α-Synuclein impairs autophagosome formation and mislocalizes Atg9 by inhibiting Rab1a.
Abstract
Parkinson’s disease (PD) is characterized pathologically by intraneuronal inclusions called Lewy bodies, largely comprised of α-synuclein. Multiplication of the α-synuclein gene locus increases α-synuclein expression and causes PD. Thus, overexpression of wild-type α-synuclein is toxic. In this study, we demonstrate that α-synuclein overexpression impairs macroautophagy in mammalian cells and in transgenic mice. Our data show that α-synuclein compromises autophagy via Rab1a inhibition and Rab1a overexpression rescues the autophagy defect caused by α-synuclein. Inhibition of autophagy by α-synuclein overexpression or Rab1a knockdown causes mislocalization of the autophagy protein, Atg9, and decreases omegasome formation. Rab1a, α-synuclein, and Atg9 all regulate formation of the omegasome, which marks autophagosome precursors.
Introduction
Parkinson’s disease (PD) is the second most common late-onset neurodegenerative disease. The muscle rigidity, tremor, and bradykinesia that are characteristic of PD patients are thought to be caused by cell death of dopaminergic neurons found in the substantia nigra. Interestingly, the cognitive and behavioral impairments seen in PD patients indicate the degeneration of other areas of the brain outside of the substantia nigra.
PD pathology is characterized by the formation of intraneuronal inclusions called Lewy bodies, which are comprised mainly of α-synuclein. Although PD is largely a sporadic disease, familial forms have revealed α-synuclein as a causal gene. Mutations in the α-synuclein gene, including point mutations and multiplications of the entire locus, have been shown to cause familial forms of PD, whereas increased severity of PD and earlier age of onset have been reported to correlate with increased α-synuclein dosage (Ross et al., 2008). Thus, wild-type α-synuclein is a likely toxic mediator in sporadic PD and is central to understanding the molecular pathogenesis of this disease.
One potential mechanism of α-synuclein toxicity has originated from elegant studies in yeast overexpressing this protein. These studies showed that wild-type α-synuclein perturbed the secretory pathway by inhibiting the activity of Rab1. Although the studies showed that Rab1 overexpression rescued α-synuclein toxicity in animal models, they did not show that the secretory pathway was affected by this protein in mammalian cells (Cooper et al., 2006; Gitler et al., 2008). Another nonmutually exclusive mechanism is that modified forms of α-synuclein appear to affect chaperone-mediated autophagy (CMA; Cuervo et al., 2004). In CMA, a proportion of cytoplasmic proteins are directly translocated into the lysosome without involvement of vesicular intermediates or autophagosomes, macroautophagy organelles. Although CMA is a very plausible contributor to pathology, it may not be the only protein degradation pathway affected, especially because the phenotype of LAMP2A/CMA-null mice is relatively benign (Massey et al., 2006). Studies have also shown a possible connection between α-synuclein and 26S proteasome dysfunction (McNaught et al., 2001, 2003; Snyder et al., 2003). α-Synuclein exists in three common forms, monomers, dimers, and protofibrils, and it is thought that an overabundance of the protofibril form inhibits ubiquitin-proteasome activity in vitro (McNaught et al., 2001, 2003; Snyder et al., 2003; Zhang et al., 2008), although these studies remain controversial, especially in vivo (Dyllick-Brenzinger et al., 2010).
Macroautophagy is the major lysosomal pathway by which cells degrade intracytoplasmic proteins. Macroautophagy, which we will henceforth call autophagy, is distinct from CMA, as macroautophagy is responsible for nonspecific, bulk degradation of cytoplasmic contents and relies on vesicular trafficking rather than direct import of substrates into lysosomes. Autophagy initiates when cells form double layered autophagosomes around a portion of cytoplasm. Autophagosomes ultimately fuse with lysosomes where their contents are degraded. This pathway, which is conserved from yeast to man, is essential for a range of normal physiological functions. Mice defective for macroautophagy die soon after birth, and neuronal knockouts of such genes cause neurodegeneration accompanied by inclusion formation (Kuma et al., 2004; Komatsu et al., 2005). Autophagy appears to impact multiple pathways relevant to neurodegeneration, as it is a key route for the degradation of a range of intracytoplasmic aggregate-prone proteins (which are a feature of most neurodegenerative diseases) and is also a disposal route for dysfunctional mitochondria (organelles implicated in many diseases, including PD). One disease-associated autophagy substrate is mutant huntingtin (associated with Huntington’s disease [HD]), and the proportions of cells with mutant huntingtin aggregates increase when autophagy is impaired (Ravikumar et al., 2002). In this sense, the percentage of cells with mutant huntingtin aggregation can serve as a sensitive indicator of autophagic substrate clearance (Klionsky et al., 2008). Therefore, we considered that autophagy may be perturbed by α-synuclein because we had previously noticed that the percentage of cells with huntingtin aggregates is enhanced when cells also overexpress α-synuclein (Furlong et al., 2000) and that this was not caused by an association between the two proteins (Furlong et al., 2000).
Results
Overexpression of wild-type α-synuclein inhibits macroautophagy
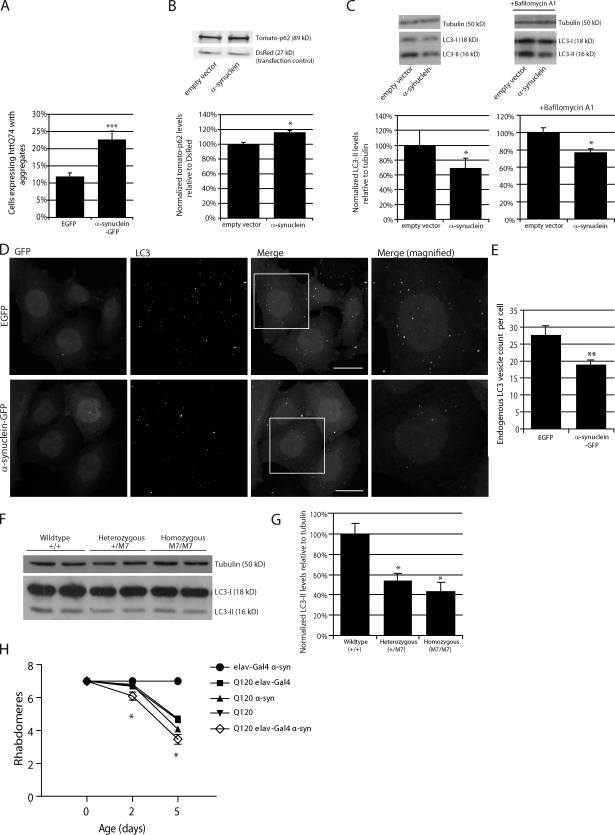
We confirmed our previous observation that α-synuclein overexpression increased the percentage of cells with huntingtin aggregates (Fig. 1 A; Furlong et al., 2000), a phenomenon which occurs when autophagy is compromised. We further observed that overexpression of α-synuclein also increased levels of another autophagy substrate, p62 (Fig. 1 B).
Figure 1.
Overexpression of α-synuclein inhibits macroautophagy. (A) Bar graph indicating the effect of α-synuclein–GFP overexpression on HA-tagged httQ74 aggregation in SKNSH, human neuroblastoma cells (odds ratio; n = 9). Note that the number of control cells with httQ74 aggregates can differ under varying experimental setups because of different cell lines, different transfection conditions (e.g., whether httQ74 is transfected along with another expression vector or siRNA), and time. Therefore, the relative change of the experimental condition is important. (B) Effect of α-synuclein overexpression on tomato-tagged p62. Tomato-p62 and DsRed (1.5:1) were transfected for the last 24 h of the experiment at low levels to observe the effect of α-synuclein overexpression on p62. DsRed was used as a transfection control for tomato-p62 (two-tailed Student’s t test; n = 3). (C) Effect of α-synuclein overexpression on LC3-II levels treated with and without bafilomycin A1. Tubulin levels demonstrate equal loading. Densitometry for a representative dataset (n = 3) with and without bafilomycin are shown (two-tailed Student’s t test). (D) Immunostaining of endogenous LC3-positive puncta in SKNSH cells overexpressing α-synuclein–GFP. The boxes in the third column are magnified in column four to show more detail of the differences in autophagosome number at high magnification. Images shown are z-stack projections. Bars, 20 µm. (E) Bar graph indicating the effect of α-synuclein overexpression on LC3 vesicle number (two-tailed Student’s t test; n = 20). (F) LC3-II levels assessed by SDS-PAGE in right hemisphere brain lysates from wild-type mice (+,+) and mice heterozygous (+/M7) and homozygous (M7/M7) for the α-synuclein transgene (M7). Representative samples are shown. (G) Quantification of LC3-II levels from F relative to tubulin by densitometry (one-way ANOVA and Bonferroni post hoc test; n = 3). (A–C, E, and G) Error bars represent SEM (*, P < 0.05; **, P < 0.01; ***, P < 0.001). (H) Neuronal expression of α-synuclein (α-syn; UAS-α-synuclein) using the elav-Gal4 driver (elav-Gal4; elavC155) significantly increased neurodegeneration of flies expressing mutant huntingtin exon-1 (Q120; gmr-Htt(exon1) Q120) in eyes (*, P < 0.02 for comparisons to all controls; paired Student’s t test; n = 5 independent experiments each based on ∼15 ommatidia from each of 10 individuals). Error bars represent SEM.
The standard means of assaying autophagy directly is to measure the levels of autophagosome-associated LC3. Processing of microtubule-associated protein 1 light chain 3B, also known as LC3, creates two forms of LC3 identifiable by immunoblotting. Initially, the LC3 precursor is cleaved to form LC3-I, a cytoplasmic, active form of the protein that does not associate with autophagosomes. Further lipidation of LC3-I creates the autophagosome-associated form, LC3-II, which is known to be a robust marker of autophagosomes (Kabeya et al., 2000). Although LC3-I can be detected by Western blotting, it is known to be largely unreliable as an indicator of autophagy because of both the instability of the protein in cell lysates and its nonassociation with autophagosomes. LC3-II levels (as a function of actin or tubulin) correlate with autophagosome number; the consensus statement on autophagy methodology is that LC3-II should be related to actin/tubulin loading controls and not to LC3-I (see Klionsky et al. [2008] for a detailed discussion; Rubinsztein et al., 2009). Alternatively, immunofluorescent staining of LC3 can be used to assay autophagosome formation.
LC3-II levels (Fig. 1 C) and the number of LC3 vesicles (Fig. 1, D and E) were decreased in cells overexpressing wild-type α-synuclein, compared with cells transfected with empty vector, and this was specific to wild-type α-synuclein as the PD-associated α-synuclein mutants A53T and A30P had no effect on LC3-II levels (Fig. S1 A). Although there was a tendency for decreased vesicle size with both α-synuclein overexpression and Rab1a knockdown (see Rab1a regulates authophagy), this was not statistically significant but was always accompanied by a significant decrease in the number of vesicles per cell. LC3-II levels can change either because of changes in the rate of LC3-II synthesis or changes in the rate of LC3-II degradation. These scenarios can be deconvoluted by assaying LC3-II levels in the presence of saturating levels of bafilomycin A1, which blocks LC3-II degradation and allows for specific assessment of LC3-II formation rates (as described in detail in Rubinsztein et al. [2009]). We confirmed that the decrease in LC3-II levels caused by α-synuclein was caused by impaired autophagosome synthesis, as LC3-II levels were also decreased in cells overexpressing α-synuclein when the cells were treated with bafilomycin A1 (Fig. 1 C). Consistent with the data, knockdown of α-synuclein increased LC3-II levels in the presence and absence of bafilomycin A1 (Fig. S1 B) and reduced the percentage of cells with huntingtin aggregates (Fig. S1 C; we used HeLa cells in this experiment as they allow more efficient siRNA silencing compared with neuronal cells). LC3-II levels were also decreased in the brains of mice overexpressing α-synuclein, suggesting that the effects we see in cell culture are present in vivo (Fig. 1, F and G). Our data suggested that the effect of the α-synuclein transgene on LC3-II levels was dose dependent in the mouse brains. We fitted a dose-dependent regression model for the effects of transgene dose (0, 1, or 2) on LC3-II levels and found statistical evidence supporting an association (P = 5.35 × 10−3). To allow for the small number of samples analyzed, we also performed a permutation test, which confirmed the association P = 0.89 × 10−3 (789/100,000 permutations had a larger test statistic than observed in the original test). It is not possible to assess whether the decrease in autophagosome numbers seen in the α-synuclein mouse brains is caused by decreased formation or increased delivery of autophagosomes to lysosomes. However, the decrease we see in LC3-II levels in the mouse brain is consistent with the definitive decrease in autophagosome formation we observe in cells with overexpression of α-synuclein.
The compound eye of Drosophila melanogaster consists of many ommatidia, each composed of eight photoreceptor neurons with light-gathering parts called rhabdomeres, seven of which can be visualized by light microscopy using the pseudopupil technique (Franceschini and Kirschfeld, 1971). Fly photoreceptors that express a mutant huntingtin fragment with 120 polyglutamine repeats exhibit photoreceptor degeneration that is not observed in flies that express the wild-type fragment with 23 polyglutamine repeats (Jackson et al., 1998). Neurodegeneration in HD flies is progressive and is associated with a decrease in the number of visible rhabdomeres in each ommatidium over time (Jackson et al., 1998). This fly model can be used to assay modulators of neurodegeneration. When autophagy is impaired, cells accumulate aggregate-prone, intracytoplasmic proteins and become more susceptible to apoptotic insults (Ravikumar et al., 2006). We have previously shown that treatment of mutant huntingtin-expressing flies with genetic or chemical inhibitors of autophagy increases neurodegeneration within the fly eye (Ravikumar et al., 2005, 2008). If α-synuclein inhibits autophagy, neurons expressing mutant huntingtin, as a model aggregate-prone toxic protein, would be predicted to degenerate more rapidly upon coexpression with α-synuclein. Consistent with our hypothesis, the toxicity of the HD mutation was enhanced by the presence of wild-type α-synuclein (Fig. 1 H). Although we cannot exclude that α-synuclein is also enhancing mutant huntingtin toxicity via autophagy-independent pathways, our cell biology data suggest that the effects on autophagy are significant contributors.
Overexpression of wild-type α-synuclein inhibits secretion in mammalian cells
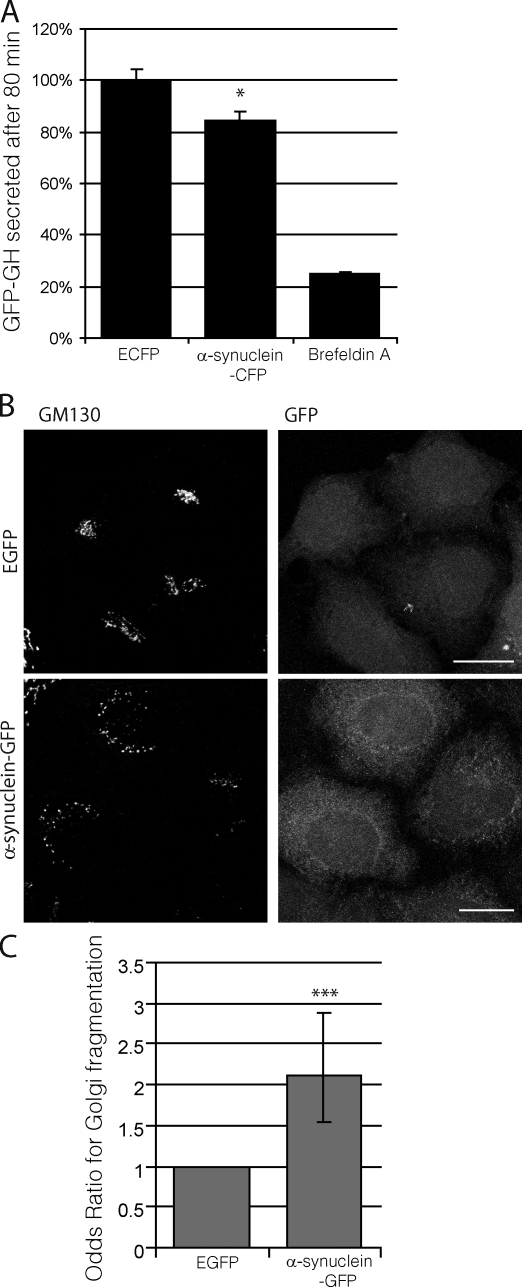
Early studies in yeast identified a potential dependence of autophagy on proper functioning of the secretory pathway (Ishihara et al., 2001; Hamasaki et al., 2003; Reggiori et al., 2004). Given the results of previous studies showing that α-synuclein inhibits secretion in yeast and in vitro by disrupting Rab1a homeostasis (Cooper et al., 2006; Gitler et al., 2008), we considered the possibility that defects in the secretory pathway caused by α-synuclein at the levels of ER to Golgi transport may underlie the autophagy defect we have observed in mammalian cells. To study the effect of α-synuclein on ER to Golgi transport in mammalian cells, we used a pharmacologically controlled secretion system that is able to sensitively measure changes in constitutive secretion. This assay showed that overexpression of wild-type α-synuclein caused a modest but reproducible inhibition of constitutive secretion in mammalian cells (Fig. 2 A). Cells overexpressing α-synuclein also showed increased Golgi fragmentation (Fig. 2, B and C), a phenotype generally caused by secretory defects (e.g., with Rab1a siRNA knockdown; Fig. S2 A). Consistent with these findings, increased Golgi fragmentation has been observed in postmortem PD brain sections in comparison with healthy controls (Fujita et al., 2006).
Figure 2.
Overexpression of α-synuclein inhibits secretion and causes Golgi fragmentation in mammalian cells. (A) Bar graph indicating the effect of α-synuclein–CFP overexpression on constitutive secretion in a HeLa-derived reporter cell line (C1). Brefeldin A, a known inhibitor of constitutive secretion, was used as a positive control (*, P < 0.05; two-tailed Student’s t test; n = 3). GH, growth hormone. (B) Immunostaining of endogenous GM130 (cis-Golgi marker) in HeLa cells overexpressing α-synuclein–GFP. Images are z-stack projections. Bars, 20 µm. (C) Quantification of α-synuclein effect on Golgi fragmentation (***, P < 0.001; odds ratio; n = 6). (A and C) Error bars represent SEM.
Rab1a regulates autophagy
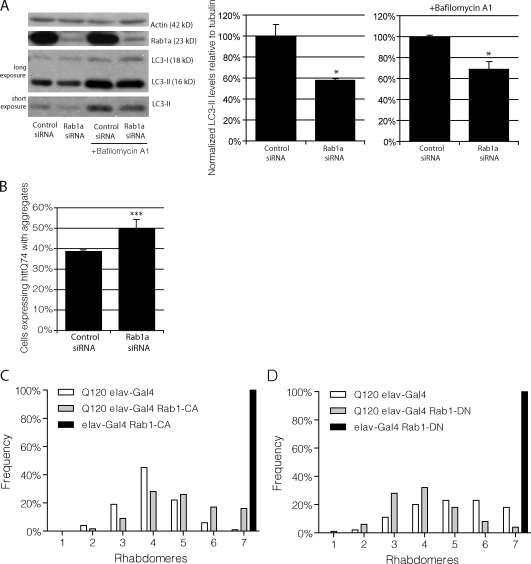
We tested whether Rab1a knockdown regulated autophagy. As there is no known specific inhibitor of Rab1a activity, we used knockdown experiments to simulate loss of Rab1a function. siRNA knockdown of Rab1a decreased LC3-II levels in the presence and absence of bafilomycin A1 (Fig. 3 A). Rab1a knockdown increased the percentage of cells with mutant huntingtin aggregates (Fig. 3 B). A constitutively active form of Rab1 suppressed mutant huntingtin toxicity in Drosophila photoreceptors (Fig. 3 C), whereas a dominant-negative Rab1 enhanced the toxicity of mutant huntingtin fragments (Fig. 3 D). Thus, Rab1a deficiency regulates autophagy and mimics the effects of α-synuclein overexpression on autophagy and related phenotypes. It is important to note that overexpression of α-synuclein does not affect Rab1a protein levels in either cells or mice brain lysates, which is consistent with an effect of α-synuclein on Rab1a activity rather than protein levels (Fig. S2 B).
Figure 3.
Knockdown of Rab1a inhibits macroautophagy. (A) LC3-II levels were assessed by SDS-PAGE after knockdown of Rab1a and treatment with bafilomycin A1 in HeLa cells. Actin levels demonstrate equal loading. Densitometry for a representative dataset (n = 3) with and without bafilomycin are shown (*, P < 0.05; Student’s t test). (B) Bar graph indicating the effect of Rab1a knockdown, as in A, on aggregation of httQ74-HA (***, P < 0.001; odds ratio; n = 9). (A and B) Error bars represent SEM. (C) Neuronal expression of constitutively active Rab1 (Rab1-CA; UASp-YFP.Rab1.Q70L) using the elav-Gal4 driver significantly suppressed neurodegeneration of 5-d-old flies expressing mutant huntingtin exon-1 (Q120; 5.0 ± 0.12 and 4.1 ± 0.06 rhabdomeres, in the presence and absence of Rab1-CA, respectively; P ≤ 0.005, by Student’s t test). Each normal fly eye contains hundreds of ommatidia with seven visible rhabdomeres. The graphs show the distributions of the numbers of rhabdomeres per ommatidium for each of the genotypes. Comparisons were performed using the paired Student’s t tests using data from five to seven independent experiments, each based on ∼10 individuals of each genotype, in which 15 ommatidia each were scored. (D) Neuronal expression of dominant-negative Rab1 (Rab1-DN; UAST-YFP.Rab1.S25N) using the elav-Gal4 driver significantly enhanced neurodegeneration of 5-d-old flies expressing mutant huntingtin exon-1 (Q120; 4.1 ± 0.1 and 5.2 ± 0.1 rhabdomeres, in the presence and absence of Rab1-DN, respectively; P < 0.02, by Student’s t test). Note that the different levels of neurodegeneration in the Q120 elav-Gal4 flies between C and D is likely because these were independent experiments, in which the duration was not identical.
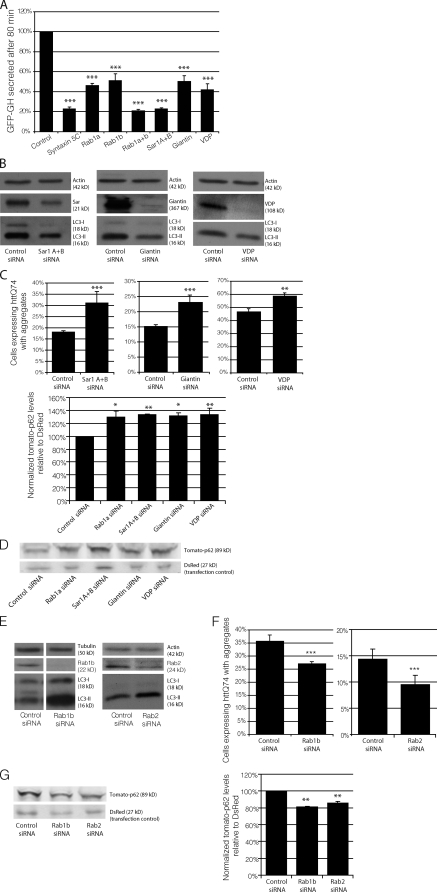
Given the role of Rab1a in secretion, we wanted to determine whether Rab1a regulates autophagy independently of general secretion or whether autophagy does in fact depend on proper functioning of the secretory pathway, as implicated in early yeast studies (Ishihara et al., 2001; Hamasaki et al., 2003; Reggiori et al., 2004). Understanding this difference may provide clues as to whether the autophagy defect caused by α-synuclein is a generic consequence of the secretory defect or whether Rab1a inhibition of autophagy can be dissociated from a general secretory defect, suggesting a specific role for Rab1a in autophagy regulation. To begin to address this issue, we used RNAi to decrease protein levels of Rab1a effectors (Giantin and VDP [vesicle-docking protein 115]), early secretory proteins critical for Rab1a secretion (Sar1A and -B), and Rab1b, an alternative isoform of Rab1, also involved in early secretion. After knockdown of these proteins, we analyzed the effects of their depletion on both secretion and autophagy to more closely look at the interplay of these two pathways. Knockdown of Rab1a, VDP, Giantin, and Sar1A and -B all partially inhibited constitutive secretion (Fig. 4 A), as measured by the percentage of protein secreted 80 min after secretion induction. Rab1b (which appears to traffic different subsets of vesicles to Rab1a; Allan et al., 2000; Alvarez et al., 2003) inhibited secretion to a similar extent as Rab1a, Giantin, and VDP. Like Rab1a, knockdown of known Rab1a effectors Giantin and VDP and Sar1A and -B decreased LC3-II levels (Fig. 4 B), increased the percentage of cells with huntingtin aggregates (Fig. 4 C), and increased p62 accumulation (Fig. 4 D). Unexpectedly, knockdown of Rab1b had the opposite effect on autophagy. Rab1b RNAi increased LC3-II (Fig. 4 E) levels, decreased the percentage of cells with huntingtin aggregates (Fig. 4 F), and decreased accumulation of p62 (Fig. 4 G).
Figure 4.
Autophagy is not dependent on all Rab proteins that regulate secretion. (A) To measure the effect of RNAi of Rab1a, Rab1b, Sar1A/B, Giantin, and VDP on constitutive secretion, HeLa-derived C1 cells were transfected with siRNA corresponding to these genes. The amount of GFP–growth hormone remaining in the cells 80 min after induction of secretion was determined using flow cytometry. As a positive control, the cells were transfected with STX5 (syntaxin5) siRNA, a known inhibitor of secretion (n = 3 independent experiments; ***, P < 0.001; multiple comparisons, one-way ANOVA, Dunnett’s multiple comparisons post hoc test). GH, growth hormone. (B) LC3-II levels were assessed by SDS-PAGE after knockdown of Sar1A and -B, Giantin, or VDP in HeLa cells. Actin levels demonstrate equal loading. (C) Bar graphs indicating the effect of knockdown, as in B, on httQ74 aggregation (**, P< 0.01; ***, P < 0.001; odds ratio; n = 9). (D) Tomato-p62 and DsRed were transfected into HeLa cells during the last 24 h of knockdown, as in B, and the effects were measured by SDS-PAGE. DsRed was used as a transfection control for tomato-p62. Quantification of tomato-p62 levels relative to DsRed is shown in the bar graph (*, P < 0.05; **, P < 0.01; Dunnett’s multiple comparison post hoc test; n = 3). Values were normalized to control values for separate experiments. (E) LC3-II levels were assessed by SDS-PAGE after knockdown of Rab1b (bands shown are from the same gel, and unrelated lanes have been omitted) or Rab2. Actin and tubulin levels demonstrate equal loading. (F) Bar graphs indicating the effect of knockdown, as in E, on the percentage of httQ74 aggregation in transfected cells (***, P < 0.001; odds ratio; n = 9). (G) Tomato-p62 and DsRed were transfected into HeLa cells during the last 24 h of knockdown, as in E, and the effects were measured by SDS-PAGE. DsRed was used as a transfection control for tomato-p62. Quantification of tomato-p62 levels relative to DsRed is shown in the bar graph (**, P < 0.01; Bonferroni’s multiple comparison post hoc test; n = 3). Values were normalized to control values for separate experiments. (A, C, D, F, and G) Error bars represent SEM.
Despite what little is known about differing functions of Rab1a and Rab1b, this clear divergence in their roles in the context of autophagy was surprising. This led us to test another functionally related Rab, Rab2. Rab2, even less characterized than Rab1b, has been shown to play a role in almost all aspects of ER–Golgi and intra-Golgi transport (Tisdale, 1999). Like Rab1b, knockdown of Rab2 increased steady-state LC3-II (Fig. 4, D and E) levels, decreased the proportion of cells with huntingtin aggregates (Fig. 4 F), and decreased p62 levels (Fig. 4 G). Although knockdown of Rab1b inhibited secretion, similar to Rab1a, it actually decreased aggregation of autophagy substrates. Interestingly, the evidence supporting an independence of autophagy function from secretory function was further corroborated by chemical data showing that the well-known autophagy inducer, trehalose (Sarkar et al., 2007), caused a partial block in secretion (Fig. S2 C). Collectively, these data provide strong evidence for a specific Rab1a-related role in the regulation of autophagy, which is independent of any effects of general secretion on autophagy. These data suggest that certain secretory routes can be impaired without decreasing autophagosome synthesis, whereas pathways regulated specifically by Rab1a and its effectors are critical for autophagy.
Rab1a is able to rescue the inhibitory effect of α-synuclein on autophagy
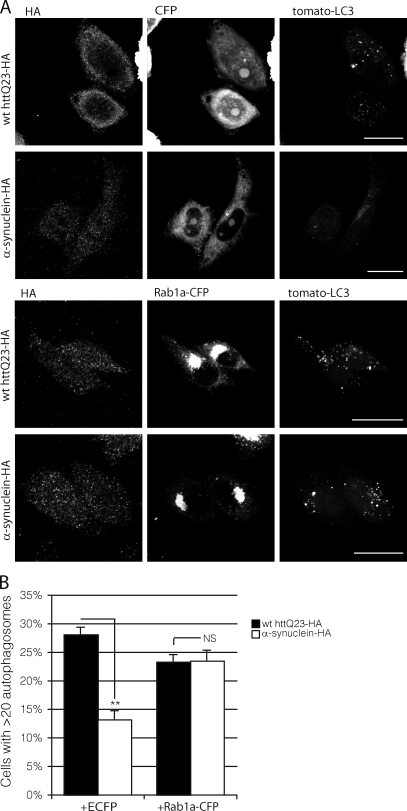
To further confirm the hypothesis that α-synuclein acts on autophagy through Rab1a, we tested and confirmed that overexpression of Rab1a rescued the loss of autophagosomes caused by α-synuclein overexpression. Overexpression of α-synuclein significantly reduced vesicle number when compared with control conditions. Coexpression of Rab1a-CFP rescued this decrease in vesicle count back to control levels (Fig. 5, A and B).
Figure 5.
Rab1a can rescue the effect of α-synuclein on macroautophagy. (A) The effect of cells expressing α-synuclein–HA with empty CFP vector (control conditions) or Rab1a-CFP (rescue conditions) on tomato-LC3 vesicles in SKNSH cells. HA-tagged wild-type (wt) huntingtin exon-1 fragment with 23 polyglutamine repeats (httQ23) was used as a control for cells expressing α-synuclein–HA in both control and rescue conditions. We have used a cut-off of 20 vesicles per cell in these experiments because the transient transfection of LC3 increases the baseline levels of vesicles (compared with endogenous LC3), and 20 vesicles is the mean observed under wild-type conditions in these cells. Bars, 20 µm. (B) Quantification of the effect of Rab1a-CFP on α-synuclein–dependent reduction of LC3 levels (**, P < 0.01; two-tailed Student’s t test; n = 3). Error bars represent SEM.
α-Synuclein and Rab1a affect the localization of the autophagy protein Atg9
We have shown that α-synuclein causes secretory and Golgi defects, consistent with Rab1a homeostasis disruption, α-synuclein and Rab1a cause similar and specific defects in autophagosome synthesis, and Rab1a can rescue the inhibitory effects of α-synuclein on autophagy. This suggests that α-synuclein inhibits autophagy via Rab1a. To identify a potential mechanism for the effects of α-synuclein and Rab1a on autophagy, we analyzed steps critical to autophagosome synthesis. Neither Rab1a knockdown nor α-synuclein overexpression affected key players in the autophagy signaling pathway such as mTOR (mammalian target of rapamycin) activity (Fig. S3 A).
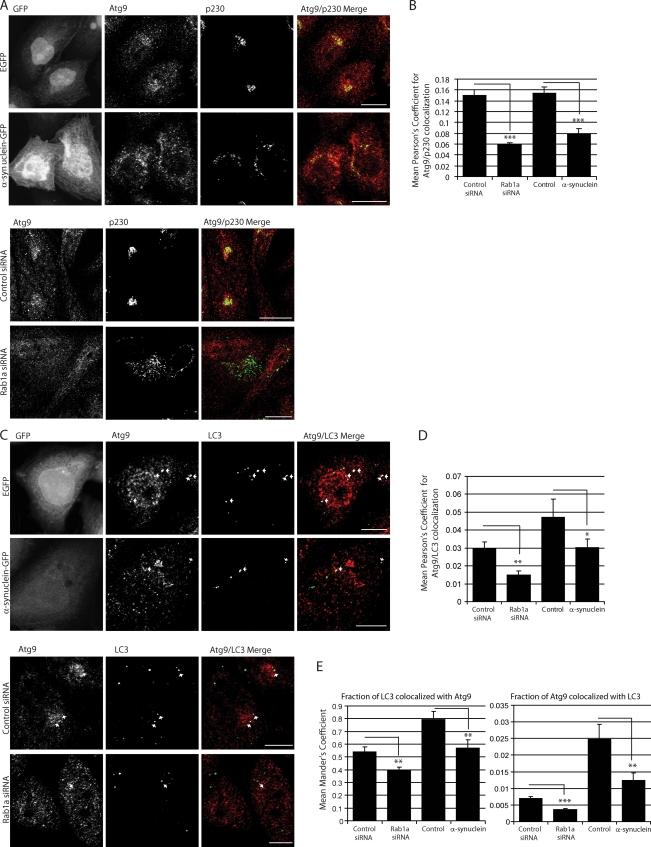
We focused next on Atg9, as it is one of the only known transmembrane autophagy proteins (implicating the secretory pathway in its transport and glycosylation) and is required for autophagosome biogenesis. Atg9 localizes to the TGN and colocalizes with a portion of LC3-positive autophagosomes. Induction of autophagy with rapamycin or starvation causes Atg9 to move away from the TGN (Fig. S3 B) to autophagosomes, causing a decrease in Atg9 colocalization with TGN markers and an increase in Atg9 colocalization with autophagosomes (Fig. S3 C; Young et al., 2006). Rab1a knockdown and α-synuclein overexpression did not overtly affect the glycosylation of Atg9 (Fig. S4 A). However, both perturbations caused Atg9 to relocate from its normal perinuclear location, where it colocalizes with the TGN marker p230, to a diffuse distribution in cells (Fig. 6 A and Fig. S4 B). This mislocalization, in cells overexpressing α-synuclein or where Rab1a was knocked down, also manifested as a lower proportion of LC3 vesicles colocalizing with Atg9 and fewer Atg9 structures colocalizing with LC3 (after controlling for decreased LC3 vesicle number through the use of Mander’s coefficients; Fig. 6, C–E). This mislocalization is not a general characteristic of autophagy inhibitors, as the normal perinuclear localization of Atg9 is unaltered in cells treated with LY290042 (a phosphatidylinositol 3-kinase inhibitor; Young et al., 2006), 3-methyladenine, or bafilomycin A1, which are all unlike rapamycin (shown as a positive control for Atg9 movement; Fig. S4 C). In addition, this characteristic alteration in Atg9 colocalization is not seen after knockdown of Rab1b nor Rab2, both of which increased the colocalization of Atg9 and LC3 (Fig. S4, D and E) and decreased Atg9 colocalization with p230. This data are in agreement with our earlier findings (Fig. 4, E–G) showing that Rab1b and Rab2 knockdown increases autophagy. Collectively, these data suggest that not only are the effects of Rab1a knockdown and α-synuclein overexpression similar and relatively specific, but that the effect of Rab1a knockdown on autophagy is not simply caused by generic disturbances in constitutive secretion or Golgi structure, as indicated by the different effects of Rab1b and Rab2.
Figure 6.
α-Synuclein and Rab1a affect the colocalization of Atg9 and LC3 vesicles. (A) Endogenous immunostaining of Atg9 and p230 in SKNSH cells with α-synuclein overexpression or Rab1a knockdown. Empty GFP vector (EGFP) was used as a control for cells expressing α-synuclein–GFP. Images shown are z-stack projections. The Atg9 panels show a clear dispersal of the perinuclear localization with α-synuclein overexpression or Rab1a knockdown. Golgi fragmentation can be seen in the p230 columns. α-Synuclein overexpression causes an increase in the disorganization of the Golgi structure, a phenotype which is amplified with Rab1a knockdown. (Magnifications of these images have been deposited in the JCB DataViewer.) (B) Quantification of Atg9 colocalization with p230 (Pearson’s coefficient; ***, P < 0.001; two-tailed Student’s t test; n = 10 from one representative experiment). (C) Immunostaining of endogenous Atg9 and LC3 in cells with α-synuclein overexpression or Rab1a knockdown. Empty GFP vector (GFP) was used as a control for cells expressing α-synuclein–GFP. Colocalization is highlighted in white on merged panel (image generated by ImageJ Colocalization plugin). Images are single confocal planes to more precisely determine colocalization. Arrows denote colocalization events. (Magnifications of these images have been deposited in the JCB DataViewer.) (D and E) Quantification of Atg9 colocalization with LC3 (Pearson’s [D] and Mander’s coefficient [E]). The Mander’s coefficient shown in the first bar graph in E more accurately predicts Atg9/LC3 colocalization when LC3 vesicle number is reduced by Rab1a knockdown or α-synuclein overexpression (*, P < 0.05; **, P < 0.01; ***, P < 0.001; two-tailed Student’s t test; n = 10). (B, D, and E) Error bars represent SEM. See related Fig. S3. Bars: (A) 20 µm; (C) 10 µm.
α-Synuclein, Rab1a, and Atg9 affect omegasome formation via Atg9
Because very little is known about Atg9 regulation of autophagosome synthesis, we looked more specifically at the effects of α-synuclein and Rab1a on the formation of the omegasome. The omegasome is a newly characterized marker that is thought to be the precursor to the forming autophagosome. Cells expressing GFP-tagged double FYVE domain–containing protein (DFCP1) allow for the visualization of forming omegasomes. The double FYVE domain present on DFCP1 binds to phosphatidylinositol-3-phosphate (PI(3)P) at the proposed site of formation of ER-associated nascent autophagosomes (Axe et al., 2008). PI(3)P plays a critical role in autophagy, and inhibition of PI(3)P synthesis blocks autophagosome formation (Kihara et al., 2001; Suzuki et al., 2007). DFCP1-GFP–positive vesicles, called omegasomes, increase after treatment with known autophagy inducers and have been shown to colocalize with LC3 and Atg5. Thus, cells expressing GFP-labeled DFCP1 allow for the visualization of early events in autophagosome formation (Axe et al., 2008).
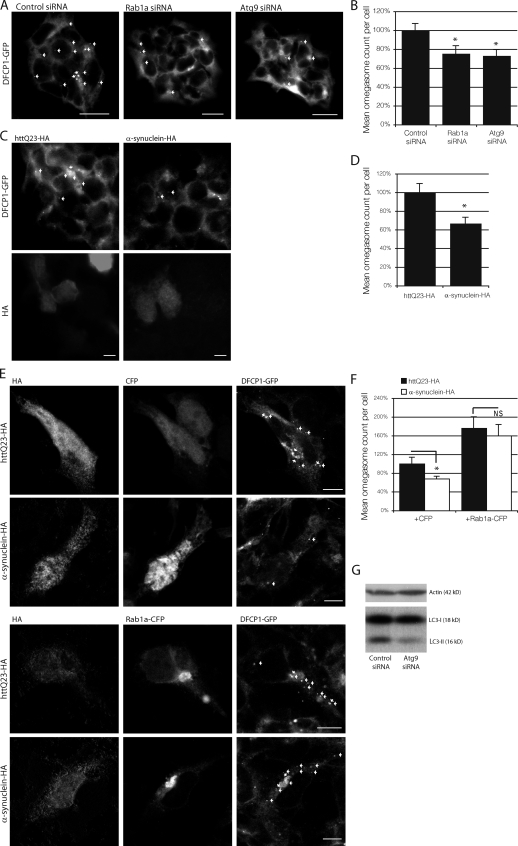
Knockdown of Rab1a and overexpression of α-synuclein reduced omegasome vesicle count under basal conditions (Fig. 7, A–D). Overexpression of Rab1a was able to rescue the deficiency in omegasome formation caused by α-synuclein overexpression and increase omegasome levels to nearly that of the control condition (Fig. 7, E and F). Interestingly, Rab1a overexpression can increase omegasome count above that of the normal basal condition but is unable to increase LC3 vesicle formation above basel levels in α-synuclein–expressing cells. The transition of the omegasome to a functional LC3-positive autophagosome may be a potential rate-limiting step that cannot be overcome simply with an increase in omegasome formation.
Figure 7.
α-Synuclein, Rab1a, and Atg9 affect omegasome and autophagosome formation. (A) The effect of Rab1a and Atg9 knockdown on omegasome formation in DFCP1-GFP HEK293, human embryonic kidney cells. (B) Quantification of A (*, P < 0.05; one-way ANOVA, Dunnett’s multiple comparison post hoc test; n = 10). (C) The effect of α-synuclein on omegasome formation in DFCP1-GFP HEK293 cells. httQ23-HA was used as a control for cells expressing α-synuclein–HA. (A and C) Arrows mark omegasomes. Images shown are single-plane images. (D) Quantification of C (*, P < 0.05; two-tailed Student’s t test; n = 10). (E) The effect of cells expressing α-synuclein–HA with empty CFP vector (control conditions) or Rab1a-CFP (rescue conditions) on omegasome formation. HA-tagged wild-type huntingtin exon-1 fragment with 23 polyglutamine repeats (httQ23) was used as a control for cells expressing α-synuclein–HA in both control and rescue conditions. Z-stack projections are shown. Arrows denote omegasomes. (F) Quantification of E (*, P < 0.05; two-tailed Student’s t test; n = 10). (B, D, and F) Error bars represent SEM. (G) Effect of Atg9 knockdown on LC3-II levels in SKNSH cell lysates. Actin was used to demonstrate equal loading. Bars: (A and C) 20 µm; (E) 10 µm.
Because α-synuclein and Rab1a appear to affect autophagy through Atg9 mislocalization, we also tested the effects of Atg9 knockdown on omegasome formation to determine whether Atg9 acts upstream of omegasome formation. Like Rab1a knockdown and α-synuclein overexpression, knockdown of Atg9 decreased omegasome and autophagosome formation under basal conditions, indicating that Atg9 acts upstream of these processes (Fig. 7, A, B, and G). Thus, these data are compatible with a model whereby α-synuclein impairs Rab1a activity, which, in turn, perturbs Atg9 function to reduce autophagosome formation at a very early point in biogenesis.
Discussion
Our data show that overexpression of α-synuclein impairs macroautophagy. This provides a mechanism whereby copy-number mutations of α-synuclein contribute to PD. Because sporadic PD is also associated with α-synuclein accumulation, our data may have much wider implications.
Our findings appear specific to wild-type α-synuclein, as the PD-associated point mutants A53T and A30P had no effect on LC3-II levels (Fig. S1 A). This may be because either A53T and A30P have no effects or have different effects on autophagy compared with the wild-type protein. Alternatively, these mutants do impair autophagy in a similar way to the wild-type protein, but their effects are masked by their known inhibition of CMA (Cuervo et al., 2004), which leads to a compensatory increase in autophagosome formation.
We have found that overexpression of wild-type α-synuclein in vitro and in vivo inhibits autophagosome synthesis, as determined by a range of different specific assays. These assays reveal a decrease in omegasome formation, a decrease in LC3-II lipidation, impaired LC3 vesicle formation, and accumulation of autophagy substrates (mutant huntingtin and p62). When viewed in the context of previous data showing that α-synuclein affects Rab1a function (Cooper et al., 2006), we were interested in determining whether α-synuclein acted via Rab1a on autophagy. We found that knockdown of Rab1a mimicked the effects seen by α-synuclein overexpression, by decreasing omegasome formation, decreasing LC3-II levels, and increasing autophagy substrates. Importantly, Rab1a expression was able to rescue the inhibitory effects of α-synuclein on both omegasome and LC3 vesicle formation.
We believe that α-synuclein and Rab1a exert their control on autophagy via a very early step in the initiation of autophagosome synthesis, as Atg9 was found to be mislocalized with α-synuclein overexpression and Rab1a knockdown. Atg9 knockdown itself reduced both omegasome formation and LC3-II levels, indicating that Atg9 controls a step in autophagosome formation upstream of omegasome formation.
These findings are important for understanding PD molecular pathogenesis and further understanding the regulation of autophagy. Our data identify a new role for Rab1a in the regulation of autophagy. The effects we observe are not simply caused by a generic impairment of ER–Golgi transport because other Rabs which mediate this step do not impair autophagy when knocked down. Furthermore, the robust autophagy inducer trehalose (Sarkar et al., 2007) appears to decrease secretion (Fig. S2 C), suggesting that bulk secretion can be uncoupled from autophagy induction. Similarly, the absence of changes in Atg9 glycosylation with Rab1a knockdown suggests that Atg9 transport, at least from the ER to the Golgi, is not overtly impaired. Our data are consistent with a model whereby other molecules trafficked via Rab1a (but not Rab2 or Rab1b) modulate Atg9 trafficking, which, in turn regulates omegasome formation.
Interestingly, in yeast, the Golgi-associated COG (conserved oligomeric Golgi) complex is important for autophagosome assembly and appears to also influence Atg9 trafficking (Yen et al., 2010). It will be informative to assess this process in mammalian cells and to also test whether specific perturbation of later steps in the secretory pathway affects autophagy in mammalian cells. However, it is important to reiterate that our data suggest that certain secretory routes (e.g., Rab1b and Rab2) can be impaired without decreasing autophagosome synthesis, whereas pathways regulated specifically by Rab1a and its effectors are critical for autophagy. Thus, either Rab1a influences autophagy via a specific Rab1a-related secretory route or may be acting via a novel, secretory-independent function of Rab1a.
Although our data suggest that specific secretion-associated components regulate autophagy, recent publications have identified an interesting situation in which autophagy regulates a specialized form of secretion. The autophagosome participates in the secretion of acyl coenzyme A–binding protein (AcbA) in starvation conditions in various species of yeast by bypassing the vacuole and fusing with the plasma membrane. This process involves the Golgi-associated proteins (GRASPs), autophagy proteins, plasma membrane SNARES, the endosomal compartments, and peroxisome activity (Duran et al., 2010; Manjithaya et al., 2010). These studies raise the possibility of a reciprocal interplay of the secretory pathway and the nonvacuole–lysosomal pathway. Although our findings involve the autophagosome–lysosome pathway and a secretory-independent function of Rab1a, it will be important to further elucidate the interaction of the secretory and autophagy pathways within mammalian cells and how a new Rab1a-specific regulation of autophagy fits into the intersection of these pathways.
The impairment of Rab1a function by overexpression of α-synuclein will have cumulative effects on both the secretory pathway, as shown in Fig. 2 (A–C) and previously (Cooper et al., 2006), and on autophagy. Although the effects we observe of α-synuclein on secretion within mammalian cells are reproducible and consistent, these effects appear to be relatively minor compared with the effects we observe of α-synuclein and Rab1a on autophagy. Cumulatively, these assays show a 30–50% decrease in autophagosome synthesis. Although this does not represent a complete block of autophagy (which would be lethal; Tsukamoto et al., 2008), a partial block of the magnitude we observed, or smaller, would be likely to manifest significant consequences over many decades in postmitotic neurons. It is of interest to note that reduction of α-synuclein levels enhances autophagy. Thus, it is likely that even small increases (or decreases) in the levels of this protein will modulate autophagy. The cumulative effects of a partial inhibition of both the secretory pathway and autophagy may explain dopamine secretion defects and proteinaceous accumulation and cell death, respectively.
Overall, this situation appears to represent a specific example of how the proteostasis of the whole cell can be disrupted by a specific aggregate-prone protein, a model which was proposed by Gidalevitz et al. (2006) in another disease context. In this case, the culprit, α-synuclein, impairs autophagy, a major route for the clearance of aggregate-prone intracytoplasmic proteins, and this in turn increases the concentration of such proteins and increases their probability of aggregation. However, the predicted outcome of reduced autophagic clearance will have pleiotropic effects. In addition to the accumulation of aggregate-prone proteins, the cells will not be as effective in clearing dysfunctional mitochondria (Twig et al., 2008) and will have increased susceptibility to certain apoptotic insults (Ravikumar et al., 2006); these are all processes that have been implicated in PD (Fig. 7 F; Cookson and van der Brug, 2008). Thus, autophagy inhibition caused by wild-type α-synuclein may provide a unifying mechanism for many of the apparently disconnected cellular pathologies in PD. It is interesting to consider that there may be compensatory regulation of CMA and macroautophagy when one of these pathways is perturbed (Martinez-Vicente et al., 2008). However, if one considers that both macroautophagy and CMA are inhibited by wild-type α-synuclein (Martinez-Vicente et al., 2008), this is likely to be particularly deleterious.
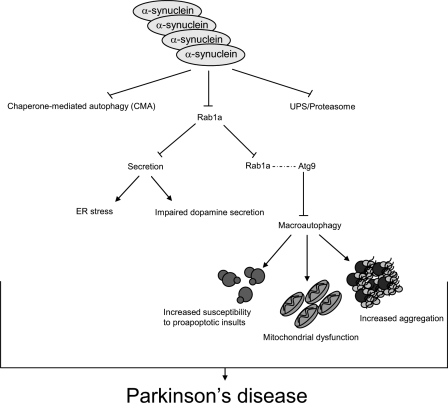
Importantly, our proposed mechanism for the inhibitory role of α-synuclein on autophagy furthers our understanding of the disease (Fig. 8). Although the inhibition of early secretion by α-synuclein may explain the dopaminergic defects seen in PD, it does not sufficiently explain the formation of inclusions and cell death characteristic of PD. In principle, multiplications of the α-synuclein locus leading to increased protein levels of α-synuclein will inhibit autophagy. The inhibition of autophagy increases accumulation of aggregate-prone proteins and sensitizes the cell to proapoptotic assaults. Increased aggregation and apoptosis are characteristic of PD, and therefore, inhibition of autophagy by α-synuclein overexpression may further explain the pathologies characteristic of PD. Collectively, the ability of α-synuclein to inhibit both autophagy and secretion may act as a potent impetus for neurodegeneration.
Figure 8.
Cumulative molecular effects of α-synuclein overexpression. Diagram illustrating the pleiotropic effects of increased intracellular levels of α-synuclein on vital intracellular pathways. The effects of α-synuclein on macroautophagy would likely cause increased susceptibility to proapoptotic insults, mitochondrial dysfunction, and increased aggregation, which are all features of PD.
Materials and methods
Cell culture
Human neuroblastoma cells (SKNSH), human cervical carcinoma cells (HeLa), and human embryonic kidney cells (HEK293) were maintained in DME supplemented with 10% fetal bovine serum, 100 U/ml penicillin/streptomycin, and 2 mM l-glutamine (Sigma-Aldrich) at 37°C and 5% CO2. DFCP1-GFP–expressing cells lines were selected in 800 µg/ml and were a gift from N.T. Ktistakis (Babraham Institute, Cambridge, England, UK).
Constructs and siRNA were transfected in OptiMeM (Invitrogen) using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s instructions using 80 nM siRNA and were compared with control siRNA (nontargeting SMARTpool; Thermo Fisher Scientific). Wild-type α-synuclein constructs were transfected at 1.5 µg per well, httQ74 constructs at 0.5 µg per well, LC3-tomato at 0.2 µg per well, Rab1a-CFP (previously characterized construct; Maier et al., 2009; gift from H. Sitte, Medical University Vienna, Vienna, Austria) at 1.5 µg per well, tomato-p62 at 0.75 µg, and DsRed at 0.5 µg per well.
Bafilomycin A1 was used at 400 nM for 4 h (Millipore). 3-methyladenine (Sigma-Aldrich) was used at 10 mM for 16 h. Rapamycin was used at 0.5 µM for 4 h. Starvation experiments were conducted in HBSS for 4 h.
Aggregate quantification
EGFP-httQ74 aggregation was detected by direct immunofluorescence, whereas httQ74-HA aggregation was detected by immunostaining (primary HA antibody [1:500; Covance] and anti–mouse Alexa Fluor–conjugated secondary antibodies [1:1,000; Invitrogen]). The proportion of transfected cells with aggregates was scored (∼200 cells per coverslip). Experiments were performed blinded and in triplicate per three independent experiments. Statistics for aggregation assays were calculated as odds ratios (the ratio of aggregate-containing nuclei in each condition) with 95% confidence intervals from the raw data. Odds ratios and p-values were determined by unconditional logistical regression analysis using the general log linear analysis in SPSS version 6.1 (SPSS Inc.).
Western blotting
Protein levels were assessed by standard SDS-PAGE techniques. Primary antibodies include anti-EGFP (Takara Bio Inc.), anti-HA (12CA5; Covance), anti-Rab1a, anti-Atg9 (both Abcam), antitubulin, and anti-LC3 (Novus Biologicals). Quantification of protein levels by Western blotting was conducted in ImageJ (National Institutes of Health) software for normal ECL development techniques and on the LI-COR Odyssey (BD).
Immunofluorescence and microscopy
Cells were grown on coverslips and fixed in 4% paraformaldehyde for 10 min and then permeabilized with −20°C methanol for 4 min. For aggregation experiments using httQ74-HA, cells were permeabilized using 0.5% Triton X-100 in PBS. 4% goat serum (Sigma-Aldrich) in PBS was used for blocking and primary and secondary buffer. Coverslips were left in primary antibody overnight at 4°C. Secondary antibodies were conjugated Alexa Fluor antibodies (Invitrogen). All primary antibodies used for Western blotting were used for immunostaining except anti-LC3 (Nanotools).
A confocal microscope (63× NA 1.4 Plan-Apochromat oil immersion lens; LSM510 META; Carl Zeiss, Inc.) along with the LSM510 version 3.2 software (Carl Zeiss, Inc.) was used for fluorescent, confocal microscopy involving immunofluorescent staining with Alexa Fluor–conjugated secondary antibodies or fluorescently tagged proteins. All confocal images were taken with a 63× oil immersion lens except for experiments involving the DFCP1-GFP HEK293 omegasome cell line, in which a 40× oil immersion lens was used. Microscopy was performed on cells fixed on coverslips. Coverslips were mounted in Prolong Gold Antifade reagent (with DAPI; Invitrogen). ImageJ and Photoshop (Adobe) were used for further analysis and processing of confocal images (the specifics of software use for colocalization analysis and image processing are discussed in the next section). A microscope (Plan-Apochromat 60× NA 1.4 oil immersion lens; Eclipse E600; Nikon) was used for aggregate counting and Golgi fragmentation quantification.
Quantification of colocalization
Colocalization of Atg9 with Golgi (p230) and LC3 was imaged using an LSM510 META confocal microscope (63× NA 1.4 Plan-Apochromat oil immersion lens) along with the LSM510 version 3.2 software after immunostaining of endogenous proteins. 10 images of a mean of five cells per image were taken per condition per experiment. Each experiment was run in at least two independent experiments. Exposure settings were unchanged throughout acquisition. Cells were imaged in z stacks, and images were analyzed by the JaCoP plugin (Bolte and Cordelières, 2006) in the ImageJ software. Pearson’s and Mander’s (original, nonthreshold) coefficients were used for reporting colocalization. Student’s t tests were performed in Excel (Microsoft) to determine statistical significance for colocalization coefficients. Images shown are z-stack projections or single planes of representative images using ImageJ and Photoshop software.
Secretion assay
The vector (pC4S1-FM4-FCS-hGH) encoding the pharmacologically regulated reporter construct was obtained from ARIAD Pharmaceuticals (Rivera et al., 2000). The construct was modified so that EGFP was cloned in-frame between the signal sequence and the first FM4 aggregation domain. The reporter construct was then subcloned into the retroviral expression vector pQXCIP (Takara Bio Inc.). A stable cell line (referred to as C1) expressing the reporter construct was generated by virally transducing HeLa cells.
For siRNA experiments, C1 cells were transfected with OnTargetPlus siRNA oligonucleotides (100 nM final concentration; Thermo Fisher Scientific) using Oligofectamine (Invitrogen) as previously described (Motley et al., 2003). For overexpression experiments, α-synuclein–CFP or CFP was transfected into C1 cells using HeLaMonster (Mirus) for 96 h and then assayed for secretion. Brefeldin A, a known inhibitor of secretion, was used at 2 µg/ml as a control for this assay.
To assess constitutive secretion, secretion of stably expressed GFP–growth hormone was initiated by incubating C1 cells with 1 µM AP21998 (ARIAD Pharmaceuticals) at 37°C in 5% CO2 for 80 min. Secretion was halted by placing cells on ice, and cells were detached from the dish by incubating them with trypsin-EDTA solution (PAA Laboratories) at 4°C for 2 h. The trypsin was quenched using DME supplemented with FCS, and the samples were then analyzed for EGFP fluorescence using a FACSCalibur flow cytometer (BD) at 0 and 80 min. Dead cells were excluded using 7-aminoactinomycin-D. For analysis of the α-synuclein overexpression experiments, which required CFP and GFP fluorescence detection, samples were analysed on a Cyan ADP (Beckman Coulter).
To measure the amount of GFP secreted, each siRNA oligonucleotide or construct was transfected in duplicate. The first sample was used for the initial time point (0 min), and the second sample was used for the final time point (80 min). The mean fluorescence of each sample was measured, and the amount of GFP remaining in the cell was calculated using FlowJo software (Tree Star, Inc.).
Statistical significance analysis was performed using Excel software for Student’s t test (overexpression experiments) and PRISM software (GraphPad Software, Inc.) for one-way analysis of variance (ANOVA; siRNA experiments). The percentage of secreted protein after 80 min was determined by dividing the GFP fluorescence at 80 min by the GFP fluorescence at 0 min. This ratio was normalized to empty vector (CFP) for α-synuclein and brefeldin A.
Golgi fragmentation
Golgi were immunostained using anti-GM130 antibody in cells overexpressing α-synuclein–GFP or EGFP. Fragmented Golgi were recognized as dispersed, noncondensed GM130 staining, whereas nonfragmented Golgi were recognized as condensed, perinuclear, and ribbon-like GM130 staining. The percentage of transfected cells with fragmented Golgi was counted in at least 200 cells of blinded experiments run in triplicate.
Drosophila strains
Pseudopupil assays were performed on progeny of the appropriate genotypes, generated by crossing virgins of the genotype elav-GAL4C155; gmr-Htt (exon1)Q1204.62 (Jackson et al., 1998) with flies expressing wild-type α-synuclein (UAS-α-syn; Feany and Bender, 2000) and with flies expressing constitutively active Rab1 (y1 P{UASp-YFP.Rab1.Q70L}CG363812c w*; Zhang et al., 2007) or dominant-negative Rab1 (y1 w*; P{UAST-YFP.Rab1.S25N}mei-S33204; Zhang et al., 2007).
As a control for the derived fly lines, virgins of the isogenic w1118 line (Ryder et al., 2004) or the pan-neural driver elav-GAL4C155 were crossed with males of each transgenic line to generate progeny to confirm that no insertions except gmr-Htt(exon1)Q120 resulted in neurodegeneration. The same virgins were crossed with males y w; gmr-Htt(exon1)Q1204.62 to assess the rate of neurodegeneration caused by mutant huntingtin.
Fly crosses and experiments were performed at 25°C. All crosses for individual experiments were performed at the same time and under the same conditions. Comparisons were performed using paired Student’s t tests using data from five to seven independent experiments, each based on ∼10 individuals of each genotype, in which 15 ommatidia each were scored. We thank M. Feany (Harvard Medical School, Boston, MA), G.R. Jackson (University of California, Los Angeles, School of Medicine, Los Angeles, CA), and the Bloomington Drosophila Stock Center for Drosophila stocks.
Mice
M7 α-synuclein mice were a gift from V.M.-Y. Lee (University of Pennsylvania, Philadelphia, PA). To generate the three possible mouse genotypes, M7 homozygous were crossed to C57BL/6J, and the F1 was intercrossed to produce the three genotypes on the F2 (+/+, M7/+, and M7/M7). F2 mice were used in all experiments. All procedures were performed in accordance with the appropriate Home Office and Local Ethical Committee approval.
F2 mice were used in all experiments. F2 mice were genotyped by real-time PCR from ear-clip DNA. α-Synuclein expression levels were verified by Western blotting. Half brains (22–23-wk-old males; n = 3 each) were homogenized in lysis buffer (0.5% Triton X-100 and 50 mM Tris HCl, pH 7.4, supplemented with complete protease inhibitor cocktail [Roche]). The homogenate was centrifuged at 13,200 rpm at 4°C. The supernatant was removed, and LC3-II and tubulin levels were assessed by SDS-PAGE. Densitometry analysis of immunoblots was performed using ImageJ software. The mean of the wild-type mice was set to 100%, and the error bars denote SEM.
For dose-dependence measures, regression analyses were performed using R. To validate the association between α-synuclein transgene dose and LC3-II levels, we used a permutation test. We randomly assigned mice to different genetic groups, tested for an association, and stored the test statistic. We repeated this 100,000 times and calculated the permutation test p-value, the proportion of test statistics larger than that observed in the original analysis.
Statistics
All data presented are reported as the mean for a representative experiment, unless otherwise stated. Three independent experiments were run for all experiments, unless otherwise stated. Statistical significance of single comparisons between groups was determined by two-tailed, unpaired, Student’s t test as performed in Excel software. Odds ratio (SPSS software; SPSS, Inc.) was used to determine significance for aggregation experiments. Statistics for aggregation assays were calculated as odds ratios (the ratio of transfected cells with aggregates in each condition) with 95% confidence intervals from the raw data. Odds ratios and p-values were determined by unconditional logistical regression analysis using the general log linear analysis in SPSS version 6.1. To simplify data presentation, bar graphs for aggregation experiments were presented as means for percent aggregation for a representative experiment with SEM of the mean. Statistical significance for comparisons between more than two groups was determined by one-way or multiple comparison ANOVA with a Bonferroni or Dunnett’s multiple comparisons post hoc test in PRISM software.
Online supplemental material
Fig. S1 shows that wild-type α-synuclein regulates autophagy. Fig. S2 shows that α-synuclein affects autophagy via inhibition of Rab1a homeostasis. Fig. S3 shows a change in Atg9 colocalization events with autophagy induction. Fig. S4 shows that Atg9 localization is important for autophagy. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201003122/DC1.
Acknowledgments
We thank H. Sitte for the Rab1a-CFP construct. We thank M. Feany, G.R. Jackson, and the Bloomington Drosophila Stock Center for Drosophila stocks and V.M.-Y. Lee for the M7 α-synuclein mice. We thank N.T. Ktistakis for sending us the DFCP1-GFP HEK293 omegasome cell line. We would also like to thank J. Cooper (University of Cambridge, Cambridge, England, UK) for help with statistical considerations and M. Gratian and M. Bowen (University of Cambridge) for technical assistance with microscopy.
The research leading to these results received funding from the Wellcome Trust (Senior Fellowship to D.C. Rubinsztein), Medical Research Council (MRC; programme grant to D.C. Rubinsztein, C.J. O’Kane, and S. Brown and Career Development Award to A.A. Peden), Wellcome Trust/MRC (Strategic Award for Neurodegeneration to D.C. Rubinsztein), Overseas Research Studentship and Cambridge Trust Overseas (award to A.R. Winslow), and the European Community’s Seventh Framework Programme (FP7/2007-2013) under grant agreement number 241791 (MEFOPA).
Footnotes
Abbreviations used in this paper:
- ANOVA
- analysis of variance
- CMA
- chaperone-mediated autophagy
- HD
- Huntington’s disease
- PD
- Parkinson’s disease
- PI(3)P
- phosphatidylinositol-3-phosphate
References
- Allan B.B., Moyer B.D., Balch W.E. 2000. Rab1 recruitment of p115 into a cis-SNARE complex: programming budding COPII vesicles for fusion. Science. 289:444–448 10.1126/science.289.5478.444 [DOI] [PubMed] [Google Scholar]
- Alvarez C., Garcia-Mata R., Brandon E., Sztul E. 2003. COPI recruitment is modulated by a Rab1b-dependent mechanism. Mol. Biol. Cell. 14:2116–2127 10.1091/mbc.E02-09-0625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axe E.L., Walker S.A., Manifava M., Chandra P., Roderick H.L., Habermann A., Griffiths G., Ktistakis N.T. 2008. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 182:685–701 10.1083/jcb.200803137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte S., Cordelières F.P. 2006. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224:213–232 10.1111/j.1365-2818.2006.01706.x [DOI] [PubMed] [Google Scholar]
- Cookson M.R., van der Brug M. 2008. Cell systems and the toxic mechanism(s) of alpha-synuclein. Exp. Neurol. 209:5–11 10.1016/j.expneurol.2007.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper A.A., Gitler A.D., Cashikar A., Haynes C.M., Hill K.J., Bhullar B., Liu K., Xu K., Strathearn K.E., Liu F., et al. 2006. Alpha-synuclein blocks ER-Golgi traffic and Rab1 rescues neuron loss in Parkinson’s models. Science. 313:324–328 10.1126/science.1129462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo A.M., Stefanis L., Fredenburg R., Lansbury P.T., Sulzer D. 2004. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science. 305:1292–1295 10.1126/science.1101738 [DOI] [PubMed] [Google Scholar]
- Duran J.M., Anjard C., Stefan C., Loomis W.F., Malhotra V. 2010. Unconventional secretion of Acb1 is mediated by autophagosomes. J. Cell Biol. 188:527–536 10.1083/jcb.200911154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyllick-Brenzinger M., D’Souza C.A., Dahlmann B., Kloetzel P.M., Tandon A. 2010. Reciprocal effects of alpha-synuclein overexpression and proteasome inhibition in neuronal cells and tissue. Neurotox. Res. 17:215–227 10.1007/s12640-009-9094-1 [DOI] [PubMed] [Google Scholar]
- Feany M.B., Bender W.W. 2000. A Drosophila model of Parkinson’s disease. Nature. 404:394–398 10.1038/35006074 [DOI] [PubMed] [Google Scholar]
- Franceschini N., Kirschfeld K. 1971. [Pseudopupil phenomena in the compound eye of drosophila]. Kybernetik. 9:159–182 10.1007/BF02215177 [DOI] [PubMed] [Google Scholar]
- Fujita Y., Ohama E., Takatama M., Al-Sarraj S., Okamoto K. 2006. Fragmentation of Golgi apparatus of nigral neurons with alpha-synuclein-positive inclusions in patients with Parkinson’s disease. Acta Neuropathol. 112:261–265 10.1007/s00401-006-0114-4 [DOI] [PubMed] [Google Scholar]
- Furlong R.A., Narain Y., Rankin J., Wyttenbach A., Rubinsztein D.C. 2000. Alpha-synuclein overexpression promotes aggregation of mutant huntingtin. Biochem. J. 346:577–581 10.1042/0264-6021:3460577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidalevitz T., Ben-Zvi A., Ho K.H., Brignull H.R., Morimoto R.I. 2006. Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science. 311:1471–1474 10.1126/science.1124514 [DOI] [PubMed] [Google Scholar]
- Gitler A.D., Bevis B.J., Shorter J., Strathearn K.E., Hamamichi S., Su L.J., Caldwell K.A., Caldwell G.A., Rochet J.C., McCaffery J.M., et al. 2008. The Parkinson’s disease protein alpha-synuclein disrupts cellular Rab homeostasis. Proc. Natl. Acad. Sci. USA. 105:145–150 10.1073/pnas.0710685105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaki M., Noda T., Ohsumi Y. 2003. The early secretory pathway contributes to autophagy in yeast. Cell Struct. Funct. 28:49–54 10.1247/csf.28.49 [DOI] [PubMed] [Google Scholar]
- Ishihara N., Hamasaki M., Yokota S., Suzuki K., Kamada Y., Kihara A., Yoshimori T., Noda T., Ohsumi Y. 2001. Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion. Mol. Biol. Cell. 12:3690–3702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson G.R., Salecker I., Dong X., Yao X., Arnheim N., Faber P.W., MacDonald M.E., Zipursky S.L. 1998. Polyglutamine-expanded human huntingtin transgenes induce degeneration of Drosophila photoreceptor neurons. Neuron. 21:633–642 10.1016/S0896-6273(00)80573-5 [DOI] [PubMed] [Google Scholar]
- Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. 2000. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19:5720–5728 10.1093/emboj/19.21.5720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara A., Noda T., Ishihara N., Ohsumi Y. 2001. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J. Cell Biol. 152:519–530 10.1083/jcb.152.3.519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D.J., Abeliovich H., Agostinis P., Agrawal D.K., Aliev G., Askew D.S., Baba M., Baehrecke E.H., Bahr B.A., Ballabio A., et al. 2008. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 4:151–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M., Waguri S., Ueno T., Iwata J., Murata S., Tanida I., Ezaki J., Mizushima N., Ohsumi Y., Uchiyama Y., et al. 2005. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 169:425–434 10.1083/jcb.200412022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuma A., Hatano M., Matsui M., Yamamoto A., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N. 2004. The role of autophagy during the early neonatal starvation period. Nature. 432:1032–1036 10.1038/nature03029 [DOI] [PubMed] [Google Scholar]
- Maier S., Reiterer V., Ruggiero A.M., Rothstein J.D., Thomas S., Dahm R., Sitte H.H., Farhan H. 2009. GTRAP3-18 serves as a negative regulator of Rab1 in protein transport and neuronal differentiation. J. Cell. Mol. Med. 13:114–124 10.1111/j.1582-4934.2008.00303.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjithaya R., Anjard C., Loomis W.F., Subramani S. 2010. Unconventional secretion of Pichia pastoris Acb1 is dependent on GRASP protein, peroxisomal functions, and autophagosome formation. J. Cell Biol. 188:537–546 10.1083/jcb.200911149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Vicente M., Talloczy Z., Kaushik S., Massey A.C., Mazzulli J., Mosharov E.V., Hodara R., Fredenburg R., Wu D.C., Follenzi A., et al. 2008. Dopamine-modified alpha-synuclein blocks chaperone-mediated autophagy. J. Clin. Invest. 118:777–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey A.C., Kaushik S., Sovak G., Kiffin R., Cuervo A.M. 2006. Consequences of the selective blockage of chaperone-mediated autophagy. Proc. Natl. Acad. Sci. USA. 103:5805–5810 10.1073/pnas.0507436103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNaught K.S., Olanow C.W., Halliwell B., Isacson O., Jenner P. 2001. Failure of the ubiquitin-proteasome system in Parkinson’s disease. Nat. Rev. Neurosci. 2:589–594 10.1038/35086067 [DOI] [PubMed] [Google Scholar]
- McNaught K.S., Belizaire R., Isacson O., Jenner P., Olanow C.W. 2003. Altered proteasomal function in sporadic Parkinson’s disease. Exp. Neurol. 179:38–46 10.1006/exnr.2002.8050 [DOI] [PubMed] [Google Scholar]
- Motley A., Bright N.A., Seaman M.N., Robinson M.S. 2003. Clathrin-mediated endocytosis in AP-2–depleted cells. J. Cell Biol. 162:909–918 10.1083/jcb.200305145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar B., Duden R., Rubinsztein D.C. 2002. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum. Mol. Genet. 11:1107–1117 10.1093/hmg/11.9.1107 [DOI] [PubMed] [Google Scholar]
- Ravikumar B., Acevedo-Arozena A., Imarisio S., Berger Z., Vacher C., O’Kane C.J., Brown S.D., Rubinsztein D.C. 2005. Dynein mutations impair autophagic clearance of aggregate-prone proteins. Nat. Genet. 37:771–776 10.1038/ng1591 [DOI] [PubMed] [Google Scholar]
- Ravikumar B., Berger Z., Vacher C., O’Kane C.J., Rubinsztein D.C. 2006. Rapamycin pre-treatment protects against apoptosis. Hum. Mol. Genet. 15:1209–1216 10.1093/hmg/ddl036 [DOI] [PubMed] [Google Scholar]
- Ravikumar B., Imarisio S., Sarkar S., O’Kane C.J., Rubinsztein D.C. 2008. Rab5 modulates aggregation and toxicity of mutant huntingtin through macroautophagy in cell and fly models of Huntington disease. J. Cell Sci. 121:1649–1660 10.1242/jcs.025726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Wang C.W., Nair U., Shintani T., Abeliovich H., Klionsky D.J. 2004. Early stages of the secretory pathway, but not endosomes, are required for Cvt vesicle and autophagosome assembly in Saccharomyces cerevisiae. Mol. Biol. Cell. 15:2189–2204 10.1091/mbc.E03-07-0479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera V.M., Wang X., Wardwell S., Courage N.L., Volchuk A., Keenan T., Holt D.A., Gilman M., Orci L., Cerasoli F., Jr, et al. 2000. Regulation of protein secretion through controlled aggregation in the endoplasmic reticulum. Science. 287:826–830 10.1126/science.287.5454.826 [DOI] [PubMed] [Google Scholar]
- Ross O.A., Braithwaite A.T., Skipper L.M., Kachergus J., Hulihan M.M., Middleton F.A., Nishioka K., Fuchs J., Gasser T., Maraganore D.M., et al. 2008. Genomic investigation of alpha-synuclein multiplication and parkinsonism. Ann. Neurol. 63:743–750 10.1002/ana.21380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein D.C., Cuervo A.M., Ravikumar B., Sarkar S., Korolchuk V., Kaushik S., Klionsky D.J. 2009. In search of an “autophagomometer”. Autophagy. 5:585–589 10.4161/auto.5.5.8823 [DOI] [PubMed] [Google Scholar]
- Ryder E., Blows F., Ashburner M., Bautista-Llacer R., Coulson D., Drummond J., Webster J., Gubb D., Gunton N., Johnson G., et al. 2004. The DrosDel collection: a set of P-element insertions for generating custom chromosomal aberrations in Drosophila melanogaster. Genetics. 167:797–813 10.1534/genetics.104.026658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S., Davies J.E., Huang Z., Tunnacliffe A., Rubinsztein D.C. 2007. Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and alpha-synuclein. J. Biol. Chem. 282:5641–5652 10.1074/jbc.M609532200 [DOI] [PubMed] [Google Scholar]
- Snyder H., Mensah K., Theisler C., Lee J., Matouschek A., Wolozin B. 2003. Aggregated and monomeric alpha-synuclein bind to the S6′ proteasomal protein and inhibit proteasomal function. J. Biol. Chem. 278:11753–11759 10.1074/jbc.M208641200 [DOI] [PubMed] [Google Scholar]
- Suzuki K., Kubota Y., Sekito T., Ohsumi Y. 2007. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 12:209–218 10.1111/j.1365-2443.2007.01050.x [DOI] [PubMed] [Google Scholar]
- Tisdale E.J. 1999. A Rab2 mutant with impaired GTPase activity stimulates vesicle formation from pre-Golgi intermediates. Mol. Biol. Cell. 10:1837–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto S., Kuma A., Murakami M., Kishi C., Yamamoto A., Mizushima N. 2008. Autophagy is essential for preimplantation development of mouse embryos. Science. 321:117–120 10.1126/science.1154822 [DOI] [PubMed] [Google Scholar]
- Twig G., Elorza A., Molina A.J., Mohamed H., Wikstrom J.D., Walzer G., Stiles L., Haigh S.E., Katz S., Las G., et al. 2008. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 27:433–446 10.1038/sj.emboj.7601963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen W.L., Shintani T., Nair U., Cao Y., Richardson B.C., Li Z., Hughson F.M., Baba M., Klionsky D.J. 2010. The conserved oligomeric Golgi complex is involved in double-membrane vesicle formation during autophagy. J. Cell Biol. 188:101–114 10.1083/jcb.200904075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young A.R., Chan E.Y., Hu X.W., Köchl R., Crawshaw S.G., High S., Hailey D.W., Lippincott-Schwartz J., Tooze S.A. 2006. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J. Cell Sci. 119:3888–3900 10.1242/jcs.03172 [DOI] [PubMed] [Google Scholar]
- Zhang J., Schulze K.L., Hiesinger P.R., Suyama K., Wang S., Fish M., Acar M., Hoskins R.A., Bellen H.J., Scott M.P. 2007. Thirty-one flavors of Drosophila rab proteins. Genetics. 176:1307–1322 10.1534/genetics.106.066761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N.Y., Tang Z., Liu C.W. 2008. alpha-Synuclein protofibrils inhibit 26 S proteasome-mediated protein degradation: understanding the cytotoxicity of protein protofibrils in neurodegenerative disease pathogenesis. J. Biol. Chem. 283:20288–20298 10.1074/jbc.M710560200 [DOI] [PubMed] [Google Scholar]