Abstract
Endoplasmic reticulum (ER) stress is a hallmark feature of secretory cells and many diseases including cancer, neurodegeneration, and diabetes. Adaptation to protein folding stress is mediated by the activation of an integrated signal transduction pathway known as the unfolded protein response (UPR). The UPR signals through three distinct stress sensors located at the ER membrane, IRE1α, ATF6 and PERK. Although PERK and IRE1α share functionally similar ER-luminal sensing domains and both are simultaneously activated in cellular paradigms of ER stress in vitro, they are selectively engaged in vivo by the physiological stress of unfolded proteins. The differences in terms of tissue-specific regulation of the UPR may be explained by the formation of distinct regulatory protein complexes. This concept is supported by the recent identification of adaptor and modulator proteins that directly interact with IRE1α. In this review we discuss recent evidence supporting a model where IRE1α signaling emerges as a highly regulated process, controlled by the formation of a dynamic scaffold onto which many regulatory components assemble.
Introduction
A number of conditions interfere with oxidative protein folding processes in the endoplasmic reticulum (ER) lumen (Ron and Walter, 2007), leading to accumulation of misfolded proteins, a cellular condition referred to as “ER stress”. Adaptation to ER stress is mediated by engagement of the unfolded protein response (UPR), an integrated signal transduction pathway that transmits information about protein folding status in the ER lumen to the nucleus to increase protein folding capacity. Conversely, cells undergo apoptosis if these mechanisms of adaptation and survival are insufficient to handle the unfolded protein load.
The occurrence of ER stress is observed in many physiological processes, especially in highly secretory cells such as plasma B lymphocytes, salivary glands and pancreatic beta cells. The high demand for efficient protein folding and secretion processes in these cells constitutes a constant source of stress initiated by the presence of large amounts of misfolded proteins that are normally generated during the protein maturation process. These folding sub products are eliminated through ER-associated degradation (ERAD), where misfolded proteins translocate to the cytosol and are degraded by the proteasome (reviewed in Vembar and Brodsky, 2008). ER stress is also triggered by conditions that alters proteostasis associated with perturbations in protein maturation, expression of certain mutant proteins, decreased chaperone function, abnormal ER calcium content or redox metabolism, altered trafficking and many others (Powers et al., 2009). As an initial response to ER stress, cells activate the UPR to decrease the unfolded protein load and recover homeostasis. In doing so, UPR signaling enforces global changes in expression of proteins related to nearly every aspect of the secretory pathway. For example, gene expression profiling has demonstrated that the UPR regulates genes involved in protein entry into the ER, folding, glycosylation, ERAD, protein quality control, redox metabolism, autophagy, lipid biogenesis, and vesicular trafficking (Figure 1A). Increasing attention has been given to the regulation of the UPR based on substantial evidence for the involvement of chronic ER stress in many diseases, including neurodegenerative conditions (Matus et al., 2008), cancer (Moenner et al., 2007), diabetes (Lipson et al., 2006), and inflammation (Todd et al., 2008), hence offering new therapeutic targets to treat these diseases.
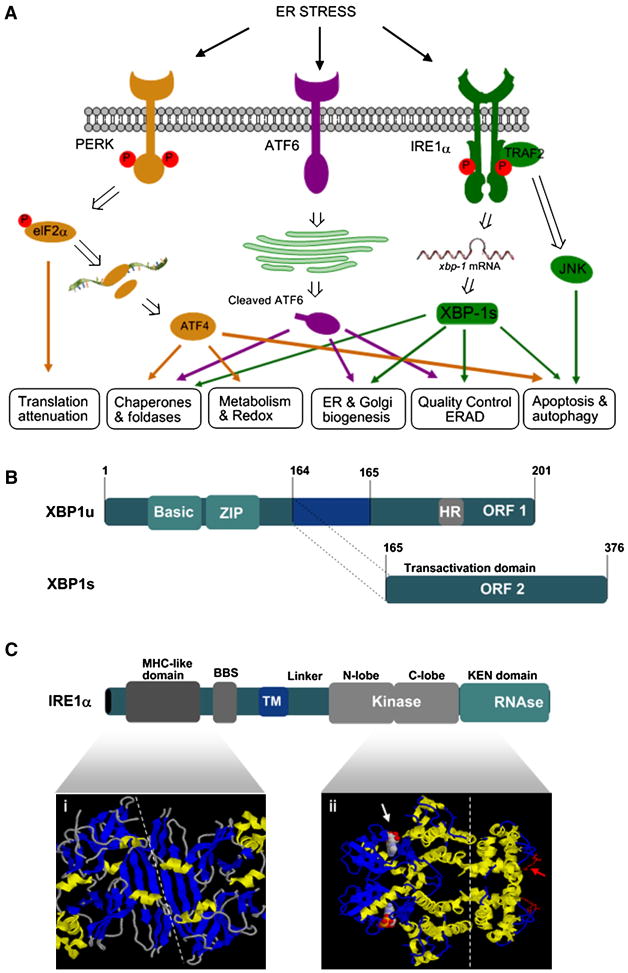
Figure 1. Essential components of the Unfolded Protein Response.
(A) UPR signaling. Accumulation of misfolded protein inside the endoplasmic reticulum (ER) lumen triggers a stress response known as UPR. There are at least three main stress sensors at the ER membrane, IRE1α, PERK, and ATF6. In cells undergoing ER stress, IRE1α auto-phosphorylates, leading to the activation of its endoribonuclease domain. This activity mediates the processing of the mRNA encoding XBP1, which is a transcriptional factor that upregulates many essential UPR genes involved in folding and protein quality control and regulates ER/Golgi biogenesis. Active IRE1α binds the adaptor protein TRAF2, triggering JNK activation, which may participate in the regulation of autophagy and apoptosis. Alternatively, activated PERK phosphorylates and inhibits translation initiator factor eIF2α, decreasing the synthesis of proteins and the overload of misfolded proteins at the ER. In addition, this event leads to the specific translation of ATF4, a transcription factor that induces the expression of genes that function in amino acid metabolism, the antioxidant response and apoptosis regulators including CHOP. A third UPR pathway is initiated by ATF6, a type II ER transmembrane protein encoding a bZIP transcriptional factor in its cytosolic domain and localized in the ER in unstressed cells. Upon ER stress induction, ATF6 is processed, increasing the expression of some ER chaperones, and ERAD-related genes. At the bottom, the cellular functions affected by each UPR-signaling branch are indicated.
(B) XBP1 splicing. Schematic representation of the unspliced and spliced forms of XBP1 (XBP1u and XBP1s, respectively). Numbers indicate amino acid positions with the initiation methionine set at 1. ORF1 and ORF2 for the C-terminal domain as well as the basic and leucine zipper (ZIP) domains are indicated. Putative hydrophobic region of XBP-1u related to targeting is also indicated.
(C) IRE1 structure. A schematic representation of the primary structure of IRE1p is presented indicating the kinase and RNAse domains. BiP-binding domain (BBD), the MHC-like domain, linker region, tramsmembrane region ™ and kinase and RNAse domains are indicated. In the bottom panel, the crystal structure of the ER luminal domain groove (MHC-I like structure) of yeast IRE1p is shown (Credle et al., 2005). The dimer interface is indicated with a white line. This structural domain of IRE1p is proposed to bind misfolded proteins to stabilize the oligomeric conformation. In addition, the three dimensional structure of the cytosolic domain of IRE1p is presented highlighting the two lobes of the kinase domain (Lee et al., 2008b). The ADP and the kinase domain are indicated with a white arrow. The KEN domain containing the RNAse activity is also shown where a red arrow indicates the putative RNAse active site (Lee et al., 2008b).
Distinct UPR signaling branches
The UPR was first characterized in yeast where a single signaling pathway governs the response to ER stress mediated by a type I transmembrane ER protein known as IRE1p (inositol-requiring transmembrane kinase/endonuclease) (Ron and Walter, 2007). In higher eukaryotes, the UPR gained complexity as it is mediated by at least three distinct UPR signaling pathways initiated by the sensors IRE1α and IRE1β, PERK (PKR-like ER kinase), and ATF6α and ATF6β (activating transcription factor 6) (Figure 1A). Activated PERK inhibits protein translation into the ER through the inactivation of the initiation factor eIF2α, alleviating ER stress by decreasing the overload of misfolded proteins. Phosphorylation of eIF2α allows the expression of ATF4 (activating transcription factor 4), a transcription factor that upregulates UPR genes that function in amino acid and redox metabolism (Harding et al., 2003), including chop/gadd153 and gadd34 (Figure 1A). A second UPR pathway is initiated by ATF6α and ATF6β, type II ER transmembrane proteins whose cytosolic domain encodes a bZIP transcriptional factor (Ron and Walter, 2007). Upon ER stress induction, ATF6 is processed at the Golgi, releasing its cytoplasmic domain which acts as a transcriptional activator controlling many UPR genes related to ERAD and folding at the ER among others (Yamamoto et al., 2007) (Figure 1). This branch of the UPR is very complex and is formed by a series of newly identified ATF6 homologues that are modulated by ER stress in specific tissues, including OASIS, CREBH, LUMAN/CREB3, CREB4, and BBF2H7 (Ron and Walter, 2007). All of these ATF6-related bZip factors are processed at the Golgi in a similar way as ATF6, but their function in the UPR is poorly characterized. Mori's group recently generated ATF6α- and ATF6β-single knockout mice, which developed normally (Yamamoto et al., 2007). However, double knockout mice are embryonic lethal, similar to the phenotype of X-Box-binding protein 1 (XBP1)(Reimold et al., 2000) or IRE1α (Urano et al., 2000) deficient mice. In contrast, PERK, ATF4, and CHOP deficient animals are viable and have varied defects in pancreatic function, metabolism, and skeletal development (Zhang et al., 2002; Harding et al., 2001; Tanaka et al., 1998; Zinszner et al., 1998).
IRE1α is the most evolutionarily conserved branch of the UPR. Nevertheless, little is known about the regulation of IRE1α activity. IRE1α is a Ser/Thr protein kinase and endoribonuclease that, upon activation, initiates the unconventional splicing of the mRNA encoding the transcriptional factor XBP1 (Ron and Walter, 2007) (Figure 1B and C). In mammalian cells, a 26 nucleotide intron of xbp1 mRNA is spliced out by activated IRE1α, leading to a shift in the codon reading frame. Translation of the new reading frame results in the conversion of XBP1 from an unspliced form of 267 amino acids to a spliced form of 371 amino acids that comprises the original N-terminal DNA binding domain plus an additional, potent transactivation domain in the C terminus (Figure 1B).
Spliced XBP1 (XBP1s) controls the upregulation of a broad spectrum of UPR-related genes involved in protein folding, protein entry to the ER, redox metabolism, ERAD and protein quality control (Lee et al., 2003b; Shaffer et al., 2004). A regulatory circuitry governed by XBP1 was interrogated by a genome-wide profiling approach. In addition to classical UPR-related genes, unexpected cell-type specific targets were identified that are linked to cell differentiation, signaling, and DNA damage pathways (Acosta-Alvear et al., 2007). A recent genetic screen systematically characterized the functional interdependencies between UPR-target genes in yeast. These factors included chaperones, glycosylation enzymes, and ERAD components as well as trafficking pathways, transcriptional regulatory networks, modulators of lipid and ion composition, and vacuolar function (Jonikas et al., 2009). The complexity of activities/processes described in this work support the concept that proteostasis emerges from the dynamic interplay between synthesis/folding, degradation and export processes. In addition, XBP1s regulates the expansion of the secretory pathway by controlling phospholipid biosynthesis and ER/Golgi biogenesis (Shaffer et al., 2004; Sriburi et al., 2004). Interestingly, XBP1 heterodimerizes with ATF6α for the induction of ER-associated degradation components (Yamamoto et al., 2007; Wu et al., 2007) and ATF6α may also modulate lipid biosynthesis and ER expansion under stress conditions (Bommiasamy et al., 2009).
Mechanism of ER stress sensing by IRE1
Role of the IRE1 ER luminal domain in its activation
It was originally proposed that, under normal conditions, the ER chaperone BiP/Grp78 binds to IRE1α or the yeast homolog IRE1p maintaining the protein in an inactive monomeric state (Bertolotti et al., 2000; Kimata et al., 2003). In ER stressed cells, BiP is released allowing IRE1α to multimerize and autophosphorylate its cytosolic domain. This phosphorylation event triggers the activation of the RNase activity, initiating XBP1 mRNA splicing and UPR responses. The functional impact of BiP association to IRE1p was addressed by mutagenesis analysis, observing that disruption of this interaction does not drastically alter the ability of IRE1p to detect protein misfolding (Kimata et al., 2004). Recently, new insights into the mechanism of IRE1α/IRE1p activation have emerged from two groups who independently solved the structure of the ER luminal domain of yeast and human IRE1 protein. Peter Walter's group speculated that misfolded proteins may directly bind to the N-terminal region of IRE1p, facilitating its oligomerization through a binding motif similar to an MHC-like groove (Figure 1C), and mutations in amino acids present in the groove or in the dimerization interface abrogated the ability of IRE1p to engage the UPR (see comparison with MHCI structure in Credle et al., 2005). Thus, misfolded proteins may be directly recognized by yeast IRE1p. The general structure of the ER stress sensing domain of IRE1p is conserved in mammals (Zhou et al., 2006). Recent in vitro studies consolidated both models for yeast IRE1p activation, suggesting that BiP first dissociates from IRE1p leading to its dimerization and cluster formation (Figure 2A, see below). In the second step, direct interaction of unfolded proteins with the stress sensing domain may orient the protein into an active IRE1p signaling cluster with full ribonuclease activity (Kimata et al., 2007).
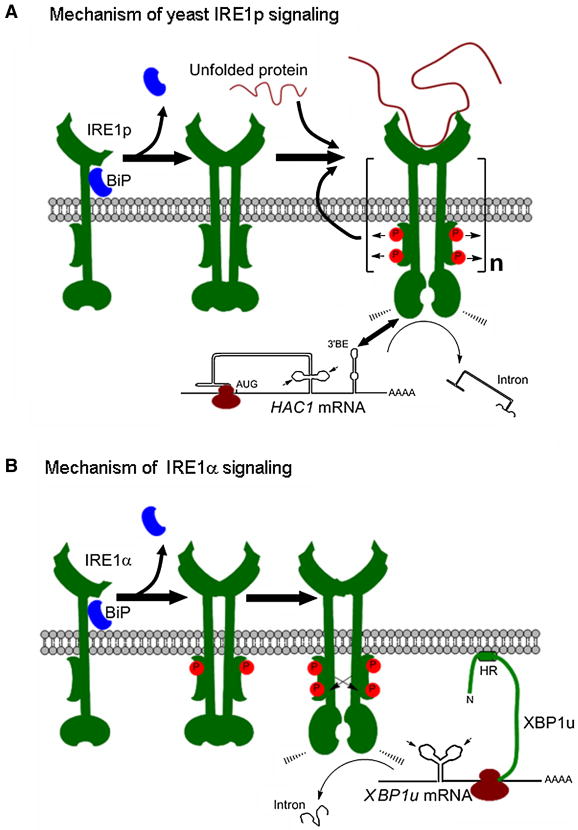
Figure 2. Mechanism of IRE1 activation in yeast and mammals.
(A) A direct recognition model proposes that unfolded proteins bind directly to the luminal domains of IRE1p, facilitating the assembly of highly ordered IRE1p clusters exemplified by the parenthesis and “n” IREp units). This may orient the cytosolic region of the dimer to form the ribonuclease active site and generation of an mRNA docking region. BiP dissociation from IRE1p may play an indirect role in unfolded-peptide loading. Oligomerization of IRE1p is essential for its auto-transphosphorylation between dimers (as indicated with arrows). IRE1p clusters recruit untranslated HAC1 mRNA contained in ribosomes (inhibited by the secondary structure of the HAC1 intron), an association which depends on structural motifs in IRE1p and the HAC1 mRNA including a bipartite element at the 3′ end (3′BE).
(B) In mammalian cells, IRE1α is maintained in a repressed state through an association with BiP. Upon ER stress BiP dissociates, leading to partial IRE1 phosphorylation and IRE1 dimerization mediated by the N-terminal ER luminal region. Dimerization triggers further phosphorylation events (auto-transphosphorylation, indicated with arrows) and activation of the RNAse domain of IRE1α. The unspliced XBP1 mRNA is translated in mammals and a hydrophobic region (HR) on the nascent peptide targets the translated XBP-1 mRNA to the ER membrane, enhancing its processing by active IRE1α. XBP-1 mRNA targeting to the ER membrane does not depend on the expression of IRE1α. In (i) and (ii) splicing sites on the XBP1 and HAC1 mRNA are indicated with an arrowhead.
Interestingly, a recent study described the generation of luminal-domain mutants of mammalian IRE1α that have low affinity for BiP, that retain significant activation even under unstressed conditions (Oikawa et al., 2009). Moreover, the luminal fragments of mammalian IRE1α did not interact with unfolded proteins in an in vitro assay (Oikawa et al., 2009) as was described previously for yeast IRE1p (Kimata et al., 2007). These data suggested that, in contrast to yeast IRE1p, the regulation of mammalian IRE1α may actually depend on the dissociation of BiP and may be independent of misfolded protein binding (Figure 2B) (Oikawa et al., 2009). This idea correlates well with the prediction that the MHC-like groove observed in the human IRE1α ER luminal domain may not be able to accommodate an unfolded protein peptide as indicated in the crystal structure (Zhou et al., 2006). In this study it was proposed that IRE1α undergoes different stages of phosphorylation, where dimerization of the ER luminal region is essential to get fully phosphorylated IRE1α and subsequent RNAse activation (Zhou et al., 2006). A similar dimer interface for the PERK-luminal region was also predicted.
The mechanisms involved in the activation of PERK and ATF6 and how these receptors sense the unfolded protein load, have not been directly investigated. Initial studies indicated that the luminal domains of both sensors bind BiP under resting conditions, and this association is lost under ER stress (Bertolotti et al., 2000; Shen et al., 2002; Chen et al., 2002). In addition, the primary sequence of the sensing domains of IRE1α and PERK are similar, and a MHC-I-like groove is also predicted to be present in PERK (see comparison of primary sequences between PERK and IRE1 in Liu et al., 2000 and Credle et al., 2005). In addition, the ER luminal domains of PERK and IRE1α are interchangeable, without affecting the rate of activation of the proteins under ER stress conditions (Liu et al., 2000). More studies are required to define the mechanisms underlying PERK and ATF6 activation.
Cluster formation by IRE1p
The crystal structure of the cytosolic domain of IRE1p was recently solved by two independent groups (Korennykh et al., 2009; Lee et al., 2008b) (Figure 1C). Korennykh and co-workers were able to visualize the architecture of IRE1p oligomers and depicted a high order rod-shaped assembly of the cytosolic domain. This polymer-like organization was critical for IRE1p signaling as demonstrated through targeted mutagenesis of the interaction interfaces between dimers (Korennykh et al., 2009). The tridimensional structure revealed a possible mechanism where oligomerization of IRE1p dimers positions the kinase domain for trans-autophosphorylation, generates the RNase active site, and creates an additional interaction surface for binding of the mRNA substrate (Korennykh et al., 2009). Oligomerization of the unphosphorylated IRE1p opens the kinase domain and positions it for trans-autophosphorylation as a second step, leading then to activation of the RNAse domain (Figure 2A). The authors also speculated that the association between multiple IRE1p polymers may underlie visible foci formation in yeast cells undergoing ER stress.
Cluster formation of IRE1α has not yet been described and early studies from David Ron's laboratory suggested that IRE1α forms mostly dimers upon activation, in contrast to PERK that multimerizes in high-molecular weight complexes upon activation (Bertolotti et al., 2000). Further studies are needed to resolve these issues regarding the similarities and differences in the mechanism of sensing ER stress in yeast and mammals.
mRNA targeting to IRE1
Recent work has provided novel insights into how the HAC1/XBP1 mRNA is recognized by IRE1p/IRE1α. Intriguing differences in the way that the XBP1 and HAC1 mRNAs are targeted to IRE1α and IRE1p, respectively were described. In unstressed yeast, most of the unspliced HAC1 mRNA is cytoplasmic and remains attached to the ribosomes in an untranslated form due to intrinsic properties of its secondary mRNA structure (Figure 2A). ER stress leads to co-localization of the HAC1 mRNA to the IRE1p clusters and this process was shown to be IRE1p-dependent (Aragon et al., 2009). HAC1 mRNA targeting to IRE1p also requires a bipartite stem loop structure in the non-translated region of the mRNA in addition to translational repression through the intron sequence to be excised. In contrast, in mammals unspliced XBP1 (XBP1u) is normally translated, and a recent report indicated that the targeting of the XBP1 mRNA to the ER membrane is dependent on the expression of XBP1u (Figure 2B). Upon translation, it was shown that XBP1u associates with membranes and recruits the XBP1 mRNA to the ER membrane through a well conserved hydrophobic region at its C-terminus (Figure 2B) that is predicted to form an α-helix. This mechanism may provide close proximity of the mRNA substrate, facilitating IRE1α-mediated splicing (Yanagitani et al., 2009). IRE1α was dispensable for ER-association of XBP-1 mRNA. In summary, it would appear that clear differences exist between IRE1α and IRE1p signaling mechanisms.
Diversity of IRE1α proximal signaling
Control of alarm signaling pathways
In mammals, in addition to catalyzing XBP1 mRNA processing, IRE1α has additional functions in cell signaling (Figure 3). The cytosolic domain of activated IRE1α binds to the adaptor protein TNFR-associated factor 2 (TRAF2), triggering the activation of the Apoptosis Signal-regulating Kinase 1 (ASK1) and cJun-N terminal kinase (JNK) pathway (Urano et al., 2000; Nishitoh et al., 2002). IRE1α also modulates other “alarm genes” such as the activation of the p38, ERK (Nguyen et al., 2004) and NF-κB pathways (Hu et al., 2006) possibly by the binding of the SH2/SH3 containing adaptor proteins Nck and a protein complex between inhibitor κB kinase (IKK)/TRAF2, respectively. However, the function of these UPR signaling branches in the context of protein misfolding is still not well understood. Activation of ASK1/JNK through the IRE1α/TRAF2 complex has been proposed to mediate at least in part apoptosis under irreversible ER stress in an analogous fashion to TNF receptor signaling (Kanda and Miura, 2004; Mauro et al., 2006). In agreement with this idea, a recent high-throughput chemical screen for inhibitors of ER stress-induced cell death revealed a crucial role of ASK1 in the process (Kim et al., 2009). These data suggest that IRE1α has a dual function in ER stress responses, (i) regulating adaptation to stress and cell survival through the control of XBP-1s expression and (ii) activation of apoptosis in cells irreversibly damaged by the activation of the JNK/ASK1 pathway.
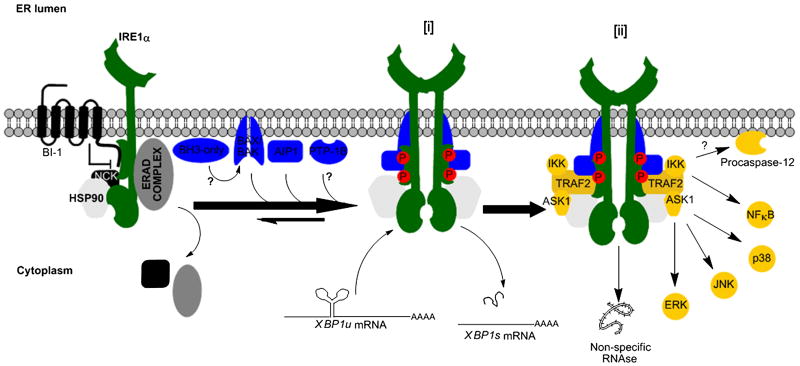
Figure 3. The IRE1α interactome.
Mammalian IRE1α signaling is initiated by the formation of a complex protein platform at the ER membrane termed the UPRosome where multiple factors assemble and modulate its activity. For example, activation of IRE1α requires the binding of accessory proteins, such as BAX, BAK, AIP1, and possibly BH3-only proteins such as PUMA and BIM (upstream of BAX/BAK), in addition to the activity of the ER-located phosphatase PTP-1B. Under chronic or prolonged ER stress, IRE1α signaling is down regulated and the ER located protein BI-1 is involved in the inactivation of IRE1α, whereas HSP90 binding decreases its turnover. In addition, active IRE1α initiates a variety of signaling responses through the binding of TRAF2 and possibly other adaptor proteins. These events trigger the activation of ASK1/JNK, ERK and p38 kinases which may regulate apoptosis and autophagy. Sequestration of IKK by IRE1α induces NF-κB signaling. In addition, a non specific RNAse activity has been described for IRE1α in flies to degrade the mRNA of proteins that have a high tendency to misfold under ER stress conditions. Inactive IRE1α interacts with components of the ERAD machinery and may regulate this process and modulators of ERK such as NCK. For simplicity, the figure separates the components that control IRE1α activation/inactivation in relation to XBP1 mRNA splicing activity [i], and the components related to the regulation of other signaling branches [ii], and this graphical separation of the complexes does not reflect a temporal dissociation between i and ii. Proteins that bids to inactive IRE1α (resting condition) are shown in gray scale colors, that regulates its activation and XBP- mRNA splicing in blue and that controls other signaling pathways in brown.
Regulation of autophagy
A new function for IRE1α was recently proposed through activation of JNK whereby it controls levels of autophagy under ER stress (Ogata et al., 2006). Autophagy is a survival pathway classically linked to adaptation to nutrient starvation. Conversely, in cells undergoing ER stress, autophagy may serve as a mechanism to eliminate damaged organelles and aggregated proteins (Levine and Kroemer, 2008). Remarkably, an initial study showed that the upregulation of autophagy under ER stress conditions is initiated by the kinase domain of IRE1α, independently of the RNAse/XBP1 signaling branch (Ogata et al., 2006). How JNK regulates autophagy is not known, but a recent report suggested that phosphorylation of the anti-apoptotic protein BCL-2 at the ER membrane by this kinase may directly affect the initiation of autophagy by modulating the activity of Beclin-1 (Pattingre et al., 2009), an essential component of the autophagy machinery (Levine and Kroemer, 2008). A genomic screen in fly cells demonstrated that knocking down UPR components, including XBP-1, increases basal autophagy levels (Arsham and Neufeld, 2009). Besides, autophagy defective cells show upregulation of essential ER chaperones (Mathew et al., 2009), suggesting a close homeostatic balance between the autophagy and UPR pathways.
Interaction with the ER-Associated degradation machinery
A well defined subset of XBP1s target genes are related to ERAD and the ER translocon, including EDEM, HERP and Sec61. Using a two-hybrid screen a recent study identified the physical interaction between the ubiquitin specific protease (USP) 14 and IRE1α. USP14 interacted with the cytoplasmic region of inactive IRE1α, and their association was inhibited by ER stress and IRE1α activation. Interestingly, a function for USP14 in ERAD was proposed, where inhibition occurs in an IRE1α-dependent manner (Nagai et al., 2009). In addition, the authors reported the association of IRE1α with essential ERAD components such as DERLIN-1, DERLIN-3, SEL1, and HRD1, suggesting that inactive IRE1α may form a macromolecular platform with the ERAD machinery (Nagai et al., 2009). The possible physiological relevance of these findings remains to be established.
Protein expression control by direct regulation of mRNA decay
A genome-wide search for substrates of the mRNA splicing activity of IRE1p revealed only HAC1 mRNA as a hit within the limits of detection, and no additional substrates were identified highlighting the specificity of the pathway (Niwa et al., 2005). Similar findings were described in a mammalian system (Nekrutenko and He, 2006). However, in insect cells, active IRE1α was proposed to control the degradation of mRNAs encoding certain ER proteins that were predicted to be difficult to fold (Hollien and Weissman, 2006). Through a complex gene profiling analysis, Weissman's group demonstrated that a subset of RNAs is selectively down-regulated during ER stress in an IRE1α-dependent and XBP1-independent manner. These effects were proposed to be a direct consequence of mRNA degradation by IRE1α ribonuclease activity (Hollien and Weissman, 2006). The authors proposed a model where selective mRNA targeting was related to misfolding propensity of certain proteins encoded by the degraded mRNA. Thus, in a dynamic way, the misfolding of a nascent protein during its translation may directly and locally activate IRE1α and its RNase domain to produce the degradation of mRNA being translated by the local ribosome. Two recent studies described the occurrence of IRE1α-dependent mRNA decay in mammalian cells (Hollien et al., 2009; Han et al., 2009). Surprisingly, although XBP1 splicing can be induced by artificial dimerization of IRE1α in the absence of ER stress, IRE1α-dependent mRNA decay was shown to require both ER stress and IRE1α activity, suggesting that these two functions of IRE1α are distinct (Hollien et al., 2009). Although the precise mechanism involved in mRNA decay is not clear, Hollien et al proposed that this novel function of IRE1α is well suited to selectively decrease the production of proteins that challenge the ER at the folding level and alleviate stress. In contrast, Han and co-workers presented evidence suggesting that endonucleolytic mediated mRNA decay of ER-localized mRNAs, including those encoding chaperones, may also culminate in apoptosis (Han et al., 2009). These complex data reinforce the concept that IRE1α controls cell fate, wherein a dual activity modulates cell survival and apoptosis in cells irreversibly damaged.
Differential regulation of UPR stress sensors: IRE1α
Recent evidence indicates that IRE1α activation is specifically regulated by a set of different proteins (co-factors and inhibitors) (Figure 3). For example, the levels of IRE1α signaling were shown to be controlled by the expression of the ER-located Protein-tyrosine phosphatase 1B (PTP-1B) (Gu et al., 2004). The absence of PTP-1B caused impaired XBP1 splicing, JNK phosphorylation, and attenuated upregulation of XBP1 target genes such as EDEM (Gu et al., 2004). Remarkably, PTP-1B deficiency did not affect PERK signaling, suggesting a specific regulation of IRE1α, and ruling out possible general effects on protein folding at the ER lumen. However, a physical association between PTP-1B and IRE1α was not evaluated, which could provide mechanistic insights about this regulation. A recent report presented evidence indicating that PTP-1B also regulates UPR signaling in vivo in diabetes models (Delibegovic et al., 2009).
IRE1α signaling is instigated by the expression of pro-apoptotic BCL-2 family members (Hetz et al., 2006). The BCL-2 family is a group of evolutionarily conserved regulators of cell death composed of both anti- and pro-apoptotic members that operate at the mitochondrial membrane to control caspase activation (Danial and Korsmeyer, 2004). Accumulating evidence indicates that, in addition to the mitochondria, members of the BCL-2 family of proteins are located at the ER membrane (reviewed in Hetz, 2007). We have described a new function for the pro-apoptotic BCL-2 family members BAX and BAK at the ER where they positively regulate the amplitude of IRE1α signaling by promoting its activation. These effects were specific for this branch of the UPR since PERK signaling was not altered by BAX and BAK deficiency (Hetz et al., 2006). This regulation was mediated by a physical association between the cytosolic domain of IRE1α and BAX/BAK and depended on the BCL-2 homology (BH) domains 1 and 3, essential motifs in the regulation of apoptosis (Figure 3). This regulation was recapitulated in vivo after challenging BAX and BAK conditional DKO mice with the ER stress agent tunicamycin (inhibitor of N-linked glycosylation), since decreased XBP1s expression and JNK phosphorylation were observed (Hetz et al., 2006).
In addition to the “multidomain” members BAX and BAK, there is another subtype of pro-apoptotic BCL-2 family members known as the “BH3-only” proteins (i.e. BIM, PUMA, and NOXA) which contain a single α-helical domain critical for apoptosis activation. In the control of apoptosis, BH3-only proteins act as upstream activators of BAX and BAK (reviewed in Youle and Strasser, 2008). A recent report indicated that the specific expression of the BH3-only proteins BIM and PUMA at the ER leads to the activation of the IRE1α/JNK pathway in a BAK-dependent manner (Klee et al., 2009). Notably, these findings were obtained in the absence of any ER stressor, suggesting that these BH3-only proteins are potent activators of the IRE1α-JNK branch acting upstream of BAX and BAK. These two BH3-only proteins have been shown to be induced by ER stress (Hetz, 2007), suggesting an interesting regulatory feedback loop. In murine cells, the proteolytic processing of the ER-resident caspase-12 has been indirectly associated to the UPR pathway by an interaction with TRAF2 and possibly with active IRE1α (Yoneda et al., 2001), but a complex between procaspase-12/TRAF2/IRE1α has not been described. Translocation of BIM from microtubules to the ER membrane has been shown to activate pro-caspase-12 processing (Morishima et al., 2004), and BAX and BAK deficient cells are resistant to pro-caspase-12 processing (Zong et al., 2003). However, the possible role of IRE1α in this process was not assessed in these two studies.
Similarly, the pro-apoptotic protein ASK1-interacting protein 1 (AIP1) was recently shown to specifically regulate and enhance IRE1α signaling (Luo et al., 2008). AIP1-deficient mice and cells derived from this mouse model displayed impaired IRE1α signaling after exposure to ER stress agents. Similar to the phenotype of PTP-1B or BAX/BAK deficient cells, the lack of AIP1 expression did not affect the PERK axis of the UPR. Structural and biochemical analyses suggested that AIP1 directly interacts with IRE1α through a domain homologous to the pleckstrin (PH domain), facilitating IRE1α dimerization and activation (Luo et al., 2008). Interestingly, the association of AIP1, and also BAX/BAK (Hetz et al., 2006), was shown to be induced by ER stress conditions. In summary, these findings suggest a novel and specific role for pro-apoptotic genes as accessory factors for the instigation of certain early UPR signaling events related to adaptation and survival (Figure 3). The possible allosteric site of IRE1α that modulates its signaling remains to be determined.
Temporal regulation of UPR signaling: Turning off IRE1α
Peter Walter's group recently reported that XBP1 mRNA splicing levels decline after prolonged ER stress, whereas PERK signaling is sustained over time (Lin et al., 2007). The authors suggested that this may serve to sensitize cells to cell death after chronic or irreversible ER stress, shutting down the pro-survival effects of IRE1α/XBP1 signaling, while enhancing the expression of the PERK-dependent pro-apoptotic factor CHOP/GADD153 and downstream expression of BIM (Puthalakath et al., 2007; Lin et al., 2009). However, the mechanism involved in the inactivation of IRE1α was not addressed. A different study provided indirect evidence suggesting that a negative regulator of IRE1α may exist. Along this line, proteasome inhibition was shown to selectively block IRE1α but not PERK activation, perhaps by the accumulation of an unknown inhibitor (Lee et al., 2003a).
Reed and colleagues initially suggested that the IRE1α pathway (and possibly ATF6) may be negatively modulated by the ER-located protein BAX inhibitor-1 (BI-1) in vivo (Bailly-Maitre et al., 2006). Under ischemic conditions, BI-1 deficient mice displayed increased expression of XBP1s and hyper-activated JNK in the liver and kidney, without affecting eIF2α phosphorylation (Bailly-Maitre et al., 2006). BI-1 is a six transmembrane containing protein functionally related to the BCL-2 family of proteins (Xu and Reed, 1998). BI-1 has no obvious homology with BCL-2-related proteins, yet it physically interacts with some members of this family (Xu and Reed, 1998; Chae et al., 2004). Further studies revealed that BI-1 homologues are present in yeast, plants, viruses and many other organisms (Chae et al., 2003; Huckelhoven, 2004) where their function remains poorly explored. Another study also suggested that BI-1 overexpression decreases activation of the UPR in vitro in classical paradigms of ER stress (Lee et al., 2007), but the mechanism underlying these observations was not directly defined.
We recently reported a direct role of BI-1 in the attenuation of IRE1α signaling. BI-1 deficient cells showed hyperactivation of IRE1α associated with increased XBP1 mRNA splicing and upregulation of XBP1s-dependent responses (Lisbona et al., 2009). Notably, attenuation of IRE1α signaling over time was markedly delayed in BI-1 deficient cells, suggesting that BI-1 has a role in the inactivation of IRE1α. The inhibition of IRE1α by BI-1 was recapitulated in vivo in BI-1 deficient mice and flies overexpressing Drosophila melanogaster BI-1 (Lisbona et al., 2009), indicating that this regulation is conserved across species. However, yeast cells deficient for the putative BI-1 homologue, the Ynl305c protein (Chae et al., 2003), did not show deficiencies in HAC1 expression, suggesting that this regulation emerged in multicellular organisms (Lisbona et al., 2009).
The formation of a protein complex between the cytosolic domain of IRE1α and BI-1 was observed. More importantly, this association was reconstituted in vitro with purified components, and BI-1 was shown to inhibit the endoribonuclease activity of IRE1α in a cell free system (Lisbona et al., 2009). The regulation of XBP1 splicing by BI-1 was mediated by its C-terminal cytosolic region, a domain previously linked with BI-1's anti-apoptotic activity (Chae et al., 2003) and to act as a pH sensor in the modulation of calcium release from the ER (Kim et al., 2008).
BI-1 and BAX/BAK's regulatory effects on XBP1 mRNA splicing were more evident when moderate to low doses of ER stressors were employed, conditions which more closely resemble a physiological state where cells are equipped to cope with injury (adaptive conditions). In agreement with this idea, BI-1 was shown to modulate immunoglobulin secretion of primary B cells (Lisbona et al., 2009), a phenomenon strictly dependent on XBP1s activity in vivo (Iwakoshi et al., 2003; Reimold et al., 2001). Randal Kaufman's group also reported that mild to low ER stress conditions evoke distinct signaling processes, where apoptosis-related events are not observed under mild ER stress conditions (Rutkowski et al., 2006).
In addition to the regulation of IRE1α activity, there is another check point that controls the stability of IRE1α, possibly affecting the amplitude of UPR responses. HSP90 was shown to interact with the cytosolic domain of IRE1α and targeting this chaperone with the inhibitors geldanamycin or 514 disrupted this complex, leading to IRE1α turnover by the proteasome (Marcu et al., 2002). HSP90 is known to regulate the stability of many protein complexes, raising the possibility that its association/dissociation with IRE1α may control the composition of the IRE1α interactome and its effects on cell signaling. Finally, Mori's group discovered that XBP1u markedly accumulates at the recovery phase of ER stress (Yoshida et al., 2006) to form a complex with XBP1s that is rapidly degraded by the proteasome. The authors proposed that this negative feedback loop helps to turn off the transcription of target genes during the recovery phase of ER stress.
Distinct roles of UPR components in organ physiology: Lessons from genetic mouse models
XBP1 is essential for the differentiation of hepatocytes, as XBP1 deficient embryos die in utero from severe liver hypoplasia and a resulting fatal anemia (Reimold et al., 2000). The first insights about the function of XBP1 in adult animals came from studies in the immune system. XBP1 was originally identified in multiple myeloma cells as a gene that is induced by Interleukin-6 treatment (Todd et al., 2008). XBP1-deficient B cells are markedly defective in antibody secretion in vivo in response to antigenic challenge, an activity later shown to be directly dependent on XBP1 splicing in stimulated B cells (Reimold et al., 2001; Iwakoshi et al., 2003). These findings provided the first link between the UPR and secretory cell function, and soon after the role of IRE1α was identified using similar experimental systems in plasma B cells (Zhang et al., 2005).
To circumvent the lethal liver phenotype of XBP-1-/- mice, we targeted an XBP1 transgene back to liver using a liver-specific promoter (Lee et al., 2005). XBP1-/-;LivXBP1 mice lacking XBP1 in all organs except the liver died shortly after birth from a severe impairment in the production of pancreatic digestive enzymes leading to hypoglycemia and death. At the cellular level, expansion of the ER was severely impaired in pancreatic exocrine cells, resulting in a complete disorganization of the ER network and decreased efficiency in zymogen granules/enzymes synthesis in the liver and salivary glands (Lee et al., 2005). These observations are consistent with a critical role of XBP1 in ER/Golgi biogenesis and phospholipid synthesis in secretory cells (Shaffer et al., 2004; Sriburi et al., 2004). Taken together with the requirement for XBP1 in plasma cell differentiation, these findings suggested that XBP1 is essential for the development of highly secretory exocrine cells.
An XBP1 conditional knockout mouse was recently generated, revealing new physiological functions of the transcription factor in diverse organs. For example, XBP1 expression in the liver is required for normal fatty acid and sterol synthesis (Lee et al., 2008a). In addition, PERK has been implicated in the regulation of lipogenic pathway in the mammary gland (Bobrovnikova-Marjon et al., 2008). Along this line, a recent study suggested that all three ER stress-sensing pathways share in protecting the organism against the deregulation of lipid metabolism upon experimental ER stress in vivo by regulating a subset of metabolic transcription factors related to lipid homeostasis (Rutkowski et al., 2008). In other studies, XBP1 deletion in intestinal epithelial cells triggered spontaneous enteritis secondary to Paneth cell dysfunction and increased susceptibility to induced colitis (Kaser et al., 2008). XBP1 polymorphisms were identified as risk factors for the human inflammatory bowel diseases Crohn's disease and ulcerative colitis (Kaser et al., 2008). ER stress has been also suggested as relevant in the occurrence of diabetes. Obesity induces ER stress in the liver, playing a central role in the development of insulin resistance and diabetes by triggering JNK activity via IRE1α and inhibition of insulin receptor signaling (Ozcan et al., 2004). Mice haploinsufficient for XBP1 showed an increased susceptibility to develop insulin resistance (Ozcan et al., 2004),
Correlative evidence suggests that activation of the UPR/IRE1α pathway may be a primary response against neurodegeneration (Matus et al., 2008). The first insights about the function of XBP1 in the nervous system came from genetic studies of human patients with bipolar disorders (Kakiuchi et al., 2003; Kato et al., 2005) and a polymorphism in the XBP1 promoter was shown to be a risk factor for this illness in the Japanese population (Kakiuchi et al., 2003). However, this findings are still under debate (Hou et al., 2004). In contrast to the drastic phenotypes described in secretory organs, the specific deletion of XBP1 in the brain was shown to have no effect on the development of the central nervous system and did not trigger any spontaneous illness or enhance the progression of a prion disease model (Hetz et al., 2008).
The phenotypes caused by defects in the PERK/eIF2α and IRE1α/XBP-1 pathways are disparate, indicating some divergence in their functions in vivo. For example, UPR-mediated translational control through eIF2α phosphorylation is not required for B lymphocyte maturation and/or plasma cell differentiation (Zhang et al., 2005). Similarly, PERK knockout mice do not show any deficiency in B cell function (Gass et al., 2007). Interestingly, PERK deficiency leads to abnormalities in the exocrine pancreas with decreased secretion of digestive enzymes, distended ER and increased apoptosis of acinar cells (Harding et al., 2001; Zhang et al., 2002), although the phenotype is modest as compared to XBP1-/-;LivXBP1 mice. Instead, PERK deficiency or lack of eIF2α phosphorylation causes progressive loss of pancreatic islet β-cells and impaired bone formation, indicating a function for the PERK pathway in β-cells and osteoblasts rather than other secretory cells (Zhang et al., 2002; Harding et al., 2001; Scheuner et al., 2005). In contrast, XBP1-/-;LivXBP1 mice do not show drastic changes at birth in the endocrine pancreas, reflected in normal levels of insulin and insulin-containing granules in β-cells. The PERK/eIF2α pathway activates a broad range of target genes, which is not surprising given that various cellular stresses converge upon the regulation of eIF2α activity. In contrast, as mentioned above, XBP1 target genes largely increase the folding capacity of the ER, trigger ER/Golgi biogenesis and improve the quality control system through ERAD regulation.
The emergence of the The UPRosome concept
The differences in cell type-specific requirements for PERK and IRE1α pathways could be attributed to specific modes of activation and/or downstream target genes regulated by each UPR branch. PERK and IRE1α share functionally similar luminal sensing domains and both are activated in cells treated in vitro with ER stress inducers. The ER stress sensing domains are, as mentioned, interchangeable between the two proteins without affecting cytosolic signaling (Liu et al., 2000). Based on the observation that PERK and IRE1α are selectively activated in vivo, it is feasible that additional mechanisms differentially regulate their activity. Based on the compelling data discussed in this review, we speculate that the specific activation of ER stress sensors in different tissue contexts may be related to the engagement of specific regulatory complexes through the association of adaptors and direct binding of modulators.
We envision a model where a complex signaling platform is assembled at the level of IRE1α to modulate its activation status in terms of signaling intensity and kinetics of activation/inactivation (Figure 3). In the context of cell fate and proteostasis, the fine tuning of UPR signaling responses is particularly relevant in life to death transitions by controlling transcriptional programs that regulate adaptation to stress or by initiating apoptosis of irreversibly damaged cells. To refer to the existence of an IRE1α signaling macromolecular complex, we have previously used the term “UPRosome” (Hetz and Glimcher, 2008). This platform initiates multiple signaling responses in a highly regulated manner, providing a mechanism for selectivity and specificity in the signaling of IRE1α (Figure 3). It remains to be determined if PERK and ATF6 are regulated in a similar manner by specific factors (i.e. UPRosome-2, UPRosome-3). It is interesting to note that a set of apoptosis-related proteins (i.e. BAX, BAK, AIP1, BI-1 and maybe PTP-1B and BH3-only proteins) interacts with IRE1α, regulating its activation status. These findings suggest a model wherein the expression of anti- and pro-apoptotic proteins at the ER membrane may determine the amplitude of UPR responses and the ability of a cell to adapt to ER injuries or to initiate apoptosis (Figure 3). It remains to be determined if distinct or dynamic IRE1α-containing complexes exist in a tissue-specific context to regulate its signaling. This model may also be useful in addressing how the transition between the catalysis of XBP-1 mRNA processing and mRNA decay occurs. Based on the fact that active IRE1α clusters in a highly organized manner, extensive biochemical characterization of the IRE1α interactome is required to determine the stoichiometry of this complex and how its protein composition evolves under conditions of mild and chronic ER stress. This characterization will be particularly relevant due to the divergent and distinct effects of the UPR in cell physiology related to the control of essential processes such as secretion, folding, autophagy, calcium signaling, organelle biogenesis, apoptosis, inflammation, and cellular differentiation. Exploration of the molecular control of fine tuning of the UPR may provide new therapeutic targets to modulate ER stress responses in the setting of diverse diseases conditions such as cancer, diabetes, autoimmunity and neurodegeneration.
Acknowledgments
We thank Dr. Diego Rodriguez for help in designing figures. This work was supported by FONDECYT no. 1070444, FONDAP grant no.15010006, High Q Foundation-CHDI, Muscular Dystrophy Association, Millennium Nucleus no. P07-048-F, ICGB grant (CH), and by NIH grant AI32412, the Harvard University Accelerator Fund and a gift from the Leila and Harold Mathers Foundation (LHG). Due to space limitations we apologize if any relevant publication for this review is not cited here.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Acosta-Alvear D, Zhou Y, Blais A, Tsikitis M, Lents NH, Arias C, Lennon CJ, Kluger Y, Dynlacht BD. XBP1 Controls Diverse Cell Type- and Condition-Specific Transcriptional Regulatory Networks. Mol Cell. 2007;27:53–66. doi: 10.1016/j.molcel.2007.06.011. [DOI] [PubMed] [Google Scholar]
- Aragon T, van AE, Pincus D, Serafimova IM, Korennykh AV, Rubio CA, Walter P. Messenger RNA targeting to endoplasmic reticulum stress signalling sites. Nature. 2009;457:736–740. doi: 10.1038/nature07641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsham AM, Neufeld TP. A genetic screen in Drosophila reveals novel cytoprotective functions of the autophagy-lysosome pathway. PLoS One. 2009;4:e6068. doi: 10.1371/journal.pone.0006068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly-Maitre B, Fondevila C, Kaldas F, Droin N, Luciano F, Ricci JE, Croxton R, Krajewska M, Zapata JM, Kupiec-Weglinski JW, Farmer D, Reed JC. Cytoprotective gene bi-1 is required for intrinsic protection from endoplasmic reticulum stress and ischemia-reperfusion injury. Proc Natl Acad Sci U S A. 2006;103:2809–2814. doi: 10.1073/pnas.0506854103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol. 2000;2:326–332. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- Bobrovnikova-Marjon E, Hatzivassiliou G, Grigoriadou C, Romero M, Cavener DR, Thompson CB, Diehl JA. PERK-dependent regulation of lipogenesis during mouse mammary gland development and adipocyte differentiation. Proc Natl Acad Sci U S A. 2008;105:16314–16319. doi: 10.1073/pnas.0808517105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommiasamy H, Back SH, Fagone P, Lee K, Meshinchi S, Vink E, Sriburi R, Frank M, Jackowski S, Kaufman RJ, Brewer JW. ATF6{alpha} induces XBP1-independent expansion of the endoplasmic reticulum. J Cell Sci. 2009;122:1626–1636. doi: 10.1242/jcs.045625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae HJ, Ke N, Kim HR, Chen S, Godzik A, Dickman M, Reed JC. Evolutionarily conserved cytoprotection provided by Bax Inhibitor-1 homologs from animals, plants, and yeast. Gene. 2003;323:101–113. doi: 10.1016/j.gene.2003.09.011. [DOI] [PubMed] [Google Scholar]
- Chae HJ, Kim HR, Xu C, Bailly-Maitre B, Krajewska M, Krajewski S, Banares S, Cui J, Digicaylioglu M, Ke N, Kitada S, Monosov E, Thomas M, Kress CL, Babendure JR, Tsien RY, Lipton SA, Reed JC. BI-1 regulates an apoptosis pathway linked to endoplasmic reticulum stress. Mol Cell. 2004;15:355–366. doi: 10.1016/j.molcel.2004.06.038. [DOI] [PubMed] [Google Scholar]
- Chen X, Shen J, Prywes R. The luminal domain of ATF6 senses endoplasmic reticulum (ER) stress and causes translocation of ATF6 from the ER to the Golgi. J Biol Chem. 2002;277:13045–13052. doi: 10.1074/jbc.M110636200. [DOI] [PubMed] [Google Scholar]
- Credle JJ, Finer-Moore JS, Papa FR, Stroud RM, Walter P. On the mechanism of sensing unfolded protein in the endoplasmic reticulum. Proc Natl Acad Sci U S A. 2005;102:18773–18784. doi: 10.1073/pnas.0509487102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116:205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- Delibegovic M, Zimmer D, Kauffman C, Rak K, Hong EG, Cho YR, Kim JK, Kahn BB, Neel BG, Bence KK. Liver-specific deletion of protein-tyrosine phosphatase 1B (PTP1B) improves metabolic syndrome and attenuates diet-induced endoplasmic reticulum stress. Diabetes. 2009;58:590–599. doi: 10.2337/db08-0913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gass JN, Jiang HY, Wek RC, Brewer JW. The unfolded protein response of B-lymphocytes: PERK-independent development of antibody-secreting cells. Mol Immunol. 2007;45:1035–43. doi: 10.1016/j.molimm.2007.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu F, Nguyen DT, Stuible M, Dube N, Tremblay ML, Chevet E. Protein-tyrosine phosphatase 1B potentiates IRE1 signaling during endoplasmic reticulum stress. J Biol Chem. 2004;279:49689–49693. doi: 10.1074/jbc.C400261200. [DOI] [PubMed] [Google Scholar]
- Han D, Lerner AG, Vande WL, Upton JP, Xu W, Hagen A, Backes BJ, Oakes SA, Papa FR. IRE1alpha Kinase Activation Modes Control Alternate Endoribonuclease Outputs to Determine Divergent Cell Fates. Cell. 2009;138:562–575. doi: 10.1016/j.cell.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Zeng H, Zhang Y, Jungries R, Chung P, Plesken H, Sabatini DD, Ron D. Diabetes mellitus and exocrine pancreatic dysfunction in perk-/- mice reveals a role for translational control in secretory cell survival. Mol Cell. 2001;7:1153–1163. doi: 10.1016/s1097-2765(01)00264-7. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, Stojdl DF, Bell JC, Hettmann T, Leiden JM, Ron D. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- Hetz C, Bernasconi P, Fisher J, Lee AH, Bassik MC, Antonsson B, Brandt GS, Iwakoshi NN, Schinzel A, Glimcher LH, Korsmeyer SJ. Proapoptotic BAX and BAK modulate the unfolded protein response by a direct interaction with IRE1alpha. Science. 2006;312:572–576. doi: 10.1126/science.1123480. [DOI] [PubMed] [Google Scholar]
- Hetz C, Glimcher L. The daily job of night killers: alternative roles of the BCL-2 family in organelle physiology. Trends Cell Biol. 2008;18:38–44. doi: 10.1016/j.tcb.2007.10.003. [DOI] [PubMed] [Google Scholar]
- Hetz C, Lee AH, Gonzalez-Romero D, Thielen P, Castilla J, Soto C, Glimcher LH. Unfolded protein response transcription factor XBP-1 does not influence prion replication or pathogenesis. Proc Natl Acad Sci U S A. 2008;105:757–762. doi: 10.1073/pnas.0711094105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz CA. ER Stress Signaling and the BCL-2 Family of Proteins: From Adaptation to Irreversible Cellular Damage. Antioxid Redox Signal. 2007;9:2345–2356. doi: 10.1089/ars.2007.1793. [DOI] [PubMed] [Google Scholar]
- Hollien J, Lin JH, Li H, Stevens N, Walter P, Weissman JS. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J Cell Biol. 2009;186:323–31. doi: 10.1083/jcb.200903014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollien J, Weissman JS. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science. 2006;313:104–107. doi: 10.1126/science.1129631. [DOI] [PubMed] [Google Scholar]
- Hou SJ, Yen FC, Cheng CY, Tsai SJ, Hong CJ. X-box binding protein 1 (XBP1) C--116G polymorphisms in bipolar disorders and age of onset. Neurosci Lett. 2004;367:232–234. doi: 10.1016/j.neulet.2004.06.012. [DOI] [PubMed] [Google Scholar]
- Hu P, Han Z, Couvillon AD, Kaufman RJ, Exton JH. Autocrine tumor necrosis factor alpha links endoplasmic reticulum stress to the membrane death receptor pathway through IRE1alpha-mediated NF-kappaB activation and down-regulation of TRAF2 expression. Mol Cell Biol. 2006;26:3071–3084. doi: 10.1128/MCB.26.8.3071-3084.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huckelhoven R. BAX Inhibitor-1, an ancient cell death suppressor in animals and plants with prokaryotic relatives. Apoptosis. 2004;9:299–307. doi: 10.1023/b:appt.0000025806.71000.1c. [DOI] [PubMed] [Google Scholar]
- Iwakoshi NN, Lee AH, Vallabhajosyula P, Otipoby KL, Rajewsky K, Glimcher LH. Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nat Immunol. 2003;4:321–329. doi: 10.1038/ni907. [DOI] [PubMed] [Google Scholar]
- Jonikas MC, Collins SR, Denic V, Oh E, Quan EM, Schmid V, Weibezahn J, Schwappach B, Walter P, Weissman JS, Schuldiner M. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science. 2009;323:1693–1697. doi: 10.1126/science.1167983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakiuchi C, Iwamoto K, Ishiwata M, Bundo M, Kasahara T, Kusumi I, Tsujita T, Okazaki Y, Nanko S, Kunugi H, Sasaki T, Kato T. Impaired feedback regulation of XBP1 as a genetic risk factor for bipolar disorder. Nat Genet. 2003;35:171–175. doi: 10.1038/ng1235. [DOI] [PubMed] [Google Scholar]
- Kanda H, Miura M. Regulatory roles of JNK in programmed cell death. J Biochem (Tokyo) 2004;136:1–6. doi: 10.1093/jb/mvh098. [DOI] [PubMed] [Google Scholar]
- Kaser A, Lee AH, Franke A, Glickman JN, Zeissig S, Tilg H, Nieuwenhuis EE, Higgins DE, Schreiber S, Glimcher LH, Blumberg RS. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134:743–756. doi: 10.1016/j.cell.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T, Kuratomi G, Kato N. Genetics of bipolar disorder. Drugs Today (Barc) 2005;41:335–344. doi: 10.1358/dot.2005.41.5.893616. [DOI] [PubMed] [Google Scholar]
- Kim HR, Lee GH, Ha KC, Ahn T, Lee BJ, Cho SG, Kim S, Seo YR, Shin YJ, Chae SW, Reed JC, Chae HJ. Bax inhibitor-1 (BI-1) is a pH-dependent regulator of Ca2+ channel activity in the endoplasmic reticulum. J Biol Chem. 2008;283:15946–55. doi: 10.1074/jbc.M800075200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I, Shu CW, Xu W, Shiau CW, Grant D, Vasile S, Cosford ND, Reed JC. Chemical biology investigation of cell death pathways activated by endoplasmic reticulum stress reveals cytoprotective modulators of ASK1. J Biol Chem. 2009;284:1593–1603. doi: 10.1074/jbc.M807308200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimata Y, Ishiwata-Kimata Y, Ito T, Hirata A, Suzuki T, Oikawa D, Takeuchi M, Kohno K. Two regulatory steps of ER-stress sensor Ire1 involving its cluster formation and interaction with unfolded proteins. J Cell Biol. 2007;179:75–86. doi: 10.1083/jcb.200704166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimata Y, Kimata YI, Shimizu Y, Abe H, Farcasanu IC, Takeuchi M, Rose MD, Kohno K. Genetic evidence for a role of BiP/Kar2 that regulates Ire1 in response to accumulation of unfolded proteins. Mol Biol Cell. 2003;14:2559–2569. doi: 10.1091/mbc.E02-11-0708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimata Y, Oikawa D, Shimizu Y, Ishiwata-Kimata Y, Kohno K. A role for BiP as an adjustor for the endoplasmic reticulum stress-sensing protein Ire1. J Cell Biol. 2004;167:445–456. doi: 10.1083/jcb.200405153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klee M, Pallauf K, Alcala S, Fleischer A, Pimentel-Muinos FX. Mitochondrial apoptosis induced by BH3-only molecules in the exclusive presence of endoplasmic reticular Bak. EMBO J. 2009;28:1757–68. doi: 10.1038/emboj.2009.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korennykh AV, Egea PF, Korostelev AA, Finer-Moore J, Zhang C, Shokat KM, Stroud RM, Walter P. The unfolded protein response signals through high-order assembly of Ire1. Nature. 2009;457:687–693. doi: 10.1038/nature07661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, Chu GC, Iwakoshi NN, Glimcher LH. XBP-1 is required for biogenesis of cellular secretory machinery of exocrine glands. EMBO J. 2005;24:4368–4380. doi: 10.1038/sj.emboj.7600903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, Iwakoshi NN, Anderson KC, Glimcher LH. Proteasome inhibitors disrupt the unfolded protein response in myeloma cells. Proc Natl Acad Sci U S A. 2003a;100:9946–9951. doi: 10.1073/pnas.1334037100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol. 2003b;23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, Scapa EF, Cohen DE, Glimcher LH. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science. 2008a;320:1492–1496. doi: 10.1126/science.1158042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GH, Kim HK, Chae SW, Kim DS, Ha KC, Mike C, Kress C, Reed JC, Kim HR, Chae HJ. Bax inhibitor-1 regulates endoplasmic reticulum stress-associated reactive oxygen species and heme oxygenase-I expression. J Biol Chem. 2007 doi: 10.1074/jbc.M700053200. [DOI] [PubMed] [Google Scholar]
- Lee KP, Dey M, Neculai D, Cao C, Dever TE, Sicheri F. Structure of the dual enzyme Ire1 reveals the basis for catalysis and regulation in nonconventional RNA splicing. Cell. 2008b;132:89–100. doi: 10.1016/j.cell.2007.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JH, Li H, Yasumura D, Cohen HR, Zhang C, Panning B, Shokat KM, Lavail MM, Walter P. IRE1 signaling affects cell fate during the unfolded protein response. Science. 2007;318:944–949. doi: 10.1126/science.1146361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JH, Li H, Zhang Y, Ron D, Walter P. Divergent effects of PERK and IRE1 signaling on cell viability. PLoS One. 2009;4:e4170. doi: 10.1371/journal.pone.0004170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipson KL, Fonseca SG, Urano F. Endoplasmic reticulum stress-induced apoptosis and auto-immunity in diabetes. Curr Mol Med. 2006;6:71–77. doi: 10.2174/156652406775574613. [DOI] [PubMed] [Google Scholar]
- Lisbona F, Rojas-Rivera D, Thielen P, Zamorano S, Todd D, Martinon F, Glavic A, Kress C, Lin JH, Walter P, Reed JC, Glimcher LH, Hetz C. BAX inhibitor-1 is a negative regulator of the ER stress sensor IRE1alpha. Mol Cell. 2009;33:679–691. doi: 10.1016/j.molcel.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CY, Schroder M, Kaufman RJ. Ligand-independent dimerization activates the stress response kinases IRE1 and PERK in the lumen of the endoplasmic reticulum. J Biol Chem. 2000;275:24881–24885. doi: 10.1074/jbc.M004454200. [DOI] [PubMed] [Google Scholar]
- Luo D, He Y, Zhang H, Yu L, Chen H, Xu Z, Tang S, Urano F, Min W. AIP1 is critical in transducing IRE1-mediated endoplasmic reticulum stress response. J Biol Chem. 2008;283:11905–11912. doi: 10.1074/jbc.M710557200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcu MG, Doyle M, Bertolotti A, Ron D, Hendershot L, Neckers L. Heat shock protein 90 modulates the unfolded protein response by stabilizing IRE1alpha. Mol Cell Biol. 2002;22:8506–8513. doi: 10.1128/MCB.22.24.8506-8513.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, Bray K, Reddy A, Bhanot G, Gelinas C, Dipaola RS, Karantza-Wadsworth V, White E. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–1075. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matus S, Lisbona F, Torres M, Leon C, Thielen P, Hetz C. The stress rheostat: an interplay between the unfolded protein response (UPR) and autophagy in neurodegeneration. Curr Mol Med. 2008;8:157–172. doi: 10.2174/156652408784221324. [DOI] [PubMed] [Google Scholar]
- Mauro C, Crescenzi E, De MR, Pacifico F, Mellone S, Salzano S, de LC, D'Adamio L, Palumbo G, Formisano S, Vito P, Leonardi A. Central role of the scaffold protein tumor necrosis factor receptor-associated factor 2 in regulating endoplasmic reticulum stress-induced apoptosis. J Biol Chem. 2006;281:2631–2638. doi: 10.1074/jbc.M502181200. [DOI] [PubMed] [Google Scholar]
- Moenner M, Pluquet O, Bouchecareilh M, Chevet E. Integrated endoplasmic reticulum stress responses in cancer. Cancer Res. 2007;67:10631–10634. doi: 10.1158/0008-5472.CAN-07-1705. [DOI] [PubMed] [Google Scholar]
- Morishima N, Nakanishi K, Tsuchiya K, Shibata T, Seiwa E. Translocation of Bim to the endoplasmic reticulum (ER) mediates ER stress signaling for activation of caspase-12 during ER stress-induced apoptosis. J Biol Chem. 2004;279:50375–50381. doi: 10.1074/jbc.M408493200. [DOI] [PubMed] [Google Scholar]
- Nagai A, Kadowaki H, Maruyama T, Takeda K, Nishitoh H, Ichijo H. USP14 inhibits ER-associated degradation via interaction with IRE1alpha. Biochem Biophys Res Commun. 2009;379:995–1000. doi: 10.1016/j.bbrc.2008.12.182. [DOI] [PubMed] [Google Scholar]
- Nekrutenko A, He J. Functionality of unspliced XBP1 is required to explain evolution of overlapping reading frames. Trends Genet. 2006;22:645–648. doi: 10.1016/j.tig.2006.09.012. [DOI] [PubMed] [Google Scholar]
- Nguyen DT, Kebache S, Fazel A, Wong HN, Jenna S, Emadali A, Lee EH, Bergeron JJ, Kaufman RJ, Larose L, Chevet E. Nck-dependent activation of extracellular signal-regulated kinase-1 and regulation of cell survival during endoplasmic reticulum stress. Mol Biol Cell. 2004;15:4248–4260. doi: 10.1091/mbc.E03-11-0851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitoh H, Matsuzawa A, Tobiume K, Saegusa K, Takeda K, Inoue K, Hori S, Kakizuka A, Ichijo H. ASK1 is essential for endoplasmic reticulum stress-induced neuronal cell death triggered by expanded polyglutamine repeats. Genes Dev. 2002;16:1345–1355. doi: 10.1101/gad.992302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa M, Patil CK, DeRisi J, Walter P. Genome-scale approaches for discovering novel nonconventional splicing substrates of the Ire1 nuclease. Genome Biol. 2005;6:R3. doi: 10.1186/gb-2004-6-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata M, Hino SI, Saito A, Morikawa K, Kondo S, Kanemoto S, Murakami T, Taniguchi M, Tanii I, Yoshinaga K, Shiosaka S, Hammarback JA, Urano F, Imaizumi K. Autophagy is activated for cell survival after ER stress. Mol Cell Biol. 2006;26:9220–9231. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa D, Kimata Y, Kohno K, Iwawaki T. Activation of mammalian IRE1alpha upon ER stress depends on dissociation of BiP rather than on direct interaction with unfolded proteins. Exp Cell Res. 2009 doi: 10.1016/j.yexcr.2009.06.009. [DOI] [PubMed] [Google Scholar]
- Ozcan U, Cao Q, Yilmaz E, Lee AH, Iwakoshi NN, Ozdelen E, Tuncman G, Gorgun C, Glimcher LH, Hotamisligil GS. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science. 2004;306:457–461. doi: 10.1126/science.1103160. [DOI] [PubMed] [Google Scholar]
- Pattingre S, Bauvy C, Carpentier S, Levade T, Levine B, Codogno P. Role of JNK1-dependent Bcl-2 phosphorylation in ceramide-induced macroautophagy. J Biol Chem. 2009;284:2719–2728. doi: 10.1074/jbc.M805920200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers ET, Morimoto RI, Dillin A, Kelly JW, Balch WE. Biological and chemical approaches to diseases of proteostasis deficiency. Annu Rev Biochem. 2009;78:959–991. doi: 10.1146/annurev.biochem.052308.114844. [DOI] [PubMed] [Google Scholar]
- Puthalakath H, O'Reilly LA, Gunn P, Lee L, Kelly PN, Huntington ND, Hughes PD, Michalak EM, Kimm-Breschkin J, Motoyama N, Gotoh T, Akira S, Bouillet P, Strasser A. ER Stress Triggers Apoptosis by Activating BH3-Only Protein Bim. Cell. 2007;129:1337–1349. doi: 10.1016/j.cell.2007.04.027. [DOI] [PubMed] [Google Scholar]
- Reimold AM, Etkin A, Clauss I, Perkins A, Friend DS, Zhang J, Horton HF, Scott A, Orkin SH, Byrne MC, Grusby MJ, Glimcher LH. An essential role in liver development for transcription factor XBP-1. Genes Dev. 2000;14:152–157. [PMC free article] [PubMed] [Google Scholar]
- Reimold AM, Iwakoshi NN, Manis J, Vallabhajosyula P, Szomolanyi-Tsuda E, Gravallese EM, Friend D, Grusby MJ, Alt F, Glimcher LH. Plasma cell differentiation requires the transcription factor XBP-1. Nature. 2001;412:300–307. doi: 10.1038/35085509. [DOI] [PubMed] [Google Scholar]
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Rutkowski DT, Arnold SM, Miller CN, Wu J, Li J, Gunnison KM, Mori K, Sadighi Akha AA, Raden D, Kaufman RJ. Adaptation to ER stress is mediated by differential stabilities of pro-survival and pro-apoptotic mRNAs and proteins. PLoS Biol. 2006;4:e374. doi: 10.1371/journal.pbio.0040374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowski DT, Wu J, Back SH, Callaghan MU, Ferris SP, Iqbal J, Clark R, Miao H, Hassler JR, Fornek J, Katze MG, Hussain MM, Song B, Swathirajan J, Wang J, Yau GD, Kaufman RJ. UPR pathways combine to prevent hepatic steatosis caused by ER stress-mediated suppression of transcriptional master regulators. Dev Cell. 2008;15:829–840. doi: 10.1016/j.devcel.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuner D, Vander MD, Song B, Flamez D, Creemers JW, Tsukamoto K, Ribick M, Schuit FC, Kaufman RJ. Control of mRNA translation preserves endoplasmic reticulum function in beta cells and maintains glucose homeostasis. Nat Med. 2005;11:757–764. doi: 10.1038/nm1259. [DOI] [PubMed] [Google Scholar]
- Shaffer AL, Shapiro-Shelef M, Iwakoshi NN, Lee AH, Qian SB, Zhao H, Yu X, Yang L, Tan BK, Rosenwald A, Hurt EM, Petroulakis E, Sonenberg N, Yewdell JW, Calame K, Glimcher LH, Staudt LM. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity. 2004;21:81–93. doi: 10.1016/j.immuni.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Shen J, Chen X, Hendershot L, Prywes R. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev Cell. 2002;3:99–111. doi: 10.1016/s1534-5807(02)00203-4. [DOI] [PubMed] [Google Scholar]
- Sriburi R, Jackowski S, Mori K, Brewer JW. XBP1: a link between the unfolded protein response, lipid biosynthesis, and biogenesis of the endoplasmic reticulum. J Cell Biol. 2004;167:35–41. doi: 10.1083/jcb.200406136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Tsujimura T, Takeda K, Sugihara A, Maekawa A, Terada N, Yoshida N, Akira S. Targeted disruption of ATF4 discloses its essential role in the formation of eye lens fibres. Genes Cells. 1998;3:801–810. doi: 10.1046/j.1365-2443.1998.00230.x. [DOI] [PubMed] [Google Scholar]
- Todd DJ, Lee AH, Glimcher LH. The endoplasmic reticulum stress response in immunity and autoimmunity. Nat Rev Immunol. 2008;8:663–674. doi: 10.1038/nri2359. [DOI] [PubMed] [Google Scholar]
- Urano F, Wang X, Bertolotti A, Zhang Y, Chung P, Harding HP, Ron D. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000;287:664–666. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Rutkowski DT, Dubois M, Swathirajan J, Saunders T, Wang J, Song B, Yau GD, Kaufman RJ. ATF6alpha optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev Cell. 2007;13:351–364. doi: 10.1016/j.devcel.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Xu Q, Reed JC. Bax inhibitor-1, a mammalian apoptosis suppressor identified by functional screening in yeast. Mol Cell. 1998;1:337–346. doi: 10.1016/s1097-2765(00)80034-9. [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Sato T, Matsui T, Sato M, Okada T, Yoshida H, Harada A, Mori K. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev Cell. 2007;13:365–376. doi: 10.1016/j.devcel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- Yanagitani K, Imagawa Y, Iwawaki T, Hosoda A, Saito M, Kimata Y, Kohno K. Cotranslational targeting of XBP1 protein to the membrane promotes cytoplasmic splicing of its own mRNA. Mol Cell. 2009;34:191–200. doi: 10.1016/j.molcel.2009.02.033. [DOI] [PubMed] [Google Scholar]
- Yoneda T, Imaizumi K, Oono K, Yui D, Gomi F, Katayama T, Tohyama M. Activation of caspase-12, an endoplastic reticulum (ER) resident caspase, through tumor necrosis factor receptor-associated factor 2- dependent mechanism in response to the ER stress. J Biol Chem. 2001;276:13935–13940. doi: 10.1074/jbc.M010677200. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Oku M, Suzuki M, Mori K. pXBP1(U) encoded in XBP1 pre-mRNA negatively regulates unfolded protein response activator pXBP1(S) in mammalian ER stress response. J Cell Biol. 2006;172:565–575. doi: 10.1083/jcb.200508145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- Zhang K, Wong HN, Song B, Miller CN, Scheuner D, Kaufman RJ. The unfolded protein response sensor IRE1alpha is required at 2 distinct steps in B cell lymphopoiesis. J Clin Invest. 2005;115:268–281. doi: 10.1172/JCI21848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, McGrath B, Li S, Frank A, Zambito F, Reinert J, Gannon M, Ma K, McNaughton K, Cavener DR. The PERK eukaryotic initiation factor 2 alpha kinase is required for the development of the skeletal system, postnatal growth, and the function and viability of the pancreas. Mol Cell Biol. 2002;22:3864–3874. doi: 10.1128/MCB.22.11.3864-3874.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Liu CY, Back SH, Clark RL, Peisach D, Xu Z, Kaufman RJ. The crystal structure of human IRE1 luminal domain reveals a conserved dimerization interface required for activation of the unfolded protein response. Proc Natl Acad Sci U S A. 2006;103:14343–14348. doi: 10.1073/pnas.0606480103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinszner H, Kuroda M, Wang X, Batchvarova N, Lightfoot RT, Remotti H, Stevens JL, Ron D. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 1998;12:982–995. doi: 10.1101/gad.12.7.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong WX, Li C, Hatzivassiliou G, Lindsten T, Yu QC, Yuan J, Thompson CB. Bax and Bak can localize to the endoplasmic reticulum to initiate apoptosis. J Cell Biol. 2003;162:59–69. doi: 10.1083/jcb.200302084. [DOI] [PMC free article] [PubMed] [Google Scholar]