Self- and hetero-association of the pro-apoptotic proteins Reaper, Hid, and Grim is required for efficient induction of the cell death program.
Abstract
Apoptosis is a specific form of cell death that is important for normal development and tissue homeostasis. Caspases are critical executioners of apoptosis, and living cells prevent their inappropriate activation through inhibitor of apoptosis proteins (IAPs). In Drosophila, caspase activation depends on the IAP antagonists, Reaper (Rpr), Head involution defective (Hid), and Grim. These proteins share a common motif to bind Drosophila IAP1 (DIAP1) and have partially redundant functions. We now show that IAP antagonists physically interact with each other. Rpr is able to self-associate and also binds to Hid and Grim. We have defined the domain involved in self-association and demonstrate that it is critical for cell-killing activity in vivo. In addition, we show that Rpr requires Hid for recruitment to the mitochondrial membrane and for efficient induction of cell death in vivo. Both targeting of Rpr to mitochondria and forced dimerization strongly promotes apoptosis. Our results reveal the functional importance of a previously unrecognized multimeric IAP antagonist complex for the induction of apoptosis.
Introduction
Apoptosis is a genetically encoded process of cell death with defined morphological features that serves to kill superfluous or unwanted cells, and abnormal regulation of this process is associated with many human diseases (Steller, 1995; Thompson, 1995; Yuan and Yankner, 2000). An evolutionarily conserved feature of apoptosis is the activation of a particular class of proteases, termed caspases (Thornberry and Lazebnik, 1998), which cleave many vital structural and regulatory proteins in the cell (Hengartner, 2000). Activation of caspases is kept in check by a conserved class of anti-apoptotic proteins, termed inhibitor of apoptosis proteins (IAPs; Reed et al., 2004; Shiozaki and Shi, 2004). IAPs can bind to both initiator and effector caspases via their BIR domains (Shi, 2002; Bergmann et al., 2003). Furthermore, many IAPs also contain a RING motif and act as E3 ubiquitin ligases to ubiquitinate cell death proteins, including caspases (Wilson et al., 2002; Tenev et al., 2005). In Drosophila, DIAP1 is strictly required to prevent caspase activation and apoptosis in virtually all somatic cells (Wang et al., 1999; Goyal et al., 2000; Lisi et al., 2000). In cells that are doomed to die, IAPs are inactivated by specific antagonists (Vucic et al., 1997; Goyal et al., 2000; Yan et al., 2004).
In Drosophila, three IAP antagonists, Reaper (Rpr), Head involution defective (Hid), and Grim are clustered together in the genome, and deleting these genes causes a severe inhibition of apoptosis (White et al., 1994; Grether et al., 1995; Chen et al., 1996). A fourth IAP antagonist, Sickle (Skl), was also identified with significant similarity to Rpr (Srinivasula et al., 2002), but due to the lack of mutants its physiological role for the induction of apoptosis is less clear. One evolutionarily conserved feature is the presence of the N-terminal IBM (IAP-binding motif), a stretch of several amino acids that interacts with the BIR domains of IAPs (Vucic et al., 1998; Shi, 2002). IAP antagonists bind IAPs and displace competitively IAP-bound caspases (Holley et al., 2002; Chai et al., 2003; Zachariou et al., 2003). Active caspases propagate a proteolytic cascade that will compromise the cell’s infrastructure and metabolism. Another aspect of IAP antagonists’ function is to stimulate IAP turnover by proteasomal degradation (Ryoo et al., 2002; Yoo et al., 2002). When expressed in human cells, Drosophila IAP antagonists preserve similar activities such as inducing cell death (McCarthy and Dixit, 1998; Haining et al., 1999) and binding and stimulating human IAP degradation (Silke et al., 2004). Humans also have IAP antagonists, among which the best characterized is Smac/Diablo (Du et al., 2000; Verhagen et al., 2000). Smac forms dimers and interacts with the BIR domains of XIAP (Wu et al., 2000), and yet the significance of dimer formation is not known. Other human IAP antagonists include HtrA2/Omi (also present in Drosophila; Hegde et al., 2002) and ARTS (Gottfried et al., 2004).
Rpr is a small protein of 65 amino acids (White et al., 1994). Previous reports have suggested that Rpr (Olson et al., 2003a), Hid (Haining et al., 1999), and Grim (Clavería et al., 2002) localize to the mitochondria. Rpr and Grim share a homologous motif outside of IBM, known as the GH3 (Grim helix 3), which is required for their mitochondrial localization (Clavería et al., 2002; Olson et al., 2003a). Disruption of this GH3 motif in Rpr not only impairs its mitochondrial translocation, but also disrupts Rpr’s ability to stimulate DIAP1 auto-ubiquitination and degradation (Freel et al., 2008).
The fact that rpr, hid, and grim share homologous IBMs and that this motif binds to specific pockets in the DIAP1 BIR domains (Wu et al., 2001; Chai et al., 2003; Yan et al., 2004) has led to the idea that the IAP antagonists have partially redundant roles. Here, we provide evidence that these proteins work together as a high-order physical complex for efficient DIAP1 inactivation. Specifically, we present a structure-function analysis of Rpr that reveals the importance of a central helical domain in dimerization, the formation of multimeric complexes with other IAP antagonists, protein localization, and the ability of Rpr to promote DIAP1 degradation.
Results
Rpr self-association is essential for its apoptotic activity
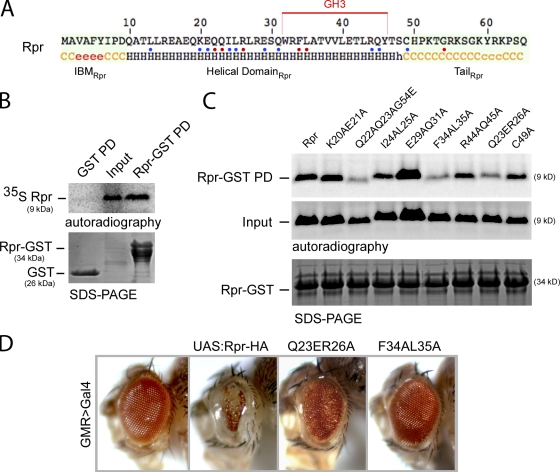
To understand how Rpr interacts with DIAP1 to induce its ubiquitination, we investigated the interaction between Rpr, DIAP1, and other related apoptosis regulator proteins. In the absence of a Rpr three-dimensional structure, we have performed a secondary structure prediction to identify structural elements in the amino acid sequence. Rpr consists of three major elements, the IBM motif (residues 1–9), a central helical domain (residues 10–48) that includes the GH3 motif (Olson et al., 2003a) and adopts an α-helical conformation, and a C-terminal unstructured tail (residues 49–65) (Fig. 1 A). Because many protein helical domains are involved in protein–protein interaction, we hypothesized that Rpr might interact with self or with other proteins through this helical domain. Supporting this idea, Rpr-GST recombinant protein was able to pull down 35S radiolabeled Rpr in vitro (Fig. 1 B). The interaction proved to be specific because control GST was not able to pull down 35S-Rpr (Fig. 1 B). Next, we set out to identify the amino acids involved in this interaction by introducing a number of point mutants that span the entire Rpr helical domain through site-directed mutagenesis (represented in Fig. 1 A). In support of our hypothesis, three mutants, Q23ER26A, F34AL35A, and Q22AQ23AG54E, were found to have reduced affinity for Rpr-GST (Fig. 1 C). Subsequently, we investigated the functional relevance of the mutations that disrupt Rpr self-association in vivo. Specifically, we generated fly transgenes in which Rpr-HA, mutant Q23ER26A, and the GH3 mutant F34AL35A were targeted to a defined genomic locus by Cre-mediated recombination (Oberstein et al., 2005) and compared their pro-apoptotic activity when expressed in developing Drosophila eyes, using the GMR>Gal4/UAS system. As previously reported, Rpr-HA–induced expression in the eyes produced severe eye ablation (White et al., 1996). On the other hand, the Q23ER26A variant had only mild apoptotic activity, whereas the GH3 mutant F34AL35A was completely inactive, yielding normal eyes (Fig. 1 D). These results support the idea that Rpr self-association is important for the protein’s apoptotic activity, and the disruption of self-association blocks protein activity in vivo.
Figure 1.
Rpr self-association and its impact on apoptotic activity. (A) Secondary structure consensus prediction of the Drosophila Rpr. Nomenclature: c, disordered; e, β-strand; h, helical. Three distinct Rpr domains are distinguishable: the IBM motif (IBMRpr), a central helical domain (Helical DomainRpr), and a C-terminal unstructured tail (TailRpr). The GH3 domain is marked with a red line above the amino acid sequence. Blue and red dots represent amino acids that were replaced by site-directed mutagenesis. Red dots represent amino acids that have an effect on the protein activities once replaced. (B) Pull-down (PD) experiment for testing the interaction between Rpr-GST and 35S-Rpr (Rpr-GST PD). As a specificity control, GST (bait) was tested for interaction with 35S-Rpr (GST PD). “Input” shows the expression of the radiolabeled Rpr and represents 10% of the protein amount used in the PD assay. “SDS-PAGE” indicates the amount of bait proteins used, as visualized by Coomassie staining. “Autoradiography” shows the radiolabeled proteins in the experiments. (C) Protein–protein interaction assay between Rpr-GST and 35S-Rpr mutants. “SDS-PAGE” indicates the amount of Rpr-GST protein used as bait. “Input” lanes indicate the autoradiography detection of the in vitro–translated 35S-Rpr mutants, for expression comparison. Each represents 10% of radiolabeled Rpr mutant amounts used in the PD assay. Rpr-GST PD is a pull-down assay, showing the binding of the individual 35S-Rpr mutants to Rpr-GST. (D) Eye images of transgenic Drosophila expressing Rpr-HA and Rpr-HA mutants Q23ER26A and the GH3 mutant F34AL35A. Genotypes: ;GMR>Gal4/+;, ;UAS:Rpr-HA/GMR>Gal4;, ;UAS:Rpr-HA Q23ER26A/GMR>Gal4;, ;UAS:Rpr-HA F34AL35A/GMR>Gal4;.
Enforced dimerization of Rpr leads to efficient cell killing
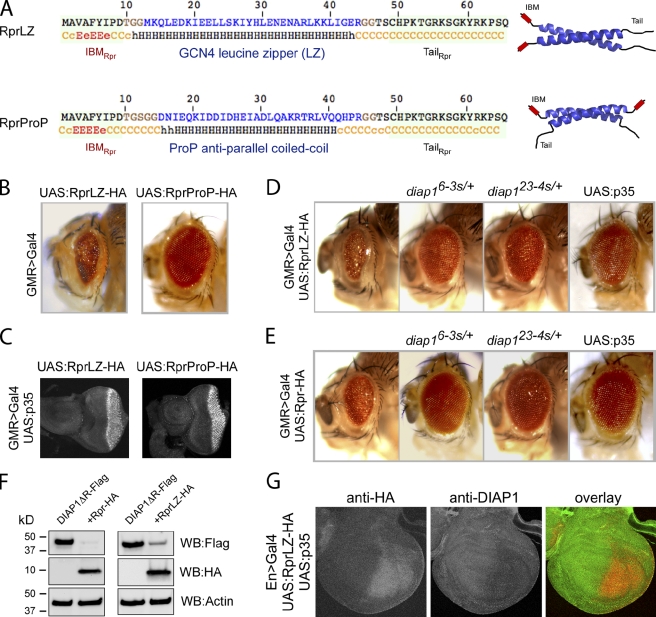
Although the above experiments indicate that Rpr self-association is required for its pro-apoptotic activity, whether it is sufficient to recapitulate Rpr’s pro-apoptotic function remained unclear. To test this, we replaced the helical domain of Rpr (residues 10–46) with well-defined dimerization domains from heterologous proteins whose three-dimensional structures have been previously determined. Specifically, we used a parallel leucine zipper (LZ) from the yeast transcription factor GCN4 (O’Shea et al., 1991) and an anti-parallel coiled-coil domain from the Escherichia coli osmosensor ProP (Zoetewey et al., 2003; Fig. 2 A). When these chimeric proteins were expressed in the fly eye using the GMR>Gal4/UAS system we found that RprLZ triggered massive cell death, as evidenced by the partially ablated eye structure (Fig. 2 B), supporting the idea that Rpr dimerization is sufficient to account for its central helical domain’s function. On the other hand, RprProP did not trigger cell death under similar conditions, despite being expressed at similar levels with RprLZ (Fig. 2 C). Next, we examined whether RprLZ induces cell death through a mechanism similar to the wild-type Rpr, namely the inhibition of DIAP1 and activation of caspases. Supporting the requirement of caspase activation, coexpression of p35, a well-established caspase inhibitor of viral origin, rescued the eye morphology caused by RprLZ (Fig. 2 D, right) as well as wild-type Rpr (Fig. 2 E, right). To test the requirement of DIAP1 inactivation, we took advantage of the diap16-3s and diap123-4s alleles, which are endogenous alleles bearing point mutations in the IBM-binding pocket of DIAP1 BIR domains, making cells resistant to Rpr-induced cell death (Goyal et al., 2000). The presence of these diap1 alleles in the background significantly suppressed apoptosis induced by the RprLZ (Fig. 2 D) as well as wild-type Rpr (Fig. 2 E). Next, we examined the ability of RprLZ to induce DIAP1 degradation. In coexpression experiments in HEK293 cells, RprLZ was able to stimulate DIAP1ΔR degradation to significant extent (Fig. 2 F, right), but lower than wild-type Rpr (Fig. 2 F, left). DIAP1ΔR was used instead of full-length DIAP1 due to its increased stability (not depicted). Ability of RprLZ to induce DIAP1 degradation was also shown in wing discs, after overexpression in the presence of p35 (Fig. 2 G). These results support the idea that RprLZ has pro-apoptotic mechanisms similar to that of wild-type Rpr.
Figure 2.
Enforced Rpr dimers kill by apoptosis in Drosophila. (A) Amino acid sequences and structural elements of Rpr dimers. RprLZ is an enforced parallel Rpr dimer where Rpr helical region (residues 10–46) was replaced with a parallel leucine zipper (GCN4), whereas RprProP is an enforced anti-parallel Rpr-dimer. LZ and ProP amino acid sequences are represented in blue. Residues in brown were inserted on both sides of each dimerization domain to preserve the same length as wild-type Rpr. IBMRpr and TailRpr are identical as in wild-type Rpr. A secondary structure prediction is represented below each sequence. Nomenclature: c, disordered; e, β-strand; h, helical. All constructs have attached a C-terminal HA tag, not represented in this diagram. To the right are schematic representations of RprLZ and RprProP with the IBMRpr (shown in red), ribbon representations of the dimerization domains LZ (PDB #2ZTA) and ProP (PDB #1R48) (shown in blue) and the TailRpr (shown in black). Note the position of the IBM motifs in RprLZ and RprProP. (B) Drosophila eye images from transgenic flies expressing RprLZ-HA or RprProP-HA. Genotypes: ;UAS:RprLZ-HA/GMR>Gal4; and ;UAS:RprProP-HA/GMR>Gal4;. (C) Eye-antennal imaginal discs from third instar transgenic larvae, expressing RprLZ-HA and RprProP-HA, stained with an anti-HA antibody. Genotype: UAS:p35/+;UAS:RprLZ-HA/GMR>Gal4; and UAS:p35/+;UAS:RprProP-HA/GMR>Gal4;. (D) Rescue of the RprLZ-HA induced eye ablation by Rpr-insensitive diap1 alleles or p35. Genotypes: ;UAS:RprLZ-HA/GMR>Gal4;, ;UAS:RprLZ-HA/GMR>Gal4;diap16-3s/+, ;UAS:RprLZ-HA/GMR>Gal4;diap123-4s/+ and UAS:p35/+;UAS:RprLZ-HA/GMR>Gal4;. (E) Rescue of the Rpr-HA induced eye ablation by Rpr-insensitive diap1 alleles or p35. Genotypes are identical to D, except that UAS:RprLZ-HA was replaced with UAS:Rpr-HA. (F) Ectopic expression of DIAP1ΔR-Flag or coexpression with Rpr-HA or RprLZ-HA in HEK293 cells, showing the ability of Rpr and RprLZ to induce DIAP1 degradation. Actin was used as a loading control. (G) Overexpression of RprLZ-HA in the presence of p35 in the posterior compartment of the wing discs and its effect on DIAP1 level. Expression of RprLZ was detected with an anti-HA antibody, whereas DIAP1 was immunostained with a rabbit anti-DIAP1 antibody.
Rpr physically interacts with other Drosophila IAP antagonists
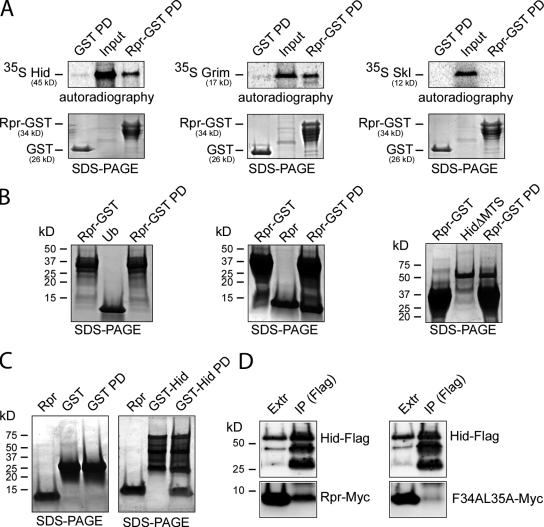
Next, we asked whether Rpr interacts with the other Drosophila IAP antagonists. Specifically, we tested potential interactions between Rpr-GST and 35S-Hid, 35S-Grim, or 35S-Skl through in vitro pull-down assays. We found that Rpr can interact with the other Drosophila IAP antagonists Hid and Grim, but not with Skl (Fig. 3 A). Under identical conditions, a control GST protein did not interact with 35S-Hid, 35S-Grim, or 35S-Skl, indicative of the specificity of the observed interactions (Fig. 3 A). The Rpr–Rpr and Rpr–Hid interactions were further confirmed by alternative pull-down experiments using Rpr-GST as “bait” and purified ubiquitin (Ub), Rpr, and HidΔMTS as “prey”. Besides confirming the specific Rpr–Rpr interaction, this experiment also indicates that Rpr–Hid interaction is not dependent on Hid’s MTS (Fig. 3 B). The interaction between Rpr and Hid was further confirmed by a reverse pull-down assay, using purified GST-Hid as bait and purified Rpr as a prey (Fig. 3 C). Moreover, we have performed competitive displacement experiments, where preformed Rpr-GST:Rpr and Rpr-GST:HidΔMTS complexes were incubated with increasing amounts of HidΔMTS or Rpr and could see displacement of proteins in these complexes (unpublished data). Next, we asked whether Rpr interacts with Hid using the same domain used for self-association. To this end, a Hid-Flag construct was cotransfected with Rpr-Myc or the GH3 mutant F34AL35A-Myc constructs in HEK293 cells followed by anti-Flag immunoprecipitation. Interestingly, Hid coimmunoprecipitated with wild-type Rpr but not with the GH3 mutant F34AL35A (Fig. 3 D). This experiment argues that Rpr uses the same domain for protein association, either with self or with Hid. Alternatively, Rpr dimers interact with Hid in an oligomeric complex, and disrupting Rpr dimer interface blocks the formation of an oligomeric complex with Hid.
Figure 3.
Rpr forms complexes with the other Drosophila IAP antagonists. (A) Pull-down assays for testing the interaction between Rpr-GST and 35S-Hid, 35S-Grim, or 35S-Skl (Rpr-GST PD). As specificity controls, pull-down experiments were performed between GST and 35S-Hid, 35S-Grim, or 35S-Skl (GST PD). “Input” lanes indicate the autoradiography detection of the in vitro–translated 35S-Hid, 35S-Grim, and 35S-Skl, for expression comparison. Each represents 10% of the radiolabeled protein amount used in the PD assay. “SDS-PAGE” shows the amount of GST or Rpr-GST used as bait. “Autoradiography” shows the phosphorimager detection of the radiolabeled proteins in the experiment. (B) Alternative pull-down experiments using purified components confirm Rpr self-association and show the interaction between Rpr and Hid lacking the mitochondrial targeting sequence. (Left) Specificity control experiment that shows the lack of interaction between Rpr-GST and ubiquitin (Rpr-GST PD). (Middle) Interaction between Rpr-GST and Rpr (Rpr-GST PD). (Right) Interaction between Rpr-GST and HidΔMTS (Rpr-GST PD). Rpr-GST (shown in the first lane of each panel) was incubated with purified ubiquitin, Rpr, or HidΔMTS (shown in the second lane of each panel). Protein complexes immobilized on glutathione Sepharose beads (Rpr-GST PD) are shown in the third lane of each panel. The proteins were separated by SDS-PAGE and visualized by Coomassie staining. (C) Reverse pull-down experiment showing the interaction between GST-Hid and Rpr. As a specificity control, GST failed to pull down Rpr. Purified Rpr protein (shown in the first lane of each panel) was incubated with either GST or GST-Hid (shown in the second lane of each panel). After incubation the complexes were pulled down using glutathione Sepharose beads (third lane of each panel). (D) Hid–Rpr and Hid–Rpr GH3 mutant F34AL35A coimmunoprecipitation experiment. Hid (anti-FLAG) does coimmunoprecipitate with Rpr (anti-Myc) from HEK293 cells (left) but not with the Rpr GH3 mutant F34AL35A (right). “Extr” represents the cell extract lane showing Hid-FLAG, Rpr-Myc, or GH3 mutant F34AL35A-Myc expression. “IP (FLAG)” represents the anti-FLAG immunoprecipitation fraction showing the level of Hid-FLAG, Rpr-Myc, or mutant levels in this fraction.
Rpr is targeted to mitochondria via interaction with Hid
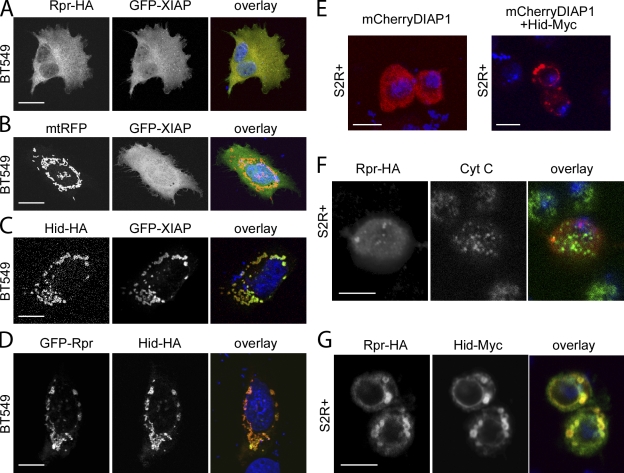
Although Rpr is known to localize to mitochondria (Olson et al., 2003a; Abdelwahid et al., 2007), prediction tools used to assess protein localization failed to identify any motifs for specific subcellular localization. This raises the possibility that Rpr localizes to the mitochondria through a novel mechanism. Thus, we decided to investigate Rpr localization by ectopic coexpression experiments in human BT549 cells, as well as in Drosophila S2R+ cells. We specifically followed the distribution of Rpr-HA, as well as XIAP, a human IAP member that is known to bind Rpr (Holley et al., 2002). Rpr-HA (Fig. 4 A) as well as GFP-fused XIAP (Fig. 4 B) was found to be spread diffusely throughout the cytoplasm. Similar experiments using Rpr-Myc and GFP-Rpr confirmed the broad distribution of Rpr in BT549 cells (unpublished data). In contrast, Hid, which has a mitochondrial targeting sequence, localizes exclusively to mitochondria and triggers GFP-XIAP translocation to this organelle (Fig. 4 C). This experiment confirms the ability of the IAP antagonist to interact with other proteins and recruit them to mitochondria. We also coexpressed GFP-Rpr and Hid in the BT549 cells and monitored any changes in the intracellular distribution of the proteins. Consistent with the ability of Rpr to bind Hid in vitro, the presence of Hid prompted GFP-Rpr distribution to change into a mitochondrial pattern (Fig. 4 D). The experiments above suggest that Rpr is not a mitochondrial protein, per se, but it is recruited to mitochondria by interaction with a mitochondrial-anchored protein. To further validate these results, we performed a similar set of experiments in Drosophila S2R+ cells. As in BT549 cells, we observed that mCherry-DIAP1 is distributed evenly throughout the cytoplasm of S2R+, but after cotransfection with Hid, mCherry-DIAP1 is translocated to mitochondria in a Hid-like pattern (Fig. 4 E). When Rpr was expressed transiently in S2R+ cells, it shows an occasional punctate staining that is only coincidental with cytochrome c (Cyt C) staining (Fig. 4 F). However, after cotransfection with Hid, Rpr’s colocalization with Hid becomes obvious (Fig. 4 G). In sum, our experiments suggest that Rpr is a soluble protein that displays a diffuse distribution throughout the cell, and coexpression with Hid leads to Rpr relocation to the mitochondria. Additionally, these experiments underline Hid’s ability to recruit DIAP1 and its human homologue XIAP to the mitochondrial membrane. To test whether the recruitment of Rpr, XIAP, and DIAP1 is indeed dependent on Hid’s MTS, we have performed coexpression experiments with HidΔMTS and Rpr, XIAP, or DIAP1. HidΔMTS localizes to the nucleus in BT549 and S2R+ cells and triggers nuclear localization of Rpr, XIAP, and DIAP1 (Fig. S1), suggesting that indeed the observed mitochondrial localization of Rpr, XIAP, and DIAP1 is dependent on mitochondrial localization of Hid. Because Rpr and Hid overexpression induce cell death, we tested whether this might induce a change in the intracellular localization of these proteins. However, addition of the caspase inhibitor zVAD-FMK did not affect localization of Rpr or Hid (Fig. S2).
Figure 4.
Rpr translocates to the mitochondria through physical interaction with Hid. (A) BT549 cells expressing Rpr-HA and GFP-XIAP. Rpr was stained with an anti-HA antibody. “Overlay” represents a composite image of Rpr (red), GFP-XIAP (green), and nuclei (DAPI, blue) staining. Bar, 20 µm. (B) BT549 cells transiently transfected with GFP-XIAP and mitochondrial RFP (mtRFP) plasmids. “Overlay” indicates GFP-XIAP (green), mitochondria (red), and nuclei (blue). Bar, 20 µm. (C) BT549 cells cotransfected with Hid-HA and GFP-XIAP plasmids. “Overlay” indicates Hid (red), GFP-XIAP (green), and nuclei (blue). Bar, 20 µm. (D) BT549 cells cotransfected with GFP-Rpr and Hid-HA plasmids. “Overlay” shows Rpr (green), Hid (red), and nuclei (blue) staining. Bar, 20 µm. (E) S2R+ Drosophila cells transiently transfected with a mCherryDIAP1 plasmid (left image) or with mCherryDIAP1 and Hid-Myc plasmids (right image). Each image shows the overlay of DIAP1 (red) and nuclei (blue) staining. Bar, 5 µm. (F) S2R+ Drosophila cell, transiently transfected with a Rpr-HA plasmid, followed by immunostaining with anti-HA and anti-Cyt C antibodies. “Overlay” indicates Rpr (red), Cyt C (green), and nuclei (blue) staining. Bar, 5 µm. (G) S2R+ Drosophila cell cotransfected with Rpr-HA and Hid-Myc plasmids. Cells were immunostained with an anti-HA antibody and an anti-Myc antibody. “Overlay” represents Rpr (green), Hid (red), and nuclei (blue) staining. Bar, 5 µm.
Functional cooperativity between Rpr and Hid
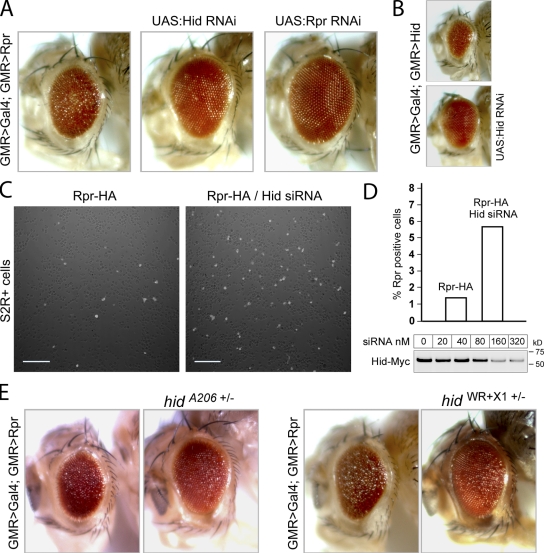
To test whether the physical interaction between Rpr and Hid has functional significance, we examined Rpr’s ability to kill cells in the absence of Hid. Although GMR>Rpr flies had rough eyes as expected, RNAi-mediated knockdown of Hid considerably suppressed this cell death phenotype of GMR>Rpr (Fig. 5 A). The cell death phenotype of GMR>Rpr is indeed caused by Rpr overexpression and could be suppressed by Rpr RNAi (Fig. 5 A). Furthermore, we could show that the used Hid RNAi line is effective at knocking down endogenous Hid because it rescues the GMR>Hid-induced eye ablation (Fig. 5 B). The effect of Hid knockdown on Rpr-induced cell death was also observed in cell culture. When Drosophila S2R+ cells were transiently transfected with a Rpr-HA plasmid in the presence of a 21-bp Hid RNA duplex (siRNA), the number of Rpr-HA–positive (5.65%), as identified by anti-HA immunostaining, were almost fourfold greater than those without Hid siRNA treatment (1.46%; Fig. 5, C and D). We interpret that depleting Hid mRNA in S2R+ cells gives these cells a better protection against Rpr-induced cell death, allowing a larger population of Rpr-positive cells to survive. The efficiency of the Hid siRNA was demonstrated by the ability to decrease the level of Hid-Myc in S2R+ cells in transient transfection experiments (Fig. 5 D). Hid’s role in Rpr-induced cell death was further validated in the heterozygous background of hid loss-of-function mutant alleles, hidA206 and hidWR+X1 (Abbott and Lengyel, 1991; Grether et al., 1995), which suppressed the degree of Rpr-induced eye ablation (Fig. 5 E). Conversely, Hid-induced cell death in Drosophila eyes is suppressed to a degree by Rpr RNAi (Fig. S3 A). These findings are in contrast to the widely recognized view that the three Drosophila IAP antagonists Rpr, Hid, and Grim induce cell death independently, are functionally redundant, and their cell death output is additive.
Figure 5.
Hid and Rpr act cooperatively to induce cell death in Drosophila. (A) Rescue of Rpr-induced eye ablation by Hid RNAi and Rpr RNAi. A rough Drosophila eye caused by overexpression of Rpr (left) is suppressed when RNAi transgenes knock down either Hid (UAS-CG5123 RNAi; middle) or Rpr (UAS-CG4319 RNAi; right). Genotypes: (left) ;GMR>Gal4/+;GMR>Rpr/+, (middle) ;GMR>Gal4/+;UAS:Hid RNAi/GMR>Rpr, (right) ;GMR>Gal4/+;UAS-Rpr RNAi/GMR>Rpr. (B) Hid-induced eye ablation (top image) is suppressed through UAS-Hid RNAi (bottom image). Genotypes: (top) GMR>Gal4/+;GMR-Hid/+;, (bottom) GMR>Gal4/+;GMR>Hid/+;UAS:Hid RNAi/+. (C) Anti-HA immunolabeling of Drosophila S2R+ cells transiently transfected with a Rpr-HA plasmid alone (left), or together with 200 nM siRNA directed against Hid mRNA (right). Bar, 100 µm. (D) Quantification of Rpr-HA–positive cells, in the absence or presence of Hid siRNA in S2R+ transient transfection experiments (top). Percentages of Rpr-positive cells were calculated by counting of at least 1,000 cells for each sample. Efficiency of Hid siRNA as assessed through anti-Myc Western blot of ectopically expressed Hid-Myc in the presence of Hid siRNA in S2R+ cells. (E) Rescue of the Rpr-induced eye ablation by the hidA206 and hidWR+X1 alleles. (First two images) Comparison of Rpr eye phenotype without or with the hidA206 allele. Genotypes: ;GMR>Gal4/+;GMR>Rpr/+ and ;GMR>Gal4/+;GMR>Rpr/hidA206. (Last two images) Rpr eye phenotype without or with hidWR+X1 allele. Genotypes: ;GMR>Gal4/+;GMR>Rpr/+ and ;GMR>Gal4/+;GMR>Rpr/hidW+RX1.
Rpr can stimulate Diap1 self-conjugation in a purified in vitro system
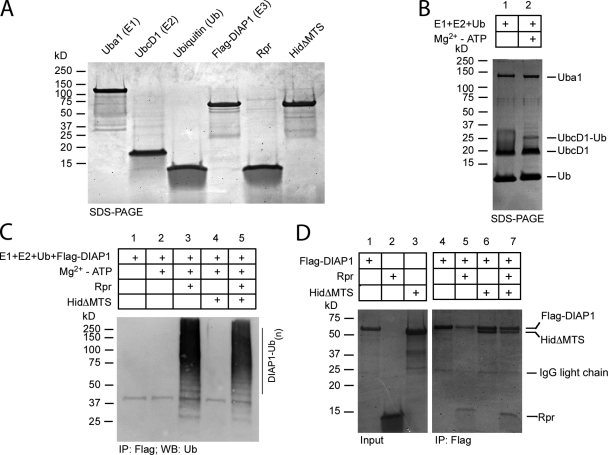
Why does Rpr kill better in the presence of Hid? To answer this question we started from the premise that both IAP antagonists act to stimulate DIAP1 auto-ubiquitination (Hays et al., 2002; Ryoo et al., 2002; Yoo et al., 2002). To this end, we purified all components involved in DIAP1 auto-ubiquitination, namely Uba1 (E1), UbcD1 (E2), ubiquitin (Ub), DIAP1, Rpr, and HidΔMTS from E. coli (Fig. 6 A). HidΔMTS entails residues 1–386. The last 24 amino acids (387–410) of Hid, which constitute the membrane-inserted mitochondrial targeting sequence, were deleted to produce protein soluble for biochemical assays. When designing this construct, we inspected Hid secondary structure to avoid terminating the protein inside a secondary structure element (unpublished data). The proteins are active, as we could reconstitute the covalent coupling of one Ub molecule on UbcD1-conjugating enzyme, in an Uba1- and Mg2+-ATP–dependent fashion (Fig. 6 B). Furthermore, by using reducing agents to break down the E2-Ub thiolesters, we confirmed the presence of the UbcD1-Ub adduct (unpublished data). We next examined DIAP1 ubiquitination in the presence of Mg2+-ATP, Rpr, and/or HidΔMTS. Although DIAP1 does not self-ubiquitinate in the presence of Mg2+-ATP (Fig. 6 C), a dramatic transfer of ubiquitin to DIAP1 could be observed when the reaction is supplemented with Rpr. Thus, we have fully reconstituted in vitro a DIAP1 auto-ubiquitinating complex from Drosophila. Because IAP antagonists bind DIAP1 with conserved motifs it is often assumed that the mechanism of DIAP1 inactivation should also be conserved. However, when HidΔMTS was added in the DIAP1 ubiquitination assay instead of Rpr, no DIAP1 auto-ubiquitination could be observed. Despite good solubility, HidΔMTS did not stimulate DIAP1 degradation. When HidΔMTS and Rpr were added together to the DIAP1 ubiquitination assay, HidΔMTS did not enhance Rpr-dependent DIAP1 ubiquitination. The inability of HidΔMTS to induce DIAP1 ubiquitination could be a result of the following reasons. First, it is possible that recombinant HidΔMTS does not reflect endogenous Hid function, perhaps due to the deletion of its C-terminal hydrophobic region. A second possibility is that Rpr and Hid have different mechanisms of DIAP1 inactivation and only Rpr induces DIAP1 ubiquitination, Hid having a different role in DIAP1 inactivation. In an attempt to address these possibilities, we examined the interactions between DIAP1, Rpr, and Hid using the purified proteins used in the ubiquitination assay. Under these conditions, HidΔMTS was able to bind DIAP1 at a roughly equimolar ratio as judged by band intensity on SDS-PAGE gel, despite HidΔMTS’s inability to stimulate DIAP1 ubiquitination (Fig. 6 D). Furthermore, we have examined the ability of HidΔMTS to form oligomers by formaldehyde cross-linking experiments. Purified HidΔMTS and Rpr appear to form oligomers under these conditions (Fig. S3 B). In addition, the interaction between Rpr and HidΔMTS was already shown in Fig. 3 B. These experiments indicate that deletion of Hid’s MTS does not block its ability to oligomerize or interact with Rpr and DIAP1. We have next examined the ability of full-length Hid or HidΔMTS to induce DIAP1ΔR degradation in HEK293 cells. Unlike Rpr (Fig. 2 F), full-length Hid and HidΔMTS do not induce DIAP1ΔR degradation (Fig. S3 C). These observations and others suggest that most probably Hid does not induce DIAP1 ubiquitination directly.
Figure 6.
DIAP1 auto-ubiquitination and interaction with Rpr and Hid. (A) SDS-PAGE gel showing E1 ubiquitin-activating enzyme Uba1 (Uba1-GST), E2 ubiquitin-conjugating enzyme UbcD1 (6His-UbcD1), 6His-ubiquitin (Ub), E3 ubiquitin ligase DIAP1 (6His-Flag-DIAP1), Rpr-His6, and HidΔMTS-His6, used in ubiquitination assays. Purification tags are not shown in the figure labeling. (B) In vitro coupling of Ub on UbcD1 (E2) in the absence (lane 1) or presence of Mg2+-ATP (lane 2). UbcD1-Ub adduct was detected by Coomassie staining. (C) In vitro DIAP1 auto-ubiquitination. Ubiquitination reactions containing E1, E2, Ub, and Flag-DIAP1, in the absence of Mg2+-ATP (lane 1) or in the presence of Mg2+-ATP (lane 2). The reaction was supplemented additionally with Rpr (lane 3), HidΔMTS (lane 4), or both (lane 5). Flag-DIAP1 was immunoprecipitated with anti-FLAG resin. Polyubiquitination species were detected in Western blot with an anti-ubiquitin antibody. (D) Coomassie-stained SDS-PAGE gel, showing the coimmunoprecipitation of Flag-DIAP1 with Rpr and HidΔMTS. “Input” shows the amount of Flag-DIAP1 (lane 1), Rpr (lane 2), or HidΔMTS (lane 3) used for co-immunoprecipitation. “IP:Flag” shows the anti-FLAG coimmunoprecipitation fractions. Lane 4 indicates the amount of Flag-DIAP1 recovered by the anti-FLAG resin. Lane 5 shows the coimmunoprecipitation of Rpr with Flag-DIAP1. Lane 6 shows the coimmunoprecipitation of HidΔMTS with Flag-DIAP1. Lane 7 shows the coimmunoprecipitation of HidΔMTS and Rpr with Flag-DIAP1.
Artificial targeting of Rpr to mitochondria stimulates its activity, and Hid promotes Rpr stability
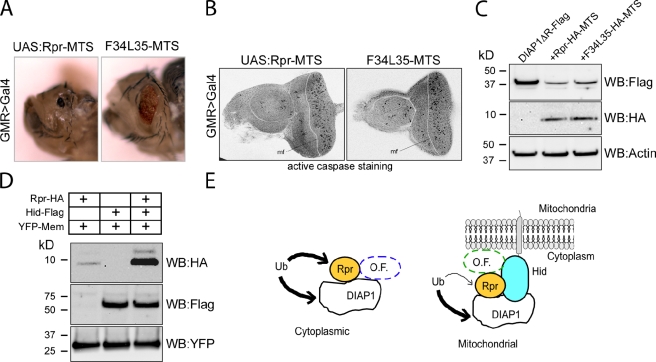
Because HidΔMTS cannot ubiquitinate DIAP1 in vitro and it does not enhance Rpr-mediated DIAP1 ubiquitination, it is possible that Hid stimulates Rpr’s activity by another mechanism. A possible scenario is that Hid recruits Rpr to the mitochondrial membrane, where Rpr is more effective in DIAP1 degradation. To test whether Rpr is more effective when present at the mitochondrial membrane, we artificially targeted Rpr at the mitochondrial membrane by appending to it the mitochondrial targeting sequence of Hid. Rpr-MTS and the GH3 mutant F34AL35A-MTS constructs were used to generate transgenic animals with the constructs inserted in the same genomic location. Rpr-MTS and F34AL35A-MTS were expressed in Drosophila eyes using the GMR>Gal/UAS system. Surprisingly, both constructs were lethal at late pupal stages at 21°C. When dissected from the pupal cases, Rpr-MTS fly eyes were reduced to a black spot and the F35AL35A-MTS fly eyes were severely affected (Fig. 7 A), underscoring the importance of Rpr’s mitochondrial targeting for its killing activity. The GH3 F34AL35A mutant, which is unable to promote eye ablation and is deficient in Hid binding, induces significant eye ablation when artificially targeted to the mitochondria, comparable to a wild-type Rpr nontargeted to the mitochondria. Next, we asked whether the eye ablation phenotype is indeed due to apoptosis. We have isolated third instar larvae eye-antennal discs expressing Rpr-MTS and F34L35-MTS in the GMR region and immunostained them with an antibody against active caspases. Consistent with the observed rough eye phenotypes, both Rpr-MTS and F34L35-MTS showed significant caspase staining in the GMR region (Fig. 7 B). Next, we examined the ability of Rpr-MTS and F34L35-MTS to induce DIAP1 degradation. After coexpression in HEK293 cells, both Rpr-MTS and F34L35-MTS induced a decrease in DIAP1ΔR level (Fig. 7 C). Expression of Rpr-MTS and F34L35-MTS in HEK293 cells was driven by cotransfection of a cmv-Gal4 driver and UAS:Rpr-MTS or UAS:F34L35-MTS constructs and was lower than that achieved with mammalian expression vectors. This might explain the incomplete DIAP1ΔR degradation. In an effort to understand how Hid enhances Rpr’s cell-killing activity, we next compared Rpr protein level in the presence or absence of Hid. When expressed in HEK293 cells, we found that Rpr’s level is in fact much higher in extracts derived from rpr and hid co-transfected cells (Fig. 7 D). On the other hand, the level of Hid did not change significantly in the presence of Rpr. As an expression, protein extraction, and loading control we have used YFP-Mem (a fluorescent marker for cell membranes), which indicates that Rpr level increase is indeed dependent on Hid’s presence. These results suggest a model where Rpr is targeted to the mitochondria by interaction with Hid, and in such a complex that potentially includes other factors, Rpr is protected against degradation (Fig. 7 E).
Figure 7.
Mitochondrial targeting enhances Rpr’s killing capacity and its stability. (A) Expression of transgenic Rpr-MTS or GH3 mutant F34L35-MTS in Drosophila eyes induces severe eye ablation. Rpr-MTS and F34AL35A-MTS constructs consist of full-length Rpr or mutant (residues 1–65), followed by an HA tag and the Hid MTS (residues 387–410). Genotypes: ;UAS:Rpr-MTS/GMR>Gal4; and ;UAS:Rpr-MTS F34AL35A/GMR>Gal4;. (B) Third instar eye-antennal discs, stained with an anti-active caspase antibody. mf line indicates the position of the morphogenetic furrow. Genotypes: ;UAS:Rpr-MTS/GMR>Gal4; and ;UAS:Rpr-MTS F34AL35A/GMR>Gal4;. (C) Rpr-MTS and F34L35-MTS induce DIAP1ΔR degradation in HEK293 cells. DIAP1ΔR was expressed alone or coexpressed with Rpr-MTS or F34L35-MTS in HEK293 cells. The level or DIAP1ΔR, Rpr, and F34L35 mutant were assessed by Western blotting. Actin was used as a loading control. (D) Hid has a stabilizing role on Rpr’s protein level in HEK293 cells. (Lane 1) Rpr protein level in HEK293 cell extracts, after transient cotransfection of a Rpr-HA and YFP-Mem constructs. (Lane 2) Hid protein level in cell extracts of HEK293 after transient cotransfection of a Hid-Flag and YFP-Mem constructs. (Lane 3) Rpr and Hid protein levels in HEK293 cell extract, after transient cotransfection with Rpr-HA, Hid-Flag, and YFP-Mem plasmids. YFP-Mem was used as a transfection control. (E) Representation of Rpr and Hid cooperative induction of cell death. Left diagram, Rpr homomers interact with DIAP1 and induce its ubiquitination in the cytoplasm. Conversely, DIAP1 induces Rpr ubiquitination. Cell death or protection from death is dependent on the balance between Rpr and DIAP1. Other factors (O.F.) such as Grim, Skl, or others could be part of this complex. (Right) Rpr forms a complex with Hid at the mitochondrial membrane. In this complex Rpr accumulates, and leads to a more efficient DIAP1 degradation, which tips the balance toward cell death. Other factors (O.F.) such as Grim or members of the Bcl2 family proteins (Debcl and Buffy) could be interacting in this mitochondrial complex.
Discussion
In this study we show that IAP antagonists undergo self-association and hetero-association that is essential for their full killing activity. Specifically, the physical association between Rpr, Hid, and Grim involves the central helical domain of Rpr. Disrupting this protein–protein interface leads to a significant loss of Rpr’s ability to induce cell death in vivo. The importance of Rpr self-association was revealed by generating enforced Rpr dimers in which the central helical domain of this protein is replaced by defined dimerization motifs. These experiments revealed that enforced parallel, but not anti-parallel dimerization of Rpr (RprLZ) can induce cell death very efficiently in transgenic Drosophila. The resulting cell death occurred by apoptosis and was rescued by the overexpression of the caspase inhibitor p35, or through Rpr-insensitive diap1 alleles. Furthermore, mutants that inhibit the self-association of Rpr have reduced pro-apoptotic activity, providing independent support for the importance of Rpr multimerization. Because an anti-parallel Rpr dimer (RprProP) was not efficiently inducing cell death in transgenic animals, it appears that the IBM motifs of multimeric Rpr have to be in a specific conformation, or at least in close proximity for efficient DIAP1 inactivation. This may occur, for example, by engaging both BIR domains of one DIAP1 molecule in a similar fashion to how SMAC can engage XIAP (Huang et al., 2003).
We also report the association of Rpr with the other IAP antagonists Grim and Hid. Hid is the only IAP antagonist that has a defined mitochondrial targeting sequence at its C terminus and is targeted to the mitochondria by itself; therefore, we focused particularly on the interaction between Rpr and Hid. Consistent with previous reports, we find that Hid consistently localizes to the mitochondria in both human and Drosophila cells. Although it has been previously reported that Rpr localizes to the mitochondria through the GH3–lipid interaction (Olson et al., 2003a; Freel et al., 2008), our results support an alternative view that Rpr’s ability to translocate to the mitochondria is an indirect consequence of associating with Hid. Specifically, in support of our model, we show that Rpr is uniformly distributed in cells when transfected alone in heterologous cells, translocating to the mitochondria only when cotransfected with Hid. We further show that the GH3 mutant F34AL35A, unlike wild-type Rpr, does not coimmunoprecipitate with Hid. This is in agreement with previous observations that a GH3 mutant failed to localize to the mitochondria in Drosophila S2 cells (Olson et al., 2003a).
Rpr induces ubiquitination of DIAP1 in vitro and in HEK293 cells. Unlike Rpr, Hid is not able to perform this function. Thus, the significance of Rpr–Hid interaction might be to bring Rpr at the mitochondrial surface to degrade DIAP1. Although both Rpr and Hid belong to the IAP antagonists family, share a conserved IBM motif, bind DIAP1, and induce cell death, their role in induction of cell death seems to be distinct. In many paradigms Hid appears to be a more potent inducer of cell death than Rpr. It is possible that the primary role of Hid is to assemble a complex at the mitochondrial membrane that recruits Rpr as one the players. The role of Rpr in this complex is to induce DIAP1 ubiquitination. Inability of Hid itself to induce DIAP1 degradation might be related to its larger size (410 amino acids) as compared with Rpr (64 amino acids) or even Grim (138 amino acids). Potentially, the bulkier Hid might interfere with conformational changes in DIAP1 or with the ubiquitin-related transfer process.
In addition, we provide evidence that Rpr is more potent at inducing apoptosis when present at the mitochondrial membrane. When Rpr was fused to the mitochondrial targeting sequence from Hid and expressed in Drosophila eyes, we observed strong cell killing and pupal lethality. Flies dissected from the pupal cases show severely ablated eyes that are reduced to black spots. Even the inactive GH3 mutant F34AL35A, when artificially targeted to the mitochondria using the Hid MTS, induces significant eye ablation. Therefore, Rpr is more potent when present at the mitochondrial membrane. We consider two possible explanations for this enhanced pro-apoptotic activity: First, Rpr may be more active at the mitochondrial surface because of increased protein stability. Consistent with this idea, cytoplasmic Rpr is not very stable (Olson et al., 2003b) and we find that Rpr accumulates to higher protein levels when the presence of Hid permits mitochondrial localization. The resulting high local concentration of Rpr may be critical for DIAP1 ubiquitination. As predicted by this model, we find that Rpr-induced cell death is less efficient when Hid is depleted by RNA knockdown. Our model is also in agreement with several previous observations. For example, it has been reported that Rpr and Hid localize to mitochondria and can induce changes of the mitochondrial ultrastructure (Abdelwahid et al., 2007). This study also showed that inhibition of Rpr localization to mitochondria significantly inhibits cell killing, and that Rpr and Hid act in concert with caspases to promote mitochondrial disruption and Cyt C release. In addition, overexpression of both rpr and hid is required to induce cell death in midline cells of the nervous system, and neither of them kills well individually (Zhou et al., 1997). This is consistent with the observation that more than one IAP antagonist is expressed and they act synergistically in the dying midline glia cells (Sonnenfeld and Jacobs, 1995; Zhou et al., 1995; Dong and Jacobs, 1997; Wing et al., 1998; Bergmann et al., 2002). Finally, Drosophila salivary gland cell death is preceded by the expression of both rpr and hid, and RNAi knockdown of hid alone is sufficient to block the death of these cells (Jiang et al., 2000; Yin and Thummel, 2004). The second, and not mutually exclusive explanation is that Rpr may be more active at the mitochondria because of local concentration of apoptosis regulators that operate at this surface. It has been previously shown that Dronc and active Drice are present at the mitochondrial membrane (Dorstyn et al., 2002), and more recently that mammalian XIAP can translocate to the mitochondrial surface in response to apoptotic stimuli (Owens et al., 2010). In addition, mitochondrial proteins involved in energy metabolism have been recently described to modulate caspase activity and cell death in Drosophila cells (Yi et al., 2007). Recently, it was shown by coimmunoprecipitation experiments in fly cell culture that Grim interacts with the Bcl-2 family proteins Debcl and Buffy (Wu et al., 2010). Thus, Rpr may be part of a higher-order complex at the mitochondria to locally regulate IAP turnover and caspase activity.
Taken together, we uncovered in this work the role of the Rpr helical domain in self-association and interaction with Hid and Grim. We revealed the mechanism of Rpr recruitment to the mitochondria by interaction with Hid. Most importantly, we provided here a new concept with respect to IAP antagonist activity in fly, which acts cooperatively by physical interaction rather than by additive cell death output.
Materials and methods
Plasmids for mammalian, insect, and bacterial expression
With the exception of GFP-Rpr, all tagged Rpr constructs used in this study had the epitopes fused to the C terminus of the Rpr coding sequence. For expression experiments in mammalian cells, Rpr-HA, Rpr-Myc, GFP-Rpr, Hid-HA, Hid-Flag, HidΔMTS-Myc (residues 1–386), GFP-XIAP, RprLZ-HA, RprProP-HA, and DIAP1ΔR-Flag (residues 1–320) were cloned into pcDNA3.1(+)Puro vector (Thomas et al., 2002). pEYFP-Mem was purchased from Takara Bio Inc. For expression in Drosophila cell culture, mCherryDIAP1, Hid-Myc, and HidΔMTS-Myc (residues 1–386) were cloned in pIE1-3 vector (EMD). For the expression of Rpr in fly cells, we generated Rpr-HA pUAST constructs where Rpr-HA sequence was flanked by Rpr’s 5′ and 3′ UTRs. We also made similar constructs with attB or pLoxP elements for targeted insertion into specific sites in the genome (Groth et al., 2004; Oberstein et al., 2005). The pLoxPUAST:Rpr-HA plasmid and its mutant variants were targeted to the [2L(38B), 13399006] site on the second chromosome (line A11; Bestgene, Inc). pUASTattB:Rpr-MTS and pUASTattB:F34AL35A-MTS were targeted to a locus on the second chromosome (line 24481, genotype: M{3xP3-RFP.attP’}ZH-22A [with M{vas-int.Dm}ZH-2A]; Bestgene, Inc). pUAST:RprLZ-HA construct was created by replacement of Rpr amino acids 10–48 in pUAST:Rpr-HA vector with the LZ-encoding DNA fragment, flanked by AgeI and SpeI restriction sites. This was achieved by the insertion of two unique restriction sites, AgeI and SpeI, at the above-mentioned positions in pUAST:Rpr-HA. pUAST:RprProP-HA was generated similarly to RprLZ by insertion of the ProP PCR fragment, between the AgeI and SpeI sites of the modified pUAST:Rpr-HA vector. pUAST:RprLZ-HA and pUAST:RprProP-HA were used to generate fly transgenes (Genetic Services, Inc.). For protein production in E. coli, the following vectors were created: pET3a:Uba1-GST, pET28b:ubiquitin, pET14b:Flag-DIAP1, pET21a:Rpr, pET21a:HidΔMTS, pET14b:UbcD1 (Ryoo et al., 2002), and pET3a:Rpr-GST (Ryoo et al., 2002). pET21a:Rpr, pET14b:Hid, pET21a:Grim, and pETDuet:Skl were used for expression in the in vitro transcription translation reactions. pET21a:Rpr construct produces Rpr with a C-terminal MGMGMHHHHHH tag. This construct was used to generate Rpr point mutants.
Protein expression and purification
For protein production we used E. coli BL21DE3 strain, transformed with appropriate plasmids. Protein production was induced with 500 µM IPTG at 25°C, overnight. Purification of GST, Rpr-GST, GST-Hid, or Uba1-GST was performed following a previously described protocol (Carrington et al., 2006; Sandu et al., 2006). Purification of 6His-UbcD1 (E2), 6His-Flag-DIAP1 (E3), Rpr-His6, HidΔMTS-His6, and 6His-ubiquitin was performed using an adapted protocol used for Ulp1 purification (Mossessova and Lima, 2000). In brief, the pellet from one liter E. coli culture was resuspended in 70 ml of lysis buffer (50 mM Tris, pH 8.0, 350 mM NaCl, 1 mM BME, 0.2% igepal, and 10 mM imidazole) and disrupted by sonication. After sonication the cell extract was cleared by centrifugation at 14,000 g for 15 min. The 6His-tagged protein was bound to Talon Metal Affinity Resin (Takara Bio Inc.) and washed with 35 ml of lysis buffer, followed by 35 ml of washing buffer (50 mM Tris, pH 8.0, 350 mM NaCl, 1 mM BME, 0.2% igepal, and 70 mM imidazole). After washing, the protein was eluted with 20 ml elution buffer (50 mM Tris, pH 8.0, 350 mM NaCl, 1 mM BME, 0.2% igepal, and 500 mM imidazole). After elution, the purified proteins were concentrated using Amicon Centricons (Millipore) and flash frozen in liquid nitrogen after addition of 10% glycerol.
Protein–protein interaction studies
Interaction of Rpr-GST with 35S-Rpr, 35S-Hid, 35S-Grim, and 35S-Skl was investigated by pull-down experiments using a protocol described previously (Sandu et al., 2006). In brief, 20 µl of a Rpr-GST bead slurry were mixed in 200 µl binding buffer (50 mM Hepes, pH 7.4, 50 mM NaCl, 0.1% NP-40, and 10% glycerol) with 10 µl of the 35S-labeled Rpr, Hid, Grim, or Skl. After 2 h nutation at 4°C, the beads were harvested by centrifugation and washed three times with 500 µl of binding buffer for 10 min at 4°C. The beads were then eluted with 15 µl SDS-PAGE loading dye at 95°C, separated on SDS-PAGE gels, and visualized by phosphorimaging of the dried gel. Interaction of Rpr with GST-Hid or GST or the interaction between Ub, Rpr, or HidΔMTS with Rpr-GST was tested in an alternative pull-down experiment. Approximately 5 µg GST-Hid, GST, or Rpr-GST linked on agarose beads were incubated with 5 µg of either Rpr-His6 (Rpr), His6-ubiquitin (Ub), or HidΔMTS-His6 (HidΔMTS) in 250 µl binding buffer (50 mM Hepes, pH 7.4, 50 mM NaCl, 0.1% NP-40, and 10% glycerol) and nutated overhead for 2 h at 4°C. The beads were washed three times with 500 µl binding buffer and denatured in SDS-sample buffer at 95°C. The samples were separated in SDS-PAGE gels, followed by Coomassie staining.
Cell culture, immunoprecipitation, immunostaining, and Western blotting
Human HEK293 and BT549 or Drosophila S2R+ cell lines were used for experiments involving expression, immunolocalization, and immunoprecipitation studies. For testing the ability of Rpr-HA, RprLZ-HA, Hid-HA, or HidΔMTS-Myc to induce DIAP1ΔR-Flag degradation, HEK293 cells were transfected with either DIAP1ΔR-Flag construct or cotransfected in equal ratios with the DIAP1ΔR-Flag construct and each one of the above-mentioned constructs. 16 h after transfection, the cells were lysed in lysis buffer (10 mM Tris, pH 8.0, 100 mM NaCl, and 0.5% NP-40) supplemented with protease inhibitor cocktail (Roche). After centrifugation at 14,000 g, cell extracts (typically 25 µg) were separated by SDS-PAGE, blotted on nitrocellulose membranes, and proteins were detected with specific antibodies. Alternatively, HEK293 cells were cotransfected with pcDNA3.1+Puro:DIAP1ΔR-Flag, pCMV-Gal4 (a gift from Brieann Fant, The Rockefeller University, New York, NY), and either pUAST:Rpr-HA-MTS or pUAST:F34L35-HA-MTS constructs. 46 h after transfection, the cells were lysed and analyzed as described above. For immunolocalization studies, Rpr-HA, Rpr-Myc, GFP-Rpr, Hid-HA, Hid-Flag, HidΔMTS-Myc, or GFP-XIAP plasmids were transfected in human BT549 cells. As a mitochondrial marker, we used an anti-Cyt C antibody (BD). Hid-FLAG and Rpr-HA or Rpr mutant complexes were immunoprecipitated with ANTI-FLAG M2 Agarose Affinity Gel (Sigma-Aldrich) after cotransfection in HEK293 cells. Detection of tagged proteins on Western blot membranes was achieved with anti-Flag HRP (Sigma-Aldrich) or anti-HA HRP (Roche) antibodies. The secondary antibodies used for immunostaining contained the following chromophores: Cy3, Cy5, Texas red (Jackson ImmunoResearch Inc.), and Alexa 488 or 546 (Invitrogen). For intracellular localization in Drosophila cell culture, proteins were expressed from the following plasmids: pIE1-3:mCherryDIAP1, pUAST:Rpr-HA, pIE1-3:Hid-Myc, pIE1-3:HidΔMTS-Myc, and pUAST-mtGFP. Actin-Gal4 plasmid was used to drive expression from pUAST vectors. Hid was stained with an anti-Myc antibody (Cell Signaling Technology) while mitochondria was detected with an anti-Cyt C antibody or mtGFP. Silencing of Hid RNA in cell culture was achieved with a 21-bp siRNA, purchased from IDT. The duplex was generated by annealing of the following two oligos: 5′-GCUCUGUGGUUUCUUCUUCTT-3′ and 5′-GAAGAAGAAACCACAGAGCTT-3′. Gene silencing was achieved according to the manufacturer’s instructions. Transfection assays in S2R+ cells were supplemented with 10 µM Q-VD-OPh. Expression level of the RprLZ or RprProP in transgenic animals was probed by immunostaining of eye imaginal discs from third instar larvae. After dissection, fixation, permeabilization, and blocking, the discs were stained with a rat anti-HA antibody (Roche), followed by an anti–rat-FITC secondary antibody. As staining controls we used a mouse anti-ELAV antibody (Developmental Studies Hybridoma Bank, Iowa City, IA), followed by anti–mouse-Cy3 secondary antibody. Detection of apoptosis in third instar eye discs was achieved by immunostaining of fixed and permeabilized discs with a cleaved caspase-3 rabbit antibody (Cell Signaling Technology), followed by an anti–rabbit-Cy3 secondary antibody.
Fluorescence microscopy
Human and Drosophila cells were grown, transfected, fixed, and stained in Lab-Tek Permanox chamber slides (Thermo Fisher Scientific). After staining, samples were mounted in Vectashield mounting medium (Vector Laboratories), sealed with nail polish and stored at 4°C until use. Images were taken at room temperature. Fluorescence images were acquired using a confocal microscope (Axioplan 2; Carl Zeiss, Inc.) equipped with a 63x/1.4 oil immersion objective lens. Images were captured and analyzed using the LSM 510 system. Images were imported into Photoshop 8.0 (Adobe) for cropping and the figures were assembled using CanvasX. Shadow/Highlight adjustment was performed for Fig. 2 G to better reveal the DIAP1 staining difference in the wing discs.
Transgenic Drosophila lines and other fly stocks
The following fly stocks were generated in this study: UAS:RprLZ-HA, UAS:RprProP-HA, LoxPUAS:Rpr-HA, LoxPUAS:Rpr-HA Q23ER26A, LoxPUAS:Rpr-HA F34AL35A, pUASTattB:Rpr-MTS, and pUASTattB:F34AL35A-MTS. The following companies performed the injections: BestGene, Inc. and Genetic Services, Inc. Additional fly stocks from the laboratory collection were used in this study: UAS:p35/;;, diap16-3s and diap123-4s alleles, hidA206 and hidWR+X1 alleles, GMR>Gal4/CyO; Sb/TM6B, GMR>Gal4; GMR>Hid/CyO; Sb/TM6B, ;GMR>Gal4/CyO; GMR>Rpr/TM6B. UAS:RNAi lines for Hid (CG5123) and Rpr (CG4319) were purchased from the Vienna Drosophila RNAi Center (Vienna, Austria). Fly crosses were incubated at 21°C.
In vitro ubiquitination assay
All proteins used in the assay are recombinant Drosophila proteins, produced and purified from E. coli as described above. Ubiquitination assay consisted of mixing together ∼3 µg E1, 3 µg E2, 3 µg ubiquitin, and 3 µg Flag-DIAP1 in a final volume of 40 µl. The reaction buffer was 25 mM Tris, pH 7.5, 50 mM NaCl, 250 µM DTT, 4 mM ATP, and 5 mM MgCl2. The effect of IAP antagonists on DIAP1 ubiquitination was tested by supplementation of the assay with ∼3 µg purified Rpr or HidΔMTS. The reaction was incubated for 40 min at 25°C followed by anti-Flag immunoprecipitation. One quarter of the DIAP1 immunoprecipitation sample was separated by SDS-PAGE and blotted on nitrocellulose membrane. The membrane was reacted with an anti-ubiquitin antibody (Sigma-Aldrich).
Structure prediction and analysis tools
Secondary structure elements of Rpr were predicted using the secondary structure consensus prediction server at PBIL (http://npsa-pbil.ibcp.fr). Various structural protein models were analyzed using Pymol Software (DeLano Scientific LLC). The design of Rpr point mutants was performed after inspection of a few structural models of the Rpr helical domain. Protein models were generated using the Protein Homology/AnalogY Recognition Engine (PHYRE; Kelley and Sternberg, 2009) and analyzed using Pymol. Residues with bulky, large side chains, oriented outwards, were chosen for mutagenesis. These residues were either hydrophobic or charged. Assuming that the surface of interaction is extensive, we chose to mutate two residues at a time. The two residues are oriented on the same side of the helix, would be part of the same interaction patch of the helix surface (charged or hydrophobic), and are positioned each on adjacent helix turns. The residues in the mutagenesis were spaced to cover the entire helical region.
Online supplemental material
Fig. S1 shows the immunolocalization experiments of Rpr-HA, GFP-XIAP, or mCherryDIAP1 with HidΔMTS-Myc. Fig. S2 shows the effect of caspase inhibitor zVAD-FMK on intracellular localization of Rpr-HA and Hid-HA. Fig. S3 shows the effect of Rpr mRNA knockdown on Hid-induced cell death in Drosophila eyes (A); shows the ability of purified Rpr and HidΔMTS to form oligomeric species after formaldehyde cross-linking (B); and the effect of Hid-HA or HidΔMTS-Myc on DIAP1ΔR-Flag degradation in HEK293 cells. Online supplemental material is available at: http://www.jcb.org/cgi/content/full/jcb.201004086/DC1.
Acknowledgments
We would like to thank members of the Steller laboratory for helpful discussions and for sharing of reagents. We thank Brieann Fant for the pCMV-Gal4 plasmid, Bill Saxton for pUAST-mtGFP plasmid and transgenic flies, and Stephen Small for pBS-LoxP-white-Lox2272 plasmid. C. Sandu thanks Chandrawatee Ramsawak for excellent technical work, and Samara Brown and Magali Susane for critical reading of the manuscript. H. Steller is an investigator of the Howard Hughes Medical Institute.
This work was supported by National Institutes of Health grant RO1GM60124 to H. Steller.
The majority of the experiments were performed by C. Sandu. H.D. Ryoo was experimentally involved in the initial phase of the project. All three authors contributed to writing the manuscript.
Footnotes
Abbreviations used in this paper:
- Cyt C
- cytochrome c
- DIAP1
- Drosophila IAP1
- Hid
- head involution defective
- IAP
- inhibitor of apoptosis protein
- IP
- immunoprecipitation
- PD
- pull-down
- Rpr
- Reaper
- Sickle
- Skl
- Western blot
- WB
References
- Abbott M.K., Lengyel J.A. 1991. Embryonic head involution and rotation of male terminalia require the Drosophila locus head involution defective. Genetics. 129:783–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelwahid E., Yokokura T., Krieser R.J., Balasundaram S., Fowle W.H., White K. 2007. Mitochondrial disruption in Drosophila apoptosis. Dev. Cell. 12:793–806 10.1016/j.devcel.2007.04.004 [DOI] [PubMed] [Google Scholar]
- Bergmann A., Tugentman M., Shilo B.Z., Steller H. 2002. Regulation of cell number by MAPK-dependent control of apoptosis: a mechanism for trophic survival signaling. Dev. Cell. 2:159–170 10.1016/S1534-5807(02)00116-8 [DOI] [PubMed] [Google Scholar]
- Bergmann A., Yang A.Y., Srivastava M. 2003. Regulators of IAP function: coming to grips with the grim reaper. Curr. Opin. Cell Biol. 15:717–724 10.1016/j.ceb.2003.10.002 [DOI] [PubMed] [Google Scholar]
- Carrington P.E., Sandu C., Wei Y., Hill J.M., Morisawa G., Huang T., Gavathiotis E., Wei Y., Werner M.H. 2006. The structure of FADD and its mode of interaction with procaspase-8. Mol. Cell. 22:599–610 10.1016/j.molcel.2006.04.018 [DOI] [PubMed] [Google Scholar]
- Chai J., Yan N., Huh J.R., Wu J.W., Li W., Hay B.A., Shi Y. 2003. Molecular mechanism of Reaper-Grim-Hid-mediated suppression of DIAP1-dependent Dronc ubiquitination. Nat. Struct. Biol. 10:892–898 10.1038/nsb989 [DOI] [PubMed] [Google Scholar]
- Chen P., Nordstrom W., Gish B., Abrams J.M. 1996. grim, a novel cell death gene in Drosophila. Genes Dev. 10:1773–1782 10.1101/gad.10.14.1773 [DOI] [PubMed] [Google Scholar]
- Clavería C., Caminero E., Martínez-A C., Campuzano S., Torres M. 2002. GH3, a novel proapoptotic domain in Drosophila Grim, promotes a mitochondrial death pathway. EMBO J. 21:3327–3336 10.1093/emboj/cdf354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong R., Jacobs J.R. 1997. Origin and differentiation of supernumerary midline glia in Drosophila embryos deficient for apoptosis. Dev. Biol. 190:165–177 10.1006/dbio.1997.8688 [DOI] [PubMed] [Google Scholar]
- Dorstyn L., Read S., Cakouros D., Huh J.R., Hay B.A., Kumar S. 2002. The role of cytochrome c in caspase activation in Drosophila melanogaster cells. J. Cell Biol. 156:1089–1098 10.1083/jcb.200111107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du C., Fang M., Li Y., Li L., Wang X. 2000. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell. 102:33–42 10.1016/S0092-8674(00)00008-8 [DOI] [PubMed] [Google Scholar]
- Freel C.D., Richardson D.A., Thomenius M.J., Gan E.C., Horn S.R., Olson M.R., Kornbluth S. 2008. Mitochondrial localization of Reaper to promote inhibitors of apoptosis protein degradation conferred by GH3 domain-lipid interactions. J. Biol. Chem. 283:367–379 10.1074/jbc.M708931200 [DOI] [PubMed] [Google Scholar]
- Gottfried Y., Rotem A., Lotan R., Steller H., Larisch S. 2004. The mitochondrial ARTS protein promotes apoptosis through targeting XIAP. EMBO J. 23:1627–1635 10.1038/sj.emboj.7600155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal L., McCall K., Agapite J., Hartwieg E., Steller H. 2000. Induction of apoptosis by Drosophila reaper, hid and grim through inhibition of IAP function. EMBO J. 19:589–597 10.1093/emboj/19.4.589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grether M.E., Abrams J.M., Agapite J., White K., Steller H. 1995. The head involution defective gene of Drosophila melanogaster functions in programmed cell death. Genes Dev. 9:1694–1708 10.1101/gad.9.14.1694 [DOI] [PubMed] [Google Scholar]
- Groth A.C., Fish M., Nusse R., Calos M.P. 2004. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 166:1775–1782 10.1534/genetics.166.4.1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haining W.N., Carboy-Newcomb C., Wei C.L., Steller H. 1999. The proapoptotic function of Drosophila Hid is conserved in mammalian cells. Proc. Natl. Acad. Sci. USA. 96:4936–4941 10.1073/pnas.96.9.4936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hays R., Wickline L., Cagan R. 2002. Morgue mediates apoptosis in the Drosophila melanogaster retina by promoting degradation of DIAP1. Nat. Cell Biol. 4:425–431 10.1038/ncb794 [DOI] [PubMed] [Google Scholar]
- Hegde R., Srinivasula S.M., Zhang Z., Wassell R., Mukattash R., Cilenti L., DuBois G., Lazebnik Y., Zervos A.S., Fernandes-Alnemri T., Alnemri E.S. 2002. Identification of Omi/HtrA2 as a mitochondrial apoptotic serine protease that disrupts inhibitor of apoptosis protein-caspase interaction. J. Biol. Chem. 277:432–438 10.1074/jbc.M109721200 [DOI] [PubMed] [Google Scholar]
- Hengartner M.O. 2000. The biochemistry of apoptosis. Nature. 407:770–776 10.1038/35037710 [DOI] [PubMed] [Google Scholar]
- Holley C.L., Olson M.R., Colón-Ramos D.A., Kornbluth S. 2002. Reaper eliminates IAP proteins through stimulated IAP degradation and generalized translational inhibition. Nat. Cell Biol. 4:439–444 10.1038/ncb798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Rich R.L., Myszka D.G., Wu H. 2003. Requirement of both the second and third BIR domains for the relief of X-linked inhibitor of apoptosis protein (XIAP)-mediated caspase inhibition by Smac. J. Biol. Chem. 278:49517–49522 10.1074/jbc.M310061200 [DOI] [PubMed] [Google Scholar]
- Jiang C., Lamblin A.F., Steller H., Thummel C.S. 2000. A steroid-triggered transcriptional hierarchy controls salivary gland cell death during Drosophila metamorphosis. Mol. Cell. 5:445–455 10.1016/S1097-2765(00)80439-6 [DOI] [PubMed] [Google Scholar]
- Kelley L.A., Sternberg M.J. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4:363–371 10.1038/nprot.2009.2 [DOI] [PubMed] [Google Scholar]
- Lisi S., Mazzon I., White K. 2000. Diverse domains of THREAD/DIAP1 are required to inhibit apoptosis induced by REAPER and HID in Drosophila. Genetics. 154:669–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy J.V., Dixit V.M. 1998. Apoptosis induced by Drosophila reaper and grim in a human system. Attenuation by inhibitor of apoptosis proteins (cIAPs). J. Biol. Chem. 273:24009–24015 10.1074/jbc.273.37.24009 [DOI] [PubMed] [Google Scholar]
- Mossessova E., Lima C.D. 2000. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol. Cell. 5:865–876 10.1016/S1097-2765(00)80326-3 [DOI] [PubMed] [Google Scholar]
- O’Shea E.K., Klemm J.D., Kim P.S., Alber T. 1991. X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science. 254:539–544 10.1126/science.1948029 [DOI] [PubMed] [Google Scholar]
- Oberstein A., Pare A., Kaplan L., Small S. 2005. Site-specific transgenesis by Cre-mediated recombination in Drosophila. Nat. Methods. 2:583–585 10.1038/nmeth775 [DOI] [PubMed] [Google Scholar]
- Olson M.R., Holley C.L., Gan E.C., Colón-Ramos D.A., Kaplan B., Kornbluth S. 2003a. A GH3-like domain in reaper is required for mitochondrial localization and induction of IAP degradation. J. Biol. Chem. 278:44758–44768 10.1074/jbc.M308055200 [DOI] [PubMed] [Google Scholar]
- Olson M.R., Holley C.L., Yoo S.J., Huh J.R., Hay B.A., Kornbluth S. 2003b. Reaper is regulated by IAP-mediated ubiquitination. J. Biol. Chem. 278:4028–4034 10.1074/jbc.M209734200 [DOI] [PubMed] [Google Scholar]
- Owens T.W., Foster F.M., Valentijn A., Gilmore A.P., Streuli C.H. 2010. Role for X-linked Inhibitor of apoptosis protein upstream of mitochondrial permeabilization. J. Biol. Chem. 285:1081–1088 10.1074/jbc.M109.072322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed J.C., Doctor K.S., Godzik A. 2004. The domains of apoptosis: a genomics perspective. Sci. STKE. 2004:re9 10.1126/stke.2392004re9 [DOI] [PubMed] [Google Scholar]
- Ryoo H.D., Bergmann A., Gonen H., Ciechanover A., Steller H. 2002. Regulation of Drosophila IAP1 degradation and apoptosis by reaper and ubcD1. Nat. Cell Biol. 4:432–438 10.1038/ncb795 [DOI] [PubMed] [Google Scholar]
- Sandu C., Morisawa G., Wegorzewska I., Huang T., Arechiga A.F., Hill J.M., Kim T., Walsh C.M., Werner M.H. 2006. FADD self-association is required for stable interaction with an activated death receptor. Cell Death Differ. 13:2052–2061 10.1038/sj.cdd.4401966 [DOI] [PubMed] [Google Scholar]
- Shi Y. 2002. Mechanisms of caspase activation and inhibition during apoptosis. Mol. Cell. 9:459–470 10.1016/S1097-2765(02)00482-3 [DOI] [PubMed] [Google Scholar]
- Shiozaki E.N., Shi Y. 2004. Caspases, IAPs and Smac/DIABLO: mechanisms from structural biology. Trends Biochem. Sci. 29:486–494 10.1016/j.tibs.2004.07.003 [DOI] [PubMed] [Google Scholar]
- Silke J., Kratina T., Ekert P.G., Pakusch M., Vaux D.L. 2004. Unlike Diablo/smac, Grim promotes global ubiquitination and specific degradation of X chromosome-linked inhibitor of apoptosis (XIAP) and neither cause apoptosis. J. Biol. Chem. 279:4313–4321 10.1074/jbc.M305661200 [DOI] [PubMed] [Google Scholar]
- Sonnenfeld M.J., Jacobs J.R. 1995. Apoptosis of the midline glia during Drosophila embryogenesis: a correlation with axon contact. Development. 121:569–578 [DOI] [PubMed] [Google Scholar]
- Srinivasula S.M., Datta P., Kobayashi M., Wu J.W., Fujioka M., Hegde R., Zhang Z., Mukattash R., Fernandes-Alnemri T., Shi Y., et al. 2002. sickle, a novel Drosophila death gene in the reaper/hid/grim region, encodes an IAP-inhibitory protein. Curr. Biol. 12:125–130 10.1016/S0960-9822(01)00657-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steller H. 1995. Mechanisms and genes of cellular suicide. Science. 267:1445–1449 10.1126/science.7878463 [DOI] [PubMed] [Google Scholar]
- Tenev T., Zachariou A., Wilson R., Ditzel M., Meier P. 2005. IAPs are functionally non-equivalent and regulate effector caspases through distinct mechanisms. Nat. Cell Biol. 7:70–77 10.1038/ncb1204 [DOI] [PubMed] [Google Scholar]
- Thomas L.R., Stillman D.J., Thorburn A. 2002. Regulation of Fas-associated death domain interactions by the death effector domain identified by a modified reverse two-hybrid screen. J. Biol. Chem. 277:34343–34348 10.1074/jbc.M204169200 [DOI] [PubMed] [Google Scholar]
- Thompson C.B. 1995. Apoptosis in the pathogenesis and treatment of disease. Science. 267:1456–1462 10.1126/science.7878464 [DOI] [PubMed] [Google Scholar]
- Thornberry N.A., Lazebnik Y. 1998. Caspases: enemies within. Science. 281:1312–1316 10.1126/science.281.5381.1312 [DOI] [PubMed] [Google Scholar]
- Verhagen A.M., Ekert P.G., Pakusch M., Silke J., Connolly L.M., Reid G.E., Moritz R.L., Simpson R.J., Vaux D.L. 2000. Identification of DIABLO, a mammalian protein that promotes apoptosis by binding to and antagonizing IAP proteins. Cell. 102:43–53 10.1016/S0092-8674(00)00009-X [DOI] [PubMed] [Google Scholar]
- Vucic D., Kaiser W.J., Harvey A.J., Miller L.K. 1997. Inhibition of reaper-induced apoptosis by interaction with inhibitor of apoptosis proteins (IAPs). Proc. Natl. Acad. Sci. USA. 94:10183–10188 10.1073/pnas.94.19.10183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vucic D., Kaiser W.J., Miller L.K. 1998. Inhibitor of apoptosis proteins physically interact with and block apoptosis induced by Drosophila proteins HID and GRIM. Mol. Cell. Biol. 18:3300–3309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.L., Hawkins C.J., Yoo S.J., Müller H.A., Hay B.A. 1999. The Drosophila caspase inhibitor DIAP1 is essential for cell survival and is negatively regulated by HID. Cell. 98:453–463 10.1016/S0092-8674(00)81974-1 [DOI] [PubMed] [Google Scholar]
- White K., Grether M.E., Abrams J.M., Young L., Farrell K., Steller H. 1994. Genetic control of programmed cell death in Drosophila. Science. 264:677–683 10.1126/science.8171319 [DOI] [PubMed] [Google Scholar]
- White K., Tahaoglu E., Steller H. 1996. Cell killing by the Drosophila gene reaper. Science. 271:805–807 10.1126/science.271.5250.805 [DOI] [PubMed] [Google Scholar]
- Wilson R., Goyal L., Ditzel M., Zachariou A., Baker D.A., Agapite J., Steller H., Meier P. 2002. The DIAP1 RING finger mediates ubiquitination of Dronc and is indispensable for regulating apoptosis. Nat. Cell Biol. 4:445–450 10.1038/ncb799 [DOI] [PubMed] [Google Scholar]
- Wing J.P., Zhou L., Schwartz L.M., Nambu J.R. 1998. Distinct cell killing properties of the Drosophila reaper, head involution defective, and grim genes. Cell Death Differ. 5:930–939 10.1038/sj.cdd.4400423 [DOI] [PubMed] [Google Scholar]
- Wu G., Chai J., Suber T.L., Wu J.W., Du C., Wang X., Shi Y. 2000. Structural basis of IAP recognition by Smac/DIABLO. Nature. 408:1008–1012 10.1038/35050012 [DOI] [PubMed] [Google Scholar]
- Wu J.W., Cocina A.E., Chai J., Hay B.A., Shi Y. 2001. Structural analysis of a functional DIAP1 fragment bound to grim and hid peptides. Mol. Cell. 8:95–104 10.1016/S1097-2765(01)00282-9 [DOI] [PubMed] [Google Scholar]
- Wu J.N., Nguyen N., Aghazarian M., Tan Y., Sevrioukov E.A., Mabuchi M., Tang W., Monserrate J.P., White K., Brachmann C.B. 2010. grim promotes programmed cell death of Drosophila microchaete glial cells. Mech. Dev. 10.1016/j.mod.2010.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan N., Wu J.W., Chai J., Li W., Shi Y. 2004. Molecular mechanisms of DrICE inhibition by DIAP1 and removal of inhibition by Reaper, Hid and Grim. Nat. Struct. Mol. Biol. 11:420–428 10.1038/nsmb764 [DOI] [PubMed] [Google Scholar]
- Yi C.H., Sogah D.K., Boyce M., Degterev A., Christofferson D.E., Yuan J. 2007. A genome-wide RNAi screen reveals multiple regulators of caspase activation. J. Cell Biol. 179:619–626 10.1083/jcb.200708090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin V.P., Thummel C.S. 2004. A balance between the diap1 death inhibitor and reaper and hid death inducers controls steroid-triggered cell death in Drosophila. Proc. Natl. Acad. Sci. USA. 101:8022–8027 10.1073/pnas.0402647101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.J., Huh J.R., Muro I., Yu H., Wang L., Wang S.L., Feldman R.M., Clem R.J., Müller H.A., Hay B.A. 2002. Hid, Rpr and Grim negatively regulate DIAP1 levels through distinct mechanisms. Nat. Cell Biol. 4:416–424 10.1038/ncb793 [DOI] [PubMed] [Google Scholar]
- Yuan J., Yankner B.A. 2000. Apoptosis in the nervous system. Nature. 407:802–809 10.1038/35037739 [DOI] [PubMed] [Google Scholar]
- Zachariou A., Tenev T., Goyal L., Agapite J., Steller H., Meier P. 2003. IAP-antagonists exhibit non-redundant modes of action through differential DIAP1 binding. EMBO J. 22:6642–6652 10.1093/emboj/cdg617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Hashimi H., Schwartz L.M., Nambu J.R. 1995. Programmed cell death in the Drosophila central nervous system midline. Curr. Biol. 5:784–790 10.1016/S0960-9822(95)00155-2 [DOI] [PubMed] [Google Scholar]
- Zhou L., Schnitzler A., Agapite J., Schwartz L.M., Steller H., Nambu J.R. 1997. Cooperative functions of the reaper and head involution defective genes in the programmed cell death of Drosophila central nervous system midline cells. Proc. Natl. Acad. Sci. USA. 94:5131–5136 10.1073/pnas.94.10.5131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoetewey D.L., Tripet B.P., Kutateladze T.G., Overduin M.J., Wood J.M., Hodges R.S. 2003. Solution structure of the C-terminal antiparallel coiled-coil domain from Escherichia coli osmosensor ProP. J. Mol. Biol. 334:1063–1076 10.1016/j.jmb.2003.10.020 [DOI] [PubMed] [Google Scholar]