Extracellular microfibrils composed of fibrillin-1 and -2 regulate bone formation through modulation of TGF-β and BMP signaling.
Abstract
Extracellular regulation of signaling by transforming growth factor (TGF)–β family members is emerging as a key aspect of organ formation and tissue remodeling. In this study, we demonstrate that fibrillin-1 and -2, the structural components of extracellular microfibrils, differentially regulate TGF-β and bone morphogenetic protein (BMP) bioavailability in bone. Fibrillin-2–null (Fbn2−/−) mice display a low bone mass phenotype that is associated with reduced bone formation in vivo and impaired osteoblast maturation in vitro. This Fbn2−/− phenotype is accounted for by improper activation of latent TGF-β that selectively blunts expression of osterix, the transcriptional regulator of osteoblast maturation, and collagen I, the structural template for bone mineralization. Cultured osteoblasts from Fbn1−/− mice exhibit improper latent TGF-β activation as well, but mature faster because of increased availability of otherwise matrix-bound BMPs. Additional in vitro evidence excludes a direct role of microfibrils in supporting mineral deposition. Together, these findings identify the extracellular microfibrils as critical regulators of bone formation through the modulation of endogenous TGF-β and BMP signaling.
Introduction
33 TGF-β–related proteins orchestrate a broad spectrum of developmental and physiological processes in mammals (Katagiri et al., 2008). Multiple mechanisms that operate within and outside the cell and at the cell surface regulate signaling by TGF-β family members in a context-specific manner (Shi and Massagué, 2003; ten Dijke and Arthur, 2007; Kang et al., 2009). Extracellular control of endogenous (local) TGF-β signaling, in particular, is emerging as an important aspect of organ formation and tissue homeostasis (Ramirez and Rifkin, 2009). TGF-β1, -2, and -3 (hereafter collectively referred to as TGF-β) are secreted either as a small latent complex (SLC) in which the bioactive homodimer remains associated with the processed propeptides (latency-associated protein [LAP]) or as a large latent complex (LLC) in which the SLC is bound to latent TGF-β–binding proteins (LTBPs) 1, 3, or 4 (Annes et al., 2003). LAP association blocks ligand interaction with TGF-β receptors, whereas LTBP binding targets the SLC to the ECM through LTBP-mediated association with fibronectin or fibrillin assemblies (microfibrils). Several extracellular molecules (many of which interact with microfibrils) and integrin receptors are involved in releasing TGF-β from the ECM, disrupting LAP-mediated latency, or inhibiting TGF-β activity. Bone morphogenetic proteins (BMPs), too, are secreted and targeted to the ECM as C-terminal disulfide cross-linked dimers that are noncovalently associated with the N-terminal propeptides (Katagiri et al., 2008). However, in contrast to TGF-β, in vitro bioactivity assays suggest that BMP prodomains generally may not confer latency to the associated dimers (Sengle et al., 2008a). Moreover, the factors and mechanisms that control BMP sequestration in and release from the ECM are less well understood than those regulating TGF-β bioavailability. Earlier characterization of a limb-patterning defect (syndactyly) in mice with inactivated fibrillin-2 gene (Fbn2) expression revealed a genetic interaction between this ECM molecule and BMP7 signaling in the forming autopods (Arteaga-Solis et al., 2001). Subsequent in vitro experiments demonstrated that BMP prodomains can directly interact with the N termini of fibrillin-1 and -2 (Sengle et al., 2008b).
Fibrillin microfibrils are ubiquitous architectural elements of the ECM that endow connective tissues with specific physical properties, either as obligatory constituents of elastic fibers or as elastin-free assemblies (Ramirez and Rifkin, 2009). Fibrillins interact with several ECM proteins during microfibril biogenesis, and many of these interactions occur at or near the same sites of LLC and BMP binding (Ramirez and Sakai, 2010). The pleiotropic manifestations of Marfan syndrome (MFS; OMIM-154700) and congenital contractural arachnodactyly (CCA; OMIM-121050), which are caused by mutations in fibrillin-1 and -2, respectively, underscore the importance of fibrillin microfibrils in the formation and function of several organ systems (Ramirez and Dietz, 2007). Moreover, the distinct phenotypes of MFS and CCA imply that fibrillin-1 and -2 have discrete functions in spite of them being the building blocks of the same matrix assemblies. Consistent with this postulate, Fbn1−/− mice do not share the limb-patterning defect of Fbn2−/− mice that was genetically associated with low BMP7 signaling (Arteaga-Solis et al., 2001; Carta et al., 2006), and aortic wall homeostasis is severely impaired only in Fbn1 mutant mice largely as a result of promiscuous TGF-β signaling (Pereira et al., 1999; Judge et al., 2004; Habashi et al., 2006). Thus, fibrillin-1 and -2 may control local TGF-β and BMP bioavailability differently, depending on the organ system, developmental stage, or physiological program.
The scope of this study was to test the aforementioned hypothesis using bone formation as an informative model system because of the following considerations. First, osteogenesis is a fairly well understood process that can be replicated in vitro using primary osteoblast cultures (Stein et al., 1990; Karsenty et al., 2009). Second, TGF-β and BMPs are abundantly stored in the bone matrix from which they are released in a timely fashion and at the appropriate concentration to regulate osteogenic differentiation (Rosen and Thies, 1992; Mundy et al., 1995; Katagiri et al., 2008). Third, Fbn1 and Fbn2 are highly expressed in the progenitor and differentiating osteoblasts of forming and adult bones (Zhang et al., 1994, 1995; Kitahama et al., 2000; Arteaga-Solis et al., 2001; Quondamatteo et al., 2002; Roman-Roman et al., 2003; Ulloa-Montoya et al., 2007). Fourth, low bone mass (osteopenia) is one of the few clinical manifestations in common between MFS and CCA patients (Ramirez and Arteaga-Solis, 2008). Our experiments demonstrate that fibrillin-2 and -1 regulate osteoblast maturation by controlling TGF-β bioavailability and calibrating TGF-β and BMP levels, respectively. Furthermore, they exclude a direct contribution of microfibrils to the formation of the organic substrate that supports mineral deposition in bone. Collectively, these findings significantly advance our understanding of the extracellular control of local TGF-β and BMP signaling in bone physiology.
Results
Loss of fibrillin-2 leads to decreased bone mass
Fbn2−/− mice are viable and fertile but proportionally smaller than wild-type (WT) littermates throughout life and irrespective of gender (Arteaga-Solis et al., 2001). Consistent with this last observation, a modest but statistically significant (P < 0.003) length reduction (4.5%) was recorded in 4-mo-old Fbn2−/− femurs compared with WT counterparts (n = 10 for each genotype). Additionally, morphometric analyses of mid-diaphyseal cross sections of 1-mo-old mutant femurs identified changes in bone shape that were appreciably more evident in 4-mo-old Fbn2−/− mice. Changes in the latter set of mutant femurs included a 15% smaller bone width (P < 0.0003) and a 25% narrower marrow cavity than WT specimens (n = 7). In contrast, 4-d-old (postnatal day [P] 4) Fbn2-null femurs were normally shaped and displayed morphologically unremarkable growth plates (unpublished data), implying a probable defect in postnatal bone modeling rather than an abnormality in endochondral bone development.
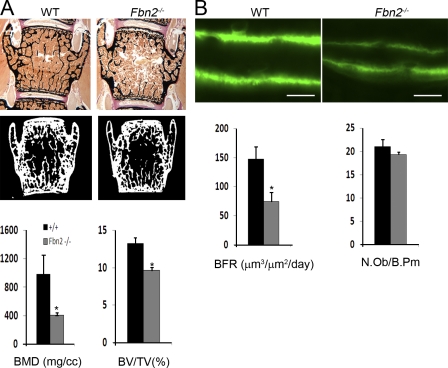
Consistent with the aforementioned prediction, microcomputed tomography (µCT) and histomorphometric analyses detected an appreciable deficit of cancellous bone in 3- and 6-mo-old mutant vertebrae. To be specific, examination of the former samples (n = 5) revealed a 58% decrease in bone mineral density (bone mineral content/total volume [TV]; P = 0.0007), a 27% decrease in bone mass (bone volume [BV]/TV; P = 0.0008; Fig. 1 A), and 52% fewer trabeculae (P < 0.003) and 66% greater intertrabecular space (P < 0.01). Parallel in vivo analyses showed a 55% reduction in bone formation rate (BFR; n = 6; P = 0.0003) associated with a seemingly normal complement of osteoblasts (number of surface osteoblasts/bone perimeter; n = 5; P = 0.24) in mutant compared with WT mice (Fig. 1 B). Altogether, these static and dynamic assessments strongly suggested that impaired bone formation is a major determinant of osteopenia in Fbn2−/− mice.
Figure 1.
Reduced bone mass and BFR in Fbn2−/− mice. (A) Representative von Kossa staining and µCT images of vertebral sections from 3-mo-old WT and Fbn2−/− male mice with histograms summarizing the µCT measurements of volumetric bone mineral density (BMD) and BV/TV in these samples. (B) Illustrative examples of dual-calcein labeling in tibiae of 3-mo-old WT and Fbn2−/− male mice with histograms summarizing BFR values and osteoblast numbers in WT and mutant samples. Error bars indicate mean ± SD, and asterisks indicate statistically significant differences (P < 0.05) between genotypes. Bars, 50 µm.
Loss of fibrillin-2 impairs osteoblast maturation
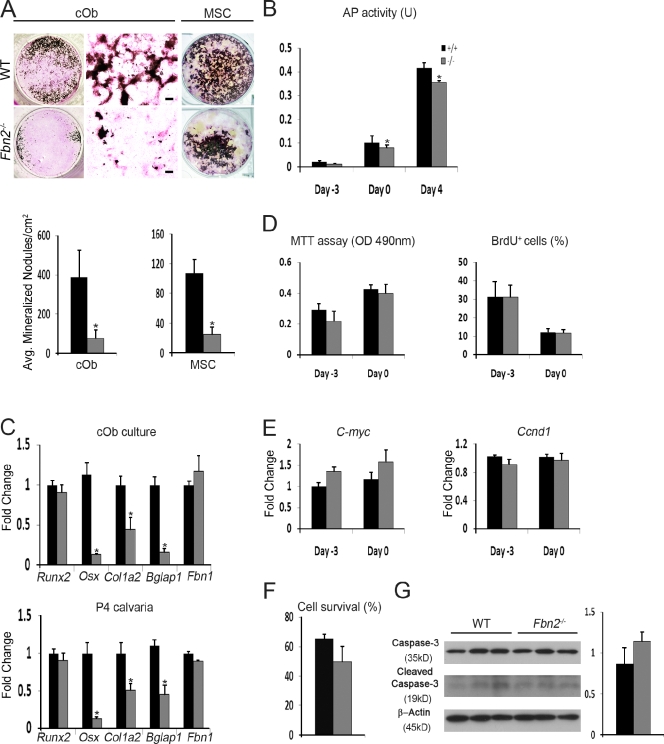
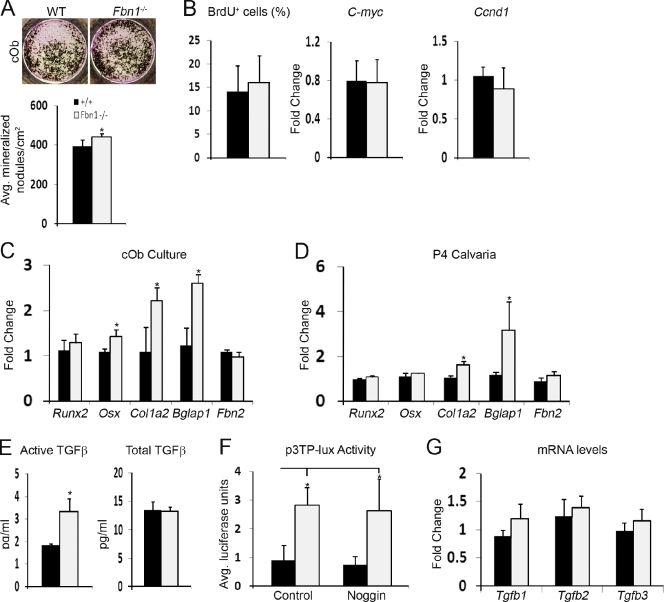
In line with the in vivo data, Fbn2-null calvarial osteoblasts (cObs) or marrow stromal cells (MSCs) cultured with osteoinductive supplements (OSs) yielded fewer and smaller mineralized nodules than the WT counterparts (Fig. 2 A). Impaired osteoblast maturation is characterized by a progressively modest reduction in AP activity (Fig. 2 B) and by an appreciable decrease in collagen deposition (as visualized by van Gieson counterstaining of mutant cOb cultures; Fig. 2 A). Quantitative real-time RT-PCR (qPCR) assays confirmed a substantial down-regulation of α2(I) collagen (Col1a2) and osteocalcin (Bglap1; a marker of terminal osteoblast differentiation) in mutant cOb in addition to excluding a compensatory up-regulation of Fbn1 (Fig. 2 C). The qPCR assays also correlated impaired maturation of Fbn2-null cOb cultures with lower than normal activity of the osterix gene (Osx), which encodes a transcriptional regulator of osteoblast differentiation (Nakashima et al., 2002), and with unremarkable levels of Runx2 mRNA, which encodes the transcriptional determinant of osteoprogenitor commitment (Fig. 2 C; Ducy et al., 1997). Identical results were obtained with RNA purified from the calvariae of Fbn2−/− newborns (Fig. 2 C). Lastly, no significant differences in cell proliferation, BrdU incorporation, and C-myc and Ccnd1 (cyclin D1) mRNA levels were noted between mutant and WT cOb cultures 3 d before and at the time of OS treatment (Fig. 2, D and E). Similar results were obtained by comparing cell survival and apoptosis of mutant and control cOb cultures (Fig. 2, F and G).
Figure 2.
Impaired maturation of Fbn2-null osteoblasts. (A) Illustrative images of von Kossa staining of neonatal cOb (left) and adult MSCs (right) along with magnified van Geison–counterstained images (middle) of cOb differentiated for 21 d after OS administration. Histograms summarize the number of mineralized nodules in WT and Fbn2-null (Fbn2−/−) cOb (n = 15) and MSC (n = 5) cultures. Bars, 200 µm. (B) AP activity of WT and Fbn2−/− cOb measured 3 d before (day −3) and 4 d after (day 4) OS administration (day 0) and normalized to total protein levels (n = 5). (C) qPCR estimates of indicated transcripts in total RNA isolated from day 4 cOb cultures (n = 4; top) or P4 calvariae of WT and Fbn2−/− mice (n = 4; bottom). (D and E) MTT and BrdU incorporation assays (D) and C-myc and Ccnd1 mRNA levels (E) at the indicated days of cOb differentiation (n = 6). (F and G) Cell survival evaluated by Trypan blue exclusion (F; n = 4) and cell apoptosis (G) measured as the fraction of cleaved caspase-3 over full-length protein (with histograms representing densitometric analyses) in WT and mutant cObs. Error bars indicate mean ± SD, and asterisks indicate statistically significant differences (P < 0.05) between genotypes.
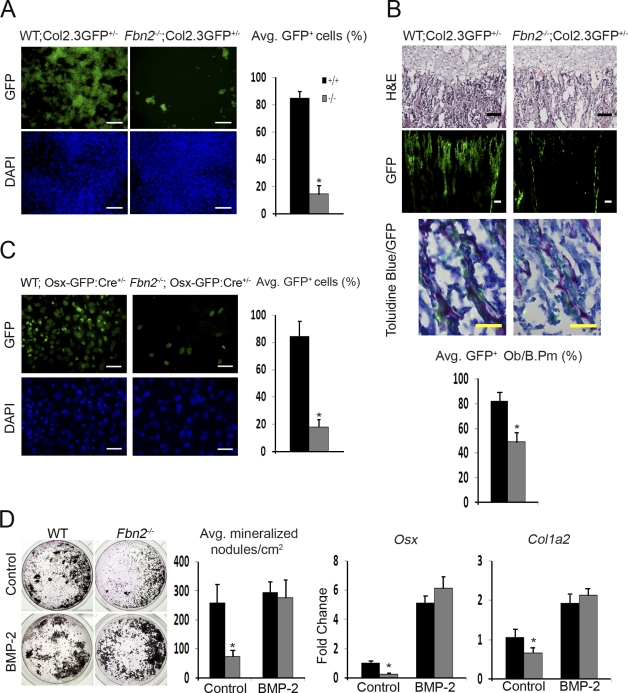
In vivo and ex vivo cell-marking experiments confirmed the aforementioned maturation defect by showing a substantial reduction in the number of GFP-positive cells in neonatal bones and cOb cultures from Fbn2−/− mice harboring the pOBCol2.3GFP transgene, which is specifically activated during osteoblast maturation (Fig. 3; Kalajzic et al., 2005). Incidentally, GFP marking confirmed that fewer surface osteoblasts are actively producing collagen I in mutant bones (Fig. 3 B). Likewise, fewer GFP-positive cells in cOb cultures from Fbn2−/− mice harboring the Osx1-GFP::Cre transgene reiterated the negative impact of the mutation on this critical regulator of osteoblast maturation (Fig. 3 C). Importantly, however, the finding that Fbn2-null cOb cultures can respond to the osteoinductive signal of exogenously added BMP2 by restoring Osx and Col1a2 activity and improving mineral nodule formation demonstrated the reversible nature of the cell defect (Fig. 3 D). Incidentally, the ∼10% greater maturation of BMP2-treated compared with untreated WT cOb was not statistically significant because of an outlier in the latter set of samples (Fig. 3 D). This point notwithstanding, we concluded that loss of fibrillin-2 delays osteoblast maturation by selectively interfering, presumably in an osterix-dependent manner, with the differentiation program that normally promotes maturation and mineralization of the bone ECM.
Figure 3.
Osterix and collagen down-regulation in Fbn2-null osteoblasts. (A and C) Illustrative images of pOBCol2.3GFP (A) or Osx-GFP::Cre transgene expression (C) in Fbn2-null and WT cOb at day 17 or 4 of differentiation, respectively, with histograms summarizing the number of GFP-positive cells (n = 3); nuclei are DAPI stained. (B) Illustrative images of tibiae of Fbn2−/− and WT mice harboring the pOBCol2.3GFP transgene that include (from top to bottom) hematoxylin/eosin staining, GFP expression, and magnified views of toluidine- and GFP-positive cells; histograms summarize cell counts in the last images (n = 3). (D) von Kossa staining after 21 d of differentiation of cOb treated with or without 100 ng/ml rhBMP2 with histograms (right) summarizing numbers of mineralized nodules and levels of Osx and Col1a2 transcripts in mutant and WT samples (n = 3). Error bars indicate mean ± SD, and asterisks indicate statistically significant differences (P < 0.05) between genotypes. Bars: (A and B [middle]) 100 µm; (B [top and bottom] and C) 50 µm.
Latent TGF-β is improperly activated in Fbn2-null osteoblast cultures
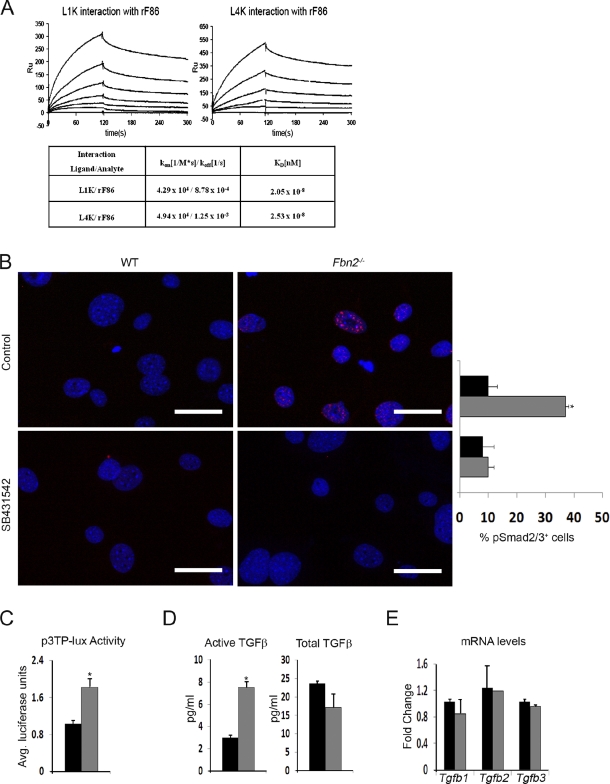
Previous evidence implicating fibrillin-1 in the extracellular control of TGF-β signaling (Isogai et al., 2003; Neptune et al., 2003) prompted us to investigate whether fibrillin-2 is involved in this regulatory process as well. To this end, interactions between fibrillin-2 and LTBPs 1 and 4 were first evaluated in vitro using surface plasmon resonance and recombinant peptides that correspond to the N-terminal segment of fibrillin-2 (rF86) and the C-terminal sequences of LTBPs 1 and 4 (L1K and L4K). The BIAcore assays established that the rF86 fragment binds the L1K and L4K peptides with the same high affinity (Kd = 20–25 nM), as previously shown for the corresponding N-terminal segment of fibrillin-1 (Fig. 4 A; Ono et al., 2009). Next, Fbn2-null cObs were found to display a greater ALK5-dependent nuclear accumulation of pSmad2 (phosphorylated Smad2) than WT cells (Fig. 4 B). Furthermore, relative levels of pSmad2/3 proteins and transcriptional activity of a transfected TGF-β–inducible plasmid (p3TP-lux) were both appreciably higher in mutant than control cells (Fig. 4 C and Fig. S1 A). Lastly, the TMLC bioassay (epithelial cells stably transfected with p3TP-lux; Abe et al., 1994) revealed more active TGF-β but nearly normal amounts of total TGF-β in Fbn2-null compared with WT cOb cultures (Fig. 4 D). The latter finding was independently supported by qPCR analyses showing normal steady-state levels of Tgf-β transcripts in Fbn2-null osteoblasts (Fig. 4 E).
Figure 4.
Fibrillin-2 controls TGF-β bioavailability. (A) Surface plasmon resonance sensograms of binding of immobilized L1K (left) and L4K (right) to rF86 at concentrations between 0 (baseline tracing) and 200 nM (top tracing). Binding was recorded as resonance units (RU), and nonspecific binding to control surface was subtracted at a molar ratio of 1:1. The table summarizes the affinity data (expressed as Kd) for interactions between the LTBPs and fibrillin-2 peptide. (B) Immunodetection of nuclear pSmad2 accumulation in day 4 cOb cultured in low serum with or without SB431542; nuclei are DAPI stained. Histograms summarize the percentage of pSmad2-positive nuclei in WT (black) and mutant (gray) cells (n = 3). (C) Transcriptional activity of p3TP-lux reporter plasmid transfected in WT or Fbn2-null cOb cultured in low serum (n = 3). (D) TMLC bioassays (n = 5) measuring active TGF-β in WT or Fbn2-null cOb cultures (left) or total TGF-β in heat-activated conditioned media of the same cultures (right). (E) qPCR estimates of Tgf-β transcripts in WT and Fbn2-null cOb (n = 3). Error bars indicate mean ± SD, and asterisks indicate statistically significant differences (P < 0.05). Bars, 50 µm.
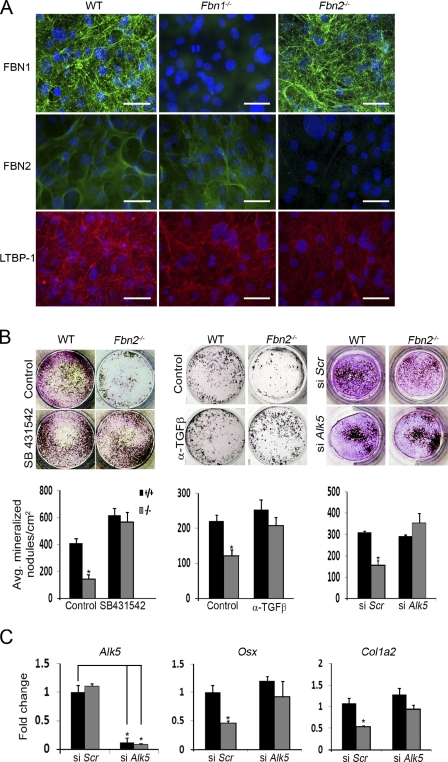
Because LTBPs target TGF-β to microfibrils (Isogai et al., 2003), we examined LTBP1 incorporation in the matrix laid down by overconfluent cOb cultures and found less immunoreactive material in Fbn2-null than WT cultures (Fig. 5 A). TMLC bioassays correlated this visual finding with an ∼47% decrease in the amount of TGF-β extracted from the matrix (relative to the amount extracted from cells) of mutant compared with WT samples (n = 3; P < 0.0001). Furthermore, qPCR analyses showed that Ltbp1 mRNA accumulation in differentiating mutant cOb (time points at days 0, 3, and 7 of osteoinduction) is less than control (∼14%) only 3 d after OS treatment (n = 3; P < 0.046). Taken at face value, these results strongly suggested that loss of fibrillin-2 promotes improper TGF-β activation mostly by impairing LLC sequestration in the osteoblast matrix. Decreased matrix incorporation of LTBP1 in the presence of robust Fbn1 expression further suggested that fibrillin-1 could not compensate for the loss of fibrillin-2 deposition in differentiating cOb cultures. This last observation is analogous to the previous finding that the BMP-dependent syndactyly of Fbn2−/− mice is not seen in Fbn1−/− mice even though both proteins are abundantly deposited in the ECM of the forming autopods (Arteaga-Solis et al., 2001; Carta et al., 2006).
Figure 5.
Elevated TGF-β signaling limits Fbn2-null osteoblast maturation. (A) Illustrative images of immunoreactive material corresponding to the indicated proteins deposited in the ECM of overconfluent WT, Fbn1-null, and Fbn2-null cOb cultures after 4 d of differentiation; nuclei are DAPI stained. Bars, 50 µm. (B) Maturation of WT and Fbn2-null cOb cultures treated with 1 µM SB431542, 300 ng/ml neutralizing pan–TGF-β antibody, or 50 µM Alk5 siRNA with histograms summarizing the number of mineralized nodules in each treatment (n = 3). (C) qPCR estimates of the indicated mRNA levels in the Alk5 silencing experiments. Error bars indicate mean ± SD, and asterisks indicate statistically significant differences (P < 0.05).
To test the postulated involvement of improper TGF-β signaling, mineral nodule formation was assessed in Fbn2-null cOb cultures that were induced to differentiate in the presence of either an ALK5 kinase inhibitor or a pan–TGF-β–neutralizing antibody or that were transfected with siRNA against Alk5 before OS treatment. Inhibition of TGF-β signaling by any of these three strategies equally improved maturation of Fbn2-null cOb (Fig. 5 B). Phenotypic rescue of Alk5-silenced mutant cOb was further associated with normalization of Osx and Col1a2 transcript levels (Fig. 5 C). These results were therefore interpreted to indicate that improper activation of latent TGF-β secondary to loss of fibrillin-2 impairs bone formation by interfering specifically with the ability of osteoblasts to assemble a mineralization-competent bone matrix. Normal TGF-β signaling in another microfibril-rich tissue (skin) and fibrillin-producing cells (dermal fibroblasts) of Fbn2−/− mice indirectly validated the specific effect of the mutation in osteoblasts and bone (unpublished data).
TGF-β and BMP signaling are both abnormally high in Fbn1-null osteoblasts
Although neonatal lethality of Fbn1−/− mice limits performing extensive analyses of bone formation (Carta et al., 2006), these mutant animals nonetheless enabled us to compare and contrast osteogenic differentiation on a matrix deficient for either fibrillin-1 or -2. Fbn1-null cOb proliferated normally but, in contrast to Fbn2-null cells, they yielded more mineral nodules than WT cultures (Fig. 6, A and B); they also displayed a modest increase in Osx expression, a significant up-regulation of Col1a2 and Bglap1, and normal Runx2 and Fbn2 activity (Fig. 6 C). In vivo levels of Col1a2 and Bglap1 (but not Osx) transcripts were appreciably higher than control, and collagen accumulation was slightly greater in mutant than WT bones (Fig. 6 D and Fig. S1 C). Moreover, AP-positive cells and mineral deposits appeared earlier and grew faster in Fbn1-null than WT cOb cultures (Fig. S1 D). Collectively, these observations were consistent with the notion that loss of fibrillin-1 accelerates osteoblast maturation.
Figure 6.
Abnormally high TGF-β activity in differentiating Fbn1-null osteoblasts. (A) Illustrative images of von Kossa–stained WT and Fbn1-null (Fbn1−/−) cOb after 21 d of differentiation with histograms summarizing the number of mineralized nodules in each sample (n = 5). (B) Cell proliferation of WT and mutant cOb at day −3 of cell culture as assessed by BrdU incorporation and qPCR quantification of C-myc and Ccnd1 transcripts (n = 3). (C and D) qPCR estimates of indicated transcripts in total RNA isolated from day 4 differentiating WT and mutant cOb cultures (C; n = 4) and from P4 WT and Fbn1−/− calvarial bones (D; n = 3). (E) TMLC bioassays (n = 5) measuring active TGF-β in WT or Fbn1-null cOb cultures (left) or total TGF-β in heat-activated conditioned media of the same cultures (right). (F) Transcriptional activity of p3TP-lux reporter plasmid transfected in WT or Fbn1-null cOb cultured in low serum with or without 1 µg/ml of noggin (n = 3). (G) qPCR estimates of TGF-β transcripts in WT and Fbn2-null cOb (n = 3). Error bars indicate mean ± SD, and asterisks indicate statistically significant differences (P < 0.05).
Similar to Fbn2-null cells, Fbn1-null cOb cultures displayed less LTBP1-immunoreactive material, more activated TGF-β, and greater p3TP-lux activity than WT cultures (Fig. 5; and Fig. 6, E and F). Normal amounts of total TGF-β and normal levels of Tgf-β transcripts further corroborated the notion that loss of fibrillin-1 deposition leads to improper activation of latent TGF-β complexes (Fig. 6, E and G). Additionally, normal responses of Fbn1-null cOb cultures to the opposing signals of recombinant TGF-β1 and BMP2 excluded possible changes of cell identity (Fig. S1 E). These findings raised the question of which factors may be responsible for overriding the negative impact of heightened TGF-β signaling on osteoblast maturation. We reasoned that BMPs were obvious candidates because they are potent osteoinductive factors that interact in vitro with fibrillin-1 (Sengle et al., 2008b).
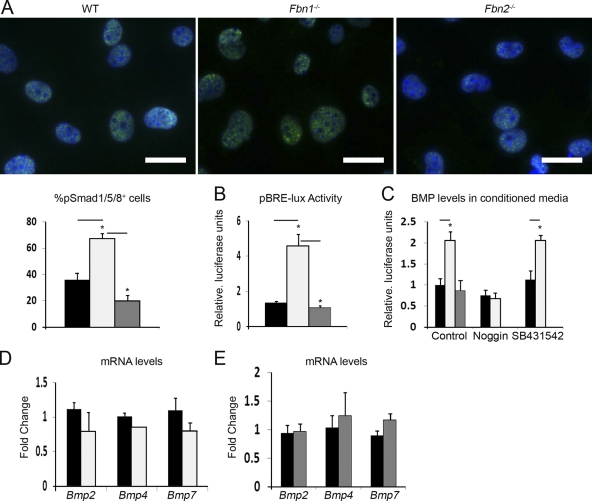
Two sets of evidence supported our hypothesis. First, immunofluorescence microscopy documented greater accumulation of pSmad1/5/8 in the nuclei of Fbn1-null cOb than in those of control or Fbn2-null cells (Fig. 7 A). These visual data were correlated with seemingly more pSmad1/5/8 proteins and higher BRE-luc activity in Fbn1-null cOb cultures compared with the WT or Fbn2-null counterparts (Fig. 7 B and Fig. S1 B). Second, BMP bioassays revealed that conditioned media from Fbn1-null cOb cultures stimulate C2C12BRA reporter cells (which harbor the BMP-inducible plasmid BRE-luc; Zilberberg et al., 2007) twofold more than control or Fbn2-null media (Fig. 7 C). Furthermore, qPCR analyses identified no significant differences in the steady-state levels of several Bmp transcripts between WT and Fbn1- or Fbn2-null cOb (Fig. 7, D and E). Lastly, ELISA assays estimated that BMP levels are appreciably higher in Fbn1-null than WT conditioned media and appreciably lower in Fbn1-null compared with WT matrices (109 vs. 81 pg/ml and 827 vs. 903 pg/ml, respectively; n = 3; P < 0.01). Collectively, these results suggested that loss of fibrillin-1 deposition impairs BMP sequestration in the ECM with the consequence of increasing BMP signaling and, thus, overriding the inhibition of osteoblast maturation by promiscuous TGF-β signaling.
Figure 7.
Fibrillin-1 regulates BMP signaling in cultured osteoblasts. (A) Immunodetection of nuclear pSmad1/5/8 accumulation in day 4 differentiating WT and Fbn1-null (Fbn1−/−) and Fbn2-null (Fbn2−/−) cOb cultured in low serum; nuclei are DAPI stained, and histograms summarize the percentage of pSmad1/5/8-positive nuclei in WT (black), Fbn1-null (white), or Fbn2-null (gray) cells (n = 3). Bars, 25 µm. (B) Transcriptional activity of pBRE-lux reporter plasmid in day 4 differentiating WT, Fbn1-null, and Fbn2-null cOb (n = 5). (C) C2Cl2BRA bioassay measuring BMP signaling in conditioned media from WT, Fbn1-null, and Fbn2-null cOb and from the first two cell cultures treated with either 1 µg/ml noggin or 1 µM SB431542 (n = 3). (D and E) qPCR estimates of Bmp transcripts in day 4 Fbn1−/− and Fbn2−/− cOb cultures, respectively (n = 3). Error bars indicate mean ± SD, and asterisks indicate statistically significant differences (P < 0.05).
Although the reason why a similar elevation of BMP signaling was not observed in Fbn2-null osteoblasts is yet to be determined, the cell culture experiments at least clarified whether or not the increases in TGF-β and BMP signaling that characterize Fbn1-null cOb are causally related to each other. BMP bioassays revealed that conditioned media from Fbn1-null cOb cultures stimulate C2C12BRA reporter cells twofold more than those from control or Fbn2-null cells and that this increase can be abrogated by adding the BMP antagonist noggin to the media, but not by using conditioned media from Fbn1-null cOb pretreated with the ALK5 inhibitor SB431542 (Fig. 7 C). Conversely, addition of noggin to Fbn1-null cOb cultures transfected with p3TP-lux did not affect expression of the TGF-β–responsive reporter plasmid (Fig. 6 F). These results therefore excluded the possibility that augmented TGF-β activity in Fbn1-null cOb cultures stimulates BMP signaling and vice versa. An implicit corollary to this conclusion is that fibrillin-1 is directly involved in calibrating the bioavailability of both TGF-β and BMP molecules during bone formation. Moreover, dysregulated TGF-β and BMP signaling in Fbn1-null bones is a tissue-specific trait in that only the former signaling abnormality characterizes the aortic media and vascular smooth cell cultures of Fbn1−/− mice, and neither of them is evident in the skin or dermal fibroblast cultures of the same mutant animals (Carta et al., 2006, 2009; unpublished data).
Fibrillin microfibrils are not structural components of the mineralization-competent matrix
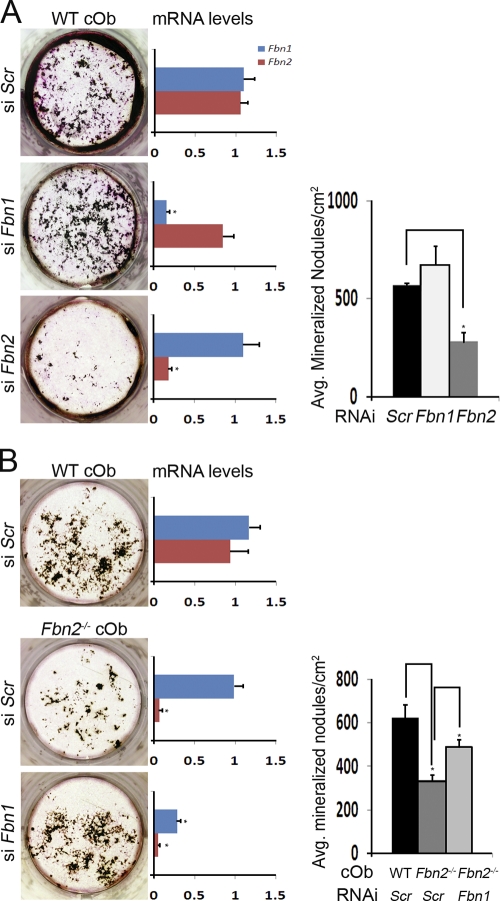
Although the aforementioned data implicated fibrillin-1 and -2 in the differential control of local TGF-β and BMP signals during bone formation, they did not formally exclude the possibility that microfibrils may also be required to form the organic substrate promoting ECM mineralization (Murshed et al., 2005). Accordingly, the mineralizing potential of the microfibril-deficient bone matrix was assessed by the siRNA approach to circumvent the problem of embryonic lethality of mice deficient for both fibrillins (Carta et al., 2006). To this end, maturation of Fbn1- or Fbn2-silenced WT cOb was first equated with the respective phenotypes of Fbn1- or Fbn2-null cells (Fig. 8 A). This finding further corroborated the cell (culture)-autonomous nature of the fibrillin mutations. Next, Fbn1 silencing was shown to improve mineral nodule formation in Fbn2-null cOb cultures (Fig. 8 B). Mineral nodule formation in cOb cultures lacking both fibrillin proteins was therefore interpreted to exclude a major structural role of microfibrils in directly supporting bone mineralization. Incidentally, little or no LTBP1-immunoreactive material in Fbn2-null cOb silenced for Fbn1 supported prior evidence indicating that fibronectin and fibrillin assemblies are sequentially involved in incorporating LTBPs in the ECM (Fig. S2; Dallas et al., 2005).
Figure 8.
Microfibrils are not a structural substrate for matrix mineralization. (A) Illustrative von Kossa–stained WT cOb cultures in which Fbn1 or Fbn2 expression was silenced by RNAi with histograms on the immediate and far right showing the number of mineralized nodules and levels of indicated transcripts (n = 3), respectively. (B) Illustrative von Kossa–stained Fbn2−/− cOb cultures in which Fbn1 was silenced with histograms on the immediate and far right, showing the number of mineralized nodules and levels of indicated transcripts, respectively (n = 3). Error bars indicate mean ± SD, and asterisks indicate statistically significant differences (P < 0.05).
Discussion
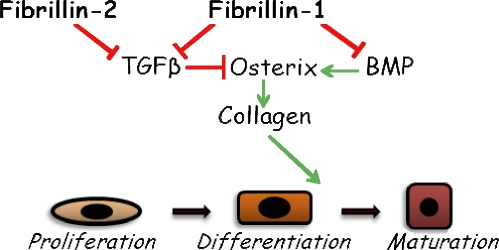
This study provides genetic validation for the long-held belief that sequential release of TGF-β and BMP ligands from the bone matrix contributes to the physiological maintenance of bone mass by balancing the pools of progenitor and mature osteoblasts (Mundy et al., 1995). It also identifies fibrillin microfibrils as the architectural elements of bone tissue that are principally involved in the storage of TGF-β and BMP complexes. Specifically, our experiments have shown that promiscuous TGF-β signaling is the cell-autonomous phenotype of primary osteoblast cultures derived from the bones of either Fbn1−/− or Fbn2−/− mice; they have established that unopposed TGF-β signaling in differentiating Fbn2-null osteoblasts selectively inhibits osterix and collagen I production; they have implied that fibrillin-1 deposition in the forming bone matrix calibrates the threshold levels of local TGF-β and BMP signals during osteoblast maturation; and they have excluded that fibrillin microfibrils are required to constitute a mineralization-competent bone matrix (Fig. 9). Two broader conclusions can be drawn from this and our previous studies of microfibril mutant mice (Arteaga-Solis et al., 2001; Neptune et al., 2003; Carta et al., 2006; Habashi et al., 2006). First, fibrillin proteins control in a different manner and with discrete cellular outcomes the bioavailability of local TGF-β and BMP ligands during tissue formation and remodeling. Second, fibrillin assemblies impart contextual specificity to TGF-β and BMP signals by either concentrating the ligands at sites of intended function in the developing embryo (positive regulation, as exemplified by syndactyly in Fbn2−/− mice) or by restricting their gradual release during postnatal tissue modeling and remodeling (negative regulation, as exemplified by bone loss in Fbn2−/− mice and aortic aneurysm formation in Fbn1 mutant mice).
Figure 9.
Model of microfibril-mediated control of osteoblast maturation. The scheme summarizes the distinct contributions of osteoblast-produced fibrillin-1 and fibrillin-2 microfibrils to osteogenic differentiation through the differential regulation of endogenous TGF-β and BMP signals that together calibrate the rate of bone formation.
Cell culture experiments have indicated that TGF-β and BMPs have both overlapping and opposing functions in bone formation. On the one hand, TGF-β stimulates the recruitment and proliferation of osteoblast progenitors but inhibits their terminal differentiation; and on the other hand, BMPs cooperate in the former process in addition to promoting osteogenic commitment of MSCs and osteoblast maturation (Alliston et al., 2008). However, albeit informative, these in vitro analyses have used exogenous stimulators or inhibitors of cell signaling and differentiation to infer the dynamics of locally released TGF-β and BMP signals during bone formation. For example, increased differentiation of C2C12 cells treated with BMP4 in the presence of an ALK5 inhibitor was interpreted to imply that endogenous TGF-β activity maintains normal bone mass by restricting the rate of osteoblast maturation through Smad-directed blockade of BMP signaling (Maeda et al., 2004). Similarly, genetic studies in mice have not directly interrogated the physiological contribution of matrix-bound TGF-β and BMP signals to bone modeling and remodeling. In this respect, Fbn-null mice are the first animal models to yield unambiguous insights into the importance of the architectural matrix in modulating the local threshold levels of TGF-β and BMP signals during osteogenic differentiation.
Dynamic changes in ECM composition accompany and influence bone formation and mineralization. Collagens I and III, fibronectin, fibrillins, and large proteoglycans predominate in the matrix of osteoprogenitor cells; as preosteoblasts cease to proliferate and begin to differentiate, collagen I production increases substantially along with continued expression of fibrillins and secretion of small proteoglycans and matricellular proteins; once fully differentiated, osteoblasts produce osteocalcin (Ramirez, 2009). Fbn2-null osteoblasts are unable to assemble a mineralization-competent (collagen-rich) ECM, conceivably because promiscuous TGF-β activity delays the emergence of osterix-producing cells. Strong support for this conclusion includes in vivo cell-marking evidence showing that Fbn2−/− bones contain significantly fewer osteoblasts expressing Col1a2 and cell culture data documenting the ability of Fbn2-null cOb to respond to TGF-β antagonism by reactivating Osx and Col1a2 expression and resuming matrix mineralization. Along the same lines, others have reported that collagen production is repressed in Osx−/− mice (which lack differentiated osteoblasts; Wang et al., 2006) and stimulated in p53−/− mice (which display Osx up-regulation and high bone mass; Nakashima et al., 2002) and that a homozygous osterix mutation causes the collagen I–related condition osteogenesis imperfecta (Lapunzina et al., 2010). Increased latent TGF-β activation in Fbn2-null cOb has no apparent effect on cell proliferation. This somewhat surprising result is at least consistent with early in vitro analyses suggesting that exogenous TGF-β modulates cOb proliferation and collagen I production through different mechanisms, which are in part influenced by ligand concentration (Centrella et al., 1987). Our finding may also reflect the involvement of other signaling pathways that are stimulated as a result of promiscuous TGF-β activity and/or a structurally impaired ECM (Carta et al., 2009). Experimental context may also reconcile the unremarkable levels of Runx2 mRNA observed in Fbn2-null cOb and the down-regulation of Runx2 transcription previously reported in TGF-β–treated osteoblastic cells (Alliston et al., 2001).
Genetic interaction between fibrillin-2 and BMP7 in the forming mouse limbs originally predicted that extracellular microfibrils might control the bioavailability of other TGF-β family members (Arteaga-Solis et al., 2001). This prediction was subsequently corroborated by in vitro binding assays showing that the prodomain of several BMPs can bind with comparable affinity to the N-terminal regions of both fibrillin-1 and -2 (Sengle et al., 2008b). Additional analyses have led to the proposal that fibrillins may act as storage scaffolds that distribute, concentrate, and confer latency to BMPs conceivably in a stage and tissue-specific manner (Gregory et al., 2005; Sengle et al., 2008a). Our finding that maturing Fbn1-null osteoblast cultures are characterized by elevated BMP signaling, less matrix-bound BMP, and normal Bmp mRNA levels is a very strong indication that fibrillin-1 microfibrils are indeed involved in sequestering BMP ligands in the bone matrix. Although heightened BMP signaling in Fbn1-null osteoblast cultures implies a context-specific mechanism that overrides the potential of fibrillin-2 to bind BMPs, it is still possible that fibrillin-2 might regulate BMP signaling earlier in osteoblastogenesis, as our study did not examine whether loss of Fbn2 expression also impairs BMP-driven osteogenic commitment. In line with evidence of competitive LTBP and fibulin binding with fibrillin-1 (Ono et al., 2009), one or more ECM molecules may compete with BMP prodomains for the N termini of fibrillins at different stages of bone matrix assembly. A similar mechanism may also explain the reason why dysregulated BMP signaling is only seen in the forming digits of Fbn2−/− mice despite abundant fibrillin-1 production (Arteaga-Solis et al., 2001). It is also interesting to note that BMP signaling is low in the developing autopods of Fbn2−/− mice (as indicated by the syndactyly phenotype of Fbn2+/−;Bmp7+/− mice; Arteaga-Solis et al., 2001) and high in the growing and remodeling bones of Fbn1 mutant mice (as inferred by the BMP bioassay of Fbn1-null osteoblasts; Fig. 7 B). Collectively, these observations therefore support the hypothesis that extracellular microfibrils can control BMP bioavailability (and by extrapolation TGF-β bioavailability) both positively or negatively depending on developmental and physiological contexts and in a fibrillin-specific manner.
Although our findings have clearly established a causal relationship between fibrillin synthesis, matrix sequestration of TGF-β and BMP complexes, and progression of osteogenic differentiation, they have also raised several important new questions about the mechanisms underlying microfibril-mediated control of local TGF-β and BMP signaling. One question is whether latent TGF-β activation in a fibrillin-deficient state may reflect greater availability of the substrate to physiologically normal levels of activators, greater stimulation of activators by a structurally abnormal matrix or a combination of both mechanisms. A corollary to this question is whether or not the same mechanism of latent TGF-β activation operates in all affected tissues of fibrillin mutant mice. This information is particularly relevant to the clinical management of organ-specific manifestations in MFS. Indeed, although systemic inhibition of TGF-β signaling (via angiotensin receptor blockade) mitigates aortic aneurysm progression in MFS mice and patients (Habashi et al., 2006; Brooke, et al., 2008), preliminary data suggest that angiotensin receptor blockade therapy is ineffective to improve osteopenia in Fbn1 mutant mice. As already noted, our study has left unresolved the important problem of how potentially equal interactions between fibrillins and TGF-β family members may impart spatiotemporal specificity to signaling events. This question is germane to the unresolved issue of how in vitro interactions between fibrillins and several other ECM proteins translate into the in vivo assembly of morphologically discrete macroaggregates. One attractive possibility is that cells may coordinate microfibril biogenesis at the plasma membrane with growth factor targeting to the ECM, as recent in vitro evidence suggests that fibronectin assemblies and cell surface receptors regulate both fibrillin polymerization and LTBP incorporation in the matrix (Dallas et al., 2005; Chen et al., 2007; Hubmacher et al., 2008; Sabatier et al., 2009). Moreover, the finding that fibrillin-2 molecules become gradually embedded within fibrillin-1 microfibrils during matrix maturation supports the notion that the dynamics of microfibril assembly may also determine the spatial distribution of signaling complexes within the ECM (Charbonneau et al., 2003, 2010). In this view, the tridimensional arrangement of fibrillin microfibrils may specify both the timely release and the optimal concentration of individual TGF-β family members and ultimately the proper behavior of resident cells, such as osteoblasts and osteoclasts during bone remodeling and fracture healing. In line with this postulate, our parallel study has implicated the fibrillins in modulating bone resorption as well through osteoblast-supported osteoclastogenesis (Nistala et al., 2010). In conclusion, this study has yielded important new insights into the extracellular control of local TGF-β and BMP signaling and implicitly, into the molecular pathophysiology of human diseases that are associated with primary or secondary deficits of the bone matrix.
Materials and methods
Histology and histomorphometry
Fbn1−/− and Fbn2−/− mice have been described previously (Arteaga-Solis et al., 2001; Carta, et al., 2006); transgenic mice pOBCol2.3GFP and Osx-GFP::Cre were provided by D. Rowe (University of Connecticut Health Center, Farmington, CT) and A. McMahon (Harvard University, Cambridge, MA), respectively (Kalajzic et al., 2005; Rodda and McMahon, 2006). Undecalcified vertebrae were embedded in methylmethacrylate, 7-µm-thick methylmethacrylate sections were stained with von Kossa, and 6-µm paraffin sections of femoral growth plates and calvariae were stained with hematoxylin/eosin and van Gieson, respectively, according to standard protocols. Lumbar vertebrae and femora of WT and mutant mice were isolated and scanned using a µCT system (eXplore Locus SP; GE Healthcare) with an isotropic voxel resolution of 9 µm in all three dimensions. All scans were performed using a density calibration phantom containing air, water, and a hydroxyapatite standard (SB3; Gammex RMI) to allow subsequent determinations of tissue mineral densities. Images were analyzed using data acquisition software (Evolver; GE Healthcare), image reconstruction software (Beam; GE Healthcare), and visualization and analysis software (Microview; GE Healthcare). To measure BFR, mice were injected with 25 mg/kg calcein 10 and 2 d before being sacrificed. Osteoblast number, BFR, and BV/TV were assessed using a microscope (DMLB; Leica) and attached color video camera (DXC-390; Sony) and the Osteomeasure analysis system (Osteometrics). Results were analyzed by an unpaired t test (Excel; Microsoft), assuming P < 0.05 as significant.
Primary osteoblast cultures
Standard protocols were used to prepare primary cultures of osteoblasts and MSCs from the calvariae or long bones of P2–4 or 7-wk-old mutant and WT mice, respectively (Bellows et al., 1986). Cells were cultured in α-MEM containing 10% FBS (HyClone) containing 1% penicillin/streptomycin (Invitrogen). Once confluent (day 0), cells were induced to differentiate by supplementing the culture medium with 50 µg/ml ascorbic acid and 10 mM β-glycerophosphate. The medium was changed every 3 d until day 21, when mineral deposits were visualized by von Kossa staining/van Geison counterstaining or by Alizarin red staining and quantified with MetaMorph imaging software (MDS Analytical Technologies). The same computer-aided quantification was used to examine cOb cultures harboring the Osx-GFP::Cre (at day 5 after induction) or pOBCol2.3GFP transgene (at day 17 after induction). In some experiments, culture media included 100 ng/ml rhBMP-2 (Wyeth Pharmaceuticals), 2 ng/ml rhTGF-β1 (R&D Systems), 1 µM SB431542 (Sigma-Aldrich) or 300 ng/ml neutralizing pan–TGF-β antibody (R&D Systems), and respective vehicle controls. Measurements of cell proliferation and viability and BrdU incorporation were performed according to standard protocols (Bonifacino et al., 2004). AP activity was measured using a commercial kit (AnaSpec). All cell images were acquired using a microscope (Eclipse TE200; Nikon) or a dissecting microscope (SMZ645; Nikon) connected to the aforementioned digital camera and processed using Photoshop (version 6.01; Adobe). Statistical analyses were performed using an unpaired t test (Excel), assuming P < 0.05 as significant.
Immunoblots and immunocytochemistry
Osteoblasts were lysed in extraction buffer (50 mM Tris, 150 mM NaCl, 5 mM EDTA, 0.5% NP-40, and 0.1% Triton X-100) supplemented with 1 mM NaF, Na2VO3, 1 mM PMSF, and 1 µg/ml aprotonin, leupeptin, and pepstatin. Cell extracts were assayed for total protein content using the BCA kit (Thermo Fisher Scientific). Proteins extracts were resolved by 10% wt/vol SDS-PAGE and transferred onto Immobilon-P membranes (Millipore). Membranes were incubated first with antibody against mouse caspase-3, Smad2/3, pSmad2/3, and pSmad1/5/8 (1:1,000 dilution; Cell Signaling Technology) or β-actin (1:10,000 dilution; Sigma-Aldrich) and subsequently with biotin-labeled anti–rabbit IgG (1:25,000 dilution; Jackson ImmunoResearch Laboratories, Inc.) and HRP-conjugated streptavidin (Millipore). Nuclear accumulation of pSmads was evaluated by immunofluorescence staining of osteoblasts at day 4 after induction that were cultured in 2% FBS and subsequently fixed and incubated with antisera against pSmad2/3 or pSmad1/5/8 at 1:200 dilution and Alexa Fluor 488 or 594 anti–rabbit IgG secondary antibody (Invitrogen). Number of pSmad-positive nuclei was calculated on multiple fields and expressed as a percentage of total cells in the visual field. Likewise, osteoblast cultures at day 4 after induction were used for fibrillin-1, fibrillin-2, and LTBP-1 immunostaining as previously described (Charbonneau et al., 2003). Nuclei were visualized with DAPI staining. Images were acquired using Axiovision software on an imaging electron microscope (Axioplan; Carl Zeiss, Inc.) with the following objectives: Fluar 10×, Plan Apochromat 40×, Plan Neo Fluar 40×, and Plan Apochromat 63× (imaging medium: air for 10, 20, and 40×; and oil for 63×).
Cell transfections and RNAi
Osteoblasts were seeded the day before transfection at a density of 13,000 cells/cm2 and cultured in 2% FBS. Cells were transiently cotransfected with 400 ng p3TP-lux or BRE-luc (provided by J. Massagué [Memorial Sloan-Kettering Cancer Center, New York, NY] and P. ten Dijke [Leiden University Medical Center, Leiden, Netherlands], respectively; Wrana et al., 1992; Logeart-Avramoglou et al., 2006) and 1 ng control plasmid SV40 Renilla luciferase (Promega) using Lipofectamine 2000 (Invitrogen). Luciferase assays were performed 24 h later and evaluated as previously described (Carta et al., 2009). WT or Fbn2-null cObs were transfected with 50 µM siRNA specific for Fbn1, Fbn2 (Thermo Fisher Scientific), or Alk5 (Invitrogen) using a nontargeting siRNA (Thermo Fisher Scientific) as a control (Carta et al., 2009). Total RNA was purified 2 d later and analyzed for gene silencing efficacy as described in the next paragraph. Parallel cell cultures were transfected once again 4 d later, and osteogenic media were added upon their reaching confluence. All experiments were performed using multiple independent samples and in triplicate. Values were analyzed using an unpaired t test, assuming P < 0.05 as significant.
RNA analyses
Total cellular or calvarial RNA was isolated using RNeasy Mini kit (QIAGEN). RNA concentration and purity were determined spectrophotometrically (NanoDrop; Thermo Fisher Scientific). Reverse transcription was performed with random hexamer primers (Invitrogen) and multiple temperature reverse transcription (AffinityScript; Agilent Technologies) using 1 µg total RNA as a template according to the manufacturer’s instructions. The cDNAs were amplified using SYBR green supermix with ROX (6-carboxy-X-rhodamine; Fermentas) on a Mastercycler ep Realplex instrument (Eppendorf). β-Actin amplification was used as an internal reference for each sample. All qPCR primer sets were purchased from SuperArray Bioscience Corporation. Thermal cycling conditions were 95°C for 10 min followed by 40 cycles consisting of 95°C for 15-s denaturation, 60°C for 30-s annealing, and 72°C for 30-s extension. Comparative quantification was performed using multiple biological replicates that were analyzed in triplicate with the same untreated WT sample designated as the calibrator across different independent experimental runs. Statistical significance was evaluated by an unpaired t test, assuming significant association at P < 0.05 compared with control samples.
TGF-β and BMP bioassays
Cells for TGF-β and BMP bioassays were provided by D. Rifkin (New York University School of Medicine, New York, NY; Abe et al., 1994; Zilberberg et al., 2007). The former bioassays used either cocultures of TMLC and osteoblasts (to measure active TGF-β) or TMLC incubated in the presence of heat-activated conditioned media collected from osteoblast cultures (to measure total TGF-β). In both cases, cells were cultured at the density of 83,000 cells/cm2 on 48-well plates in serum-free DME containing 0.1% BSA, and TGF-β activity was assessed 16 h later by measuring luciferase activity in cell lysates with a luminometer (TD-20; Turner Designs) as described previously (Abe et al., 1994). Resulting reporter gene values (normalized to cell number) were converted into amount of TGF-β (pg/ml) by comparing them with relative luciferase units of TMLC treated with increasing doses of rhTGF-β1. BMP bioassays were similarly performed by measuring relative luciferase units of C2C12BRA cells incubated with cOb-conditioned media as previously described (Zilberberg et al., 2007). To evaluate matrix-bound TGF-β, cells were washed twice with ice-cold PBS and removed using a lysis buffer (0.5% sodium deoxycholate and 10 mM Tris–HCl buffer, pH 8.0, containing 150 mM NaCl and 1% NP-40). Cell debris were removed with ice-cold PBS, and total TGF-β (activated by adding α-MEM containing 0.1% BSA and heating the tissue culture plate at 80°C for 15 min) was evaluated using the TLMC bioassay. To evaluate matrix-bound BMPs, cells were washed twice with PBS and removed using a lysis buffer (0.5% Triton X-100 and 20 mM ammonium hydroxide, pH 7.0). After removing cellular debris with ice-cold PBS, matrix-bound BMPs were solubilized into 10 mM Tris–HCl buffer containing 2 M urea, 2% SDS, and 10% glycerol, pH 6.8. Diluted samples were assayed using the BMP Quantikine kit (R&D Systems). Both bioassays were performed with multiple samples in triplicate and evaluated using an unpaired t test (Excel). Significant association was defined when P < 0.05 compared with control.
Surface plasmon resonance–binding assays
Binding analyses were performed using a BIAcoreX (BIAcore AB) and previously described recombinant LTBP1 (L1K), LTBP4 (L4K), and fibrillin-2 (rF86) peptides (Isogai et al., 2003; Ono et al., 2009). Peptides L1K and L4K were covalently coupled to CM5 sensor chips (research grade) using the amine coupling kit following the manufacturer’s instructions (BIAcore AB). Binding responses caused by analyte interaction with the surface-coupled ligand were normalized by subtracting the background binding to plain control flow cells. Binding assays were performed at 25°C in 10 mM Hepes buffer, pH 7.4, containing 0.15 M NaCl, 2 mM EDTA, and 0.005% vol/vol P20 surfactant (HBS-EP buffer; BIAcore AB). Peptide rF86 was diluted in HBS-EP buffer and injected at several concentrations and different flow rates over immobilized LTBP peptides. The surface was regenerated with a pulse of 10 mM glycine, pH 1.7. Kinetic constants were calculated by nonlinear fitting (1:1 interaction model with mass transfer) to the association and dissociation curves according to the manufacturer’s instructions (BIAevaluation software version 3.0; BIAcore AB). Apparent equilibrium dissociation constants (Kd) were calculated as the ratio of kd/ka.
Online supplemental material
Fig. S1 shows immunoblots of pSmad2/3 and pSmad1/5/8 proteins in fibrillin-deficient osteoblasts, collagen accumulation in Fbn1-null bones, AP and mineral nodule formation at different stages of Fbn1-null osteoblast maturation, and mineralization of and fibrillin-deficient osteoblast cultures treated with rhBMP2 or rhTGF-β1. Fig. S2 shows LTBP1 immunostaining in Fbn2-null cOb silenced for Fbn1. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201003089/DC1.
Acknowledgments
We are indebted to Drs. Juan Massagué, Andrew McMahon, Daniel Rifkin, David Rowe, and Peter ten Dijke for sharing key reagents, Drs. Ernesto Canalis, Theresa Guise, Nicola Partridge, and Marian Young for experimental advices, and Drs. Harry Dietz and Daniel Rifkin for insightful discussions. We also thank Ms. Catherine Liu and Maria del Solar for excellent technical assistance and Ms. Karen Johnson for organizing the manuscript.
This work was supported by grants from the National Institutes of Health (AR42044 and AR49698), the Shriners Hospitals for Children, and the National Marfan Foundation.
Footnotes
Abbreviations used in this paper:
- BFR
- bone formation rate
- BMP
- bone morphogenetic protein
- BV
- bone volume
- CCA
- congenital contractural arachnodactyly
- cOb
- calvarial osteoblast
- LAP
- latency-associated protein
- LLC
- large latent complex
- LTBP
- latent TGF-β–binding protein
- µCT
- microcomputed tomography
- MFS
- Marfan syndrome
- MSC
- marrow stromal cell
- OS
- osteoinductive supplement
- qPCR
- quantitative real-time RT-PCR
- SLC
- small latent complex
- TV
- total volume
- WT
- wild type
References
- Abe M., Harpel J.G., Metz C.N., Nunes I., Loskutoff D.J., Rifkin D.B. 1994. An assay for transforming growth factor-β using cells transfected with a plasminogen activator inhibitor-1 promoter-luciferase construct. Anal. Biochem. 216:276–284 10.1006/abio.1994.1042 [DOI] [PubMed] [Google Scholar]
- Alliston T., Choy L., Ducy P., Karsenty G., Derynck R. 2001. TGF-β-induced repression of CBFA1 by Smad3 decreases cbfa1 and osteocalcin expression and inhibits osteoblast differentiation. EMBO J. 20:2254–2272 10.1093/emboj/20.9.2254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alliston T., Piek E., Derynck R. 2008. TGF-β family signaling in skeletal development, maintenance and disease. The TGF-β Family. Derynck R., Miyazono K., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: 667–723 [Google Scholar]
- Annes J.P., Munger J.S., Rifkin D.B. 2003. Making sense of latent TGFbeta activation. J. Cell Sci. 116:217–224 10.1242/jcs.00229 [DOI] [PubMed] [Google Scholar]
- Arteaga-Solis E., Gayraud B., Lee S.Y., Shum L., Sakai L.Y., Ramirez F. 2001. Regulation of limb patterning by extracellular microfibrils. J. Cell Biol. 154:275–281 10.1083/jcb.200105046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellows C.G., Aubin J.E., Heersche J.N., Antosz M.E. 1986. Mineralized bone nodules formed in vitro from enzymatically released rat calvaria cell populations. Calcif. Tissue Int. 38:143–154 10.1007/BF02556874 [DOI] [PubMed] [Google Scholar]
- Bonifacino J.S., Dasso M., Harford J.B., Lippincott-Schwartz J., Yamada K.M. 2004. Short Protocols in Cell Biology Science: a Compendium of Methods from Current Protocols in Cell Biology. J. Wiley & Sons Inc, Hoboken, NJ: 826 pp. [Google Scholar]
- Brooke B.S., Habashi J.P., Judge D.P., Patel N., Loeys B., Dietz H.C., III 2008. Angiotensin II blockade and aortic-root dilation in Marfan’s syndrome. N. Engl. J. Med. 358:2787–2795 10.1056/NEJMoa0706585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carta L., Pereira L., Arteaga-Solis E., Lee-Arteaga S.Y., Lenart B., Starcher B., Merkel C.A., Sukoyan M., Kerkis A., Hazeki N., et al. 2006. Fibrillins 1 and 2 perform partially overlapping functions during aortic development. J. Biol. Chem. 281:8016–8023 10.1074/jbc.M511599200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carta L., Smaldone S., Zilberberg L., Loch D., Dietz H.C., Rifkin D.B., Ramirez F. 2009. p38 MAPK is an early determinant of promiscuous Smad2/3 signaling in the aortas of fibrillin-1 (Fbn1)-null mice. J. Biol. Chem. 284:5630–5636 10.1074/jbc.M806962200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centrella M., McCarthy T.L., Canalis E. 1987. Transforming growth factor β is a bifunctional regulator of replication and collagen synthesis in osteoblast-enriched cell cultures from fetal rat bone. J. Biol. Chem. 262:2869–2874 [PubMed] [Google Scholar]
- Charbonneau N.L., Dzamba B.J., Ono R.N., Keene D.R., Corson G.M., Reinhardt D.P., Sakai L.Y. 2003. Fibrillins can co-assemble in fibrils, but fibrillin fibril composition displays cell-specific differences. J. Biol. Chem. 278:2740–2749 10.1074/jbc.M209201200 [DOI] [PubMed] [Google Scholar]
- Charbonneau N.L., Jordan C.D., Keene D.R., Lee-Arteaga S., Dietz H.C., Rifkin D.B., Ramirez F., Sakai L.Y. 2010. Microfibril structure masks fibrillin-2 in postnatal tissues. J. Biol. Chem. 285:20242–20251 10.1074/jbc.M109.087031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Sivakumar P., Barley C., Peters D.M., Gomes R.R., Farach-Carson M.C., Dallas S.L. 2007. Potential role for heparan sulfate proteoglycans in regulation of transforming growth factor-β (TGF-β) by modulating assembly of latent TGF-β-binding protein-1. J. Biol. Chem. 282:26418–26430 10.1074/jbc.M703341200 [DOI] [PubMed] [Google Scholar]
- Dallas S.L., Sivakumar P., Jones C.J.P., Chen Q., Peters D.M., Mosher D.F., Humphries M.J., Kielty C.M. 2005. Fibronectin regulates latent transforming growth factor-β (TGF β) by controlling matrix assembly of latent TGF β-binding protein-1. J. Biol. Chem. 280:18871–18880 10.1074/jbc.M410762200 [DOI] [PubMed] [Google Scholar]
- Ducy P., Zhang R., Geoffroy V., Ridall A.L., Karsenty G. 1997. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 89:747–754 10.1016/S0092-8674(00)80257-3 [DOI] [PubMed] [Google Scholar]
- Gregory K.E., Ono R.N., Charbonneau N.L., Kuo C.L., Keene D.R., Bächinger H.P., Sakai L.Y. 2005. The prodomain of BMP-7 targets the BMP-7 complex to the extracellular matrix. J. Biol. Chem. 280:27970–27980 10.1074/jbc.M504270200 [DOI] [PubMed] [Google Scholar]
- Habashi J.P., Judge D.P., Holm T.M., Cohn R.D., Loeys B.L., Cooper T.K., Myers L., Klein E.C., Liu G., Calvi C., et al. 2006. Losartan, an AT1 antagonist, prevents aortic aneurysm in a mouse model of Marfan syndrome. Science. 312:117–121 10.1126/science.1124287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubmacher D., El-Hallous E.I., Nelea V., Kaartinen M.T., Lee E.R., Reinhardt D.P. 2008. Biogenesis of extracellular microfibrils: Multimerization of the fibrillin-1 C terminus into bead-like structures enables self-assembly. Proc. Natl. Acad. Sci. USA. 105:6548–6553 10.1073/pnas.0706335105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isogai Z., Ono R.N., Ushiro S., Keene D.R., Chen Y., Mazzieri R., Charbonneau N.L., Reinhardt D.P., Rifkin D.B., Sakai L.Y. 2003. Latent transforming growth factor β-binding protein 1 interacts with fibrillin and is a microfibril-associated protein. J. Biol. Chem. 278:2750–2757 10.1074/jbc.M209256200 [DOI] [PubMed] [Google Scholar]
- Judge D.P., Biery N.J., Keene D.R., Geubtner J., Myers L., Huso D.L., Sakai L.Y., Dietz H.C. 2004. Evidence for a critical contribution of haploinsufficiency in the complex pathogenesis of Marfan syndrome. J. Clin. Invest. 114:172–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalajzic I., Staal A., Yang W.P., Wu Y., Johnson S.E., Feyen J.H., Krueger W., Maye P., Yu F., Zhao Y., et al. 2005. Expression profile of osteoblast lineage at defined stages of differentiation. J. Biol. Chem. 280:24618–24626 10.1074/jbc.M413834200 [DOI] [PubMed] [Google Scholar]
- Kang J.S., Liu C., Derynck R. 2009. New regulatory mechanisms of TGF-β receptor function. Trends Cell Biol. 19:385–394 10.1016/j.tcb.2009.05.008 [DOI] [PubMed] [Google Scholar]
- Karsenty G., Kronenberg H.M., Settembre C. 2009. Genetic control of bone formation. Annu. Rev. Cell Dev. Biol. 25:629–648 10.1146/annurev.cellbio.042308.113308 [DOI] [PubMed] [Google Scholar]
- Katagiri T., Suda T., Miyazono K. 2008. The bone morphogenetic proteins. The TGF-β Family. Derynck R., Miyazono K., Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 121–149 [Google Scholar]
- Kitahama S., Gibson M.A., Hatzinikolas G., Hay S., Kuliwaba J.L., Evdokiou A., Atkins G.J., Findlay D.M. 2000. Expression of fibrillins and other microfibril-associated proteins in human bone and osteoblast-like cells. Bone. 27:61–67 10.1016/S8756-3282(00)00292-1 [DOI] [PubMed] [Google Scholar]
- Lapunzina P., Aglan M., Temtamy S., Caparrós-Martín J.A., Valencia M., Letón R., Martínez-Glez V., Elhossini R., Amr K., Vilaboa N., Ruiz-Perez V.L. 2010. Identification of a frameshift mutation in Osterix in a patient with recessive osteogenesis imperfecta. Am. J. Hum. Genet. 87:110–114 10.1016/j.ajhg.2010.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logeart-Avramoglou D., Bourguignon M., Oudina K., Ten Dijke P., Petite H. 2006. An assay for the determination of biologically active bone morphogenetic proteins using cells transfected with an inhibitor of differentiation promoter-luciferase construct. Anal. Biochem. 349:78–86 10.1016/j.ab.2005.10.030 [DOI] [PubMed] [Google Scholar]
- Maeda S., Hayashi M., Komiya S., Imamura T., Miyazono K. 2004. Endogenous TGF-β signaling suppresses maturation of osteoblastic mesenchymal cells. EMBO J. 23:552–563 10.1038/sj.emboj.7600067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundy G.R., Boyce B., Hughes D., Wright K., Bonewald L., Dallas S., Harris S., Ghosh-Choudhury N., Chen D., Dunstan C., et al. 1995. The effects of cytokines and growth factors on osteoblastic cells. Bone. 17(2, Suppl):71S–75S 10.1016/8756-3282(95)00182-D [DOI] [PubMed] [Google Scholar]
- Murshed M., Harmey D., Millán J.L., McKee M.D., Karsenty G. 2005. Unique coexpression in osteoblasts of broadly expressed genes accounts for the spatial restriction of ECM mineralization to bone. Genes Dev. 19:1093–1104 10.1101/gad.1276205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K., Zhou X., Kunkel G., Zhang Z., Deng J.M., Behringer R.R., de Crombrugghe B. 2002. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 108:17–29 10.1016/S0092-8674(01)00622-5 [DOI] [PubMed] [Google Scholar]
- Neptune E.R., Frischmeyer P.A., Arking D.E., Myers L., Bunton T.E., Gayraud B., Ramirez F., Sakai L.Y., Dietz H.C. 2003. Dysregulation of TGF-β activation contributes to pathogenesis in Marfan syndrome. Nat. Genet. 33:407–411 10.1038/ng1116 [DOI] [PubMed] [Google Scholar]
- Nistala H., Lee-Arteaga S., Smaldone S., Siciliano G., Ramirez F. 2010. Extracellular microfibrils modulate osteoblast-supported osteoclastogenesis by restricting TGF beta stimulation of RANKL production. J. Biol. Chem. 10.1074/jbc.M110.125328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono R.N., Sengle G., Charbonneau N.L., Carlberg V., Bächinger H.P., Sasaki T., Lee-Arteaga S., Zilberberg L., Rifkin D.B., Ramirez F., et al. 2009. Latent transforming growth factor β-binding proteins and fibulins compete for fibrillin-1 and exhibit exquisite specificities in binding sites. J. Biol. Chem. 284:16872–16881 10.1074/jbc.M809348200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira L., Lee S.Y., Gayraud B., Andrikopoulos K., Shapiro S.D., Bunton T., Biery N.J., Dietz H.C., Sakai L.Y., Ramirez F. 1999. Pathogenetic sequence for aneurysm revealed in mice underexpressing fibrillin-1. Proc. Natl. Acad. Sci. USA. 96:3819–3823 10.1073/pnas.96.7.3819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quondamatteo F., Reinhardt D.P., Charbonneau N.L., Pophal G., Sakai L.Y., Herken R. 2002. Fibrillin-1 and fibrillin-2 in human embryonic and early fetal development. Matrix Biol. 21:637–646 10.1016/S0945-053X(02)00100-2 [DOI] [PubMed] [Google Scholar]
- Ramirez F. 2009. Extracellular matrix in the skeleton. The Skeletal System. Pourquié O., editor Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: 341–353 [Google Scholar]
- Ramirez F., Arteaga-Solis E. 2008. Marfan syndrome and related disorders. Primer on the Metabolic Bone Diseases and Disorders of Mineral Metabolism. Seventh Edition Rosen C.J., editor American Society for Bone and Mineral Research Publications, Washington, D.C. 450–454 [Google Scholar]
- Ramirez F., Dietz H.C. 2007. Marfan syndrome: from molecular pathogenesis to clinical treatment. Curr. Opin. Genet. Dev. 17:252–258 10.1016/j.gde.2007.04.006 [DOI] [PubMed] [Google Scholar]
- Ramirez F., Rifkin D.B. 2009. Extracellular microfibrils: contextual platforms for TGFbeta and BMP signaling. Curr. Opin. Cell Biol. 21:616–622 10.1016/j.ceb.2009.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez F., Sakai L.Y. 2010. Biogenesis and function of fibrillin assemblies. Cell Tissue Res. 339:71–82 10.1007/s00441-009-0822-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodda S.J., McMahon A.P. 2006. Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development. 133:3231–3244 10.1242/dev.02480 [DOI] [PubMed] [Google Scholar]
- Roman-Roman S., Garcia T., Jackson A., Theilhaber J., Rawadi G., Connolly T., Spinella-Jaegle S., Kawai S., Courtois B., Bushnell S., et al. 2003. Identification of genes regulated during osteoblastic differentiation by genome-wide expression analysis of mouse calvaria primary osteoblasts in vitro. Bone. 32:474–482 10.1016/S8756-3282(03)00052-8 [DOI] [PubMed] [Google Scholar]
- Rosen V., Thies R.S. 1992. The BMP proteins in bone formation and repair. Trends Genet. 8:97–102 [DOI] [PubMed] [Google Scholar]
- Sabatier L., Chen D., Fagotto-Kaufmann C., Hubmacher D., McKee M.D., Annis D.S., Mosher D.F., Reinhardt D.P. 2009. Fibrillin assembly requires fibronectin. Mol. Biol. Cell. 20:846–858 10.1091/mbc.E08-08-0830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengle G., Ono R.N., Lyons K.M., Bächinger H.P., Sakai L.Y. 2008a. A new model for growth factor activation: type II receptors compete with the prodomain for BMP-7. J. Mol. Biol. 381:1025–1039 10.1016/j.jmb.2008.06.074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengle G., Charbonneau N.L., Ono R.N., Sasaki T., Alvarez J., Keene D.R., Bächinger H.P., Sakai L.Y. 2008b. Targeting of bone morphogenetic protein growth factor complexes to fibrillin. J. Biol. Chem. 283:13874–13888 10.1074/jbc.M707820200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Massagué J. 2003. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 113:685–700 10.1016/S0092-8674(03)00432-X [DOI] [PubMed] [Google Scholar]
- Stein G.S., Lian J.B., Owen T.A. 1990. Bone cell differentiation: a functionally coupled relationship between expression of cell-growth- and tissue-specific genes. Curr. Opin. Cell Biol. 2:1018–1027 10.1016/0955-0674(90)90151-4 [DOI] [PubMed] [Google Scholar]
- ten Dijke P., Arthur H.M. 2007. Extracellular control of TGFbeta signalling in vascular development and disease. Nat. Rev. Mol. Cell Biol. 8:857–869 10.1038/nrm2262 [DOI] [PubMed] [Google Scholar]
- Ulloa-Montoya F., Kidder B.L., Pauwelyn K.A., Chase L.G., Luttun A., Crabbe A., Geraerts M., Sharov A.A., Piao Y., Ko M.S.H., et al. 2007. Comparative transcriptome analysis of embryonic and adult stem cells with extended and limited differentiation capacity. Genome Biol. 8:R163 10.1186/gb-2007-8-8-r163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Kua H.Y., Hu Y., Guo K., Zeng Q., Wu Q., Ng H.H., Karsenty G., de Crombrugghe B., Yeh J., Li B. 2006. p53 functions as a negative regulator of osteoblastogenesis, osteoblast-dependent osteoclastogenesis, and bone remodeling. J. Cell Biol. 172:115–125 10.1083/jcb.200507106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrana J.L., Attisano L., Cárcamo J., Zentella A., Doody J., Laiho M., Wang X.F., Massagué J. 1992. TGF β signals through a heteromeric protein kinase receptor complex. Cell. 71:1003–1014 10.1016/0092-8674(92)90395-S [DOI] [PubMed] [Google Scholar]
- Zhang H., Apfelroth S.D., Hu W., Davis E.C., Sanguineti C., Bonadio J., Mecham R.P., Ramirez F. 1994. Structure and expression of fibrillin-2, a novel microfibrillar component preferentially located in elastic matrices. J. Cell Biol. 124:855–863 10.1083/jcb.124.5.855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Hu W., Ramirez F. 1995. Developmental expression of fibrillin genes suggests heterogeneity of extracellular microfibrils. J. Cell Biol. 129:1165–1176 10.1083/jcb.129.4.1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberberg L., ten Dijke P., Sakai L.Y., Rifkin D.B. 2007. A rapid and sensitive bioassay to measure bone morphogenetic protein activity. BMC Cell Biol. 8:41 10.1186/1471-2121-8-41 [DOI] [PMC free article] [PubMed] [Google Scholar]