WRKY33 functions downstream of pathogen-responsive MPK3 and MPK6 in reprogramming the expression of camalexin biosynthetic genes; this drives the metabolic flow to camalexin production in Arabidopsis challenged by pathogens. Biochemical and genetic analyses demonstrate that the phosphorylation of WRKY33 by MPK3/MPK6 plays an important role in the process.
Abstract
Plant sensing of invading pathogens triggers massive metabolic reprogramming, including the induction of secondary antimicrobial compounds known as phytoalexins. We recently reported that MPK3 and MPK6, two pathogen-responsive mitogen-activated protein kinases, play essential roles in the induction of camalexin, the major phytoalexin in Arabidopsis thaliana. In search of the transcription factors downstream of MPK3/MPK6, we found that WRKY33 is required for MPK3/MPK6-induced camalexin biosynthesis. In wrky33 mutants, both gain-of-function MPK3/MPK6- and pathogen-induced camalexin production are compromised, which is associated with the loss of camalexin biosynthetic gene activation. WRKY33 is a pathogen-inducible transcription factor, whose expression is regulated by the MPK3/MPK6 cascade. Chromatin immunoprecipitation assays reveal that WRKY33 binds to its own promoter in vivo, suggesting a potential positive feedback regulatory loop. Furthermore, WRKY33 is a substrate of MPK3/MPK6. Mutation of MPK3/MPK6 phosphorylation sites in WRKY33 compromises its ability to complement the camalexin induction in the wrky33 mutant. Using a phospho-protein mobility shift assay, we demonstrate that WRKY33 is phosphorylated by MPK3/MPK6 in vivo in response to Botrytis cinerea infection. Based on these data, we conclude that WRKY33 functions downstream of MPK3/MPK6 in reprogramming the expression of camalexin biosynthetic genes, which drives the metabolic flow to camalexin production in Arabidopsis challenged by pathogens.
INTRODUCTION
Plant recognition of pathogen-associated molecular patterns (PAMPs) or pathogen-derived effector proteins triggers massive changes in gene expression, cellular metabolism, and eventually induced resistance (Staskawicz et al., 1995; Dangl and Jones, 2001; Nürnberger and Scheel, 2001; Martin et al., 2003; Ausubel, 2005; Boller, 2005). One of the earliest signaling events after plant sensing of invading pathogens is the activation of mitogen-activated protein kinases (MAPKs) (Tena et al., 2001; Zhang and Klessig, 2001; Ichimura et al., 2002; Nakagami et al., 2005). Arabidopsis thaliana has three stress/pathogen-responsive MAPKs: MPK3, MPK6, and MPK4. MPK3 and MPK6 function together in a single MAPK cascade because they share common upstream kinases, are coactivated, and are functionally redundant (Asai et al., 2002; Ren et al., 2002, 2008; Wang et al., 2008). MPK3 and MPK6 are orthologous to tobacco (Nicotiana tabacum) WIPK and SIPK, respectively (Zhang and Klessig, 2001; Ichimura et al., 2002; Ren et al., 2002). In tobacco, SIPK and WIPK share a common upstream MAPKK, Nt MEK2 (Yang et al., 2001). There are two Nt MEK2 orthologs in Arabidopsis, MKK4 and MKK5 (Ren et al., 2002). Arabidopsis MPK4 forms another independent MAPK cascade with upstream MKK1/MKK2 and MEKK1 (Petersen et al., 2000; Suarez-Rodriguez et al., 2007; Qiu et al., 2008a).
Loss- and gain-of-function studies provide genetic evidence supporting a positive role of the MPK3/MPK6 cascade in signaling plant disease resistance (Yang et al., 2001; Asai et al., 2002; Jin et al., 2003; Kroj et al., 2003; del Pozo et al., 2004; Menke et al., 2004; Beckers et al., 2009). Identification of the first plant MAPK substrate revealed that MPK3/MPK6 regulate ethylene production by phosphorylating a subset of ACC synthase (ACS) isoforms (Liu and Zhang, 2004; Joo et al., 2008; Han et al., 2010). Ethylene plays important roles in plant defense (Broekaert et al., 2006; van Loon et al., 2006). Recently, ERF104, an ethylene response factor, was shown to be a MPK6 substrate that plays important roles in plant resistance to a nonadapted bacterial pathogen (Bethke et al., 2009). The MPK3/MPK6 cascade is also involved in defense gene activation, reactive oxygen species generation, and hypersensitive response–like cell death (Ren et al., 2002; Kroj et al., 2003; Kim and Zhang, 2004; Liu et al., 2007). The importance of MAPK signaling in plant–pathogen interactions is also supported by studies of bacterial effectors, several of which target plant MAPK cascades (Zhang et al., 2007; Cui et al., 2010).
Induction of antimicrobial phytoalexins is an integral part of plant disease resistance (VanEtten et al., 1989; Hammerschmidt, 1999; Dixon, 2001). Evidence supporting a positive role of phytoalexins in plant disease resistance comes from studies of both pathogens and plants. Disruption of pathogen genes that encode enzymes known to detoxify phytoalexins can lead to loss of pathogenicity, and the virulence of a pathogen on a specific host sometimes coevolves with the generation of enzymes that are capable of degrading plant phytoalexins (VanEtten et al., 1989; Morrissey and Osbourn, 1999). In addition, mutations of plant genes in the phytoalexin biosynthetic and regulatory pathways, which result in reduced phytoalexin biosynthesis, can lead to increased susceptibility of plants to pathogens (Thomma et al., 1999; Ferrari et al., 2003, 2007; Nafisi et al., 2007; Ren et al., 2008). In recent years, the biosynthetic pathways of a number of phytoalexins have been fully elucidated, and it has been demonstrated that phytoalexin induction is associated with the activation of genes encoding enzymes in the biosynthetic pathways (Hammerschmidt, 1999; Dixon, 2001). However, the signal transduction pathway(s) leading to the activation of these genes are mostly unclear.
We previously reported that the pathogen-responsive MPK3/MPK6 cascade plays a positive role in regulating the biosynthesis of camalexin (3-thiazol-2’-yl-indole; Tsuji et al., 1992), the major phytoalexin in Arabidopsis (Ren et al., 2008). Activation of the MPK3/MPK6 cascade leads to coordinated upregulation of multiple genes encoding enzymes in the camalexin biosynthetic pathway, including CYP71A13, which converts indole-3-acetaldoxime to indole-3-acetonitrile, and PAD3, which encodes another P450 enzyme (CYP71B15) that carries out the last step of camalexin biosynthesis (Zhou et al., 1999; Schuhegger et al., 2006; Nafisi et al., 2007; Böttcher et al., 2009). We also hypothesized that MPK3/MPK6 are likely to phosphorylate a transcription factor or factors, which is/are directly responsible for activating the expression of camalexin biosynthetic genes (Ren et al., 2008).
In our search for the transcription factor(s) downstream of MPK3/MPK6 in Arabidopsis or their orthologous WIPK/SIPK in tobacco, we identified WRKY transcription factors, including Arabidopsis WRKY33, as potential downstream targets based on their gene activation in the gain-of-function GVG-Nt-MEK2DD plants (Kim and Zhang, 2004; Wan et al., 2004). Later, it was shown that WRKY33 expression is highly induced in Arabidopsis treated with PAMPs or infected by pathogens and that wrky33 mutants are more susceptible to Botrytis cinerea and to Alternaria brassicicola (Zheng et al., 2006; Lippok et al., 2007). In the same studies, it was also demonstrated that WRKY33 is nuclear localized and that it binds to the W-box cis-element. More recently, WRKY33 was shown to be essential for the induction of camalexin biosynthesis in Arabidopsis infected with Pseudomonas syringae, and WRKY33 directly binds to the PAD3 promoter (Qiu et al., 2008b).
In this report, we demonstrate that the WRKY33 transcription factor functions downstream of MPK3/MPK6 in activating the expression of camalexin biosynthetic genes. In the wrky33 mutant background, both the gain-of-function MPK3/MPK6- and B. cinerea–induced camalexin production are compromised, which is associated with the loss of activation of camalexin biosynthetic genes. WRKY33 is a pathogen-inducible transcription factor, whose expression is regulated by the MPK3/MPK6 cascade. In addition, WRKY33 is a substrate of MPK3/MPK6. Using a phospho-protein mobility shift assay, we show that WRKY33 is phosphorylated by MPK3/MPK6 in vivo in response to B. cinerea infection. Furthermore, mutation of MPK3/MPK6 phosphorylation sites in WRKY33 compromises its ability to complement the deficiency of camalexin induction in the wrky33 mutant. These results demonstrate that WRKY33 acts downstream of MPK3/MPK6 in reprogramming the expression of camalexin biosynthetic genes, which drives the metabolic flow to camalexin production in Arabidopsis infected by pathogens.
RESULTS
WRKY33 Is Essential for Gain-of-Function GVG-Nt-MEK2DD– and B. cinerea–Induced Camalexin Biosynthesis
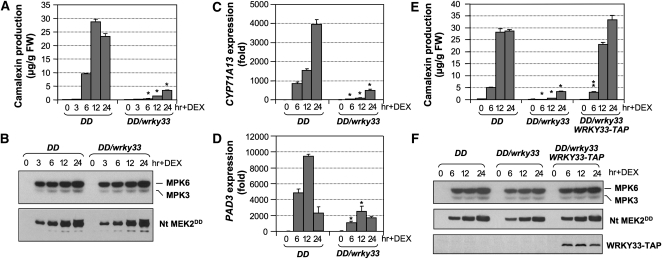
Using a gel mobility shift assay, we identified WRKY transcription factors as potential targets of SIPK/WIPK in tobacco defense response (Kim and Zhang, 2004). To identify the specific WRKY(s) involved, we took a genetic approach in Arabidopsis by crossing the dexamethasone (DEX)-inducible promoter-driven constitutively active Nt MEK2DD transgene (GVG-Nt-MEK2DD, abbreviated as DD) (Yang et al., 2001; Ren et al., 2002) into different wrky mutant backgrounds. Our initial efforts were focused on WRKY members, including WRKY6, WRKY33, WRKY40, and WRKY53, whose expressions are induced in the DD plants after DEX treatment (Wan et al., 2004) (Y. Liu and S. Zhang, unpublished data). Known MPK3/MPK6-regulated defense responses, including defense gene activation, ethylene induction, and camalexin production (Kim and Zhang, 2004; Liu and Zhang, 2004; Ren et al., 2008), were monitored in the DD/wrky double mutants. As shown in Figure 1A, DD-induced camalexin production was blocked in the wrky33, but not wrky6, wrky40, or wrky53, background, suggesting that WRKY33 is downstream of MPK3/MPK6 in regulating camalexin biosynthesis. DD expression and MPK3/MPK6 activation after DEX treatment were not affected in the DD/wrky33 double mutant (Figure 1B). Compromised camalexin induction was associated with the loss of activation of camalexin biosynthetic genes, including CYP71A13 and PAD3 (Figures 1C and 1D), consistent with our previous report that gene activation is involved in MPK3/MPK6-induced camalexin biosynthesis (Ren et al., 2008).
Figure 1.
Induction of Camalexin Biosynthesis in the Gain-of-Function GVG-Nt-MEK2DD Transgenic Plants (DD) Is Dependent on the WRKY33 Transcription Factor.
(A) Mutation of WRKY33 inhibited camalexin biosynthesis in DD seedlings. Two-week-old DD and DD/wrky33 seedlings were treated with DEX (1 μM final concentration). Camalexin accumulation was measured at indicated times. Error bars indicate se (n = 3). FW, fresh weight.
(B) Normal DD induction (bottom) and MPK3/MPK6 activation (top) in DD/wrky33 seedlings. Flag-tagged DD protein was detected by immunoblot analysis using an anti-Flag antibody. MPK6 and MPK3 activation were determined by an in-gel kinase assay using MBP as a substrate.
(C) and (D) Activation of camalexin biosynthetic genes, including CYP71A13 (C) and PAD3 (D), was compromised in the wrky33 background. Transcript levels were determined by real-time qPCR. Error bars indicate se (n = 3).
(E) Complementation of wrky33 mutation by a native WRKY33 promoter-driven WRKY33-TAP construct. Error bars indicate se (n = 3).
(F) Induction of WRKY33-TAP protein in DD/WRKY33-TAP/wrky33 plants after MPK3/MPK6 activation. Total protein extracts prepared from seedlings shown in (E) were subjected to an in-gel kinase assay using MBP as a substrate (top), and immunoblot analyses using anti-Flag antibody to detect Flag-tagged DD protein (middle) and anti-IgG-HRP conjugate to detect the TAP-tagged WRKY33 (bottom). Statistically different data groups at a specific time point (P value < 0.05) are indicated using different numbers of asterisks (0 to 2) vertically placed above the columns in the graphs.
Two wrky33 mutant alleles were analyzed, which gave similar results. In addition, we transformed a WRKY33 native promoter driven tandem-affinity purification (TAP)-tagged WRKY33 construct (WRKY33-TAP) into the DD/wrky33 background. As shown in Figure 1E, this construct fully rescued camalexin induction in the DD/wrky33 plants. Again, the induction of the DD protein and the MPK3/MPK6 activation were the same in plants with different genotypes (Figure 1F, top and middle). Previously, we reported that WRKY33 expression is highly induced by MPK3/MPK6 activation (Wan et al., 2004). With the TAP tag, we examined the WRKY33 protein levels before and after MPK3/MPK6 activation. As shown in Figure 1F (bottom), the WRKY33 protein was undetectable before MPK3/MPK6 activation and accumulated to high levels after MPK3/MPK6 activation, consistent with the activation of WRKY33 gene expression in DD plants after DEX treatment (Wan et al., 2004).
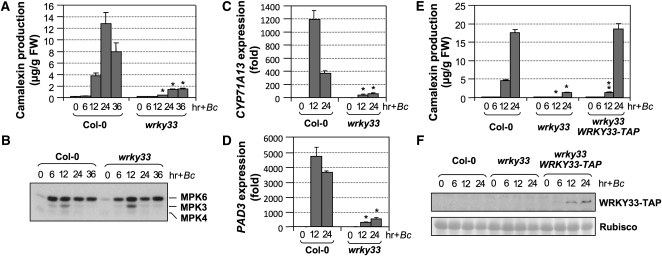
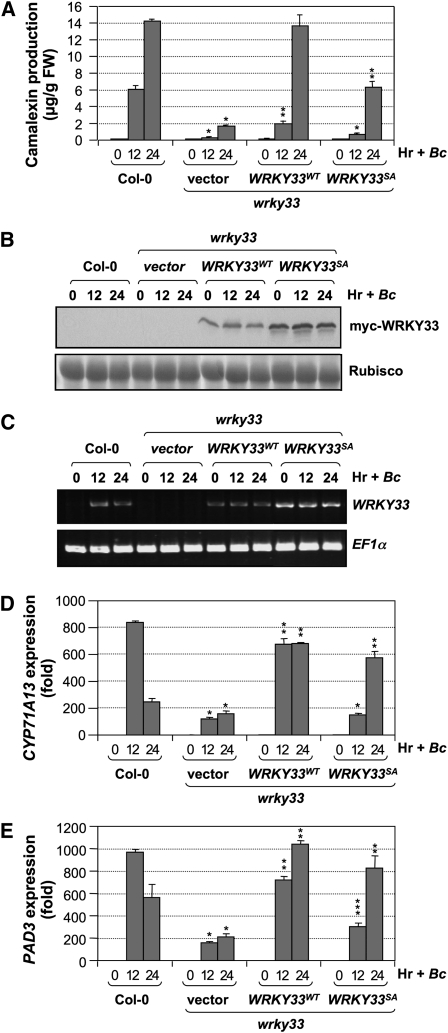
WRKY33 is also essential for B. cinerea–induced camalexin biosynthesis. Camalexin induction in the wrky33 mutants was compromised (Figure 2A), which was associated with the greatly reduced activation of CYP71A13 and PAD3 gene expression (Figures 2C and 2D). The activation of MPK3/MPK6 was not affected in the wrky33 mutants (Figure 2B), consistent with a function of WRKY33 downstream of MPK3/MPK6. The WRKY33-TAP transgene fully rescued the deficiency of wrky33 at 24 h after B. cinerea inoculation (Figure 2E). Again, the WRKY33-TAP protein was absent before pathogen infection and was induced by B. cinerea infection (Figure 2F), which was associated with the activation WRKY33 at transcriptional level (see Supplemental Figure 1 online).
Figure 2.
WRKY33 Is Essential to Camalexin Induction in Arabidopsis after B. cinerea Infection.
(A) Mutation of WRKY33 compromised B. cinerea–induced camalexin biosynthesis. Two-week-old wild-type (Col-0) and wrky33 seedlings were inoculated with B. cinerea spores, and camalexin accumulation was measured at indicated times. Error bars indicate se (n = 3). FW, fresh weight.
(B) MPK3/MPK6 activation in the wrky33 mutant was not affected. MAPK activation in these seedlings was determined by an in-gel kinase assay using MBP as a substrate.
(C) and (D) Activation of camalexin biosynthetic genes, including CYP71A13 (C) and PAD3 (D), was compromised in the wrky33 mutant. Transcript levels were determined by real-time qPCR. Error bars indicate se (n = 3).
(E) Complementation of wrky33 mutation by a native WRKY33 promoter-driven WRKY33-TAP construct. Error bars indicate se (n = 3).
(F) Induction of WRKY33-TAP protein in WRKY33-TAP/wrky33 plants by B. cinerea. Total protein extracts prepared from seedlings shown in (E) were subjected to immunoblot analyses using a goat anti-IgG-HRP conjugate to detect the TAP-tagged WRKY33 (top). Equal amounts (10 μg) were loaded to each lane and were confirmed by Ponceau S staining (bottom). Statistically different data groups at a specific time point (P value < 0.05) are indicated using different numbers of asterisks (0 to 2) vertically placed above the columns in the graphs.
The Pathogen-Responsive MPK3/MPK6 Cascade Is Involved in WRKY33 Gene Activation
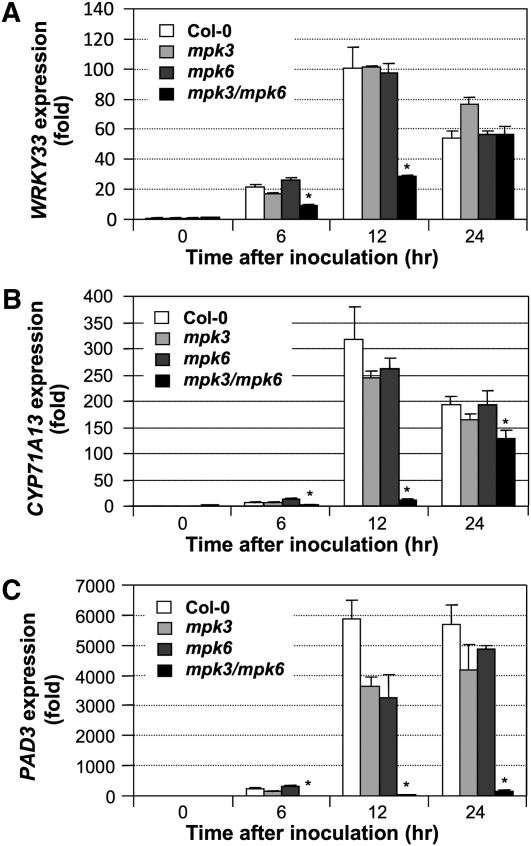
WRKY33 is a PAMP/pathogen-responsive WRKY transcription factor that is essential to B. cinerea resistance (Zheng et al., 2006; Lippok et al., 2007). Gain-of-function activation of MPK3/MPK6 is sufficient to activate WRKY33 expression (Wan et al., 2004), suggesting that the MPK3/MPK6 cascade might be involved in PAMP/pathogen-induced WRKY33 gene activation. To provide loss-of-function evidence, we examined WRKY33 expression in B. cinerea–infected wild type (Columbia-0 [Col-0]), single mpk3 or mpk6 mutants, and the rescued mpk3 mpk6 double mutant (Wang et al., 2007; Ren et al., 2008). The conditionally rescued mpk3 mpk6 double mutant was obtained by transforming a DEX-inducible promoter-driven MPK6 (GVG-MPK6) into mpk3−/−/mpk6+/− plants. When the T3 mpk3−/−/mpk6+/−/GVG-MPK6+/+ plants began to flower, DEX was sprayed every other day to rescue the embryo lethality of the mpk3−/−/mpk6−/−/GVG-MPK6+/+ zygotes. Progenies with mpk3−/−/mpk6−/−/GVG-MPK6+/+ genotype, which still show developmental defects (Wang et al., 2007), were called rescued mpk3 mpk6 double mutants and were used for experiments.
As shown in Figure 3A, WRKY33 activation was not affected in the single mpk3 or mpk6 mutants. However, in the rescued mpk3 mpk6 double mutant, WRKY33 induction was compromised and much delayed, suggesting that the MPK3/MPK6 cascade is required for full induction of WRKY33 expression. The residual activation of WRKY33 in the mpk3 mpk6 double mutant suggests that other pathways are also involved in B. cinerea–induced WRKY33 expression. It is also possible that the basal level expression of the GVG-MPK6 transgene might be able to compensate the mutant to a certain extent, although this is unlikely since we detected little/no activity from the transgenic MPK6 by the in-gel kinase activity assay in the rescued mpk3 mpk6 double mutant after B. cinerea infection (Ren et al., 2008; Han et al., 2010).
Figure 3.
WRKY33 Induction after B. cinerea Infection Is Dependent on the MPK3/MPK6 Cascade.
(A) Induction of WRKY33 gene expression was compromised in the rescued mpk3 mpk6 double mutant. Wild-type (Col-0), mpk3, mpk6, and mpk3 mpk6 seedlings were inoculated with B. cinerea. Samples were collected at indicated times. Induction of the WRKY33 transcript was quantified by real-time qPCR. Error bars indicate se (n = 3).
(B) and (C) Activation of camalexin biosynthetic genes, including CYP71A13 (B) and PAD3 (C), was compromised in the mpk3 mpk6 mutant. Error bars indicate se (n = 3). Asterisks above the columns indicate the data sets that are statistically different from those without asterisk at a specific time point (P value < 0.05).
In Vitro Phosphorylation of WRKY33 by MPK3 and MPK6
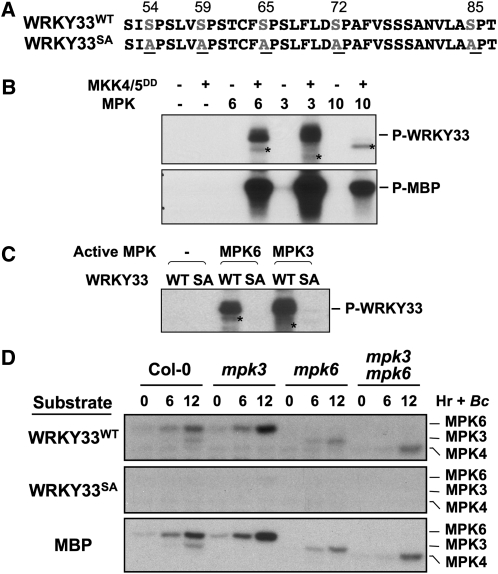
Despite a partial blockage of WRKY33 gene activation, camalexin induction in the mpk3 mpk6 double mutant was almost completely inhibited (Ren et al., 2008). Associated with it, induction of CYP71A13 and PAD3 expression was also abolished (Figures 3B and 3C). This result suggests that MPK3/MPK6 might regulate WRKY33 at additional levels. It is possible that phosphorylation by MPK3/MPK6 is required to fully activate the de novo synthesized WRKY33 protein. In the absence of MPK3 and MPK6, residual induction of WRKY33 is unable to fully activate the expression of downstream camalexin biosynthetic genes, resulting in compromised camalexin induction. An examination of WRKY33 protein sequence revealed a cluster of five potential MAPK phosphorylation sites (Ser-54, Ser-59, Ser-65, Ser-72, and Ser-85) in the N terminus of the WRKY33 protein (Figure 4A). As a result, we prepared a His-tagged recombinant WRKY33 protein for in vitro MAPK phosphorylation assays.
Figure 4.
In Vitro Phosphorylation of WRKY33 by MPK3 and MPK6.
(A) Putative MAPK phosphorylation sites in the N terminus of WRKY33 and the loss-of-phosphorylation WRKY33 mutant with all five Ser mutated to Ala (WRKY33SA).
(B) Phosphorylation of WRKY33 in vitro by the activated MPK3 and MPK6 but not MPK10. Reactions with various components omitted (−) were used as controls. The asterisks in the top panel indicate the phosphorylation of MAPKs by MKK4DD/MKK5DD.
(C) Mutation of MAPK phosphorylation sites abolished the phosphorylation of WRKY33 by MPK3 and MPK6. Recombinant WRKY33WT (WT) and WRKY33SA (SA) were incubated with activated MPK3 and MPK6 as in (B). Phosphorylated WRKY33 was visualized by autoradiography after gel electrophoresis.
(D) Phosphorylation of WRKY33WT, but not WRKY33SA, by the native MAPKs extracted from seedlings treated with B. cinerea. Total extracts were prepared from wild-type (Col-0), mpk3, mpk6, and mpk3 mpk6 seedlings infected with B. cinerea. MAPK activities were detected by an in-gel kinase assay using recombinant WRKY33WT (top), recombinant WRKY33SA (middle), and MBP (bottom) as substrates.
As shown in Figure 4B (top), activated recombinant MPK3 and MPK6 strongly phosphorylated WRKY33. By contrast, MPK10, a closely related homolog of MPK3 and MPK6, failed to do so. All three MAPKs were able to phosphorylate myelin basic protein (MBP), demonstrating that all were active (Figure 4B, bottom). Without activation by the constitutively active MKK4DD/MKK5DD, neither MPK3 nor MPK6 was able to phosphorylate WRKY33 (Figure 4B), confirming the importance of phosphorylation activation of MPK3/MPK6 by its upstream MKK4/MKK5. In the autoradiogram, the phosphorylation labeling of MAPKs by MKK4DD/MKK5DD was evident (Figure 4B, top). When all five Ser residues were mutated to Ala (WRKY33SA), the protein could no longer be phosphorylated by MPK3/MPK6 (Figure 4C).
In addition to recombinant MAPKs, we also analyzed the phosphorylation of WRKY33 by the native MAPKs. In this assay, recombinant WRKY33WT or WRKY33SA protein was embedded in an SDS-PAGE gel instead of MBP. Phosphorylation of the embedded WRKY33 was determined by an in-gel kinase assay using total protein extracts from Col-0, mpk3, mpk6, and mpk3 mpk6 seedlings treated with B. cinerea, which activates MPK3/MPK6 and MPK4 cascades (Ren et al., 2008; Han et al., 2010). As shown in Figure 4D, identical kinase activity patterns were observed when WRKY33WT and MBP were used as the substrates. By contrast, no kinase activity was detected when WRKY33SA was embedded in the gel. The loss of kinase bands in their respective mutants confirmed the MAPK identities. In addition to MPK3 and MPK6, we also detected the activity of MPK4 in assays using WRKY33WT and MBP (but not WRKY33SA) as substrates, suggesting that MPK4 is able to phosphorylate WRKY33 in vitro on the same Ser residues. Similar to our previous reports (Ren et al., 2008; Han et al., 2010), higher levels of MPK4 activity were observed in the mpk3 mpk6 double mutant after B. cinerea infection (Figure 4D). This could be a result of higher MPK4 protein levels in the mpk3 mpk6 double mutant (Han et al., 2010). It is also possible that MPK4 cascade and MPK3/MPK6 cascade share common upstream components after the sensing of B. cinerea. The loss of MPK3/MPK6 cascade leads to a higher signaling strength feeding into the MPK4 cascade.
MPK3/MPK6 Phosphorylation Sites in WRKY33 Are Essential for Its Full Activity in Vivo
To determine the importance of WRKY33 phosphorylation by MPK3/MPK6 in vivo, we investigated the ability of WRKY33SA in complementing the wrky33 mutant phenotype. In this experiment, we used the constitutive 35S promoter–driven WRKY33WT and WRKY33SA constructs so that the regulation of WRKY33 at the phosphorylation level by MPK3/MPK6 could be studied separately from its regulation at the transcriptional level. A four-copy myc tag (4myc) was added to the N terminus for easy detection of the WRKY33 protein. More than 30 independent lines of each construct were analyzed to determine the induction of camalexin production after B. cinerea infection. We found that WRKY33SA lines had lower levels of camalexin induction than WRKY33WT lines with similar levels of expression, and none of the WRKY33SA lines showed full rescue of the wrky33 mutant. The highest level of rescue was ~50% at 24 h after inoculation (Figure 5A). By contrast, many WRKY33WT-rescued lines were obtained. Examination of WRKY33 protein levels revealed that even the lines with partial complementation expressed WRKY33SA at a higher level than the WRKY33WT line with full rescue (Figures 5A and 5B). This result revealed that WRKY33SA was less efficient in complementing the wrky33 mutant. The higher WRKY33SA protein level was associated with a higher level of gene expression (Figure 5C). In the wild-type (Col-0) control, induction of WRKY33 expression was evident. In both transgenic lines, transcripts were constitutively expressed because of the 35S promoter. In the vector/wrky33 control, no WRKY33 transcript was detectable, demonstrating the specificity of the RT-PCR reaction. RT- PCR with a primer pair that spans the whole open reading frame was used to examine WRKY33 transgene expression in the wrky33 background because wrky33 mutant alleles still produce nonfunctional transcripts (Zheng et al., 2006). We tried several pairs of quantitative PCR (qPCR) primers, and all of them amplified the cDNAs from the mutated gene transcripts. We found that the native promoter driven WRKY33SA construct also failed to fully complement the camalexin induction in the wrky33 mutant background (see Supplemental Figure 2 online).
Figure 5.
MPK3/MPK6 Phosphorylation Sites in WRKY33 Are Important for Its Function in Vivo.
(A) Loss-of-phosphorylation WRKY33 can only partially complement the wrky33 mutant. Myc epitope-tagged WRKY33WT and WRKY33SA under the control of the constitutive 35S promoter were transformed into the wrky33 mutant. An empty vector was used as a negative control. Camalexin accumulation was determined at indicated times after B. cinerea inoculation, and seedlings were collected for protein and RNA preparations. Error bars indicate se (n = 3). FW, fresh weight.
(B) Expression of WRKY33SA at a higher level despite a lower level of complementation. Levels of WRKY33 protein in samples collected in (A) were determined by immunoblot analysis using an anti-myc epitope antibody (top). Equal loading was confirmed by Ponceau S staining (bottom).
(C) Higher WRKY33SA protein levels were associated with higher transcript levels. Levels of WRKY33 transcript in samples collected in (A) were examined by RT-PCR using a primer pair that did not amplify WRKY33 with the T-DNA insertion (top). EF1α was used to show equal inputs of cDNA templates (bottom). Twenty-five cycles of PCR were performed.
(D) and (E) WRKY33SA is less efficient in supporting the B. cinerea–induced activation of camalexin biosynthetic genes, including CYP71A13 (D) and PAD3 (E). Transcript levels were determined by real-time qPCR. Error bars indicate se (n = 3). Statistically different data groups at a specific time point (P value < 0.05) are indicated using different numbers of asterisks (0 to 3) vertically placed above the columns in the graphs.
Loss-of-phosphorylation mutant WRKY33SA was also less efficient in complementing the activation of camalexin biosynthetic genes in the wrky33 mutant (Figures 5D and 5E). The induction of CYP71A13 and PAD3 gene expression was much lower at 12 h after B. cinerea inoculation in the WRKY33SA/wrky33 seedlings in comparison to that in the wild-type control (Col-0) and WRKY33WT/wrky33 seedlings. The much-delayed induction of camalexin biosynthetic genes is likely to hamper the accumulation enzyme activities, resulting in the lower camalexin production in WRKY33SA/wrky33 seedlings (Figure 5A). Once the cell death sets in at the later stage of the infection process, the cell will eventually have a reduced metabolic capacity and may lose the ability to produce camalexin.
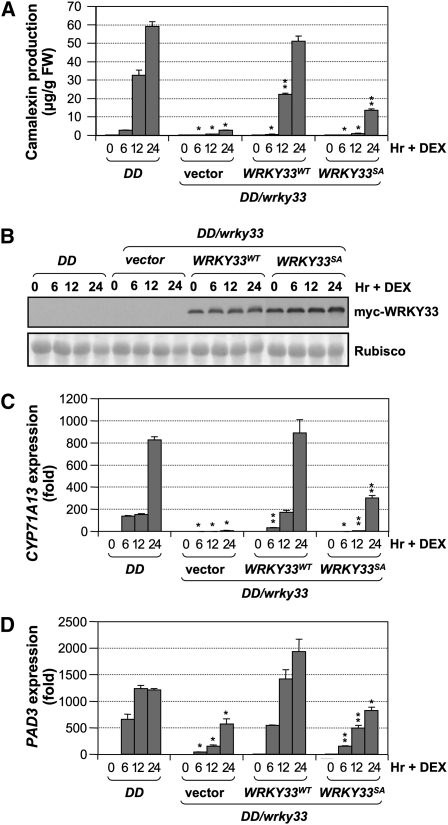
To determine the importance of WRKY33 phosphorylation in camalexin induction in the gain-of-function DD plants, we crossed the transgenic lines shown in Figure 5A (homozygous vector/wrky33, WRKY33WT/wrky33, and WRKY33SA/wrky33) with DD/wrky33 to generate DD/vector/wrky33, DD/WRKY33WT/wrky33, and DD/WRKY33SA/wrky33. Large numbers of crosses were performed to obtain enough F1 seeds for experiments. They were homozygous for wrky33 and heterozygous for DD and WRKY33WT or WRKY33SA. Camalexin accumulation after DEX treatment in these lines was compared. As shown in Figure 6A, WRKY33WT was able to fully complement the loss of endogenous WRKY33. However, WRKY33SA could only partially rescue wrky33. Immunoblot analysis using an anti-myc antibody revealed that WRKY33SA expressed at a higher level than WRKY33WT (Figure 6B), ruling out the possibility that the partial complementation by the WRKY33SA transgene was a result of lower expression. Again, the lower efficiency of WRKY33SA in complementing the DD-induced camalexin production in the wrky33 mutant (Figure 6A) was associated with the compromised induction of camalexin biosynthetic genes, including CYP71A13 and PAD3 (Figures 6C and 6D). Based on these data, we can conclude that WRKY33SA, a loss-of-phosphorylation mutant, cannot achieve the full activity of WRKY33WT in activating the expression of camalexin biosynthetic genes, highlighting the importance of WRKY33 phosphorylation by MPK3/MPK6 in camalexin induction.
Figure 6.
MPK3/MPK6 Phosphorylation Sites in WRKY33 Are Also Required for Camalexin Induction in the Gain-of-Function DD Seedlings.
(A) Loss-of-phosphorylation WRKY33 can only partially complement the camalexin induction in DD/wrky33 mutant. The same transgenic lines shown in Figure 5 were crossed to DD/wrky33 lines to generate DD/wrky33/4myc-WRKY33WT, DD/wrky33/4myc-WRKY33SA, and vector control lines. Camalexin accumulation was monitored at indicated times after DEX (1 μM) treatment, and seedlings were collected for protein preparations. Error bars indicate se (n = 3).
(B) Partial complementation by WRKY33SA was not a result of a lower expression level. Levels of WRKY33 protein in the transgenic lines were determined in samples collected in (A) using an anti-myc antibody (top). Equal loading was confirmed by Ponceau S staining (bottom).
(C) and (D) WRKY33SA is less efficient in supporting the MPK3/MPK6-induced activation of camalexin biosynthetic genes, including CYP71A13 (C) and PAD3 (D). Transcript levels were determined by real-time qPCR. Error bars indicate se (n = 3). Statistically different data groups at a specific time point (P value < 0.05) are indicated using different numbers of asterisks (0 to 2) vertically placed above the columns in the graphs.
Phosphorylation of WRKY33 by MPK3/MPK6 in Vivo
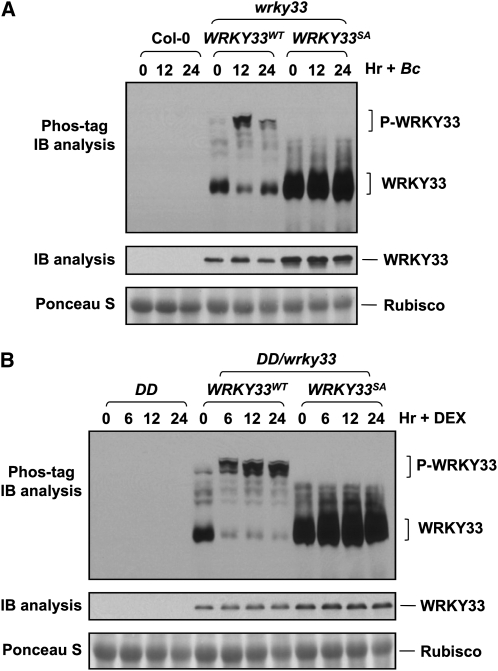
The genetic evidence above demonstrates that MPK3/MPK6 phosphorylation sites in the N terminus of WRKY33 are important for the full induction of camalexin biosynthesis in plants challenged by B. cinerea or in the gain-of-function DD plants (Figures 5 and 6). To provide direct evidence that WRKY33 is phosphorylated by MPK3/MPK6 in vivo, we used the Phos-tag mobility shift assay, in which the binding of phospho-proteins to the Phos-tag reagent in the SDS-PAGE gel matrix slows down their movement (Bethke et al., 2009). Protein extracts from WRKY33WT/wrky33 and WRKY33SA/wrky33 plants treated with B. cinerea were first separated in a Phos-tag SDS-PAGE gel, and 4myc-tagged WRKY33 was detected by immunoblot analysis. Extracts from the wild type (Col-0) were used as a negative control to determine the specificity of the anti-myc immunoblot analysis. As shown in Figure 7A, upshift of 4myc-tagged WRKY33WT was observed after B. cinerea infection, which was associated with a decrease in unphosphorylated WRKY33WT protein. Such upshift was absent in the extracts from WRKY33SA/wrky33 plants, demonstrating the phosphorylation of WRKY33 on the five MAPK phosphorylation sites after B. cinerea infection. Total 4myc-tagged WRKY33 proteins were determined by regular immunoblot (Figure 7A, middle). Equal loading of protein was double confirmed by staining of nitrocellulose membrane with Ponceau S (Figure 7A, bottom).
Figure 7.
In Vivo Phosphorylation of WRKY33 by MPK3/MPK6.
(A) WRKY33 becomes phosphorylated in seedlings infected with B. cinerea. Protein extracts from wild-type (Col-0), wrky33/4myc-WRKY33WT, and wrky33/4myc-WRKY33SA seedlings treated with B. cinerea for different times were separated in an SDS-PAGE gel with Phos-tag reagent. After being transferred to a nitrocellulose membrane, myc-tagged WRKY33WT and WRKY33SA proteins were detected by an anti-myc antibody (top). A regular immunoblot (IB) was done at the same time to detect total WRKY33 protein (middle). Equal loading was confirmed by Ponceau S staining (bottom).
(B) WRKY33 phosphorylation in gain-of-function DD seedlings after DEX treatment. Protein extracts from DD, DD/wrky33/4myc-WRKY33WT, and DD/wrky33/4myc-WRKY33SA seedlings treated with DEX (1 μM) at various times were separated in a Phos-tag SDS-PAGE gel. After being transferred to nitrocellulose membranes, WRKY33WT and WRKY33SA proteins were detected by an anti-myc antibody (top). A regular immunoblot was done at the same time to detect the total WRKY33 protein (middle). Equal loading was confirmed by Ponceau S staining (bottom).
To demonstrate the phosphorylation of WRKY33 by MPK3/MPK6, we analyzed the phosphorylation status of WRKY33 in the DD background. Protein extracts from DD, DD/WRKY33WT/wrky33, and DD/WRKY33SA/wrky33 plants treated with DEX for different times were subjected to Phos-tag mobility shift assays. Within 6 h after DEX treatment, the majority of the WRKY33WT protein was phosphorylated, as indicated by the upshift of the 4myc-tagged WRKY33. Associated with this, the amount of unphosphorylated protein decreased (Figure 7B, top). By contrast, no such upshift of WRKY33SA was observed, demonstrating again that the phosphorylation was on the MAPK phosphorylation sites. Based on these data, we conclude that WRKY33 is phosphorylated after MPK3/MPK6 activation and that the phosphorylation is dependent on the MAPK phosphorylation sites in the N terminus of WRKY33. Combined with the genetic evidence that the MPK3/MPK6 phosphorylation sites are required for the complementation of the wrky33 mutant phenotype, we can conclude that MPK3/MPK6 phosphorylation of WRKY33 is important to the activation of camalexin biosynthetic genes.
MPK3/MPK6 Phosphorylation of WRKY33 Does Not Alter Its DNA Binding Activity
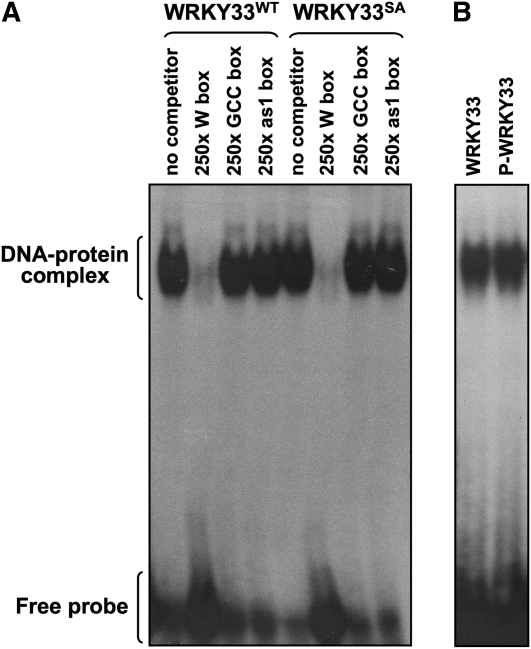
Phosphorylation of a transcription factor by a kinase may change the DNA binding activity of the transcription factor. To determine whether phosphorylation of WRKY33 by MPK3/MPK6 alter its W-box binding activity, we performed an electrophoresis mobility shift assay (EMSA). As shown in Figure 8, wild-type WRKY33 (WRKY33WT) and loss-of-phosphorylation WRKY33 mutant (WRKY33SA) had similar DNA binding activity to W-box. Inclusion of unlabeled W-box, but not GCC-box or as-1 box, in the binding reaction effectively competed the binding of WRKY33 to the 32P-labeled W-box probe, demonstrating the specificity of W-box binding activity of WRKY33 protein. This experiment also demonstrated that the mutation of the five Ser residues in the MPK3/MPK6 phosphorylation sites to Ala residues does not interfere with the DNA binding activity of WRKY33.
Figure 8.
Phosphorylation of WRKY33 Does Not Alter Its DNA Binding Ability to the W-Box cis-Element.
(A) EMSA was performed using freshly prepared recombinant WRKY33WT or WRKY33SA protein and 32P-labeled W-box probe. The specificity of W-box binding activity was demonstrated by competition assay using 250-fold excess unlabeled W-box, GCC-box, or as1-box DNAs.
(B) Phosphorylation of WRKY33 does not enhance its W-box binding activity. Freshly prepared recombinant WRKY33WT was phosphorylated using the activated MPK3 and MPK6 (equal mixture). A control reaction without MPK3/MPK6 was processed side-by-side. The W-box binding activity of the phosphorylated and unphosphorylated (from the control reaction) was determined by EMSA as in (A).
To determine the effect of MPK3/MPK6 phosphorylation on the DNA binding activity of WRKY33, we first phosphorylated WRKY33 using the activated MPK3/MPK6. Since both MPK3 and MPK6 phosphorylate WRKY33 on the same sites (Figure 4), we used an equal mixture of MPK3 and MPK6 as the enzyme. A control reaction without the addition of MPK3/MPK6 was set side-by-side, which was used as unphosphorylated WRKY33. EMSA revealed no difference in the W-box binding activity between the phosphorylated WRKY33 and unphosphorylated WRKY33 (Figure 8B). This finding suggests that the phosphorylation of WRKY33 by MPK3/MPK6 might affect the transactivation activity rather than the W-box binding activity of WRKY33. This is consistent with the fact that MPK3/MPK6 phosphorylation sites in WRKY33 are far away from the DNA binding domain (Zheng et al., 2006), making it unlikely that the phosphorylation will change its DNA binding activity. In addition, we found that WRKY33 is constitutively associated with the chromatin since extraction buffer without SDS was unable to extract WRKY33 from cells.
Binding of WRKY33 to Its Own Promoter Suggests Potential Self-Activation of Transcription
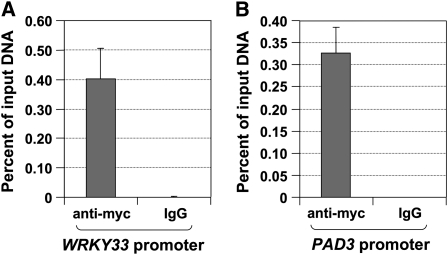
Increase in the levels of WRKY33 protein, which is associated with WRKY33 gene activation (Figures 1F, 2F, and 3A), is likely to play an important role in the activation of downstream genes during plant defense response. It was demonstrated that a cluster of W-box in the promoter of WRKY33 is involved in the WRKY33 gene activation in plants infected by pathogens or after PAMP treatment (Lippok et al., 2007). Furthermore, it was shown in the same report that WRKY transcription factors interact with these W-boxes in vivo based on chromatin immunoprecipitation (ChIP) assay. However, since an antibody against all/most WRKY transcription factors was used in the ChIP assay, the identity of the WRKY remains unknown. To determine whether WRKY33 is involved in regulating its own expression, we performed ChIP-qPCR assay to see whether WRKY33 binds to its own promoter. As shown in Figure 9A, WRKY33 promoter was greatly enriched with an anti-myc antibody that immunoprecipitates the 4myc-tagged WRKY33 transgene product. By contrast, IgG control failed to pull down WRKY33 promoter DNA.
Figure 9.
WRKY33 Binds to Its Own Promoter and the Promoter of PAD3 in Vivo.
ChIP-qPCR analysis was performed using DD/4myc-WRKY33WT plants generated from the cross of wrky33/4myc-WRKY33WT with DD lines. Input chromatin was isolated from 2-week-old seedlings 12 h after DEX treatment. Epitope-tagged WRKY33-chromatin complex was immunoprecipitated with an anti-myc antibody. A control reaction was processed side-by-side using mouse IgG. ChIP- and input-DNA samples were quantified by real-time qPCR using primers specific to the promoters of WRKY33 (A) and PAD3 (B) genes. The ChIP results are presented as percentage of input DNA. Error bars indicate se (n = 3).
To further validate the ChIP-qPCR experiment, we quantified the enrichment of the PAD3 promoter. Our results indicated that WRKY33 is likely to target PAD3 genes directly and is involved in the upregulation of PAD3 expression (Figures 1D and 2D). As shown in Figure 9B, anti-myc antibody effectively enriched the PAD3 promoter DNA, while the control IgG failed to do so. This result is consistent with previous finding that WRKY33 directly interacts with PAD3 promoter (Qiu et al., 2008b).
DISCUSSION
Induction of phytoalexins in plants after pathogen invasion is an integral part of induced plant disease resistance (VanEtten et al., 1989; Glazebrook et al., 1997; Hammerschmidt, 1999; Morrissey and Osbourn, 1999; Dixon, 2001). The biosynthetic pathways of a number of phytoalexins have been fully defined. However, the signaling pathways regulating their biosynthesis are largely unclear. Our previous study demonstrated that the Arabidopsis MPK3/MPK6 cascade is an important regulatory pathway controlling camalexin biosynthesis in Arabidopsis (Ren et al., 2008). The activation of MPK3/MPK6 leads to the upregulation of the expression of camalexin biosynthetic genes, implicating the involvement of downstream transcription factors. In this report, we demonstrate that WRKY33 is a key component downstream of MPK3/MPK6 in the pathogen-induced camalexin biosynthesis. In wrky33 mutants, both the gain-of-function MPK3/MPK6- and the pathogen-induced camalexin productions are compromised, which is associated with the loss of activation of camalexin biosynthetic genes. Genetic analysis revealed that the MAPK phosphorylation sites in WRKY33 are important for its full function/activity in vivo. Phospho-protein mobility shift assays allowed us to demonstrate the in vivo phosphorylation of WRKY33 by MPK3/MPK6 after B. cinerea infection. Taken together, we can conclude that WRKY33, a novel MPK3/MPK6 substrate, plays an essential role in the transcriptional activation of camalexin biosynthetic genes and camalexin induction in Arabidopsis in response to pathogen infection.
Dual-Level Regulation of WRKY33 by the MPK3/MPK6 Cascade in Plant Defense Response
Expression of many WRKY genes is highly induced by stresses, especially pathogen-related stimuli (Dong et al., 2003; Pandey and Somssich, 2009; Rushton et al., 2010). However, the signaling pathways and downstream transcription factors are unknown. Gain-of-function activation of MPK3/MPK6 was sufficient to induce WRKY33 expression and WRKY33 protein accumulation (Figure 1F) (Wan et al., 2004), suggesting that the MPK3/MPK6 cascade might be involved in pathogen-induced WRKY33 expression. In this report, we provide loss-of-function evidence to support this conclusion. In the mpk3 mpk6 double mutant, B. cinerea–induced WRKY33 induction was compromised (Figure 3A). However, the induction of WRKY33 was not completely inhibited but rather reduced greatly, especially at earlier time points. This delayed induction of WRKY33 (Figure 3A) was associated with the blockage of camalexin induction (Ren et al., 2008). Based on these results, we conclude that, although the MPK3/MPK6 cascade plays important roles in regulating WRKY33 expression, other signaling pathways are also involved. It is also possible that the downstream signaling process involved in the induction of WRKY33 expression can still be triggered in the absence of MPK3/MPK6. Another scenario is that, in the rescued mpk3 mpk6 double mutant, the basal level expression of MPK6 gene from the leaky DEX-inducible promoter might be able to partially compensate the mutant, although this is unlikely since we detected little/no activity from the transgenic MPK6 by the in-gel kinase activity assay in the rescued mpk3 mpk6 double mutant after B. cinerea infection (Ren et al., 2008; Han et al., 2010).
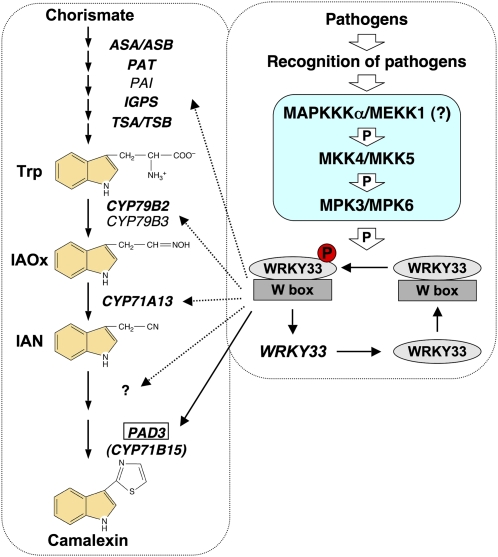
W-box, the WRKY binding site, exists at high frequencies in WRKY promoters, which led to the hypothesis that WRKY transcription factors can autoactivate their own expression (Dong et al., 2003; Pandey and Somssich, 2009; Rushton et al., 2010). The WRKY33 promoter has three W-box cis-elements, which are required for efficient pathogen- or PAMP-triggered gene activation (Lippok et al., 2007). In the same report, it was also shown that the rapid induction of WRKY33 is independent of de novo protein synthesis, suggesting the involvement of posttranslational regulation of a preexisting factor. In this report, we demonstrate that WRKY33 is a substrate of the stress/pathogen-inducible MPK3/MPK6, and phosphorylation of WRKY33 is likely to promote the transactivation activity of WRKY33. It is tempting to speculate that the phosphorylation of the basal-level WRKY33 protein might be involved in turning on WRKY33 expression, forming a positive feedback regulation loop. In support of this hypothesis, we found that WRKY33 interacts directly with the W-boxes in the promoter of WRKY33 based on ChIP-qPCR assay (Figure 9). The activation of WRKY33 at both transcriptional and posttranscriptional levels eventually drives the high-level activation of camalexin biosynthetic genes and the induction of camalexin biosynthesis, as depicted in our working model (Figure 10).
Figure 10.
A Model Depicting the Involvement of WRKY33 Downstream of MPK3/MPK6 Cascade in Regulating Camalexin Biosynthesis in Plants Challenged by Pathogens.
A simplified camalexin biosynthetic pathway and its regulatory pathway are placed in separate rectangular boxes with dashed outlines. Genes whose expressions are induced by pathogen infection and MPK3/MPK6 activation are marked by bold, italic font. Arrows with solid lines are used to connect WRKY33 and its direct target genes based on ChIP-qPCR analysis. WRKY33 binds constitutively to the W-box cis-elements. Upon phosphorylation by MPK3/MPK6, WRKY33 is able to activate the expression of its target genes, including WRKY33, forming a potential positive feedback regulatory loop downstream of MPK3/MPK6 cascade. The activation of WRKY33 at both transcriptional and posttranslational levels eventually drives the high-level activation of camalexin biosynthetic genes and the induction of camalexin biosynthesis. One arrow may represent multiple steps because of unknown components.
[See online article for color version of this figure.]
Camalexin Biosynthetic Enzymes and Key Regulators of the Biosynthetic Pathway Are Required for Plant Fungal Resistance
Camalexin induction plays important roles in Arabidopsis resistance to fungal pathogens. Mutation of key enzymes in the camalexin biosynthetic pathway compromises resistance (Glazebrook et al.,1997; Thomma et al., 1999; Zhou et al., 1999; Ferrari et al., 2003, 2007; Nafisi et al., 2007). Mutation of the Arabidopsis WRKY33 gene also causes enhanced susceptibility to the necrotrophic fungal pathogens B. cinerea and A. brassicicola. Ectopic overexpression of WRKY33, on the other hand, increases resistance to these two necrotrophic fungal pathogens (Zheng et al., 2006). Previously, we found that the MPK3/MPK6 cascade regulates camalexin biosynthesis and that mutation of MPK3 also compromises plant resistance to B. cinerea (Ren et al., 2008). In this report, we demonstrate that WRKY33 is downstream of the MPK3/MPK6 cascade in activating the expression of camalexin biosynthetic genes in response to B. cinerea infection. They function together in one regulatory pathway to control the expression of camalexin biosynthetic genes (Figure 10). It is likely that the compromised fungal resistance in the wrky33 mutant is, at least in part, due to the lack of camalexin induction.
Plant sensing of B. cinerea invasion triggers long-lasting activation of MPK3/MPK6, which regulates WRKY33 at both transcriptional and posttranslational levels by direct phosphorylation of WRKY33 at the N-terminal Ser residues. At present, the PAMPs/effectors in B. cinerea and the sensors/receptors in Arabidopsis that trigger the long-lasting activation of the MPK3/MPK6 cascade are unknown. One of the known PAMPs from B. cinerea, cell wall–derived polysaccharide elicitors, only activates MPK3/MPK6 transiently (Han et al., 2010), which is not associated with the induction of camalexin (data not shown). Prolonged activation of MPK3/MPK6 leads to the coordinated high-level induction of multiple genes in the camalexin biosynthetic pathway (Ren et al., 2008), which drives the metabolic flow from primary metabolism to the formation of camalexin, a secondary metabolite.
Is MPK4 Involved in Camalexin Induction in Arabidopsis Challenged by B. cinerea?
Our finding that WRKY33 is essential to camalexin biosynthesis is consistent with a previous report (Qiu et al., 2008b). However, more research is needed to reconcile how WRKY33 is regulated. Qiu et al. (2008b) conclude that MPK4 regulates WRKY33 by sequestering it in the MPK4/MKS1 complex in the absence of pathogens. After sensing an invading pathogen, the activation of MPK4 phosphorylates MKS1 (but not WRKY33), which releases WRKY33 from the complex so it can activate gene expression (Qiu et al., 2008b). In the conditional gain-of-function DD Arabidopsis plants, no MPK4 activation was detectable (Figure 1B) (Liu and Zhang, 2004; Ren et al., 2008). However, WRKY33-dependent camalexin induction was normal, suggesting that MPK4 activation is not essential to the WRKY33-dependent activation of camalexin induction. Furthermore, in the rescued mpk3 mpk6 double mutant, high levels of MPK4 protein and activation was detected (Figure 4D) (Ren et al., 2008; Han et al., 2010), but camalexin induction by B. cinerea infection was compromised (Ren et al., 2008), suggesting that MPK4 activation is not sufficient to support the camalexin induction. Finally, we analyzed camalexin induction in the mpk4 mutant and found no difference in the camalexin induction between the mpk4 mutant and its wild-type control after B. cinerea infection (see Supplemental Figure 3 online). Based on these results, we conclude that MPK4 is not required for camalexin induction in Arabidopsis after B. cinerea infection. It is possible that MPK4 has differential roles in camalexin induction in response to different pathogens; for example, MPK4 is not required for the camalexin induction by a fungal pathogen (this study) but is involved in camalexin induction by a bacterial pathogen (Qiu et al., 2008b).
The Signaling Specificity of Multifunctional MPK3/MPK6 Is Conferred by Their Diverse Substrates
MPK3/MPK6 are involved in many different processes, including induction of ethylene biosynthesis in plants under stress (Kim et al., 2003; Liu and Zhang, 2004; Joo et al., 2008; Han et al., 2010), camalexin induction (Ren et al., 2008; this report), stomatal development (Wang et al., 2007; Lampard et al., 2008), flower petal abscission (Cho et al., 2008), and ovule development (Wang et al., 2008). It appears that their multifunctionality and signaling specificity are conferred by their ability to phosphorylate different substrates. Four MPK3/MPK6 substrates have been reported with functional data (Liu and Zhang, 2004; Lampard et al., 2008; Bethke et al., 2009; this report). A subset of ACS isoforms, the rate-limiting enzyme in the ethylene biosynthetic pathway, can be directly phosphorylated by MPK3 and MPK6, which stabilize the ACS protein and lead to ethylene induction (Liu and Zhang, 2004; Joo et al., 2008; Han et al., 2010). In the stomatal pathway, phosphorylation of SPEECHLESS, a basic helix-loop-helix transcription factor involved in stomatal initiation, negatively regulates stomatal development (Lampard et al., 2008). ERF104, a member of the ethylene response factor (ERF) transcription factor family, forms a complex with MPK6. Upon MPK6 activation by flg22 PAMP treatment, ERF104 is released from the complex so it can access its target genes (Bethke et al., 2009).
Phosphorylation of WRKY33 by MPK3/MPK6 enhances its activity in promoting the expression of downstream camalexin biosynthetic genes. Different from ACS2/ACS6, accumulation of WRKY33 protein in plants after B. cinerea infection or in gain-of-function DD plants after DEX treatment is a result of transcriptional activation (Figures 1F and 2F) but not of protein stabilization. WRKY33 expressed under the 35S promoter showed no change in protein levels after B. cinerea infection or in DD background after DEX treatment (Figures 5B and 6B). As a result, MPK3/MPK6 are capable of regulating their substrates at different levels, including transcriptional activity, protein stability, and protein complex formation. In addition, the expression pattern of MPK3/MPK6 substrates can also affect signaling specificity. ACS2/ACS6, ERF104, and WRKY33 are expressed in most tissues, which is consistent with the general stress/defense responses. By contrast, SPEECHLESS is expressed only in cells about to enter the stomatal lineage, which confers the specific role of MPK3/MPK6 in plant stomatal development (Wang et al., 2007; Lampard et al., 2008). Research aimed at identifying additional MPK3/MPK6 substrates will reveal the molecular mechanisms underlying the complex roles of MPK3/MPK6 in plant growth, development, and response to environment and/or pathogens.
METHODS
Plant Growth, Treatments, Camalexin Measurement, and Statistical Analysis
Arabidopsis thaliana plants were grown under a 14-h light cycle (100 μE m−2 s−1) at 22°C. Seedlings were grown in 20-mL gas chromatography vials with 6 mL of half-strength Murashige and Skoog liquid medium in a growth chamber under continuous light as described before (Ren et al., 2008). Two-week-old seedlings were used for experiments. Seedlings were collected at various time points after the addition of DEX or inoculation of Botrytis cinerea spores (4.0 × 105 spores per vial). Procedures for B. cinerea maintenance and spore preparation were as previously described (Ren et al., 2008; Han et al., 2010).
Camalexin production by Arabidopsis seedlings was determined using a previously described method (Tsuji et al., 1992; Glazebrook and Ausubel, 1994) with slight modification (Ren et al., 2008). Briefly, camalexin accumulation in the culture medium, which reflects its production in the seedlings, was quantified by fluorospectrometry with a standard curve established using known concentrations of camalexin.
At least two independent repetitions were performed for experiments with multiple time points. For single time point experiments, at least three independent repetitions were done. Results from one of the independent repeats that gave similar results were shown. n = 3 indicates independent biological samples from one of the repeats. Student’s t test was used to determine whether the difference between two groups of data at a specific time point is statistically significant (P < 0.05). Statistically different data groups are indicated using different number of asterisks (0 to 3) vertically placed above the columns in the graphs.
Mutant Lines and Generation of Transgenic Plants
Mutant alleles of mpk3-1 (SALK_151594) and mpk6-2 (Salk_073907) were used for experiments (Liu and Zhang, 2004; Wang et al., 2007). The generation of rescued mpk3 mpk6 double mutant was detailed by Wang et al. (2007). All mutants used in this study are in the Col-0 background. Two T-DNA insertion mutant alleles, wrky33-1 (SALK_006603) and wrky33-2 (GABI_324B11), were described previously (Zheng et al., 2006). Both wrky33-1 and wrky33-2 alleles were used for experiments demonstrating the requirement of WRKY33 in B. cinerea– and DD-induced camalexin production (Figures 1A to 1D and 2A to 2D). Similar results were obtained and results using wrky33-1 were shown. Complementation experiments using WRKY33-TAP, WRKY33WT, and WRKY33SA transgenes (Figures 1E, 2E, and 5 to 7) were done in only wrky33-2 background because wrky33-1 acquired kanamycin resistance from the T-DNA insertion. For crosses, wrky33 mutations were followed by PCR genotyping, and the DD transgene was followed by hygromycin resistance. F3 double homozygous seedlings were used for experiments unless stated otherwise.
For the generation of the native promoter-driven WRKY33-TAP (PWRKY33:WRKY33-TAP) construct, an ~1.3-kb DNA fragment of the WRKY33 promoter was PCR amplified from Arabidopsis genomic DNA using primers 5′-ATCAAGCTTCCACATATCGTGCAATAAGAAACT-3′ and 5′-ATCGAGCTCACGAAAAATGGAAGTTTGTTTTATAAAAGA-3′. The promoter sequence was digested with HindIII/SacI and inserted into the same restriction sites to replace the cauliflower mosaic virus 35S promoter of plant transformation/expression vector pOCA30 (Chen and Chen, 2002). The full-length WRKY33 cDNA was PCR amplified using primers 5′-ATCGAGCTCTATATGGACAATAGCAGAACCAGACA-3′ and 5′-ATCGGATCCGGGCATAAACGAATCGAAAAATG-3′ and fused with the TAP tag as previously described (Xing and Chen, 2006). The WRKY33-TAP construct was then subcloned behind the WRKY33 promoter in pOCA30. The constructs were verified by DNA sequencing.
To generate loss-of-phosphorylation WRKY33 mutant (WRKY33SA), we PCR amplified the wild-type WRKY33 cDNA and cloned it into pBluescript II KS vector. Mutations were introduced by QuickChange site-directed mutagenesis (Stratagene) and confirmed by sequencing. Primers used for mutagenesis were as follows: 12AAF1, 5′-ctccttcttcaatctctatcGctccttctcttgtcGctccttccacttgtttc-3′; 34AAF1, 5′-ctccttccacttgtttcGCtccctctctttttctcgatGcccctgcttttgtctcc-3′; 5AF1, 5′-ctctgctaacgttctagctGctccaaccacaggagc-3′, and their complementary primers. Mutated nucleotides are marked with uppercase letters. WRKY33SA with all five Ser residues mutated to Ala residues was generated by three successive mutagenesis steps.
To generate the 35S:4myc-WRKY33WT and 35S:4myc-WRKY33SA constructs, we amplified the wild-type and WRKY33SA cDNA fragments using primers 5′-GGAATTCCATATGGCTGCTTCTTTTCTTACAATGGACAATAGCAGAACCAGACA-3′ and 5′-GACTAGTTCAGGGCATAAACGAATCG-3′ and cloned the PCR fragment into a modified pBlueScript II KS vector with a 4myc epitope tag coding sequence at the 5′-end. The WRKY33 cDNA with a 4myc epitope tag coding sequence was then moved into the SpeI/XhoI sites of a modified pBI121 vector.
All generated binary vectors were transformed into Agrobacterium trumefaciens strain GV3101. Arabidopsis transformation was performed by the floral dip procedure (Clough and Bent, 1998), and transformants were identified by screening for kanamycin or hygromycin resistance. Independent lines with expression of tagged WRKY33 were identified based on RNA gel blots and/or immunoblotting analyses. From these transformants, those with a single copy of T-DNA insertion (based on the 3:1 segregation of antibiotic/herbicide resistance in T2 progeny) were isolated, and homozygote transgenic plants were further identified in the progeny based on segregation of antibiotic resistance.
Preparation of Recombinant WRKY33 Proteins and in Vitro Phosphorylation Assay
WRKY33WT and WRKY33SA cDNA was PCR amplified using primers (5′-GGAATTCATGGCTGCTTCTTTTCTTACAATG-3′ and 5′-CCGCTCGAGTCAGGGCATAAACGAATCG-3′) and ligated into pET32a (+) vector in frame. The constructs were transformed into Escherichia coli strain BL21 (DE3). Recombinant protein expression was induced with 0.25 mM isopropylthio-β-galactoside for 3 h at 28°C. His-tagged proteins were purified using nickel columns and dialysis to 20 mM Tris-HCl, pH 7.5, overnight at 4°C.
The in vitro phosphorylation assay was performed as previously described (Liu and Zhang, 2004; Han et al., 2010). In brief, recombinant WRKY33 proteins were mixed with activated MPK3, MPK6, MPK4, and MPK10 (20:1 substrate enzyme ratio) in the kinase reaction buffer (20 mM HEPES, pH 7.5, 10 mM MgCl2, and 1 mM DTT) with 25 μM ATP and [γ-32P]-ATP (1 μCi per reaction). The reactions were stopped by the addition of SDS sample buffer after 30 min. Phosphorylated WRKY33 was visualized by autoradiography after being resolved in a 10% SDS-PAGE gel.
DNA-Protein EMSA
Synthetic DNA oligonucleotide (5′-CGTTGACCGTTGACCGAGTTGACTTTTTA-3′) with three W-boxes (underlined) was used as probe. DNA probe labeling and gel mobility shift assays were performed as previously described (Kim and Zhang, 2004). Briefly, two complementary strands of the oligonucleotides were annealed and then labeled at the 5′-end using T4 polynucleotide kinase. The 32P-labeled DNA probe was purified using Bio-Spin column (Bio-Rad). Freshly prepared recombinant WRKY33WT or WRKY33SA protein (1 μg) was incubated with 20,000 to 50,000 cpm of DNA probe (2 pmole) for 30 min at room temperature in binding buffer (20 mM HEPES, pH 7.9, 0.1 μg/μL herring sperm DNA, 0.5 mM DTT, 0.1 mM EDTA, and 50 mM KCl) in the presence or absence of unlabeled competitor DNA. The resulting protein-DNA complexes were resolved in 5% nondenaturing polyacrylamide gel in half-strength TBE buffer. Following electrophoresis, the gel was dried onto 3MM paper and exposed to x-ray film. For testing the effect of MAPK phosphorylation on the DNA binding activity of WRKY33, recombinant WRKY33WT was first incubated with the activated MPK3 and MPK6 (equal mix) in the kinase reaction buffer with 50 μM ATP for 60 min at room temperature before performing the EMSA assay.
Protein Extraction, Immunoblot Analysis, and in-Gel Kinase Assay
Proteins for in-gel kinase assay and immunoblot detection of Flag-tagged DD were extracted as previously described (Zhang and Klessig, 1997). For detection of tagged WRKY33 proteins, total proteins were extracted using 3 volumes (v/w) of SDS-loading buffer without bromophenol blue dye (Joo et al., 2008). The concentration of protein was determined using the Bio-Rad protein assay kit with BSA as the standard. Immunoblot detection of tagged transgene products was performed as previously described (Liu and Zhang, 2004). Antibody against the myc-epitope tag was purchased from Millipore, and the anti-IgG-horseradish peroxidase (HRP) conjugate used to detect the TAP-tagged WRKY33 was purchased from Sigma-Aldrich. MBP, WRKY33, or WRKY33SA recombinant protein was used as the substrate for the in gel-kinase assay (Zhang and Klessig, 1997; Liu and Zhang, 2004).
Mobility Shift Assay to Detect in Vivo Phosphorylated Proteins
Phos-tag reagent (NARD Institute) was used for the phospho-protein mobility shift assay to detect in vivo phosphorylated WRKY33 protein. Proteins (10 μg) were separated in a 10% SDS-PAGE gel containing 100 μmol/L Phos-tag and 200 μM MnCl2. After proteins were transferred to a nitrocellulose membrane, 4myc-tagged WRKY33 was detected using the anti-myc antibody (Millipore).
qPCR Analysis
Total RNA was extracted using TRIzol reagent (Invitrogen). After DNase treatment, 1 μg of total RNA was used for reverse transcription. qPCR analysis was performed using an Optican 2 real-time PCR machine (MJ Research) as previously described (Ren et al., 2008). After normalization to an EF-1α control, the relative levels of gene expression were calculated. The primer pairs (forward and backward) used for qRT-PCR were EF1α (At5g60390, 5′-TGAGCACGCTCTTCTTGCTTTCA-3′ and 5′-GGTGGTGGCATCCATCTTGTTACA-3′), CYP71A13 (At2g30770, 5′-GGGTAGAGGCTGGACCAAAT-3′ and 5′-ACAACCGAAGATGGAAATGC-3′), CYP71B15 (PAD3, At3g26830, 5′-GGTACGGGATAAATCTCTATGA-3′ and 5′-AGATACAGTCGATGAACCTAC-3′), WRKY33 (At 2g38470, 5′-GTGATATTGACATTCTTGACGA-3′ and 5′-GATGGTTGTGCACTTGTAGTA-3′), and WRKY33-TAP transgene (5′-AACAACGAAACGCCTTCATC-3′ and 5′-CGGAATTCGCGTCTACTTTC-3′).
WRKY33 transgene expression in the wrky33 mutant background was examined by RT-PCR using a primer pair (5′-TTCAGTCCCTCTCTTTTTCTCGAT-3′ and 5′-GGTCTCCTCGTTTGGTTCTTC-3′) that spans the whole open reading frame because nonfunctional transcripts are still produced from the mutated wkry33 gene (Zheng et al., 2006). Equal cDNA input was confirmed by PCR using EF1α control (5′-GATGGTCAGACCCGTGAGCACG-3′ and 5′-CAGTCTCAACACGTCCCACTGGC-3′). Twenty-five cycles of PCR were performed.
ChIP-qPCR Analysis
F1 plants generated from the cross of wrky33/4myc-WRKY33 and DD lines were used for ChIP assay. Two-week-old seedlings treated with 1 μM DEX for 12 h were processed as described (Kaufmann et al., 2010). Chromatin was isolated from 0.8 g of frozen tissue and sonicated with a Bioruptor sonicator (15 s on and 15 s off cycles, medium-energy settings) for 6 min. Immunoprecipitation was performed by incubating chromatin with 2 μg of anti-myc antibody (Millipore) or mouse IgG (negative control) for 1 h at 4°C. The protein-chromatin immunocomplexes were captured using Protein G-Dynal magnetic beads (Invitrogen). After Proteinase K digestion, the immunoprecipitated DNA was purified using ChIP DNA Clean and Concentrator kit (Zymo Research). Immunoprecipitated DNA and input DNA were analyzed by qPCR using primers specific for the promoter regions of PAD3 and WRKY33. The primer pairs (forward and backward) used for ChIP-qPCR were PAD3 (5′-TGTTCATGCACTTCGTCTCG-3′ and 5′-CTTCACTGACCGAGCTAACAAA-3′) and WRKY33 (5′-TTTTTGAGCAAGAGCCAAGAAT-3′ and 5′-GGCTCAATGCTTTCATCATCTT-3′) that flank the W-boxes in the promoters. The ChIP results are presented as percentage of input DNA.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: MPK3 (At3g45640), MPK6 (At2g43790), MKK4 (At1g51660), MKK5 (At3g21220), EF1α (At5g60390), CYP71A13 (At2g30770), CYP71B15 (PAD3, At3g26830), and WRKY33 (At 2g38470).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Induction of WRKY-TAP Expression in WRKY33-TAP/wrky33 Plants Infected by B. cinerea.
Supplemental Figure 2. MPK3/MPK6 Phosphorylation Sites in WRKY33 Are Required for Full Complementation of wrky33 Mutation.
Supplemental Figure 3. B. cinerea–Induced Camalexin Biosynthesis Is Not Affected by MPK4 Mutation.
Acknowledgments
We thank Melody Kroll for proofreading the manuscript. This work was supported by National Science Foundation Grants MCB-0543109 and IOS-0743957 to S.Z. and IOS-0958066 to Z.C.
References
- Asai T., Tena G., Plotnikova J., Willmann M.R., Chiu W.-L., Gomez-Gomez L., Boller T., Ausubel F.M., Sheen J. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983 [DOI] [PubMed] [Google Scholar]
- Ausubel F.M. (2005). Are innate immune signaling pathways in plants and animals conserved? Nat. Immunol. 6: 973–979 [DOI] [PubMed] [Google Scholar]
- Beckers G.J.M., Jaskiewicz M., Liu Y., Underwood W.R., He S.Y., Zhang S., Conrath U. (2009). Mitogen-activated protein kinases 3 and 6 are required for full priming of stress responses in Arabidopsis thaliana. Plant Cell 21: 944–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethke G., Unthan T., Uhrig J.F., Pöschl Y., Gust A.A., Scheel D., Lee J. (2009). Flg22 regulates the release of an ethylene response factor substrate from MAP kinase 6 in Arabidopsis thaliana via ethylene signaling. Proc. Natl. Acad. Sci. USA 106: 8067–8072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T. (2005). Peptide signalling in plant development and self/non-self perception. Curr. Opin. Cell Biol. 17: 116–122 [DOI] [PubMed] [Google Scholar]
- Böttcher C., Westphal L., Schmotz C., Prade E., Scheel D., Glawischnig E. (2009). The multifunctional enzyme CYP71B15 (PHYTOALEXIN DEFICIENT3) converts cysteine-indole-3-acetonitrile to camalexin in the indole-3-acetonitrile metabolic network of Arabidopsis thaliana. Plant Cell 21: 1830–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broekaert W.F., Delauré S.L., De Bolle M.F.C., Cammue B.P.A. (2006). The role of ethylene in host-pathogen interactions. Annu. Rev. Phytopathol. 44: 393–416 [DOI] [PubMed] [Google Scholar]
- Chen C., Chen Z. (2002). Potentiation of developmentally regulated plant defense response by AtWRKY18, a pathogen-induced Arabidopsis transcription factor. Plant Physiol. 129: 706–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S.K., Larue C.T., Chevalier D., Wang H., Jinn T.-L., Zhang S., Walker J.C. (2008). Regulation of floral organ abscission in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 105: 15629–15634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cui H., Wang Y., Xue L., Chu J., Yan C., Fu J., Chen M., Innes R.W., Zhou J.-M. (2010). Pseudomonas syringae effector protein AvrB perturbs Arabidopsis hormone signaling by activating MAP kinase 4. Cell Host Microbe 7: 164–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl J.L., Jones J.D.G. (2001). Plant pathogens and integrated defence responses to infection. Nature 411: 826–833 [DOI] [PubMed] [Google Scholar]
- del Pozo O., Pedley K.F., Martin G.B. (2004). MAPKKKalpha is a positive regulator of cell death associated with both plant immunity and disease. EMBO J. 23: 3072–3082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon R.A. (2001). Natural products and plant disease resistance. Nature 411: 843–847 [DOI] [PubMed] [Google Scholar]
- Dong J., Chen C., Chen Z. (2003). Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol. Biol. 51: 21–37 [DOI] [PubMed] [Google Scholar]
- Ferrari S., Galletti R., Denoux C., De Lorenzo G., Ausubel F.M., Dewdney J. (2007). Resistance to Botrytis cinerea induced in Arabidopsis by elicitors is independent of salicylic acid, ethylene, or jasmonate signaling but requires PHYTOALEXIN DEFICIENT3. Plant Physiol. 144: 367–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari S., Plotnikova J.M., De Lorenzo G., Ausubel F.M. (2003). Arabidopsis local resistance to Botrytis cinerea involves salicylic acid and camalexin and requires EDS4 and PAD2, but not SID2, EDS5 or PAD4. Plant J. 35: 193–205 [DOI] [PubMed] [Google Scholar]
- Glazebrook J., Ausubel F.M. (1994). Isolation of phytoalexin-deficient mutants of Arabidopsis thaliana and characterization of their interactions with bacterial pathogens. Proc. Natl. Acad. Sci. USA 91: 8955–8959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J., Zook M., Mert F., Kagan I., Rogers E.E., Crute I.R., Holub E.B., Hammerschmidt R., Ausubel F.M. (1997). Phytoalexin-deficient mutants of Arabidopsis reveal that PAD4 encodes a regulatory factor and that four PAD genes contribute to downy mildew resistance. Genetics 146: 381–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerschmidt R. (1999). Phytoalexins: What have we learned after 60 years? Annu. Rev. Phytopathol. 37: 285–306 [DOI] [PubMed] [Google Scholar]
- Han L., Li G.J., Yang K.Y., Mao G., Wang R., Liu Y., Zhang S. (2010). Mitogen-activated protein kinase 3 and 6 regulate Botrytis cinerea-induced ethylene production in Arabidopsis. Plant J. 64: 114–127 [DOI] [PubMed] [Google Scholar]
- Ichimura K., et al. ; MAPK Group (2002). Mitogen-activated protein kinase cascades in plants: A new nomenclature. Trends Plant Sci. 7: 301–308 [DOI] [PubMed] [Google Scholar]
- Jin H., Liu Y., Yang K.-Y., Kim C.Y., Baker B., Zhang S. (2003). Function of a mitogen-activated protein kinase pathway in N gene-mediated resistance in tobacco. Plant J. 33: 719–731 [DOI] [PubMed] [Google Scholar]
- Joo S., Liu Y., Lueth A., Zhang S. (2008). MAPK phosphorylation-induced stabilization of ACS6 protein is mediated by the non-catalytic C-terminal domain, which also contains the cis-determinant for rapid degradation by the 26S proteasome pathway. Plant J. 54: 129–140 [DOI] [PubMed] [Google Scholar]
- Kaufmann K., Muiño J.M., Østerås M., Farinelli L., Krajewski P., Angenent G.C. (2010). Chromatin immunoprecipitation (ChIP) of plant transcription factors followed by sequencing (ChIP-SEQ) or hybridization to whole genome arrays (ChIP-CHIP). Nat. Protoc. 5: 457–472 [DOI] [PubMed] [Google Scholar]
- Kim C.Y., Liu Y., Thorne E.T., Yang H., Fukushige H., Gassmann W., Hildebrand D., Sharp R.E., Zhang S. (2003). Activation of a stress-responsive mitogen-activated protein kinase cascade induces the biosynthesis of ethylene in plants. Plant Cell 15: 2707–2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C.Y., Zhang S. (2004). Activation of a mitogen-activated protein kinase cascade induces WRKY family of transcription factors and defense genes in tobacco. Plant J. 38: 142–151 [DOI] [PubMed] [Google Scholar]
- Kroj T., Rudd J.J., Nürnberger T., Gäbler Y., Lee J., Scheel D. (2003). Mitogen-activated protein kinases play an essential role in oxidative burst-independent expression of pathogenesis-related genes in parsley. J. Biol. Chem. 278: 2256–2264 [DOI] [PubMed] [Google Scholar]
- Lampard G.R., Macalister C.A., Bergmann D.C. (2008). Arabidopsis stomatal initiation is controlled by MAPK-mediated regulation of the bHLH SPEECHLESS. Science 322: 1113–1116 [DOI] [PubMed] [Google Scholar]
- Lippok B., Birkenbihl R.P., Rivory G., Brümmer J., Schmelzer E., Logemann E., Somssich I.E. (2007). Expression of AtWRKY33 encoding a pathogen- or PAMP-responsive WRKY transcription factor is regulated by a composite DNA motif containing W box elements. Mol. Plant Microbe Interact. 20: 420–429 [DOI] [PubMed] [Google Scholar]
- Liu Y., Ren D., Pike S., Pallardy S., Gassmann W., Zhang S. (2007). Chloroplast-generated reactive oxygen species are involved in hypersensitive response-like cell death mediated by a mitogen-activated protein kinase cascade. Plant J. 51: 941–954 [DOI] [PubMed] [Google Scholar]
- Liu Y., Zhang S. (2004). Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell 16: 3386–3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G.B., Bogdanove A.J., Sessa G. (2003). Understanding the functions of plant disease resistance proteins. Annu. Rev. Plant Biol. 54: 23–61 [DOI] [PubMed] [Google Scholar]
- Menke F.L.H., van Pelt J.A., Pieterse C.M.J., Klessig D.F. (2004). Silencing of the mitogen-activated protein kinase MPK6 compromises disease resistance in Arabidopsis. Plant Cell 16: 897–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey J.P., Osbourn A.E. (1999). Fungal resistance to plant antibiotics as a mechanism of pathogenesis. Microbiol. Mol. Biol. Rev. 63: 708–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nafisi M., Goregaoker S., Botanga C.J., Glawischnig E., Olsen C.E., Halkier B.A., Glazebrook J. (2007). Arabidopsis cytochrome P450 monooxygenase 71A13 catalyzes the conversion of indole-3-acetaldoxime in camalexin synthesis. Plant Cell 19: 2039–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagami H., Pitzschke A., Hirt H. (2005). Emerging MAP kinase pathways in plant stress signalling. Trends Plant Sci. 10: 339–346 [DOI] [PubMed] [Google Scholar]
- Nürnberger T., Scheel D. (2001). Signal transmission in the plant immune response. Trends Plant Sci. 6: 372–379 [DOI] [PubMed] [Google Scholar]
- Pandey S.P., Somssich I.E. (2009). The role of WRKY transcription factors in plant immunity. Plant Physiol. 150: 1648–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen M., et al. (2000). Arabidopsis map kinase 4 negatively regulates systemic acquired resistance. Cell 103: 1111–1120 [DOI] [PubMed] [Google Scholar]
- Qiu J.-L., et al. (2008b). Arabidopsis MAP kinase 4 regulates gene expression through transcription factor release in the nucleus. EMBO J. 27: 2214–2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J.-L., Zhou L., Yun B.-W., Nielsen H.B., Fiil B.K., Petersen K., Mackinlay J., Loake G.J., Mundy J., Morris P.C. (2008a). Arabidopsis mitogen-activated protein kinase kinases MKK1 and MKK2 have overlapping functions in defense signaling mediated by MEKK1, MPK4, and MKS1. Plant Physiol. 148: 212–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D., Liu Y., Yang K.-Y., Han L., Mao G., Glazebrook J., Zhang S. (2008). A fungal-responsive MAPK cascade regulates phytoalexin biosynthesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 105: 5638–5643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D., Yang H., Zhang S. (2002). Cell death mediated by MAPK is associated with the hydrogen peroxide production in Arabidopsis. J. Biol. Chem. 277: 559–565 [DOI] [PubMed] [Google Scholar]
- Rushton P.J., Somssich I.E., Ringler P., Shen Q.J. (2010). WRKY transcription factors. Trends Plant Sci. 15: 247–258 [DOI] [PubMed] [Google Scholar]
- Schuhegger R., Nafisi M., Mansourova M., Petersen B.L., Olsen C.E., Svatos A., Halkier B.A., Glawischnig E. (2006). CYP71B15 (PAD3) catalyzes the final step in camalexin biosynthesis. Plant Physiol. 141: 1248–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staskawicz B.J., Ausubel F.M., Baker B.J., Ellis J.G., Jones J.D.G. (1995). Molecular genetics of plant disease resistance. Science 268: 661–667 [DOI] [PubMed] [Google Scholar]
- Suarez-Rodriguez M.C., Adams-Phillips L., Liu Y., Wang H., Su S.-H., Jester P.J., Zhang S., Bent A.F., Krysan P.J. (2007). MEKK1 is required for flg22-induced MPK4 activation in Arabidopsis plants. Plant Physiol. 143: 661–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tena G., Asai T., Chiu W.-L., Sheen J. (2001). Plant mitogen-activated protein kinase signaling cascades. Curr. Opin. Plant Biol. 4: 392–400 [DOI] [PubMed] [Google Scholar]
- Thomma B.P.H.J., Nelissen I., Eggermont K., Broekaert W.F. (1999). Deficiency in phytoalexin production causes enhanced susceptibility of Arabidopsis thaliana to the fungus Alternaria brassicicola. Plant J. 19: 163–171 [DOI] [PubMed] [Google Scholar]
- Tsuji J., Jackson E.P., Gage D.A., Hammerschmidt R., Somerville S.C. (1992). Phytoalexin accumulation in Arabidopsis thaliana during the hypersensitive reaction to Pseudomonas syringae pv syringae. Plant Physiol. 98: 1304–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanEtten H.D., Matthews D.E., Matthews P.S. (1989). Phytoalexin detoxification: Importance for pathogenicity and practical implications. Annu. Rev. Phytopathol. 27: 143–164 [DOI] [PubMed] [Google Scholar]
- van Loon L.C., Geraats B.P.J., Linthorst H.J.M. (2006). Ethylene as a modulator of disease resistance in plants. Trends Plant Sci. 11: 184–191 [DOI] [PubMed] [Google Scholar]
- Wan J., Zhang S., Stacey G. (2004). Activation of a mitogen-activated protein kinase pathway in Arabidopsis by chitin. Mol. Plant Pathol. 5: 125–135 [DOI] [PubMed] [Google Scholar]
- Wang H., Liu Y., Bruffett K., Lee J., Hause G., Walker J.C., Zhang S. (2008). Haplo-insufficiency of MPK3 in MPK6 mutant background uncovers a novel function of these two MAPKs in Arabidopsis ovule development. Plant Cell 20: 602–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Ngwenyama N., Liu Y., Walker J.C., Zhang S. (2007). Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19: 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing D., Chen Z. (2006). Effects of mutations and constitutive overexpression of EDS1 and PAD4 on plant resistance to different types of microbial pathogens. Plant Sci. 171: 251–262 [Google Scholar]
- Yang K.-Y., Liu Y., Zhang S. (2001). Activation of a mitogen-activated protein kinase pathway is involved in disease resistance in tobacco. Proc. Natl. Acad. Sci. USA 98: 741–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., et al. (2007). A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-induced immunity in plants. Cell Host Microbe 1: 175–185 [DOI] [PubMed] [Google Scholar]
- Zhang S., Klessig D.F. (1997). Salicylic acid activates a 48-kD MAP kinase in tobacco. Plant Cell 9: 809–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Klessig D.F. (2001). MAPK cascades in plant defense signaling. Trends Plant Sci. 6: 520–527 [DOI] [PubMed] [Google Scholar]
- Zheng Z., Qamar S.A., Chen Z., Mengiste T. (2006). Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 48: 592–605 [DOI] [PubMed] [Google Scholar]
- Zhou N., Tootle T.L., Glazebrook J. (1999). Arabidopsis PAD3, a gene required for camalexin biosynthesis, encodes a putative cytochrome P450 monooxygenase. Plant Cell 11: 2419–2428 [DOI] [PMC free article] [PubMed] [Google Scholar]