A reservoir of Atg9-containing vesicles and tubules provides the initial membranes necessary for autophagophore formation in yeast.
Abstract
Eukaryotes use the process of autophagy, in which structures targeted for lysosomal/vacuolar degradation are sequestered into double-membrane autophagosomes, in numerous physiological and pathological situations. The key questions in the field relate to the origin of the membranes as well as the precise nature of the rearrangements that lead to the formation of autophagosomes. We found that yeast Atg9 concentrates in a novel compartment comprising clusters of vesicles and tubules, which are derived from the secretory pathway and are often adjacent to mitochondria. We show that these clusters translocate en bloc next to the vacuole to form the phagophore assembly site (PAS), where they become the autophagosome precursor, the phagophore. In addition, genetic analyses indicate that Atg1, Atg13, and phosphatidylinositol-3-phosphate are involved in the further rearrangement of these initial membranes. Thus, our data reveal that the Atg9-positive compartments are important for the de novo formation of the PAS and the sequestering vesicle that are the hallmarks of autophagy.
Introduction
The conserved catabolic pathway of autophagy is essential to generate an internal pool of nutrients that permit cells to survive during prolonged periods of starvation (Mizushima et al., 2008). Recent studies have revealed that because of its ability to eliminate unwanted structures, autophagy also participates in development, cellular differentiation, degradation of aberrant structures, and life span extension, as well as protecting against pathogens and tumors (Levine and Deretic, 2007; Levine and Kroemer, 2008; Mizushima et al., 2008). As a result, this pathway plays a relevant role in the pathophysiology of neurodegenerative, cardiovascular, muscular, and autoimmune diseases.
The general mechanism of autophagy involves the sequestration of the cytoplasmic cargo into large double-membrane vesicles called autophagosomes that fuse with lysosomes/vacuoles to provide their contents with access to the hydrolytically active interior of these organelles, thus allowing for the cargo’s degradation. The current model of autophagosome biogenesis is that they are formed by expansion and successive sealing of a small membrane cisterna termed the phagophore or isolation membrane (Reggiori and Klionsky, 2005). The phagophore appears to be generated at a specialized site called the preautophagosomal structure or the phagophore assembly site (PAS; Xie and Klionsky, 2007). Most of the studies about the PAS have been done in the yeast Saccharomyces cerevisiae, where only one of these structures is present in each cell and it is found in proximity to the vacuole (Xie and Klionsky, 2007). These investigations have revealed that the autophagy-related (Atg) proteins, the components of the conserved machinery mediating autophagosome biogenesis, assemble according to a hierarchical order and form this specialized site (Suzuki et al., 2007). Despite the apparent importance of the PAS, very little is known about this site at the molecular level, including the origin of the nucleating membrane and the subsequent rearrangements that allow it to form a sequestering compartment.
Most of the Atg proteins are cytosolic and transiently associate with the PAS by interacting with other Atg components and/or lipids. Atg9 is the only conserved integral membrane protein that is essential for autophagosome formation (Lang et al., 2000; Noda et al., 2000; Young et al., 2006). In yeast, Atg9 localizes to several puncta dispersed throughout the cytoplasm, some of which colocalize with mitochondria markers; however, it remains unclear whether they are part of the outer membrane of this organelle or adjacent to it (Reggiori et al., 2004a, 2005). Atg9 cycles between these peripheral puncta (hereafter referred as the Atg9 reservoirs), which do not contain detectable levels of most other conserved Atg proteins, and the PAS, which supports the notion that Atg9 is involved in the delivery of membrane necessary for the formation of the sequestering vesicles (Reggiori et al., 2004a). In addition, Atg9 is one of the first Atg components to be localized to the PAS (Suzuki et al., 2007). In atg9Δ cells, nearly all the other Atg proteins fail to be recruited to the PAS, making its role crucial in the assembly of the Atg machinery.
The key organizational function of Atg9 is also exemplified by the fact that Atg9 transport to the PAS is regulated during the yeast cytoplasm-to-vacuole targeting (Cvt) pathway, a biosynthetic selective type of autophagy devoted to the delivery into the vacuole of cytosolic oligomers mostly formed by the precursor vacuolar protease aminopeptidase I (prApe1; Klionsky et al., 1992). After oligomerization, the prApe1 complex binds to its receptor, Atg19, to form the Cvt complex, which is subsequently recruited near the vacuole surface in an Atg11-dependent step (Shintani et al., 2002). Atg11 also binds Atg9 and thus mediates the simultaneous movement of both the Cvt complex and Atg9 (Shintani et al., 2002; Shintani and Klionsky, 2004; He et al., 2006). This regulated relocation triggers the recruitment of the Atg proteins to the PAS and subsequent sequestration of the Cvt complex into a double-membrane Cvt vesicle (Shintani and Klionsky, 2004).
Despite the important progress made in understanding autophagy, the study of the precise function of these factors as well as the mechanism of this pathway has been hampered by the lack of information regarding the membrane dynamics during the autophagosome formation process. To gain insights into this crucial question, we have studied the biogenesis of the yeast PAS by combining a recently developed immunoelectron microscopy (IEM) protocol (Griffith et al., 2008) with yeast genetics. We show that the PAS originates from Atg9-positive clusters of vesicles and tubules that we called Atg9 reservoirs. Translocation of one or more reservoirs in proximity to the vacuole, together with the successive recruitment of other Atg proteins, generates the PAS. Hence, fusion of the tubulovesicular membranes of the Atg9 reservoirs leads to the formation of the phagophore necessary for the subsequent biogenesis of a double-membrane vesicle. Thus, our results suggest the de novo formation of the PAS from vesicles and tubules and highlight the crucial role of Atg9 in this process.
Results
Atg9 localizes to tubulovesicular clusters adjacent to mitochondria
It has been proposed that Atg9 is one of the factors mediating the transport of lipid bilayers to the PAS (Reggiori et al., 2004a). Consequently, the identification of the membranes positive for Atg9 is crucial to understand the nature and biogenesis of the PAS, and its role in autophagosome formation. Fluorescence microscopy has shown that Atg9 resides in numerous puncta, some of them partially colocalizing with or near mitochondria (Reggiori et al., 2005; Reggiori and Klionsky, 2006). This approach, however, does not make it possible to determine the nature of the Atg9 compartments and their structural relationship with mitochondria. To unveil their morphology and acquire mechanistic insights into their contribution to PAS formation, we analyzed the localization and trafficking of Atg9 at the ultrastructural level using a recently developed IEM procedure (Griffith et al., 2008). As the cellular levels of endogenous Atg9 are too low for IEM detection, we created a functional C-terminal GFP fusion protein (Atg9-GFP), which behaves like the endogenous one (Fig. 1).
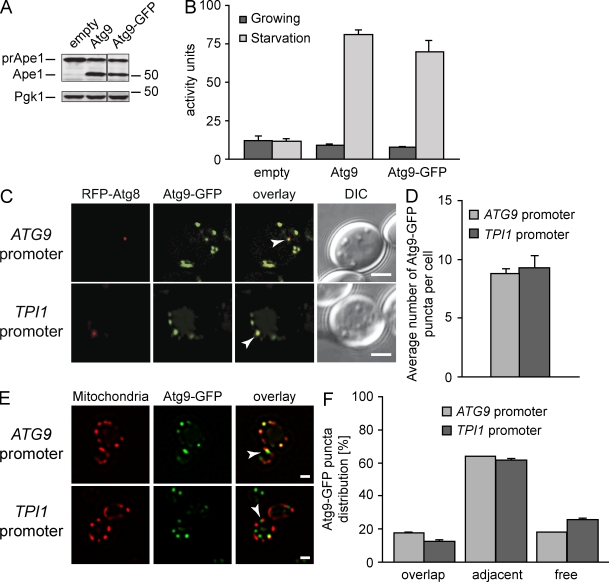
Figure 1.
The Atg9-GFP construct for IEM analyses. (A) The prApe1 processing is normal in Atg9-GFP–expressing cells. The atg9Δ (JCK007) mutant transformed with either an empty vector (pRS416), a plasmid carrying the ATG9 gene (pJK1-2416), or the ATG9-GFP fusion (pATG9GFP416) was grown in YPD medium to log phase before analyzing prApe1 maturation by immunoblotting. The cytosolic protein Pgk1 was used as a loading control. Mr is indicated in kilodaltons. The black line indicates that intervening lanes have been spliced out. (B) Autophagy is normal in the presence of Atg9-GFP. The atg9Δ (FRY300) cells expressing Pho8Δ60 and transformed with the same plasmids described in A were shifted from YPD medium (dark gray bars) to SD-N medium (light gray bars) for 4 h. Autophagy induction was determined by a Pho8 activity assay. (C and D) Atg9-GFP has a normal distribution, and one of the Atg9-GFP-containing puncta is the PAS. Strains expressing Atg9-GFP under the control of the ATG9 promoter (FRY162) or the TPI1 promoter (MMY067) were transformed with a plasmid (promRFPATG8415) carrying the PAS protein marker RFP-Atg8. (C) Transformants were cultured to log phase and imaged. The number of Atg9-GFP puncta per cell was counted (D) and error bars represent the standard deviation of the mean. Arrowheads highlight colocalizations. (E and F) Part of Atg9-GFP distributes to mitochondria. The strains analyzed in C were transformed with the pmitoDsRed415 plasmid expressing mitochondria-targeted RFP and imaged (E). Arrowheads highlight Atg9 puncta adjacent to mitochondria. Determination of Atg9-GFP puncta distribution on mitochondria (F) was determined as described in Materials and methods, and error bars represent the standard deviation of the mean. DIC, differential interference contrast. Bar, 2 µm.
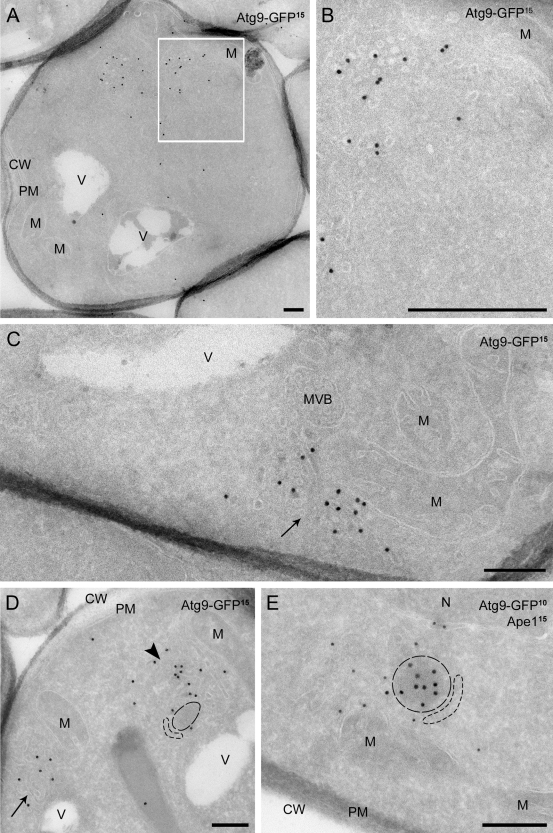
Wild-type cells expressing Atg9-GFP were grown in rich medium and prepared for IEM. The vast majority of Atg9-GFP was present in clusters of vesicular (mean diameter of 30–40 nm) and tubular structures (Fig. 2, A–D; and Fig. S1). The Atg9 clusters (defined in Materials and methods) were found in 53% of the cells, and their numbers vary from one to three, with occasionally four, per cell section (Table I). The gold particles corresponding to Atg9-GFP decorated the cytosolic surface of these membranes, in agreement with Atg9 topology that predicts GFP fused to the C terminus to be on the cytosolic face of lipid bilayers (He et al., 2006). Interestingly, the Atg9 clusters were occasionally seen surrounding a circular electron-dense cytoplasmic structure with a diameter of 100–150 nm (Figs. 2 D and S1, A [arrowhead] and E). Double immunogold labeling for GFP and Ape1 identified it as the Cvt complex (Figs. 2 E and S1 B; Baba et al., 1997). When more than one Atg9 cluster was observed in a cell section, only one of them was associated with a Cvt complex (Tables I and II; 1% of the cell sections are Cvt complex positive). In contrast, the Cvt complex was associated with an Atg9 cluster in 80% of the cells where the former was detected, reflecting the close association between these two structures (He et al., 2006).
Figure 2.
IEM analysis of wild-type cells expressing Atg9-GFP. Atg9-GFP (MMY067) cells were grown to log phase and processed for IEM as described in Materials and methods. Preparations shown in A–D were immunogold labeled only for GFP, whereas those presented in E were double immunogold labeled for GFP and Ape1. (A) Overview of a labeled cell section. (B) Insets of A illustrating an Atg9-positive cluster of vesicles and tubules. (C) Atg9-containing membranous arrangements (arrow) adjacent to mitochondria (≤ 50 nm distance). (D) Micrograph showing a GFP-labeled cell with two Atg9 clusters (marked by an arrow and an arrowhead), one of which (arrowhead) associated with an electron-dense structure with a 100–150 nm diameter (broken lines). (E) The circular electron-dense structures found in close proximity to the Atg9 clusters and labeled for Ape1 are Cvt complexes (broken lines). The dashed cisterna emphasizes the Cvt complex surface not associated with Atg9-containing membranes. D and E are also shown in Fig. S1 (A and B) without broken lines for clarity, while additional examples of Atg9 clusters are presented in Fig. S1, C–H. The size of the gold particles is indicated on the top of each picture. CW, cell wall; M, mitochondria; MVB, multivesicular body; N, nucleus; PM, plasma membrane; V, vacuole. Bars, 200 nm.
Table I.
Frequency of the Atg9-GFP containing structures and the Cvt complex
| Criteria | Strain background | ||
| WT | atg11Δ | atg1Δ | |
| Atg9-GFP–containing membranous clusters/cell section | |||
| 0 | 47 ± 1.8 | 57 ± 2.3 | 84 ± 1.2 |
| 1 | 28 ± 1.3 | 25 ± 0.8 | 14 ± 0.7a |
| 2 | 19 ± 0.9 | 16 ± 0.8 | 2 ± 0.2a |
| 3 | 5 ± 0.8 | 2 ± 1.1 | 0 |
| ≥4 | 1 ± 0.2 | 0 | 0 |
| Cvt complex/cell section | |||
| 0 | 99 ± 0.1 | 99.7 ± 0.3 | 95 ± 0.8 |
| 1 | 1 ± 0.4 | 0.3 ± 0.1a | 5 ± 0.8a |
| 2 | 0 | 0 | 0 |
| 3 | 0 | 0 | 0 |
| ≥4 | 0 | 0 | 0 |
Counting and statistical analyses were carried out as described in Materials and methods. All the results are expressed in percentages ± the standard error of the mean.
P ≤ 0.05.
Table II.
Relative subcellular distribution of the Atg9-GFP containing structures and Cvt complex
| Criteria/cell type | Location | |||
| Adjacent to mitochondria | Perivacuolar | Perinuclear | Other cytoplasmic | |
| Atg9-GFP–containing membranous clusters | ||||
| WT | 57.0 ± 0.2 | 3.8 ± 0.05 | 4.9 ± 0.05 | 34.3 ± 0.1 |
| atg11Δ | 50 ± 0.4 | 4 ± 0.3 | 2 ± 0.03 | 44 ± 0.1 |
| atg1Δ | 17.5 ± 0.1 | 37.5 ± 0.1 | 7.5 ± 0.05 | 37.5 ± 0.2 |
| Cvt complexes associated with Atg9-GFP–containing membranous clusters | ||||
| WT | 58.8 ± 0.3 | 2.0 ± 0.1 | 3.9 ± 0.2 | 35.3 ± 0.3 |
| atg11Δ | ND | ND | ND | ND |
| atg1Δ | 14.3 ± 0.2 | 57.1 ± 0.2 | 12.2 ± 0.5 | 16.4 ± 0.8 |
| Free Cvt complexes | ||||
| WT | 2.2 ± 0.2 | 0 | 0 | 97.8 ± 0.3 |
| atg11Δ | 15.4 ± 1.1 | 0 | 1.2 ± 0.8 | 83.4 ± 1.5 |
| atg1Δ | ND | ND | ND | ND |
Counting and statistical analyses were carried out as described in Materials and methods. All the results are expressed in percentages ± the standard error of the mean. ND, not determined because this structure is not present or very rare in the indicated strain.
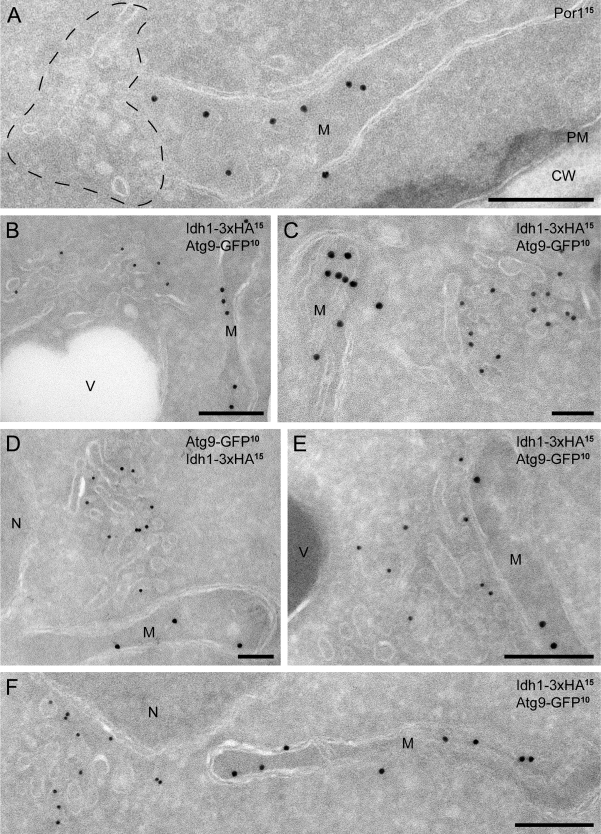
In agreement with the fluorescence microscopy data (Reggiori et al., 2005; He et al., 2006; Reggiori and Klionsky, 2006), the Atg9 clusters were predominantly adjacent to mitochondria (57%, Table II), but they were never seen continuous with the outer membrane of this organelle. To reinforce the notion that these clusters are separated from mitochondria, we immunolocalized Por1, a component of the mitochondrial outer membrane. The Por1 was found exclusively on the surface of mitochondria and was absent from the Atg9-containing clusters (Fig. 3 A). An identical result was also obtained with Idh1, another mitochondrial protein marker (Fig. 3, B–F).
Figure 3.
Mitochondrial protein markers do not localize to the Atg9 clusters. (A) Ultrathin cryosections obtained from a wild-type (MMY067) strain were immunogold labeled only for Por1 because the anti-Por1 antibody does not work in double labeling. The broken line highlights an Atg9 cluster. (B–F) Cells expressing both Atg9-GFP and the mitochondrial marker Idh1-3xHA (MOY003) were processed for IEM, and cryo-sections were double immunogold labeled for GFP and HA. CW, cell wall; M, mitochondria; N, nucleus; PM, plasma membrane; V, vacuole. Bars, 200 nm.
In parallel, we addressed whether higher levels of Atg9-GFP were altering the Atg9 reservoirs morphology. When we analyzed an untransformed wild-type strain by IEM, we detected identical clusters adjacent to mitochondria, albeit with a reduced size (Fig. S2 A). To determine if these clusters showed similar biophysical properties in wild-type and Atg9-GFP–overexpressing cells, cell extracts were centrifuged at 13,000 g. Under these conditions, most of the endogenous Atg9 was found in the low-speed supernatant (S13) fraction and only a small amount in the pellet (P13) fraction (Fig. S2 B; Reggiori et al., 2004b, 2005). The reduction in the amount of Atg9 in the P13 fraction in comparison to previously published data (Reggiori et al., 2005) may represent more efficient spheroplast lysis. Using the same approach for cells overproducing Atg9-GFP, we found this fusion protein in the S13 fraction (Fig. S2 B). This result indicates that higher levels of Atg9 change the volume of the Atg9 clusters to accommodate more protein but that they do not alter their biophysical properties. Similar enlargements have been reported for other organelles like the ER, where overproduction of a resident protein leads to ER expansion without altering its physiology (Wright et al., 1988).
We concluded that the Atg9-containing compartments consist of a cluster of vesicles and tubules that are often in the vicinity of mitochondria, but not connected with them.
Atg9 resides in a novel compartment originating from the secretory system
The proximity of the Atg9 clusters adjacent to, but distinct from, mitochondria raised the question of their biogenesis. As Atg9 is a transmembrane protein, it could be inserted into membranes either in the mitochondria or in the ER.
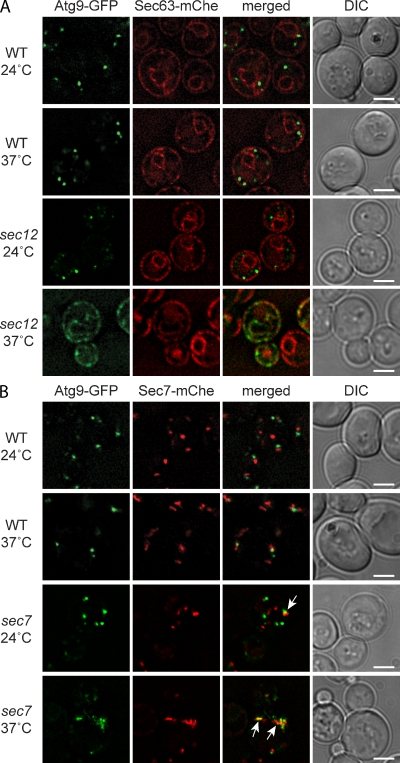
To follow the Atg9 biosynthetic route, Atg9-GFP expression was put under the control of the galactose-inducible GAL1 promoter. Transfer into a galactose-containing medium for 2 h allowed us to detect Atg9 clusters (Fig. 4). No intermediate structures, e.g., mitochondria or ER, were visualized even at earlier time points, probably due to the rapid transport of Atg9 after synthesis. To further explore whether Atg9 is transported through the secretory pathway, we used a strain carrying a thermosensitive allele of SEC12, which blocks ER exit (Barlowe and Schekman, 1993). At a permissive temperature, the localization pattern of Atg9-GFP was indistinguishable from that observed in wild-type cells (Fig. 4 A). At a restrictive temperature, in contrast, the transport block between the ER and Golgi apparatus resulted in the accumulation of newly synthesized Atg9 in the ER, as revealed by colocalization with the specific protein marker Sec63 (Fig. 4 A; Deshaies et al., 1991). This transport block was reversible, which indicates that Atg9-GFP was not amassed in a terminal structure but rather accumulated in a transport intermediate (unpublished data). To examine the role of the Golgi complex in Atg9 transport, we used a thermosensitive sec7 allele, which blocks protein traffic from this organelle but does not affect localization of the protein (Franzusoff and Schekman, 1989; Jackson and Casanova, 2000). At 24°C, Atg9-GFP was again normally distributed to several cytoplasmic puncta, and those were only rarely positive for the late Golgi compartment protein marker Sec7 (Fig. 4 B; Losev et al., 2006). In contrast, at 37°C, newly synthesized Atg9-GFP was present in circular structures, often positive for Sec7ts tagged with Discosoma red fluorescent protein (dsRed), which occasionally had an elongated conformation (Fig. 4 B, arrows). These structures are likely Berkeley bodies, aberrant Golgi generated as a result of the sec7 sorting defect (Novick et al., 1980). Again, the transport block was reversible, which indicates that Atg9-GFP was not accumulated in a terminal structure (unpublished data). We concluded that Atg9 is translocated into the ER and reaches its final destination, the Atg9 clusters, probably after passing through the Golgi.
Figure 4.
Atg9 is transported through part of the secretory pathway. (A) Atg9 is translocated into the ER. The wild-type (MMY126) and sec12 (MMY129) cells expressing Sec63-mChe-V5 and carrying the pGalATG9GFP416 plasmid were grown in SMD at 24°C before being transferred into a galactose-containing medium. Cultures were subsequently split and separately incubated at either 24°C or 37°C for 2 h before imaging. No fluorescence signal was detected when cells were grown in the presence of glucose (not depicted). (B) Atg9 passes through the Golgi before reaching its final destination. Wild-type (MMY125) and sec7 (MMY127) cells expressing genomically mChe-V5–tagged Sec7 and Sec7ts, respectively, and transformed with the pGalATG9GFP416 plasmid were analyzed as in A. Arrows highlight the colocalization between Atg9-GFP and Sec7ts-mChe in sec7ts cells at 37°C. DIC, differential interference contrast. Bars, 2 µm.
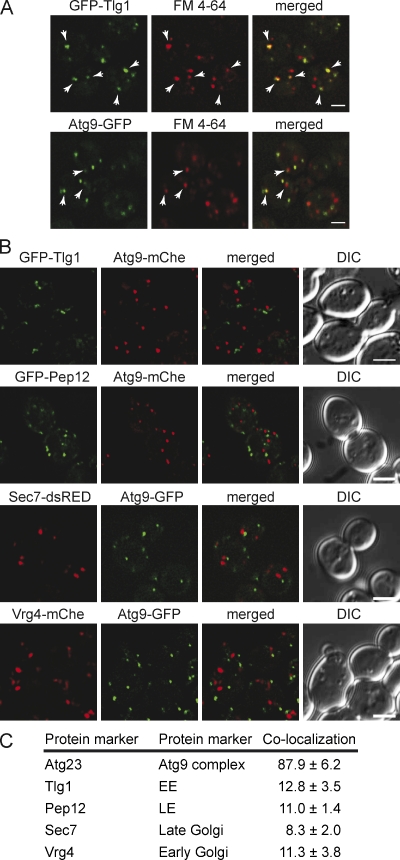
As a post-Golgi organelle, the Atg9 clusters could be endosomes. To test this possibility, we used FM 4-64, a lipophilic dye that, after associating with the plasma membrane, passes through endosomes before reaching the vacuole (Vida and Emr, 1995). At 4°C, endocytosis is inhibited and FM 4-64 exclusively associates with the plasma membrane. Transfer of FM 4-64–loaded cells to RT allows the internalization of this dye in a time-dependent manner (Sipos et al., 2004). After 10 min at RT, FM 4-64 reached early endosomes (EE; Sipos et al., 2004), as demonstrated by the extensive colocalization with the specific protein marker Tlg1 (Fig. 5 A; Holthuis et al., 1998). Several Atg9 clusters were faintly positive for FM 4-64 at the same internalization time point (Fig. 5 A, arrows).
Figure 5.
Atg9 concentrates into a novel organelle. (A) Atg9-GFP colocalizes with some FM 4-64-positive puncta shortly after endocytosis. The strains expressing genomically GFP-tagged Tlg1 (FRY360) or Atg9 (FRY162) were grown to log phase and then exposed to FM 4-64 as described in Materials and methods. Arrowheads highlight the colocalization between FM 4-64 and GFP-Tlg1 or Atg9-GFP. (B) Atg9 does not colocalize with endosomal and Golgi protein markers. The Atg9-GFP Vrg4-mChe-V5 (FRY340), Atg9-GFP Sec7-dsRED (FRY341), GFP-Tlg1 Atg9-mChe-V5 (FRY342), and GFP-Pep12 Atg9-mChe-V5 (FRY344) strains were grown to log phase before being fixed and imaged. Bars, 2 µm. (C) The colocalization experiments shown in B were statistically evaluated as described in Materials and methods. Atg23 was used as a positive control for colocalization because forming a complex with Atg9 (Reggiori et al., 2004a). DIC, differential interference contrast.
The fact that the Atg9 clusters are connected with the endocytic and secretory systems prompted us to investigate whether known protein markers of these compartments localized on Atg9 clusters. We created strains expressing Atg9-GFP together with dsRed- or Cherry-tagged Tlg1 (EE), Pep12 (late endosomes [LE]), Vrg4 (early Golgi), Sec7 (late Golgi), and Atg23 (colocalizes with Atg9; Holthuis et al., 1998; Losev et al., 2006) under the control of the endogenous promoters. As expected, Atg23 displayed substantial colocalization with Atg9 (Fig. 5 C). In contrast, the tagged endosomal and Golgi proteins did not (Fig. 5, B and C).
Collectively, our results show that the Atg9 clusters represent a novel compartment originating from the secretory pathway and able to exchange at least lipids with the endocytic system.
The Atg9 reservoirs and the PAS have a similar morphological organization
Atg9 is a highly dynamic protein that shuttles between the Atg9 reservoirs and the PAS (Reggiori et al., 2004a; Geng et al., 2008). Therefore, one of the observed Atg9 clusters described could represent the PAS. In wild-type cells it is not possible to distinguish the PAS from the Atg9 reservoirs. Therefore, we capitalized on certain characteristics of the Cvt pathway for two reasons: (1) it uses primarily the same Atg machinery as autophagy, and (2) specific gene deletions can modulate Atg9 trafficking. For instance, Atg11 is essential for both the association between Atg9 and the Cvt complex, and their transport near the vacuole in growing conditions, where they recruit the Atg proteins (Fig. S3 A; He et al., 2006). As a consequence, no PAS is formed in the atg11Δ strain (Fig. S3 B; Shintani and Klionsky, 2004), and this allows exclusive visualization of the Atg9 reservoirs.
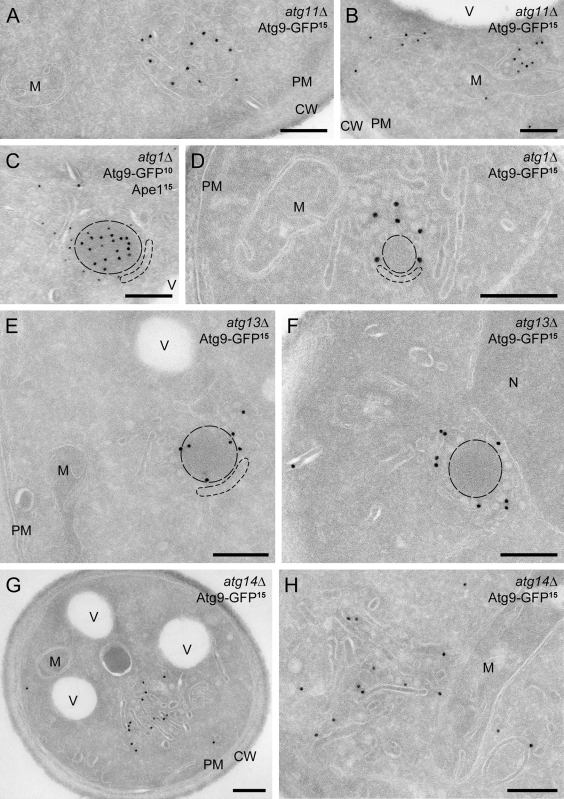
IEM analysis of the atg11Δ cells revealed that the Atg9 clusters adjacent to mitochondria are the Atg9 reservoirs (Fig. 6, A and B; Fig. S4, A–C; and Table II). The Atg9-positive structures were found in 43% of the cell sections, and the subcellular distribution of these compartments was identical to that observed in wild-type cells, which shows that the Atg9 reservoirs are the main site of Atg9 concentration (Table II). As expected, the Atg9 reservoirs were no longer associated with Cvt complexes in agreement with the described phenotype of the atg11Δ mutant (Fig. S3 A; He et al., 2006).
Figure 6.
Atg9-GFP distribution in various atg mutants. The atg11Δ (MMY069; A and B), atg1Δ (MMY068; C and D), atg13Δ (MMY070; E and F), and atg14Δ (MMY071; G and H) strains were grown and processed as described in Fig. 2. Cryosections were immunolabeled for GFP alone (A, B, and D–J) or in combination with Ape1 (C). (A and B) The Atg9 reservoirs observed in atg11Δ cells. (C and D) The PAS accumulated in the atg1Δ knockout. (E and F) The PAS present in the atg13Δ strain displays a Cvt complex surrounded by numerous small vesicles. (G and H) Enrichment of tubular membranes at the PAS of the atg14Δ mutant. Cvt complexes are highlighted with broken lines. D–F are also shown in Fig. S4 (E, F, I, and J) without dashed lines for clarity, while additional examples are presented in Fig. S4 (A–D, G, H, and K–Q). CW, cell wall; M, mitochondria; N, nucleus; PM, plasma membrane; V, vacuole. Bars, 200 nm.
We then used a similar genetic approach to capture the PAS and analyze its ultrastructure. Deletion of ATG genes blocks PAS assembly at a precise stage (Suzuki et al., 2007). Under these conditions, this specialized site is identified by fluorescence microscopy as a perivacuolar punctum positive for the Cvt complex and for several Atg proteins, the array of which depends on the knocked out gene (Suzuki et al., 2007). Atg1 is one of the first proteins to be recruited to the PAS, and consequently, an early formation intermediate accumulates when it is absent (Suzuki et al., 2007). Crucially, Atg9 is exclusively localized at the PAS in atg1Δ cells because it cannot recycle to the reservoirs (Fig. S3; Reggiori et al., 2004a), providing an excellent tool to capture the forming PAS. Remarkably, the PAS appeared as a single cluster of Atg9-containing vesicles and tubules, very similar to the Atg9 reservoirs observed in wild-type and atg11Δ cells (Fig. 6, C and D; and Fig. S4, D–H). A main difference was that this single cluster was detected in fewer cells (Table I; 16% of the cell sections in comparison with 53% and 43% for wild-type and atg11Δ, respectively) and was no longer located in close proximity to the mitochondria, but was mostly adjacent to the vacuole (Table II; 37% of the time), which reinforces the notion that it corresponds to the PAS (Suzuki et al., 2007). Furthermore, this cluster was associated with the Cvt complex in 94% of the cases, which confirms its PAS identity (Fig. 6, C and D; and Fig. S4, E–H).
To corroborate the conclusions drawn from the examination of the atg1Δ mutant, we next analyzed the PAS in atg13Δ and atg14Δ cells, where the PAS biogenesis is also blocked at an early assembly stage (Suzuki et al., 2007) and Atg9 is accumulated at this site (Fig. S3; Reggiori et al., 2004a). Similar to atg1Δ cells, the number of Atg9 reservoirs was strongly reduced in these mutants, and when detected, they were often in association with the Cvt complex and proximal to the vacuole, further suggesting that this is indeed the PAS (Fig. 6, E–H; and Fig. S4, I–Q). Strikingly, the PAS observed in the absence of ATG13 and ATG14 displayed a slightly different morphology from that of atg1Δ cells. In the atg13Δ knockout (Fig. 6, E and F; and S4, I–M), the PAS was comprised almost entirely of small vesicular profiles with a diameter of ∼25–30 nm, whereas in the atg14Δ mutant, more and longer tubular profiles were present in this structure, which in general appeared to be larger (Fig. 6, G and H; and Fig. S3, N–Q).
Together, our data strongly support the concept that the PAS is a vesicular and tubular cluster with morphology very similar to Atg9 reservoirs. The data also indicate that double-membrane vesicles are formed through a process that initially requires the fusion/remodeling of these membranes through the direct or indirect functions of Atg1, Atg13, and Atg14.
The Atg9 reservoirs participate in the generation of the PAS
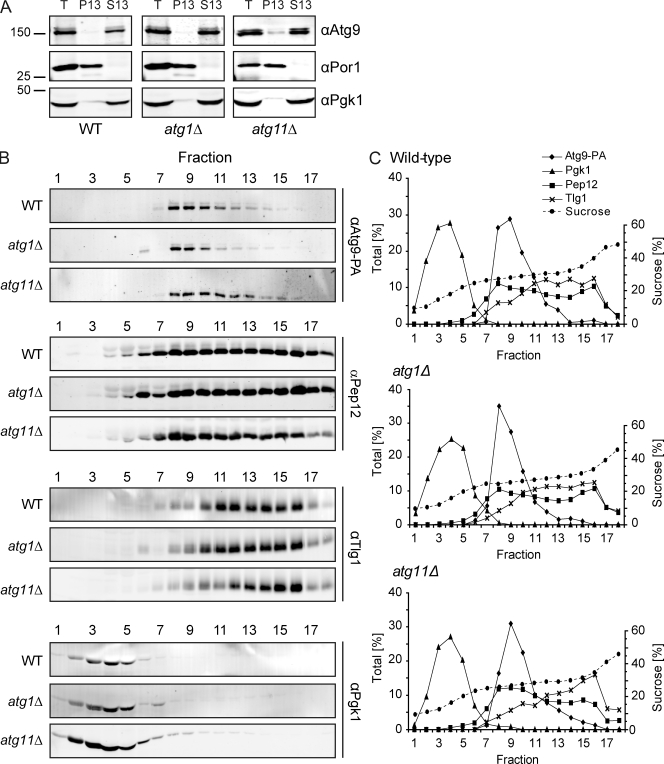
The strong morphological similarity between the Atg9 reservoirs and the PAS suggests that the latter originates from the former. If so, these two structures are expected to have similar biochemical properties. To test this, we fractionated intracellular membranes of wild-type, atg11Δ, and atg1Δ strains. Cell extracts were first centrifuged at 13,000 g. In all three strains, most of the Atg9 was found in the low speed S13 fraction and only a small amount in the pellet P13 fraction (Figs. 7 A and S2 B; Reggiori et al., 2004b, 2005). The S13 supernatant fraction was then separated on a sucrose step-density gradient (Reggiori et al., 2004b). From wild-type cells, Atg9 fractionated in a single peak that was for the most part distinct from EE and LE (Fig. 7, B and C). Importantly, the Atg9 reservoirs and the PAS that accumulated in the atg11Δ and atg1Δ mutant, respectively, are also present in the same fractions (Fig. 7, B and C). This finding indicates that the Atg9 reservoirs and the PAS have almost identical densities, which supports the notion that the PAS could be derived from the Atg9 reservoirs.
Figure 7.
Subcellular fractionation of Atg9 clusters in wild-type, atg11Δ, and atg1Δ backgrounds. (A) The Atg9-PA (FRY172), Atg9-PA atg11Δ pep4Δ (FRY250), and Atg9-PA atg1Δ pep4Δ (FRY196) strains were grown to log phase, converted to spheroplasts, and lysed, then cell extracts were centrifuged at 13,000 g for 15 min. The cell extract (T), the supernatant (S13), and the pellet (P13) fractions were then separated by SDS-PAGE, and Atg9 distribution was analyzed by Western blotting using anti-PA antibodies. Efficient lysis and correct fractionation were assessed by verifying the partitioning of cytosolic Pgk1 and mitochondrial Por1. Mr is indicated in kilodaltons. (B) The S13 supernatants were fractionated on sucrose step-density gradients, and the fractions were analyzed by Western blotting using antisera against PA (for Atg9-PA), Pep12, Tlg1, and Pgk1. (C) Quantification of the immunoblots.
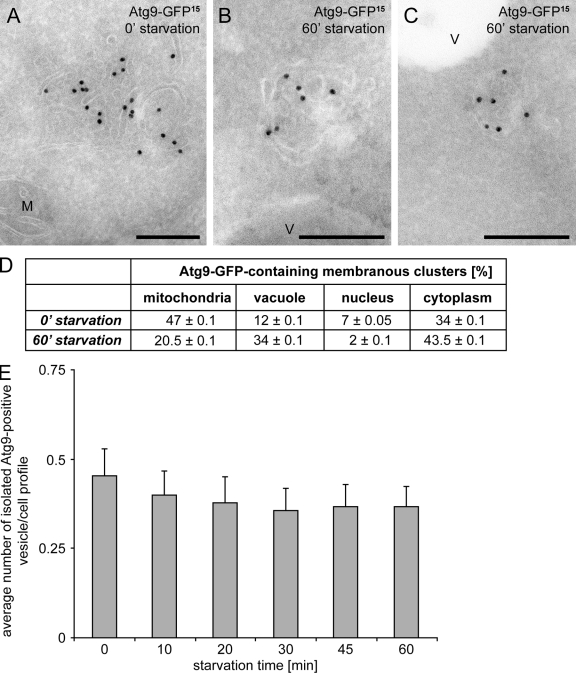
Two possible models could explain how the PAS is generated from the Atg9 reservoirs. In the first, reservoirs translocate en bloc toward the vacuole, whereas in the second model, single vesicles derived from one or more reservoirs assemble near the vacuole. To determine how the PAS is generated from the Atg9 reservoirs, we took advantage of the fact that Atg9 transport to the PAS requires the Cvt complex in growing conditions, but not when autophagy is induced (Shintani and Klionsky, 2004). In the ape1Δ atg1Δ double-knockout cells maintained in growing conditions, therefore, Atg9 remains locked in the Atg9 reservoirs, whereas it accumulates at the PAS when these cells are starved (Shintani and Klionsky, 2004). To determine how Atg9 is transported from the Atg9 reservoirs to the PAS, we followed the relocalization of Atg9 in the ape1Δ atg1Δ mutant by IEM over time (up to 1 h) after autophagy induction by nitrogen starvation. At the early time points, the only Atg9-positive structures we identified were clusters of vesicles and tubules; these corresponded to the Atg9 reservoirs, as they were often found, in 47% of the cases, adjacent to the mitochondria (Fig. 8, A and D). Before starvation (time 0 min), 40% of the cell profiles were positive for 1–3 Atg9 reservoirs. In contrast, 60 min after the induction of autophagy, only 14% of the cell profiles were positive for Atg9 clusters and only one of them was observed per cell section. The Atg9-positive compartments were mostly found close to the vacuole after 60 min (Fig. 8, B–D). This observation is in line with the design of the experiment, which predicts an atg11Δ-equivalent phenotype (43% of cell profiles positive for 1–3 Atg9 clusters) before autophagy induction and an atg1Δ phenotype (16% of cell profiles positive for one Atg9 compartment) after starving the cells. The observed phenotypes were not caused by a structural reorganization of the Atg9 clusters, e.g., fusion and fission, because the mean surface section of the Atg9 clusters is very similar at the 0 and 60 min time points (0.121 and 0.139 mm2, respectively; t test assessment of the individual measurements revealed no significant difference between the values; P = 0.09). Importantly, we did not observe a significant increase in the total number of isolated Atg9-positive vesicles in the cytoplasm, which supports the notion that Atg9 reservoirs translocate en bloc (Fig. 8 E).
Figure 8.
The Atg9 reservoirs move en bloc to become the PAS. The ape1Δ atg1Δ (MMY078) double mutant was grown to log phase before being nitrogen starved to induce autophagy. Cell aliquots were collected after 0, 10, 20, 30, 45, and 60 min. They were then processed for IEM, and cryosections were immunogold labeled for GFP (A–C). (A) An Atg9 reservoir adjacent to a mitochondrion observed at the 0 min time point. (B and C) Atg9 clusters close to the vacuole limiting membrane detected after 60 min of starvation. The gold particles size is indicated on the top of each picture. M, mitochondria; V, vacuole. Bars, 200 nm. (D) Relative subcellular distribution of the Atg9-GFP clusters in the ape1Δ atg1Δ cells before and after induction of autophagy for 60 min. Statistical analyses were performed as described in Materials and methods. (E) The number of isolated Atg9-positive vesicles in the cytoplasm does not increase after triggering Atg9 relocalization in the ape1Δ atg1Δ mutant by autophagy induction. 100 cell profiles were randomly selected and the number of isolated Atg9-positive vesicles per cell profile was established. Results in D and E are expressed in percentages ± the standard error of the mean (error bars).
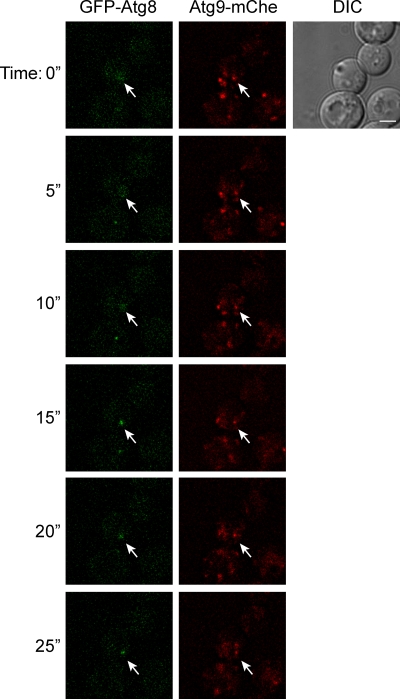
To further demonstrate that the Atg9 reservoirs are the PAS precursor, we analyzed the evolution of this compartment by time-lapse fluorescence microscopy. At present, the primary distinguishing feature of the PAS is that most of the Atg proteins associate at least transiently with this site. Thus, the idea behind this experiment is that an Atg9 reservoir should acquire the rest of the conserved Atg proteins as it becomes a PAS. To test this hypothesis, we generated a strain expressing higher levels of Atg9-monomeric cherry (mChe) to avoid rapid bleaching of the red fluorescent protein, allowing longer recording intervals. In addition, ATG8 was genomically tagged with GFP in the same cells. We selected Atg8 as a PAS protein marker because this factor is the last Atg protein to be recruited at this site (Suzuki et al., 2007), and consequently, its presence at the PAS reflects the complete assembly of the Atg machinery. The engineered cells were then grown to logarithmic (log) phase before being transferred in SD-N medium for 20 min and imaged. Nitrogen starvation was used to induce autophagy because under these conditions, double-membrane vesicles form at a higher frequency, increasing the chance of capturing PAS biogenesis. As shown in Fig. 9 and Video 1, this approach allowed us to observe Atg9 reservoirs becoming the PAS; GFP-Atg8 was seen to move from a cytosolic location and ultimately colocalized with a reservoir. Importantly, the same result was obtained with cells expressing endogenous Atg9-GFP and carrying a plasmid expressing mChe-Atg8 (Video 2). These results demonstrate that the PAS originates from the Atg9 reservoirs, and consequently, this latter compartment supplies at least part of the phagophore membrane. They also further support the notion that the Atg9 reservoirs translocate en bloc to become the PAS.
Figure 9.
Live cell imaging of an Atg9 reservoir becoming the PAS. Atg9-mChe GFP-Atg8 (MMY120) cells were grown to log phase and transferred to SD-N medium for 20 min before being imaged as described in Materials and methods. Sequential images acquired with a time lapse of 5 s are shown. The arrow highlights the Atg9 reservoirs that ultimately colocalize with Atg8 in the process of becoming the PAS. The complete movie reconstruction is presented in Video 1. An identical result was obtained with cells expressing endogenous Atg9-GFP (FRY172) and carrying the pCumCheV5ATG8415 plasmid, which expresses mChe-Atg8 (Video 2). DIC, differential interference contrast. Bar, 2 µm.
Discussion
To shed light on the long-standing issue in autophagy of how autophagosomes are generated, we investigated PAS biogenesis using Atg9 as a protein marker. We discovered that the Atg9 reservoirs are clusters of vesicles and tubules that are often adjacent to mitochondria but not continuous with them (Fig. 2, Fig. S1, and Tables I and II). The reason for the intimate connection between these two organelles remains unknown, and their proximity could be simply due to both organelles being associated with actin cables. Nevertheless, it is now clear that the Atg9 reservoirs are not directly generated from mitochondria. First, we never observed the membrane of the vesicles/tubules comprising this compartment in continuity with the mitochondrial outer membrane. Second, Atg9 was not detected on the surface of this organelle, and conversely, the mitochondrial protein markers Por1 and Idh1 were not found localizing to the Atg9 reservoirs (Fig. 2, B–E; Fig. 3; and Fig. S1). Finally, our studies about Atg9 biosynthesis show that this protein is translocated into the ER and reaches its final location via part of the secretory pathway (Fig. 4). Our results could appear to contradict published studies that suggested Atg9 localized at the mitochondria (Reggiori et al., 2005; He et al., 2006; Reggiori and Klionsky, 2006; Yen et al., 2007). In these studies, the mitochondrial localization of Atg9 was only hypothetical due to the resolution limits of fluorescence microscopy, and thus these previous analyses could not exclude the possibility that Atg9 is in a structure adjacent to mitochondria. Moreover, only a small fraction of Atg9-containing membranes (Fig. 6 A) are found associated with mitochondria in subcellular fractionation experiments (Reggiori et al., 2005). Consequently, our data are not inconsistent with the literature, but rather explain previous observations and solve the discordance about Atg9 localization. It has been reported that the sec12 mutation causes an Atg9 redistribution from preexisting Atg9 reservoirs to the mitochondria surface (Reggiori and Klionsky, 2006). The Atg9 localization to mitochondria, however, was probably artifactual due to this organelle autofluorescence (unpublished data).
Based on our data, the Atg9 reservoirs emerge as a new organelle, and newly synthesized Atg9 is delivered to these sites through part of the secretory pathway (Fig. 4). Because of the only partial colocalization between newly synthesized Atg9 and the late Golgi protein marker Sec7 in sec7 cells (Fig. 4 B), it remains unclear from which Golgi compartment Atg9 is exiting. In addition, it cannot be excluded that Atg9 passes through another organelle, and from there mediates the formation of the Atg9 reservoirs. These issues are currently being investigated in our laboratory. The Atg9 reservoirs are accessible to endocytic material, indicating that they are able to exchange materials with the endocytic system (Fig. 5 A). Atg9, however, shows minimal colocalization, and only modestly fractionates with protein markers of the EE, LE, and Golgi, which indicates that this protein concentrates in a unique organelle (Fig. 5, B and C; and Fig. 7 C; Noda et al., 2000; Reggiori et al., 2004b; Yen et al., 2007). In support of this, the disruption of the endocytic system with specific deletions such as that of VPS4 leads to the concentration of endosomal proteins and several late Golgi factors into an abnormal, large LE adjacent to the vacuole (Odorizzi et al., 1998, 2003) without affecting Atg9 distribution or autophagy efficiency (Epple et al., 2003; Reggiori et al., 2004b; Reggiori and Klionsky, 2006).
It has been postulated that Atg9 is involved in supplying the nascent autophagosomes with lipid bilayers (Reggiori et al., 2004a). Here, we show that the PAS originates from at least one of the Atg9 reservoirs (Figs. 8 and 9). Thus, we argue that the initial membranes of the PAS and by extension of double-membrane vesicles are derived from the Atg9 reservoirs. We cannot exclude the possibility that during the expansion of the phagophore, additional lipid bilayers are obtained from a different source, e.g., from the ER, mitochondria, or Golgi (Hayashi-Nishino et al., 2009; Geng et al., 2010; Hailey et al., 2010; van der Vaart et al., 2010).
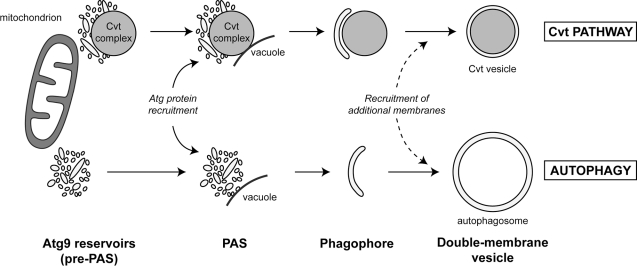
In addition to the similar morphology between the Atg9 reservoirs and the PAS (Figs. 1, 6, S1, and S4), one of our unpredicted discoveries has been the en bloc translocation of the reservoirs to form the PAS next to the vacuole. Our time-lapse fluorescence microscopy showed that the Atg machinery can be recruited to a single Atg9 reservoir (Fig. 9 and Videos 1 and 2). When the Atg9 movement was triggered from the reservoirs to the PAS, we did not observe an increase in the number of isolated Atg9-containing vesicles and tubules in the cytoplasm (Fig. 8). These data support the notion that the Atg9 reservoirs move as clusters. This observation fits with our previous studies showing Atg9 present in cytoplasmic clusters (Reggiori et al., 2004a, 2005) and the demonstration that Atg9 self-interacts (Reggiori et al., 2005; He et al., 2008). An alternative model is that the PAS is generated by a small cluster of vesicles and/or tubules that results from the fragmentation of an Atg9 reservoir, but we do not consider this likely because we have never seen such a scission event during the live-cell imaging experiments, and small clusters comprising less than six vesicular and/or tubular profiles have only rarely been observed in our IEM preparations (see Materials and methods). All together, our data allow us to postulate a model where at least one Atg9 reservoir acts as a pre-PAS and where a change in localization of this compartment determines the biogenesis of the PAS (Fig. 10). The movement of an Atg9 reservoir in close proximity to the vacuole triggers the hierarchical recruitment of the remaining Atg proteins that mediate the rearrangement of these vesicles and tubules into what becomes the phagophore. It remains to be determined which factors on the vacuole limiting membrane or adjacent to it induce the Atg machinery assembly. Nonetheless, at this time we cannot conclusively rule out alternative models, and this hypothesis has to be experimentally demonstrated in future.
Figure 10.
Model for the role of the Atg9 reservoirs in double-membrane vesicle formation. The Atg9 reservoirs, which often are adjacent to mitochondria, act as a pre-PAS. Association with the prApe1 oligomer in nutrient-rich conditions (Cvt pathway) and probably cellular signals during starvation (autophagy) induces the translocation of one or more Atg9 reservoirs into close proximity with the vacuole. This relocalization event triggers the recruitment of the rest of the Atg proteins to a reservoir, leading to the formation of the PAS. Successive fusion of the tubulovesicular membranes composing the PAS and possibly acquisition of additional membrane from other Atg9 reservoirs and/or other sources creates a double-membrane vesicle.
The mammalian orthologue of Atg9 (mAtg9) cycles between the TGN and LE; and after autophagy stimulation, it relocalizes to autophagosomal membranes (Young et al., 2006). All the proteins encoded by the genes knocked out in our study possess orthologues, and some of them have also been implicated in mAtg9 trafficking (Young et al., 2006; Itakura et al., 2008; Chan et al., 2009). Consequently, by extension, our results suggest that the origin of the initial autophagosomal membranes in higher eukaryotes could be the TGN and/or LE, and that the Atg9-positive membranes lead to the formation of the mammalian PAS after relocalization to the cell periphery, but this hypothesis remains to be experimentally tested.
The current hypothesis for double-membrane vesicle formation is that Atg proteins assemble at the PAS, and at this site, they mediate the formation of a phagophore that in turn expands into an autophagosome. Two main models have been proposed for the generation of phagophores: first, the generation by emergence from a defined organelle (which is supported by the recent observation according to which mammalian autophagosomes are generated in close proximity to the ER or mitochondria; Hayashi-Nishino et al., 2009; Hailey et al., 2010), and second, de novo formation by fusion of vesicles. Our results clearly support the model where the initial events of double-membrane vesicle biogenesis in yeast involve the de novo fusion of vesicles and tubules. One observation in support of de novo formation is that we have detected the Atg9-containing clusters of vesicles and tubules always positioned on one side of the Cvt complex, never completely surrounding this circular structure (Fig. 2, D and E; Fig. 6, C–F, cisternae circled with broken lines; Fig. S1, A–D and E; and Fig. S4, E–M). It is consequently tempting to imagine that the initial fusions generate a small cisterna, e.g., the phagophore. Our data do not exclude the possibility that after these early events, the completion of autophagosomes entails the acquisition of additional membranes through a different mechanism.
What would be the molecular basis for the fusion events? It has recently been shown that lipidated Atg8 mediates the tethering and hemifusion of membranes in vitro (Nakatogawa et al., 2007). Our observations indicate that the early fusion events during double-membrane vesicle biogenesis probably do not require these Atg8 functions. In the atg1Δ and atg13Δ mutants, lipidated Atg8 is present at the PAS (Fig. S1, C–H; Suzuki et al., 2007), but our micrographs clearly illustrate that the Atg9-containing vesicles and tubules are not hemifused (Fig. 6, C–F; and Fig. S4, E, F, and H–M). In atg14Δ cells, moreover, Atg8 is not associated with the PAS (Fig. S3, C–H; Suzuki et al., 2007), but the Atg9-positive membranes appear to have undergone some fusion, and this is exemplified by the presence of larger tubular profiles (Fig. 6, G and H; and Fig. S4, N–Q). Our data are consistent with the observation that the tethering and hemifusion properties of lipidated Atg8 do not play a role before the expansion of the autophagosomal membranes (Nakatogawa et al., 2007; Sou et al., 2008). Because Sec18/NSF and SNARE proteins have so far not been implicated in double-membrane vesicle formation (Ishihara et al., 2001; Reggiori et al., 2004b), future studies will have to address which factors carry out these initial fusion events. The Atg proteins are the most likely candidates because deletion of some of them leads to the formation of a PAS with different membrane rearrangement (Fig. 6, C–H; Fig. S2; and Fig. S4, D–Q). Our observations show that the protein composition of the PAS dictates the extent of fusion events (Fig. 6, G and H; and Fig. S4, N–Q) and possibly fission events (Fig. 6, E and F; and Fig. S4, C, D, and N–Q) occurring at this site. Atg14 is involved in the recruitment of the kinase complex that generates the PAS pool of phosphatidylinositol-3-phosphate (Obara et al., 2006). Consequently, our results also indicate that the membrane rearrangements occurring at the PAS could be directly influenced by the lipid composition.
In conclusion, our work has revealed that the PAS is formed from a preexisting cluster of Atg9-containing vesicles and tubules whose composition is unique and from which atypical fusion events generate the double-membrane sequestering vesicles. This discovery provides the knowledge essential to perform further studies on the function of Atg proteins. Understanding the role of Atg proteins in rearranging and fusing membranes will be crucial to unveil the molecular mechanism of autophagy, which in turn will be essential to understand the contribution of this pathway in physiological and pathological situations.
Materials and methods
Plasmids
To create the ATG9-GFP fusion under the control of a TPI1 promoter (pATG9GFP416), ATG9 was amplified by PCR and cloned as a HindIII–XmaI fragment into the pSNA3416 plasmid digested with HindIII–AgeI (Reggiori and Pelham, 2001). The 3′ primer used for this PCR reaction introduced a Gly-Ala-Gly-Ala-Gly-Ala-Gly protein linker between Atg9 and GFP. The integration vector pATG9GFP406 was generated by swapping TPI1-ATG9-GFP as a XhoI–SacI fragment into a pRS406 plasmid (Sikorski and Hieter, 1989). This construct leads to levels of Atg9-GFP eight times higher when compared with endogenous Atg9 as assessed by Western blotting. The plasmid expressing the Atg9-GFP chimera under the control of the GAL1 promoter (pGalATG9416) was made by excising the TPI1 promoter from the pATG9GFP416 vector with XhoI–HindIII and replacing it with that of GAL1, which was obtained by PCR from genomic DNA. The integration vector pATG9mCheV5403, which leads to the expression of Atg9-mChe-V5 under the control of a TPI1 promoter, was created as follows: first, PCR-amplified mCHE-V5 was cloned into the pSNA3416 plasmid as an AgeI–KpnI fragment, resulting in the pSNA3mCheV5416 plasmid. The SNA3 gene was then excised from this plasmid with HindIII and AgeI, and replaced by PCR-amplified ATG9 digested with HindIII and XmaI to produce the pATG9mCheV5416 plasmid. The integration vector pATG9mCheV5403 was finally made by swapping TPI1-ATG9-mCHE-V5 as a XhoI–SacI fragment into a pRS403 plasmid (Sikorski and Hieter, 1989). As with the Atg9-GFP chimera (Fig. 1), the TPI1 promoter-driven Atg9-mChe fusion protein is also functional.
The plasmids expressing monomeric RFP-Atg8 under the control of the authentic promoter (promRFPATG8415) were created in two steps. First, GFP was excised from the pRS316GFP–AUT7 plasmid (a gift of Y. Ohsumi, National Institute for Basic Biology, Okazaki, Japan; Suzuki et al., 2001) with BamHI and replaced by the gene coding for RFP flanked by BamHI sites. The promoter and gene fusions were then shuttled into the pRS415 vector (Sikorski and Hieter, 1989). The plasmid expressing mChe-V5-Atg8 under the control of the CUP1 promoter (pCumCheV5ATG8415) was created by excising and replacing GFP in the pCuGFPATG8415 vector (Kim et al., 2002; Suzuki et al., 2001, 2007), with PCR-generated mCHE-V5 digested with SpeI–XmaI.
The plasmid pmitoDsRed415 was generated by digesting the pADHmitoDsRED (Meeusen and Nunnari, 2003) vector with NotI–HindIII and cloning the resulting ADH1 promoter-mitoDsRED fragment into the pRS415 vector using the same restriction enzymes.
Template plasmids for the PCR-based integration of the mCherry-V5 tag (pFA6a-mCherry-V5-TRP1, pFA6a-mCherry-V5-HIS3, and pFA6a-mCherry-V5-KanMX) at the 3′ end of genes were created as follows: the GFP gene was excised with PacI and AscI from the pFA6a-GFP-TRP1, pFA6a-GFP-HIS3, and pFA6a-GFP-KanMX vectors (Longtine et al., 1998), respectively, and replaced with the PCR-amplified mCherry-V5.
Plasmids pRS416, pJK1-2416, and those used for the genomic integration of Sec7-DsRED, GFP-Atg8, and RFP-Ape1 have been described previously (Sikorski and Hieter, 1989; Noda et al., 2000; Suzuki et al., 2001; Reggiori et al., 2004a; Losev et al., 2006). The plasmid for the genomic integration of Sec7-DsRED was a gift of B. Glick (University of Chicago, Chicago, IL).
Strains
The S. cerevisiae strains used in this study are listed in Table III. For gene disruptions, the APE1, ATG1, ATG9, ATG11, ATG13, and ATG14 coding regions were replaced with the Escherichia coli kanr, the S. cerevisiae TRP1, or the Saccharomyces pombe HIS5 genes using PCR primers containing ∼60 bases of identity to the regions flanking the open reading frame.
Table III.
Strains used in this study
| Name | Genotype | Reference/origin |
| AFM69-1A | MATa sec7-4 his3,11-15 leu2-3,112 ura3-1 | Reggiori et al., 2004b |
| BY4742 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | Euroscarf |
| FBY217 | MATα sec12-4 his3 leu2 ura3 trp1 ade2 | Reggiori et al., 2004b |
| FRY162 | SEY6210 ATG9-GFP::HIS5S.p. | Reggiori et al., 2005 |
| FRY172 | SEY6210 ATG9-PA::TRP1 pep4Δ::LEU2 | Reggiori et al., 2004a |
| FRY196 | SEY6210 ATG9-PA::TRP1 atg1Δ::URA3 pep4Δ::LEU2 | Reggiori et al., 2004a |
| FRY250 | SEY6210 ATG9-PA::TRP1 atg11Δ::URA3 pep4Δ::LEU2 | This study |
| FRY300 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 pho13Δ::KAN pho8::PHO8Δ60 atg9Δ0 | This study |
| FRY340 | SEY6210 ATG9-GFP::TRP1 VRG4-mCHE-V5::HIS5 S.p. | This study |
| FRY341 | SEY6210 ATG9-GFP::TRP1 SEC7-dsRED::URA3 | This study |
| FRY342 | SEY6210 GFP-TLG1 ATG9-mCHE-V5::TRP1 | This study |
| FRY344 | SEY6210 GFP-PEP12 ATG9-mCHE-V5::TRP1 | This study |
| FRY360 | SEY6210 GFP-TLG1 | This study |
| JCK007 | SEY6210 atg9Δ::HIS3 | Noda et al., 2000 |
| MMY067 | SEY6210 atg9Δ::KAN TPI1-ATG9-GFP::URA3 | This study |
| MMY068 | SEY6210 atg9Δ::KAN atg1Δ::HIS5 S.p. TPI1-ATG9-GFP::URA3 | This study |
| MMY069 | SEY6210 atg9Δ::KAN atg11Δ::HIS5 S.p. TPI1-ATG9-GFP::URA3 | This study |
| MMY070 | SEY6210 atg9Δ::KAN atg13Δ::HIS5 S.p. TPI1-ATG9-GFP::URA3 | This study |
| MMY071 | SEY6210 atg9Δ::KAN atg14Δ::HIS5 S.p. TPI1-ATG9-GFP::URA3 | This study |
| MMY072 | SEY6210 atg9Δ::KAN TPI1-ATG9-GFP::URA3 RFP-APE1::LEU2 | This study |
| MMY073 | SEY6210 atg9Δ::KAN atg1Δ::HIS5 S.p TPI1-ATG9-GFP::URA3 RFP-APE1::LEU2 | This study |
| MMY074 | SEY6210 atg9Δ::KAN atg11Δ::HIS5 S.p TPI1-ATG9-GFP::URA3 RFP-APE1::LEU2 | This study |
| MMY075 | SEY6210 atg9Δ::KAN atg13Δ::HIS5 S.p TPI1-ATG9-GFP::URA3 RFP-APE1::LEU2 | This study |
| MMY076 | SEY6210 atg9Δ::KAN atg14Δ::HIS5 S.p TPI1-ATG9-GFP::URA3 RFP-APE1::LEU2 | This study |
| MMY078 | SEY6210 atg9Δ::KAN atg1Δ::HIS5 S.p. ape1Δ::HIS5 S.p. TPI1-ATG9-GFP::URA3 | This study |
| MMY120 | SEY6210 TPI1-ATG9-mCHE-V5::HIS3 | This study |
| MMY125 | BY4742 SEC7-mCHE-V5::KAN | This study |
| MMY126 | BY4742 SEC63-mCHE-V5::KAN | This study |
| MMY127 | MATa sec7-4-mCHE-V5::KAN his3,11-15 leu2-3,112 ura3-1 | This study |
| MMY129 | MATα sec12-4 SEC63-mChe-V5::HIS3 leu2 ura3 trp1 ade2 | This study |
| MOY003 | SEY6210 atg9Δ::KAN TPI1-ATG9-GFP::URA3 IDH1-3xHA::HIS3 | This study |
| SEY6210 | MATα ura3-52 leu2-3, 112 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 mel GAL | Robinson et al., 1988 |
PCR-based integration of GFP, mCHE-V5, and a triple HA tag at the 3′ end of ATG9, IDH1, SEC63, SEC7, and VRG4 was used to generate strains expressing C-terminal fusion proteins under the control of the native promoters. The template for integration was pFA6a-3xHA-HIS5, pFA6a-GFP-TRP1, pFA6a-mCherry-V5-TRP1, pFA6a-mCherry-V5-HIS3, and pFA6a-mCherry-V5-HIS3 (Longtine et al., 1998; and see Plasmids). PCR-based integration of GFP at the 5′ end of TLG1 and PEP12 followed by Cre recombinase-mediated excision of the auxotrophic marker was used to generate strains expressing N-terminal fusion proteins under the control of their native promoters (Gauss et al., 2005). The PCR template for integration was pOM43.
Western blotting using specific antibodies recognizing the proteins encoded by the deleted genes, analysis of the Ape1 processing, and PCR verification were used to confirm all deletions and integrations, and the functionality of all the genomic fusions.
Media
Yeast cells were grown in rich (YPD; 1% yeast extract, 2% peptone, and 2% glucose) or synthetic minimal media (SMD; 0.67% yeast nitrogen base, 2% glucose, and amino acids and vitamins as needed). Starvation experiments were conducted by adding 200 ng/ml of rapamycin or by transferring cells into a nitrogen starvation medium (SD-N; 0.17% yeast nitrogen base without amino acids and ammonium sulfate, and 2% glucose).
Fluorescence microscopy
Yeast cells were grown in the appropriate medium before imaging. Endosomes were stained with FM 4-64 as described previously (Sipos et al., 2004). In brief, 1 ml of log phase cultures was centrifuged and resuspended in 100 µl of ice-cold medium containing 20 µM of FM 4-64 (Invitrogen). After 15 min on ice, cells were washed once with ice-cold medium without FM 4-64 and then resuspended in the same solution before being spotted on a cover slide. Preparations were successively imaged at RT over a 10-min period.
Fluorescence signals were captured with a fluorescence microscope system (DeltaVision RT; (Applied Precision) with a 100× objective lens, equipped with a CoolSNAP HQ camera (Photometrix). Images were generated by collecting a stack of 20 pictures with focal planes 0.20 µm apart to cover the entire volume of a yeast cell (4–5 µm) and by successively deconvolving and analyzing them with the softWoRx software (Applied Precision). A single focal plane is shown at each time point.
To perform statistical analyses of the colocalization degree between fluorescence signals, mild conditions were used to fix cells without destroying the fluorescent proteins (Reggiori et al., 2005). The three-dimensional projection of the picture stacks allow for visualizing all the Atg9-positive puncta present in each cell. Subsequently, the number of these puncta colocalizing with the fluorescent organelle protein marker present in the same cells was determined. Percentages represent how many Atg9-positive puncta colocalize with a specific organelle protein marker. In total, ∼200–350 Atg9-positive puncta were analyzed for each time point, and this counting was repeated twice with samples from two independent experiments. Standard deviations between these two countings were calculated. To determine the number of Atg9-positive compartments per cell in strains expressing endogenous or overexpressed Atg9-GFP, the number of green puncta per cell was counted in 100 randomly chosen cells. Estimation of colocalization between endogenous or overexpressed Atg9-GFP puncta and mitochondria labeled with mitochondria-targeted DsRed was also performed in 100 randomly chosen cells. Three different categories of Atg9-GFP puncta were found. Those completely colocalizing with mitochondria were defined as overlapping and those adjacent or apposed to mitochondria were defined as adjacent, whereas those not in proximity to mitochondria were defined as free.
The time-lapse experiments were performed by imaging the same cells every 5 s for 4 min and 5 s (50 pictures). At each time point, images were generated by collecting a stack of three pictures with focal planes 0.20 µm apart and by successively deconvolving them.
IEM
Cells were fixed, embedded in gelatin, and cryo-sectioned as described previously (Griffith et al., 2008). Sections were then immuno-labeled using rabbit anti-GFP (Abcam) and anti-Ape1 antisera (a gift from I. Sandoval and M. Mazón, Universidad Autonoma de Madrid, Madrid, Spain), mouse anti-Por1 (Invitrogen), and anti-HA antibodies (a gift of G. Bu, Washington University in St. Louis, St. Louis, MO), and goat anti-GFP antibody (Rockland Immunochemicals, Inc.) follow by protein A (PA)-gold detection. The specificity of the antigenic reaction has been controlled for each single or double immunogold labeling that has been performed (Fig. S5). Sections were viewed in an electron microscope (1200 EX; JEOL).
Statistical analysis of IEM data
The presence of gold particles associated with membranes comprising at least six vesicular and/or tubular profiles with a distance no greater than 10–15 nm defined an Atg9 cluster, whereas labeled single vesicles at a distance >100 nm from a membranous structure, vesicle, or organelle, were defined as isolated vesicles. Membranes comprising 2–5 vesicular and/or tubular profiles have not been included in the statistical analyses because they were very rarely observed. A gold particle was assigned to a compartment when no further than 25 nm away from its limiting membrane. The number of Atg9 clusters and Cvt complexes per cell section were counted in wild-type, atg11Δ, and atg1Δ strains by randomly analyzing 250 cell profiles per strain over three independent experiments. This counting was also used to determine the number of cell sections positive for an Atg9 cluster or a Cvt complex.
The relative distribution of Atg9-GFP tubulovesicular clusters was assessed by randomly analyzing 150 sampled profiles from at least four distinct grids. These structures (adjacent or not to a Cvt complex) were designated as being associated with a well-defined organelle (nucleus, vacuole, or mitochondria) if within 50 nm from its limiting membrane. Structures further away were defined as cytoplasmic. Estimation of the average surface section of the Atg9 clusters was performed by analyzing 30 micrographs per condition using the point-hit method (Rabouille, 1999).
Subcellular fractionation
Separation of membranes present in low-speed supernatant fractions on sucrose gradients were performed as described previously (Reggiori et al., 2004b). In brief, 500 OD600 equivalents of spheroplasts were lysed in 5 ml of ice-cold hypo-osmotic buffer (50 mM K2HPO4, pH 7.5, 200 mM sorbitol, and 1 mM EDTA) containing freshly added Complete protease inhibitors (Roche) and 2 mM PMSF by 20 aspirations through a syringe. Cell lysates were centrifuged twice at 500 g for 5 min, and 4 ml of the supernatant fractions (T) were subjected to centrifugation at 13,000 g for 15 min to be separated into supernatant (S13) and pellet (P13) fractions. The S13 fractions (2 ml) were subsequently loaded on a top of sucrose step gradients and centrifuged at 170,000 g for 16 h. All fractions were mixed with 4× Sample buffer, heated at 37°C for 5 min, and resolved by SDS-PAGE followed by Western blotting using antibodies or serum against GFP (Roche), V5 (Invitrogen), Tlg1 (a gift from H. Pelham, Medical Research Council Laboratory of Molecular Biology, Cambridge, England, UK), Por1, Pgk1, and Pep12 (all from Invitrogen).
Pho8 activity assay
4 OD600 of cells in log phase or nitrogen-starved for 4 h were harvested by centrifugation, washed with 1 ml of ice-cold water, and resuspended in 400 µl of Lysis buffer (20 mM Pipes, pH 6.8, 0.5% Triton X-100, 50 mM KCl, 100 mM potassium acetate, 10 mM MgSO4, 10 µM ZnSO4, and 2 mM PMSF). The same volume of glass beads was added and cells were disrupted by agitation for 5 min. After pelleting of cell debris at 13,000 g for 5 min, 100 µl of lysate were mixed with 400 µl of reaction buffer (250 mM Tris-HCl, pH 8.5, 0.4% Triton TX-100, 10 mM MgSO4, 10 µM ZnSO4, and 1.25 mM p-nitrophenyl phosphate; Sigma-Aldrich) prewarmed at 37°C. Incubation was performed at 37°C for 20 min and stopped by adding 500 µl of 1 M glycine buffer, pH 11.0. Samples were then centrifuged at 13,000 g for 2 min, and 1 ml of supernatant was taken to measure the absorbance at OD420. Protein concentration in the lysate was determinate with a Dc Protein Assay kit (Bio-Rad Laboratories). Pho8 activity was calculated as (1,000 × OD420)/(min × [prot]), where min = 20 min and [prot] = protein concentration in the lysate expressed in mg/ml. Results were expressed in a plot as the percentage of the activity measured for the wild-type strain starved for nitrogen, with error bars representing the standard deviation from three experiments.
Ape1 analysis by Western blotting
2.5 OD600 of cells grown in YPD to log phase were collected by centrifugation, and proteins were precipitated with 400 µl of ice-cold 10% trichloroacetic acid for 30 min. After spinning the samples for 5 min, pellets were washed with ice-cold acetone. Pellets were subsequently air dried, resuspended in 100 µl of sample buffer (80 mM Tris-HCl, pH 6.8, 2% SDS, 8.7% glycerol, 2.5% 2-mercaptoethanol, and 0.05% bromophenol blue), and boiled for 5 min. Aliquots of 20 µl were loaded on 8% SDS-PAGE gels, and after Western blotting, membranes were probed with the anti-Ape1 antibodies. The anti-Ape1 polyclonal antiserum was generated by injecting the synthetic peptide CKHWRSVYDEFGEL corresponding to amino acid residues 502–514 of Ape1 in New Zealand white rabbits (New England Peptide).
Online supplemental material
Fig. S1 shows Fig. 2 (D and E) without dashed lines as well as additional examples of Atg9 clusters in wild-type cells. Fig. S2 illustrates that Atg9 overexpression leads to an expansion of the membranes containing this protein without altering their biophysical properties. Fig. S3 shows the subcellular distribution of Atg9 in various mutant strains. Fig. S4 presents Fig. 6 (C–F) without dashed lines and ultrastructural views of Atg9 clusters in various mutant strains. Fig. S5 displays the immunogold labeling controls. Videos 1 and 2 show an Atg9 reservoir becoming the PAS. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200912089/DC1.
Acknowledgments
The authors thank G. Bu, B. Glick, H. Pelham, Y. Ohsumi, M. Mazón and I. Sandoval for reagents; C. Rabouille and J. Klumperman for critical reading of the manuscript; and R. Scriwanek and M. van Peski for assistance with the figure preparation.
F. Reggiori is supported by the Netherlands Organization for Health Research and Development (ZonMW-VIDI) and by the Utrecht University (High Potential grant). D.J. Klionsky is supported by Public Health Service grant GM53396 from the National Institutes of Health.
Footnotes
Abbreviations used in this paper:
- Atg
- autophagy related
- Cvt
- cytoplasm-to-vacuole targeting
- dsRed
- Discosoma red fluorescent protein
- EE
- early endosomes
- IEM
- immunoelectron microscopy
- LE
- late endosomes
- mChe
- monomeric Cherry
- PA
- protein A
- PAS
- phagophore assembly site
References
- Baba M., Osumi M., Scott S.V., Klionsky D.J., Ohsumi Y. 1997. Two distinct pathways for targeting proteins from the cytoplasm to the vacuole/lysosome. J. Cell Biol. 139:1687–1695 10.1083/jcb.139.7.1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlowe C., Schekman R. 1993. SEC12 encodes a guanine-nucleotide-exchange factor essential for transport vesicle budding from the ER. Nature. 365:347–349 10.1038/365347a0 [DOI] [PubMed] [Google Scholar]
- Chan E.Y.W., Longatti A., McKnight N.C., Tooze S.A. 2009. Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domains using an Atg13-independent mechanism. Mol. Cell. Biol. 29:157–171 10.1128/MCB.01082-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies R.J., Sanders S.L., Feldheim D.A., Schekman R. 1991. Assembly of yeast Sec proteins involved in translocation into the endoplasmic reticulum into a membrane-bound multisubunit complex. Nature. 349:806–808 10.1038/349806a0 [DOI] [PubMed] [Google Scholar]
- Epple U.D., Eskelinen E.-L., Thumm M. 2003. Intravacuolar membrane lysis in Saccharomyces cerevisiae. Does vacuolar targeting of Cvt17/Aut5p affect its function? J. Biol. Chem. 278:7810–7821 10.1074/jbc.M209309200 [DOI] [PubMed] [Google Scholar]
- Franzusoff A., Schekman R. 1989. Functional compartments of the yeast Golgi apparatus are defined by the sec7 mutation. EMBO J. 8:2695–2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauss R., Trautwein M., Sommer T., Spang A. 2005. New modules for the repeated internal and N-terminal epitope tagging of genes in Saccharomyces cerevisiae. Yeast. 22:1–12 10.1002/yea.1187 [DOI] [PubMed] [Google Scholar]
- Geng J., Baba M., Nair U., Klionsky D.J. 2008. Quantitative analysis of autophagy-related protein stoichiometry by fluorescence microscopy. J. Cell Biol. 182:129–140 10.1083/jcb.200711112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng J., Nair U., Yasumura-Yorimitsu K., Klionsky D.J. 2010. Post-golgi sec proteins are required for autophagy in Saccharomyces cerevisiae. Mol. Biol. Cell. 21:2257–2269 10.1091/mbc.E09-11-0969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith J., Mari M., De Mazière A., Reggiori F. 2008. A cryosectioning procedure for the ultrastructural analysis and the immunogold labelling of yeast Saccharomyces cerevisiae. Traffic. 9:1060–1072 10.1111/j.1600-0854.2008.00753.x [DOI] [PubMed] [Google Scholar]
- Hailey D.W., Rambold A.S., Satpute-Krishnan P., Mitra K., Sougrat R., Kim P.K., Lippincott-Schwartz J. 2010. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 141:656–667 10.1016/j.cell.2010.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi-Nishino M., Fujita N., Noda T., Yamaguchi A., Yoshimori T., Yamamoto A. 2009. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat. Cell Biol. 11:1433–1437 10.1038/ncb1991 [DOI] [PubMed] [Google Scholar]
- He C., Song H., Yorimitsu T., Monastyrska I., Yen W.-L., Legakis J.E., Klionsky D.J. 2006. Recruitment of Atg9 to the preautophagosomal structure by Atg11 is essential for selective autophagy in budding yeast. J. Cell Biol. 175:925–935 10.1083/jcb.200606084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C., Baba M., Cao Y., Klionsky D.J. 2008. Self-interaction is critical for Atg9 transport and function at the phagophore assembly site during autophagy. Mol. Biol. Cell. 19:5506–5516 10.1091/mbc.E08-05-0544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holthuis J.C., Nichols B.J., Dhruvakumar S., Pelham H.R.B. 1998. Two syntaxin homologues in the TGN/endosomal system of yeast. EMBO J. 17:113–126 10.1093/emboj/17.1.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara N., Hamasaki M., Yokota S., Suzuki K., Kamada Y., Kihara A., Yoshimori T., Noda T., Ohsumi Y. 2001. Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion. Mol. Biol. Cell. 12:3690–3702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E., Kishi C., Inoue K., Mizushima N. 2008. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol. Biol. Cell. 19:5360–5372 10.1091/mbc.E08-01-0080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson C.L., Casanova J.E. 2000. Turning on ARF: the Sec7 family of guanine-nucleotide-exchange factors. Trends Cell Biol. 10:60–67 10.1016/S0962-8924(99)01699-2 [DOI] [PubMed] [Google Scholar]
- Kim J., Huang W.-P., Stromhaug P.E., Klionsky D.J. 2002. Convergence of multiple autophagy and cytoplasm to vacuole targeting components to a perivacuolar membrane compartment prior to de novo vesicle formation. J. Biol. Chem. 277:763–773 10.1074/jbc.M109134200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D.J., Cueva R., Yaver D.S. 1992. Aminopeptidase I of Saccharomyces cerevisiae is localized to the vacuole independent of the secretory pathway. J. Cell Biol. 119:287–299 10.1083/jcb.119.2.287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang T., Reiche S., Straub M., Bredschneider M., Thumm M. 2000. Autophagy and the cvt pathway both depend on AUT9. J. Bacteriol. 182:2125–2133 10.1128/JB.182.8.2125-2133.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B., Deretic V. 2007. Unveiling the roles of autophagy in innate and adaptive immunity. Nat. Rev. Immunol. 7:767–777 10.1038/nri2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B., Kroemer G. 2008. Autophagy in the pathogenesis of disease. Cell. 132:27–42 10.1016/j.cell.2007.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A., III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961 [DOI] [PubMed] [Google Scholar]
- Losev E., Reinke C.A., Jellen J., Strongin D.E., Bevis B.J., Glick B.S. 2006. Golgi maturation visualized in living yeast. Nature. 441:1002–1006 10.1038/nature04717 [DOI] [PubMed] [Google Scholar]
- Meeusen S., Nunnari J. 2003. Evidence for a two membrane-spanning autonomous mitochondrial DNA replisome. J. Cell Biol. 163:503–510 10.1083/jcb.200304040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Levine B., Cuervo A.M., Klionsky D.J. 2008. Autophagy fights disease through cellular self-digestion. Nature. 451:1069–1075 10.1038/nature06639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatogawa H., Ichimura Y., Ohsumi Y. 2007. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 130:165–178 10.1016/j.cell.2007.05.021 [DOI] [PubMed] [Google Scholar]
- Noda T., Kim J., Huang W.-P., Baba M., Tokunaga C., Ohsumi Y., Klionsky D.J. 2000. Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. J. Cell Biol. 148:465–480 10.1083/jcb.148.3.465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P., Field C., Schekman R. 1980. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 21:205–215 10.1016/0092-8674(80)90128-2 [DOI] [PubMed] [Google Scholar]
- Obara K., Sekito T., Ohsumi Y. 2006. Assortment of phosphatidylinositol 3-kinase complexes—Atg14p directs association of complex I to the pre-autophagosomal structure in Saccharomyces cerevisiae. Mol. Biol. Cell. 17:1527–1539 10.1091/mbc.E05-09-0841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odorizzi G., Babst M., Emr S.D. 1998. Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell. 95:847–858 10.1016/S0092-8674(00)81707-9 [DOI] [PubMed] [Google Scholar]
- Odorizzi G., Katzmann D.J., Babst M., Audhya A., Emr S.D. 2003. Bro1 is an endosome-associated protein that functions in the MVB pathway in Saccharomyces cerevisiae. J. Cell Sci. 116:1893–1903 10.1242/jcs.00395 [DOI] [PubMed] [Google Scholar]
- Rabouille C. 1999. Quantitative aspects of immunogold labeling in embedded and nonembedded sections. Methods Mol. Biol. 117:125–144 [DOI] [PubMed] [Google Scholar]
- Reggiori F., Klionsky D.J. 2005. Autophagosomes: biogenesis from scratch? Curr. Opin. Cell Biol. 17:415–422 10.1016/j.ceb.2005.06.007 [DOI] [PubMed] [Google Scholar]
- Reggiori F., Klionsky D.J. 2006. Atg9 sorting from mitochondria is impaired in early secretion and VFT-complex mutants in Saccharomyces cerevisiae. J. Cell Sci. 119:2903–2911 10.1242/jcs.03047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Pelham H.R.B. 2001. Sorting of proteins into multivesicular bodies: ubiquitin-dependent and -independent targeting. EMBO J. 20:5176–5186 10.1093/emboj/20.18.5176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Tucker K.A., Stromhaug P.E., Klionsky D.J. 2004a. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev. Cell. 6:79–90 10.1016/S1534-5807(03)00402-7 [DOI] [PubMed] [Google Scholar]
- Reggiori F., Wang C.-W., Nair U., Shintani T., Abeliovich H., Klionsky D.J. 2004b. Early stages of the secretory pathway, but not endosomes, are required for Cvt vesicle and autophagosome assembly in Saccharomyces cerevisiae. Mol. Biol. Cell. 15:2189–2204 10.1091/mbc.E03-07-0479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Shintani T., Nair U., Klionsky D.J. 2005. Atg9 cycles between mitochondria and the pre-autophagosomal structure in yeasts. Autophagy. 1:101–109 10.4161/auto.1.2.1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J.S., Klionsky D.J., Banta L.M., Emr S.D. 1988. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol. Cell. Biol. 8:4936–4948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T., Klionsky D.J. 2004. Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J. Biol. Chem. 279:29889–29894 10.1074/jbc.M404399200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T., Huang W.-P., Stromhaug P.E., Klionsky D.J. 2002. Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev. Cell. 3:825–837 10.1016/S1534-5807(02)00373-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R.S., Hieter P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 122:19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipos G., Brickner J.H., Brace E.J., Chen L., Rambourg A., Kepes F., Fuller R.S. 2004. Soi3p/Rav1p functions at the early endosome to regulate endocytic trafficking to the vacuole and localization of trans-Golgi network transmembrane proteins. Mol. Biol. Cell. 15:3196–3209 10.1091/mbc.E03-10-0755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sou Y.S., Waguri S., Iwata J., Ueno T., Fujimura T., Hara T., Sawada N., Yamada A., Mizushima N., Uchiyama Y., et al. 2008. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol. Biol. Cell. 19:4762–4775 10.1091/mbc.E08-03-0309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Kirisako T., Kamada Y., Mizushima N., Noda T., Ohsumi Y. 2001. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 20:5971–5981 10.1093/emboj/20.21.5971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Kubota Y., Sekito T., Ohsumi Y. 2007. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 12:209–218 10.1111/j.1365-2443.2007.01050.x [DOI] [PubMed] [Google Scholar]
- van der Vaart A., Griffith J., Reggiori F. 2010. Exit from the golgi is required for the expansion of the autophagosomal phagophore in yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 21:2270–2284 10.1091/mbc.E09-04-0345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vida T.A., Emr S.D. 1995. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 128:779–792 10.1083/jcb.128.5.779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright R., Basson M., D’Ari L., Rine J. 1988. Increased amounts of HMG-CoA reductase induce “karmellae”: a proliferation of stacked membrane pairs surrounding the yeast nucleus. J. Cell Biol. 107:101–114 10.1083/jcb.107.1.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Klionsky D.J. 2007. Autophagosome formation: core machinery and adaptations. Nat. Cell Biol. 9:1102–1109 10.1038/ncb1007-1102 [DOI] [PubMed] [Google Scholar]
- Yen W.L., Legakis J.E., Nair U., Klionsky D.J. 2007. Atg27 is required for autophagy-dependent cycling of Atg9. Mol. Biol. Cell. 18:581–593 10.1091/mbc.E06-07-0612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young A.R.J., Chan E.Y.W., Hu X.W., Köchl R., Crawshaw S.G., High S., Hailey D.W., Lippincott-Schwartz J., Tooze S.A. 2006. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J. Cell Sci. 119:3888–3900 10.1242/jcs.03172 [DOI] [PubMed] [Google Scholar]