Cdc48/p97/VCP plays a ubiquitin-independent role during autophagosome formation in S. cerevisiae.
Abstract
The molecular details of the biogenesis of double-membraned autophagosomes are poorly understood. We identify the Saccharomyces cerevisiae AAA–adenosine triphosphatase Cdc48 and its substrate-recruiting cofactor Shp1/Ubx1 as novel components needed for autophagosome biogenesis. In mammals, the Cdc48 homologue p97/VCP and the Shp1 homologue p47 mediate Golgi reassembly by extracting an unknown monoubiquitinated fusion regulator from a complex. We find no requirement of ubiquitination or the proteasome system for autophagosome biogenesis but detect interaction of Shp1 with the ubiquitin-fold autophagy protein Atg8. Atg8 coupled to phosphatidylethanolamine (PE) is crucial for autophagosome elongation and, in vitro, mediates tethering and hemifusion. Interaction with Shp1 requires an FK motif within the N-terminal non–ubiquitin-like Atg8 domain. Based on our data, we speculate that autophagosome formation, in contrast to Golgi reassembly, requires a complex in which Atg8 functionally substitutes ubiquitin. This, for the first time, would give a rationale for use of the ubiquitin-like Atg8 during macroautophagy and would explain why Atg8-PE delipidation is necessary for efficient macroautophagy.
Introduction
Macroautophagy sequesters superfluous cytosol and organelles into double-membraned autophagosomes, which finally fuse with lysosomes for degradation. Despite identification of >30 Atg (autophagy) genes, the molecular mechanism of autophagosome biogenesis is poorly understood (Mizushima et al., 2008; Farré et al., 2009; Nakatogawa et al., 2009). All proposed models predict elongation and final closure or cisternal assembly of double-membraned precursors during autophagosome formation (Axe et al., 2008; Longatti and Tooze, 2009). Previous studies assumed that the autophagic machinery mediates the required membrane fusions independent from SNAREs (Reggiori et al., 2004) and identified the ubiquitin-like protein Atg8 as a key component, especially for elongation of the forming autophagosome (Nakatogawa et al., 2007; Xie et al., 2008). Atg8 is coupled by a ubiquitin-like conjugation system to phosphatidylethanolamine (PE; Ichimura et al., 2000). In vitro Atg8-PE induced liposome tethering and hemifusion (Nakatogawa et al., 2007). However, it remained open how Atg8 mediates membrane fusion and why macroautophagy needs a ubiquitin-like protein.
Autophagosome biogenesis is morphologically reminiscent of nuclear envelope expansion and postmitotic Golgi reassembly. The mammalian AAA-ATPase p97/VCP is a multifunctional enzyme in the ubiquitin–proteasome pathway; for example, in ER-associated degradation (ERAD) and ubiquitin fusion degradation (for reviews see Jentsch and Rumpf, 2007; Meyer and Popp, 2008). Some experiments suggested that p97 only handles ubiquitinated proteins (Dalal et al., 2004); others reported a direct function in membrane fusion (for reviews see Jentsch and Rumpf, 2007; Meyer and Popp, 2008). p97 mediates multiple functions by interacting with numerous ubiquitin-binding adaptors. During Golgi regrowth, p97 binds via p47 to an unknown monoubiquitinated fusion regulator, which prevents untimely SNARE pairing. As an ATPase, p97 then segregates the ubiquitin conjugate from the SNARE to allow fusion. Finally, deubiquitination by the cysteine proteinase VCIP135 is essential (Kondo et al., 1997; Uchiyama et al., 2002; Wang et al., 2004). In this paper, we identify Cdc48 and Shp1/Ubx1, the yeast homologues of p97 and p47, as novel components of autophagosome biogenesis in Saccharomyces cerevisiae. We found no requirement of ubiquitination or the proteasome system for macroautophagy but show interaction of Atg8, which is dependent on an FK motif in its non–ubiquitin-like N-terminal helical domain (NHD), with Shp1. Based on our data, we speculate that S. cerevisiae autophagosome formation uses a protein complex analogous to that mediating mammalian nuclear envelope growth and Golgi reassembly with the distinction that Atg8 replaces ubiquitin. The cysteine proteinase Atg4 would then be equivalent to VCIP135. Our model would explain why efficient macroautophagy requires the ubiquitin-fold Atg8- and Atg4-dependent delipidation of Atg8-PE.
Results and discussion
Cdc48 and its cofactor Shp1/Ubx1 are essential for macroautophagy and micronucleophagy
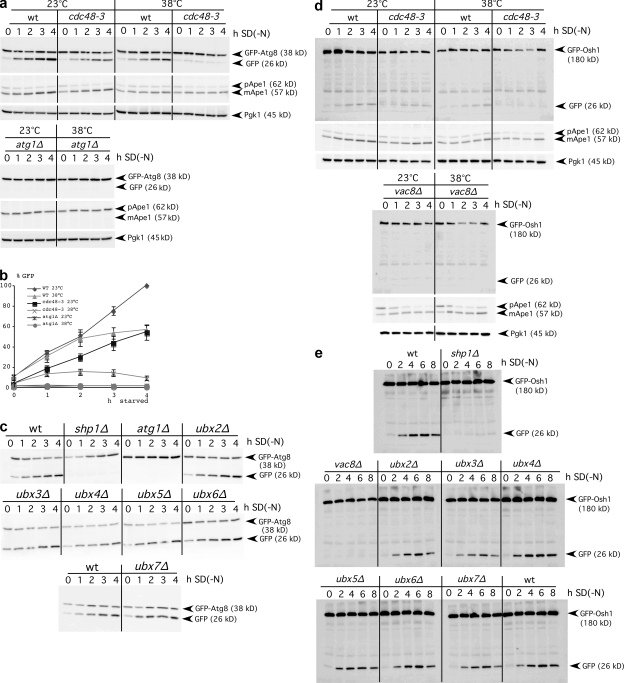
Cdc48 is essential for viability; we thus used temperature-sensitive cdc48-3 mutant cells (Latterich et al., 1995). We measured macroautophagy with a standard assay (Meiling-Wesse et al., 2002; Cheong and Klionsky, 2008). In addition to elongation of growing autophagosome membranes, Atg8 is involved in cargo recognition. Accordingly, macroautophagy selectively targets part of GFP-Atg8 to vacuoles, where degradation yields proteolysis-resistant GFP. Increasing GFP levels in immunoblots therefore reflects the macroautophagic rate. At the permissive temperature, starved cdc48-3 cells showed normal macroautophagy, and shift to nonpermissive 38°C severely blocked macroautophagy (Fig. 1, a and b). Cellular survival was unaffected at 38°C. To exclude strain-dependent effects, we repeated the experiment in another genetic background (unpublished data). At 23 or 38°C, no free GFP appeared in autophagy-deficient atg1Δ cells (Fig. 1, a and b).
Figure 1.
Macroautophagy and PMN require Cdc48 and Shp1. (a) GFP levels from GFP-Atg8 degradation reflect the autophagic rate. S. cerevisiae cells grown stationary at 23°C were starved at 23 or 38°C and analyzed in immunoblots with antibodies to GFP (top), proaminopeptidase I (middle), and Pgk1 as a loading control (bottom). (b) Quantification of GFP levels, mean and SD, from at least three experiments. (c) Immunoblot measurement of macroautophagy in ubx mutants at 30°C. (d) GFP levels from breakdown of the PMN marker GFP-Osh1 reflect the PMN rate. Cells were treated as in panel a. (e) Measurement of the PMN rate, as in panel d, in ubx mutants at 30°C.
Cdc48/p97 is expected to extract proteins from protein complexes or membranes during membrane fusions and other processes (for reviews see Jentsch and Rumpf, 2007; Meyer and Popp, 2008). To mediate its divergent roles, it associates with numerous substrate-recruiting and -processing cofactors (Jentsch and Rumpf, 2007; Schuberth and Buchberger, 2008). The Ubx domain proteins are Cdc48/p97 regulators involved in substrate recruitment (Schuberth et al., 2004). S. cerevisiae has seven Ubx proteins, with Shp1/Ubx1 being the mammalian p47 homologue. The GFP-Atg8 degradation assay showed block of starvation-induced macroautophagy in shp1Δ cells but not in cells lacking any other Ubx protein (Fig. 1 c). As a second assay for nonselective macroautophagy, we expressed 3-phosphoglycerate kinase (Pgk1) fused to GFP (Pgk1-GFP) and followed with immunoblot generation of GFP by proteolysis. The lack of GFP in atg1Δ cells confirmed autophagy dependence of GFP formation. shp1Δ cells were defective in the macroautophagic breakdown of this cytosolic marker (Fig. S1 a). During starvation with the proteinase B inhibitor PMSF, autophagic bodies accumulate in the vacuoles of wild-type, but not of autophagy-deficient, cells. Light microscopy showed that shp1Δ cells failed to accumulate autophagic bodies in the vacuole, further supporting a defect in autophagosome formation or their vacuolar fusion (Fig. S1 b).
We next assessed the requirement of Cdc48 and Shp1 for selective autophagy. Piecemeal microautophagy of the nucleus (PMN) requires the core Atg proteins (Krick et al., 2008). It occurs at nucleus–vacuole junctions formed by the interaction of Vac8, Nvj1, Tsc13, and Osh1 (Roberts et al., 2003). The micronucleophagic rate can be monitored in immunoblots after generation of proteolysis-resistant GFP through vacuolar breakdown of GFP-Osh1 (Krick et al., 2008). cdc48-3 cells at the nonpermissive temperature and shp1Δ cells, but not other ubx mutants, showed defective PMN (Fig. 1, d and e). The cytoplasm to vacuole targeting (Cvt) pathway, as a selective macroautophagic pathway, delivers proaminopeptidase I to the vacuole under nonstarvation conditions. In shp1Δ and cdc48-3 cells, mature aminopeptidase I formed even at nonpermissive temperature (Fig. 1, a and d; and Fig. S1, a and c).
Functionality of the Cvt pathway in shp1Δ and cdc48-3 cells seems surprising at first glance, because, as shown subsequently, Atg8 interacts with Shp1. However, for unknown reasons, the requirement of Atg8 differs between the Cvt pathway and macroautophagy. In Atg8-deficient cells the Cvt pathway is blocked, whereas few aberrantly small autophagosomes still form during starvation induction of macroautophagy (Abeliovich et al., 2000; Chang and Huang, 2007). Indeed, Atg8 is crucial for control of autophagosomal size (Nakatogawa et al., 2007; Xie et al., 2008). Because few autophagosomes are sufficient for proaminopeptidase I transport (Suzuki et al., 2002), we anticipate either that Cdc48 and Shp1 predominantly affect elongation of autophagosomes or that few aberrant autophagosomes even form without their action.
Macroautophagy does not depend on the ubiquitin–proteasome system
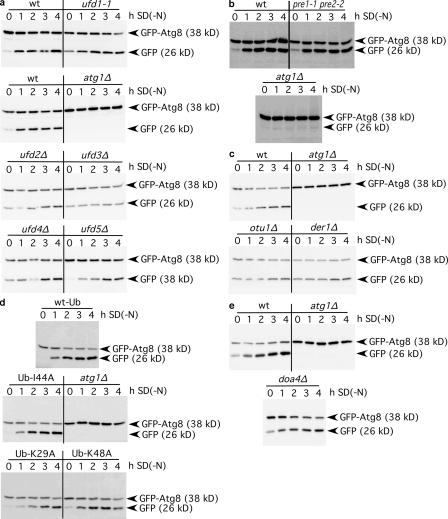
To our knowledge, no precise molecular function was assigned to Shp1; only slight effects on proteasomal degradation of ubiquitinated proteins were reported (Schuberth et al., 2004). We analyzed whether the role of Cdc48 and Shp1 in macroautophagy requires the ubiquitin–proteasome system and respective Cdc48 cofactors. Ufd1 is a crucial substrate-recruiting Cdc48 cofactor (Ye et al., 2001; Jentsch and Rumpf, 2007). In agreement with mutually exclusive binding of Shp1 and Ufd1 to Cdc48, no macroautophagy defect was detectable in ufd1-1 mutants (Fig. 2 a). Macroautophagy was also normal in cells lacking the substrate-processing cofactors Ufd2 and Ufd3 (Fig. 2 a), the ubiquitin ligase Ufd4, the proteasome regulator Ufd5 (Fig. 2 a), the ERAD component Der1 (Fig. 2 c), and the proteasome-deficient pre1-1 pre2-2 cells (Fig. 2 b). Mammalian VCIP135 is distantly related to yeast Otu1, another Cdc48 substrate-processing cofactor. otu1Δ cells showed normal macroautophagy (Fig. 2 c). These data argue against the need for the ubiquitin–proteasome system in the macroautophagic function of Cdc48 and Shp1. Indeed, overexpression of mutated ubiquitin K48A, which is unable to form polyubiquitin via lysine-48 that is recognized by the proteasome (Chau et al., 1989; Sloper-Mould et al., 2001), did not inhibit macroautophagy (Fig. 2 d). Overexpression of mutated ubiquitin I44A suppressed Golgi reassembly, which is consistent with the proposed extraction of a monoubiquitinated fusion regulator from the membrane by the p97–p47 complex (Wang et al., 2004). However, overexpression of ubiquitin I44A did not affect macroautophagy (Fig. 2 d). To further analyze the requirement of ubiquitination, we used cells lacking the deubiquitinating enzyme Doa4. In doa4Δ cells, processes requiring monoubiquitination, such as the multivesicular bodies pathway, are also affected. doa4Δ cells in two genetic backgrounds showed efficient macroautophagy (Fig. 2 e). In sum, macroautophagy requires Cdc48 and Shp1 independent of ubiquitination and proteasomal degradation.
Figure 2.
The ubiquitin–proteasome system is dispensable for macroautophagy. The autophagic rate was determined as in Fig. 1. Mutants defective in the Cdc48 substrate-recruiting cofactor Ufd1, the substrate-processing cofactors Ufd2 and Ufd3, the ubiquitin-ligase Ufd4, the proteasome regulator Ufd5 (a), the proteasome (b), the deubiquitinating cofactor Otu1 (c), the ERAD component Der1 (c), and the deubiquitinating enzyme Doa4 (e) show normal macroautophagy rates. Overexpression of ubiquitin-K29A, -K48A, and -I44A did not interfere with macroautophagy (d).
Shp1 affects autophagosome biogenesis
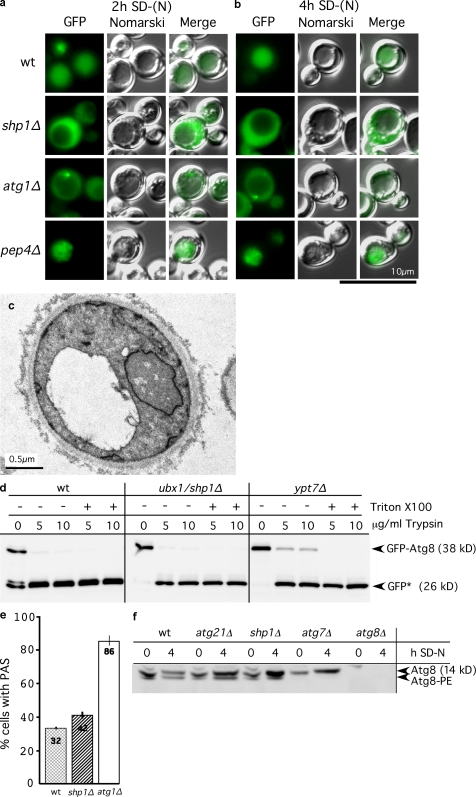
Next, we examined at which step Shp1 affects macroautophagy. The last step is intravacuolar lysis of autophagic bodies dependent on vacuolar acidification and proteinases. Light and electron microscopy showed no vacuolar accumulation of autophagic bodies in starved shp1Δ cells (Fig. 3, a–c). Fluorescence microscopy further demonstrated that, in contrast to wild-type cells, the autophagic cargo GFP-Atg8 did not reach the vacuole in shp1Δ cells (Fig. 3, a and b). GFP-Atg8–positive autophagosomes did not accumulate in the cytosol and were also not detected in shp1Δ cells by electron microscopy (unpublished data). The presence of mature carboxypeptidase Y (Fig. S1 c) in starved shp1Δ cells ruled out that disturbed vacuolar proteolysis caused the GFP-Atg8 degradation defect. Shp1 thus affects either biogenesis of autophagosomes or their vacuolar fusion. We distinguished between these possibilities in a protease protection experiment with spheroplasts hypotonically lysed under conditions that preserved the integrity of autophagosomes. Vacuolar fusion of autophagosomes requires Ypt7. Accordingly, the part of GFP-Atg8 enclosed in autophagosomes is protease protected in ypt7Δ, but not in wild-type, cells because of the rapid vacuolar fusion of autophagosomes (Fig. 3 d). The absence of protease-protected GFP-Atg8 in starved shp1Δ cells indicated defective autophagosome biogenesis or closure (Fig. 3 d). Many S. cerevisiae Atg proteins colocalize at the pre-autophagosomal structure (PAS), the site of autophagosome biogenesis. However, strong cytosolic staining masked detection of Cdc48 and Shp1 at the PAS in direct and indirect fluorescence microscopy. Because Shp1 is dispensable for proaminopeptidase I maturation, it may function in elongation of the isolation membrane, a role proposed for Atg8 (Nakatogawa et al., 2007; Xie et al., 2008). We thus examined whether Shp1 affects localization or lipidation of Atg8. Upon starvation, 42% of shp1Δ and 32% of wild-type cells showed GFP-Atg8–positive PAS punctae (Fig. 3 e), indicating normal Atg8 PAS recruitment. Also, Atg8-PE was formed in shp1Δ cells, and compared with wild type, the Atg8 level was slightly increased, most likely as a result of the autophagic defect (Fig. 3 f).
Figure 3.
Shp1 affects autophagosome biogenesis. (a and b) Fluorescence microscopy of starved cells showed defective vacuolar uptake of GFP-Atg8. No GFP-Atg8–positive autophagosomes accumulated in the cytosol. (c) Electron microscopy of starved shp1Δ cells showed no vacuolar accumulation of autophagic bodies. (d) Lysed spheroplasts of starved cells were trypsin digested with and without detergent. Immunoblots with GFP antibodies showed proteolysis-resistant GFP-Atg8 (inside autophagosomes) in ypt7Δ but not in wild-type and shp1Δ cells. GFP-Atg8 breakdown yields GFP*. (e) Cells with a GFP-Atg8–positive PAS punctum were scored in fluorescence microscopy. The mean and SD of two experiments are shown, with >200 cells analyzed per strain. (f) To analyze Atg8-PE, extracts were separated in 6 M urea SDS-PAGE and immunoblotted with anti-Atg8.
Shp1 interacts with Atg8 and Cdc48
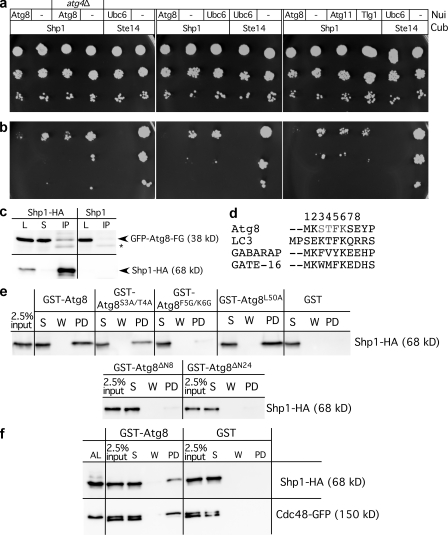
We used the split-ubiquitin system to test interaction of Atg8 with Shp1. We fused the baits with the Nui (N-terminal ubiquitin half) and preys with its Cub (C-terminal domain), followed by a modified Ura3. Protein interaction restores ubiquitin and leads to Ura3 degradation. Slower growth without uracil thus indicates protein interaction (Müller and Johnsson, 2008). Clear interaction between Shp1 and Atg8 was detected (Fig. 4, a and b). No significant interaction was observed between Shp1 and Atg11, Tlg1, or Ubc6 (Fig. 4, a and b). In atg4Δ cells deficient in Atg8 lipidation, interaction between Atg8 and Shp1 was only rather weak but reproducible (Fig. 4, a and b), pointing to the possibility that Shp1 preferentially interacts with Atg8-PE. In wild-type cells, we expected only transient interaction of Atg8 with Shp1 and Cdc48 as a result of Atg8 delipidation by Atg4. Consistently, only little Atg8 coimmunoprecipitated with chromosomally expressed Shp1-HA (Fig. S2 a). To stabilize the complex, we deleted ATG4 and expressed Atg8-FG. Atg4 has two functions: it removes the C-terminal arginine from Atg8-FGR to allow lipidation, and it delipidates Atg8-PE (Nakatogawa et al., 2009). Therefore, in atg4Δ cells, Atg8-FGR is unlipidated, whereas Atg8-FG is permanently lipidated. Coimmunoprecipitation of Shp1-HA chromosomally expressed in atg4Δ cells showed a clear interaction with GFP-Atg8-FG (Fig. 4 c). No clear precipitation of GFP-Atg8-FGR was detectable (unpublished data). This suggests that Shp1 might preferentially interact with Atg8-PE.
Figure 4.
Interaction of Atg8, Shp1, and Cdc48. The split-ubiquitin system proposed preferential interaction of Shp1 with lipidated Atg8. Protein interaction results in slower growth on medium lacking uracil. Dilutions were dropped on medium with (a) and without (b) uracil. Ste14-Cub/Nui-Ubc6, positive; Ste14-Cub/pRS314, negative control. (c) Shp1-HA from lysates (L) of starved atg4Δ cells expressing GFP-Atg8-FG were immunoprecipitated with HA antibodies. S, supernatant; IP, immunoprecipitate. Immunoblots with GFP (top) and HA antibodies (bottom) are shown. *, cross-reaction. (d) Atg8 N terminus aligned with mammalian homologues. (e) GST fusions on beads were incubated with lysates of cells chromosomally expressing Shp1-HA with native promoter. (f) As in panel e, GST fusions were incubated with lysate from cells chromosomally expressing Shp1-HA and Cdc48-GFP with native promoters. AL, alkaline lysis; W, wash, PD, bound. Immunoblots with HA (top) or GFP antibodies (bottom). The Cdc48-GFP double band might be a result of proteolysis.
We confirmed this interaction in a pull down with GST-Atg8. Incubation of crude extracts of S. cerevisiae cells chromosomally expressing Shp1-HA from its own promoter resulted in strong binding of Shp1-HA to GST-Atg8 but not to GST (Fig. 4 e). Pgk1 did not bind to GST-Atg8 or GST, confirming selective interaction (unpublished data). In mass spectrometry of the pull-down eluates, Cdc48, but not Shp1, was identified (unpublished data). The binding of Shp1 to Escherichia coli–expressed nonlipidated GST-Atg8 might reflect the binding of Shp1 to unlipidated Atg8. However, we favor an alternate explanation. Lipidation induces Atg8 oligomerization, and mutations impairing oligomerization affect liposome tethering and phagophore elongation (Nakatogawa et al., 2007). In vitro Atg8 oligomerization is enhanced at 10-µM concentrations. We therefore propose that the ∼20-µM concentration of GST-Atg8 on beads could mimic oligomerization and thus lipidation. Atg8 contains a C-terminal ULD and a 24 amino acid NHD that is absent in ubiquitin. Crystallography of Atg8 bound to a peptide of the cargo receptor Atg19 showed a closed conformation in which the NHD buries part of the ULD (Noda et al., 2008). During this study, the nuclear magnetic resonance solution structure of Atg8 (Schwarten et al., 2010) unraveled flexibility in the NHD. The structure with the lowest target function showed the first eight amino acids in an open conformation projected away from the ULD. Speculatively, oligomerization releases the NHD from the ubiquitin-like domain (ULD) and induces the open conformation (Nakatogawa et al., 2007). Truncation of the NHD affected autophagosomal elongation and reduced the macroautophagic rate to ∼70% for Atg8-ΔN8 lacking the first helix of the NHD and to ∼60% for Atg8-ΔN24 lacking the complete NHD (Nakatogawa et al., 2007; Fig. S2 b). We thus analyzed its relevance for interaction with Shp1 by incubating immobilized GST-Atg8-ΔN8 and GST-Atg8-ΔN24 with extracts from cells chromosomally expressing Shp1-HA. We observed no binding (Fig. 4 e). Putatively, the NHD might help to discriminate ubiquitin from Atg8. Comparison of the first eight amino acids of Atg8 with its mammalian homologues LC3, γ-aminobutyrate type A receptor-associated protein, and GATE-16 showed that amino acids 5 and 6 are strongly conserved (Fig. 4 d). We generated an Atg8-F5G/K6G mutant that was unable to bind Shp1-HA (Fig. 4 e). We also generated an Atg8-S3A/T4A mutant that still effectively bound Shp1-HA (Fig. 4 e). We thus identified the FK motif within the Atg8 NHD as essential for Shp1 binding. Nakatogawa et al. (2007) reported that an Atg8-L50A mutant in the ULD was lipidated and showed increased multimerization and liposome clustering but almost no formation of autophagic bodies. As a control, we also generated an Atg8-L50A mutant and found normal binding to Shp1 (Fig. 4 e), supporting the crucial role of the NHD for Shp1 binding. To demonstrate the existence of a ternary Atg8, Shp1, and Cdc48 complex, we used GST-Atg8 and extracts of cells chromosomally expressing Cdc48-GFP and Shp1-HA with native promoters (Fig. 4 f).
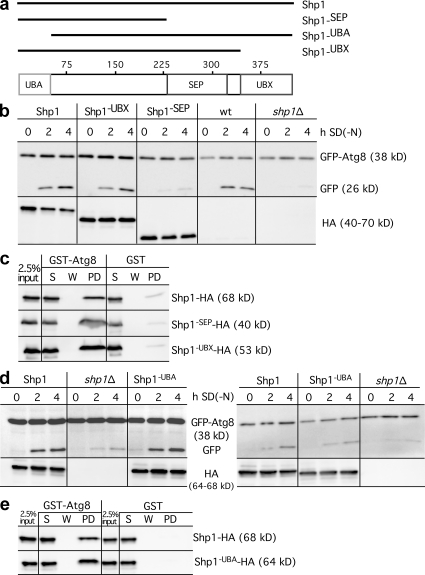
We next analyzed the relevance of Shp1 domains for interaction with Atg8. Shp1 contains a ubiquitin-associated domain (UBA) involved in ubiquitin binding, an SEP (Shp1, eyes-closed, p47) domain involved in p47 trimerization, and a Cdc48/p97-binding Ubx domain (Fig. 5 a). BS1 or SHP box is a second p97 binding site of p47 at the end of the SEP domain. Because of the second Cdc48 binding site, BS1 deletion of the Ubx domain alone did not block macroautophagy (Fig. 5 b). Accordingly, deletion of the SEP and UBX domain, which removes both Cdc48 binding sites, severely inhibited autophagy (Fig. 5 b). The C-terminally truncated Shp1 variants were chromosomally integrated. The N-terminally truncated Shp1 species were on plasmids with the CUP1 promoter. Deletion of the UBA domain had no obvious effect on autophagy, irrespective of induction with exogenous Cu2+ (Fig. 5 d, left) or, when grown in normal medium, containing traces of Cu2+ (Fig. 5 d, right). All truncated Shp1 variants interacted with Atg8 (Fig. 5, c and e). This suggests that Atg8 binding requires the domain between the UBA and the SEP domain. We thus expect that the Atg8-FK motive, which, as part of the NHD, is absent in ubiquitin, interacts with this Shp1 domain.
Figure 5.
Relevance of Shp1 domains for Atg8 interaction. (a) Shp1 domains and generated Shp1 variants. (b) The autophagic rate is measured as in Fig. 1 a with cells chromosomally expressing Shp1 truncations with native promoter. GFP, top; HA antibodies, bottom. (c and e) As in Fig. 4 e, GST-Atg8 beads were incubated with extracts of cells expressing chromosomally (c) or plasmid-encoded (e) truncated HA-tagged Shp1 variants. S, supernatant; W, wash; PD, bound. (d) Autophagic activity of Shp1-UBA-HA with the CUP1 promoter. Left, induction with 50 µM Cu2+; right, medium without exogenous Cu2+.
Defects in the secretory pathway affect autophagosome biogenesis (Ishihara et al., 2001; Reggiori et al., 2004). However, because Atg8 does not affect sorting via the ER and Golgi, the complex of Atg8 with Cdc48-Shp1 cannot have such an indirect effect on macroautophagy. Our data further suggest that the autophagic function of Shp1 requires neither the ubiquitin–proteasome system nor the Shp1 UBA domain. In addition, Atg8 mutants with impaired tethering and hemifusion, including Atg8-ΔN24, showed unaltered PAS localization (Nakatogawa et al., 2007), leading to the conclusion that Atg8 mediates phagophore elongation at the PAS (Nakatogawa et al., 2007). Most recently, LC3 was shown to mediate phagophore elongation, whereas the γ-aminobutyrate type A receptor-associated protein/GATE-16 subfamily most likely mediates autophagosome sealing (Weidberg et al., 2010).
In analogy to Golgi reassembly, we speculate that Atg8 may act as a fusion regulator, which must be extracted from a complex with a fusion mediator by the AAA-ATPase Cdc48 and its adaptor Shp1. In this hypothetical model, deubiquitination by the cysteine proteinase VCIP135 would be reflected by Atg8 delipidation by the cysteine proteinase Atg4, which is needed for efficient macroautophagy. Indeed, GATE-16, a mammalian Atg8 homologue, interacts with the SNARE GOS-28 (Müller et al., 2002). Because another study detected no SNAREs at the PAS (Reggiori et al., 2004), further work is needed to clarify whether small amounts of SNAREs that escaped detection are involved in autophagosome elongation or whether unknown components, probably Atg proteins, take over their role. While finishing our study, two papers reported that mammalian p97/VCP mutants of patients suffering from the multisystem degenerative disease IBMPFD (inclusion body myopathy, Paget disease of bone, and frontotemporal dementia) cause macroautophagic defects by disturbing autophagosome maturation (Ju et al., 2009; Tresse et al., 2010). One study suggests that p97 might selectively affect autophagic degradation of ubiquitinated proteins (Tresse et al., 2010). Both studies do not provide insights into the molecular function of p97 during macroautophagy but underline the medical relevance of macroautophagy.
Materials and methods
Antibodies
Generation of anti–proApe I was described in Barth and Thumm (2001). Anti-GFP antibodies were purchased from Roche. Anti-Pgk1 and anti–carboxypeptidase Y were purchased from Invitrogen, and anti-HA was obtained from Santa Cruz Biotechnology, Inc.
Strains
YYH3/PM373 MATa ura3-52 leu2-3,-112 his4-519 ade1-100 ufd1-1 and its wild type are described in Johnson et al. (1995). Other ufd and ubx deletions were obtained from Euroscarf. pre1-1 pre2-2 cells were described in Heinemeyer et al. (1993). MATa leu2 pep4Δ::URA3 cdc48-3 cells (Latterich et al., 1995) were crossed with WCG4-α (Thumm et al., 1994) or BY4741 (Euroscarf). Ascospores with the cdc48-3 allele were selected. atg deletions are in the WCG background.
Autophagic rate measurement
The GFP-Atg8 breakdown assay is described in Meiling-Wesse et al. (2002). Measurement of PMN was described in Krick et al. (2008). For these assays, cells expressing either GFP-Atg8 or GFP-Osh1 from plasmids were grown to stationary phase and then were shifted to SD-(N) medium (1.7% yeast nitrogen base without amino acid, ammonium sulfate, and 2% glucose). Samples were analyzed by Western blotting using a GFP antibody and secondary horseradish peroxidase–conjugated goat anti–mouse (Dianova) with the ECL or ECL Plus detection kit (GE Healthcare) and an LAS-3000 imaging system (Fujifilm). Temperature-sensitive strains were grown at 23°C to stationary phase and shifted to SD-(N) at either 23 or 38°C. To measure the GFP-Atg8 or GFP-Osh1 breakdown in cells expressing mutated ubiquitin, the plasmids pRS315-GFP-Atg8 and pRS315-GFP-Osh1 were constructed. An insert containing GFP-ATG8 under control of the ATG8 promoter was isolated from pGFP-Atg8 (Suzuki et al., 2001) by Not1–Sal1 digestion and ligated into the Not1–Sal1 cut pRS315. GFP-Osh1 under control of the PHO5 promoter was isolated from pRS416-GFP-Osh1 by Sal1–Nae1 digest and ligated in Sal1–Nae1-digested pRS315. Each of the generated plasmids was cotransformed with plasmids expressing the ubiquitin variants from the CUP1 promoter (Sloper-Mould et al., 2001). For the degradation assay, stationary cells were induced in selection medium with 100 µM CuSO4 for 2 h at 30°C. Then, cells were shifted to SD-(N) medium at 30°C, and samples corresponding to 2 OD600 were taken and analyzed by immunoblotting as described.
Fluorescence microscopy
An Axioskope 2 with a digital AxioCam MRm camera, a 100× 1.45 NA oil Plan-Fluar objective, and AxioVision software (release 4.5; Carl Zeiss, Inc.) were used. The images were processed with Photoshop CS4 (Adobe) or Canvas X (ACD Systems International, Inc.). Fluorescent pictures were taken with the GFP filter set.
Proteinase protection
According to Meiling-Wesse et al. (2002), 60 OD600 of stationary cells in the BY4741 background carrying a GFP-Atg8 plasmid was starved in SD-(N) medium for 4 h at 30°C. Cells were harvested, washed once with DTT buffer (10 mM Tris-sulfate, pH 9.4, and 10 mM DTT), and resuspended as a 2-OD/ml suspension in DTT buffer. The suspension was incubated at 30°C for 15 min while shaking. Cells were then harvested and resuspended as a 10-OD/ml suspension in SP buffer (1 M sorbitol and 20 mM Pipes, pH 6.8). 1.2 mg Zymolyase 20T (Seikagaku) was added, and samples were incubated for 25 min at 30°C. The resulting spheroplasts were washed with SP buffer and hypotonically lysed using PS200 buffer (20 mM Pipes, pH 6.8, 200 mM sorbitol, and 5 mM MgCl2). After two preclearing steps, 300-µl aliquots were treated with trypsin sequencing grade (Roche) in the presence or absence of Triton X-100 for 30 min at 30°C. The reaction was stopped by TCA precipitation. The protein pellets were washed twice with acetone and resuspended in 50 µl of Laemmli buffer. Samples were analyzed by immunoblotting as described.
GST pull down
Atg8ΔN8 and Atg8ΔN24 were amplified with GST-ΔN8ATG8 forward, GST-ΔN8ATG8 reverse, GST-ΔN24ATG8 forward, and GST-ΔN8ATG8 reverse with pGFP-Atg8 as a template. Oligonucleotides are listed in Table S1. Products and pGEX-4T-3 were digested with BamH1–Xho1 and ligated. GST-Atg8 was mutagenized with the Quikchange II kit (Agilent Technologies). nATG8 S3A/T4A forward and nATG8 S3A/T4A reverse, nATG8 F5G/K6G forward and nATG8 F5G/K6G reverse, or nATG8 L50A forward and nATG8 L50A reverse and GST-Atg8 as a template were used.
HA was integrated after SHP1 in WCG4-α and in cells chromosomally expressing Cdc48-GFP (Invitrogen; Janke et al., 2004) with S2 HA-Nat and S3 HA-Nat. SHP1-UBX-HA and SHP1-SEP-HA were made in WCG4-α with cSHP1-UBX-HA S3 or cSHP1-SEP-HA S3 and S2 HA-Nat and pYM17 as a template.
CUP1::SHP1 plasmids, pRS313, and pYM-N1 (CUP1) digested with SacI–EcoRI were ligated to pRS313-CUP1. SHP1-HA (GST-SHP1-Eco and pCUP1-SHP1-HA Xho) and SHP1-UBA-HA (CUP1-SHP1-UBA Eco and plasmid pCUP1-SHP1-HA Xho) were amplified with SHP1-HA::natNT2 chromosomal DNA. Products and pRS313-CUP1 were digested with EcoRI–XhoI and ligated.
40 OD600 of stationary cells was glass-bead lysed in cold PBS, 0.5% Triton X-100, and protease inhibitors. 2 OD600 of cleared lysate was removed, and the rest was incubated for 4 h at 4°C with beads carrying equal amounts of GST fusions. After bead sedimentation, 2 OD600 was removed. Beads were washed three times with PBS, 0.5% Triton X-100, and protease inhibitors and twice with PBS, 250 mM NaCl, and protease inhibitors. Washing buffer corresponding to 2 OD600 was taken. Beads were resuspended in 50 µl Laemmli. For immunoblotting, 10 µl of samples and anti-HA was used.
Coimmunoprecipitation
WCG4-α cells chromosomally expressing Shp1-HA with its own promoter and cells additionally deleted for ATG4 were used. ATG4 was deleted with an amplified product of Atg4-His forward and Atg4-His reverse and pFa6a-His3MX6. Strains were transformed with GFP-Atg8 or GFP-Atg8-FG (Suzuki et al., 2001).
Cells were starved for 4 h in SD-(N), and then 40 OD600 of cells was glass-bead lysed in PBS, 0.5% Triton X-100, and protease inhibitors. After centrifugation, 5% of the lysate was taken. After 2 h at 4°C with 0.6 µg HA antibody, 2.5 mg protein A Sepharose CL-4B in PBS was added to the rest. After 2 h at 4°C, beads were sedimented, and 2 OD600 of supernatant was taken and washed three times with lysis and twice with wash buffer (PBS and 250 mM NaCl). Wash buffer corresponding to 2 OD600 was taken. Finally, beads were resuspended in Laemmli and immunoblotted using double eluate volume.
Split-ubiquitin assay
1 OD600 of cells was diluted 1:10, 1:100, 1:1,000, and 1:10,000 on to plates with uracil and on plates without uracil but with 250 µM methionine and 100 µM CuSO4 (Laser et al., 2000).
In SEY6210, ATG4 was deleted using S1 Atg4 ko NAT and S2 Atg4 ko NAT (Janke et al., 2004). For MET25-SHP1-Cub-RURA3, SHP1 was amplified with Shp1-Cub forward and Shp1-Cub reverse, cut with Cla1–Sal1, and ligated in MET25-Cub-RURA3 (provided by F. Reggiori [University Medical Centre Utrecht, Utrecht, The Netherlands] and N. Johnsson [Universitaet Ulm, Ulm, Germany]). For CUP1-Nui-ATG8, ATG8 was amplified with Nui-Atg8 forward and Nui-Atg8 reverse, cut with BamH1–Xho1, and ligated in pRS314-Nui-PCUP1-Nub (provided by N. Johnsson; Wittke et al., 1999). CUP1-Nui-ATG11 and CUP1-Nui-Tlg1 were obtained from F. Reggiori.
Electron microscopy
Cells starved for 4 h in SD-(N) were permanganate fixed and embedded in Epon (Epple et al., 2003). An electron microscope (JEM1200EX-II; JEOL) was used.
Online supplemental material
Fig. S1 shows the defect of shp1Δ cells in unselective starvation-induced macroautophagy. Fig. S2 shows coimmunoprecipitation of Shp1-HA and GFP-Atg8 in wild-type cells and proaminopeptidase I maturation in cells expressing N-terminally truncated Atg8 variants. Table S1 shows oligonucleotides used in this study. Online supplemental materials are available at http://www.jcb.org/cgi/content/full/jcb.201002075/DC1.
Acknowledgments
We thank L. Hicke, T. Rapoport, F. Madeo, H. Urlaub, N. Johnsson, F. Reggiori, T. Prick, and D. Wolf for strains, plasmids, and help. We further thank the Electron Microscopy Unit of the Institute of Biotechnology, University of Helsinki for providing laboratory facilities.
This study was supported by the Deutsche Forschungsgemeinschaft.
Footnotes
Abbreviations used in this paper:
- Cvt
- cytoplasm to vacuole targeting
- ERAD
- ER-associated degradation
- NHD
- N-terminal helical domain
- PAS
- pre-autophagosomal structure
- PE
- phosphatidylethanolamine
- Pgk1
- 3-phosphoglycerate kinase
- PMN
- piecemeal microautophagy of the nucleus
- SEP
- Shp1, eyes-closed, p47
- ULD
- ubiquitin-like domain
References
- Abeliovich H., Dunn W.A., Jr., Kim J., Klionsky D.J. 2000. Dissection of autophagosome biogenesis into distinct nucleation and expansion steps. J. Cell Biol. 151:1025–1034 10.1083/jcb.151.5.1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axe E.L., Walker S.A., Manifava M., Chandra P., Roderick H.L., Habermann A., Griffiths G., Ktistakis N.T. 2008. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 182:685–701 10.1083/jcb.200803137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth H., Thumm M. 2001. A genomic screen identifies AUT8 as a novel gene essential for autophagy in the yeast Saccharomyces cerevisiae. Gene. 274:151–156 10.1016/S0378-1119(01)00614-X [DOI] [PubMed] [Google Scholar]
- Chang C.-Y., Huang W.-P. 2007. Atg19 mediates a dual interaction cargo sorting mechanism in selective autophagy. Mol. Biol. Cell. 18:919–929 10.1091/mbc.E06-08-0683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau V., Tobias J.W., Bachmair A., Marriott D., Ecker D.J., Gonda D.K., Varshavsky A. 1989. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science. 243:1576–1583 10.1126/science.2538923 [DOI] [PubMed] [Google Scholar]
- Cheong H., Klionsky D.J. 2008. Biochemical methods to monitor autophagy-related processes in yeast. Methods Enzymol. 451:1–26 10.1016/S0076-6879(08)03201-1 [DOI] [PubMed] [Google Scholar]
- Dalal S., Rosser M.F.N., Cyr D.M., Hanson P.I. 2004. Distinct roles for the AAA ATPases NSF and p97 in the secretory pathway. Mol. Biol. Cell. 15:637–648 10.1091/mbc.E03-02-0097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epple U.D., Eskelinen E.L., Thumm M. 2003. Intravacuolar membrane lysis in Saccharomyces cerevisiae. Does vacuolar targeting of Cvt17/Aut5p affect its function? J. Biol. Chem. 278:7810–7821 10.1074/jbc.M209309200 [DOI] [PubMed] [Google Scholar]
- Farré J.-C., Krick R., Subramani S., Thumm M. 2009. Turnover of organelles by autophagy in yeast. Curr. Opin. Cell Biol. 21:522–530 10.1016/j.ceb.2009.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemeyer W., Gruhler A., Möhrle V., Mahé Y., Wolf D.H. 1993. PRE2, highly homologous to the human major histocompatibility complex-linked RING10 gene, codes for a yeast proteasome subunit necessary for chrymotryptic activity and degradation of ubiquitinated proteins. J. Biol. Chem. 268:5115–5120 [PubMed] [Google Scholar]
- Ichimura Y., Kirisako T., Takao T., Satomi Y., Shimonishi Y., Ishihara N., Mizushima N., Tanida I., Kominami E., Ohsumi M., et al. 2000. A ubiquitin-like system mediates protein lipidation. Nature. 408:488–492 10.1038/35044114 [DOI] [PubMed] [Google Scholar]
- Ishihara N., Hamasaki M., Yokota S., Suzuki K., Kamada Y., Kihara A., Yoshimori T., Noda T., Ohsumi Y. 2001. Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion. Mol. Biol. Cell. 12:3690–3702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C., Magiera M.M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E., Knop M. 2004. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 21:947–962 10.1002/yea.1142 [DOI] [PubMed] [Google Scholar]
- Jentsch S., Rumpf S. 2007. Cdc48 (p97): a “molecular gearbox” in the ubiquitin pathway? Trends Biochem. Sci. 32:6–11 10.1016/j.tibs.2006.11.005 [DOI] [PubMed] [Google Scholar]
- Johnson E.S., Ma P.C., Ota I.M., Varshavsky A. 1995. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J. Biol. Chem. 270:17442–17456 10.1074/jbc.270.29.17442 [DOI] [PubMed] [Google Scholar]
- Ju J.-S., Fuentealba R.A., Miller S.E., Jackson E., Piwnica-Worms D., Baloh R.H., Weihl C.C. 2009. Valosin-containing protein (VCP) is required for autophagy and is disrupted in VCP disease. J. Cell Biol. 187:875–888 10.1083/jcb.200908115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo H., Rabouille C., Newman R., Levine T.P., Pappin D., Freemont P., Warren G. 1997. p47 is a cofactor for p97-mediated membrane fusion. Nature. 388:75–78 10.1038/40411 [DOI] [PubMed] [Google Scholar]
- Krick R., Muehe Y., Prick T., Bremer S., Schlotterhose P., Eskelinen E.-L., Millen J., Goldfarb D.S., Thumm M. 2008. Piecemeal microautophagy of the nucleus requires the core macroautophagy genes. Mol. Biol. Cell. 19:4492–4505 10.1091/mbc.E08-04-0363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laser H., Bongards C., Schüller J., Heck S., Johnsson N., Lehming N. 2000. A new screen for protein interactions reveals that the Saccharomyces cerevisiae high mobility group proteins Nhp6A/B are involved in the regulation of the GAL1 promoter. Proc. Natl. Acad. Sci. USA. 97:13732–13737 10.1073/pnas.250400997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latterich M., Fröhlich K.U., Schekman R. 1995. Membrane fusion and the cell cycle: Cdc48p participates in the fusion of ER membranes. Cell. 82:885–893 10.1016/0092-8674(95)90268-6 [DOI] [PubMed] [Google Scholar]
- Longatti A., Tooze S.A. 2009. Vesicular trafficking and autophagosome formation. Cell Death Differ. 16:956–965 10.1038/cdd.2009.39 [DOI] [PubMed] [Google Scholar]
- Meiling-Wesse K., Barth H., Thumm M. 2002. Ccz1p/Aut11p/Cvt16p is essential for autophagy and the cvt pathway. FEBS Lett. 526:71–76 10.1016/S0014-5793(02)03119-8 [DOI] [PubMed] [Google Scholar]
- Meyer H., Popp O. 2008. Role(s) of Cdc48/p97 in mitosis. Biochem. Soc. Trans. 36:126–130 10.1042/BST0360126 [DOI] [PubMed] [Google Scholar]
- Mizushima N., Levine B., Cuervo A.M., Klionsky D.J. 2008. Autophagy fights disease through cellular self-digestion. Nature. 451:1069–1075 10.1038/nature06639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J., Johnsson N. 2008. Split-ubiquitin and the split-protein sensors: chessman for the endgame. Chembiochem. 9:2029–2038 10.1002/cbic.200800190 [DOI] [PubMed] [Google Scholar]
- Müller J.M., Shorter J., Newman R., Deinhardt K., Sagiv Y., Elazar Z., Warren G., Shima D.T. 2002. Sequential SNARE disassembly and GATE-16-GOS-28 complex assembly mediated by distinct NSF activities drives Golgi membrane fusion. J. Cell Biol. 157:1161–1173 10.1083/jcb.200202082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatogawa H., Ichimura Y., Ohsumi Y. 2007. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 130:165–178 10.1016/j.cell.2007.05.021 [DOI] [PubMed] [Google Scholar]
- Nakatogawa H., Suzuki K., Kamada Y., Ohsumi Y. 2009. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat. Rev. Mol. Cell Biol. 10:458–467 10.1038/nrm2708 [DOI] [PubMed] [Google Scholar]
- Noda N.N., Kumeta H., Nakatogawa H., Satoo K., Adachi W., Ishii J., Fujioka Y., Ohsumi Y., Inagaki F. 2008. Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes Cells. 13:1211–1218 10.1111/j.1365-2443.2008.01238.x [DOI] [PubMed] [Google Scholar]
- Reggiori F., Wang C.W., Nair U., Shintani T., Abeliovich H., Klionsky D.J. 2004. Early stages of the secretory pathway, but not endosomes, are required for Cvt vesicle and autophagosome assembly in Saccharomyces cerevisiae. Mol. Biol. Cell. 15:2189–2204 10.1091/mbc.E03-07-0479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts P., Moshitch-Moshkovitz S., Kvam E., O’Toole E., Winey M., Goldfarb D.S. 2003. Piecemeal microautophagy of nucleus in Saccharomyces cerevisiae. Mol. Biol. Cell. 14:129–141 10.1091/mbc.E02-08-0483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuberth C., Buchberger A. 2008. UBX domain proteins: major regulators of the AAA ATPase Cdc48/p97. Cell. Mol. Life Sci. 65:2360–2371 10.1007/s00018-008-8072-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuberth C., Richly H., Rumpf S., Buchberger A. 2004. Shp1 and Ubx2 are adaptors of Cdc48 involved in ubiquitin-dependent protein degradation. EMBO Rep. 5:818–824 10.1038/sj.embor.7400203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarten M., Stoldt M., Mohrlüder J., Willbold D. 2010. Solution structure of Atg8 reveals conformational polymorphism of the N-terminal domain. Biochem. Biophys. Res. Commun. 395:426–431 10.1016/j.bbrc.2010.04.043 [DOI] [PubMed] [Google Scholar]
- Sloper-Mould K.E., Jemc J.C., Pickart C.M., Hicke L. 2001. Distinct functional surface regions on ubiquitin. J. Biol. Chem. 276:30483–30489 10.1074/jbc.M103248200 [DOI] [PubMed] [Google Scholar]
- Suzuki K., Kirisako T., Kamada Y., Mizushima N., Noda T., Ohsumi Y. 2001. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 20:5971–5981 10.1093/emboj/20.21.5971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Kamada Y., Ohsumi Y. 2002. Studies of cargo delivery to the vacuole mediated by autophagosomes in Saccharomyces cerevisiae. Dev. Cell. 3:815–824 10.1016/S1534-5807(02)00359-3 [DOI] [PubMed] [Google Scholar]
- Thumm M., Egner R., Koch B., Schlumpberger M., Straub M., Veenhuis M., Wolf D.H. 1994. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 349:275–280 10.1016/0014-5793(94)00672-5 [DOI] [PubMed] [Google Scholar]
- Tresse E., Salomons F.A., Vesa J., Bott L.C., Kimonis V., Yao T.-P., Dantuma N.P., Taylor J.P. 2010. VCP/p97 is essential for maturation of ubiquitin-containing autophagosomes and this function is impaired by mutations that cause IBMPFD. Autophagy. 6:217–227 10.4161/auto.6.2.11014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama K., Jokitalo E., Kano F., Murata M., Zhang X., Canas B., Newman R., Rabouille C., Pappin D., Freemont P., Kondo H. 2002. VCIP135, a novel essential factor for p97/p47-mediated membrane fusion, is required for Golgi and ER assembly in vivo. J. Cell Biol. 159:855–866 10.1083/jcb.200208112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Satoh A., Warren G., Meyer H.H., Wang Y. 2004. VCIP135 acts as a deubiquitinating enzyme during p97-p47–mediated reassembly of mitotic Golgi fragments. J. Cell Biol. 164:973–978 10.1083/jcb.200401010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidberg H., Shvets E., Shpilka T., Shimron F., Shinder V., Elazar Z. 2010. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 29:1792–1802 10.1038/emboj.2010.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittke S., Lewke N., Müller S., Johnsson N. 1999. Probing the molecular environment of membrane proteins in vivo. Mol. Biol. Cell. 10:2519–2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Nair U., Klionsky D.J. 2008. Atg8 controls phagophore expansion during autophagosome formation. Mol. Biol. Cell. 19:3290–3298 10.1091/mbc.E07-12-1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Meyer H.H., Rapoport T.A. 2001. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 414:652–656 10.1038/414652a [DOI] [PubMed] [Google Scholar]