This study examines the regulation of SOMNUS (SOM), which is a key negative regulator of seed germination. ABI3 was found to regulate SOM expression together with PIL5, a previously identified regulator of SOM. PIL5 and ABI3, which interact to form a complex, regulate SOM expression independently in maturing seeds, but collaboratively in imbibed seeds.
Abstract
A previous study showed that SOMNUS (SOM), which encodes a C3H-type zinc finger protein, is a key negative regulator of seed germination that acts downstream of PHYTOCHROME INTERACTING FACTOR3-LIKE5 (PIL5). However, it was not determined if PIL5 is the sole regulator of SOM expression. Public microarray data suggest that the expression of SOM mRNA is regulated also by ABSCISIC ACID INSENSITIVE3 (ABI3), another key regulator of seed germination. By analyzing abi3 mutants and ABI3 overexpression lines, we show here that ABI3 activates the expression of SOM mRNA collaboratively with PIL5 in imbibed seeds. Chromatin immunoprecipitation analysis coupled with electrophoretic mobility shift assay indicate that ABI3 activates the expression of SOM mRNA by directly binding to two RY motifs present in the SOM promoter in vivo, which is further supported by the greatly decreased expression of a reporter gene driven by a SOM promoter bearing mutated RY motifs. At the protein level, the ABI3 protein interacts with the PIL5 protein. The ABI3-PIL5 interaction, however, does not affect targeting of ABI3 and PIL5 to SOM promoters. Taken together, our results indicate that ABI3 and PIL5 collaboratively activate the expression of SOM mRNA by directly binding to and interacting with each other at the SOM promoter.
INTRODUCTION
The decision for a seed to germinate at a given time and space is determined by seed developmental status and environmental conditions. In Arabidopsis thaliana, freshly harvested seeds display seed dormancy, a property that inhibits germination even in favorable environmental conditions (Finch-Savage and Leubner-Metzger, 2006). Abscisic acid (ABA) plays important roles in both initiating and maintaining seed dormancy, as shown by the shallow seed dormancy of ABA synthetic mutants, such as aba2, and the deep seed dormancy of ABA catabolic mutants, such as a cyp707a triple mutant (Léon-Kloosterziel et al., 1996; Okamoto et al., 2010). Mutations in some of the ABA signaling genes, such as ABSCISIC ACID INSENSITIVE3 (ABI3), which encodes a DNA binding protein that acts as a positive component of ABA signaling, also disrupts seed dormancy (Lopez-Molina et al., 2002). Other components, such as DELAY OF GERMINATION1 and HISTONE MONOUBIQUITINATION1, have been identified as being important regulators of seed dormancy, but their relationship with ABA signaling has not been determined (Bentsink et al., 2006; Liu et al., 2007).
Freshly harvested seeds eventually lose dormancy when they are sufficiently dried or have undergone stratification. It is not clear how seed dormancy breaks, but, once broken, nondormant seeds germinate if environmental conditions are favorable. Among various environmental factors, light and temperature play important roles in the decision to germinate. A seed must monitor and integrate various environmental conditions into cellular processes in order for germination to occur. Ultimately, favorable conditions activate germination-promoting hormone signaling, such as gibberellic acid (GA) signaling, and repress germination-inhibiting hormone signaling, such as ABA signaling. The molecular pathways involved in monitoring environmental conditions and integrating them with hormonal signaling are currently being actively investigated (Finkelstein et al., 2008; Holdsworth et al., 2008; Seo et al., 2009).
Phytochrome-mediated light signaling provides a good model system of how environmental conditions are perceived and integrated into internal cellular processes. Inside seed cells, phytochromes perceive red and far-red spectra of light. In Arabidopsis, phytochrome B (phyB), which is the major phytohcrome present in dry seeds, perceives red light and promotes seed germination. Phytochrome A (phyA), which accumulates during seed imbibition, also perceives light and promotes seed germination. Unlike phyB, however, phyA perceives both very low fluences of all spectra of visible light and prolonged far-red light (Shinomura et al., 1996). Other minor phytochromes, such as phyE, also play a role in promoting seed germination in imbibed seeds (Hennig et al., 2002). When imbibed seeds perceive light, activated phytochromes enter the nucleus and transmit the light signal, partly by destabilizing a germination-inhibiting, phytochrome-interacting bHLH transciption factor, called PIL5 (also known as PIF1) (Oh et al., 2004, 2006). The central role of PIL5 in mediating phytohcrome signaling can be inferred from the 100% germination frequency of the pil5 mutant, even if phytochrome B is inactivated by far-red light. In addition, microarray analysis indicates that all genes in imbibed seeds that are differentially expressed in response to red light are either directly or indirectly regulated by PIL5 (Oh et al., 2009). Thus, the destabilization of PIL5 by activated phytohcromes serves to release the repression of seed germination imposed by PIL5 and allows seeds to germinate.
PIL5 inhibits seed germination both by coordinating various hormone signaling cascades and by inhibiting cell wall loosening in imbibed seeds (Oh et al., 2009). Chromatin immunoprecipitation analysis coupled with microarray analysis shows that PIL5 regulates the expression of 166 genes by directly binding to their promoters. These direct target genes include cell wall loosening genes, such as expansin and xyloglucan endotransglycosylase genes, and various hormone signaling genes, such as GA signaling genes (GA INSENSITIVE [GAI] and REPRESSOR OF GA [RGA]), ABA signaling genes (ABI5 and ABI3), auxin signaling genes (AUXIN RESPONSE FACTOR18 [ARF18] and INDOLE ACETIC ACID-INDUCED PROTEIN16 [IAA16]), cytokinin signaling genes (CYTOKININ RESPONSE FACTOR1 [CRF1], CRF2, and CRF3), a JA signaling gene (JAZ1), and a BR signaling gene (BES1-INTERACTING MYC-LIKE PROTEIN2). The direction of expression indicates that PIL5 inhibits seed germination partly by activating the expression of GAI and RGA, which inhibit seed germination as negative GA signaling components, and of ABI3 and ABI5, which also inhibit seed germination as positive ABA signaling components. The role of other signaling components in seed germination is not clearly defined. However, the repression of JAZ1 and IAA16, negative signaling components of JA and auxin signaling, respectively, and the activation of ARF8, a positive auxin signaling component, are also consistent with the inhibitory role of PIL5 in seed germination. PIL5 also modulates hormone signaling by regulating the metabolism of various hormones. For example, PIL5 decreases the level of GA by repressing GA synthetic genes (GA3ox1 and GA3ox2) and activating a GA catabolic gene (GA2ox1), while it increases the ABA level by activating ABA synthetic genes (ABA DEFICIENT1 [ABA1], NINE-CIS-EPOXYCAROTENOID DIOXYGENASE6 [NCED6], and NCED9) and repressing an ABA catabolic gene (CYP707A2). The reciprocal regulation of hormone synthetic and catabolic genes can also be seen in auxin metabolism, as PIL5 activates the expression of genes involved in auxin synthesis (NITRILASE1 [NIT1], NIT3, ALDEHYDE OXIDASE1, and AMIDASE1) but represses the expression of a gene involved in auxin catabolism (DWARF IN LIGHT1). Curiously, PIL5 does not bind to the promoters of most genes involved in hormone metabolism, suggesting that PIL5 indirectly regulates these metabolic genes through other direct target genes. However, the degree of separation between PIL5 and genes involved in hormone metabolism is not known. Furthermore, genetic networks operating under PIL5 to regulate seed germination have not yet been identified. An analysis of the genes directly targeted by PIL5 might be useful to identify these genetic networks.
As a positive ABA signaling component, ABI3 also regulates various aspects of seed development (Giraudat et al., 1992). It belongs to a plant-specific B3 domain protein family that includes ABI3, FUSCA3 (FUS3), and LEAFY COTYLEDON2 (LEC2) in Arabidopsis and VIVIPAROUS1 (VP1) in maize (Zea mays). ABI3 contains four domains, dubbed A, B1, B2, and B3, which are named after the activation domain and the three basic amino acid clusters contained in it (Bies-Etheve et al., 1999). Among them, the B3 domain is known to bind to the RY motif (CATGCA) in vitro (Suzuki et al., 1997; Mönke et al., 2004; Braybrook et al., 2006), while the B1 and B2 domains interact with a set of bZIP transcription factors, including ABI5, bZIP10, bZIP25, and TRAB1 (Hobo et al., 1999; Nakamura et al., 2001; Lara et al., 2003), that bind to either the G-box element (CACGTG) or ABA response elements (ABREs) (Choi et al., 2000). Consistent with its binding to the RY motif and its interaction with bZIP factors, promoters of many ABI3-regulated genes contain RY motifs coupled with a G-box or ABREs. Upon binding to these promoters either directly or indirectly, ABI3 regulates the expression of genes involved in seed dormancy, seed germination, desiccation tolerance, the accumulation of seed storage proteins, and the breakdown of chlorophyll molecules in developing seeds. The genetic networks operating under ABI3 to regulate these processes are not fully understood.
We previously showed that SOMNUS (SOM) is a key direct target of PIL5 that inhibits seed germination downstream of PIL5 (Kim et al., 2008). SOM encodes a C3H-type zinc finger protein of unknown function. The expression analysis suggests that SOM inhibits seed germination partly by activating the expression of ABA synthetic genes and by inhibiting the expression of GA synthetic genes downstream of PIL5 but not by activating the expression of GAI and RGA in imbibed seeds. In this study, we further investigated how the expression of SOM is regulated by ABI3 and PIL5. We found that ABI3 directly binds to RY motifs present in the SOM promoter and activates the expression of SOM independently of PIL5 in maturing seeds but collaboratively with PIL5 in imbibed seeds. The interaction between ABI3 and PIL5 at the protein level further indicates that these two key regulators form a complex that regulates the expression of SOM in imbibed seeds.
RESULTS
ABI3 Regulates the Expression of SOM
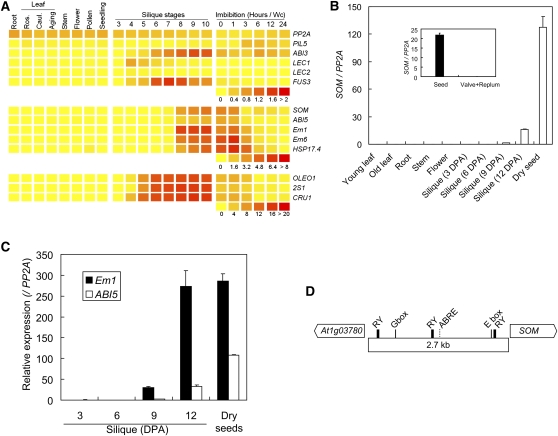
SOM inhibits seed germination by regulating genes downstream of PIL5 that are involved in both GA and ABA metabolism in imbibed seeds (Oh et al., 2007). We further investigated how the expression of SOM is regulated. The public microarray data compiled using the BAR HeatMapper tool suggested that SOM is a seed-specific gene whose expression increases during seed maturation (Figure 1A). The timing of SOM expression is slightly later than that of ABI3 or FUS3 but similar to that of ABI5 or Em1 during seed maturation. Unlike ABI3 and ABI5, PIL5 is expressed at a low level throughout seed maturation (Figure 1A).
Figure 1.
SOM Is Highly Expressed during Seed Maturation.
(A) Public micorarray data showing the seed-specific expression of SOM. The expression levels are visualized by BAR HeatMapper. Numbers beneath the heat map indicate the relative expression levels, and the higher expression levels are indicated by more reddish colors. Ros., rosette; Caul., cauline; Aging, aging rosette. Hours/Wc, imbibition hours in continuous white light.
(B) Seed-specific expression of SOM during seed maturation. The SOM mRNA expression levels were quantified by quantitative RT-PCR and are indicated as relative expression levels compared with PP2A mRNA (SOM/PP2A) (sd, n = 3). An inset indicates the relative expression levels of SOM mRNA in seeds and other tissues (valve and replum) that are separated from the 12-DPA silique.
(C) Relative expression of Em1 and ABI5 mRNAs during seed maturation (sd, n = 3).
(D) A diagram of the SOM promoter showing the RY motifs. G-box (CACGTG), PIL5-associated E-box (CANNTG), and ABRE (CACGTA) are also indicated.
Experimental analysis further supports that SOM is a seed-specific gene. We determined the expression levels of SOM in various tissues. SOM was expressed at very low levels in leaf, root, stem, and flower tissues but at high levels in seeds. During seed maturation, SOM began to be detected in siliques at 9 d postanthesis (DPA), and its expression increased beyond this time point (Figure 1B). The expression pattern of SOM during seed maturation was similar to that of ABI5 and Em1, two seed-specific genes that are known to be regulated by ABI3 (Figure 1C). When siliques were separated into developing seeds and other parts (valve and replum), SOM was detected only in the developing seeds but not in the valve and replum (Figure 1B, inset), further supporting the seed-specific expression of SOM.
We analyzed the upstream region of SOM and noticed that there were three RY motifs, in addition to other motifs, such as an ABRE and E-boxes, in the 2.7-kb intergenic region ranging from the transcription start site of SOM to its upstream neighboring gene (At1g03780) (Figure 1D). The three RY motifs are 0.3, 1.4, and 2.4 kb away from the transcription start site of SOM. Since the RY motif is bound by ABI3 or its homologs, the presence of RY motifs in its promoter suggests that SOM is regulated by ABI3 or related B3-domain proteins during seed maturation.
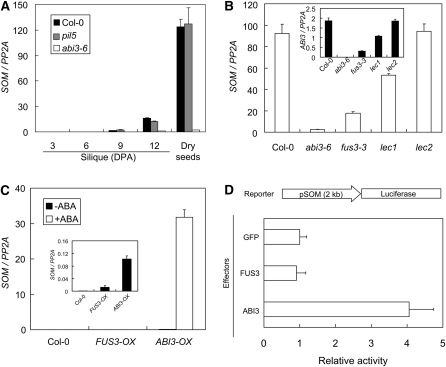
To experimentally test whether ABI3 regulates the expression of SOM during seed maturation, we determined the SOM transcript levels in an abi3 mutant. Since PIL5 regulates the expression of SOM in imbibed seeds, we first tested if PIL5 also regulates the expression of SOM during seed maturation. As in the wild type, however, the expression level of SOM increased during seed maturation and reached a similar level in the pil5 mutant (Figure 2A), indicating that PIL5 does not play a significant role in regulating the expression of SOM during seed maturation. In contrast with the pil5 mutant, the expression level of SOM remained low in the abi3 mutants during seed maturation and also in dry seeds (Figure 2A). These results indicate that ABI3 regulates the expression of SOM during seed maturation.
Figure 2.
ABI3 Regulates the Expression of SOM.
(A) Decreased expression of SOM mRNA (SOM/PP2A) in the abi3-6 mutant but not in the pil5 mutant seeds.
(B) Relative expression of SOM mRNA (SOM/PP2A) in freshly harvested abi3-6 and other related mutant seeds. An inset indicates the relative expression levels of ABI3 mRNA (ABI3/PP2A) in abi3-6 and its related mutant seeds.
(C) Increased expression of SOM mRNA (SOM/PP2A) in leaves of ABI3-OX. An inset indicates a magnified view of the relative expression levels of SOM mRNA in the absence of ABA treatment.
(D) Transient expression assay showing the activation of the firefly luciferase reporter gene linked to the SOM promoter by ABI3 but not by FUS3 or GFP in Arabidopis protoplasts. Error bars are sd (n = 3).
Seed maturation is known to be regulated by three B3-domain proteins (i.e., ABI3, FUS3, and LEC2) and a HAP3 subunit of CCAAT binding protein (LEC1) (Giraudat et al., 1992; Luerssen et al., 1998; Raz et al., 2001; Stone et al., 2001). To investigate if these other proteins also regulate the expression of SOM, we determined the expression levels of SOM transcript in abi3, fus3, lec1, and lec2 mutant seeds. The abi3 mutant seeds expressed the lowest level of SOM mRNA. The fus3 and lec1 mutant seeds also expressed lower levels of SOM than the wild type, while the lec2 mutant expressed a similar level of SOM as the wild type (Figure 2B). Since ABI3 levels were lower in the fus3 and lec1 mutants than in the wild type (Figure 2B, inset), the reduced expression of SOM in the fus3 and lec1 mutants could be due to the lower levels of ABI3 in these mutants.
To further show that ABI3 is the major regulator of SOM, we generated transgenic lines expressing either ABI3 or FUS3 under the cauliflower mosaic virus (CaMV) 35S promoter. Transgenic lines expressing FUS3 exhibited inflorescences with flowers that opened prematurely and short and thick siliques. For the analysis, we selected homozygous transgenic lines that expressed similar levels of ABI3 and FUS3 (ABI3-OX and FUS3-OX) (see Supplemental Figure 1 online). We determined the expression levels of the SOM transcript in transgenic leaves to avoid the complex interlocked transcriptional regulation among ABI3 and other seed maturation genes in the seeds (To et al., 2006). SOM mRNA was expressed at higher levels in ABI3-OX but only slightly higher levels in FUS3-OX (Figure 2C, inset). When the plants were subjected to ABA treatment, the SOM expression level increased dramatically only in the ABI3-OX line (Figure 2C), supporting the notion that ABI3 activates the expression of SOM. ABI3 also activates the expression of luciferase in a transient expression assay using Arabidopsis protoplasts and a 2-kb SOM promoter linked to the luciferase gene (Figure 2D). Taken together, these results suggest that the seed-specific expression of SOM is caused by the seed-specific expression of ABI3.
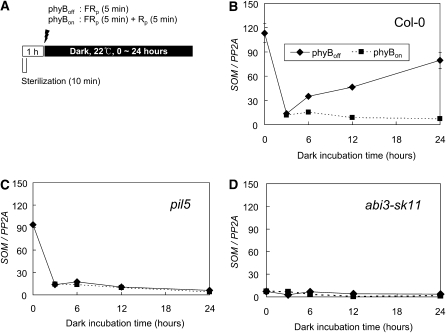
ABI3 is also required for the expression of SOM mRNA in imbibed seeds. When imbibed, the level of SOM transcript decreased drastically in the beginning of seed imbibition (after 3 h of seed imbibition) regardless of the light conditions. The initial decrease in SOM transcript is in agreement with the initial decrease in the majority of seed-specific mRNAs during seed imbibition. After imbibition, however, the expression of SOM depended on the light conditions. If a red light pulse was administered at the beginning of seed imbibition (phyBon), the SOM transcript level remained low, whereas it increased if a far-red pulse was given (phyBoff) (Figures 3A and 3B). This light-dependent expression of SOM in imbibed seeds, as reported before (Kim et al., 2008), was regulated by PIL5, as the SOM transcript levels remained low in the pil5 mutant, regardless of the light conditions (Figure 3C). The SOM transcript levels remained even lower in the abi3-sk11 mutant, regardless of the light conditions (Figure 3D). These results indicate that ABI3 is necessary for the high expression of SOM mRNA also in imbibed seeds.
Figure 3.
ABI3 Regulates the Light-Dependent Expression Levels of SOM mRNA in Imbibed Seeds.
(A) A diagram showing the light treatment schemes. Seeds were treated with either a far-red pulse (FRp, 2.8 μmol·m−2·s−1 for 5 min) or a far-red pulse followed by a red pulse (Rp, 18.7 μmol·m−2·s−1 for 5 min) 1 h after the start of seed sterilization, before transferring the seeds to the dark.
(B) Light-dependent expression of SOM mRNA (SOM/PP2A) during imbibition of wild-type seeds (sd, n = 3).
(C) Abolition of light-dependent expression of SOM mRNA (SOM/PP2A) during imbibition of pil5 mutant seeds (sd, n = 3).
(D) Decreased expression of SOM mRNA (SOM/PP2A) during imbibition of abi3-sk11 mutant seeds (sd, n = 3).
ABI3 Regulates the Expression of SOM by Directly Binding to RY Motifs in the SOM Promoter in Vivo
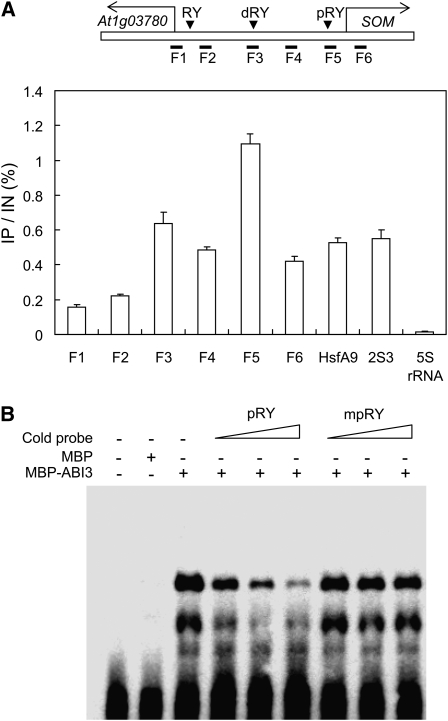
The presence of RY motifs in the SOM promoter suggested that ABI3 regulates the expression of SOM by directly binding to its promoter. To determine if ABI3 directly binds to the SOM promoter in vivo, we performed chromatin immunoprecipitation (ChIP) analysis using imbibed seeds of a transgenic line expressing flag-tagged ABI3 under the control of the CaMV 35S promoter. The flag-tagged ABI3 was functional, as it rescued the green desiccation-intolerant seed phenotype of the abi3-6 mutant. After the ChIP with anti-flag antibody, the enrichments of specific SOM promoter fragments in the immunoprecipitates were determined by six primer pairs (F1 to F6; Figure 4A). A primer pair that amplifies the genomic region around the 5S rRNA gene was included as a negative control, and two primer pairs that amplify two seed-specific gene promoters containing RY motifs (HsfA9 and 2S3) were also included as potential ABI3 target sites (Kagaya et al., 2005; Kotak et al., 2007).
Figure 4.
ABI3 Directly Binds to the SOM Promoter through RY Motifs Both in Vivo and in Vitro.
(A) ChIP analysis showing the direct binding of ABI3 to the SOM promoter in vivo. Top diagram, the SOM promoter and three RY motifs (a proximal RY motif [pRY], a distal RY motif [dRY], and an RY motif near At1g03780 [RY]). Seeds imbibed for 12 h under phyBoff conditions were used for ChIP analysis. F1 to F6 indicate genomic DNA fragments around the SOM promoter tested for enrichment by quantitative PCR (sd, n = 3). IP indicates immunoprecipitated DNA, and IN indicates input DNA.
(B) EMSA analysis showing the direct binding of ABI3 to pRY in the SOM promoter in vitro. MBP-ABI3, ABI3 protein fused to MBP; cold probe, nonlabeled wild-type pRY (pRY) or mutated pRY (mpRY). The increased concentration of competitors (5×, 10×, and 20×) is indicated by triangles.
The ChIP-enriched SOM promoter fragments ranging from F3 to F5 fragments, with the strongest enrichment being at the F5 fragment (Figure 4A). The HsfA9 and 2S3 promoters were also enriched to a greater extent than the negative control, indicating that ABI3 binds directly to the promoters of SOM, HsfA, and 2S3 in vivo. We noticed that the region of the SOM promoter that was enriched by ABI3 is relatively wide (>1 kb), as if ABI3 binds to a RY motif (pRY) at 0.3 kb upstream of the transcription start site in addition to a RY motif (dRY) at 1.4 kb upstream. However, not all fragments containing an RY motif were enriched. A fragment (F1) near an RY motif at 2.4 kb upstream of the transcription start site was not enriched much by ABI3, indicating that ABI3 binds to only some RY motifs.
To determine if ABI3 can directly bind to an RY motif present in the SOM promoter, we performed electrophoretic mobility shift assay (EMSA) analysis using a proximal RY motif–containing double-stranded oligomer from the F5 fragment region. Recombinant ABI3 fused to maltose binding protein (MBP-ABI3) bound to the oligomer, and its binding was effectively out-competed by nonlabeled oligomer (Figure 4B). Unlike unlabeled oligomer, however, a double-stranded oligomer containing a mutated RY motif (mpRY) did not compete for the binding to MBP-ABI3. Taken together, these results indicate that ABI3 binds to the SOM promoter through RY motifs and activates the expression of SOM.
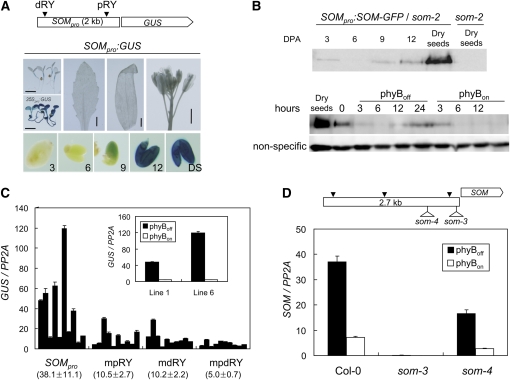
To investigate the role of these RY motifs further in vivo, we cloned a 2-kb region of the SOM promoter, fused it to a reporter gene (β-glucuronidase [GUS]), and generated transgenic lines (SOMpro:GUS). The 2-kb SOM promoter expressed the GUS reporter gene in seeds and mildly in hypocotyls, vegetative leaves, cauline leaves, and flowers (Figure 5A). When developing embryos were dissected out, we observed that GUS expression began to be detected in embryos 9 DPA and was strong in dry seeds, consistent with the expression pattern of endogenous SOM. Unlike the SOM promoter, the CaMV 35S promoter drove expression of the GUS reporter gene throughout seedlings (Figure 5A). The expression of SOM-green fluorescent protein (GFP) under the same 2-kb SOM promoter also increased dramatically in developing seeds. In imbibed seeds, the SOM-GFP protein level decreased initially but increased again when phyB was in an inactive state, consistent with the expression pattern of endogenous SOM transcript in this condition (Figure 5B). These results indicate that the 2-kb region of the SOM promoter linked to a reporter gene functionally substitutes the endogenous SOM promoter.
Figure 5.
RY Motifs Are Necessary for the High Expression of SOM.
(A) Seed-specific expression driven by a 2-kb region of the SOM promoter. SOMpro:GUS indicates various tissues and organs of transgenic plants harboring SOMpro:GUS, while 35Spro:GUS indicates transgenic plants harboring 35Spro:GUS. Numbers and DS on figures indicate DPA and dry seeds, respectively. Bar = 5 mm.
(B) Immunoblot analysis showing the increased expression of SOM-GFP protein by the SOM promoter during seed maturation and imbibition. hours, hours after imbibition.
(C) Decreased expression of GUS reporter transcripts (GUS/PP2A) by the SOM promoter with mutations in their RY motifs. Numbers indicate average ± sd (n = 10). Seeds imbibed for 24 h under phyBoff condition were used for analysis. The inset shows the light-dependent expression of the GUS transcript in two transgenic lines. Error bars are sd (n = 3).
(D) Decreased expression of SOM mRNA (SOM/PP2A) in the som-4 allele. Seeds imbibed for 12 h were used for analysis. Triangles indicate RY motifs in the SOM promoter (sd, n = 3).
To determine the role of RY motifs in regulating the expression of SOM, the 2-kb SOM promoter bearing mutations in two RY motifs, either individually or together, was fused to a GUS reporter gene and the respective transgenic lines were generated (mpRY:GUS, mdRY:GUS, and mpdRY:GUS). For each promoter construct, 10 randomly chosen independent transgenic lines were established, and the activities of promoters were determined by measuring the GUS transcript levels in the imbibed seeds. As expected, different transgenic lines expressed different levels of GUS transcript; thus, we determined the average expression levels of 10 transgenic lines. The average expression level of GUS mRNA driven by wild-type 2-kb SOM promoter was 38 relative to that of PP2A (Figure 5C). When a distal or a proximal RY motif was mutated, the average relative expression levels decreased to 10, whereas when both RY motifs were mutated, the average relative expression level decreased further to 5, indicating that two RY motifs contribute similarly to the high expression of SOM. Consistent with the view that both RY motifs contribute to SOM expression, the relative expression level of SOM was reduced when a T-DNA fragment was inserted between the proximal and distal RY motifs (som-4 allele), whereas the level was further reduced when a T-DNA fragment was inserted between the proximal RY motif and the transcription start site (som-3 allele) (Figure 5D). Taken together, these results indicate that both RY motifs are required for the high expression of SOM.
ABI3 Interacts with PIL5 to Potentiate the Expression of SOM in Imbibed Seeds
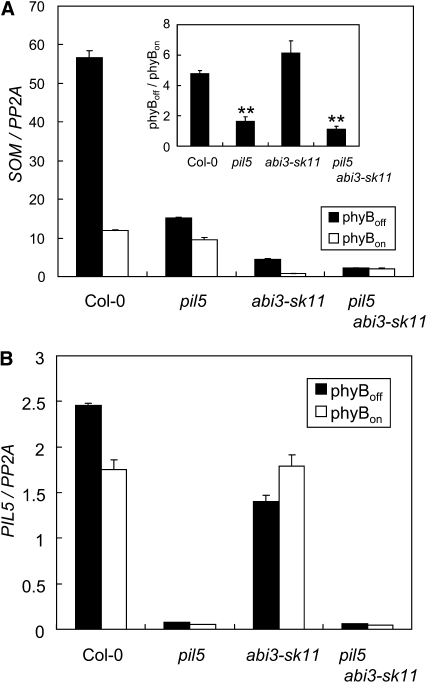
Our analysis indicates that the expression of SOM mRNA is regulated by both ABI3 and PIL5 in imbibed seeds. In imbibed seeds, public microarray data show that PIL5 is expressed comparably in both tissues, while ABI3 is expressed higher in endosperm. Consistent with the requirement of both ABI3 and PIL5 for the expression of SOM, SOM is also expressed higher in endosperms than in embryos of imbibed seeds (see Supplemental Figure 2 online). We further investigated the role of ABI3 and PIL5 in regulating the expression of SOM by comparing the expression levels of SOM in various mutants in imbibed seeds. In the wild type, the expression levels of SOM transcript were ~5-fold higher under the phyBoff condition than the phyBon condition (Figure 6A). In the pil5 mutant, the expression levels of SOM transcript were similarly low, regardless of the light conditions, consistent with a previous report that showed that PIL5 mediates light signaling to regulate the expression of SOM mRNA (Kim et al., 2008). In the abi3 mutants, the expression levels of SOM mRNA were greatly reduced relative to the wild type, indicating that ABI3 is also necessary for the high expression of SOM (Figure 6A). However, the SOM mRNA was still expressed 6-fold higher in FR-treated seeds than in R-treated imbibed seeds of the abi3 mutant, indicating that ABI3, although necessary for the high expression of SOM, is dispensable for the light-dependent expression of SOM mRNA. This reduced expression of SOM in the abi3 mutant was not due to the lower levels of PIL5 transcript (Figure 6B). Taken together, these results suggest that PIL5 regulates the light-dependent expression of SOM mRNA even in the absence of ABI3, while ABI3 collaborates with PIL5 to increase the expression of SOM mRNA in imbibed seeds.
Figure 6.
PIL5 Requires ABI3 to Activate the Expression of SOM Efficiently.
(A) The expression levels of SOM mRNA (SOM/PP2A) in the wild type, the pil5 and abi3-sk11 mutants, and the abi3-sk11 pil5 double mutant. Seeds were imbibed for 12 h. The inset indicates the fold difference in SOM mRNA expression between the phyBoff and phyBon conditions (sd, n = 3). Student’s t test: **P < 0.01.
(B) Expression levels of PIL5 mRNA (PIL5/PP2A) in the wild type, the pil5 and abi3-sk11 mutants, and the pil5 abi3-sk11 double mutant (sd, n = 3).
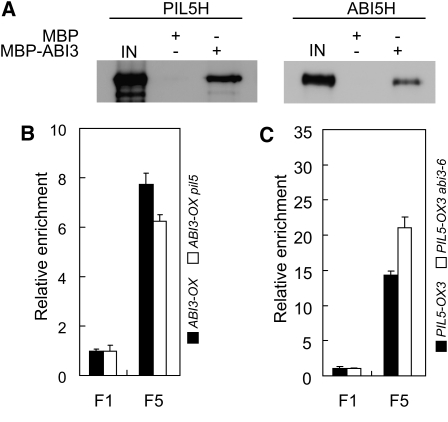
Numerous transcription factors have been shown to regulate the expression of target genes collaboratively by forming a complex on the promoters of the target genes. For example, ABI3 has been shown to interact with a subset of bZIP transcription factors, including ABI5, bZIP10, and bZIP25, in Arabidopsis (Nakamura et al., 2001; Lara et al., 2003). In maize, VP1, an ABI3 homolog, also interacts with TRAB1, a bZIP transcription factor (Hobo et al., 1999). We investigated if ABI3 and PIL5 interact with each other at the protein level. We detected the interaction by an in vitro binding assay using MBP-tagged ABI3 and His-tagged PIL5. As a positive control, we included His-tagged ABI5, a known ABI3-interacting transcription factor. When MBP-ABI3 was precipitated, it coprecipitated PIL5 (Figure 7A), whereas MBP alone did not coprecipitate PIL5, indicating that ABI3 interacts with PIL5. Consistent with a previous report, MBP-ABI3 but not MBP alone coprecipitated ABI5. The results indicate that ABI3 interacts with various transcription factors, including PIL5 and ABI5, at the protein level.
Figure 7.
ABI3 and PIL5 Interact at the Protein Level.
(A) In vitro binding assay showing the protein–protein interaction between ABI3 and PIL5. MBP-ABI3, MBP-fused ABI3 protein; PIL5H, His-tagged PIL5 protein; ABI5H, His-tagged ABI5 protein; IN, 5% of input proteins.
(B) ChIP assay showing the similar enrichments of SOM promoter fragments by ABI3-flag both in the ABI3-OX and ABI3-OX pil5 lines. F1 and F5 are as indicated in Figure 4A. The values shown are immunoprecipitated DNA/input DNA relative to F1. Error bars are sd (n = 3).
(C) ChIP assay showing that PIL5-myc enriches SOM promoter fragments to a similar extent both in the PIL5-OX3 and PIL5-OX3 abi3-6 lines. F1 and F5 are as indicated in Figure 4A. The values shown are immunoprecipitated DNA/input DNA relative to F1 (sd, n = 3).
The physical interaction between ABI3 and PIL5 may suggest that they help each other to bind to the SOM promoter. Alternatively, they may bind to the SOM promoter independently but interact with each other to activate the expression of SOM mRNA. The light-dependent expression of SOM mRNA in the abi3 mutant suggested that PIL5 binds to the SOM promoter even in the absence of ABI3 (Figure 6A). We further determined if ABI3 and PIL5 affect the binding of each other to the SOM promoter in vivo. ChIP analysis was used to determine the binding of ABI3 and PIL5 to the SOM promoter in vivo in the presence or absence of PIL5 or ABI3. The ChIP analysis showed that ABI3 binds to the SOM promoter similarly both in ABI3-OX and ABI3-OX/pil5 (Figure 7B), while PIL5 also binds to the SOM promoter similarly both in PIL5-OX and PIL5-OX/abi3 (Figure 7C), indicating that ABI3 and PIL5 bind to the SOM promoter even in the absence of each other. Taken together, these results indicate that PIL5 and ABI3 do not drastically affect each other’s ability to bind to the SOM promoter in vivo. Rather, they suggest that they interact with each other to activate the expression of SOM.
DISCUSSION
We show that ABI3 binds to the SOM promoter through two RY motifs using ChIP analysis and EMSA. Binding of ABI3 to both RY motifs was necessary to activate the expression of SOM mRNA fully, as mutations in either of two RY motifs greatly reduced the expression of a reporter gene. We analyzed the relationship between ABI3 and PIL5, which also regulates the expression of SOM by directly binding to its promoter (Kim et al., 2008). We found that ABI3 and PIL5 regulate the expression of SOM either independently or collaboratively. In maturing seeds, ABI3, but not PIL5, was necessary for the high expression of SOM mRNA, whereas, in imbibed seeds, PIL5 mediated light-dependent expression of SOM mRNA and ABI3 potentiated the ability of PIL5 to increase the expression level of SOM mRNA. At the protein level, ABI3 interacts with PIL5. This protein–protein interaction, however, did not drastically affect the targeting of ABI3 to the SOM promoter, indicating that ABI3 and PIL5 interact with each other to regulate SOM transcription collaboratively in imbibed seeds. Since ABI3 and PIL5 are key signaling components of ABA signaling and light signaling (Lopez-Molina et al., 2002; Oh et al., 2007), respectively, our results suggest that the SOM promoter integrates ABA and light signaling to regulate seed germination.
ABI3 Is Necessary for the High Expression of SOM Both in Maturing and Imbibed Seeds, but PIL5 Is Necessary Only in Imbibed Seeds
Our expression analysis shows that ABI3 is necessary for the activation of SOM mRNA both in maturing seeds and in imbibed seeds. However, PIL5 was necessary only in the imbibed seeds, but not in the maturing seeds. What determines this stage-specific role of PIL5 is not clear. A few different possible explanations may account for the stage specificity. First, maturing seeds may express other PIF family members that activate the expression of SOM mRNA redundantly with PIL5. Public microarray data indicate that PIL5 is expressed at lower levels than other PIFs (PIF5 and PIF6) in maturing seeds, while PIL5 is a dominant PIF in imbibed seeds (see Supplemental Figure 3 online). If other PIFs could activate the expression of SOM redundantly with PIL5, the mutation in PIL5 would be phenotypically manifested more severely in imbibed seeds than in maturing seeds. Second, other transcription factors that are known to interact with ABI3 may functionally substitute for PIL5 in maturing seeds. The same public microarray data indicate that ABI5, EEL, DPBF2, and AREB3 are expressed at high levels in maturing seeds (see Supplemental Figure 3 online) (Bensmihen et al., 2005). Since these bZIP transcription factors are known to activate the expression of seed storage-specific protein genes in maturing seeds, it is possible that these bZIP factors bind to the SOM promoter and activate the expression of SOM in maturing seeds. Alternatively, stage-specific modification or cofactors that enable ABI3 to activate the expression of SOM mRNA alone may account for the dispensability of PIL5 in maturing seeds. Further analysis is needed to clarify why PIL5 is dispensable for the activation of SOM mRNA in maturing seeds.
ABI3 Binds to the SOM Promoter through RY Motifs to Activate the Expression of SOM mRNA
A previous study showed that SOM is a C3H-type zinc finger protein that inhibits seed germination downstream of PIL5 partly by activating ABA biosynthesis and inhibiting GA biosynthesis in imbibed seeds (Oh et al., 2007; Kim et al., 2008). We further investigated how the expression of SOM is regulated in seeds. Public microarray data indicated that SOM is highly expressed in maturing seeds, which is reminiscent of the expression pattern of ABI3-regulated genes, such as ABI5 (Figure 1A). The presence of RY motifs, which serve as a binding site of B3-domain proteins, such as ABI3, FUS3, and LEC2, further suggested that SOM is regulated by ABI3 or its homologs. Consistent with this notion, the expression level of SOM was drastically reduced in abi3 and weakly reduced in fus3. Transgenic lines overexpressing ABI3 and FUS3 showed that the expression of SOM mRNA is activated mainly by ABI3. Apparently, ABA strongly enhanced the expression of SOM by ABI3. Taken together, our results suggest that the high expression of SOM in maturing seeds is caused by the seed-specific expression of ABI3 and the concomitant increase in ABA level during seed maturation.
Our ChIP analysis coupled with EMSA indicated that ABI3 directly binds to two regions of the SOM promoter through RY motifs and activates the expression of SOM mRNA. The binding of ABI3 to both RY motifs was necessary for the high expression of SOM mRNA, as mutations in either one of the RY motifs greatly decreased the expression levels of a reporter gene. As in the SOM promoters, multiple RY motifs are also found in the promoters of many seed storage protein genes, including OLEOSIN, CRUCIFERIN, and 2S, suggesting that promoters of ABI3-regulated genes recruit multiple copies of ABI3. Interestingly, an insertion of a 4.5-kb T-DNA fragment from pBIN-pROK2 between two of the RY motifs also reduced the expression of SOM mRNA. Since the insertion of the T-DNA fragment increases the distance between the distal RY motif and the transcription start site, the reduced expression may indicate that ABI3 must be in relatively close proximity to the transcription start site to activate the transcription. Alternatively, the insertion of T-DNA may disrupt the molecular interaction among proteins that bind to the proximal region and proteins that bind to distal regions of the SOM promoter.
The ChIP analysis further indicates that ABI3 binds to only a subset of RY motifs in vivo. A genomic region we scanned by ChIP analysis included three RY motifs. Among these RY motifs, only two were highly enriched, and one was not enriched by ABI3. The enrichment was not correlated with the distance from the transcription start site, as two enriched RY motifs were 0.3 and 1.4 kb upstream of the SOM transcription start site, respectively, while one nonenriched RY motif was 0.3 kb upstream of the transcription start site of At1g03780 (Figure 1D). These results indicate that ABI3 binds to only a subset of RY motifs in vivo and imply that in vivo ABI3 binding sites will have to be determined experimentally, such as by ChIP analysis. Binding of the transcription factor to its target appeared to be more selective under in vivo conditions than in vitro, and this seems to be a rather general phenomenon, instead of an exception. A recent ChIP-Chip analysis showed that PIL5 binds to only a subset of G-box elements in vivo (Oh et al., 2009). Similarly, a bZIP transcription factor, HY5, binds to only a subset of G-box elements (Lee et al., 2007). The molecular mechanisms that determine how the in vivo binding sites are selected for each transcription factor are not known.
ABI3 and PIL5 Interact and Activate the Expression of SOM mRNA Collaboratively in Imbibed Seeds
ABI3 and PIL5 collaboratively activated the expression of SOM mRNA. The expression of SOM mRNA was greatly reduced in the abi3 mutant. Mutations in RY motifs greatly decreased the expression of reporter genes, supporting the hypothesis that ABI3 activates the expression of SOM. Nevertheless, the expression of SOM mRNA was still repressible by red light, as long as PIL5 was present, indicating that PIL5 mediates light signaling to activate the expression of SOM mRNA, while ABI3 enhances the expression level.
Although the molecular mechanisms underlying the ability of ABI3 to enhance the activity of PIL5 are not known, the interaction between ABI3 and PIL5 at the protein level may contribute to this collaboration. ABI3 and PIL5 may form a protein complex that activates the transcription more efficiently. Alternatively, ABI3 and PIL5 may help each other bind to the SOM promoter.
Our data indicate that ABI3 and PIL5 do not drastically affect each other’s targeting to the SOM promoter, implying that the synergism comes after their targeting to the promoter, rather than during the targeting itself (Figures 6 and 7). Our ChIP analysis indicates that ABI3 enriches the SOM promoter and that this enrichment is of a similar magnitude in the wild type and the pil5 mutant. PIL5 also enriched the SOM promoter, and this enrichment was similar in the wild type and the abi3 mutants. These results indicate that ABI3 and PIL5 can bind to their target sites in the absence of each other. Our results are similar to a previous report showing that ABRE binding factors bind to the maize rab28 promoter even in the absence of VP1 (Busk and Pagès, 1997), while they are different to another report showing that ABI3 slightly enhances the binding between bZIP10/OPAQUE2 and the G-box element in vitro (Lara et al., 2003). Since a slight increase or decrease in binding affinity is difficult to resolve in our ChIP assay, the results do not exclude the possibility that ABI3 mildly affects the ability of PIL5 or other interacting bZIP factors to bind to DNA. Therefore, even though ABI3 and PIL5 do not drastically affect each other’s binding to SOM promoter, further analysis is needed to determine if they affect DNA binding activity more subtly.
The direct interaction between ABI3 and PIL5 indicates that three germination-inhibiting transcription factors (PIL5, ABI3, and ABI5) form a functional module, both through transcriptional regulation and protein–protein interaction, to inhibit seed germination. At the transcriptional level, PIL5 directly binds to promoters of ABI3 and ABI5 and activates the expression of their mRNAs, while ABI3 activates the expression of ABI5 mRNA. At the protein level, ABI3 interacts with both its regulator (PIL5) and its regulatee (ABI5), which leads to the synergistic activation of their target gene expression. This type of functional module that is composed of interconnected transcription factors is likely to be useful to regulate the expression of wide sets of genes either independently, in combination with each other or with other proteins as well, or interdependently. Further studies are needed to decipher which target genes are regulated by these three factors independently, in combination, or interdependently.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana plants were grown in a growth room with a 16-h-light/8-h-dark cycle at 22 to 24°C. T-DNA insertion lines (abi3-sk11, som-4, lec1, and lec2) were obtained from the Salk Institute (Salk_023411, CS806302, Salk_131219, and CS873714, respectively), and insertion sites of abi3-sk11 and som-4 were determined by amplifying and sequencing the flanking regions. The lec1 and lec2 mutants were confirmed by monitoring anthocyanin accumulation and desiccation intolerance in seeds. To generate ABI3-OX and FUS3-OX plants, full-length ABI3 and FUS3 cDNAs were amplified, cloned into binary vectors, and transformed into Arabidopsis, and then homozyogous lines were selected (primers are presented in Supplemental Table 1 online). For transgenic lines harboring the GFP-tagged mini-gene construct of SOM (SOMpro:SOM-GFP) in the som-2 background, SOM cDNA and its promoter region were sequentially cloned into the pbGFP1 binary vector and transformed into the som-2 mutant. Homozygous lines were selected. For mutagenesis of the RY motifs in the SOM promoter, inverse PCR was performed with mutated primer sets (see Supplemental Table 1 online) using a SOM promoter-containing plasmid as a template. Mutated SOM promoters were confirmed by sequencing, cloned into a pbGG4 binary vector for promoter-reporter construction, and transformed into Columbia-0 (Col-0). Ten independent transgenic lines showing the 3:1 segregation ratio were used to monitor the expression of GUS mRNA during imbibition. The abi3-6 and fus3-3 mutant seeds were kind gifts from F. Parcy at Centre National de la Recherche Scientifique, France. All the mutants used in this study (som-2, som-3, som-4, pil5-1, PIL5-OX3, lec1, lec2, fus3-3, abi3-sk11, and abi3-6) are in the Col-0 background. Desiccation-intolerant mutants were maintained by sowing seeds harvested from green siliques.
Quantitative and Qualitative Gene Expression Analysis
For mRNA expression analysis, total RNAs were extracted either from dried seeds, imbibed seeds, or transgenic leaves using a Spectrum plant total RNA kit (Sigma-Aldrich) and converted to cDNAs using MMLV-RTase (Promega) according to the manufacturers’ protocols. Transcript levels of each mRNA were determined by real-time PCR and normalized with the level of PP2A mRNA using the delta Ct method (Czechowski et al., 2005; Schmittgen and Livak, 2008). Amplification log curves of each primer set were confirmed to be parallel to that of PP2A amplification to validate the comparisons among samples and primer sets before use. Real-time PCR was performed with a protocol (45 cycles, each cycle consisting of 95°C/58°C/72°C for 20 s each) in an iCycler iQ5 cycler (Bio-Rad). We tested more than three different seed batches to confirm the gene expression patterns and presented representative results in each figure.
Expression browser and Heat-Mapper tools provided by BAR (The Bio-Array Resource for Arabidopsis Functional Genomics; http://bbc.botany.utoronto.ca/) were employed to show the heat map of gene expression patterns.
To determine the tissue-specific expression of SOM by histochemical GUS reporter assay, seed coats, which inhibit penetration of X-Gluc into seeds, were carefully pinched off the mature or imbibed seeds using forceps. Decoated seed embryos were stained for GUS activity after having undergone fixing in 90% acetone for 30 min, as described previously (Jefferson et al., 1987).
For the effector activity assay of ABI3 and FUS3 using Arabidopsis protoplasts, mesophyll protoplasts were prepared from fully mature leaves as described previously (Yoo et al., 2007). Briefly, leaf slices were dipped in cell wall digestion solution (10 mM MES, pH 5.7, 1.5% cellulase R10, 0.4% macerozyme R10, 0.4 M mannitol, 20 mM KCl, 10 mM CaCl2, and 0.1% [w/v] BSA) and incubated for 3 h with mild shaking. One hundred microliters of protoplasts (2 × 104) was used for each transfection. After incubation for 1 d, luciferase activity was measured using a Dual Luciferase assay kit (Promega).
For the analysis of protein levels, samples were ground in protein extraction buffer (100 mM NaH2PO4, 8 M urea, and 10 mM TrisCl, pH 8.0) using Tissue-lyzer (Qiagen). Tissue lysates were cleared by centrifugation at 20,000g for 10 min and subjected to immunoblotting as described previously (Park et al., 2004).
In Vitro Binding Assay
For the in vitro binding assay between PIL5 and ABI3, recombinant MBP-ABI3 protein was produced in Escherichia coli at 25°C using the pMAL-c2X-ABI3 expression vector and purified using amylose-resin following the manufacturer’s protocol (NEB) (see Supplemental Table 1 online for primer sets). His-tagged PIL5 and ABI5 (PIL5H and ABI5H) were produced as described previously (Oh et al., 2007). MBP-resin and MBP-ABI3-resin were incubated with purified PIL5H and ABI5H at 4°C for 2 h, sedimented by spin down, and washed five times. MBP resin was used as an MBP control as well as a nonspecific binding control. Then, 1× PBS with 0.1% Nonidet P-40, 0.5 mM DTT, 10% glycerol, 1 mM PMSF, and Protease inhibitor cocktail (Roche) was used for binding and washing. PIL5H and ABI5H bound to MBP-ABI3 were visualized by protein blotting using anti-His antibody.
ChIP Analysis and EMSA
ChIP was performed essentially as described previously, except that three separate steps (the elution of the DNA–protein complex from resin, the reversal of cross-linking, and the digestion with proteinase K) were combined into one step in this study (Kim et al., 2008). Immunoprecipitated DNA was eluted from resin in 200 μL of elution buffer containing 20 mM Tris-HCl, pH 7.5, 5 mM EDTA, 50 mM NaCl, 1% SDS, and 50 μg/mL proteinase K at 68°C for 2 h with rotary mixing. Residual DNA was further eluted by subsequent incubation in 100 μL of elution buffer at 68°C for 5 min. Eluted DNA was purified with a PCR purification kit (Solgent) and subjected to real-time PCR. For ChIP of flag-tagged ABI3, anti-flag antibody-conjugated resin (Sigma-Aldrich) was used, while anti-myc Ab (Cell Signaling) and protein A–conjugated resin (Upstate) was used for ChIP of myc-tagged PIL5. For EMSA, previously described procedures were followed, except that 5′ biotin-labeled probes were synthesized by chemical modification (Bioneer).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: PIL5 (At2g20180), ABI3 (At3g24650), SOM (At1g03790), LEC1 (At1g21970), LEC2 (At1g28300), FUS3 (At3g26790), ABI5 (At2g36270), Em1 (At3g51810), Em6 (At2g40170), 2S3 (At4g27160), RGA (At2g01570), HsfA9 (At5g54070), 5S rRNA (At3g41979), and PP2A (At1g13320).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Expression Levels of ABI3 and FUS3 Transgenes.
Supplemental Figure 2. Tissue-Specific Expression of ABI3, PIL5, and SOM.
Supplemental Figure 3. Expression of PIFs and bZIPs during Seed Maturation and Imbibition.
Supplemental Table 1. Primers Used in This Study.
Acknowledgments
This work was supported in part by grants from the National Research Foundation of Korea (2007-0056949 and 2009-0077873) to G.C.
References
- Bensmihen S., Giraudat J., Parcy F. (2005). Characterization of three homologous basic leucine zipper transcription factors (bZIP) of the ABI5 family during Arabidopsis thaliana embryo maturation. J. Exp. Bot. 56: 597–603 [DOI] [PubMed] [Google Scholar]
- Bentsink L., Jowett J., Hanhart C.J., Koornneef M. (2006). Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc. Natl. Acad. Sci. USA 103: 17042–17047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bies-Etheve N., da Silva Conceicao A., Giraudat J., Koornneef M., Léon-Kloosterziel K., Valon C., Delseny M. (1999). Importance of the B2 domain of the Arabidopsis ABI3 protein for Em and 2S albumin gene regulation. Plant Mol. Biol. 40: 1045–1054 [DOI] [PubMed] [Google Scholar]
- Braybrook S.A., Stone S.L., Park S., Bui A.Q., Le B.H., Fischer R.L., Goldberg R.B., Harada J.J. (2006). Genes directly regulated by LEAFY COTYLEDON2 provide insight into the control of embryo maturation and somatic embryogenesis. Proc. Natl. Acad. Sci. USA 103: 3468–3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busk P.K., Pagès M. (1997). Protein binding to the abscisic acid-responsive element is independent of VIVIPAROUS1 in vivo. Plant Cell 9: 2261–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H., Hong J., Ha J., Kang J., Kim S.Y. (2000). ABFs, a family of ABA-responsive element binding factors. J. Biol. Chem. 275: 1723–1730 [DOI] [PubMed] [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M.K., Scheible W.R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch-Savage W.E., Leubner-Metzger G. (2006). Seed dormancy and the control of germination. New Phytol. 171: 501–523 [DOI] [PubMed] [Google Scholar]
- Finkelstein R., Reeves W., Ariizumi T., Steber C. (2008). Molecular aspects of seed dormancy. Annu. Rev. Plant Biol. 59: 387–415 [DOI] [PubMed] [Google Scholar]
- Giraudat J., Hauge B.M., Valon C., Smalle J., Parcy F., Goodman H.M. (1992). Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4: 1251–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig L., Stoddart W.M., Dieterle M., Whitelam G.C., Schäfer E. (2002). Phytochrome E controls light-induced germination of Arabidopsis. Plant Physiol. 128: 194–200 [PMC free article] [PubMed] [Google Scholar]
- Hobo T., Kowyama Y., Hattori T. (1999). A bZIP factor, TRAB1, interacts with VP1 and mediates abscisic acid-induced transcription. Proc. Natl. Acad. Sci. USA 96: 15348–15353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdsworth M.J., Bentsink L., Soppe W.J. (2008). Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol. 179: 33–54 [DOI] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. (1987). GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagaya Y., Okuda R., Ban A., Toyoshima R., Tsutsumida K., Usui H., Yamamoto A., Hattori T. (2005). Indirect ABA-dependent regulation of seed storage protein genes by FUSCA3 transcription factor in Arabidopsis. Plant Cell Physiol. 46: 300–311 [DOI] [PubMed] [Google Scholar]
- Kim D.H., Yamaguchi S., Lim S., Oh E., Park J., Hanada A., Kamiya Y., Choi G. (2008). SOMNUS, a CCCH-type zinc finger protein in Arabidopsis, negatively regulates light-dependent seed germination downstream of PIL5. Plant Cell 20: 1260–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotak S., Vierling E., Bäumlein H., von Koskull-Döring P. (2007). A novel transcriptional cascade regulating expression of heat stress proteins during seed development of Arabidopsis. Plant Cell 19: 182–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara P., Oñate-Sánchez L., Abraham Z., Ferrándiz C., Díaz I., Carbonero P., Vicente-Carbajosa J. (2003). Synergistic activation of seed storage protein gene expression in Arabidopsis by ABI3 and two bZIPs related to OPAQUE2. J. Biol. Chem. 278: 21003–21011 [DOI] [PubMed] [Google Scholar]
- Lee J., He K., Stolc V., Lee H., Figueroa P., Gao Y., Tongprasit W., Zhao H., Lee I., Deng X.W. (2007). Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19: 731–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léon-Kloosterziel K.M., Gil M.A., Ruijs G.J., Jacobsen S.E., Olszewski N.E., Schwartz S.H., Zeevaart J.A., Koornneef M. (1996). Isolation and characterization of abscisic acid-deficient Arabidopsis mutants at two new loci. Plant J. 10: 655–661 [DOI] [PubMed] [Google Scholar]
- Liu Y., Koornneef M., Soppe W.J. (2007). The absence of histone H2B monoubiquitination in the Arabidopsis hub1 (rdo4) mutant reveals a role for chromatin remodeling in seed dormancy. Plant Cell 19: 433–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L., Mongrand S., McLachlin D.T., Chait B.T., Chua N.H. (2002). ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J. 32: 317–328 [DOI] [PubMed] [Google Scholar]
- Luerssen H., Kirik V., Herrmann P., Miséra S. (1998). FUSCA3 encodes a protein with a conserved VP1/AB13-like B3 domain which is of functional importance for the regulation of seed maturation in Arabidopsis thaliana. Plant J. 15: 755–764 [DOI] [PubMed] [Google Scholar]
- Mönke G., Altschmied L., Tewes A., Reidt W., Mock H.P., Bäumlein H., Conrad U. (2004). Seed-specific transcription factors ABI3 and FUS3: Molecular interaction with DNA. Planta 219: 158–166 [DOI] [PubMed] [Google Scholar]
- Nakamura S., Lynch T.J., Finkelstein R.R. (2001). Physical interactions between ABA response loci of Arabidopsis. Plant J. 26: 627–635 [DOI] [PubMed] [Google Scholar]
- Oh E., Kang H., Yamaguchi S., Park J., Lee D., Kamiya Y., Choi G. (2009). Genome-wide analysis of genes targeted by PHYTOCHROME INTERACTING FACTOR 3-LIKE5 during seed germination in Arabidopsis. Plant Cell 21: 403–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Kim J., Park E., Kim J.I., Kang C., Choi G. (2004). PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell 16: 3045–3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Yamaguchi S., Hu J., Yusuke J., Jung B., Paik I., Lee H.S., Sun T.P., Kamiya Y., Choi G. (2007). PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell 19: 1192–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Yamaguchi S., Kamiya Y., Bae G., Chung W.I., Choi G. (2006). Light activates the degradation of PIL5 protein to promote seed germination through gibberellin in Arabidopsis. Plant J. 47: 124–139 [DOI] [PubMed] [Google Scholar]
- Okamoto M., et al. (2010). Genome-wide analysis of endogenous abscisic acid-mediated transcription in dry and imbibed seeds of Arabidopsis using tiling arrays. Plant J. 62: 39–51 [DOI] [PubMed] [Google Scholar]
- Park E., Kim J., Lee Y., Shin J., Oh E., Chung W.I., Liu J.R., Choi G. (2004). Degradation of phytochrome interacting factor 3 in phytochrome-mediated light signaling. Plant Cell Physiol. 45: 968–975 [DOI] [PubMed] [Google Scholar]
- Raz V., Bergervoet J.H., Koornneef M. (2001). Sequential steps for developmental arrest in Arabidopsis seeds. Development 128: 243–252 [DOI] [PubMed] [Google Scholar]
- Schmittgen T.D., Livak K.J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3: 1101–1108 [DOI] [PubMed] [Google Scholar]
- Seo M., Nambara E., Choi G., Yamaguchi S. (2009). Interaction of light and hormone signals in germinating seeds. Plant Mol. Biol. 69: 463–472 [DOI] [PubMed] [Google Scholar]
- Shinomura T., Nagatani A., Hanzawa H., Kubota M., Watanabe M., Furuya M. (1996). Action spectra for phytochrome A- and B-specific photoinduction of seed germination in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 93: 8129–8133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone S.L., Kwong L.W., Yee K.M., Pelletier J., Lepiniec L., Fischer R.L., Goldberg R.B., Harada J.J. (2001). LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc. Natl. Acad. Sci. USA 98: 11806–11811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M., Kao C.Y., McCarty D.R. (1997). The conserved B3 domain of VIVIPAROUS1 has a cooperative DNA binding activity. Plant Cell 9: 799–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- To A., Valon C., Savino G., Guilleminot J., Devic M., Giraudat J., Parcy F. (2006). A network of local and redundant gene regulation governs Arabidopsis seed maturation. Plant Cell 18: 1642–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572 [DOI] [PubMed] [Google Scholar]