Summary
T cell fate is associated with mutually exclusive expression of CD4 or CD8 in helper and cytotoxic T cells, respectively. How expression of one locus is temporally coordinated with repression of the other has been a long-standing enigma, though we know RUNX transcription factors activate the Cd8 locus, silence the Cd4 locus, and repress the Zbtb7b locus (encoding the transcription factor ThPOK), which is required for CD4 expression. Here we found that nuclear organization was altered by interplay among members of this transcription factor circuitry: RUNX binding mediated association of Cd4 and Cd8 whereas ThPOK binding kept the loci apart. Moreover, targeted deletions within Cd4 modulated CD8 expression and pericentromeric repositioning of Cd8. Communication between Cd4 and Cd8 thus appears to enable long-range epigenetic regulation to ensure that expression of one excludes the other in mature CD4 or CD8 single-positive (SP) cells.
Graphical Abstract
Highlights
► Association of Cd4 and Cd8 genes enables coordinate gene regulation ► Cd4 and Cd8 associate with one another in CD8-expressing cells ► RUNX transcription factor mediates association of Cd4 and Cd8 genes ► Targeted deletions within Cd4 modulate expression and nuclear location of Cd8
Introduction
Cell fate is determined by complex patterns of gene expression that are often mediated by a surprisingly limited number of transcription factors. Within a particular lineage, key factors can both upregulate and repress the expression of different target genes, which can number in the hundreds and be scattered throughout the genome. How are these activities coordinated? Given that tissue-specific expression profiles can be accompanied by tissue-specific patterns of locus conformation (Roldán et al., 2005; Sayegh et al., 2005; Skok et al., 2007) or nuclear location (near the nuclear periphery, pericentromeric heterochromatin [Brown et al., 1999], or within a chromosome territory [Chambeyron and Bickmore, 2004]), it is reasonable to ask whether higher-order nuclear organization might be involved in coordinating this simultaneous expression and repression.
Lymphocyte development provides an attractive model system for investigating whether there is a correlation between cell fate decisions and the spatial organization of the nucleus, because developmental stages and effector cell functions are clearly differentiated by the expression of cell surface glycoproteins that mark lineage commitment (and transcription of whose loci must therefore be carefully orchestrated). T lymphocytes express both CD4 and CD8 during development, but mature helper T cells express only CD4 protein while mature cytotoxic T cells express only CD8 (Kioussis and Ellmeier, 2002). Both populations of T cells arise from common thymic precursors that are propelled through a series of developmental stages by recombination of variable, diversity, and joining gene segments (V(D)J) that eventually form a unique antigen receptor. Recombination begins at the earliest stage of development in double-negative (DN) thymocytes, which express neither CD4 nor CD8. Productive V(D)J rearrangement of one allele leads to assembly and expression of the pre-T cell receptor (pre-TCR) on the surface of the cell; signaling through the pre-TCR promotes differentiation to the double-positive (DP) stage, in which the cells express both CD4 and CD8. DP cells then enter a transitional stage during which CD8 expression diminishes (CD4+CD8lo) before finally becoming either CD4+ helper T cells or CD8+ cytotoxic T cells (sometimes referred to as CD4 or CD8 single-positive [SP] cells, respectively). The CD4 and CD8 coreceptors are regulated to ensure mutually exclusive expression (and complementary repression).
How is this coregulation achieved? Two lines of evidence led us to suspect that nuclear organization might play a role. First, it is increasingly apparent that transcriptional status is related to chromosomal positioning (Chubb et al., 2006; Iborra et al., 1996; Osborne et al., 2004; Ragoczy et al., 2006). Second, members of the RUNX family of transcription factors are important in governing cell fate decisions in developing T lymphocytes (Collins et al., 2009)—and they are known to have a role in nuclear organization as well (Stein et al., 2007). RUNX1 and RUNX3, which are expressed at different stages of T cell development, both activate and repress expression of the Cd4 and Cd8 loci in a complementary fashion. How such exquisite control is achieved, however, has been difficult to ascertain with molecular genetic and biochemical approaches.
To test the notion that higher-order nuclear organization might facilitate epigenetic regulation of these loci, we used 3-dimensional DNA fluorescent in situ hybridization (FISH) to examine the interplay between the Cd4 and Cd8 loci during T cell development in wild-type mice and a variety of mutant lines.
Results
Cd4 and Cd8 Associate with One Another in CD8-Expressing Murine Cells
Cd4 and Cd8 are located on chromosome 6 in the mouse, separated by a distance of 53.3 megabases (Mb). We used the two BAC probes RP23-121J20 and RP23-139M18, which cover the Cd4 and Cd8 loci, respectively, to follow the nuclear localization of these loci during T cell development (Figure S1A available online). 3D DNA FISH and confocal microscopy were carried out as previously described (Roldán et al., 2005) in sorted thymocyte populations from wild-type mice (Figure S1B). It is important to point out that, in our analyses, we sorted cells in different stages of thymocyte development by expression of CD4 and CD8 as well as a number of other developmental markers (Figure S1C). We measured the distance between the center of mass of the Cd4 and Cd8 signals in individual cells by using Image J software (Figure 1A). Interallelic distances were displayed as cumulative frequency curves. A left shift in the cumulative frequency curve indicates closer distances, as shown by the distribution. The statistical significance of the difference between distributions was calculated by the two-sample Kolmogorov-Smirnov (KS) test (Figure 1B).
Figure 1.
Cd4 and Cd8 Associate in Murine CD8-Expressing Cells
(A) Confocal microscopy sections showing a range of distances between the Cd4 and Cd8 loci. Scale bars represent 1 μm.
(B) Association of Cd4-Cd8 in DP T cells compared to B cells (top); association of Cd4-Cd8 compared to Tcrb-Lrig1 in DP T cells (bottom). The separation of signals is plotted as a cumulative frequency of association. Association of Cd4-Cd8 in DP cells was increased compared to B cells and higher than association of Tcrb-Lrig1 in DP cells. The Kolmogorov-Smirnov test was used for statistical analysis. At least three independent experiments were performed for each data set. n = 238–264 alleles.
(C) Association of Cd4-Cd8 in developing T cell populations and statistical analysis between specified stages. Association is increased in DP and CD8 SP cells. n = 166–238 alleles.
(D) Images of Cd4 and Cd8 and their individual chromosome 6 territories in DP cells.
Two controls were used for these experiments: we measured the distance between these same loci in a different cell type, namely B cells, and we measured the distance between two different loci, Tcrb and Lrig1, which are also located on chromosome 6 and separated by a similar distance (52.9 Mb) (Figure S1A). We observed significantly reduced association between Cd4 and Cd8 alleles in splenic B cells compared to DP cells (p = 1.67e-11) (Figure 1B). To examine the control loci Tcrb and Lrig1, we used the two BAC probes RP24-365F23 and RP23-148M10, respectively (Figure S1A). Association between Tcrb and Lrig1 was significantly lower than between Cd4 and Cd8 in DP cells (Figure 1B; p = 2.44e-10). We used the distribution of the frequency of association of Cd4 and Cd8 in a control cell type (splenic B cells) and of Tcrb and Lrig1 in DP cells to provide the background measurement of association against which the distribution in developing thymocytes could be compared.
At the earliest (DN) stage of development, when T cells express neither CD4 nor CD8, we found significantly decreased association of Cd4 and Cd8 compared to DP T cells (Figure 1C; p = 3.37e-14). As DP cells differentiate to the CD4+CD8lo transitional stage, the loci dissociated somewhat (p = 3.12e-07), but Cd4 and Cd8 achieved close association again in CD8 SP cells. In CD4 SP thymocytes, the two loci moved farther apart than in the transitional CD4+CD8lo stage and were significantly more separated compared to CD8 SP cells (p = 1.43e-10). Cd4 and Cd8 association therefore correlated well with CD8 expression, whether the cells were isolated from thymus, peripheral spleen, or lymph node (data not shown).
In the same subsets of sorted thymocytes, we examined the positions of the Cd4 and Cd8 loci relative to pericentromeric heterochromatin (PCH), a repressive subcompartment of the nucleus. This was carried out as described previously (Merkenschlager et al., 2004), with a labeled γ-satellite repeat probe to identify PCH regions. Association of Cd4 and Cd8 was scored if the signals were juxtaposed or overlapping with PCH. At the DN stage, approximately 50% of Cd4 and 30% of Cd8 alleles were associated with PCH (Figure S1D and Table S1). In DP cells, which express both proteins, ∼25% of Cd4 and ∼35% of Cd8 alleles were located at PCH. In CD4+CD8lo and CD4 SP cells, a large proportion of Cd8 alleles were repositioned to PCH (up to 70%), consistent with its diminished expression. Likewise, in CD8 SP cells, ∼65% of Cd4 alleles were repositioned to pericentromeric regions.
Taken together these data indicate that Cd4 and Cd8 associate specifically in T lineage cells (in contrast to B cells) and that the greatest degree of close interaction occurs in DP and CD8 SP cells. Furthermore, in agreement with what has been published, we found that repositioning of each locus to PCH inversely correlated with the expression of the coreceptor at that developmental stage (Delaire et al., 2004; Merkenschlager et al., 2004).
Cd4 and Cd8 Association Occurs Predominantly in cis
The association we observed between the Cd4 and Cd8 loci could be occurring either in cis (between the two alleles on the same chromosome) or in trans (between the two alleles on separate chromosomes). To determine which is the case, we measured the distance between the two chromosome territories and analyzed whether Cd4 and Cd8 were positioned closer on the same, or different, chromosomes in DP and CD8 SP sorted T cells. We used a chromosome paint that hybridizes to chromosome 6 in addition to the two BAC probes RP23-121J20 and RP23-139M18 (Figure 1D). In 40%–60% of DP or CD8 SP cells, the two chromosome 6 territories were separated by >1 μm, indicating that any interaction between Cd4 and Cd8 was occurring predominantly in cis on the same chromosome. Even when the two territories were separated by <1 μm, it was still possible to assign each Cd4 and Cd8 locus to its respective chromosome territory in most cells. In only a small population of cells (10% of DP cells and 2% of CD8 SP cells) was it difficult to determine which territory the loci belonged to and whether Cd4 and Cd8 were closer on the same or different chromosomes. Thus, interaction between Cd4 and Cd8 occurs predominantly between loci located on the same chromosome.
The E8I and E8II Enhancers Promote Cd8 Transcription and Cd4-Cd8 Association
Having established that Cd4-Cd8 association occurs in Cd8-expressing cells, we turned our attention to regulatory elements in the Cd8 locus to determine how Cd8 transcription affects the relationship between the two loci. CD8+ T cells express a heterodimer of CD8α and CD8β chains that are governed by at least five enhancer elements (E8I to E8V; Figure S2A) that drive expression of CD8α and CD8β in a developmentally regulated manner (Ellmeier et al., 1997, 1998; Hostert et al., 1997, 1998), although genetic analysis of these enhancer elements indicates overlapping and redundant roles in regulating CD8 expression. We first made use of mice lacking the E8I enhancer (Ellmeier et al., 1998), which is active in CD8 SP thymocytes but not DP cells (Ellmeier et al., 1997). The location of Cd8 enhancers within the Cd8 locus is shown in Figure S2A. DNA FISH and confocal microscopy analysis of sorted thymocyte populations from E8I-deficient mice (Figure S2B) showed wild-type levels of Cd4-Cd8 association in DP thymocytes (data not shown) but reduced association in CD8 SP cells (p = 7.19e-06 compared to wild-type CD8 SP) (Figure 2A). Consistent with this finding, Cd8 transcription was decreased in E8I-deficient CD8 SP cells but not DP cells (Figure 2B) and surface CD8 expression in all E8I-deficient thymocyte subsets was 25% lower than in wild-type (Ellmeier et al., 1998). These data are consistent with the notion that Cd8 transcription promotes association of Cd4 and Cd8 loci, but it is also possible that E8I mediates the association by recruiting factors to the Cd8 locus that promote its interaction with Cd4 in CD8 SP cells.
Figure 2.
The E8I and E8II Enhancers Promote Cd8 Transcription and Cd4-Cd8 Association
(A) Cd4-Cd8 association in wild-type and E8I-deficient CD8 SP cells (E8I Δ/Δ). Association is decreased in E8I-deficient CD8 SP compared to wild-type cells. n = 230–264 alleles.
(B) Cd8a RNA expression in wild-type and E8I-deficient DP and CD8 SP cells. Standard error bars were calculated from three independent experiments.
(C) Flow cytometry analysis of wild-type and E8IE8II double-mutant DP cells (E8I Δ/Δ E8II Δ/Δ) (TCRβintCD24+).
(D) Cd4-Cd8 association in wild-type and E8IE8II double-mutant DP cells. Statistical analyses are between specified genotypes. Association is lower in CD8+ and CD8lo E8IE8II double-mutant than in wild-type cells. Confocal microscopy sections of Cd4-Cd8 distances are representative of each genotype. Scale bars represent 1 μm. n = 204–248 alleles.
(E) RT-PCR analysis of Cd8a (top) or Cd4 (bottom) expression in wild-type and E8IE8II double-mutant DP cells. Standard error bars were calculated from two independent experiments.
(F) Cd4 or Cd8 association with pericentromeric heterochromatin in wild-type and E8IE8II double-mutant DP cells. Cd8 recruitment is higher in double-mutant than in wild-type control cells.
To explore this further, we took advantage of the variegated CD8 expression in mice doubly deficient for E8I and E8II (Ellmeier et al., 2002). These mice lose expression of CD8 in approximately one-third of their DP stage thymocytes while retaining wild-type amounts of surface expression in the remaining two-thirds (Figure 2C; Ellmeier et al., 2002). By gating on these populations, we were able to sort E8IE8II double-mutant DP cells that are CD8-expressing and E8IE8II double-mutant DP cells that have reduced CD8 expression (referred to here as CD8+ E8IE8II and CD8lo E8IE8II double-mutant DP cells, respectively; Figure S2C). DNA FISH and confocal microscopy showed decreasing amounts of Cd4-Cd8 association with reduced CD8 expression: CD8+ E8IE8II double-mutant DP cells showed significantly less Cd4-Cd8 association than did wild-type (p = 9.13e-05) and CD8lo E8IE8II double-mutant DP cells showed even lower levels of Cd4-Cd8 association (p = 3.33e-08 compared to wild-type) (Figure 2D). Consistent with these results, RT-PCR analysis demonstrated that Cd8 transcription was lower than wild-type in CD8+ E8IE8II and almost abolished in CD8lo E8IE8II double-mutant DP cells (Figure 2E).
Repositioning of Cd8 to PCH was probably affected both by deletion of these enhancer elements and by the reduction in transcription. We observed increased positioning of Cd8 to PCH in the CD8+ E8IE8II double-mutant DP cells; about 52% of the double-mutant cells had at least one allele associated with PCH in CD8+ DP, versus 35% in wild-type DP cells. This repositioning was even greater (65%) in CD8lo E8IE8II double-mutant DP cells (p = 2.60e-02 for CD8+ E8IE8II double-mutant DP compared to wild-type; p = 6.90e-07 for CD8lo E8IE8II double-mutant DP compared to wild-type; Figure 2F; Table S2). Pericentromeric localization of Cd8 correlates with epigenetic differences between CD8lo E8IE8II double-mutant DP cells and CD8+ E8IE8II double-mutant DP because the Cd8 locus in the CD8lo cells has an epigenetic “off” state (Bilic et al., 2006).
Decreased Cd8 transcription therefore correlates with diminished association of Cd4 and Cd8 and increased repositioning of Cd8 alleles to PCH. It is not clear, however, whether Cd8 transcription is a cause or a consequence of the euchromatic location of Cd8 and the increased association of Cd4-Cd8. Both of these effects could be mediated by the presence of trans-acting factors recruited to the Cd8 locus by E8I and E8II.
Cd4-Cd8 Association Requires the RUNX Binding Partner CBFβ
To determine whether trans-acting proteins are involved in mediating the Cd4-Cd8 association, we focused on members of the RUNX family, which are known to bind both the Cd4 and Cd8 loci and have an important role in governing cell fate decisions in developing T lymphocytes (Collins et al., 2009). RUNX1 and RUNX3 are expressed at different stages of T cell development: RUNX1 binds the Cd4 silencer element in DN cells to suppress Cd4 expression (Taniuchi et al., 2002a; Zou et al., 2001), and loss of RUNX1 or CBFβ (which stabilizes the interaction of RUNX proteins with DNA) or deletion of the Cd4 silencer allows Cd4 expression in DN cells (Leung et al., 2001; Taniuchi et al., 2002b). RUNX3 expression is activated at later stages of T cell development, and in addition to binding to the Cd4 silencer to prevent Cd4 derepression, it has a crucial role in activating Cd8 expression in transitional CD4+CD8lo and CD8 SP cells (Egawa et al., 2007). RUNX proteins bind along the Cd8 locus, most prominently to E8I (specifically in CD8 SP cells) and to E8II in DP cells as well as E8IV in all thymocytes (Sato et al., 2005). Thus, RUNX1 and RUNX3 simultaneously regulate expression of the Cd4 and Cd8 loci in an opposite manner.
To test the hypothesis that RUNX proteins are involved in bringing the Cd4 and Cd8 loci together, we used conditional Cbfb-deleted mice in which the gene encoding CBFβ, the requisite heterodimeric binding partner of all RUNX proteins, is conditionally deleted in all thymocytes from the DN stage onward by crossing to Lck-cre mice, which permits transition to the DP stage but results in blocked T cell development beyond the DP stage (Figure 3A; Egawa et al., 2007). Our DNA FISH and confocal analyses of sorted thymocyte populations from CBFβ-deficient mice (Figure S3) revealed that Cd4-Cd8 come into close contact at a lower frequency in DP cells from these mice (p = 5.35e-07; Figures 3B and 3C). This indicates a role for RUNX proteins in mediating association between the two loci.
Figure 3.
Cd4-Cd8 Association Requires the RUNX Binding Partner CBFβ
(A) Flow cytometry analysis of wild-type and CBFβ-deficient thymocytes (conditional CbfbF/F crossed to Lck-cre).
(B) Cd4-Cd8 association in wild-type and CBFβ-deficient DN and DP T cells, including statistical analysis. Association is lower in CBFβ-deficient DP cells than in wild-type counterparts. n = 228–264 alleles.
(C) Confocal microscopy sections of Cd4-Cd8 distances representative of wild-type or CBFβ-deficient DP cells. Scale bars represent 1 μm.
See also Figure S3.
ThPOK Inhibits the Association of Cd4 and Cd8
The zinc finger transcription factor ThPOK, which has been shown to be required for CD4+ T cell lineage commitment (He et al., 2005; Sun et al., 2005), binds to the Cd4 silencer to prevent CD4 silencing in CD4-fated thymocytes (Muroi et al., 2008). In CD4-fated cells, ThPOK expression is increased from the basal post-positive selection level, and ThPOK binds to both the Zbtb7b (gene encoding ThPOK) silencer and the Cd4 silencer, where it is thought to antagonize RUNX function and prevent the Zbtb7b and Cd4 loci from being silenced (Muroi et al., 2008; Wildt et al., 2007). ThPOK has also been implicated in repressing Cd8 expression (Jenkinson et al., 2007), and peripheral CD8+ T cells transduced with a retroviral vector expressing Zbtb7b have significantly lower Cd8 transcription than either empty vector-transduced CD8+ T cells or those transduced with a Zbtb7b retrovirus carrying the HD mutation (which leads to a defect in the generation of CD4+ T helper cells) (Figure 4A; Dave et al., 1998). Furthermore, ThPOK-deficient mice show increased expression of RUNX3 in CD4+CD8lo transitional cells (Egawa and Littman, 2008), which could exert an effect on Cd4-Cd8 association.
Figure 4.
ThPOK Inhibits Cd4-Cd8 Association
(A) Cd8a RNA expression in peripheral CD8+ T cells transduced with empty pMIGR, pMIGR.ThPOK, or pMIGR.ThPOK.HD.
(B) Flow cytometry analysis of wild-type and Zbtb7bhd/hd mature SP cells.
(C) RUNX3 staining in wild-type CD8 cells and wild-type or Zbtb7bhd/hd CD4 SP cells. Scale bars represent 1 μm.
(D) Cd4-Cd8 association in wild-type and Zbtb7bhd/hd CD4 SP cells. Association is higher in Zbtb7bhd/hd CD4 SP than in wild-type cells. n = 206–218 alleles.
(E) Cd4 recruitment to pericentromeric heterochromatin in wild-type and Zbtb7bhd/hd CD4 SP cells. Recruitment is higher in Zbtb7bhd/hd cells.
(F) RT-PCR analysis of Cd4 or Cd8a expression in wild-type and Zbtb7bhd/hd CD4 SP cells. Standard error bars were calculated from three independent experiments.
(G) Cd4-Cd8 association in wild-type and ThPOK transgenic DP cells. Cd4-Cd8 association is lower in ThPOK transgenic DP cells. n = 316–356 alleles.
We generated Zbtb7bhd/hd mice (Figures S4A–S4D), sorted the few remaining CD4 SP thymocytes (Figure 4B; Figure S4E), and used a RUNX3-specific antibody for immunofluorescence. Wild-type CD4 SP cells expressed very little RUNX3 protein compared to wild-type CD8 SP cells, but Zbtb7bhd/hd CD4 SP cells expressed levels of RUNX3 equivalent to wild-type CD8 SP cells (Figure 4C). The increase in RUNX3 expression in CD4 SP cells from Zbtb7bhd/hd mice significantly increased interaction between Cd4 and Cd8 (p = 3.15e-03 compared to wild-type CD4 SP cells) (Figure 4D). Furthermore, we observed a significant rise in the percentage of Cd4 alleles positioned at PCH (from 40% in wild-type CD4 SP cells to almost 70% in Zbtb7bhd/hd CD4 SP cells, p = 1.14e-05; Figure 4E; Table S3). In line with our previous observations, the increased frequency in Cd4-Cd8 association was accompanied by an increase in Cd8 transcription (Figure 4F). These data suggest that one of the functions of ThPOK could be to inhibit the reassociation of the Cd4 and Cd8 loci in CD4-fated CD4+CD8lo and CD4 SP thymocytes, thereby preventing Cd4 silencing.
To test the idea that ThPOK could separate Cd4 and Cd8, we compared the frequency of association of these loci in DP cells from wild-type and ThPOK transgenic mice (Sun et al., 2005). These mice express a wild-type form of the protein by using human CD2-based regulatory elements that drive expression as early as the DP stage of development (where the endogenous locus is not normally transcribed). Although the presence of the transgene does not substantially alter the total number of thymocytes, it directs cells toward the CD4 lineage and impairs CD8 development; there are virtually no CD8 SP cells in these mice (Sun et al., 2005). We found a significant decrease in the frequency of Cd4-Cd8 association in DP cells where ThPOK is prematurely expressed (p = 1.40e-08) (Figure 4G and for sort strategy see Figure S4F). Together these experiments indicate that ThPOK negatively regulates association of Cd4 and Cd8 as well as commitment to the CD8 lineage.
The Cd4 Proximal Enhancer Inhibits Cd4-Cd8 Association after Positive Selection
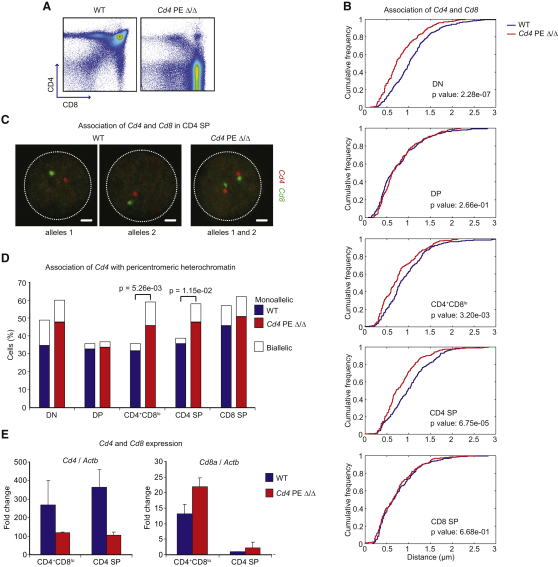
The experiments we have described above indicate that the transcription factor RUNX could bring Cd4 and Cd8 together to streamline their regulation (Schoenfelder et al., 2010). It is also possible that Cd4 and Cd8 could exert a more direct influence over each other. If so, alterations of key regulatory elements within the Cd4 locus would translate into changes in Cd8 regulation. To address this question, we made use of gene-targeted mice. Cd4 expression is regulated by a silencer element and at least one stage-specific enhancer element (Chong et al., 2010; Kioussis and Ellmeier, 2002). The proximal enhancer Cd4 PE, located 13 Kb upstream of the Cd4 start site, is absolutely required for transcription, and therefore expression, of Cd4 in DP thymocytes (Chong et al., 2010). The position of this enhancer is diagrammed in Figure S5A. After positive selection in Cd4 proximal enhancer (PE)-deficient mice, CD4-expressing single-positive thymocytes and CD4+ peripheral T cells were detected, albeit at reduced numbers, and levels of CD4 expression were comparable to wild-type mice, suggesting that one or more putative enhancer elements rescue Cd4 expression (Figure 5A and data not shown). DNA FISH and confocal microscopy analysis of sorted thymocyte populations from Cd4 PE-deficient mice (Figure S2B) revealed that the Cd4 PE, and therefore Cd4 transcription, is not required for either the Cd4-Cd8 association at the DP stage or for the repositioning away from PCH, because the degree of Cd4-Cd8 association and pericentromeric localization were comparable to wild-type (Figures 5B–5D; Table S4).
Figure 5.
The Cd4 Proximal Enhancer Inhibits Cd4-Cd8 Association
(A) Flow cytometry analysis of wild-type and Cd4 PE-deficient thymocytes (Cd4 PE Δ/Δ).
(B) Cd4-Cd8 association in wild-type and Cd4 PE-deficient cells, including statistical analysis. Association is higher in Cd4 PE-deficient DN, CD4+CD8lo, and CD4 SP cells than in wild-type cells. n = 196–286 alleles.
(C) Confocal microscopy sections of Cd4-Cd8 distances representative of each genotype. Scale bars represent 1 μm.
(D) Recruitment of Cd4 to pericentromeric heterochromatin in wild-type and Cd4 PE-deficient cells. Recruitment is higher in Cd4 PE-deficient DN, CD4+CD8lo and CD4 SP than in wild-type cells.
(E) RT-PCR analysis of Cd4 or Cd8a expression in wild-type and Cd4 PE-deficient cells. Standard error bars were calculated from three independent experiments.
In contrast to wild-type cells, however, Cd4-Cd8 association in Cd4 PE-deficient mice occurred at a higher frequency in DN cells (p = 2.28e-07 compared to wild-type controls) and remained high in both CD4+CD8lo and CD4 SP cells after positive selection (p = 3.20e-03 and p = 6.75e-05 for CD4+CD8lo and CD4 SP cells, respectively, compared to the appropriate wild-type controls) (Figure 5B). Deletion of the Cd4 PE also increased the frequency with which the Cd4 locus relocated to PCH beyond the DP stage; in both CD4+CD8lo and CD4 SP cells from Cd4 PE-deficient mice, Cd4 association with PCH reached the same levels as in CD8 SP cells (59% in Cd4 PE-deficient cells compared to 36% in wild-type CD4+CD8lo cells, p = 5.26e-03; 58% in Cd4 PE-deficient cells compared to 39% in wild-type CD4 SP cells, p = 1.15e-02; Figure 5D; Table S4). In order to correlate these positional changes with the transcriptional state of each locus, we set aside a subset of each sorted thymocyte population for real-time RT-PCR analysis. Cd4 transcription was virtually abolished in DP cells from Cd4 PE-deficient mice (data not shown) and substantially reduced in CD4+CD8lo and CD4 SP cells (Figure 5E), consistent with the surface expression of CD4 on these thymocyte subsets. These data demonstrate that Cd4 transcription is not required for Cd4 to associate with Cd8 at the DP stage and further suggest that, in the absence of robust transcription after positive selection, the Cd4 locus remains associated with the Cd8 locus. Cd4-Cd8 pairs were more frequently located at PCH, with Cd4 positioned close to these regions, whereas Cd8 remained euchromatic, i.e., the two loci were not equivalently associated with this repressive compartment. Furthermore, Cd8 expression was slightly increased in DP and CD4+CD8lo cells. These results are consistent with results in wild-type cells above: association of the two loci correlates with CD8 expression.
The Cd4 Silencer Mediates Cd4-Cd8 Association
To explore the role of the Cd4 silencer on Cd4-Cd8 association and the coordinate regulation of the two loci, we next analyzed the organization of these genes in sorted thymocyte populations from wild-type and Cd4 silencer-deficient mice. The position of the silencer within the Cd4 locus is shown in Figure S6A (Sawada et al., 1994). Germline deletion of the Cd4 silencer (sil) allows Cd4 derepression in DN thymocytes and CD8 lineage T cells (Taniuchi et al., 2002b); flow cytometry analysis shows a lack of non-CD4-expressing cells in both these populations (Figure 6A).
Figure 6.
The Cd4 Silencer Mediates Cd4-Cd8 Association
(A) Flow cytometry analysis of wild-type and Cd4 sil-deficient thymocytes (Cd4 sil Δ/Δ).
(B) Cd4-Cd8 association in wild-type and Cd4 sil-deficient cells, including statistical analysis. Association is lower in Cd4 sil-deficient DP, CD4+CD8lo, and CD8 SP than in wild-type cells. n = 210–340 alleles.
(C) Confocal microscopy sections of Cd4-Cd8 distances representative of each genotype. Scale bars represent 1 μm.
(D) Cd8 recruitment to pericentromeric heterochromatin in wild-type and Cd4 sil-deficient cells. Recruitment is higher in Cd4 sil-deficient DP and CD8 SP cells than in wild-type cells.
DNA FISH was performed on sorted thymocyte populations from wild-type and Cd4 sil-deficient mice (Figure S6B) as described previously, and cells were analyzed by confocal microscopy. Surprisingly, the Cd4-Cd8 association in Cd4 sil-deficient DP cells did not increase beyond the level observed in the DN thymocyte population and remained low at all subsequent stages of development (p = 5.48e-13 in DP cells, p = 3.51e-03 in CD4+CD8lo cells, and p = 2.62e-07 in CD8 SP cells, compared to the appropriate wild-type controls; Figure 6B). Confocal sections showing decreased association of Cd4 and Cd8 in DP and CD8 SP cells from Cd4 sil-deficient mice are shown in Figure 6C. These data indicate that the Cd4 silencer region is required to mediate the close association of Cd4 and Cd8 in both DP and CD8 SP thymocytes.
Absence of the Cd4 silencer did not appear to affect localization of Cd4 at PCH, but, surprisingly, Cd8 alleles were significantly repositioned to PCH at the DP stage (51% in Cd4 sil-deficient cells compared to 32% in wild-type DP cells, p = 5.00e-03) and subsequent stages of development, most notably in CD8 SP cells (60% in Cd4 sil-deficient cells compared to 45% in wild-type CD8 SP cells, p = 3.26e-03; Figure 6D; Table S5). Loss of a regulatory element on the Cd4 locus thus results in a long-range epigenetic effect on the Cd8 locus. Consistent with results described above, the decreased association of Cd4 and Cd8 and the increased repositioning of Cd8 to PCH correlated with slightly decreased Cd8 transcription in DP and CD8 SP cells (data not shown).
CD4 and CD8 Associate in Human Peripheral CD8+ T Cells but Not Peripheral CD4+ T Cells or B Cells
Unlike the murine loci, CD4 and CD8 are located on different chromosomes in humans. To test whether association between CD4 and CD8 is conserved between the two species despite this difference, we measured association between the CD4 and CD8 loci, which in human cells are located on chromosomes 12 and 2, respectively. If colocalization serves an important role in coordinating the expression of these coreceptors, we would expect that the two loci should be associated more frequently in CD8-expressing T cells. We performed 3D DNA FISH and confocal microscopy analysis on peripheral CD4+ and CD8+ T cells as well as on B cells sorted from human peripheral blood cells (Figure S7A). For this experiment we used two BAC probes, RP11-101F21 and CTD-2291B5, which hybridize to CD4 and CD8 on chromosomes 12 and 2 (Figure 7A).
Figure 7.
CD4 and CD8 Also Associate in Human CD8-Expressing Cells
(A) 3D DNA FISH on human sorted CD8+ T cells, CD4+ T cells, and B cells.
(B) CD4-CD8 association in B and T cells, including statistical analysis between specified cell types. Association in CD8+ T cells is increased compared to CD4+ T and B cells. n = 196–206 alleles.
See also Figure S7.
As expected (because the two loci are located on different chromosomes) CD4-CD8 interallelic distances are much larger in human cells than in murine cells. Nonetheless, we observed closer association of CD4 and CD8 in human peripheral CD8+ T cells than in either peripheral CD4+ T cells or B cells (p = 7.64e-04 and p = 1.20e-03, respectively; Figure 7B; Table S6). These data indicate that cross-talk between the Cd4 and Cd8 loci is conserved between species, underscoring the importance of this relationship in regulating expression of the two loci (Figure S7B).
Discussion
Recent studies have begun to reveal the complex interplay between nuclear organization, chromatin architecture, and gene expression. Several lines of evidence from the current study link Cd4-Cd8 association with Cd8 transcription. First, we observed that Cd4 and Cd8 closely associated only in wild-type thymocytes that express CD8 (DP and CD8 SP, but not DN, CD4+CD8lo, or CD4 SP). Second, where Cd4-Cd8 association was decreased (i.e., in CD8+ and CD8lo E8IE8II double-mutant DP cells, E8I-deficient CD8 SP cells, and Cd4 sil-deficient DP and CD8 SP cells), Cd8 transcription was also decreased to varying degrees. Third, when there was prolonged association between Cd4 and Cd8 (in Cd4 PE-deficient CD4+CD8lo cells, Cd4 PE-deficient CD4 SP cells, and Zbtb7bhd/hd CD4 SP cells), Cd8 transcription increased. Thus, the loss of a regulatory element (Cd4 PE) in one locus (Cd4) can influence the transcriptional status of a distant locus (Cd8), presumably through their physical association. Similarly, loss of the Cd4 sil on the Cd4 locus exerts an influence on the Cd8 locus, increasing the frequency with which the latter is positioned at PCH.
Beyond elucidating the genetic requirements for Cd4-Cd8 association, we also wanted to gain insight into whether transcription factors known to be involved in T cell lineage commitment could be involved in mediating the association between Cd4 and Cd8. For this, we made use of mice with a conditional CbfbF/F allele crossed to Lck-cre to delete CBFβ at the early DN stage. We found that Cd4-Cd8 association was substantially reduced in the CBFβ-deficient DP cells, despite equivalent levels of Cd4 and Cd8 transcription, suggesting that association of the two loci could occur at sites where RUNX is enriched in the nucleus. Loss of ThPOK leads to elevated expression of RUNX3 in CD4-fated thymocytes (Egawa and Littman, 2008), and overexpression of ThPOK in peripheral CD8+ T cells decreases CD8 expression. We therefore predicted that loss of ThPOK might prolong the Cd4-Cd8 association in CD4 SP cells from Zbtb7bhd/hd mice. This is indeed what we observed, indicating that binding of ThPOK to the Cd4 locus could be a mechanism for keeping the two loci separate. Furthermore, the loss of Cd4-Cd8 association in these cells was accompanied by a substantial increase in the localization of Cd4 to pericentromeric heterochromatin and a concomitant decrease in Cd4 transcription. Conversely premature expression of ThPOK in DP cells led to separation of Cd4 and Cd8.
Is association between Cd4 and Cd8 a cause or consequence of Cd8 transcription? We believe it may be both, in the same way that changes in location of loci relative to pericentromeric heterochromatin are likely to be both a cause and a consequence of changes in gene activation and repression. As with most epigenetic correlations, this is a chicken-and-egg situation and we cannot pinpoint the initiating event.
These studies allow us to put forth the following model. The Cd4 and Cd8 loci come into close proximity in DP thymocytes. After positive selection, all thymocytes pass through a CD4+CD8lo transitional stage in which Cd8 transcription decreases and it moves to pericentromeric regions, disrupting the Cd4-Cd8 association. In CD4-fated cells, ThPOK binds to the Cd4 silencer, preventing it from interacting again with the Cd8 locus. In CD8-fated cells, RUNX3 mediates the reassociation of Cd4 and Cd8 by binding to the Cd4 silencer and the Cd8 locus, predominantly within E8I. Thus, RUNX-mediated Cd4-Cd8 association silences the Cd4 locus, repositioning it to repressive pericentromeric heterochromatin.
Although it has been known for some time that chromosomal interactions can exert an effect on gene expression in trans in Drosophila (transvection) (Lewis, 1985) and possibly plants (paramutation) (Stam, 2009), there are still only a few instances in which association of alleles is known to exert epigenetic control in mammals. Two examples involve the pairing of homologous alleles: X inactivation (Bacher et al., 2006; Xu et al., 2006) and allelic exclusion (Hewitt et al., 2009). Heterologous association between different loci has been noted in developing B cells as well: one immunoglobulin light chain (Igk) allele transiently associates with one immunoglobulin heavy chain (Igh) allele at pericentromeric regions, inducing a change in nuclear location and a conformational change within the Igh locus to prevent ongoing recombination (Hewitt et al., 2008). Similarly, association of different loci has been shown to occur in T cell subsets: the Ifng locus interacts with the Il4 locus just prior to commitment to either the Th1 or Th2 cell lineage, which express either IFN-γ or IL-4, respectively. The association of Ifng and Il4 could facilitate the coordinate regulation of these loci in the differentiated CD4+ T cell subsets (Spilianakis et al., 2005) but no trans acting factors that could mediate the association have been identified. Clearly this is an underexplored area of epigenetic regulation.
Our findings add to a growing body of evidence that nuclear architecture plays a dynamic role in regulating gene expression (Fraser and Bickmore, 2007). That association of Cd4-Cd8 is conserved in both mouse and humans, despite being located on different chromosomes in the latter, underscores the importance of this mechanism for regulating CD4 and CD8 coreceptor expression. Undoubtedly, a fuller understanding of the mechanism of Cd4-Cd8 association will yield insight into how these coreceptors are regulated during T cell development and how long-range chromosomal interactions control gene expression.
Experimental Procedures
Mice
C57Bl/6 mice were purchased from Jackson Laboratories or Taconic. Cd4 PE-deficient (Chong et al., 2010), Cd4 sil-deficient (Zou et al., 2001), CbfbF/F (Naoe et al., 2007), E8I-deficient (Ellmeier et al., 1998), E8IE8II double-mutant (Ellmeier et al., 2002), Lck-cre (Lee et al., 2001), and ThPOK transgenic (Sun et al., 2005) mice have previously been described. Mice were housed in SPF conditions at the Skirball animal facility at NYU School of Medicine. Experiments were performed in accordance with approved protocols for the NYU Institutional Animal Care and Usage Committee (IACUC).
Generation of Zbtb7bhd/hd Mice
A targeting vector was created with a neomycin selection cassette and Zbtb7b exons 2 and 3 mutated at position 389 from arginine to glycine (assembled from BAC RP23-126P10) and flanked by loxP sites (Figure S4A). An XbaI site was inserted for Southern blot screening. The construct was targeted into E14 129Sv embryonic stem cells. Breeding of derived mice with EIIA-cre mice (cre expression in early embryos) gave progeny with the mutant allele. Mice were backcrossed onto C57Bl/6 for at least four generations and screened by Southern blot and PCR (Figures S4B–S4D). Primer sequences for targeting vectors and genotyping are available upon request.
Flow Cytometry Analysis and Cell Sorting
Analyses and sorting were performed on an LSRII or FACSAria, respectively (both BD Biosciences; Figures S1–S7). Antibodies to mouse antigens were as follows: CD24 FITC (clone M1/69, BD, 1:1000 dilution), CD69 PE (H1.2F3, BD, 1:100), TCRβ APC (H57-597, eBiosciences, 1:500), CD4 Alexa Fluor 700 (GK1.5, eBioscience, 1:1000), CD8α PE-Cy7 (53-6.7, eBioscience, 1:1000), CD44 PE-Cy5.5 (IM7, eBioscience, 1:500), CD25 PE-Cy7 (PC61.5, eBioscience, 1:500), and Thy1.2 FITC (CD90.2, clone 30-H12, BD, 1:500). Antibodies to human antigens were as follows: CD4 Pacific Blue (RPA-T4, BD, 1:50), CD8a FITC (RPA-T8, eBioscience, 1:50), CD3 APC-Cy7 (UCHT1, eBioscience, 1:50), CD45RA PE (HI100, eBioscience, 1:20), CD45RO APC (UCHL1, eBioscience, 1:20), and CD19 FITC (HIB19, eBioscience, 1:50).
T and B Cell Isolation from Human Peripheral Blood Mononuclear Cells
Adult PBMCs were isolated (Manel et al., 2008) and depleted of CD14+ cells (autoMACS Pro). CD4+ and CD8+ T cells and B cells were purified by flow cytometry as CD3+CD4+CD45RA+CD45RO– or CD3+CD8+CD45RA+CD45RO– and CD3–CD19+, respectively (Figure S7).
Purification of Resting Splenic B Cells
This was carried out as described (Skok et al., 2001).
Three-Dimensional DNA FISH
Sorted cells were washed in PBS, attached to glass coverslips coated with poly-L-lysine, and fixed for DNA FISH as described (Skok et al., 2001). Bacterial artificial chromosome (BAC) probes and the γ-satellite probe (Skok et al., 2001) were labeled by nick translation with ChromaTide Alexa Fluor 488-5-dUTP, 594-5-dUTP (Invitrogen), or dUTP-indodicarbocyanine (Cy5; GE Healthcare).
Cd4-Cd8 DNA FISH Combined with Chromosome 6 Paint
Cells were dropped onto poly-L-lysine coated slides, incubated in 0.075 M KCl (10 min), fixed in cold methanol/acetic acid 3:1 (2× 10 min), and dehydrated in an ethanol series. RNaseA treatment (100 μg/ml, 1 hr) and dehydration preceded denaturation (70% formamide/2× SSC, 75°C, 3 min), dehydration, and probe hybridization (overnight, 37°C, humid chamber). Slides were rinsed in 50% formamide/2× SSC (3×, 45°C) and 1× SSC (3×, 60°C) and mounted in Prolong Gold (Invitrogen) with 1.5 μg/ml DAPI. FITC-labeled chromosome 6 paint (Cambio) was prepared in 7.5 μl hybridization buffer. Cd4 and Cd8 probes were resuspended in 7.5 μl hybridization buffer. Probes were mixed just prior to hybridization.
Immunofluorescence
Cells were adhered to poly-L-lysine-coated coverslips, fixed (2% paraformaldehyde/PBS, 10 min), permeabilized (0.4% Triton/PBS, 5 min), and blocked (2.5% BSA/0.1% Tween/10% goat serum/PBS, 30 min). RUNX3 detection used a rabbit RUNX3 antibody (Egawa et al., 2007) (1:50,000 in blocking solution, 1 hr). Cells were rinsed (0.2% BSA/0.1% Tween-20/PBS) and incubated in goat anti-rabbit Alexa 488 (Invitrogen, 1:500 in blocking solution, 1 hr). Coverslips were rinsed (0.1% Tween-20/PBS) and mounted in Prolong Gold with DAPI.
Microscopy and Analysis
Optical sections of 80 nm x-y pixel size and separated by 0.3 μm were acquired by confocal laser scanning microscopy (Leica SP5, 100×/1.4 oil objective). Only cells with signals from both alleles (typically >95%) were analyzed with Leica software. At least three independent experiments were performed (n = 166 to 356 alleles for Cd4-Cd8 association, see Supplemental Tables for one representative experiment of PCH analysis). Distances between the center of the Cd4 and Cd8 signals was measured with Image J software.
The empirical interallelic distance distributions were compared to test whether they had been drawn from the same underlying continuous distribution. The statistical significance of pair-wise distributions' dissimilarity was assessed with the nonparametric two-sample Kolmogorov-Smirnov (KS) test (Massey, 1951). The reported p values were calculated with MATLAB 7.9 (The MathWorks Inc., Natick, MA).
Association of Cd4 and Cd8 with pericentromeric domains was scored if the loci signals were juxtaposed or overlapping with γ-satellite signals. Statistical significances for PCH localization were calculated with χ2 test (Campbell, 1989). Yates' correction was applied when any category had less than 10 observations. Each data set was paired with the most relevant stage, genotype, or cell type.
RT-PCR
RNA was extracted with TRIZOL (Invitrogen). Reverse transcription was performed with Superscript III (Invitrogen), cDNA analyzed in triplicate with Quantitect Multiplex PCR Mix (QIAGEN) for Taqman probes or iQ SYBR Green Supermix (BioRad) in the iCycler (BioRad), and normalized to beta-actin (Actb). Primer sequences: Actb: 5′-GCTCTGGCTCCTAGCACCAT, 3′-GCCACCGATCCACACAGAGT, probe: FAM-TCAAGATCATTGCTCCTCCTGAGCGC-TAMRA; Cd4: 5′-GACTGACCCTGAAGCAGGAG, 3′-CTGTCTGGTTCACCCCTCTG; Cd8a: 5′-CACAGGAGCCGAAAGCGT, 3′-GGGCTTGCCTTCCTGTCTG. Standard error bars were calculated from two to four independent experiments.
Acknowledgments
We would like to thank J. de Nooij and T. Jessell for RUNX3 antibody (Kramer et al., 2006). We also thank members of the J.A.S. and D.R.L. labs for thoughtful discussions and critical comments on the manuscript. This work was supported by a Leukemia and Lymphoma Scholar Award, an NIH 1R01GM086852 grant, and a WT project grant (WT 085096) (J.A.S.). A.C. and D.R.L. were supported by funds from the Howard Hughes Medical Institute and from the Helen and Martin Kimmel Center for Biology and Medicine. S.L.H. is supported by a Fellow Scholar Award from the American Society of Hematology and M.M. is supported by NSF IGERT grant 0333389. M.M.W.C. was funded sequentially by a Postdoctoral Fellowship from the Cancer Research Institute and a Senior Fellowship from the Helen and Martin Kimmel Center for Stem Cell Biology. A.E.C. and D.J.B. are supported by a BBSRC project grant (Biotechnology and Biological Sciences Research Council). Work in the lab of W.E. is supported by the Austrian Science Fund (FWF; P19930). Work in the lab of R.B. is supported by the Intramural Research Program of the National Cancer Institute, Center for Cancer Research, NIH. M.S. is an Irvington Institute Fellow of the Cancer Research Institute.
Supplemental Information
References
- Bacher C.P., Guggiari M., Brors B., Augui S., Clerc P., Avner P., Eils R., Heard E. Transient colocalization of X-inactivation centres accompanies the initiation of X inactivation. Nat. Cell Biol. 2006;8:293–299. doi: 10.1038/ncb1365. [DOI] [PubMed] [Google Scholar]
- Bilic I., Koesters C., Unger B., Sekimata M., Hertweck A., Maschek R., Wilson C.B., Ellmeier W. Negative regulation of CD8 expression via Cd8 enhancer-mediated recruitment of the zinc finger protein MAZR. Nat. Immunol. 2006;7:392–400. doi: 10.1038/ni1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K.E., Baxter J., Graf D., Merkenschlager M., Fisher A.G. Dynamic repositioning of genes in the nucleus of lymphocytes preparing for cell division. Mol. Cell. 1999;3:207–217. doi: 10.1016/s1097-2765(00)80311-1. [DOI] [PubMed] [Google Scholar]
- Campbell R. Cambridge University Press; Cambridge, UK: 1989. Statistics for Biologists. [Google Scholar]
- Chambeyron S., Bickmore W.A. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 2004;18:1119–1130. doi: 10.1101/gad.292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong M.M., Simpson N., Ciofani M., Chen G., Collins A., Littman D.R. Epigenetic propagation of CD4 expression is established by the Cd4 proximal enhancer in helper T cells. Genes Dev. 2010;24:659–669. doi: 10.1101/gad.1901610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chubb J.R., Trcek T., Shenoy S.M., Singer R.H. Transcriptional pulsing of a developmental gene. Curr. Biol. 2006;16:1018–1025. doi: 10.1016/j.cub.2006.03.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins A., Littman D.R., Taniuchi I. RUNX proteins in transcription factor networks that regulate T-cell lineage choice. Nat. Rev. Immunol. 2009;9:106–115. doi: 10.1038/nri2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dave V.P., Allman D., Keefe R., Hardy R.R., Kappes D.J. HD mice: a novel mouse mutant with a specific defect in the generation of CD4(+) T cells. Proc. Natl. Acad. Sci. USA. 1998;95:8187–8192. doi: 10.1073/pnas.95.14.8187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaire S., Huang Y.H., Chan S.W., Robey E.A. Dynamic repositioning of CD4 and CD8 genes during T cell development. J. Exp. Med. 2004;200:1427–1435. doi: 10.1084/jem.20041041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egawa T., Littman D.R. ThPOK acts late in specification of the helper T cell lineage and suppresses Runx-mediated commitment to the cytotoxic T cell lineage. Nat. Immunol. 2008;9:1131–1139. doi: 10.1038/ni.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egawa T., Tillman R.E., Naoe Y., Taniuchi I., Littman D.R. The role of the Runx transcription factors in thymocyte differentiation and in homeostasis of naive T cells. J. Exp. Med. 2007;204:1945–1957. doi: 10.1084/jem.20070133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellmeier W., Sunshine M.J., Losos K., Hatam F., Littman D.R. An enhancer that directs lineage-specific expression of CD8 in positively selected thymocytes and mature T cells. Immunity. 1997;7:537–547. doi: 10.1016/s1074-7613(00)80375-1. [DOI] [PubMed] [Google Scholar]
- Ellmeier W., Sunshine M.J., Losos K., Littman D.R. Multiple developmental stage-specific enhancers regulate CD8 expression in developing thymocytes and in thymus-independent T cells. Immunity. 1998;9:485–496. doi: 10.1016/s1074-7613(00)80632-9. [DOI] [PubMed] [Google Scholar]
- Ellmeier W., Sunshine M.J., Maschek R., Littman D.R. Combined deletion of CD8 locus cis-regulatory elements affects initiation but not maintenance of CD8 expression. Immunity. 2002;16:623–634. doi: 10.1016/s1074-7613(02)00309-6. [DOI] [PubMed] [Google Scholar]
- Fraser P., Bickmore W. Nuclear organization of the genome and the potential for gene regulation. Nature. 2007;447:413–417. doi: 10.1038/nature05916. [DOI] [PubMed] [Google Scholar]
- He X., He X., Dave V.P., Zhang Y., Hua X., Nicolas E., Xu W., Roe B.A., Kappes D.J. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature. 2005;433:826–833. doi: 10.1038/nature03338. [DOI] [PubMed] [Google Scholar]
- Hewitt S.L., Farmer D., Marszalek K., Cadera E., Liang H.E., Xu Y., Schlissel M.S., Skok J.A. Association between the Igk and Igh immunoglobulin loci mediated by the 3′ Igk enhancer induces ‘decontraction’ of the Igh locus in pre-B cells. Nat. Immunol. 2008;9:396–404. doi: 10.1038/ni1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt S.L., Yin B., Ji Y., Chaumeil J., Marszalek K., Tenthorey J., Salvagiotto G., Steinel N., Ramsey L.B., Ghysdael J. RAG-1 and ATM coordinate monoallelic recombination and nuclear positioning of immunoglobulin loci. Nat. Immunol. 2009;10:655–664. doi: 10.1038/ni.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hostert A., Tolaini M., Roderick K., Harker N., Norton T., Kioussis D. A region in the CD8 gene locus that directs expression to the mature CD8 T cell subset in transgenic mice. Immunity. 1997;7:525–536. doi: 10.1016/s1074-7613(00)80374-x. [DOI] [PubMed] [Google Scholar]
- Hostert A., Garefalaki A., Mavria G., Tolaini M., Roderick K., Norton T., Mee P.J., Tybulewicz V.L., Coles M., Kioussis D. Hierarchical interactions of control elements determine CD8alpha gene expression in subsets of thymocytes and peripheral T cells. Immunity. 1998;9:497–508. doi: 10.1016/s1074-7613(00)80633-0. [DOI] [PubMed] [Google Scholar]
- Iborra F.J., Pombo A., Jackson D.A., Cook P.R. Active RNA polymerases are localized within discrete transcription “factories” in human nuclei. J. Cell Sci. 1996;109:1427–1436. doi: 10.1242/jcs.109.6.1427. [DOI] [PubMed] [Google Scholar]
- Jenkinson S.R., Intlekofer A.M., Sun G., Feigenbaum L., Reiner S.L., Bosselut R. Expression of the transcription factor cKrox in peripheral CD8 T cells reveals substantial postthymic plasticity in CD4-CD8 lineage differentiation. J. Exp. Med. 2007;204:267–272. doi: 10.1084/jem.20061982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kioussis D., Ellmeier W. Chromatin and CD4, CD8A and CD8B gene expression during thymic differentiation. Nat. Rev. Immunol. 2002;2:909–919. doi: 10.1038/nri952. [DOI] [PubMed] [Google Scholar]
- Kramer I., Sigrist M., de Nooij J.C., Taniuchi I., Jessell T.M., Arber S. A role for Runx transcription factor signaling in dorsal root ganglion sensory neuron diversification. Neuron. 2006;49:379–393. doi: 10.1016/j.neuron.2006.01.008. [DOI] [PubMed] [Google Scholar]
- Lee P.P., Fitzpatrick D.R., Beard C., Jessup H.K., Lehar S., Makar K.W., Pérez-Melgosa M., Sweetser M.T., Schlissel M.S., Nguyen S. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 2001;15:763–774. doi: 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- Leung R.K., Thomson K., Gallimore A., Jones E., Van den Broek M., Sierro S., Alsheikhly A.R., McMichael A., Rahemtulla A. Deletion of the CD4 silencer element supports a stochastic mechanism of thymocyte lineage commitment. Nat. Immunol. 2001;2:1167–1173. doi: 10.1038/ni733. [DOI] [PubMed] [Google Scholar]
- Lewis E.B. Regulation of the genes of the bithorax complex in Drosophila. Cold Spring Harb. Symp. Quant. Biol. 1985;50:155–164. doi: 10.1101/sqb.1985.050.01.021. [DOI] [PubMed] [Google Scholar]
- Manel N., Unutmaz D., Littman D.R. The differentiation of human T(H)-17 cells requires transforming growth factor-beta and induction of the nuclear receptor RORgammat. Nat. Immunol. 2008;9:641–649. doi: 10.1038/ni.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey F.J. The Kolmogorov-Smirnov test for goodness of fit. J. Am. Stat. Assoc. 1951;253:1951. [Google Scholar]
- Merkenschlager M., Amoils S., Roldan E., Rahemtulla A., O'connor E., Fisher A.G., Brown K.E. Centromeric repositioning of coreceptor loci predicts their stable silencing and the CD4/CD8 lineage choice. J. Exp. Med. 2004;200:1437–1444. doi: 10.1084/jem.20041127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muroi S., Naoe Y., Miyamoto C., Akiyama K., Ikawa T., Masuda K., Kawamoto H., Taniuchi I. Cascading suppression of transcriptional silencers by ThPOK seals helper T cell fate. Nat. Immunol. 2008;9:1113–1121. doi: 10.1038/ni.1650. [DOI] [PubMed] [Google Scholar]
- Naoe Y., Setoguchi R., Akiyama K., Muroi S., Kuroda M., Hatam F., Littman D.R., Taniuchi I. Repression of interleukin-4 in T helper type 1 cells by Runx/Cbf beta binding to the Il4 silencer. J. Exp. Med. 2007;204:1749–1755. doi: 10.1084/jem.20062456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne C.S., Chakalova L., Brown K.E., Carter D., Horton A., Debrand E., Goyenechea B., Mitchell J.A., Lopes S., Reik W., Fraser P. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat. Genet. 2004;36:1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- Ragoczy T., Bender M.A., Telling A., Byron R., Groudine M. The locus control region is required for association of the murine beta-globin locus with engaged transcription factories during erythroid maturation. Genes Dev. 2006;20:1447–1457. doi: 10.1101/gad.1419506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roldán E., Fuxa M., Chong W., Martinez D., Novatchkova M., Busslinger M., Skok J.A. Locus ‘decontraction’ and centromeric recruitment contribute to allelic exclusion of the immunoglobulin heavy-chain gene. Nat. Immunol. 2005;6:31–41. doi: 10.1038/ni1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Ohno S., Hayashi T., Sato C., Kohu K., Satake M., Habu S. Dual functions of Runx proteins for reactivating CD8 and silencing CD4 at the commitment process into CD8 thymocytes. Immunity. 2005;22:317–328. doi: 10.1016/j.immuni.2005.01.012. [DOI] [PubMed] [Google Scholar]
- Sawada S., Scarborough J.D., Killeen N., Littman D.R. A lineage-specific transcriptional silencer regulates CD4 gene expression during T lymphocyte development. Cell. 1994;77:917–929. doi: 10.1016/0092-8674(94)90140-6. [DOI] [PubMed] [Google Scholar]
- Sayegh C.E., Jhunjhunwala S., Riblet R., Murre C. Visualization of looping involving the immunoglobulin heavy-chain locus in developing B cells. Genes Dev. 2005;19:322–327. doi: 10.1101/gad.1254305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfelder S., Sexton T., Chakalova L., Cope N.F., Horton A., Andrews S., Kurukuti S., Mitchell J.A., Umlauf D., Dimitrova D.S. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat. Genet. 2010;42:53–61. doi: 10.1038/ng.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skok J.A., Brown K.E., Azuara V., Caparros M.L., Baxter J., Takacs K., Dillon N., Gray D., Perry R.P., Merkenschlager M., Fisher A.G. Nonequivalent nuclear location of immunoglobulin alleles in B lymphocytes. Nat. Immunol. 2001;2:848–854. doi: 10.1038/ni0901-848. [DOI] [PubMed] [Google Scholar]
- Skok J.A., Gisler R., Novatchkova M., Farmer D., de Laat W., Busslinger M. Reversible contraction by looping of the Tcra and Tcrb loci in rearranging thymocytes. Nat. Immunol. 2007;8:378–387. doi: 10.1038/ni1448. [DOI] [PubMed] [Google Scholar]
- Spilianakis C.G., Lalioti M.D., Town T., Lee G.R., Flavell R.A. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435:637–645. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- Stam M. Paramutation: A heritable change in gene expression by allelic interactions in trans. Mol Plant. 2009;2:578–588. doi: 10.1093/mp/ssp020. [DOI] [PubMed] [Google Scholar]
- Stein G.S., Lian J.B., van Wijnen A.J., Stein J.L., Javed A., Montecino M., Choi J.Y., Vradii D., Zaidi S.K., Pratap J., Young D. Organization of transcriptional regulatory machinery in nuclear microenvironments: Implications for biological control and cancer. Adv. Enzyme Regul. 2007;47:242–250. doi: 10.1016/j.advenzreg.2006.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G., Liu X., Mercado P., Jenkinson S.R., Kypriotou M., Feigenbaum L., Galéra P., Bosselut R. The zinc finger protein cKrox directs CD4 lineage differentiation during intrathymic T cell positive selection. Nat. Immunol. 2005;6:373–381. doi: 10.1038/ni1183. [DOI] [PubMed] [Google Scholar]
- Taniuchi I., Osato M., Egawa T., Sunshine M.J., Bae S.C., Komori T., Ito Y., Littman D.R. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell. 2002;111:621–633. doi: 10.1016/s0092-8674(02)01111-x. [DOI] [PubMed] [Google Scholar]
- Taniuchi I., Sunshine M.J., Festenstein R., Littman D.R. Evidence for distinct CD4 silencer functions at different stages of thymocyte differentiation. Mol. Cell. 2002;10:1083–1096. doi: 10.1016/s1097-2765(02)00735-9. [DOI] [PubMed] [Google Scholar]
- Wildt K.F., Sun G., Grueter B., Fischer M., Zamisch M., Ehlers M., Bosselut R. The transcription factor Zbtb7b promotes CD4 expression by antagonizing Runx-mediated activation of the CD4 silencer. J. Immunol. 2007;179:4405–4414. doi: 10.4049/jimmunol.179.7.4405. [DOI] [PubMed] [Google Scholar]
- Xu N., Tsai C.L., Lee J.T. Transient homologous chromosome pairing marks the onset of X inactivation. Science. 2006;311:1149–1152. doi: 10.1126/science.1122984. [DOI] [PubMed] [Google Scholar]
- Zou Y.R., Sunshine M.J., Taniuchi I., Hatam F., Killeen N., Littman D.R. Epigenetic silencing of CD4 in T cells committed to the cytotoxic lineage. Nat. Genet. 2001;29:332–336. doi: 10.1038/ng750. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.