcliffe P.J. (2004). Oxygen sensing by HIF hydroxylases. Nat. Rev. Mol. Cell Biol. 5: 343–354 [DOI] [PubMed] [Google Scholar]
- Shendure J. (2008). The beginning of the end for microarrays? Nat. Methods 5: 585–587 [DOI] [PubMed] [Google Scholar]
- Shikanai T., Müller-Moulé P., Munekage Y., Niyogi K.K., Pilon M. (2003). PAA1, a P-type ATPase of Arabidopsis, functions in copper transport in chloroplasts. Plant Cell 15: 1333–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siaut M., Cuiné S., Cagnon C., Fessler B., Nguyen M., Carrier P., Beyly A., Beisson F., Triantaphylidès C., Li-Beisson Y., Peltier G. (2011). Oil accumulation in the model green alga Chlamydomonas reinhardtii: characterization, variability between common laboratory strains and relationship with starch reserves. BMC Biotechnol. 11: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J.C., Denny R., Dorschel C., Gorenstein M.V., Li G.Z., Richardson K., Wall D., Geromanos S.J. (2006b). Simultaneous qualitative and quantitative analysis of the Escherichia coli proteome: A sweet tale. Mol. Cell. Proteomics 5: 589–607 [DOI] [PubMed] [Google Scholar]
- Silva J.C., Gorenstein M.V., Li G.Z., Vissers J.P., Geromanos S.J. (2006a). Absolute quantification of proteins by LCMSE: A virtue of parallel MS acquisition. Mol. Cell. Proteomics 5: 144–156 [DOI] [PubMed] [Google Scholar]
- Sommer F., Kropat J., Malasarn D., Grossoehme N.E., Chen X., Giedroc D.P., Merchant S.S. (2010). Two metal-responsive domains in the nutritional copper senson in Chlamydomonas. Plant Cell 12: 4098–4113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M., Diekhans M., Baertsch R., Haussler D. (2008). Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 24: 637–644 [DOI] [PubMed] [Google Scholar]
- Swem D.L., Swem L.R., Setterdahl A., Bauer C.E. (2005). Involvement of SenC in assembly of cytochrome c oxidase in Rhodobacter capsulatus. J. Bacteriol. 187: 8081–8087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka R., Tanaka A. (2007). Tetrapyrrole biosynthesis in higher plants. Annu. Rev. Plant Biol. 58: 321–346 [DOI] [PubMed] [Google Scholar]
- Terauchi A.M., Lu S.F., Zaffagnini M., Tappa S., Hirasawa M., Tripathy J.N., Knaff D.B., Farmer P.J., Lemaire S.D., Hase T., Merchant S.S. (2009). Pattern of expression and substrate specificity of chloroplast ferredoxins from Chlamydomonas reinhardtii. J. Biol. Chem. 284: 25867–25878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terauchi A.M., Peers G., Kobayashi M.C., Niyogi K.K., Merchant S.S. (2010). Trophic status of Chlamydomonas reinhardtii influences the impact of iron deficiency on photosynthesis. Photosynth. Res. 105: 39–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ’t Hoen P.A., Ariyurek Y., Thygesen H.H., Vreugdenhil E., Vossen R.H., de Menezes R.X., Boer J.M., van Ommen G.J., den Dunnen J.T. (2008). Deep sequencing-based expression analysis shows major advances in robustness, resolution and inter-lab portability over five microarray platforms. Nucleic Acids Res. 36: e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusnády G.E., Simon I. (2001). The HMMTOP transmembrane topology prediction server. Bioinformatics 17: 849–850 [DOI] [PubMed] [Google Scholar]
- Vijayan P., Browse J. (2002). Photoinhibition in mutants of Arabidopsis deficient in thylakoid unsaturation. Plant Physiol. 129: 876–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki H., Hayashi M., Fukazawa M., Kobayashi Y., Shikanai T. (2009). SQUAMOSA Promoter Binding Protein-Like7 is a central regulator for copper homeostasis in Arabidopsis. Plant Cell 21: 347–361 [DOI] [PMC free article] [PubMed] [Google Scholar]