HMG-CoA reductase has a key role in the regulation of the mevalonate pathway for isoprenoid biosynthesis and is modulated by many diverse endogenous and environmental stimuli. In this work, protein phosphatase 2A emerges as a positive and negative multilevel regulator of plant HMG-CoA reductase during normal development and in response to a variety of stress conditions.
Abstract
Plants synthesize a myriad of isoprenoid products that are required both for essential constitutive processes and for adaptive responses to the environment. The enzyme 3-hydroxy-3-methylglutaryl-CoA reductase (HMGR) catalyzes a key regulatory step of the mevalonate pathway for isoprenoid biosynthesis and is modulated by many endogenous and external stimuli. In spite of that, no protein factor interacting with and regulating plant HMGR in vivo has been described so far. Here, we report the identification of two B′′ regulatory subunits of protein phosphatase 2A (PP2A), designated B′′α and B′′β, that interact with HMGR1S and HMGR1L, the major isoforms of Arabidopsis thaliana HMGR. B′′α and B′′β are Ca2+ binding proteins of the EF-hand type. We show that HMGR transcript, protein, and activity levels are modulated by PP2A in Arabidopsis. When seedlings are transferred to salt-containing medium, B′′α and PP2A mediate the decrease and subsequent increase of HMGR activity, which results from a steady rise of HMGR1-encoding transcript levels and an initial sharper reduction of HMGR protein level. In unchallenged plants, PP2A is a posttranslational negative regulator of HMGR activity with the participation of B′′β. Our data indicate that PP2A exerts multilevel control on HMGR through the five-member B′′ protein family during normal development and in response to a variety of stress conditions.
INTRODUCTION
The enzyme 3-hydroxy-3-methylglutaryl CoA reductase (HMGR) catalyzes the first committed step of the mevalonate (MVA) pathway for isoprenoid biosynthesis. In plants, this pathway provides precursors for a wide variety of isoprenoid products that are required for very diverse functions, including sterols for membrane biogenesis, sesquiterpenoid phytoalexins and steroid glycoalkaloids for defense, brassinosteroids and cytokinins for control of growth and development, farnesyl and geranyl groups for protein prenylation, dolichols for protein glycosylation, and ubiquinone for respiration (Stermer et al., 1994; Chappell, 1995). All known plant HMGR variants are targeted primarily to the endoplasmic reticulum and have the same topology in the membrane (Campos and Boronat, 1995). The diverged N-terminal region and the conserved catalytic domain are located in the cytosol, whereas only a short stretch of amino acids connecting the two transmembrane segments is in the lumen. This is consistent with the cytosol being the only site for MVA biosynthesis in plant cells (Campos and Boronat, 1995).
In all plant species studied so far, HMGR is encoded by a multigenic family. In Arabidopsis thaliana, two genes (HMG1 and HMG2) encode three HMGR isoforms (HMGR1S, HMGR1L, and HMGR2) (Enjuto et al., 1994; Lumbreras et al., 1995). HMGR1S and HMGR1L proteins derive from the HMG1 gene and are identical in sequence, but the 1L isoform has an N-terminal extension of 50 amino acid residues. The analysis of a null HMG1 mutant (hmg1-1) has confirmed the essential role of this gene (Suzuki et al., 2004). The hmg1-1 plants show dwarfism, early senescence, and male sterility. By contrast, disruption of HMG2 does not affect the phenotype nor the fertility of the plant under normal growth conditions (Ohyama et al., 2007). The HMGR1S transcript is found in all tissues but at fairly higher levels during the first stages of development and in inflorescences (Enjuto et al., 1994). By contrast, HMGR1L and HMGR2 transcripts are detected only in seedlings, roots, and inflorescences and are about 10 times less abundant than the HMGR1S mRNA (Enjuto et al., 1995; Lumbreras et al., 1995). These observations suggest a housekeeping role for HMGR1S and a more specialized function for HMGR1L and HMGR2.
Plant HMGR has a key regulatory role in the MVA pathway, critical not only for normal plant development, but also for the adaptation to demanding environmental conditions. Consistent with this notion, plant HMGR is modulated by myriad endogenous signals and external stimuli, such as phytohormones, calcium, calmodulin, light, blockage of isoprenoid biosynthesis, chemical challenge, wounding, elicitor treatment, and pathogen attack (Stermer et al., 1994; Rodríguez-Concepción et al., 2011). It has been proposed that the major changes in HMGR activity would be determined at the transcriptional level, whereas the posttranslational control would allow a finer and faster adjustment (Chappell, 1995). Whereas transcriptional modulation of HMGR has been demonstrated in many plant systems, evidence of posttranslational regulation is still scarce (Rodríguez-Concepción et al., 2011). The membrane domain of plant HMGR exerts negative regulation on the catalytic domain, thus limiting phytosterol biosynthesis (Harker et al., 2003). Arabidopsis HMGR1 can be phosphorylated in cell-free extracts by Brassica oleracea and Spinacea oleracea SnF1-related kinase (SnRK1) activity at a conserved Ser residue (Ser-577 in HMGR1S) (Dale et al., 1995; Sugden et al., 1999). This modification completely inactivates HMGR activity and therefore could determine the flux through the MVA pathway. In fact, higher phytosterol levels were obtained in tobacco (Nicotiana tabacum) seeds when a variant of Arabidopsis HMGR1S without the phosphorylation site was constitutively expressed (Hey et al., 2006). However, the gain in seed phytosterol content was only one-tenth of that obtained in seeds or leaves that constitutively expressed a single copy of the HMGR catalytic domain (Harker et al., 2003). Thus, present evidence indicates that negative regulation of plant HMGR activity relies more on the presence of the N-terminal domain than on phosphorylation at the conserved Ser residue.
Ser/Thr protein phosphatase 2A (PP2A) is a structurally conserved heterotrimeric enzyme composed of a scaffolding/regulatory A subunit, also termed PR65, a regulatory B-type subunit, and a catalytic C subunit (Mayer-Jaekel and Hemmings, 1994; Luan, 2003). The A and C subunits constitute the core of the holoenzyme, whereas the B-type subunit is more variable. Numerous studies performed in animal cells indicate that the B-type subunit determines the intracellular location and substrate specificity of the PP2A holoenzyme (Virshup, 2000; Janssens and Goris, 2001; Sontag, 2001). The targets of B-type subunits are therefore potential substrates for PP2A. So far, three distinct B-type protein families, named B, B′, and B′′, have been confirmed as components of PP2A holoenzymes (Luan, 2003; DeLong, 2006).
To investigate the posttranslational regulation of Arabidopsis HMGR, we searched for proteins that interact specifically with the cytosolic N-terminal region. We identified two Arabidopsis B′′ subunits of PP2A, designated B′′α and B′′β, that bind HMGR1L and HMGR1S, but not HMGR2. We found that PP2A is not only a posttranslational negative regulator of HMGR activity and protein levels, but also a positive regulator of HMG1 transcript levels. Whereas B′′β plays a role in the posttranscriptional repression of HMGR in unchallenged seedlings, B′′α modulates HMGR transcript, protein, and activity levels in response to salt challenge. Our data suggest that the multilevel control of HMGR is a major role of the five-member B′′ protein family in Arabidopsis.
RESULTS
Identification of B′′α and B′′β as HMGR-Interacting Proteins
To identify proteins that interact with the N-terminal region of Arabidopsis HMGR, we conducted a yeast two-hybrid screening. A cDNA fragment encoding the N-terminal region of HMGR1L (NT1L) was cloned in plasmid pAS2-1 to obtain a translational fusion with the C terminus of the GAL4 binding domain (BD-NT1L). The chimeric construct was used to screen 1.4 × 106 clones of a pACT library prepared from Arabidopsis 3-d-old etiolated seedlings (Kim et al., 1997). The three Arabidopsis HMGR isoforms (HMGR1S, HMGR1L, and HMGR2) are represented in the transcript population of this developmental stage (Enjuto et al., 1994; Lumbreras et al., 1995). The screening produced two positive clones encoding B′′ subunits of PP2A fused to the C terminus of the GAL4 activation domain (AD-B′′α and AD-B′′β). The corresponding plasmids led to a very strong interaction in two different genetic backgrounds, as estimated both by the extent of growth in the selection medium without His and the intensity of blue staining after detection of β-galactosidase activity with X-gal (Figure 1B; see Supplemental Figure 1A online). Clone pACT-B′′α contains an open reading frame of 1614 bp that encodes a polypeptide of 538 amino acid residues (protein B′′α) with an estimated molecular mass of 62.3 kD. Clone pACT-B′′β(88-536) encodes a truncated B′′β variant, lacking the first 87 amino acid residues. A cDNA with the corresponding entire coding sequence (1608 bp) was isolated by PCR (see Methods for details). The complete protein has 536 amino acid residues and an estimated molecular mass of 62.0 kD. The two-hybrid analysis did not show interaction between the B′′ variants and the catalytic domain of HMGR1 or other isoprenoid biosynthetic enzymes nor between these preys or the NT1L bait and appropriate negative controls (see Supplemental Figures 1A and 1B online).
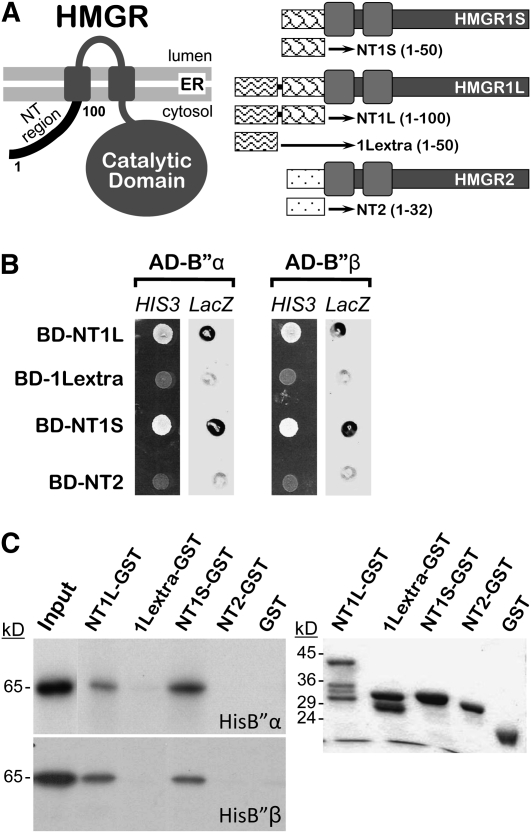
Figure 1.
Interaction of HMGR1L and HMGR1S with B′′α and B′′β.
(A) Schematic representation and topology in the membrane of Arabidopsis HMGR isoforms, depicting N-terminal fragments used in the two-hybrid analysis. The amino acid positions delimiting protein fragments are indicated in parentheses.
(B) Two-hybrid analysis in yeast. Cells from strain Y190 were cotransformed with a pACT2 derivative encoding AD-B′′α or AD-B′′β and a pAS2-1 derivative encoding BD-NT1L, BD-1Lextra, BD-NT1S, or BD-NT2. Interaction between the assayed partners was confirmed by the occurrence of growth on selective medium without His (HIS3 lanes) and β-galactosidase activity (LacZ lanes).
(C) In vitro GST pull-down analysis. Equivalent amounts of radiolabeled in vitro–synthesized HisB′′α or HisB′′β were incubated with the bait NT1L-GST, 1Lextra-GST, NT1S-GST, or NT2-GST or subjected directly to electrophoresis (Input). Fluorograms of the radiolabeled products retained by the indicated resins are shown on the left, whereas a Coomassie blue–stained gel with the GST fusions used in the assay is shown on the right.
To analyze if B′′α and B′′β recognize only HMGR1L or also the other Arabidopsis HMGR isoforms, we performed two-hybrid and glutathione S-transferase (GST) pull-down analyses. cDNAs coding for the N-terminal region of HMGR1S (NT1S), the extra region of HMGR1L not present in HMGR1S (1Lextra), or the N-terminal region of HMGR2 (NT2) were cloned in frame with the sequence coding for the GAL4 binding domain to obtain constructs that encode the chimeric products BD-NT1S, BD-NT2, and BD-1Lextra (Figures 1A and 1B). The two-hybrid experiments showed that B′′α and B′′β interact with NT1S and NT1L but not with NT2 or 1Lextra (Figure 1B). This was further confirmed by GST pull-down analysis (Figure 1C). In vitro–synthesized B′′α and B′′β with an appended His tag (HisB′′α and HisB′′β) were specifically retained in matrices containing NT1L-GST or NT1S-GST but not in 1Lextra-GST or NT2-GST resins. It should be noted that C-terminal instead of N-terminal GST fusions were used in these assays. This was originally intended to preserve the N-terminal position of the HMGR sequence, but the results indicate that the sequence itself and not its position (N-terminal or internal) is relevant. On the one hand, efficient binding of B′′ proteins was similarly observed using N-terminal GST fusions of NT1L or NT1S (GST-NT1L and GST-NT1S, respectively; see Supplemental Figure 1C online). On the other, NT1L-GST and NT1S-GST were retained with similar yields (Figure 1C), although NT1L represents a natural extension of NT1S (N-terminal regions of HMGR1L and HMGR1S, respectively). It can be concluded that the binding site for B′′α and B′′β is located in the region of 50 amino acid residues common to NT1S and NT1L and that the original N terminus is not required for the interaction. Altogether, our results provide evidence for direct binding between B′′ proteins and the N-terminal region of HMGR1L and HMGR1S.
B′′α and B′′β Are Calcium Binding Regulatory Subunits of PP2A
Databank searches and phylogenetic analysis revealed that the two proteins identified in the two-hybrid screening are similar to regulatory B′′ subunits of PP2A (Figure 2A). As predicted from this observation, B′′α and B′′β (named AtB′′α and AtB′′β in Figure 2A) interact with Arabidopsis PR65 subunit (Figure 2B). B′′α, B′′β , and three other products encoded in the Arabidopsis genome form a family of five members (Figure 2A). A cDNA encoding B′′α (corresponding to gene At5g44090) was isolated previously (Hendershot et al., 1999). The other members of the group were named accordingly: B′′β (At5g28850), B′′γ (At5g28900), B′′δ (At1g54450), and B′′ε (At1g03960). Fifty-eight percent of the amino acid residues are identical in the five proteins. The most distant sequences of the group (B′′δ and B′′ε) still share 65.8% identical (78.3% similar) residues, whereas B′′β and B′′γ are 99.6% identical. The five B′′ proteins from Arabidopsis, together with two other sequences from rice (Oryza sativa), form a compact clade (Figure 2A, group I), closely related to a broader group of animal B′′ PP2A subunits (group II). These two clades are distantly related to the Arabidopsis TONNEAU2 protein, which defines a diverged class of B′′ PP2A subunit, with plant and animal representatives (Figure 2A, group III) (Camilleri et al., 2002).
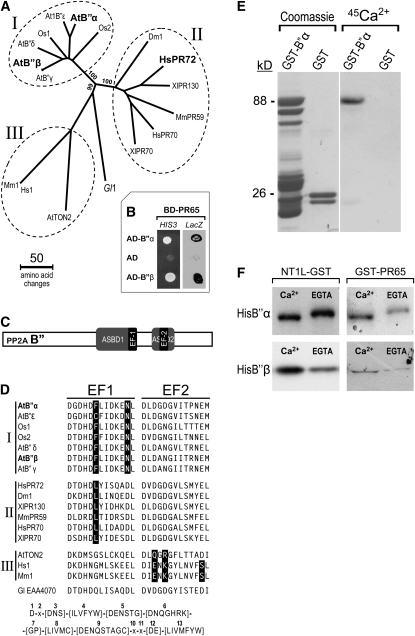
Figure 2.
Characterization of the B′′ PP2A Protein Family.
(A) Phylogenetic analysis of eukaryotic B′′ PP2A subunits. The branching points leading to three groups were highly significant, since they occurred in 99 or 100% of 1000 bootstrap replicates, as indicated. The first two letters of the sequence names refer to the organism. At, Arabidopsis thaliana; Dm, Drosophila melanogaster; Gl, Giardia lamblia; Hs, Homo sapiens; Mm, Mus musculus; Os, Oryza sativa; Xl, Xenopus laevis. A text file of the sequence alignment used in this analysis is available as Supplemental Data Set 1 online.
(B) Two-hybrid analysis. Yeast cells from strain Y190 were cotransformed with a pAS2-1 derivative encoding BD-PR65 and either control pACT2 plasmid encoding AD or pACT2 derivatives encoding AD fused to B′′α or B′′β. Growth on selective medium without His (HIS3 lane) or staining after β-galactosidase assay (LacZ lane) indicates positive interaction. BD-PR65 corresponds to the A2 (pDF1) variant of PR65.
(C) Schematic representation of the B′′ PP2A subunit. Positions of ASBD and EF-hand motifs are indicated, keeping proportionality with that of the actual primary sequence.
(D) Comparison of the EF-hand motifs. Sequences corresponding to the two EF-hand motifs of B′′ subunits were aligned and compared with the EF-hand consensus (PROSITE entry PS00018) shown at the bottom. EF1 and EF2 correspond to positions 330 to 342 and 403 to 515 of B′′α, and 327 to 339 and 400 to 412 of B′′β, respectively. Residues that do not fit the consensus are represented in white over black background. B′′ sequences were classified in three groups, as indicated on the left, according to divergence of EF1 and EF2 from consensus. The first two letters of the sequence names refer to the organism as in Figure 2A.
(E) Ca2+ binding assay. GST-B′′α and GST produced in E. coli were partially purified by glutathione affinity chromatography. Shown are a Coomassie blue–stained gel of the eluted fractions (left) and an autoradiogram of equivalent samples (right) after electroblotting and incubation of the PVDF membrane with 45Ca2+.
(F) Ca2+-dependent in vitro binding and band shift assays. Radiolabeled in vitro–synthesized HisB′′α or HisB′′β were incubated with matrices containing NT1L-GST or GST-PR65 bait in the presence of 5 mM Ca2+ or 5 mM EGTA. Equivalent volumes of eluted samples were subjected to SDS-PAGE (10% acrylamide) and fluorography. GST-PR65 corresponds to the A3 (pDF2) variant of PR65.
It was shown that a human regulatory B′′ subunit of PP2A (B′′/PR72, named HsPR72 in Figure 2A) is a Ca2+-calcium binding protein and that interaction with the cation is mediated by a typical tandem of EF-hand motifs (EF1 and EF2) (Janssens et al., 2003). Sequence alignment revealed that the pair of EF-hand motifs is also present in the B′′ subunits from plants (Figure 2D). These motifs and the two domains required for interaction with the PR65 subunit (ASBD1 and ASBD2) (Li and Virshup, 2002) are in a core region (Figure 2C) that shows the highest conservation between animal and plant B′′ PP2A subunits. In the core region, HsPR72 (residues 192 to 403) and Arabidopsis B′′α (residues 232 to 442) are 52.4% identical (73.6% similar). Interestingly, classification of B′′ sequences according to divergence of the two EF-hand motifs from the consensus resulted in a three-group clustering, which is identical to that obtained after phylogenetic analysis of full-length sequences (cf. Figures 2A and 2D). This tight correlation suggests that variations in the Ca2+ binding motifs occurring during evolution have been important in the functional specialization of PP2A B′′ subclasses.
To test whether Arabidopsis B′′α is a Ca2+ binding protein, an Escherichia coli extract containing partially pure GST-B′′α was electrophoresed under denaturing conditions and transferred to a polyvinylidene fluoride (PVDF) membrane. As observed in Figure 2E, the GST-B′′α chimera bound 45Ca2+. No calcium was bound to GST or other proteins of the extract. To examine whether the availability of calcium influences the binding capacity of B′′ isoforms, we performed GST pull-down analysis with the NT1L and PR65 targets as baits. Isotopically labeled HisB′′α and HisB′′β were incubated with NT1L-GST or GST-PR65 matrices in the presence of calcium or EGTA, and the retained samples were subjected to gel electrophoresis and fluorography. Interestingly, the interaction of B′′β with NT1L or PR65 and the interaction of B′′α with PR65 increased in the presence of extra calcium ions, compared with the binding in the presence of the calcium chelator (Figure 2F). By contrast, the absence of free calcium ions (presence of EGTA) did not limit the association between B′′α and NT1L. In spite of the denaturing conditions, the transient exposure to calcium increased the electrophoretic mobility of the B′′ variants, particularly in the case of HisB′′α (Figure 2F). As reported for other proteins (Klee et al., 1979), the band shift is indicative of a conformational change in Arabidopsis B′′ due to Ca2+ binding.
PP2A Is a Posttranslational Negative Regulator of HMGR
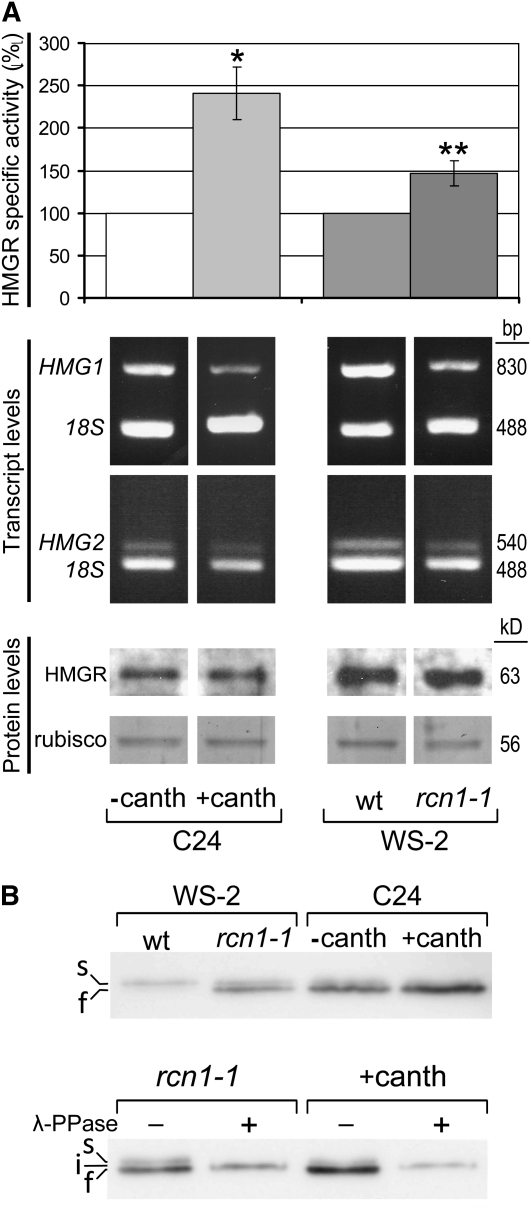
The observation that Arabidopsis HMGR interacts with regulatory subunits of PP2A prompted us to examine whether HMGR is under control of PP2A. We first measured the HMGR-specific activity in seedlings grown in the presence or absence of cantharidin (canth), a commonly used Ser-Thr phosphatase inhibitor that has the highest affinity for PP2A (one order of magnitude lower Kd for PP2A than PP1) (Li et al., 1993). The pharmacological block of PP2A consistently led to an increase of HMGR activity (Figure 3A, top). To confirm the regulation of HMGR by PP2A, we compared the rcn1-1 mutant with its parental line Wassilewskija-2 (WS-2). This mutant is defective in the major isoform of the regulatory A subunit of PP2A and has a 50% reduction in PP2A activity (Deruère et al., 1999). As shown in Figure 3A, the knockout of RCN1 gave rise to a 45% increase of HMGR activity. Therefore, both the genetic and pharmacological approaches indicate a negative regulation of HMGR by PP2A. To characterize the type of regulation, we measured HMGR transcript and protein levels in the samples used for the HMGR activity assays. As observed in Figure 3A, the genetic and pharmacological blockage of PP2A did not cause an increase, but rather a slight decrease, in the HMG1 and HMG2 transcript levels and did not affect appreciably the amount of HMGR protein. The augmented HMGR activity cannot be explained by increases in mRNA or protein levels and therefore seems exclusively due to a posttranslational regulation. To test whether the phosphorylation status of HMGR depends on PP2A, we first submitted aliquots of the above protein samples to electrophoresis under conditions that allowed discrimination of closely migrating bands (Figure 3B). As observed in the top gel of Figure 3B, both the genetic and pharmacological blocks of PP2A led to accumulation of a faster-migrating band recognized by the anti-CD1-i antibody (cf. lane WS-2_rcn1-1 with lane WS-2_wt and lane C24_+canth with lane C24_−canth), suggesting that this band could correspond to a phosphorylated form of HMGR. It should be noted, however, that the ratio between the phosphorylated and nonphosphorylated HMGR bands was different in the untreated WS-2 and C24 plants (cf. lanes WS-2_wt and C24_−canth). When WS-2_rcn1-1 and C24_+canth extracts were treated with phosphatase prior to electrophoresis, the two HMGR bands were replaced by a single band of intermediate migration (bottom gel of Figure 3B, band i). This supports that the faster-migrating band (named f in Figure 3B) corresponds to a phosphorylated HMGR variant. The slower-migrating band (named s) also seems to be phosphorylated. Hence, our data suggest that two different forms of phosphorylated HMGR may exist in Arabidopsis and that the dephosphorylation of one of them depends on PP2A.
Figure 3.
Posttranslational Regulation of HMGR by PP2A.
(A) HMGR transcript, protein, and activity levels. Arabidopsis C24, WS-2 wild-type (wt), and WS-2 rcn1-1 mutant seedlings were grown for 3 to 4 weeks under short-day conditions in MS medium alone or in MS medium containing 10 μM cantharidin as indicated. The HMGR activity (top panel) is represented as the percentage with respect to the C24 (three experiments) or WS-2 wild-type sample (five experiments), indicating the average values and the corresponding sd. The HMGR-specific activity (pmol HMG-CoA/min*mg) was 2.33 ± 0.88 for C24 and 1.99 ± 0.34 for WS-2 wild type. Asterisks indicate levels of statistical significance as determined for Student’s t test: *P < 3 10−3 for +canth versus −canth; **P < 3 10−4 for rcn1-1 versus the wild type. The HMG1 and HMG2 transcript levels (middle panel) were estimated by quantitative PCR and agarose gel electrophoresis using the amplification product of 18S rRNA (18S) as an internal reference. The protein levels (bottom panel) were estimated by immunoblot with the anti-CD1-i antibody using the Coomassie blue–stained Rubisco band of the same filter as a normalization reference. The sizes of the PCR products (pb) or proteins (kD) are indicated on the right.
(B) Phosphorylation status of HMGR. The above protein samples were subjected to SDS-PAGE (top gel) or incubated in the presence (+) or absence (−) of protein phosphatase from phage λ (λ-PPase), as indicated, prior to SDS-PAGE (bottom gel). Electrophoresis was performed in 9% acrylamide mini gels with 8 mA constant current. HMGR was detected by immunoblot with the anti-CD1-i antibody. s, i, f: slow-, intermediate-, and fast-migrating HMGR bands.
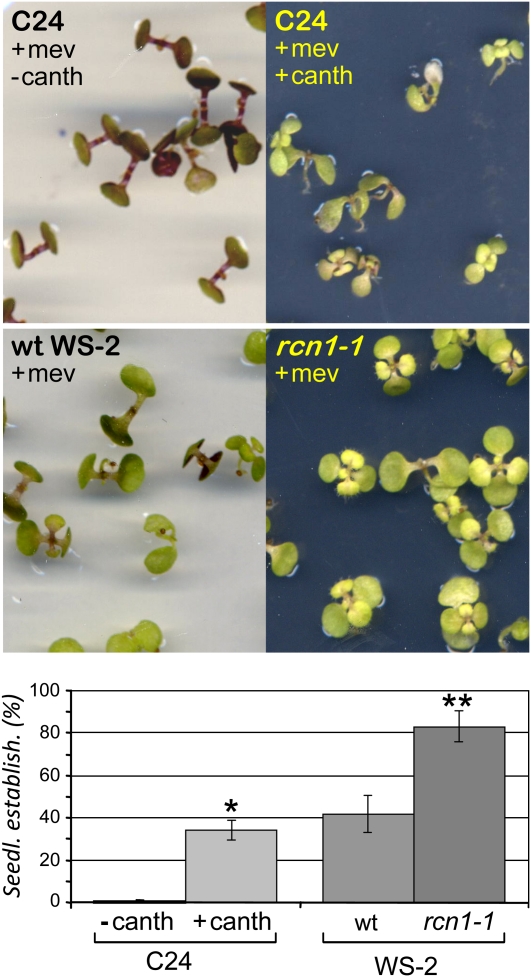
The above results show PP2A-dependent variations of HMGR activity but do not illustrate whether the observed changes have physiological consequences in vivo. To analyze whether HMGR is effectively modulated by PP2A in the plant, we arrested development with mevinolin, a specific HMGR inhibitor, and tried to release this blockage by modulating HMGR activity through additional pharmacological or genetically induced inhibition of PP2A. In the presence of 5 μM mevinolin, development of wild-type Arabidopsis seedlings is arrested at the two cotyledon stage (Figure 4, C24 +mev −canth). A severe stress in these seedlings is also evidenced by the accumulation of reddish anthocyanin. It has been shown that the percentage of seedlings that develop true leaves (achieve seedling establishment) in the presence of mevinolin is a direct function of the HMGR activity level (Rodríguez-Concepción et al., 2004). Cantharidin is toxic to the plant, but its addition allowed a significant proportion of the C24 seedlings to overcome the mevinolin-induced developmental block (Figure 4, C24 +mev +canth, notice the leaf curling and the smaller plant size in the presence of the two drugs). The mevinolin treatment affected WS-2 germinating plants less severely than those of C24 (Figure 4). This is due to the presence of the phyD-1 mutation in the WS-2 ecotype (Aukerman et al., 1997), which causes an increase in HMGR activity (Rodríguez-Concepción et al., 2004). The seedling establishment rate was also higher in the rcn1-1 mutant than in its genetic background WS-2 (Figure 4). It can be concluded that both the pharmacological and genetic blocks of PP2A cause an increase of HMGR activity, which is observed in the vitro assays (Figure 3A) and in planta (Figure 4). In these cases, HMGR is regulated by PP2A posttranslationally.
Figure 4.
Regulation of HMGR by PP2A in Planta.
Arabidopsis C24, WS-2 wild-type (wt), and WS-2 rcn1-1 mutant seedlings were grown for 15 d under long-day conditions in MS medium containing 5 μM mevinolin or 5 μM mevinolin and 10 μM cantharidin as indicated. The resistance to mevinolin is represented in the bottom panel as the percentage of seedling establishment (Seedl. establish.). The bars indicate the mean values and the corresponding sd of three independent experiments, with at least 60 plants per experimental condition. Asterisks indicate levels of statistical significance as determined for Student’s t test: *P < 2 10−4 for +canth versus −canth; **P < 10−6 for rcn1-1 versus the wild type.
Posttranscriptional Regulation of HMGR by the B′′β Gene
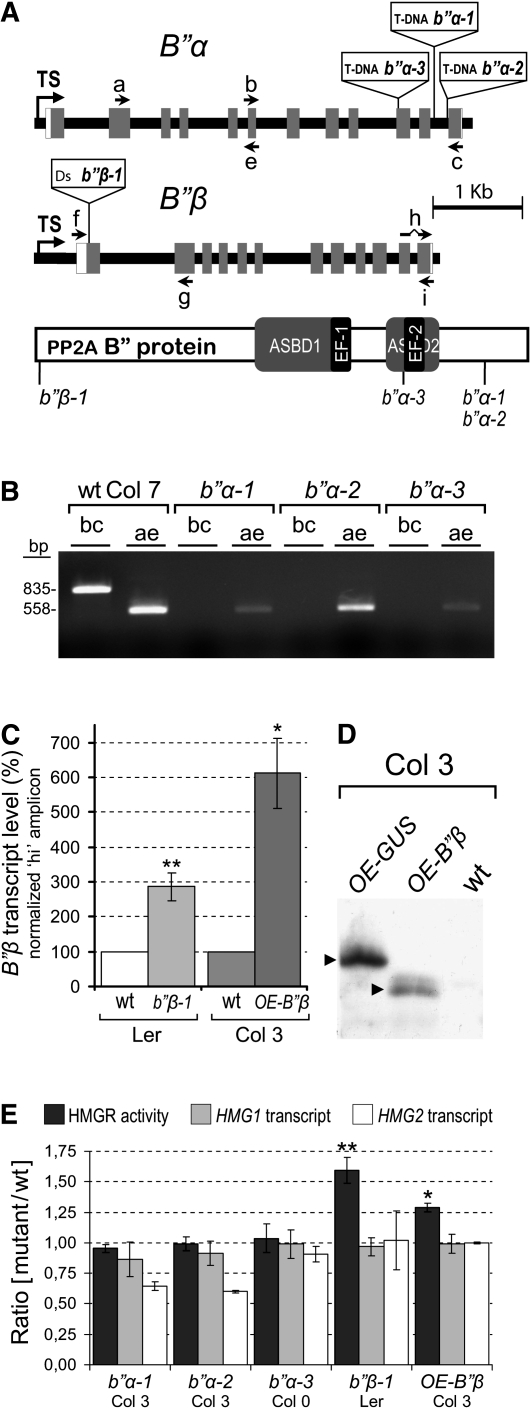
To determine whether the B′′ PP2A subunits play a role in the regulation of HMGR by PP2A, we analyzed several B′′α and B′′β Arabidopsis mutants. Three independent T-DNA insertion lines of B′′α (designated b′′α-1, b′′α-2, and b′′α-3) and a transposon Ds insertion line of B′′β (designated b′′β-1) were studied. In b′′α-1 and b′′α-2 mutants, the T-DNA is located in the eleventh intron of the B′′α gene (Figure 5A). In the b′′α-3 mutant, the T-DNA is inserted just upstream of the sequence encoding the EF-2 and therefore disrupts the conserved ASBD2 core element (Figure 5A). In the b′′β-1 mutant, the Ds element interrupts the B′′β gene nine residues downstream of the ATG start codon but 15 and 54 residues upstream, respectively, of two in-frame ATG triplets (Figure 5A). None of the Arabidopsis B′′ mutants showed phenotypic alterations with respect to the corresponding genetic background under the sterile conditions at the seedling stage.
Figure 5.
Characterization of b′′α and b′′β Mutants.
(A) The disrupted B′′ genes. Gene structure is drawn to scale, but the inserted T-DNA (8.0 kb) and Ds transposable element (6.6 kb) are only indicated. Black line, gene-flanking sequences and introns; white boxes, transcribed nontranslated regions; gray boxes, coding exons; TS, transcription start site. Arrows a to i show the annealing position of primers. The crooked line of primer h represents a missing intron sequence. A protein scheme with disruption points corresponding to the above insertions is shown at the bottom. The conserved ASBD and EF-hand motifs are indicated.
(B) RT-PCR analysis of b′′α-1, b′′α-2, and b′′α-3 mutants. The bc and ae primer sets were used to detect the corresponding B′′α transcript regions in total RNA from B′′α mutant and wild-type (wt) control seedlings. The size of the amplicons (bp) is indicated to the left of the agarose gel.
(C) B′′β transcript levels in b′′β-1 and OE-B′′β mutants. Total RNA was extracted from b′′β-1, OE-B′′β , and the corresponding wild-type seedlings, grown in MS medium for 14 d under long-day conditions. The B′′β transcript level was measured by qRT-PCR with the hi primer set using the At4g26410 transcript as a normalization reference (Czechowski et al., 2005). The graph shows the average ± sd percentage of normalized B′′β transcript of mutant with respect to the wild type from four (b′′β-1) or three (OE-B′′β) independent culture and sample processing experiments. Asterisks indicate levels of statistical significance as determined for Student’s t test: *P < 2 10−3 for OE-B′′β versus wild-type Col-3; **P < 2 10−4 for b′′β-1 versus wild-type Ler.
(D) Immunodetection of the HA-B′′β protein in OE-B′′β plants. A segregating population of OE-B′′β, a transgenic control OE-GUS and their parental wild-type line were analyzed by immunoblot with the anti-HA 3F10 monoclonal antibody. Samples correspond to total protein from supernatant fraction of 14-d-old seedlings. On the left, arrowheads indicate the position of epitope-tagged HA-B′′β and HA-GUS proteins corresponding to OE-B′′β and OE-GUS plants, respectively.
(E) HMGR activity and transcript levels. Arabidopsis b′′α-1, b′′α-2, b′′α-3, b′′β-1, and OE-B′′β mutant seedlings, together with the corresponding wild-type controls (Col-7, Col-7, Col-0, Ler, and Col-3, respectively), were grown for 15 d under long-day conditions in MS medium. After collection and freezing, sample aliquots were taken for HMGR-specific activity and transcript level determination. HMG1 and HMG2 transcript levels were estimated by qRT-PCR using the GAPDH transcript for normalization, as previously described (Nieto et al., 2009). The graph represents the HMGR-specific activity (black) and the relative HMG1 (gray) and HMG2 (white) transcript levels as the average ratio ± sd between the mutants and the corresponding wild-type control from at least three independent culture and sample processing experiments. The HMGR-specific activity (pmol HMG-CoA/min*mg) was 8.62 ± 0.81 for wild-type Col-7, 9.78 ± 1.44 for wild-type Col-0, 16.55 ± 2.39 for wild-type Ler, and 8.38 ± 1.62 for wild-type Col-3. The asterisks indicate levels of statistical significance as determined for Student’s t test: *P < 0.02 for OE-B′′β versus wild-type Col-3; **P < 0.002 for b′′β-1 versus wild-type Ler.
As a first step to characterize the mutants, we examined the presence of B′′α and B′′β transcripts by RT-PCR. A set of two primers flanking the insertion site was used to confirm the absence of the complete transcript (oligonucleotides b, c for B′′α; and f, g for B′′β) (Figures 5A and 5B; see Supplemental Figure 2 online). A second amplicon, positioned either upstream (oligonucleotides a, e for B′′α) or downstream (oligonucleotides h, i for B′′β) of the corresponding insertion site (Figure 5A), was devised to determine if that part of the transcript was present (ae and hi amplicons, respectively). As observed in Figure 5B, the ae amplicon is still detected in the three b′′α insertion mutants. Therefore, none of them is null at the transcript level. However, the b′′α-3 mutant is likely a knockout in what concerns protein function, since the hypothetical peptide that might result from the truncated B′′α transcript (amino acid residues 1 to 397 of B′′α) is not able to interact with PR65 nor with HMGR in the two-hybrid system (see Supplemental Figure 3 online). As observed in Figure 5C, much higher amounts of the hi amplicon were obtained by real-time quantitative RT-PCR (qRT-PCR) in the b′′β-1 Ds insertion line, with respect its wild-type reference. Therefore, line b′′β-1 is a B′′β transcript overexpression mutant.
The HMGR transcript and activity levels of the b′′α and b′′β mutants were compared with those of the corresponding parental lines in 15-d-old seedlings grown under sterile conditions. As observed in Figure 5E, the disruption of B′′α does not affect HMGR activity nor HMG1 transcript levels as indicated by a ratio of mutant to wild type close to 1. Although a 10 to 40% reduction of HMG2 transcript occurs in the b′′α mutants (Figure 5E), no major effect on the total HMGR activity is observed, possibly because HMG2 is expressed at a much lower rate than HMG1 (Enjuto et al., 1995). The b′′β-1 mutant shows higher HMGR activity than its parental line (Figure 5E), as confirmed by seedling establishment assays (see Supplemental Figure 4 online). The rise of HMGR activity in the b′′β-1 mutant occurred without a significant increase in HMGR transcript levels (Figure 5E), suggesting posttranscriptional regulation by B′′β. To confirm this hypothesis, we examined HMGR transcript and activity levels in an independent B′′β overexpressing line (OE-B′′β) generated by a transgenic approach. In this line, B′′β expression is under control of the cauliflower mosaic virus 35S promoter, and the resulting protein is tagged with the Hemagglutinin epitope (HA) (see Methods for details). As shown in Figure 5C, OE-B′′β had ~6 times higher B′′β transcript levels than did the corresponding wild-type line. The HA-B′′β protein was detected by immunoblot with an HA-specific antibody (see Methods for details) (Figure 5D). Similarly to the b′′β-1 mutant, OE-B′′β contains higher HMGR activity than its parental line, without a change in the relative levels of HMG1 and HMG2 transcripts (Figure 5E). These data indicate posttranscriptional regulation of HMGR by B′′β, which is consistent with the observed posttranslational regulation by PP2A.
B′′α Negatively Regulates Root Growth in Response to Salt
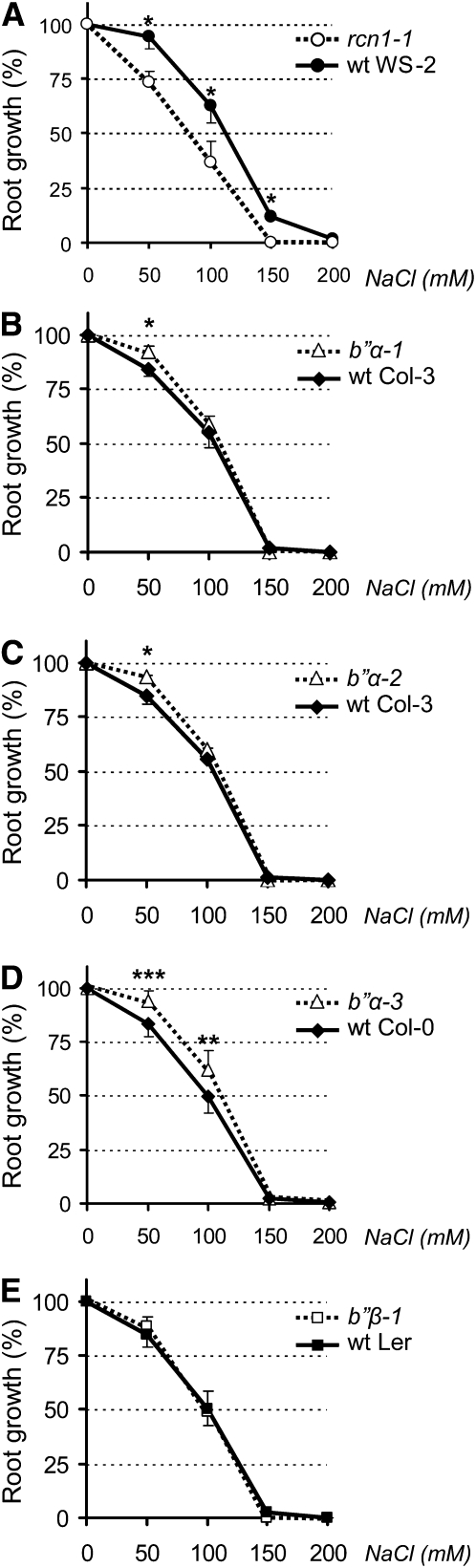
We observed that disruption of B′′α does not affect HMGR in seedlings grown in standard sterile medium. Therefore, we wondered whether B′′α could have a regulatory role but only under particular conditions. Since the Arabidopsis RCN1 and PP2Ac-2 genes, encoding scaffolding and catalytic PP2A subunits, have been shown to regulate root growth in response to several challenges (Pernas et al., 2007; Blakeslee et al., 2008), we decided to test the root response to salt in the B′′ disruption mutants. Four-day-old seedlings grown in half-concentrated Murashige and Skoog (MS) medium were transferred to plates containing the same medium and different concentrations of NaCl (Figure 6). The seedlings were grown for six additional days in the vertical position and the length of the newly developed main root was measured. As previously reported (Blakeslee et al., 2008), salt stress inhibits the growth of the root in seedlings and this inhibition is more drastic in rcn1-1 than in the wild type (Figure 6A). By contrast, the b′′α-1, b′′α-2, and b′′α-3 mutants developed a longer main root than did the corresponding wild type at several salt concentrations (Figures 6B to 6D). The effect of B′′α disruption on root growth was more evident at 50 mM than at 100 mM NaCl (Figures 6B to 6D). On the other hand, the disruption of B′′β had no effect on the inhibition of root growth by the challenging condition (Figure 6E). Thus, under salt stress, although RCN1 is a positive regulator of root growth, B′′α behaves as a negative regulator and B′′β seems neutral.
Figure 6.
Effect of NaCl on the Root Growth of PP2A Mutants.
Mutant rcn1-1 (A), b′′α-1 (B), b′′α-2 (C), b′′α-3 (D), and b′′β-1 (E) Arabidopsis seedlings (dotted lines) grown for 4 d in half-concentrated MS were transferred, together with the corresponding wild-type (wt) control (black lines), to plates containing the same medium and the indicated concentrations of NaCl. After six additional days, the new growth of the main root was measured and expressed as the percentage with respect to the growth in the absence of NaCl. The graphs show the average and sd of three ([A] to [C] and [E]) or seven (D) independent experiments, with 10 plants per experimental condition. For clarity, only the positive (mutant) or negative (wild type) sd is shown. Asterisks indicate levels of statistical significance as determined by paired Student’s t test: *P < 0.05, **P < 0.005, and ***P < 0.0005 for the indicated mutant versus the corresponding wild-type line.
HMGR Transcript, Protein, and Activity Levels Are Modulated in Response to Salt
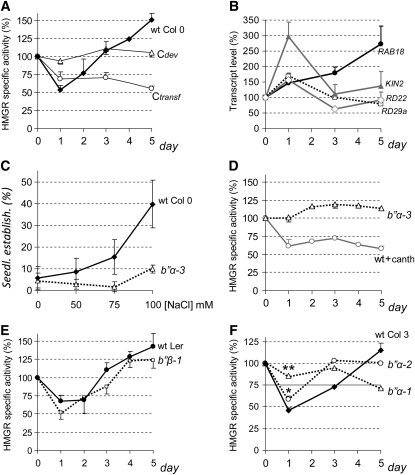
Since B′′α was involved in the plant response to salt, we decided to examine whether its interacting partner HMGR is influenced by the challenging condition. Arabidopsis wild-type Columbia-0 (Col-0) seedlings were grown for 2 weeks on polyester filters layered on half-concentrated MS medium and then the filters were transferred to new plates containing the same medium supplemented with 50 mM NaCl. This treatment did not affect the seedling phenotype, but the HMGR activity decreased to 50% 1 d after transfer and then increased during the next 4 d to levels 50% above the starting value (Figure 7A, wt Col 0). These changes were not observed in nontransferred seedlings (Figure 7A, Cdev), so they were not induced by normal development. However, the transfer to fresh plates containing half concentrated MS without NaCl supplement caused a reduction of HMGR activity that did not recover over time (Figure 7A, Ctransf). This was a bit surprising because the transferred plants were not even touched. The process, done under sterile conditions, involved only opening of the original and destination plates and did not last longer than a minute. Remarkably, the transfer affected the HMGR activity measured in the seedlings 24 h later and the effect remained at least several days. The initial decrease of HMGR activity was less intense when plants were transferred to MS alone than when they were transferred to salt-containing medium (cf. Ctransf and wt Col 0 at day 1 in Figure 7A), maybe denoting a less intense challenging condition. However, these observations left still open the question of whether Arabidopsis HMGR could respond to salt treatment devoid of the other challenging stimuli. To confirm that the plants were detecting and responding to salt challenge, we first measured the relative levels of RAB18, RD29a, RD22, and KIN2 transcripts, known to accumulate under salt stress (Nylander et al., 2001; Song et al., 2009), in the same samples used for the above experiments. As observed in Figure 7B, the abundance of all four transcripts rose during the first day after transfer, confirming the salt stress condition, but, whereas RAB18 expression kept increasing until the fifth day, RD29a, RD22, and KIN2 transcripts dropped almost to their starting levels. To demonstrate that Arabidopsis HMGR can be modulated in response to salt challenge alone, we performed seedling establishment assays in the presence of statin and increasing concentrations of NaCl. In these in vivo assays, vernalization, germination, and growth occurred in a single plate, so without a transfer step. As observed in Figure 7C, higher salt concentrations led to a higher proportion of wild-type Col-0 seedlings developing true leaves. This is in full agreement with the increase of HMGR activity observed at long term when plants are transferred to salt-containing medium (Figure 7A, wt Col 0). We conclude that the HMGR activity increases in Arabidopsis seedlings in response to salt, without another stress condition, such as mechanical injury or short-time desiccation that may occur upon transfer.
Figure 7.
Temporal Profile of HMGR Activity in Seedlings Subjected to Salt Stress.
Mutant and wild-type (wt) Arabidopsis seedlings were grown for 2 weeks under long-day conditions on polyester filters layered over half-concentrated MS. The filters were transferred to plates containing the same medium supplemented with 50 mM NaCl or 50 mM NaCl and 10 μM cantharidin (+canth) and growth was continued. Samples were collected and frozen at the indicated times. Cdev is a development control of nontransferred wild-type Col-0 plants. Ctransf is a control of wild-type Col-0 plants transferred to half-concentrated MS medium without NaCl.
(A) and (D) to (F) Average percentage of HMGR-specific activity of the different genotypes (wild type, black lines; mutant, dotted lines) and treatments (+canth, gray line) with respect to the corresponding starting values. Calculations were made from at least three independent culture and sample processing experiments. For clarity, only the positive (wild type) or negative (mutant) sd is shown. The HMGR-specific activity (pmol HMG-CoA/min*mg) at day 0 was 9.78 ± 1.44 for wild-type Col-0, 15.02 ± 1.63 for wild-type Col-3, 13.23 ± 4.51 for b′′α-1, 11.63 ± 2.36 for b′′α-2, 9.32 ± 0.73 for b′′α-3, 16.55 ± 2.39 for wild-type Ler, and 25.52 ± 1.78 for b′′β-1.
(B) qRT-PCR analysis of RAB18, RD29a, RD22, and KIN2 marker transcripts in wild-type Col-0 seedlings transferred to NaCl-containing plates using the At4g26410 transcript as a normalization reference. The graph shows the average percentage and sd of the relative transcript level with respect to the corresponding starting values in two independent assays.
(C) Wild-type Col-0 and b′′α-3 plants were germinated and grown for 16 d on MS medium containing 4 μM lovastatin or 5 μM mevinolin and the indicated concentrations of NaCl; the graph represents the average and sd of seedling establishment percentage in four independent experiments, with 60 plants per experimental condition.
(F) Asterisks indicate levels of statistical significance of mutant versus the wild type as determined by Student’s t test: **P < 10−12 for b′′α-1; *P < 10−4 for b′′α-2.
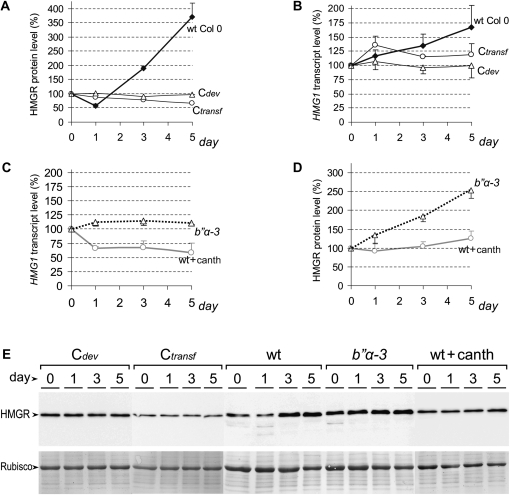
To determine if the variations of HMGR activity were caused by transcriptional or posttranslational mechanisms, we measured HMGR protein levels by immunoblotting and HMG1 transcript levels by real-time PCR. The profile of HMGR protein level in wild-type Col-0 (Figure 8A; see also Figure 8E, wt section), consisting of a decrease and a subsequent increase, paralleled that of the activity (Figure 7A) but with a notable difference. Whereas the HMGR protein level rose 3.5 times at day 5 of treatment (Figure 8A), the HMGR activity increased only 1.5 times (Figure 7A), suggesting posttranslational repression of the enzyme. The accumulation of HMGR protein was likely due to activation of transcription, since it was paralleled by an increase in HMG1 transcript level (cf. Figures 8A and 8B). However, the initial decrease in HMGR activity and protein level (Figures 7A and 8A) was simultaneous with a rise in HMG1 transcript (Figure 8B). A closer examination of the HMGR protein pattern helped to solve the inconsistency. The severe reduction in the band corresponding to entire HMGR at day 1 after challenge correlated with the presence of two faster migrating weak bands (Figure 8E, wt section, lane 1) that may correspond to degradation products. The signs of degradation disappeared after the first day of challenge (Figure 8E, wt section, lanes 3 and 5). Changes in protein stability, together with the steady increase in transcript level, may support the decrease and subsequent rise in HMGR activity.
Figure 8.
Temporal Profile of HMGR Transcript and Protein Levels in Seedlings Subjected to Salt Stress.
HMGR transcript and protein levels were determined in the same samples used in Figure 7. HMG1 transcript levels were measured by qRT-PCR using the At4g26410 transcript as a normalization reference (Czechowski et al., 2005). HMGR protein level was estimated by quantification of the anti-CD1-i signal after immunoblot. The chemiluminescence HMGR band was normalized to the Coomassie blue–stained Rubisco band of the same immunoblot filter.
(A) to (D) The graphs show the average percentage of HMGR protein level ([A] and [D]) and relative transcript level ([B] and [C]) of the different genotypes (wild type [wt], black lines; mutant, dotted lines) and treatments (+canth, gray line) with respect to the corresponding starting values. Calculations were made from at least three independent culture and sample processing experiments. For clarity, only the positive (wild type) or negative (mutant) sd is shown.
(E) Representative chemiluminescence (top) and Coomassie blue–stained (bottom) electrophoretic patterns used in HMGR protein level determinations.
No significant variations of HMGR transcript and protein levels occurred in the developmental control (Cdev in Figures 8A and 8B). In the transfer control, however, the HMG1 transcript increased 35% at day 1 and subsequently returned to the starting levels (Figure 8B, Ctransf). The HMG1 transcript was not paralleled by the HMGR protein that decreased steadily to reach 67% of the starting level at day 5 (Figure 8A, Ctransf). We conclude that the transfer to new plates without NaCl induced changes in HMGR transcript, protein, and activity levels, but the corresponding profiles were clearly different from those of the transfer to salt-containing plates.
PP2A and B′′α Are Involved in the Multilevel Response of HMGR to Salt
The above results indicate HMGR is subjected to transcriptional and posttranscriptional regulation in response to salt, and we therefore decided to test whether PP2A and B′′ subunits play a role in this process. To that purpose, we transferred b′′α-1, b′′α-2, b′′α-3, and b′′β-1 mutant seedlings, together with the appropriate wild-type controls to plates containing 50 mM NaCl and also wild-type Col-0 seedlings to plates containing 50 mM NaCl and 10 μM cantharidin and studied HMGR transcript, protein, and activity levels in these assays. The exposure to cantharidin slightly delayed growth and development and occasionally produced reddish color in the seedlings, denoting the stress condition, but the plants remained viable. In salt-treated plants, the partial block of PP2A with the drug caused a reduction, instead of an increase, in HMG1 transcript level that did not recover over time (cf. Figure 8C with 8B). The long-lasting reduction in HMG1 transcript, occurring upon PP2A block by cantharidin, was paralleled by a similar decrease in HMGR activity (cf. Figure 8C with 7D). However, the HMGR protein level showed nearly no loss and increased slightly over time (Figure 8D, wt+canth). This could be explained by a higher stability of the HMGR protein, since no sign of degradation was observed in the HMGR protein pattern of cantharidin-treated seedlings (Figure 8E, wt+canth section). Our data suggest that PP2A is required for the removal of HMGR protein occurring under salt stress and that a portion of the HMGR rescued by a blockage of the phosphatase becomes inactive.
Very similar HMGR activity profiles were observed in b′′β-1 mutant and wild-type Landsberg erecta (Ler) seedlings transferred to salt-containing plates (Figure 7E), indicating that the overexpression of B′′β does not affect the HMGR response to salt. By contrast, the disruption of B′′α disturbed the multilevel response of HMGR to this condition. In the b′′α-1 and b′′α-2 mutants, the HMGR activity profile was quite different from that of the corresponding wild type (Figure 7F). The initial decrease of activity was significantly affected, as indicated by Student’s t tests at day 1 (Figure 7F). The b′′α-3 mutant shows a more severe alteration of the HMGR response to salt, further confirming the role of B′′α. In this mutant, the salt challenge did not induce a steady increase in the HMG1 transcript level as in the wild type (cf. Figures 8B and 8C). The flat profile of HMGR activity in the b′′α-3 mutant is in sharp contrast with the crooked profile of the wild type (cf. Figures 7A and 7D). Also, in the seedling establishment assays, the HMGR activity of the b′′α-3 mutant did not increase in response to salt, as in the wild type (Figure 7C). Both HMGR activity and HMG1 transcript profiles in the b′′α-3 mutant are well above those of the cantharidin-treated wild-type seedlings (Figures 7D and 8C), indicating that the pharmacological block of PP2A exerted a more drastic action on HMGR. The HMGR protein profile of the b′′α-3 mutant showed a steady increase from the day of transfer, without a minimum occurring at day 1 as in cantharidin-treated and cantharidin-untreated wild-type samples (Figures 8A and 8D). In agreement, no sign of degradation was observed in the HMGR protein pattern of the b′′α-3 mutant at day 1 or later after transfer (Figure 8E, section b′′α-3). The accumulation of HMGR protein in the b′′α-3 mutant during the 5 d after transfer (Figure 8D) was more pronounced that the rise in HMGR activity (Figure 7D), indicating once again posttranslational negative regulation. On the whole, these results indicate multilevel and multistep modulation of HMGR by PP2A in response to salt challenge.
DISCUSSION
Control of HMGR by PP2A
Our data show that PP2A is a posttranslational negative regulator of HMGR in Arabidopsis plants. We base this conclusion on the increase of HMGR activity detected-by in vitro assays and seedling establishment in vivo assays, occurring in response to pharmacological or genetic block of PP2A. The modulation of HMGR1S and HMGR1L by PP2A was suggested by their specific interaction with the B′′ regulatory subunits and was confirmed by the rise of HMGR activity in the rcn1 mutant. The comparison between phosphatase-treated and untreated wild-type, cantharidin, and rcn1-1 extracts indicated that dephosphorylation by PP2A, not simply the presence of interacting PP2A complexes, is involved in the negative regulation. Altogether, our results implicate a PP2A-dependent dephosphorylation event in the repression of HMGR. Previous work showed that plant HMGR can be inactivated by SnRK1-mediated phosphorylation at a conserved Ser residue of the catalytic domain (Ser-577 in Arabidopsis HMGR1S) (Dale et al., 1995). The covalent modification by PP2A should occur in another position, since dephosphorylation at the SnRK1 site would lead to HMGR activation. In agreement, our electrophoretic patterns suggested that Arabidopsis HMGR may exist in two different phosphorylated forms. We showed posttranslational repression of HMGR by PP2A in the absence of challenge other than the block of the phosphatase. However, in both wild-type and b′′α-3 seedlings submitted to long-term salt stress, HMGR protein increased much more than HMGR activity. So, our results suggest that negative regulation of HMGR by dephosphorylation might occur under stress conditions as well.
The control of HMGR by PP2A is not restricted to posttranslational dephosphorylation. PP2A regulates HMG1 transcript levels both under stress and nonstress conditions. HMG1 transcript accumulation and HMGR protein level are modulated by B′′α and PP2A in response to salt. Our results suggest that Arabidopsis HMGR protein level is determined, at least in part, by protein degradation. This type of regulation has been proposed in potato (Solanum tuberosum) as a part of the normal developmental program and in response to darkness (Korth et al., 2000). The immunodetection of HMGR in the lytic vacuole of Arabidopsis cotyledon cells might suggest developmental regulation via degradation of this enzyme also in Arabidopsis (Leivar et al., 2005). Whereas HMGR degradation might be triggered by direct interaction with B′′α, additional factors and a more intricate pathway could be required to induce HMG1 expression. In our salt stress conditions, both the HMG1 and RAB18 transcripts showed a sustained accumulation over several days, whereas the RD29a, RD22, and KIN2 transcripts peaked and returned to their starting values. From this group, only the HMG1 and RAB18 transcripts seem appropriate as markers of long-lasting response to salt in Arabidopsis.
The HMGR activity of Arabidopsis seedlings transferred to salt-containing plates decreased by 50% in 1 d and increased to 150% of the original level in the next 4 d. Our data indicate that these variations mostly occur because of the salt stress condition but also suggest that other challenging factors, as the physical manipulation of the plant or the short exposure to nonsaturated water atmosphere may influence the observed response. The salt-induced modulation of HMGR activity was finely characterized by in vitro assays and confirmed by seedling establishment in vivo assays. Whereas the reduction in HMGR activity correlated with a decrease in HMGR protein level, the increase paralleled accumulation of HMG1 transcript. We may speculate that the initial decrease in HMGR obeys to the need for a blockage of certain biosynthetic routes to redirect photoassimilates and energy for the urgent synthesis of adaptive compounds and that the Arabidopsis seedling requires a surplus of isoprenoid products at the long term to cope with the challenging condition. However, in this scenario, it is unclear why the increase in HMGR protein levels is accompanied by a lower rise in HMGR activity, suggesting simultaneous posttranslational repression. Another role of HMGR required for the adaptation to the stress condition, perhaps a structural one, would reconcile this apparent contradiction. Interestingly, a similar biphasic profile of HMGR activity (decrease and subsequent increase) has been reported in two other plant systems exposed to different challenges. In tobacco BY-2 cell cultures subjected to block with mevinolin, the apparent HMGR activity decreased by 50% in ~4 h and subsequently increased 13-fold over the initial value in 20 h (Hemmerlin et al., 2003). In potato tubers, wounding induced a 15% decrease in HMGR activity, followed by a 4-fold increase that peaked 1 h after challenge (Yang et al., 1991). In this system, the decrease in HMGR activity was simultaneous with a sharp increase in HMGR transcript levels, as we observe in Arabidopsis. The coincident biphasic response of HMGR activity to challenging conditions might suggest underlying control by PP2A and B′′ regulatory subunits also in the tobacco and potato systems. The simultaneous, but opposite, changes in HMGR transcript and protein levels may occur at different rates, depending on the plant system and the type and severity of the stress condition. Thus, the final effect in HMGR activity at a given time after challenge may be quite variable. This may explain why the stress-induced positive and negative control of HMGR by PP2A was overlooked in the past. Our observation that the mere transfer of Arabidopsis seedlings to new plates without additional salt alters HMGR transcript, protein, and activity levels is relevant to this respect. We found that the pharmacological or genetic block of PP2A suppressed not only the salt-induced HMGR response but also the transfer-induced response. Thus, our results favor that PP2A might be involved in the modulation of HMGR under a variety of challenging conditions.
We have identified regulatory genes for HMGR that operate both under stress and normal development conditions. B′′α modulates HMGR in Arabidopsis seedlings upon transfer to salt-containing medium. B′′β overexpression does not affect the HMGR response to this challenge but stimulates HMGR activity when seedlings are grown in sterile conditions without noticeable stress. Since b′′β-1 and OE- B′′β accumulate high levels of B′′β transcript, they might behave as dominant-negative mutants with respect to PP2A. The excess of B′′β protein could out-compete B′′-containing PP2A heterotrimers that would otherwise bind and repress HMGR. In agreement with this, a B′′β protein variant labeled with an HA epitope was detected in the OE- B′′β mutant, and higher HMGR activity is present in b′′β-1 and OE- B′′β lines. However, further work is required to define the mechanism by which B′′β overexpression positively regulates HMGR.
The five Arabidopsis B′′ genes are represented by full-length or partial cDNA sequences in the GenBank database, so all of them are expressed. However, the B′′α (34% of the EST entries), B′′β (29%), and B′′γ (27%) transcripts are much more abundant than those encoding B′′δ (4%) or B′′ε (6%). According to the ATH1 22k microarray data accessible at the Genevestigator facility (https://www.genevestigator.com/gv/index.jsp), the levels of the Arabidopsis B′′ transcripts remain quite constant in different organs and throughout development. Interestingly, the microarray data suggest that the abundance of the B′′α transcript increases in response to salt, abscisic acid (ABA), or mildew and decreases in response to potyvirus infection. Since B′′γ is nearly identical to B′′β (only two conservative changes in 536–amino acid residue sequence), it will most likely bind HMGR1L and HMGR1S. Thus, the multilevel control of HMGR is likely a major role for the Arabidopsis B′′ protein family. On the other hand, B′′α and B′′β are 67.4% identical (76.9% similar) (i.e., nearly as divergent as the most distant members B′′δ and B′′ε; 64.0% identical and 74.7% similar). Therefore, it would not be surprising if all five B′′ isoforms interact with HMGR. The redundancy in the binding to HMGR underlines the importance of the Arabidopsis B′′ protein family. With this shared biochemical function, the different B′′ variants might have specific roles in the modulation of HMGR and maybe other targets during normal development and in response to a variety of stress and nonstress conditions. For instance, in the sterile medium, we observed that no gene can substitute for B′′α in regulating the multilevel response of HMGR to salt.
HMGR, PP2A, and the Signal Transduction Network
PP2A is a major phosphatase that accounts for ~25% of the total protein phosphatase activity in crude homogenates from several plants (MacKintosh and Cohen, 1989). It is found in most plant tissues and in many subcellular locations, including the nucleus, cytosol, and membranes, and in insoluble fractions (Smith and Walker, 1996). A considerable number of biochemical, pharmacological, and genetic studies have shown that PP2A is involved in the control of constitutive plant processes, such as cell cycle, metabolism, ion channel fluxes, growth and development, pollination and seed germination, and in the defense against many biotic or abiotic challenges (Smith and Walker, 1996; Luan, 2003; DeLong, 2006). PP2A has been proposed to modulate several metabolic processes in vascular plants, including sucrose synthesis, quinate metabolism, nitrogen assimilation, and nocturnal CO2 fixation (Siegl et al., 1990; MacKintosh et al., 1991; MacKintosh, 1992; Dong et al., 2001), but its involvement in isoprenoid biosynthesis had not been established. The broad distribution and functional diversity likely correspond to a multiplicity of PP2A holoenzymes (each formed by one A, one B-type, and one C subunit) reflected at the genomic level. In Arabidopsis, for instance, three genes code for A subunits, 17 for B-type subunits, and five for C subunits, which could theoretically form up to 255 different PP2A heterotrimers (Zhou et al., 2004). However, little is known about biochemical, structural, and regulatory properties of plant PP2A holoenzymes, and our knowledge on particular PP2A subunits at a protein level is also very limited and sparse. A definite role for PP2A subunits in plant growth and development has been uncovered only in the cases of Arabidopsis TONNEAU2 (subunit B′′) involved in the organization of the cortical cytoskeleton (Camilleri et al., 2002) and Arabidopsis RCN1 (subunit A). The knockout of RCN1 deregulates several phytohormone signaling processes, such as auxin transport and responsiveness to ABA and ethylene and seedling phototropism (Garbers et al., 1996; Kwak et al., 2002; Larsen and Cancel, 2003; Tseng and Briggs, 2010). It was proposed that RCN1 might contribute to the crosstalk between auxin and ABA signaling pathways that would be modulated as well by accompanying regulatory and catalytic subunits of PP2A (Kwak et al., 2002). The characterization of B-type regulatory PP2A subunits is particularly relevant, since they are expected to provide specificity to PP2A action. Here, we report that the B′′α PP2A subunit is involved in the regulation of root growth under salt stress. The data suggest that the multilevel control of HMGR by B′′α and PP2A is part of the plant response to this challenging condition. It is worth noting that this response is produced at a moderate stress (50 mM NaCl in half concentrated MS medium; 83 mM total salt) and without any noticeable alteration in the seedling phenotype. Thus, the salt-induced modulation of HMGR is not a desperate plant response but seems to contribute successfully to surmounting the adverse condition.
Plant PP2A activity or particular PP2A subunits have been defined as positive or negative transducers of a variety of environmental or hormonal signals in different plant systems (Monroy et al., 1998; Kwak et al., 2002; Larsen and Cancel, 2003; He et al., 2004; Pernas et al., 2007), but only two PP2A targets have been identified so far (Tseng and Briggs, 2010; Tang et al., 2011). Arabidopsis HMGR could be a PP2A target, since we have shown that its phosphorylation status is altered upon blockage of the phosphatase. Since HMGR catalyzes a key regulatory step of the isoprenoid biosynthetic route, the control of this enzyme by PP2A may be considered as an end point of signal transduction pathways. It has been shown that distinct light perception pathways involving cryptochrome and phytochrome photoreceptors contribute to downregulation of HMGR (Rodríguez-Concepción et al., 2004). We observed that the rcn1-1 mutation further increases HMGR activity in homozygous phyD-1 plants defective in phytochrome D (WS-2 ecotype). This additive effect supports that HMGR inhibition in Arabidopsis is mediated by two separate signal transduction pathways that involve phytochrome D and PP2A, respectively. As mentioned above, PP2A participates in ABA and auxin signaling, and exogenous addition of these factors affects HMGR activity (Russell and Davidson, 1982; Nishi and Tsuritani, 1983). Future investigations should help clarify to what extent modulation of HMGR by phytohormones depends on PP2A and whether this is part of the response to biotic or abiotic challenge.
METHODS
Plant Material and Growth Conditions
The rcn1-1 mutant (Garbers et al., 1996) was a kind gift of A. Delong (Brown University, Providence, RI). Mutants b′′α-1 and b′′α-2 (available at The Arabidopsis Information Resource under names SAIL_139_A03 and SAIL 76_G08, respectively) were obtained from Syngenta and mutants b′′α-3 (SALK_081091) and b′′β-1 (GS_5_88433) from the Nottingham Arabidopsis Stock Centre.
Plants were vernalized at 4°C for 3 d and grown in MS medium (Murashige and Skoog, 1962) at 22°C under long-day (16 h light/8 h darkness) or short-day (8 h ligh/16 h darkness) conditions. In the assays of Figures 7 and 8, germination and growth was done on polyester filter over half-concentrated MS medium. Seedlings were harvested during the last hour of the dark period to standardize the effect of photoperiod on HMGR levels. For harvesting, plates were removed from the growth chamber individually. After plate opening, seedlings were immediately frozen in liquid nitrogen and stored at −80°C until analysis.
For root growth assays (Figure 6), seeds were germinated in vertical plates containing half-concentrated MS and 1% agar. After 4 d under long-day conditions, 10 mutant and 10 of the corresponding wild-type plants at a similar developmental stage were transferred to 12 × 12-cm square plates containing the same medium and different concentrations of NaCl. Root tips were aligned to a marked reference. Growth was continued for six additional days with plates in a vertical position. The average of the main root new growth was measured and calculated as a percentage of that obtained on the plate without salt.
Generation and Analysis of the OE-B′′β Transgenic Line
The B′′β cDNA was amplified by PCR with primers 25F-XhoI and 25R-XhoI (see Supplemental Table 1 online) to generate XhoI flanking sites that allowed cloning into the SalI site of vector pMenchu (Ferrando et al., 2000). The new construct encoded a B′′β variant with an HA epitope at its N terminus. A blunt-ended NotI-NotI fragment of this construct, including the HA-B′′β-coding sequence, was cloned between blunt ended EcoRI and SacI sites of vector pPCV812 (Koncz et al., 1994). The resulting binary plasmid was introduced into Agrobacterium tumefaciens GV3101 (pMP90RK) strain (Koncz and Schell, 1986), which was then used to transform Arabidopsis thaliana Col-3 plants (generation T0) by the floral dip method (Clough and Bent, 1998). Transformants (generation T1) were selected with 40 μg/mL Hygromycin B in MS medium. Self-fertilization of one of the transformants rendered a segregating pool of T2 seeds that constituted the OE-B′′β line.
The presence of the HA-B′′β protein in the OE-B′′β line was examined by immunoblot analysis. Entire 14-d-old seedlings, grown under short-day conditions, were homogenized in liquid nitrogen and mixed with prechilled extraction buffer (2 μL per mg) containing 50 mM Tris-HCl, pH 7.6, 10% (v/v) glycerol, 1 mM EDTA, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride (PMSF), 10 μg/mL aprotinin, 0.5 μg/mL leupeptin, 3 μg/mL E64, and Protease Inhibitor Cocktail for plant cell and tissue extracts (Sigma-Aldrich; 20 μL/mL). After two centrifugations at 200g, 4°C for 10 min, 40 μg protein of the remaining supernatant were resolved by SDS-PAGE and transferred to a PVDF membrane. Proteins containing the HA epitope were detected by chemiluminescence with anti-HA high-affinity rat monoclonal antibody, clone 3F10 (1:1000 dilution; Roche Applied Science) and secondary rabbit anti-rat, peroxidase-conjugated serum (1:3000 dilution; Dako).
Cloning of the B′′β cDNA
As a first step to clone a cDNA encoding the entire B′′β, we amplified the 5′ part of the sequence missing in the original pACT-B′′β(88-536) clone. A linear PCR reaction, performed with the pACT library as a template and a single antisense primer (25GSP1), was followed by a nested exponential PCR performed with a sense primer (pACTBF) complementary to a sequence of the pACT vector and an antisense primer (25GSP2) complementary to a sequence of the B′′β insert. The resulting products were cloned, sequenced, and compared with the corresponding genomic sequence. We could then design the sense primer 25totF and the antisense primer 25totR to amplify a full-length B′′β cDNA from the pACT library. This cDNA was cloned in vector pGEM-T Easy (Promega) to obtain plasmid #6 of Supplemental Table 2 online. The identity and integrity of the final cDNA was confirmed by sequencing. Primers are listed in Supplemental Table 1 online.
Two-Hybrid Screening and Analysis
The two-hybrid screening and subsequent analysis was performed with the GAL4-based Matchmaker Two-Hybrid System 2 (Clontech). Translational fusions to the GAL4 activation domain (AD) were constructed in vector pACT or pACT2. Translational fusions to the GAL4 binding domain (BD) were constructed in vector pAS2-1. Yeast strain CG-1945 was transformed with the pAS2-1 derivative encoding the N-terminal region of HMGR1L (BD-NT1L) and subsequently retransformed with the pACT library derived from 3-d-old etiolated Arabidopsis seedlings (Kim et al., 1997). This phage library (reference CD4-22) was obtained from the ABRC and converted to its plasmid form (pACT) in the laboratory. From the 1.4 × 106 clones screened, those expressing AD fusion proteins that interact with BD-NT1L were identified as growing colonies in plates containing selection medium that did not contain Leu, Trp, and His (SC-Leu-Trp-His) but had 5 mM 3-amino-1,2,4-aminotriazole. The pACT-derived plasmids of positive clones were isolated to reconfirm interactions in yeast strain Y190 by growth on selection medium (SC-Leu-Trp-His+25 mM 3-amino-1,2,4-aminotriazole) and β-galactosidase filter-lifting assay.
Two-hybrid analyses were performed with the following chimeric proteins that contained the indicated Arabidopsis proteins or protein fragments: AD-B′′α, entire B′′α (GenBank AAD45158); AD-B′′α(1-538), residues 1 to 538 of B′′α; AD-B′′α(1-397), residues 1 to 397 of B′′α (AAD45158); AD-B′′β, entire B′′β (NP_568509); AD-B′′β(88-536), residues 88 to 536 of B′′β; BD-CD1, residues 166 to 592 of HMGR1S (P14891), equivalent to residues 216 to 642 of HMGR1L (AAR83122); BD-HMGS, entire 3-hydroxy-3-methylglutaryl CoA synthase (P54873); BD-NT1L, residues 1 to 100 of HMGR1L (AAR83122); BD-1Lextra, residues 1 to 50 of HMGR1L; BD-MVK, entire MVA kinase (P46086); BD-ΔNtFPS1, residues 26 to 342 of farnesyl-diphosphate synthase 1 (AAB07264); BD-NT1S, residues 1 to 50 of HMGR1S (P14891); BD-NT2, residues 1 to 32 of HMGR2 (P43256); BD-PR65, entire A2 variant of PR65 (NP_189208); and BD-SQS1, entire squalene synthase 1 (P53799). The cloning strategy for the different constructs is indicated in Supplemental Table 2 online. The EST clone G5C6T7 (pDF1) encoding the A2 variant of PR65 was obtained from the ABRC (http://www.Arabidopsis.org/).
Protein Expression and GST Binding Assays
Constructs encoding fusion proteins GST-B′′α, GST-NT1L, and GST-PR65 (A3 variant; NP_172790) were generated in vectors pGEX-4T-1 or pGEX-5X-2 (Amersham Biosciences). Constructs encoding NT1L-GST, 1Lextra-GST, NT1S-GST, and NT2-GST were generated in vector pET-23d(+) (Novagen) by a two-step cloning procedure. Introduction of the sequence encoding NT1L, 1Lextra, NT1S, or NT2 was followed by cloning of the GST-coding sequence. Constructs encoding His-tagged proteins were generated in vectors pRSET C or pET-28a(+).The cloning procedures are indicated in Supplemental Table 2 online.
Expression and binding of GST fusion proteins were performed as described (Ausubel et al., 1987), with minor modifications. GST chimeras were produced in Escherichia coli BL21 cells (pGEX-4T-1 and pGEX-5X-2 derivatives) or in E. coli BL21(DE3)pLysS cells (pET-23d+ derivatives). Cultures were grown at 37°C but shifted to 22°C after they reached 0.4 to 0.6 OD600 and 0.4 mM isopropyl-β-d-thiogalactopyranoside was added. Bacterial cells were resuspended in 10 mL of ice-cold lysis buffer composed of 10 mM NaPO4, pH 7.4, 154 mM NaCl, 1 mM PMSF, 10 μg/mL aprotinin, 0.5 μg/mL leupeptin, 3 μg/mL E64, and 50 μL/mL bacterial protease inhibitor cocktail (Sigma-Aldrich) and lysed by sonication. The lysate was supplemented with 1% (v/v) Triton X-100 and centrifuged at 12,500g for 10 min at 4°C. After addition of 500 μL of a 50% slurry of glutathione-agarose beads (Sigma-Aldrich) to the supernatant, the mixture was incubated with gentle shaking for 30 min at 4°C. The agarose beads were washed four times with a buffer containing 10 mM NaPO4, pH 7.4, 154 mM NaCl, and 1% (v/v) Triton X-100 and resuspended in 250 μL of lysis buffer.
The 45Ca2+ binding to GST derivatives (Figure 2E) was performed as described (Maruyama et al., 1984). [35S]-Radiolabeled HisB′′α, HisB′′β(88-536), and HisB′′β proteins were synthesized in vitro from the corresponding plasmids with the Transcription and Translation Coupled Reticulocyte System (Promega). Three to five micrograms of GST fusion proteins in Agarose beads were mixed with an additional 20 μL of glutathione-agarose slurry and added to 400 μL of buffer BBB (50 mM Tris-HCl, pH 7.5, 150 mM KCl, 10% [v/v] glycerol, 1 mM MgCl2, and 1% [v/v] Triton X-100), containing 1 mM PMSF, 10 μg/mL aprotinin, 0.5 μg/mL leupeptin, 3 μg/mL E64, and 10 mg/mL BSA. After the final addition of 1 to 5 μL of the in vitro translation samples, the mixtures were incubated for 1 h at 4°C with gentle rotation. Beads were recovered by centrifugation and washed three times with 1 mL of buffer BBB. When indicated (Figure 2F), buffer BBB contained additional 5 mM Ca2+ or 5 mM EGTA at both binding and washing steps. Proteins were released from the resin by boiling in 15 μL of electrophoresis sample buffer and analyzed by denaturing polyacrylamide gel electrophoresis and fluorography.
Determination of Transcript Levels
Total RNA was extracted from frozen samples with the RNeasy plant mini kit (Qiagen), including the DNase I digestion step. Two micrograms of the RNA were reverse transcribed with the High Capacity RNA-to-cDNA kit (Applied Biosystems). In the experiments of Figure 3A, quantitative PCR was performed with the QuantumRNA 18S Internal Standards kit (Ambion). Transcript HMG1 was amplified with oligonucleotides H1.3F and H1.2R for 25 cycles using a 4:6 ratio of normal 18S rRNA primers to competimers. Transcript HMG2 was amplified during 30 cycles, with oligonucleotides H2.4F and H2.3R and a 2:8 primers to competimers ratio. Linearity in the amplification of the HMG1 and HMG2 transcripts and the 18S rRNA was confirmed with ethidium bromide staining after PCR using different volumes of the corresponding cDNA templates. In the experiments of Figures 5C, 5E, 8B, and 8C, transcripts were quantitated by qRT-PCR. In these assays, the reverse transcription mix was diluted 1:100 with water, and a 4 μL aliquot was analyzed in triplicate, using the Power SyBR Green Master Mix and an ABI PRISM 7700 sequence detection system (Applied Biosystems). The PCR thermal profile consisted of 40 cycles with 30 s at 94°C, 40 s at 61°C, and 45 s at 72°C, preceded by 10 min at 95°C. Relative quantification of the target transcripts was obtained according to the threshold cycle (CT) 2−ΔΔCT method (Livak and Schmittgen, 2001) using the At4g26410 transcript as a normalization reference, since it shows very stable levels under a variety of biotic and abiotic stress conditions (Czechowski et al., 2005). Amplification efficiencies of target and reference transcripts were close to 1 and approximately equal, as required for the 2−ΔΔCT method. In preliminary standard curve assays, linearity between CT and the logarithm of cDNA sample volume was obtained over a range of four orders of magnitude dilution for both HMG1 and At4g26410 transcripts. In Arabidopsis nontreated seedlings, these transcripts have very similar levels, since their CT values do not differ more than one cycle. Melting curves were examined to confirm the absence of unspecific amplification products. The primer sets and the amplicon size for the different transcripts were as follows: At4g26410, primers 264F* and 264R (81 bp); HMG1, 764F* and 764R (85 bp); B′′β, h* and i (96 bp); RAB18, 18F* and 18R; RD29a, 29aF1and 29aR1* (95 bp); RD22, 22F* and 22R (85 bp); KIN2, K2F* and K2R (84 bp). The asterisk denotes the presence of an exon-exon junction in the primer sequence. Primers are listed in Supplemental Table 1 online.
Determination of HMGR Protein Levels
HMGR protein levels were determined by immunoblot using a rabbit polyclonal antibody raised against the catalytic domain of Arabidopsis HMGR1 (CD1) that also recognizes the HMGR2 isoform (Leivar et al., 2005). Occasional cross-reaction of the serum with non-HMGR proteins of the Arabidopsis electrophoretic pattern suggested contamination of the original CD1 immunogen with E. coli proteins. To prevent unspecific signals, we immunosubtracted the rabbit anti-CD1 serum with E. coli BL21[DE3] protein extract prepared according to the same protocol previously applied to obtain the CD1 immunogen (Dale et al., 1995) but omitting the Affi-Gel Blue chromatography step. Ten milligrams of purified E. coli proteins were coupled at 75% efficiency to a HiTrap NHS-activated HP column (GE Healthcare) to obtain the immunosubtracting resin. The new antibody preparation (anti-CD1-i) was used at 1:1000 dilution and the secondary antibody (horseradish peroxidase anti-rabbit IgG, Santa Cruz) at 1:10,000. For immunoblot analyses, each well in 10% acrylamide SDS gels was loaded with 5 μg of total protein. Immunoblot images were developed with ECL+Plus (GE Healthcare) for 1 to 10 min in a LAS-3000 luminescent image analyzer (Fujifilm). After exposure, PDVF membranes were stained for 1 min with 1% (w/v) Coomassie Brilliant Blue and photographed with the image analyzer. The intensity of the chemiluminescence band detected with the anti-CD1-i antibody was normalized to the intensity of the Coomassie-stained large subunit ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) band after triplicate quantification of both with the Multi Gauge software (Fujifilm).
Digestion with λ Phosphatase
Protein extracts were prepared from Arabidopsis seedlings as for the determination of HMGR activity. Sixty micrograms of protein were incubated for 7 min at 30°C with 800 units of λppase in 50 μL of incubation mix containing 40 mM HEPES-KOH, pH 7.2, 100 mM sucrose, 50 mM KCl, 0.2% (v/v) Triton X-100, 30 mM MnCl2, 10 mM DTT, 60 μg/mL PMSF, 75 μg/mL aprotinin, 21 μg/mL E64, 100 μg/mL leupeptin, and 9 μg/mL pepstatin A. Digestion was stopped by addition of 50 μL twice-concentrated electrophoresis buffer and immediate boiling.
Determination of HMGR-Specific Activity
Entire seedlings (250 to 400 mg) were homogenized in liquid nitrogen and mixed with prechilled extraction buffer (2 to 3 μL per mg) containing 40 mM HEPES-KOH, pH 7.2, 100 mM sucrose, 50 mM KCl, 0.2% (v/v) Triton X-100, 10 mM DTT, 100 μg/mL PMSF, 15 μg/mL aprotinin, 10 μg/mL E64, 20 μg/mL leupeptin, and 1.5 μg/mL pepstatin A (antiproteolytics from Sigma-Aldrich) and either 5 mM CaCl2 (Figure 3A) or 5 mM EGTA (Figures 5E, 7A, 7D, 7E, and 7F). The slurry was centrifuged twice at 200g, 4°C for 10 min. The HMGR activity of the final supernatant was determined radiometrically as described (Bach et al., 1986) with the following modifications. The standard assay system consisted of 208 mM Tris-HCl, pH 7.2, 50 mM EDTA, 10 mM DTT, 0.5 mg/mL BSA, 2.5 mM NADP, 50 mM glucose-6-phosphate, 8.3 mU yeast glucose-6-phosphate dehydrogenase, 33.3 μM HMG-CoA, and 14.6 μM (1295 Bq) [3-14C]-HMG-CoA (Amersham Biosciences) in a final volume of 42 μL. Bands of labeled mevalonolactone were cut from the thin-layer chromatographic plastic sheet and counted after localizing them by exposure to a phosphor imager screen. Protein concentration was determined with the Protein Assay reagent (Bio-Rad) and BSA as a standard. Radioactive incorporation into mevalonolactone was measured in duplicate aliquots from the original extract and protein concentration in quadruplicate to determine the corresponding HMGR-specific activity.
Phylogenetic Analysis
Phylogenetic analyses were conducted with the program MEGA version 2.1 (Kumar et al., 2001) using the neighbor-joining algorithm and the number of amino acid differences as a distance. Those sites in which at least one of the sequences displayed a deletion were not considered. As an outgroup, we chose a B′′-like sequence from Giardia lamblia, a protist that represents the earliest diverging lineage in the eukaryotic line of descent in both small subunit rRNA- and protein-based phylogenetic trees (Hashimoto et al., 1994). The bootstrap procedure (Felsenstein, 1985) with 1000 replicates was applied to determine the consistency of the analysis.
Accession Numbers
The cDNA sequence data from this article can be found in the GenBank database under accession numbers AY462121 (B′′α, At5g44090), AY462122 (B′′β, At5g28850), AY063937 (B′′γ, At5g28900), NM_104323 (B′′δ, At1g54450), and NM_202028 (B′′ε, At1g03960). The input protein sequences of Figure 2A are available at the GenBank database under the following accession numbers: AtB′′α (AAD45158), AtB′′β (NP_568509), AtB′′γ (NP_198242), AtB′′δ (NP_175847) and AtB′′ε (AAO42077), AtTON2 (AAG35792), Dm1 (AAG22156), Gl1 (XP_001705580), Hs1 (NP_060387), HsPR70 (NP_037371), HsPR72 (AAB02614), Mm1 (BAA95061), MmPR59 (AAC98973), Os1 (AAK13162), Os2 (BAD34016), XlPR70 (AAK98642), and XlPR130 (AAK98643).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Specific Interaction of B′′α and B′′β(88-538) with the N-Terminal Region of HMGR1L.
Supplemental Figure 2. RT-PCR Analysis of the b′′β-1 Mutant.
Supplemental Figure 3. Two-Hybrid Analysis of B′′α(1-397).
Supplemental Figure 4. Seedling Establishment Analysis of the b′′β-1 Mutant.
Supplemental Table 1. PCR Primers.
Supplemental Table 2. Plasmid Constructs.
Supplemental Data Set 1. Text File of the Sequence Alignment Used for the Phylogenetic Analysis in Figure 2A.
Acknowledgments
We thank A. Delong (Brown University), Syngenta, the Nottingham Arabidopsis Stock Centre, and the ABRC for seed stocks and cDNA clones. We thank Gabriela Puntes for the genotyping of the b′′β-1 mutant, Patricia Lorden and Roser Adalid from excellent technical assistance, Carme Caelles for advice in the phosphatase treatments, and Victoria Lumbreras and Víctor González for helpful discussions. We also thank the staff of the Installació Radiactiva of the Faculty of Biology, Serveis Cientificotècnics, and Serveis de Camps Experimentals of the University of Barcelona for their help in plant culture, DNA sequencing, real-time PCR analysis, and HMGR activity assays. This work was supported by grants of the Spanish Ministerio de Ciencia e Innovación (BMC2003-03450 and BFU2006-14655 to N.C., BFU2006-00544 and BIO2009-06984 to A.F., and BIO2009-09523 to A.B., all of them including European Regional Development Funds), the Spanish Consolider-Ingenio 2010 Program (CSD2007-00036 Centre for Research in Agrigenomics), and the Generalitat de Catalunya (2009SGR0026). P.L. and M.A. were recipients of predoctoral fellowships from the Comissió Interdepartamental de Recerca i Innovació Tecnològica and the Spanish Ministerio de Educación Cultura y Deporte, respectively.
References
- Aukerman M.J., Hirschfeld M., Wester L., Weaver M., Clack T., Amasino R.M., Sharrock R.A. (1997). A deletion in the PHYD gene of the Arabidopsis Wassilewskija ecotype defines a role for phytochrome D in red/far-red light sensing. Plant Cell 9: 1317–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel F.M., Brent R., Kingston R.E., Moore D.D., Seidman J.G., Smith J.A., Struhl K. (1987). Current Protocols in Molecular Biology. (New York: John Wiley and Sons; ). [Google Scholar]
- Bach T.J., Rogers D.H., Rudney H. (1986). Detergent-solubilization, purification, and characterization of membrane-bound 3-hydroxy-3-methylglutaryl-coenzyme A reductase from radish seedlings. Eur. J. Biochem. 154: 103–111 [DOI] [PubMed] [Google Scholar]
- Blakeslee J.J., Zhou H.W., Heath J.T., Skottke K.R., Barrios J.A.R., Liu S.Y., DeLong A. (2008). Specificity of RCN1-mediated protein phosphatase 2A regulation in meristem organization and stress response in roots. Plant Physiol. 146: 539–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camilleri C., Azimzadeh J., Pastuglia M., Bellini C., Grandjean O., Bouchez D. (2002). The Arabidopsis TONNEAU2 gene encodes a putative novel protein phosphatase 2A regulatory subunit essential for the control of the cortical cytoskeleton. Plant Cell 14: 833–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos N., Boronat A. (1995). Targeting and topology in the membrane of plant 3-hydroxy-3-methylglutaryl coenzyme A reductase. Plant Cell 7: 2163–2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell J. (1995). Biochemistry and molecular biology of the isoprenoid biosynthetic pathway in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46: 521–547 [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M.K., Scheible W.R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale S., Arró M., Becerra B., Morrice N.G., Boronat A., Hardie D.G., Ferrer A. (1995). Bacterial expression of the catalytic domain of 3-hydroxy-3-methylglutaryl-CoA reductase (isoform HMGR1) from Arabidopsis thaliana, and its inactivation by phosphorylation at Ser577 by Brassica oleracea 3-hydroxy-3-methylglutaryl-CoA reductase kinase. Eur. J. Biochem. 233: 506–513 [DOI] [PubMed] [Google Scholar]
- DeLong A. (2006). Switching the flip: Protein phosphatase roles in signaling pathways. Curr. Opin. Plant Biol. 9: 470–477 [DOI] [PubMed] [Google Scholar]
- Deruère J., Jackson K., Garbers C., Söll D., Delong A. (1999). The RCN1-encoded A subunit of protein phosphatase 2A increases phosphatase activity in vivo. Plant J. 20: 389–399 [DOI] [PubMed] [Google Scholar]
- Dong L.Y., Ermolova N.V., Chollet R. (2001). Partial purification and biochemical characterization of a heteromeric protein phosphatase 2A holoenzyme from maize (Zea mays L.) leaves that dephosphorylates C4 phosophoenolpyruvate carboxylase. Planta 213: 379–389 [DOI] [PubMed] [Google Scholar]
- Enjuto M., Balcells L., Campos N., Caelles C., Arró M., Boronat A. (1994). Arabidopsis thaliana contains two differentially expressed 3-hydroxy-3-methylglutaryl-CoA reductase genes, which encode microsomal forms of the enzyme. Proc. Natl. Acad. Sci. USA 91: 927–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjuto M., Lumbreras V., Marín C., Boronat A. (1995). Expression of the Arabidopsis HMG2 gene, encoding 3-hydroxy-3-methylglutaryl coenzyme A reductase, is restricted to meristematic and floral tissues. Plant Cell 7: 517–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. (1985). Confidence limits on phylogenetics: an approach using the bootstrap. Evolution 39: 783–791 [DOI] [PubMed] [Google Scholar]
- Ferrando A., Farràs R., Jásik J., Schell J., Koncz C. (2000). Intron-tagged epitope: A tool for facile detection and purification of proteins expressed in Agrobacterium-transformed plant cells. Plant J. 22: 553–560 [DOI] [PubMed] [Google Scholar]
- Garbers C., DeLong A., Deruére J., Bernasconi P., Söll D. (1996). A mutation in protein phosphatase 2A regulatory subunit A affects auxin transport in Arabidopsis. EMBO J. 15: 2115–2124 [PMC free article] [PubMed] [Google Scholar]
- Harker M., Holmberg N., Clayton J.C., Gibbard C.L., Wallace A.D., Rawlins S., Hellyer S.A., Lanot A., Safford R. (2003). Enhancement of seed phytosterol levels by expression of an N-terminal truncated Hevea brasiliensis (rubber tree) 3-hydroxy-3-methylglutaryl-CoA reductase. Plant Biotechnol. J. 1: 113–121 [DOI] [PubMed] [Google Scholar]
- Hashimoto T., Nakamura Y., Nakamura F., Shirakura T., Adachi J., Goto N., Okamoto K.-I., Hasegawa M. (1994). Protein phylogeny gives a robust estimation for early divergences of eukaryotes: phylogenetic place of a mitochondria-lacking protozoan, Giardia lamblia. Mol. Biol. Evol. 11: 65–71 [DOI] [PubMed] [Google Scholar]
- He X.H., Anderson J.C., del Pozo O., Gu Y.Q., Tang X.Y., Martin G.B. (2004). Silencing of subfamily I of protein phosphatase 2A catalytic subunits results in activation of plant defense responses and localized cell death. Plant J. 38: 563–577 [DOI] [PubMed] [Google Scholar]
- Hemmerlin A., Hoeffler J.F., Meyer O., Tritsch D., Kagan I.A., Grosdemange-Billiard C., Rohmer M., Bach T.J. (2003). Cross-talk between the cytosolic mevalonate and the plastidial methylerythritol phosphate pathways in tobacco bright yellow-2 cells. J. Biol. Chem. 278: 26666–26676 [DOI] [PubMed] [Google Scholar]
- Hendershot J.D., III, Esmon C.A., Lumb J.E., Rundle S.J. (1999). Identification and characterization of sequences encoding a 62 kD B′′ regulatory subunit of Arabidopsis thaliana protein phosphatase 2A (Accession No. AF165429) (PGR 99-125). Plant Physiol. 121: 31110577163 [Google Scholar]
- Hey S.J., Powers S.J., Beale M.H., Hawkins N.D., Ward J.L., Halford N.G. (2006). Enhanced seed phytosterol accumulation through expression of a modified HMG-CoA reductase. Plant Biotechnol. J. 4: 219–229 [DOI] [PubMed] [Google Scholar]
- Janssens V., Goris J. (2001). Protein phosphatase 2A: A highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem. J. 353: 417–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens V., Jordens J., Stevens I., Van Hoof C., Martens E., De Smedt H., Engelborghs Y., Waelkens E., Goris J. (2003). Identification and functional analysis of two Ca2+-binding EF-hand motifs in the B′′/PR72 subunit of protein phosphatase 2A. J. Biol. Chem. 278: 10697–10706 [DOI] [PubMed] [Google Scholar]
- Kim J., Harter K., Theologis A. (1997). Protein-protein interactions among the Aux/IAA proteins. Proc. Natl. Acad. Sci. USA 94: 11786–11791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klee C.B., Crouch T.H., Krinks M.H. (1979). Calcineurin: A calcium- and calmodulin-binding protein of the nervous system. Proc. Natl. Acad. Sci. USA 76: 6270–6273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C., Martini N., Szabados L., Hrouda M., Bachmair A., Schell J. (1994). Specialized vectors for gene tagging and expression studies. In Plant Molecular Biology Manual, Gelvin S.B., Schilperoort R.A., Verma D.P.S., eds (Dordrecht, The Netherlands: Kluwer; ), pp. 1–22 [Google Scholar]
- Koncz C., Schell J. (1986). The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 204: 383–396 [Google Scholar]
- Korth K.L., Jaggard D.A.W., Dixon R.A. (2000). Developmental and light-regulated post-translational control of 3-hydroxy-3-methylglutaryl-CoA reductase levels in potato. Plant J. 23: 507–516 [DOI] [PubMed] [Google Scholar]
- Kumar S., Tamura K., Ingrid B., Jakobsen I.B., Nei M. (2001). MEGA2: Molecular Evolutionary Genetics Analysis Software. (Tempe, AZ: Arizona State University; ). [DOI] [PubMed] [Google Scholar]
- Kwak J.M., Moon J.H., Murata Y., Kuchitsu K., Leonhardt N., DeLong A., Schroeder J.I. (2002). Disruption of a guard cell-expressed protein phosphatase 2A regulatory subunit, RCN1, confers abscisic acid insensitivity in Arabidopsis. Plant Cell 14: 2849–2861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen P.B., Cancel J.D. (2003). Enhanced ethylene responsiveness in the Arabidopsis eer1 mutant results from a loss-of-function mutation in the protein phosphatase 2A A regulatory subunit, RCN1. Plant J. 34: 709–718 [DOI] [PubMed] [Google Scholar]
- Leivar P., González V.M., Castel S., Trelease R.N., López-Iglesias C., Arró M., Boronat A., Campos N., Ferrer A., Fernàndez-Busquets X. (2005). Subcellular localization of Arabidopsis 3-hydroxy-3-methylglutaryl-coenzyme A reductase. Plant Physiol. 137: 57–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Virshup D.M. (2002). Two conserved domains in regulatory B subunits mediate binding to the A subunit of protein phosphatase 2A. Eur. J. Biochem. 269: 546–552 [DOI] [PubMed] [Google Scholar]
- Li Y.M., MacKintosh C., Casida J.E. (1993). Protein phosphatase 2A and its [3H]cantharidin/[3H]endothall thioanhydride binding site. Inhibitor specificity of cantharidin and ATP analogues. Biochem. Pharmacol. 46: 1435–1443 [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Luan S. (2003). Protein phosphatases in plants. Annu. Rev. Plant Biol. 54: 63–92 [DOI] [PubMed] [Google Scholar]
- Lumbreras V., Campos N., Boronat A. (1995). The use of an alternative promoter in the Arabidopsis thaliana HMG1 gene generates an mRNA that encodes a novel 3-hydroxy-3-methylglutaryl coenzyme A reductase isoform with an extended N-terminal region. Plant J. 8: 541–549 [DOI] [PubMed] [Google Scholar]
- MacKintosh C. (1992). Regulation of spinach-leaf nitrate reductase by reversible phosphorylation. Biochim. Biophys. Acta 1137: 121–126 [DOI] [PubMed] [Google Scholar]
- MacKintosh C., Coggins J., Cohen P. (1991). Plant protein phosphatases. Subcellular distribution, detection of protein phosphatase 2C and identification of protein phosphatase 2A as the major quinate dehydrogenase phosphatase. Biochem. J. 273: 733–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKintosh C., Cohen P. (1989). Identification of high levels of type 1 and type 2A protein phosphatases in higher plants. Biochem. J. 262: 335–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama K., Mikawa T., Ebashi S. (1984). Detection of calcium binding proteins by 45Ca autoradiography on nitrocellulose membrane after sodium dodecyl sulfate gel electrophoresis. J. Biochem. 95: 511–519 [DOI] [PubMed] [Google Scholar]
- Mayer-Jaekel R.E., Hemmings B.A. (1994). Protein phosphatase 2A—A ‘ménage à trois’. Trends Cell Biol. 4: 287–291 [DOI] [PubMed] [Google Scholar]
- Monroy A.F., Sangwan V., Dhindsa R.S. (1998). Low-temperature signal transduction during cold acclimation: Protein phosphatase 2A as an early target for cold-inactivation. Plant J. 13: 653–660 [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and bio-assays with tobacco tissue cultures. Physiol. Plant. 15: 473–497 [Google Scholar]
- Nieto B., Forés O., Arró M., Ferrer A. (2009). Arabidopsis 3-hydroxy-3-methylglutaryl-CoA reductase is regulated at the post-translational level in response to alterations of the sphingolipid and the sterol biosynthetic pathways. Phytochemistry 70: 53–59 [DOI] [PubMed] [Google Scholar]
- Nishi A., Tsuritani I. (1983). Effect of auxin on the metabolism of mevalonic acid in suspension-cultured carrot cells. Phytochemistry 22: 399–401 [Google Scholar]
- Nylander M., Svensson J., Palva E.T., Welin B.V. (2001). Stress-induced accumulation and tissue-specific localization of dehydrins in Arabidopsis thaliana. Plant Mol. Biol. 45: 263–279 [DOI] [PubMed] [Google Scholar]
- Ohyama K., Suzuki M., Masuda K., Yoshida S., Muranaka T. (2007). Chemical phenotypes of the hmg1 and hmg2 mutants of Arabidopsis demonstrate the in-planta role of HMG-CoA reductase in triterpene biosynthesis. Chem. Pharm. Bull. (Tokyo) 55: 1518–1521 [DOI] [PubMed] [Google Scholar]
- Pernas M., García-Casado G., Rojo E., Solano R., Sánchez-Serrano J.J. (2007). A protein phosphatase 2A catalytic subunit is a negative regulator of abscisic acid signalling. Plant J. 51: 763–778 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Concepción M., Campos N., Ferrer A., Boronat A. (2011). Biosynthesis of isoprenoid precursors in Arabidopsis. In Isoprenoid Synthesis and Function in Plants and Microorganisms: New Concepts and Experimental Approaches, Bach T.J., Rohmer M., Gerhenzon J., eds (New York: Springer: ), in press [Google Scholar]
- Rodríguez-Concepción M., Forés O., Martinez-García J.F., González V., Phillips M.A., Ferrer A., Boronat A. (2004). Distinct light-mediated pathways regulate the biosynthesis and exchange of isoprenoid precursors during Arabidopsis seedling development. Plant Cell 16: 144–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell D.W., Davidson H. (1982). Regulation of cytosolic HMG-CoA reductase activity in pea seedlings: Contrasting responses to different hormones, and hormone-product interaction, suggest hormonal modulation of activity. Biochem. Biophys. Res. Commun. 104: 1537–1543 [DOI] [PubMed] [Google Scholar]
- Siegl G., MacKintosh C., Stitt M. (1990). Sucrose-phosphate synthase is dephosphorylated by protein phosphatase 2A in spinach leaves. Evidence from the effects of okadaic acid and microcystin. FEBS Lett. 270: 198–202 [DOI] [PubMed] [Google Scholar]
- Smith R.D., Walker J.C. (1996). Plant protein phosphatases. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47: 101–125 [DOI] [PubMed] [Google Scholar]
- Song H., Zhao R., Fan P., Wang X., Chen X., Li Y. (2009). Overexpression of AtHsp90.2, AtHsp90.5 and AtHsp90.7 in Arabidopsis thaliana enhances plant sensitivity to salt and drought stresses. Planta 229: 955–964 [DOI] [PubMed] [Google Scholar]
- Sontag E. (2001). Protein phosphatase 2A: The Trojan horse of cellular signaling. Cell. Signal. 13: 7–16 [DOI] [PubMed] [Google Scholar]
- Stermer B.A., Bianchini G.M., Korth K.L. (1994). Regulation of HMG-CoA reductase activity in plants. J. Lipid Res. 35: 1133–1140 [PubMed] [Google Scholar]
- Sugden C., Donaghy P.G., Halford N.G., Hardie D.G. (1999). Two SNF1-related protein kinases from spinach leaf phosphorylate and inactivate 3-hydroxy-3-methylglutaryl-coenzyme A reductase, nitrate reductase, and sucrose phosphate synthase in vitro. Plant Physiol. 120: 257–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M., Kamide Y., Nagata N., Seki H., Ohyama K., Kato H., Masuda K., Sato S., Kato T., Tabata S., Yoshida S., Muranaka T. (2004). Loss of function of 3-hydroxy-3-methylglutaryl coenzyme A reductase 1 (HMG1) in Arabidopsis leads to dwarfing, early senescence and male sterility, and reduced sterol levels. Plant J. 37: 750–761 [DOI] [PubMed] [Google Scholar]
- Tang W., et al. (2011). PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat. Cell Biol. 13: 124–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng T.S., Briggs W.R. (2010). The Arabidopsis rcn1-1 mutation impairs dephosphorylation of Phot2, resulting in enhanced blue light responses. Plant Cell 22: 392–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virshup D.M. (2000). Protein phosphatase 2A: A panoply of enzymes. Curr. Opin. Cell Biol. 12: 180–185 [DOI] [PubMed] [Google Scholar]
- Yang Z.B., Park H.S., Lacy G.H., Cramer C.L. (1991). Differential activation of potato 3-hydroxy-3-methylglutaryl coenzyme A reductase genes by wounding and pathogen challenge. Plant Cell 3: 397–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H.W., Nussbaumer C., Chao Y., DeLong A. (2004). Disparate roles for the regulatory A subunit isoforms in Arabidopsis protein phosphatase 2A. Plant Cell 16: 709–722 [DOI] [PMC free article] [PubMed] [Google Scholar]