This work shows that in the phytopathogenic fungus Ustilago maydis, the genetic pathway involved in the transmission of signals required for the response to DNA damage is also used to control the specific dikaryotic cell cycle that predominates proliferation in planta.
Abstract
In the phytopathogenic fungus Ustilago maydis, the dikaryotic state dominates the period of growth occurring during the infectious phase. Dikaryons are cells in which two nuclei, one from each parent cell, share a single cytoplasm for a period of time without undergoing nuclear fusion. In fungal cells, maintenance of the dikaryotic state requires an intricate cell division process that often involves the formation of a structure known as the clamp connection as well as the sorting of one of the nuclei to this structure to ensure that each daughter dikaryon inherits a balance of each parental genome. Here, we describe an atypical role of the DNA damage checkpoint kinases Chk1 and Atr1 during pathogenic growth of U. maydis. We found that Chk1 and Atr1 collaborate to control cell cycle arrest during the induction of the virulence program in U. maydis and that Chk1 and Atr1 work together to control the dikaryon formation. These findings uncover a link between a widely conserved signaling cascade and the virulence program in a phytopathogen. We propose a model in which adjustment of the cell cycle by the Atr1-Chk1 axis controls fidelity in dikaryon formation. Therefore, Chk1 and Atr1 emerge as critical cell type regulators in addition to their roles in the DNA damage response.
INTRODUCTION
The sexual life cycle starts with fusion of two different haploid cells, proceeds to the formation of a diploid, and ends with meiosis to generate four haploid cells (Chen et al., 2007). However, the way in which haploid cells are brought together and the predominance of the haploid or diploid phase varies between species. In fungal cells, mating, the process equivalent to fertilization, brings together two haploid nuclei in the same cytoplasm. In some species of fungi, this process is followed by nuclear fusion, resulting in a diploid nucleus that enters meiosis immediately (as occurs in the fission yeast Schizosaccharomyces pombe) or that is maintained and proliferates in the diploid state (as in budding yeast Saccharomyces cerevisiae). However, for a large group of fungi, mating results in a dikaryon, a cell in which the two nuclei, one from each parent cell, share a single cytoplasm for a period of time without undergoing nuclear fusion. The dikaryon stage is typical in the life cycles of many fungal species primarily in the Basidiomycota, a large group that includes mushrooms, bracket fungi, and many phytopathogenic fungi, such as the maize pathogen Ustilago maydis.
Establishment and maintenance of dikaryotic growth in Basidiomycete fungi is controlled by information specified at the Mating Type (MAT) loci, specialized regions of fungal genomes akin to the sex chromosomes of larger eukaryotes (Lee et al., 2010). Although the specific contribution of the MAT locus components to dikaryon formation varies among species characterized thus far, a central element common to all of them is the activation of a specific transcriptional cascade controlled by a heterodimeric homeodomain transcription factor, with components derived from the MAT locus of each parent. Dikaryotic maintenance involves an intricate cell division process to ensure that each dikaryon inherits a balance of each parental genome. In many dikaryotic basidiomycetes (with some exceptions, such as various Uredinomycetes; for instance, see Ikeda et al., 2003) cell division involves the formation of a specialized structure known as the clamp connection as well as the sorting of one of the nuclei to this structure. In this way, nuclear division occurs in a synchronous and independent fashion in two distinct subcellular compartments. Once nuclear division is finished, the clamp connection is resolved to reconstitute the dikaryotic status of daughter cells (Brown and Casselton, 2001; Gladfelter and Berman, 2009). Studies with Coprinopsis cinerea and Schizophyllum commune indicated that the MAT genes encoding homeodomain transcription factors govern nuclear pairing as well as clamp formation (Casselton and Olesnicky, 1998; Kües, 2000); therefore, a connection between MAT genes and cell cycle control is predicted, although the details behind these connections are largely unknown.
In U. maydis, the dikaryotic state dominates the period of growth occurring during the infectious phase. This state is dependent upon the MAT-encoded homeoprotein called the b-complex, whose subunits, bW and bE, are provided by each mating partner (Feldbrügge et al., 2004; Brefort et al., 2009). During induction of the virulence program in U. maydis, an infectious dikaryotic hypha is produced on the plant surface as a result of mating of a pair of compatible haploid budding cells. The infectious hypha or filament is composed of a single dikaryotic cell that is arrested in the cell cycle at G2 phase (Mielnichuk et al., 2009), and its formation is dependent on synthesis of the b-complex. The arrest is transient, and eventually the filament manages to enter the plant tissue, where it starts to proliferate while maintaining its dikaryotic status. Since mutations that abolish this transient cell cycle arrest also impair dikaryon formation (Mielnichuk et al., 2009), it is thought that b-induced cell cycle arrest is required for dikaryon establishment in U. maydis. In our efforts to describe the mechanisms behind this transient cell cycle arrest, we recently reported that the widely conserved Chk1 protein kinase is required for this cell cycle arrest (Mielnichuk et al., 2009). Chk1 is better known as a key signal transducer within the DNA damage response cascade (Chen and Sanchez, 2004) in a broad range of eukaryotes, including U. maydis (Pérez-Martín, 2009). Here, we investigated the response of Chk1 to b-complex formation. We present findings showing that Chk1 is activated through phosphorylation by the conserved upstream activating kinase Atr1 and that the Atr1-Chk1 regulatory axis serves in maintenance of dikaryotic status in planta. Because Chk1 controls cell cycle progression, we propose that b-complex–mediated activation of the Atr1-Chk1 axis is part of the mechanism responsible of coordination during a dikaryotic cell cycle.
RESULTS
b-Dependent Cell Cycle Arrest Requires Chk1 Activating Phosphorylation
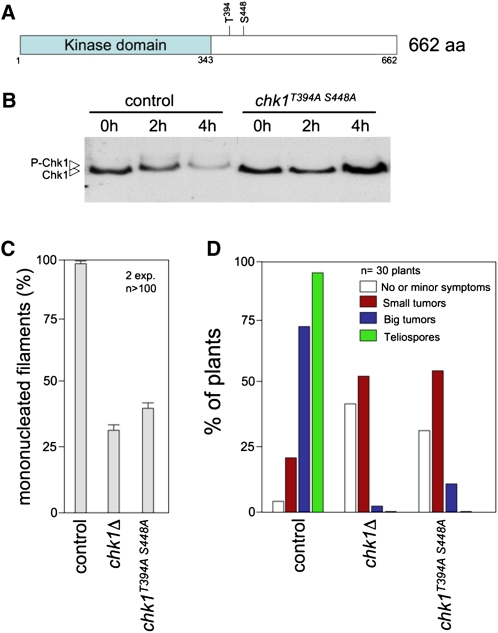
Assembly of the heterodimeric b-complex during dikaryon formation in U. maydis is concomitant with transient cell cycle arrest as well as with accumulation of phosphorylated forms of Chk1 and translocation of Chk1 into the nucleus, two hallmarks of Chk1 activation (Mielnichuk et al., 2009). Previous research (Pérez-Martín, 2009) established that in response to DNA damage, Chk1 activation results in G2 cell cycle arrest and involves phosphorylation at two residues (Thr-394 and Ser-448) located in the regulatory domain (Figure 1A). Mutant isoforms containing Ala in place of these residues could not be activated in response to DNA damage signals (Pérez-Martín, 2009). To determine whether these phosphorylation sites were also important during the cell cycle arrest associated with dikaryon formation, we measured the ability of these phosphorylation refractory Chk1 mutants to support the b-induced cell cycle arrest. For this, a chk1T394A S448A mutant allele tagged with the T7 epitope was integrated at the native locus in AB33 cells, a haploid strain that carries compatible (i.e., able to dimerize) bE and bW genes under the control of the inducible nar1 promoter that is induced by the addition of nitrate to medium (Brachmann et al., 2001). As a control, a T7-tagged wild-type chk1 allele was used. Upon induction, Chk1T394A S448A protein did not show the reduced electrophoretic mobility observed with wild-type Chk1 protein after induction of heterodimeric b protein (Figure 1B). Moreover, the cells carrying the nonphosphorylatable Chk1 allele were impaired in cell cycle arrest just as were cells deleted for chk1 (Figure 1C). We also analyzed the ability of compatible haploid cells carrying the chk1T394A S448A allele to infect plants and found that the nonphosphorylatable mutant showed similar virulence defects as those observed in cells lacking Chk1 protein (Figure 1D). In summary, these results show that the chk1T394A S448A mutant phenocopies all defects observed in the chk1 null mutant, implying that phosphorylation of Chk1 at these residues is integral to the mechanism of b-dependent activation of Chk1.
Figure 1.
A chk1 Allele Refractory to Phosphorylation Mimics the chk1Δ Loss-of-Function Mutation with Respect to b-Dependent Filament Cell Cycle Arrest and in Planta Proliferation.
(A) Scheme of Chk1 showing the kinase domain and two phosphorylatable residues required for Chk1 activation. aa, amino acids.
(B) In vivo phosphorylation of Chk1 during b-dependent filamentation. AB33-derived cells carrying an endogenous chk1-T7 (control, UCS31) or the chk1T394A S448A allele (UMP183) were incubated in inducing conditions (MM-NO3) for the indicated time (in hours). Protein extracts were immunoprecipitated with a commercial anti-T7 antibody, and immunoprecipitates were subjected to SDS-PAGE and immunoblotted with anti-T7 antibody.
(C) AB33 (control, UCS31) and derived strains lacking the chk1 gene (chk1Δ, UMP114) or carrying the chk1T394A S448A allele (UMP183) were incubated in inducing conditions (MM-NO3). Cells were stained with 4′,6-diamidino-2-phenylindole to detect nuclei. The percentage of cells producing mononucleated filaments (i.e., cell cycle arrested) after 24 h of incubation is shown graphically. The graph shows the result from two independent experiments, counting more than 100 cells each. Means and sds are shown.
(D) Quantification of symptoms in maize plants after 14 d after infection with wild-type (control, UCM350xUCM520), chk1Δ (UMP122xUMP129), or chk1T394A S448A (UMP190xUMP191) mutant crosses.
Atr1 Is Required in the Response to DNA Damage in U. maydis
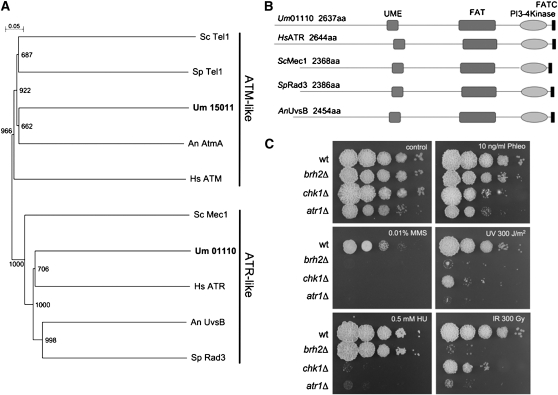
In other model systems, Chk1 phosphorylation in response to DNA damage is performed by two phosphatidylinositol 3-kinase-related kinases (PIKKs), ATM and ATR (Zhou and Elledge, 2000; Nyberg et al., 2002). Putative orthologs of these two PIKKs are present in U. maydis genome (Figure 2A; see Supplemental Data Set 1 online). The genes encoding Atm1 (Um15011) and Atr1 (Um01110) were identified as entries (noted in parentheses) in the manually annotated Munich Information Center for Protein Sequences U. maydis database (see http://mips.helmholtz-muenchen.de/genre/proj/ustilago/). To further analyze the relationships between these putative kinases and Chk1, gene deletion of the corresponding genes was attempted. Constructions containing a hygromycin resistance cassette flanked by regions located upstream and downstream of the atr1 and atm1 open reading frames were transformed in haploid cells. Only mutants lacking the atr1 gene were obtained. As this result suggested that atm1 was essential, a diploid/meiotic analysis protocol was employed. We successfully inactivated one atm1 allele in the diploid FBD11 strain, replacing it with the hygromycin resistance cassette, generating the atm1Δ null allele. After sporulation, we analyzed the meiotic progeny of this strain, and we found no hygromycin resistant cells, indicating that atm1 was an essential gene. We decided to focus on the characterization of Atr1. Sequence comparisons of U. maydis Atr1 with orthologs from different organisms revealed the presence of conserved domains, including the PIKK-specific domains FAT and FATC (Bosotti et al., 2000), suggesting that U. maydis Atr1 is a bona fide Atr1 ortholog (Figure 2B). Consistently, we found that cells lacking Atr1 were extremely sensitive to several genotoxic insults: UV irradiation, which induces pyrimidine dimers in DNA; hydroxyurea (HU), which inhibits ribonucleotide reductase and therefore affects replication by depletion of deoxynucleotide triphosphates, causing replication fork stalling and collapse; methyl methanesulfonate, which induces DNA alkylation; phleomycin, a radiomimetic drug that causes double-strand breaks in DNA; and ionizing radiation (IR), which generates double-strand breaks (Figure 2C; see Supplemental Figure 2 online). As comparisons we used mutant cells lacking the chk1 gene as well as cells lacking brh2, which encodes a BRCA2-like protein required for homology-directed recombinational repair (Kojic et al., 2002).
Figure 2.
U. maydis Atr1.
(A) Dendrogram of characterized Atr1 and Atm1-like proteins. The tree was created by the distance-based minimum evolution method, based on 1000 replicates. Bootstrap values are given, and branching points and the scale bar denote substitutions per site. The proteins used were Homo sapiens ATR (CAA70298.1) and ATM (AAB38309.1); S. pombe Rad3 (SPBC216.05) and Tel1 (SPCC23B6.03c); S. cerevisiae Mec1 (YBR136W) and Tel1 (YBL088C); Aspergillus nidulans AtmA (AN0038.2) and UvsB (AN6975.2); and U. maydis Um15011 (Atm1) and Um01110 (Atr1).
(B) Schematic representation of the domain architecture of Atr1 proteins in different organisms. Boxes represent UME (UVSB PI-3 kinase/MEI-4/ESR1 conserved domain), FAT (FRAP/ATM/TRRAP conserved domain), the PI3-kinase domain, and FATC (FRAP/ATM/TOR C-terminal region), all of them conserved domains in ATR-like kinases.
(C) Sensitivity of cells lacking atr1 gene (UCS9) in comparison to wild-type (wt; UCM350), chk1Δ (UMP122), and brh2Δ (UCM565) cells to different chemicals as well as IR and UV irradiation. A 108 cells/mL cell suspension and a series of 10-fold dilutions were spotted (2 μL per spot) onto agar plates containing the indicated drugs (HU, hydroxyurea; Phleo, phleomycin; MMS, methyl methanesulfonate). For UV ad IR sensitivity, cells were irradiated at the indicated dose after being spotted. The spots were photographed after incubation for 2 d.
Atr1 Phosphorylates and Activates Chk1 in Response to DNA Damage in U. maydis
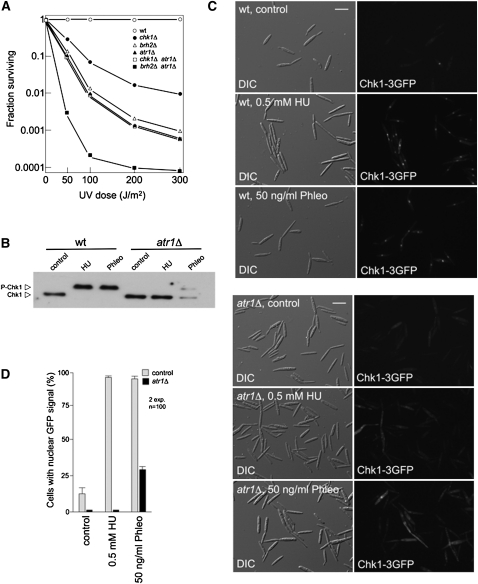
To define relationships between atr1 and chk1, we constructed double mutants of chk1 and atr1 and analyzed sensitivity to UV. As control for this epistasis analysis, we also constructed an atr1 brh2 double mutant strain. Our genetic results suggest that atr1 acts in the same pathway as chk1, since double chk1 atr1 mutants showed the same sensitivity as atr1 mutant, while brh2 seems to work in a distinct pathway than atr1 since their effects were additive (Figure 3A).
Figure 3.
Atr1 Is Required for Activation of Chk1 in Response to DNA Damage in U. maydis.
(A) Epistasis analysis between chk1, atr1, and brh2. Survival curves against UV irradiation of the indicated single and double mutants were obtained. Cells were grown to late log phase, adjusted to a density of 2 × 107 cells per mL, and irradiated with UV. Survival was determined by counting colonies visible after incubation for 2 to 3 d. wt, wild type.
(B) In vivo phosphorylation of Chk1 in response to agents that induce DNA damage depends on Atr1. Wild-type (UMP162) and atr1Δ (UMP207) cells carrying an endogenous Chk1-T7 allele were grown with no treatment (control) or in the presence of 0.5 mM HU or 50 ng/mL phleomycin (Phleo) for 6 h. Protein extracts were immunoprecipitated with a commercial anti-T7 antibody, and immunoprecipitates were subjected to SDS-PAGE and immunoblotted with anti-T7 antibody.
(C) Atr1 is required to localize Chk1 at the nucleus. Cell images of wild-type (UMP111) and atr1Δ (UCS15) strains carrying a Chk1-3GFP fusion protein after 3 h of incubation in the presence of HU or phleomycin (Phleo). DIC, differential interference contrast. Bar = 10 μm.
(D) Quantification of the cell response to DNA damage as the percentage of cells carrying a clear nuclear GFP fluorescence signal. The graph shows the result from two independent experiments, counting more than 100 cells each. Means and sds are shown.
We also analyzed whether Atr1 was required for the accumulation of phosphorylated forms of Chk1 in response to DNA damage agents. Our current view of DNA damage–dependent Chk1 activation in U. maydis is that there are two main signals to be detected by DNA surveillance systems: DNA double-strand breaks (induced here by phleomycin treatment) and single-strand DNA tracts as a hallmark of replication stress (caused by HU treatment). We challenged cultures of wild-type and atr1 mutant strains carrying the Chk1-T7 allele with HU or phleomycin and found that absence of Atr1 abolished the mobility shift in the presence of HU, while in the presence of phleomycin, the shift was only partially abrogated. Distinct behavior in Chk1 phosphorylation with respect to these two DNA damaging agents was previously noted (Pérez-Martin, 2009), suggesting that these two stimuli are transmitted by different signaling pathways, and our results support the idea that at least one of them seems to be totally dependent on Atr1.
Given that phosphorylation seemed to be required for Chk1 activation and that in response to DNA damage Chk1 accumulated in the nucleus, we also analyzed the ability of Chk1 to translocate to the nucleus in response to DNA damage in the absence of Atr1. We examined the subcellular localization of green fluorescent protein (GFP)-tagged Chk1 in the presence of either HU or phleomycin (Figure 3C). While control cells showed a clear nuclear accumulation of the fluorescent signal in the presence of these DNA damaging agents, in the absence of Atr1, GFP-tagged Chk1 failed to accumulate in the nucleus. This response was dramatic in the presence of HU but somewhat less obvious in the presence of phleomycin, mirroring the above phosphorylation results (Figure 3D).
Atr1-Dependent Phosphorylation of Chk1 Is Required for b-Dependent Cell Cycle Arrest
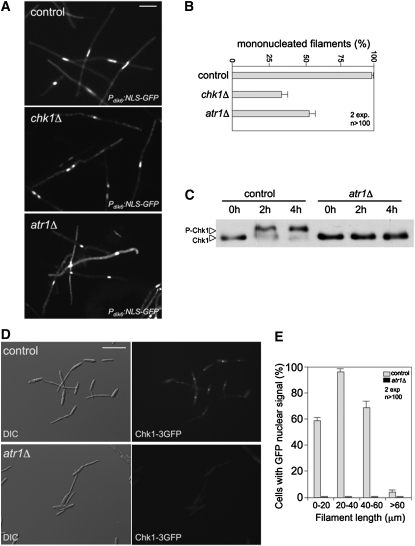
We analyzed whether Atr1 was required for b-induced cell cycle arrest. For this, we deleted the atr1 gene in strains expressing the homeoprotein b-complex heterodimer (AB33 background). We observed that deletion of atr1 in this genetic background resulted in elongated cells. To distinguish the b-induced filaments from such a cell population background, the haploid strains used expressed a GFP fusion to a nuclear localization signal under control of the dik6 promoter, which is specifically activated by the b-complex heterodimer (Flor-Parra et al., 2006). In this way, only cells expressing the b-dependent program produced a fluorescent nuclear signal. We found that in the control strain, almost the totality of the b-expressing cell population carried a single nucleus, while chk1Δ and atr1Δ filaments frequently carried more than two nuclei (Figures 4A and 4B), which was consistent with a defect in the ability to arrest entry into mitosis. These results suggest that most likely Atr1 is the PIKK kinase involved in the activation of Chk1 in response to b-induction.
Figure 4.
Atr1 Is Required for b-Dependent Cell Cycle Arrest.
(A) Cell images of control (UCS20) and a derived strain lacking the chk1 gene (chk1Δ, UMP112) or atr1 (atr1Δ, UCS21) incubated for 8 h in inducing conditions (MM-NO3). Strains carried a NLS-GFP fusion under control of the b-dependent dik6 promoter to detect the nucleus. Bar = 15 μm.
(B) Percentage of cells producing mononucleated filaments (i.e., cell cycle arrested) after 24 h of incubation. The graph shows the result from two independent experiments, counting more than 100 cells each. Means and sds are shown.
(C) Dependence on atr1 for in vivo phosphorylation of Chk1 during b-dependent filamentation. Control (UCS31) and atr1Δ (UCS32) AB33-derived cells carrying an endogenous chk1-T7 allele were incubated in MM-NO3 for the indicated time (in hours). Protein extracts were immunoprecipitated with a commercial anti-T7 antibody, and immunoprecipitates were subjected to SDS-PAGE and immunoblotted with anti-T7 antibody.
(D) Atr1 is required to localize Chk1 at the nucleus in response to b-induction. Cell images of control (UMP133) and atr1Δ strain (UMP208) carrying a Chk1-3GFP fusion protein after 6 h in inducing conditions (MM-NO3). Bar = 30 μm.
(E) Distribution of cells showing nuclear GFP accumulation in function of cell length. Quantification is the result of measurement of two independent experiments, counting more than 100 cells each. Mean and sd are shown.
To gain additional support for this idea, we investigated whether Atr1 was required for the accumulation of phosphorylated forms of Chk1 in response to the induction of the b-dependent program. Consistently, we found that there was no Chk1 phosphorylation in cells lacking Atr1 after b-induction (Figure 4C). In addition, given that phosphorylation seemed to be required for Chk1 activation and nuclear accumulation, the relationships between phosphorylation and nuclear localization in response to the induction of the b-dependent program were also investigated. When AB33-derived cells carrying a Chk1-GFP fusion were induced to produce filaments, we observed a clear GFP signal accumulation in the nucleus (Figure 4D). As we already noted, Chk1 activations seems to be a transient response and the nuclear GFP signal disappears in long filaments (Mielnichuk et al., 2009). To quantify this apparently transient response, we measured cells (n = 100 cells, two independent experiments) producing filaments of different length and plotted against the presence or not of a nuclear GFP signal (Figure 4E), confirming that only shorter filaments (i.e., early stages) showed nuclear accumulation of Chk1. When similar measures were performed in AB33-derived cells carrying a Chk1-GFP fusion but lacking the atr1 gene, we found that no cells showed accumulation of nuclear GFP signal at any stage (Figures 4D and 4E).
Atr1 and Chk1 Have Roles during Pathogenic Growth in Planta
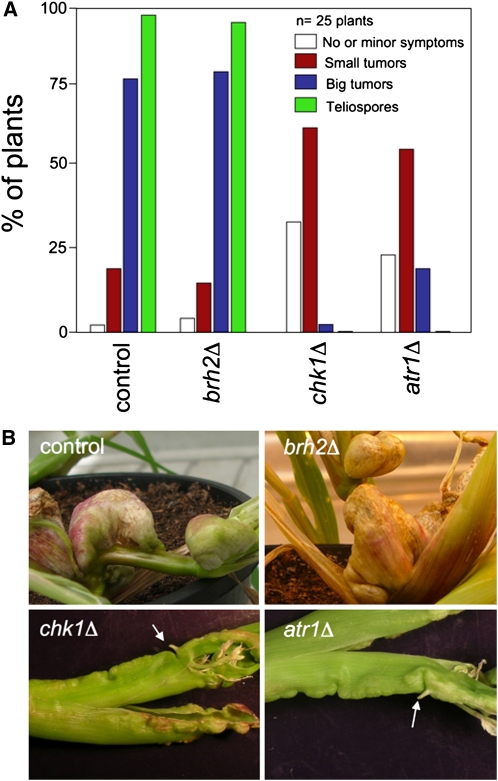
U. maydis infection of maize results in anthocyanin pigment production by the plant and the formation of tumors that are filled with proliferating fungal cells that eventually differentiate into black teliospores (Banuett and Herskowitz, 1996). We tested strains defective either in Chk1 or Atr1 for pathogenicity during the infection process. We found that >25% of the plants showed no or minor symptoms such as chlorosis (Figure 5A). This result can be easily attributed to impaired formation of a functional infective filament, as a consequence of impaired cell cycle arrest during dikaryon establishment (Mielnichuk et al., 2009). However, once plants were infected with these mutant strains, they rarely showed big tumors, and even in these rare occasions, no teliospores were found in these tumors. A striking feature observed in tumors induced by chk1Δ and atr1Δ strains is the development of small, shootlike structures (Figure 5B). Such structures were described previously in plants infected by U. maydis mutants exhibiting overactivation of cAMP cascade and were attributed to a defective fungal development inside the plant (Krüger et al., 2000).
Figure 5.
Atr1 and Chk1 Are Required for Full Virulence in U. maydis.
(A) Quantification of symptoms in maize plants after 14 d after infection with crosses of wild-type (control, UCM350xUCM520), brh2Δ (UCM565xUCM575), chk1Δ (UMP122xUMP129), and atr1Δ (UCS9xUCS10) strains.
(B) Morphology of tumors caused by wild-type (control) and compatible combinations of brh2Δ, chk1Δ, and atr1Δ strains. Representative tumors were photographed 16 d after infection. Arrows mark shoot-like structures.
These results strongly suggest roles of Chk1 and Atr1 beyond the initial steps of infection. A possible explanation for these results might be in the compromised ability of the mutant cells to deal with DNA damage occurring during proliferation inside the plant in response to the plant defense system (i.e., reactive oxygen and/or nitrogen species). However, this explanation seems unlikely since the DNA damage–sensitive brh2 mutant cells complete the life cycle at levels similar to wild-type cells (Figure 5A). This suggests that the Atr1-Chk1 axis has some additional role in the pathogenic process beyond signaling repair of DNA damage.
Chk1 and Atr1 Are Required to Maintain the Dikaryotic Growth
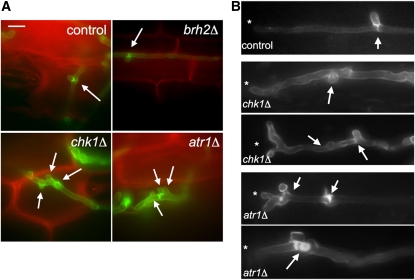
We tested whether proliferation inside the plant was affected by the disruption of Atr1-Chk1. Infected plant tissue was stained with Alexa-Fluor–labeled wheat germ agglutinin (WGA-AF488), a lectin that binds to chitin, enabling detection of fungal cell walls, and with propidium iodide, to visualize plant membranes (Doehlemann et al., 2009). U. maydis grows as a dikaryon during its pathogenic state. Previous work (Scherer et al., 2006) showed that clamp-like structures were involved in the distribution of nuclei in hyphae and thereby in the ability to proliferate inside the plant. Consistently, we found that in wild-type and brh2Δ filaments, single clamp-like structures were placed at the position of septum formation. However, atr1Δ and chk1Δ mutants were impaired in proliferation, and hyphae showed aberrant formation and variable distribution of clamp-like structures (Figures 6A and 6B). To quantify these defects, individual hyphae were counted and sorted in relation to the number of clamp-like structures observed at the three more apical septa (Figure 7A). Wild-type and brh2Δ filaments showed the expected distribution of one clamp-like structure per septum. However, filaments defective in chk1 or atr1 showed a clear deviation from this pattern, with apical septa that carry more than one clamp-like structure. Similar defects in clamp-like formation were observed in plants infected with compatible haploid cells carrying the chk1T394A S448A allele (see Supplemental Figures 1 and 3 online).
Figure 6.
Atr1 and Chk1 Are Required for Normal in Planta Proliferation of Hyphae.
(A) Images of plant tissues 2 d after infection with the indicated crosses of fungal cells. Hyphae (stained by WGA-AF488; green) grow intracellularly in epidermal cells of maize (stained by PI; red). Arrows mark clamp-like connections. Bar = 10 μm.
(B) Examples of aberrant hyphal morphologies found in chk1Δ and atr1Δ filaments in planta. Asterisk marks the hyphal tip. Arrows denoted aberrant clamp-like structures.
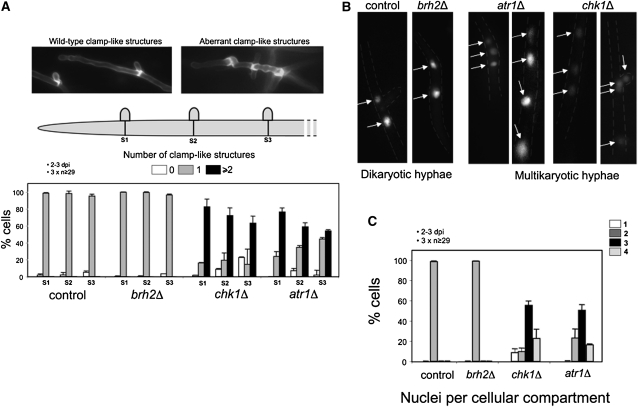
Figure 7.
Atr1 and Chk1 Are Required for Dikaryon Maintenance in Planta.
(A) Distribution of clamp-like structures in mutant cells. Infected plant tissue (obtained 2 to 3 d after infection [dpi]) with the indicated strains was stained with WGA-AF488 to detect hyphae and PI to mark maize cells. Then individual hyphae were counted and sorted in relation to the number of clamp-like structures observed at the three more apical septa (S1, S2, and S3). Control (wild type) and brh2Δ filaments showed the expected distribution of one clamp-like structure per septum. Filaments defective in chk1 or atr1 showed a clear deviation of this pattern. The graph shows the result from three independent experiments, counting more than 29 filaments each. Means and sds are shown. Top images exemplify typical wild-type (left) or mutant (right) hyphae.
(B) Examples of wild-type and the indicated mutant hyphae expressing a nuclear localized 3xGFP to detect nuclei. Wild-type hyphae contain two nuclei per cell as observed for brh2Δ. The atr1Δ and chk1Δ hyphae contain multiple nuclei per cell compartment. The distribution of the nuclei with respect to the clamp-like structures is given in the lateral illustrations.
(C) Quantification of the number of nuclei per cellular compartment in hyphae from infected plant tissue with the indicated strains. The graph shows the result from three independent experiments, counting more than 29 filaments each. Means and sds are shown.
Since formation of clamp-like structures is directly related to the process of nuclear division, we wondered whether nuclear distribution was also affected by the disruption of Atr1 or Chk1. To visualize fungal nuclei in infected plants, we used strains carrying a triple GFP gene fused to a nuclear localization sequence under the control of the constitutive promoter Ptef1 (Flor-Parra et al., 2006). While wild-type and brh2Δ filaments carried two nuclei per cellular compartment, atr1Δ and chk1Δ mutants carried variable combinations from 1 to 4 (Figures 7B and 7C). These observations suggest that Chk1 and Atr1 have roles in appropriate nuclear segregation and therefore that in their absence, dikaryotic cells replicate aberrantly, most likely causing the observed proliferation defects in planta.
DISCUSSION
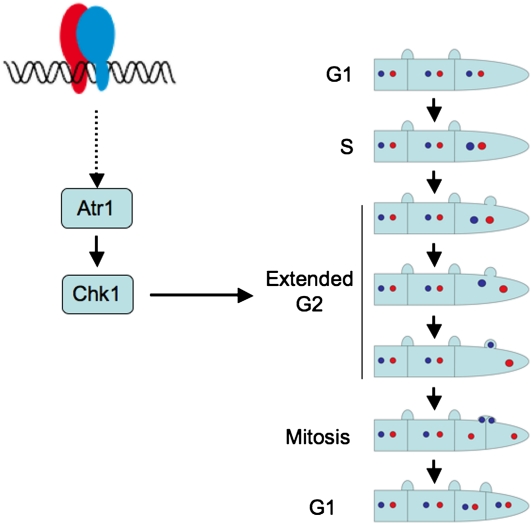
Dikaryon cell division involves nuclear migration and sorting of the divided nuclei to ensure that each daughter cell inherits a balance of each parental genome (Casselton, 2002). This cell cycle relies on a synchronized nuclear division and the development of clamp connections, a specialized projection formed close to the position of the future septum formation. One nucleus enters and divides in the developing clamp cell, whereas the other divides in the main hypha, with the result that mitosis occurs in two distinct cell compartments. It is not known in which cell cycle stage these processes take place, but G2 phase is most likely, as it happens after nuclear DNA duplication and before mitosis. Moreover, the movement of one of the nuclei into the clamp cell is reminiscent of the movement of the diploid nucleus into the daughter bud during vegetative cell division in U. maydis, a process occurring at the G2/M transition (Straube et al., 2005). Such a nuclear ballet most likely will need an extended G2 period to proceed. One of the roles of the Atr1-Chk1 axis during DNA damage response is to provide an elongated G2 phase to cells to allow time to repair DNA and resolve other problems before mitosis starts (Toettcher et al., 2009). On this basis, we propose that the role of the Atr1-Chk1 axis during the dikaryotic cell cycle is to synchronize nuclei as well as to provide an extended window during G2 phase to enable the above-mentioned process (Figure 8). It could be well that in the absence of the Atr1-Chk1 axis, the nuclear cycle, clamp cell formation, and cytokinesis occur asynchronously resulting in the defects such as observed in mutant hyphae inside the plant.
Figure 8.
Working Model of the Role That Atr1-Chk1 Play during Pathogenic Growth of U. maydis inside the Plant.
During dikaryon cell division, one nucleus enters and divides in the developing clamp cell, whereas the other divides in the main hypha, with the result that mitosis occurs in two distinct cell compartments. Most likely these processes take place during G2 phase. Since such a nuclear ballet will need an extended G2 period, we propose that the role of Atr1 and Chk1 during dikaryotic cell cycle is to synchronize nuclei as well as provide an extended window during G2 phase to allow the above-mentioned process.
Since b proteins are required for synchronization between cell cycle and cytokinesis (Wahl et al., 2010), we propose that in normal conditions it is the timely activation of the Atr1-Chk1 axis by the b homeodomain protein that produces accurate control of cell cycle. We have no clue about how the b protein activates this cascade. Our preliminary attempts to correlate expression of compatible b alleles with an increase in chk1 or atr1 transcription were negative (data not shown). Chk1 activation is linked to DNA damage in vegetative cells, and our results indicated that Atr1 is also required for the response to DNA damage. Therefore, it is tempting to speculate that b-induction could be associated with some class of DNA damage that triggers this kinase cascade during b-dependent filament formation. It is worth noting that b proteins activate the transcription of a gene encoding a putative DNA polymerase β (Brachmann et al., 2001), which belongs to the family X of DNA polymerases that are described to be involved in a number of DNA repair processes (Ramadan et al., 2004). On the other hand, several lines of evidence suggest that it is unlikely that activation of the Atr1-Chk1 cascade during b-induction is related to DNA damage. First, in previous studies using the formation of Rad51 foci as reporter for active DNA repair, there was no evidence for DNA damage associated with the induction of the infective hyphae (Mielnichuk et al., 2009). Second, we observed no defect in the ability to arrest the cell cycle (see Supplemental Figure 4 online) or to infect plants (Figure 5A) in cells lacking Brh2, which is required for DNA repair by homologous recombination (Kojic et al., 2002). Finally, preliminary research indicated that cells lacking the PolX polymerase were able to arrest cell cycle at levels comparable to wild-type cells (see Supplemental Figure 4 online). Two recent reports showed that activation of the DNA damage response cascade can be triggered in the absence of DNA damage by stable association of elements of the cascade with chromatin (Bonilla et al., 2008; Soutoglou and Misteli, 2008). Whether a similar mechanism could explain our observations in U. maydis will require additional research.
There are increasing numbers of reports indicating that the ability of the DNA damage response cascade to modulate cell cycle progression can be used during developmental processes. Some of these processes were linked with limited DNA damage as happens in B cell differentiation (Sherman et al., 2010) or even in the absence of apparent DNA damage, such as in the midblastula transition in Drosophila melanogaster embryos (Sibon et al., 1997) or in the asynchronous division at two-cell-stage Caenorhabditis elegans embryos (Brauchle et al., 2003). The surprising finding that a regulatory cascade involved in DNA damage responses plays a role in a fungal developmental process mirrors these previous results and reinforces the emerging idea that checkpoint cascades may have roles beyond cell surveillance by virtue of their ability to interact with cell cycle machinery.
METHODS
Ustilago maydis Genetic Methods
U. maydis strains are listed in Table 1. Manipulations with U. maydis, culture methods, gene disruption and gene transfer procedures, survival after DNA damage, and plant infections have been described previously (Castillo-Lluva and Pérez-Martín, 2005; Flor-Parra et al., 2007; Mielnichuk and Pérez-Martín, 2008; Mielnichuk et al., 2009). Protein extracts, immunoprecipitations, and immunoblot analysis were performed as described previously (García-Muse et al., 2004; Sgarlata and Pérez-Martín, 2005). To detect the phosphorylated forms of Chk1, T7-tagged Chk1 proteins were immunoprecipitated using an anti-T7 antibody (Sigma-Aldrich) from cell extracts and subjected to SDS-PAGE in 8% acrylamide/0.1% bisacrylamide, pH 9.2, gels. Blots were incubated with anti-T7-horseradish peroxidase (Sigma-Aldrich) and visualized using enhanced chemiluminescence (Renaissance; Perkin-Elmer).
Table 1.
Strains Used in This Work
| Strain | Relevant Genotype | Reference |
| UCM350 | a1b1 | Kojic et al. (2002) |
| UCM520 | a2b2 | Kojic et al. (2002) |
| UMP122 | a1b1 chk1Δ | Pérez-Martín (2009) |
| UMP129 | a2b2 chk1Δ | Mielnichuk et al. (2009) |
| UCS9 | a1b1 atr1Δ | This work |
| UCS10 | a2b2 atr1Δ | This work |
| UCM565 | a1b1 brh2Δ | Kojic et al. (2002) |
| UCM575 | a2b2 brh2Δ | Kojic et al. (2002) |
| UCS22 | a1b1 atr1Δ chk1Δ | This work |
| UCS27 | a1b1 atr1Δ brh2Δ | This work |
| AB33 | a2 Pnar1:bW2 Pnar1:bE1 | Brachmann et al. (2001) |
| UCS31 | a2 Pnar1:bW2 Pnar1:bE1 chk1-T7 | This work |
| UMP114 | a2 Pnar1:bW2 Pnar1:bE1 chk1Δ | Mielnichuk et al. (2009) |
| UCS32 | a2 Pnar1:bW2 Pnar1:bE1 chk1-T7 atr1Δ | This work |
| UMP183 | a2 Pnar1:bW2 Pnar1:bE1 chk1-T7T394A S448A | This work |
| UMP190 | a1b1 chk1-T7T394A S448A | This work |
| UMP191 | a2b2 chk1-T7T394A S448A | This work |
| UMP162 | a1b1 chk1-T7 | This work |
| UMP207 | a1b1 atr1Δ chk1-T7 | This work |
| UMP111 | a1b1 chk1-3GFP | Pérez-Martín (2009) |
| UCS15 | a1b1 atr1Δ chk1-3GFP | This work |
| UCS20 | a2 Pnar1:bW2 Pnar1:bE1 Pdik6:NLS-3GFP | This work |
| UMP112 | a2 Pnar1:bW2 Pnar1:bE1 Pdik6:NLS-3GFP chk1Δ | Mielnichuk et al. (2009) |
| UCS21 | a2 Pnar1:bW2 Pnar1:bE1 Pdik6:NLS-3GFP atr1Δ | This work |
| UMP196 | a1 b1 Ptef1:NLS-3GFP | This work |
| UMP197 | a1 b1 Ptef1:NLS-3GFP brh2Δ | This work |
| UMP198 | a1 b1 Ptef1:NLS-3GFP atr1Δ | This work |
| UMP199 | a1 b1 Ptef1:NLS-3GFP chk1Δ | This work |
| UMP133 | a2 Pnar1:bW2 Pnar1:bE1 chk1-3GFP | Mielnichuk et al. (2009) |
| UMP208 | a2 Pnar1:bW2 Pnar1:bE1 chk1-3GFP atr1Δ | This work |
Null mutants were constructed by replacing the entire open reading frames with cassettes expressing resistance to antibiotics by standard methodology (Brachmann et al., 2004). Briefly, a pair of DNA fragments flanking the atr1 open reading frame were amplified and ligated to a gene cassette encoding hygromycin resistance and flanked by SfiI sites. The 5′ fragment spans from nucleotide −1967 to nucleotide −29 (considering the adenine in the ATG as nucleotide +1), and it was produced by PCR amplification using the primers ATR1-22 (5′-TTAATTAAGCAGATCCACTGCTGAACGGGTTTC-3′) and ATR1-3 (5′-GGTGGCCATCTAGGCCTTCCTTAGGCTTGGACACTGGAGATCAGT-3′). The flanking 3′ fragment was obtained after PCR amplification with primers ATR1-4 (5′-ATAGGCCTGAGTGGCCACGGTTTGCAGCTGCATACAGTAGGATAT-3′) and ATR1-23 (5′-TTAATTAAGGAACTCATCAGCGTGTGGAACCGA-3′) and spans from nucleotide +7912 to nucleotide +9962.
Microscopy
To visualize fungal hyphae in plant by wheat germ agglutinin-Alexa fluor 488 (WGA-AF488; Invitrogen) and propidium iodide (PI) (Sigma-Aldrich), samples 2 to 3 d after infection of maize (Zea mays) leaves were incubated in staining solution (1 μg/mL PI, 10 μg/mL WGA-AF488, and 0.02% Tween 20) for 30 min and washed in 1× PBS, pH 7.4 (Doehlemann et al., 2009). Analysis of the infection stages was done using a Deltavision wide-field microscope (Applied Precision). Image deconvolution was performed using z-series of between 10 and 15 focal planes, acquired at 0.5-μm intervals. Image processing was performed using Adobe Photoshop CS2 and Canvas 8.0 (Deneba)
To quantify the clamp-like structures in fungal hyphae, plant leaves 2 to 3 d after infections were analyzed. Individual hyphae where the hyphal tip as well as the penetration points could be observed were analyzed, quantifying the number of clamp-like structures. To quantify the number of nuclei per cellular compartment, maize leaves 2 to 3 d after infections were analyzed. Only nuclei in individual hyphae were quantified.
Sequence Analysis
Alignments were made with ClustalW (Thompson et al., 1997). Phylogenetic dendrograms were constructed using MEGA 2.1 (Kumar et al., 2001) with the minimum evolution or maximum parsimony algorithm and gap deletion option.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under accession numbers JF690671 (Atm1) and JF690672 (Atr1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Phosphorylation of Chk1 Is Required for Normal in Planta Proliferation of Hyphae.
Supplemental Figure 2. Complementation of chk1 and atr1 Deletion Mutants.
Supplemental Figure 3. Single Phosphorylation Mutants in Chk1 Showed Wild-Type Phenotypes with Respect to Cell Cycle Arrest and Infection.
Supplemental Figure 4. Absence of Brh2 or PolX Does Not Affect the Ability of the b-Complex to Arrest Cell Cycle.
Supplemental Data Set 1. Sequence Alignment of Atr- and Atm-Like PI3K Kinases.
Acknowledgments
We thank Lorraine Symington (Columbia University) for stimulating discussions. C.d.S. was supported by a Formación de Personal Investigador contract. This work was supported by a Grant from the Spanish Government (BIO2008-04054). W.K.H. received grant support from National Institutes of Health Grant GM042482.
References
- Banuett F., Herskowitz I. (1996). Discrete developmental stages during teliospore formation in the corn smut fungus, Ustilago maydis. Development 122: 2965–2976 [DOI] [PubMed] [Google Scholar]
- Bonilla C.Y., Melo J.A., Toczyski D.P. (2008). Colocalization of sensors is sufficient to activate the DNA damage checkpoint in the absence of damage. Mol. Cell 30: 267–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosotti R., Isacchi A., Sonnhammer E.L. (2000). FAT: A novel domain in PIK-related kinases. Trends Biochem. Sci. 25: 225–227 [DOI] [PubMed] [Google Scholar]
- Brachmann A., König J., Julius C., Feldbrügge M. (2004). A reverse genetic approach for generating gene replacement mutants in Ustilago maydis. Mol. Genet. Genomics 272: 216–226 [DOI] [PubMed] [Google Scholar]
- Brachmann A., Weinzierl G., Kämper J., Kahmann R. (2001). Identification of genes in the bW/bE regulatory cascade in Ustilago maydis. Mol. Microbiol. 42: 1047–1063 [DOI] [PubMed] [Google Scholar]
- Brauchle M., Baumer K., Gönczy P. (2003). Differential activation of the DNA replication checkpoint contributes to asynchrony of cell division in C. elegans embryos. Curr. Biol. 13: 819–827 [DOI] [PubMed] [Google Scholar]
- Brefort T., Doehlemann G., Mendoza-Mendoza A., Reissmann S., Djamei A., Kahmann R. (2009). Ustilago maydis as a pathogen. Annu. Rev. Phytopathol. 47: 423–445 [DOI] [PubMed] [Google Scholar]
- Brown A.J., Casselton L.A. (2001). Mating in mushrooms: Increasing the chances but prolonging the affair. Trends Genet. 17: 393–400 [DOI] [PubMed] [Google Scholar]
- Casselton L.A. (2002). Mate recognition in fungi. Heredity 88: 142–147 [DOI] [PubMed] [Google Scholar]
- Casselton L.A., Olesnicky N.S. (1998). Molecular genetics of mating recognition in basidiomycete fungi. Microbiol. Mol. Biol. Rev. 62: 55–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo-Lluva S., Pérez-Martín J. (2005). The induction of the mating program in the phytopathogen Ustilago maydis is controlled by a G1 cyclin. Plant Cell 17: 3544–3560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E.H., Grote E., Mohler W., Vignery A. (2007). Cell-cell fusion. FEBS Lett. 581: 2181–2193 [DOI] [PubMed] [Google Scholar]
- Chen Y., Sanchez Y. (2004). Chk1 in the DNA damage response: Conserved roles from yeasts to mammals. DNA Repair (Amst.) 3: 1025–1032 [DOI] [PubMed] [Google Scholar]
- Doehlemann G., van der Linde K., Assmann D., Schwammbach D., Hof A., Mohanty A., Jackson D., Kahmann R. (2009). Pep1, a secreted effector protein of Ustilago maydis, is required for successful invasion of plant cells. PLoS Pathog. 5: e1000290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldbrügge M., Kämper J., Steinberg G., Kahmann R. (2004). Regulation of mating and pathogenic development in Ustilago maydis. Curr. Opin. Microbiol. 7: 666–672 [DOI] [PubMed] [Google Scholar]
- Flor-Parra I., Castillo-Lluva S., Pérez-Martín J. (2007). Polar growth in the infectious hyphae of the phytopathogen Ustilago maydis depends on a virulence-specific cyclin. Plant Cell 19: 3280–3296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flor-Parra I., Vranes M., Kämper J., Pérez-Martín J. (2006). Biz1, a zinc finger protein required for plant invasion by Ustilago maydis, regulates the levels of a mitotic cyclin. Plant Cell 18: 2369–2387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Muse T., Steinberg G., Perez-Martín J. (2004). Characterization of B-type cyclins in the smut fungus Ustilago maydis: Roles in morphogenesis and pathogenicity. J. Cell Sci. 117: 487–506 [DOI] [PubMed] [Google Scholar]
- Gladfelter A., Berman J. (2009). Dancing genomes: Fungal nuclear positioning. Nat. Rev. Microbiol. 7: 875–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K., Nakamura H., Matsumoto N. (2003). Mycelial incompatibility operative in pairings between single basidiospore isolates of Helicobasidium mompa. Mycol. Res. 107: 847–853 [DOI] [PubMed] [Google Scholar]
- Kojic M., Kostrub C.F., Buchman A.R., Holloman W.K. (2002). BRCA2 homolog required for proficiency in DNA repair, recombination, and genome stability in Ustilago maydis. Mol. Cell 10: 683–691 [DOI] [PubMed] [Google Scholar]
- Krüger J., Loubradou G., Wanner G., Regenfelder E., Feldbrügge M., Kahmann R. (2000). Activation of the cAMP pathway in Ustilago maydis reduces fungal proliferation and teliospore formation in plant tumors. Mol. Plant Microbe Interact. 13: 1034–1040 [DOI] [PubMed] [Google Scholar]
- Kües U. (2000). Life history and developmental processes in the basidiomycete Coprinus cinereus. Microbiol. Mol. Biol. Rev. 64: 316–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Tamura K., Jakobsen I.B., Nei M. (2001). MEGA2: Molecular evolutionary genetics analysis software. Bioinformatics 17: 1244–1245 [DOI] [PubMed] [Google Scholar]
- Lee S.C., Ni M., Li W., Shertz C., Heitman J. (2010). The evolution of sex: A perspective from the fungal kingdom. Microbiol. Mol. Biol. Rev. 74: 298–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mielnichuk N., Pérez-Martín J. (2008). 14-3-3 regulates the G2/M transition in the basidiomycete Ustilago maydis. Fungal Genet. Biol. 45: 1206–1215 [DOI] [PubMed] [Google Scholar]
- Mielnichuk N., Sgarlata C., Pérez-Martín J. (2009). A role for the DNA-damage checkpoint kinase Chk1 in the virulence program of the fungus Ustilago maydis. J. Cell Sci. 122: 4130–4140 [DOI] [PubMed] [Google Scholar]
- Nyberg K.A., Michelson R.J., Putnam C.W., Weinert T.A. (2002). Toward maintaining the genome: DNA damage and replication checkpoints. Annu. Rev. Genet. 36: 617–656 [DOI] [PubMed] [Google Scholar]
- Pérez-Martín J. (2009). DNA-damage response in the basidiomycete fungus Ustilago maydis relies in a sole Chk1-like kinase. DNA Repair (Amst.) 8: 720–731 [DOI] [PubMed] [Google Scholar]
- Ramadan K., Shevelev I., Hübscher U. (2004). The DNA-polymerase-X family: Controllers of DNA quality? Nat. Rev. Mol. Cell Biol. 5: 1038–1043 [DOI] [PubMed] [Google Scholar]
- Scherer M., Heimel K., Starke V., Kämper J. (2006). The Clp1 protein is required for clamp formation and pathogenic development of Ustilago maydis. Plant Cell 18: 2388–2401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgarlata C., Pérez-Martín J. (2005). Inhibitory phosphorylation of a mitotic cyclin-dependent kinase regulates the morphogenesis, cell size and virulence of the smut fungus Ustilago maydis. J. Cell Sci. 118: 3607–3622 [DOI] [PubMed] [Google Scholar]
- Sherman M.H., Kuraishy A.I., Deshpande C., Hong J.S., Cacalano N.A., Gatti R.A., Manis J.P., Damore M.A., Pellegrini M., Teitell M.A. (2010). AID-induced genotoxic stress promotes B cell differentiation in the germinal center via ATM and LKB1 signaling. Mol. Cell 39: 873–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibon O.C., Stevenson V.A., Theurkauf W.E. (1997). DNA-replication checkpoint control at the Drosophila midblastula transition. Nature 388: 93–97 [DOI] [PubMed] [Google Scholar]
- Soutoglou E., Misteli T. (2008). Activation of the cellular DNA damage response in the absence of DNA lesions. Science 320: 1507–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straube A., Weber I., Steinberg G. (2005). A novel mechanism of nuclear envelope break-down in a fungus: Nuclear migration strips off the envelope. EMBO J. 24: 1674–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. (1997). The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toettcher J.E., Loewer A., Ostheimer G.J., Yaffe M.B., Tidor B., Lahav G. (2009). Distinct mechanisms act in concert to mediate cell cycle arrest. Proc. Natl. Acad. Sci. USA 106: 785–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl R., Zahiri A., Kämper J. (2010). The Ustilago maydis b mating type locus controls hyphal proliferation and expression of secreted virulence factors in planta. Mol. Microbiol. 75: 208–220 [DOI] [PubMed] [Google Scholar]
- Zhou B.B., Elledge S.J. (2000). The DNA damage response: Putting checkpoints in perspective. Nature 408: 433–439 [DOI] [PubMed] [Google Scholar]