Abstract
Background
While numerous SNPs in Chromosome 9p21 have been associated with coronary artery disease (CAD) and incident MI in Caucasians, there are limited and conflicting reports on the association of this locus with prognosis in Caucasians with existing CAD and no reports in blacks or Hispanics. We investigated the hypothesis that 9p21 polymorphisms are associated with increased risk for adverse cardiovascular outcomes in patients with documented CAD.
Methods and Results
We studied the association of 155 chromosome 9p21 SNPs with adverse outcomes among hypertensive CAD patients of multiple races/ethnicities in INVEST GENES (the INternational VErapamil SR Trandolapril STudy GENetic Substudy, n= 1,460, n = 5,979 for 2 SNPs) and with replication testing of 4 SNPs in the INFORM (INvestigation oF Outcomes from acute coronary syndRoMe; n=714) study of acute coronary syndrome (ACS) patients. In INVEST, the haplotype comprised of the risk allele for the widely reported 9p21 SNPs was associated with better prognosis in Caucasians (OR, 95% CI: 0.72, 0.57–0.92, p = 0.0085) but not blacks (1.21, 0.68–1.24, p = 0.52) or Hispanics (0.96, 0.65–1.44, p=0.86). A less commonly reported LD block was associated with worse prognosis in INVEST among both Caucasians (OR=1.52 (1.20–1.93), p = 0.0006) and African Americans (OR = 4.11 (1.55–10.88), p = 0.004).
Conclusions
Our findings suggest previously reported chromosome 9p21 SNPs, which predict incident CAD, are not associated with higher risk for adverse outcomes in patients with established CAD. The less commonly reported LD block warrants further investigation.
Keywords: chromosome 9p21, cardiovascular outcomes, genetic polymorphisms, INVEST, INFORM
INTRODUCTION
Epidemiological and experimental evidence indicate that coronary artery disease (CAD) arises from a combination of genetic and environmental factors. Four genome-wide association studies in 2007 independently identified the chromosomal locus 9p21 as being a susceptibility locus for CAD or myocardial infarction (MI) among Caucasians.1–4 Subsequent studies have replicated these findings and also found association of this locus with stroke and aortic aneurysm in Northern European and North American Caucasian populations1, 5–9 and East Asian populations.10, 11 A recent meta-analysis included 22 reports with 47 distinct datasets and found a statistically significant association between 9p21 SNPs and risk for coronary heart/artery disease or MI that varied by age at disease onset, with odds ratios higher at earlier ages (< 55 years) of disease onset.12 The chromosome 9p21 represents the most replicated locus for CAD and MI in Caucasians to date, with rs1333049 being the strongest SNP, tagging multiple SNPs in high linkage disequilibrium in this region. The risk allele is common with an allele frequency of almost 50%, and is associated with a 20% to 30% increased risk per risk allele. While the association of the 9p21 region with risk for developing CAD is well defined, the association of this region with adverse outcomes in patients with established CAD is unclear. Four recent studies have reported conflicting results about the association of 9p21 SNPs with clinical outcomes in Caucasian patients with existing CAD. One reported the CAD/MI risk-allele [C] of rs1333049 was associated with higher risk for recurrent MI and mortality after acute coronary syndrome (ACS),13 two found no association with subsequent mortality in CAD patients14, 15, and the fourth found a trend for decreased risk for incident MI per C allele in CAD patients.16 We were unable to identify reports on such associations with prognosis in individuals of African descendant or Hispanic ethnicity. Additionally, hypertension is the most prevalent CAD risk factor and its control clearly reduces risk; yet no reports have addressed the possible contribution of blood pressure on these genetic associations.
We sought to determine whether the risk alleles of the 9p21 SNPs, previously associated with increased risk of incident CAD and MI, are also associated with increased risk for adverse cardiovascular (CV) outcomes such as death, nonfatal MI or nonfatal stroke among elderly hypertensive patients with established stable CAD. We utilized data from the INternational VErapamil SR Trandolapril study (INVEST) clinical trial, which included a diverse population with well controlled BP, and we tested this hypothesis in Caucasians, African Americans and Hispanics. We sought to replicate our findings in a distinct cohort of ACS patients from the INvestigation oF Outcomes from acute coronary syndRoMes study (INFORM). This question is particularly important given that the exact molecular mechanism by which the chromosome 9p21 region increases risk of incident CAD is not yet fully understood (and may have different implications in different patient populations) and currently, there is no direct way to pharmacologically modulate the risk imposed by the chromosome 9p21 locus. Further, direct-to-consumer genetics companies are offering genotyping of this locus to patients, so an understanding of how well the previously noted associations apply to different patient populations is important.
METHODS
INVEST-GENES Participants
The INternational VErapamil SR Trandolapril STudy (INVEST) compared adverse cardiovascular outcomes in 22,576 hypertensive CAD patients randomized to two different antihypertensive treatment strategies.17, 18 Details of the methods and main outcomes were previously reported.17, 18 Briefly, patients 50 years or older with CAD, and essential hypertension requiring drug therapy were eligible. Patients were randomly assigned to a verapamil SR- or an atenolol-based treatment strategy. Details on addition of study drugs and dose titration are published elsewhere.17, 18 The INVEST GENEtic Substudy (INVEST-GENES) cohort consisted of 5,979 patients from 213 sites in the United States who provided genomic DNA samples and additional written informed consent for genetic studies.
INVEST-GENES case-control samples
The INVEST-GENES case-control sample, a subset of INVEST-GENES cohort, included all 304 patients (cases) who experienced a primary outcome event during the trial (first occurrence of all cause death, nonfatal MI or nonfatal stroke) and frequency-matched controls (who did not experience death, MI or stroke), matched on age (decades), gender and race/ethnicity (defined as self-identified race/ethnicity as recorded by the physician).
Outcomes
The primary outcome was the first occurrence of all-cause mortality, nonfatal MI or nonfatal stroke. Secondary outcomes were the individual components of the primary outcome. Events were adjudicated by an independent committee that was blinded to treatment strategy.17
INFORM Participants (Replication cohort)
INFORM is a prospective cohort study of ACS patients admitted to two Kansas City, MO, medical centers between March 2001 and October 2002.19 Standard definitions were used to diagnose ACS patients with either MI or unstable angina. MI patients were defined by an elevated troponin value in the setting of symptoms or electrocardiographic changes (both ST-segment elevation and non–ST-segment elevation changes) consistent with an MI. Unstable angina was diagnosed if the patient had a negative troponin blood test and any one of the following: new-onset angina (<2 months) of at least class III of the Canadian Cardiovascular Society Classification, prolonged (>20 minutes) rest angina, recent (<2 months) worsening of angina, or angina that occurred within 2 weeks of an MI.19
Outcomes
The primary endpoint of INFORM was the composite of first occurrence of all-cause mortality or cardiac rehospitalization. The secondary outcomes were the individual components of the primary outcomes. Cardiac rehospitalization was assessed by patient interview and included hospitalization for MI, heart failure, chest pain, planned percutaneous coronary intervention or planned coronary artery bypass grafting. Mortality assessment was made using the Social Security Administration Death Master File (http://www.ntis.gov/products/ssadmf.asp).
Genotyping
Genotyping details are shown in the supplemental material. Briefly, we obtained genotype data for 153 SNPs in 9p21 region on the INVEST-GENES case-control samples from the gene-centric Human CVD 50K SNP array (Illumina). We also genotyped rs2383207 and rs10757278 in INVEST-GENES cohort; and rs2383207, rs10757278, rs7049105 and rs2157719 in INFORM cohort.
STATISTICAL ANALYSES
To minimize the potential for population stratification in our racially and ethnically diverse population in INVEST, all analyses were conducted separately by race/ethnicity and adjusted for the first 2 eigenvectors in the principal component analysis (PCA) performed on a set of LD-pruned SNPs using EIGENSTRAT software20. The analysis in Caucasian individuals was considered primary and analyses in the Hispanic and African-American individuals were considered secondary given that we were under-powered in these groups and did not have a replication cohort for the Hispanic group. Hardy-Weinberg equilibrium was tested for each racial/ethnic group using χ2 analysis with one degree of freedom. Linkage disequilibrium (r2) between the SNPs in each racial/ethnic group was estimated using Haploview.21 Haplotypes were constructed for each race/ethnicity group using PHASE v2.1.22
For the INVEST-GENES case-control samples, the association of the 153 SNPs in the Ch9p21 region and primary outcome was assessed using logistic regression within each race/ethnicity group with and without adjusting for age, gender, PCA (first 2 eigenvectors), treatment strategy diabetes, history of MI and heart failure, average on-treatment blood pressure and smoking. For all genotypes, the risk allele (defined as published risk allele or minor allele for those not published before) homozygotes were coded as 2, heterozygotes as 1 and non-risk allele homozygotes as 0. Odds ratios per risk allele and 95% confidence interval were calculated. To be considered statistically significant for this analysis, the p values for single SNPs needed to be <0.00032 (0.05/155). Case-control association analysis of these SNPs were performed in PLINK.23 For haplotype analysis, the p values needed to be <0.0056 (0.05/9, where 9 is the number of haplotypes with frequency>1% in Caucasians) to be considered statistically significant.
For the SNPs tested in the INVEST GENES and INFORM cohorts, Cox Proportional Hazard modeling was performed to assess the association of the SNPs and the time to adverse cardiovascular outcomes adjusting for pre-specified variables that were shown to be associated with outcomes. Unadjusted hazard ratios were also reported. In INVEST, models were adjusted for age, gender, treatment strategy, diabetes, history of heart failure and history of MI, average on-treatment blood pressure, hyperlipidemia and smoking. In INFORM, models were adjusted for age, gender, ACS type, revascularization strategy (medical management, percutaneous coronary intervention, or coronary artery bypass graft), hypertension, diabetes, history of heart failure, hyperlipidemia and history of MI. The hazard ratios represent risk per risk allele (as defined as previous CAD risk allele or minor allele if not published previously). The assumption of the proportional hazards was tested in each Cox regression model using Schoenfeld residuals.24 We defined statistical significance as a p value of <0.0125 for replication in INFORM along with point estimates in the same direction as in INVEST-GENES.23
RESULTS
INVEST GENES Participants' Baseline Characteristics
The INVEST-GENES cohort consists of an ethnically diverse (40% Caucasians, 47% Hispanics and 12.5% African Americans), elderly population (mean age of 66±10 years) of patients with hypertension and documented CAD (23.2% with history of MI, 74.4% had diagnosis of classic angina pectoris). The average follow up was 2.8 years, with a total of 16,946 person-years of follow up. In INVEST GENES, 5907 (98.8%) patients had genotype data on at least one 9p21 SNP. The baseline characteristics and medical history of these patients are shown in supplemental Table S1.
INVEST-GENES case-control samples (n=1460) included 858 Caucasians (58.8%), 212 blacks (14.5%) and 390 Hispanics (26.7%) elderly patients (age: 70.5±9.3 years) with hypertension and documented CAD (54% with history of MI, 64% had diagnosis of classic angina pectoris). There were significant differences in baseline demographics (such as age, gender, body mass index and baseline blood pressure) and medical history (such as percentage of patients with history of MI, heart failure, angina and diabetes) between Caucasians, African Americans and Hispanic patients, all p values <0.0001.
9p21 SNP association with INVEST-GENES primary outcome
We analyzed the association of all 153 SNPs in 9p21 region on the CVD SNP array with the INVEST primary outcome in the 1,460 case-control samples and 2 SNPs (rs2383207 and rs10757278) in the entire 5,979 INVEST-GENES cohort. The 8 SNPs with significant association (p<0.00032 in Caucasians) with the primary outcome are shown in Table 1. None of these SNPs reached statistical significance in African Americans or Hispanics, 2 SNPs in AA and 1 SNP in Hispanics that trended toward significance (p<0.01) and are shown in Table 1. The 6 SNPs previously reported to be associated with risk for developing CAD/MI in Caucasians (rs10757274, rs2891168, rs2383206, rs2383207, rs10757278 and rs1333049) are included in Table 2 as well and highlighted in bold, even though they did not meet the threshold of statistical significance used for the novel SNPs from the CVD SNP array in INVEST GENES Caucasians. The genotype and allele frequencies of these SNPs are shown in Supplemental Table S3.
Table 1.
CVD SNP array SNPs associated with INVEST primary outcome.
| Caucasians (n=858) | African Americans (212) | Hispanics (n=390) | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Positiona | Alleles | Minor allele | Risk alleleb | MAF | OR per risk allele | 95% CI | p value | MAF | OR per risk allele | 95% CI | p value | MAF | OR per risk allele | 95% CI | p value |
| rs7044859 | 22008781 | [T/A] | A§ | A | 0.48 | 0.63 | 0.50–0.81 | 0.00024 | 0.34 | 0.65 | 0.38–1.10 | 0.11 | 0.36 | 0.99 | 0.67–1.47 | 0.98 |
| rs10757264 | 22009732 | [A/G] | A | A | 0.49 | 1.66 | 1.28–2.14 | 0.00013 | 0.33 | 1.51 | 0.87–2.64 | 0.15 | 0.34 | 1.24 | 0.81–1.90 | 0.33 |
| rs10965212 | 22013795 | [T/A] | T | T | 0.49 | 1.60 | 1.25–2.05 | 0.00017 | 0.32 | 1.54 | 0.88–2.68 | 0.13 | 0.34 | 1.09 | 0.73–1.62 | 0.68 |
| rs7049105¶ | 22018801 | [A/G] | A | A | 0.49 | 1.61 | 1.25–2.06 | 0.00018 | 0.32 | 1.67 | 0.94–2.98 | 0.08 | 0.34 | 1.10 | 0.74–1.62 | 0.64 |
| rs10965215 | 22019445 | [A/G] | A | A | 0.49 | 0.63 | 0.49–0.81 | 0.00030 | 0.37 | 0.88 | 0.51–1.49 | 0.63 | 0.44 | 0.77 | 0.52–1.14 | 0.19 |
| rs564398 | 22019547 | [A/G] | G | G | 0.39 | 1.53 | 1.20–1.95 | 0.0006 | 0.08 | 3.57 | 1.41–9.03 | 0.0072 | 0.2400 | 0.96 | 0.60–1.54 | 0.86 |
| rs2157719¶ | 22023366 | [A/G] | G | G | 0.4 | 1.58 | 1.24–2.01 | 0.00020 | 0.08 | 3.57 | 1.41–9.03 | 0.0072 | 0.2400 | 1.00 | 0.64–1.59 | 0.98 |
| rs2151280 | 22024719 | [C/T] | T | T | 0.49 | 0.63 | 0.49–0.81 | 0.00025 | 0.36 | 0.90 | 0.52–1.56 | 0.72 | 0.44 | 0.76 | 0.52–1.12 | 0.17 |
| rs16905613 | 22070363 | [A/G] | G | G | 0.0081 | 0.10 | 0.03–0.36 | 0.00020 | 0.00 | na | 0.00 | na | ||||
| rs10757274 | 22086055 | [A/G] | A | G | 0.5 | 0.76 | 0.60–0.98 | 0.031 | 0.0310 | 1.19 | 0.70–2.00 | 0.53 | 0.46 | 0.88 | 0.59–1.30 | 0.51 |
| rs2891168 | 22088619 | [A/G] | G | G | 0.5 | 0.76 | 0.60–0.96 | 0.024 | 0.29 | 0.76 | 0.59–0.96 | 0.06 | 0.46 | 0.88 | 0.60–1.32 | 0.55 |
| rs2383206 | 22105026 | [A/G] | A | G | 0.47 | 0.78 | 0.61–0.98 | 0.035 | 0.46 | 1.32 | 0.80–2.17 | 0.28 | 0.44 | 0.85 | 0.58–1.23 | 0.39 |
| rs2383207¶ | 22105959 | [A/G] | A | G | 0.46 | 0.75 | 0.58–0.96 | 0.022 | 0.14 | 0.69 | 0.38–1.25 | 0.22 | 0.31 | 0.96 | 0.68–1.35 | 0.81 |
| rs10757278¶ | 22114477 | [A/G] | G | G | 0.49 | 0.80 | 0.63–1.02 | 0.078 | 0.25 | 1.96 | 1.27–3.12 | 0.003 | 0.47 | 0.96 | 0.69–1.32 | 0.81 |
| rs1333049 | 22115503 | [C/G] | C | C | 0.49 | 0.78 | 0.61–0.99 | 0.038 | 0.29 | 1.28 | 0.76–2.13 | 0.35 | 0.47 | 1.01 | 0.68–1.49 | 0.96 |
| rs7853656 | 22134530 | [G/T] | G | G | 0.18 | 0.64 | 0.45–0.90 | 0.014 | 0.29 | 0.60 | 0.30–1.18 | 0.13 | 0.19 | 0.29 | 0.14–0.57 | 0.00034 |
Highlighted in boldface are the 6 SNPs that were mostly commonly reported as associated with CAD and MI in the literature.
Abbreviations: OR: odds ratio adjusted for age, gender, PCA, diabetes, history of MI, heart failure, treatment strategy, average on-treatment blood pressure and smoking; CI: confidence interval.
genome build 36.3;
risk allele as previously published or as the minor allele if not previously published;
T is the minor allele in African Americans and Hispanics;
G is minor for African Americans.
indicates SNPs that were included in replication analysis in INFORM
Table 2.
Risk for INVEST primary outcome and rs2383207 and rs10757278
| Caucasians (n = 2364) | African Americans (n = 736) | Hispanics (n = 2770) | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Event/N, % | Unadjusted HR | p value | Adjusted HR† | p value | Event/N, % | Unadjusted HR | p value | Adjusted HR† | p value | Event/N, % | Unadjusted HR | p value | Adjusted HR† | p value | ||
| rs2383207 | AA | 48/444, 10.8% | 0.74(0.59–0.92) | 0.0058 | 0.75(0.60–0.93) | 0.0083 | 2/13, 15.4% | 0.74(0.41–1.31) | 0.3 | 0.69(0.38–1.25) | 0.22 | 9/269, 3.4% | 0.92(0.65–1.30) | 0.63 | 1.02(0.71–1.46) | 0.91 |
| (risk allele: G) | AG | 79/1170, 3.8% | 9/153, 5.9% | 31/1182, 2.6% | ||||||||||||
| GG | 41/693, 5.9% | 31/549, 5.7% | 32/1266, 2.5% | |||||||||||||
| rs10757278 | AA | 56/588, 9.5% | 0.80(0.65–0.98) | 0.036 | 0.81(0.66–1.0) | 0.05 | 21/461, 4.6% | 1.85(1.19–2.86) | 0.006 | 1.95(1.25–3.02) | 0.003 | 23,787, 2.9% | 0.94(0.69–1.30) | 0.71 | 0.96(0.69–1.33) | 0.8 |
| (risk allele: G) | AG | 81/1142, 7.1% | 13/214, 6.1% | 34/1320, 2.6% | ||||||||||||
| GG | 36/567, 6.4% | 7/43, 16.3% | 16/614, 2.6% | |||||||||||||
hazard ratio per risk allele (A for both SNPs), adjusted for age, gender, treatment strategy, diabetes, history of myocardial infarction, heart failure, average on-treatment blood pressure, smoking and hyperlipidemia.
Linkage disequilibrium and haplotype analysis of the 9p21 SNPs
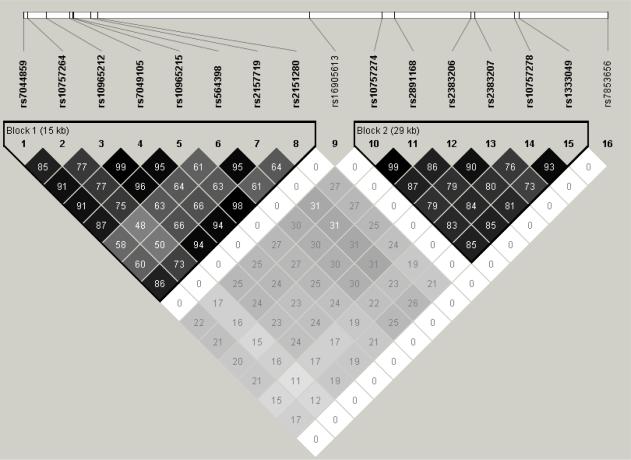
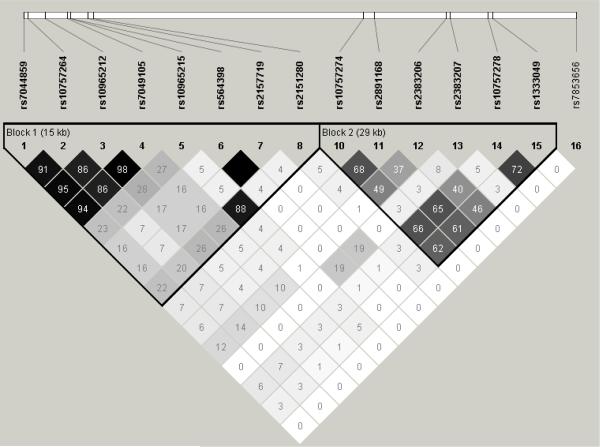
We explored the linkage disequilibrium (LD) among the 16 top SNPs described above, including the 6 most reported 9p21 SNPs from the literature in Haploview and constructed haplotypes for each race/ethnic group. In Caucasian patients, these SNPs formed 2 distinct LD blocks, with the 8 strongest SNPs in this analysis in one block (block 1) and the 6 mostly reported SNPs in another block (block 2) (Figure 1). As expected, the LD of the SNPs in each block was the strongest in Caucasians, weakest in African Americans and intermediate in Hispanics.
Figure 1.
LD (r2) of significant chromosome 9p21 SNPs on the Illumina CVD SNP array in Caucasian (A), African Americans (B) and Hispanics (C).
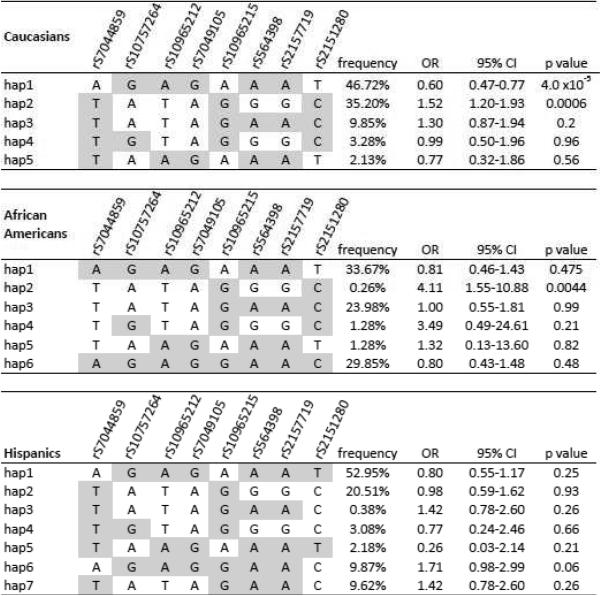
In LD block 1, which consists of the 8 strongest SNPs, the two most frequent haplotypes in Caucasians (hap1 and hap 2) accounted for 82% of the Caucasian chromosomes, 73% of the Hispanic chromosomes, and 34% of the African American chromosomes (Figure 2A). Hap1 of block 1 was associated with 40% better prognosis (lower risk for primary outcome) in Caucasians with OR=0.60 and 95% confidence interval of 0.47–0.77 (p=4.0*10−5), but not in African Americans (OR=0.81, 0.46–1.43, p=0.475) or Hispanics (0.80, 0.55–1.17, p=0.25). The second most frequent haplotype, Hap2, with complementary alleles to Hap1, was associated with worse prognosis (higher risk for primary outcome) among both Caucasians (OR=1.52, 95% confidence interval, 1.20–1.93, p=0.0006) and African Americans (OR=4.11, 1.55–10.88, p=0.004), but not Hispanics (OR=0.98, 0.59–1.62, p=0.93) (Figure 2A).
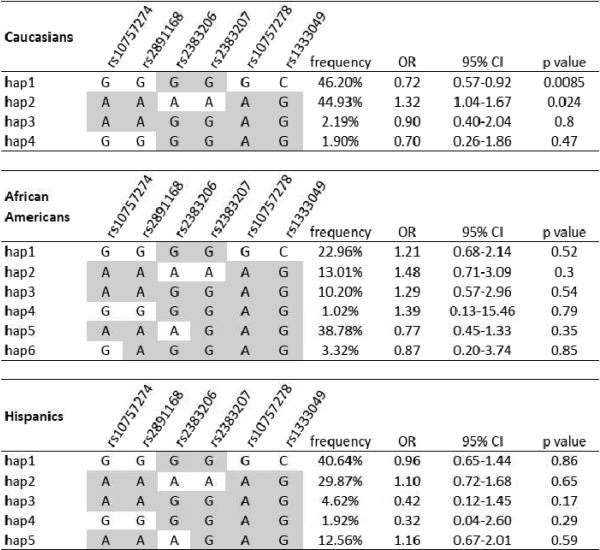
Figure 2.
Haplotype analyses of the 2 LD blocks in 9p21 region in Caucasians, African Americans and Hispanics. (highlighted in gray are the major alleles in each population). A. Block 1: less commonly reported haplotype block B. Block 2: the haplotype block containing the 6 most widely reported SNPs
In LD block 2 containing the 6 9p21 SNPs from the literature (rs2383206, rs2383207, rs10757278, rs1333049, rs10757274 and rs2891168), the 2 most frequent haplotypes (hap1: GGGGGC and hap2: AAAAAG) accounted for over 91% of the Caucasian chromosomes, 71% of the Hispanics chromosomes and 36% of the African American chromosomes (Figure 2B). In Caucasian patients, block 2 hap1 (containing risk alleles from the incident CAD/MI literature) was trended toward better prognosis (OR= 0.72, 95% CI, 0.57–0.92, p=0.0085), while block 2 hap2, with complementary alleles, trended towards worse prognosis (OR=1.32, 95% CI, 1.04–1.67, p=0.024). There was no evidence of such associations in African Americans and Hispanics. We did not observe any differences in 9p21 associations between randomized treatment strategies (ie atenolol- versus verapamil-based treatment strategy) (treatment*SNP interaction p=0.86).
Rs2383207 and rs10757278 Associations with Primary and Secondary Outcomes in INVEST GENES Cohort
As described in the supplemental methods section, the two most widely reported SNPs from the body of literature surrounding the chromosome 9p21 region and CAD/MI risk association, rs2383207 and rs10757278, were not captured on the CVD SNP array and were genotyped separately in the entire INVEST-GENES cohort. The event rates and unadjusted and adjusted hazard ratios (HRs) for the primary outcome in each race/ethnic group of rs2383207 and rs10757278 are shown in Table 2. Neither of these SNPs reached statistical significance in any of the three populations when the association with INVEST primary outcome were tested. The HRs for prognosis among the risk alleles were directionally opposite compared to the published HRs for CAD/MI risk in Caucasians for both SNPs, with the well-documented G alleles showing a trend for protection against adverse cardiovascular outcome The adjusted hazard ratios and 95% confidence intervals for rs2383207 and rs10757278 G alleles were 0.75 (0.60–0.93), p=0.0083 and 0.81 (0.66–1.0), p=0.05, respectively.
Replication of Associations in INFORM
The INFORM genetics cohort consisted of DNA samples from 573 Caucasian and 141 African American patients with a mean age of 60±13 years, 37% female, 28% with ST elevation MI, 30% with non-ST elevation MI and 42% with unstable angina (Supplemental Table S2). Given the strong LD within the two haplotype blocks, to replicate the findings in INVEST, INFORM samples were genotyped for 4 variants: rs7049105 and rs2157719 from LD block 1 and rs2383207 and 10757278 from LD block 2. Of the INFORM cohort, 557 Caucasians and 126 African Americans had genotype data on at least one SNP. The genotype and allele frequencies of these 4 SNPs in INFORM are shown in Supplemental Table S3. The associations of the composite outcome with these 4 SNPs are shown in Table 3. None of the 4 SNPs reached statistical significance for replication, confirming absence of increased risk associated with the risk alleles as defined in the literature. Consistent with findings in the INVEST-GENES case-control and cohort, the G allele of rs2383207 was associated with a trend toward lower risk for the composite outcome of death or cardiac rehospitalization in INFORM Caucasians (adjusted HR: 0.81(0.64–1.02), p=0.07) and African Americans (adjusted HR: 0.64(0.37–1.12), p=0.12) (Table 3). The association of rs10757278 also showed the same trend in the Caucasian INFORM patients with the G allele of this SNP being associated with a trend toward lower risk of the composite outcome (adjusted HR: 0.75(0.58–0.95), p =0.019). For Block 1 SNPs rs7049105 and rs2157719, the effect sizes in Caucasians were comparable to those in INVEST-GENES and in the same direction, with adjusted HR of 1.17(0.92–1.49), p=0.22 and 1.16 (0.91–1.49), p=0.24 respectively (Table 3), although they did not achieve statistical significance.
Table 3.
Risk for INFORM primary outcome and rs2383207, rs10757278, rs7049105 and rs2157719
| INFORM Caucasians (n=557) | INFORM African Americans (n=126) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Event/N, % | Unadjusted HR | p value | Adjusted HR† | p value | Event/N, % | Unadjusted HR | p value | Adjusted HR† | p value | ||
| rs7049105 (risk allele: A) | AA | 41/108, 38.0% | 1.20(0.95–1.53) | 0.13 | 1.17(0.91–1.49) | 0.22 | 1/6, 16.7% | 0.95(0.50–1.81) | 0.88 | 0.75(0.36–1.54) | 0.43 |
| AG | 55/230, 23.9% | 12/41/29.3% | |||||||||
| GG | 35/127, 27.6% | 12/46, 26.1% | |||||||||
|
| |||||||||||
| rs2157719 (risk allele: G) | AA | 45/181, 24.9% | 1.22(0.96–1.56) | 0.41 | 1.16(0.91–1.49) | 0.24 | 18/73, 24.7% | 1.51(0.56–4.08) | 0.41 | 1.40(0.48–4.05) | 0.54 |
| AG | 63/230, 27.4% | 5/14, 35.7% | |||||||||
| GG | 26/69, 37.7% | - | |||||||||
|
| |||||||||||
| rs2383207 (allele: G) | AA | 32/95, 33.7% | 0.80(0.63–1.01) | 0.6 | 0.81(0.64–1.02) | 0.07 | 32/95, 33.7% | 0.61(0.37–1.0) | 0.05 | 0.64(0.37–1.12) | 0.12 |
| AG | 62/218, 28.4% | 7/12, 58.3% | |||||||||
| GG | 36/152, 23.7% | 16/76, 21.1% | |||||||||
|
| |||||||||||
| rs10757278 (allele: G) | AA | 35/106, 33.0% | 0.75(0.58–0.94) | 0.017 | 0.75(0.59–0.95) | 1.09 | 19/57, 33.3% | 0.49(0.23–1.03) | 0.06 | 0.49(0.22–1.09) | 0.08 |
| AG | 67/226, 30.0% | 8/36, 22.2% | |||||||||
| GG | 26/132, 19.7% | 0/6, 0% | |||||||||
adjusted for age, gender, heart failure, hypertension, diabetes, hyperlipidemia, ACS type, ACS treatment strategy (medical management, percutaneous intervention, coronary artery bypass grafting)
DISCUSSION
The chromosome 9p21 region is the most replicated risk locus for CAD. However, whether this chromosomal region is also associated with prognosis in those with existing CAD is not well understood. Using two similar but independent cohorts, our data suggest that the 9p21 alleles that were previously associated with higher risk for development of CAD/MI are not associated with higher risk for long term adverse outcomes among patients with established CAD. Instead, in both of our populations, the previously identified CAD/MI risk alleles trended toward protection from adverse outcomes among patients with established CAD. A less commonly reported LD block (block 1) was more strongly associated with prognosis in INVEST among both Caucasians and African Americans than the more commonly reported LD block (block 2).
Specifically, the SNPs located in block 2 including the 6 most often reported 9p21 risk alleles: rs10757274 (G), rs2891168 (G), rs2383206 (G), rs2383207 (G), rs10757278 (G) and rs1333049 (C) were associated with trends toward improved prognosis in Caucasians with documented CAD in INVEST GENES Caucasians and African Americans. Similarly, in INFORM Caucasians and African Americans (with 2 of the 6 SNPs being genotyped in INFORM), the direction of association in INFORM was consistent with those in INVEST GENES and p values trended toward significance. Thus both study populations strongly suggest that the traditional risk alleles for CAD are not associated with additional prognostic risk in those with established CAD. In fact, the data suggest these alleles may be protective, as it relates to long term adverse cardiovascular outcomes.
Previous reports may provide some insight into why we did not observe an association between chromosome 9p21 and higher risk for adverse outcomes among patients with established CAD and if anything, saw results in the opposite direction from the widely replicated associations with CAD/MI risk. First is the study by Horne et al in which they too saw no association with MI compared to individuals with CAD/no MI. They confirmed these findings in 7 European case-control sets again showing no association with MI when compared to those with CAD/no MI. In fact, the ORs in this study were all less than 1 (0.9, with 95% CI of 0.81–0.99), consistent with our finding.16 Second is a recent study by Dandona et al in which investigators found that rs1333049 [C allele] of the chromosome 9p21 region was associated with greater atherosclerotic burden rather than with the likelihood for plaque rupture, with no association with MI when disease severity was accounted for.15 Unfortunately we do not have angiographic data in all patients to evaluate the association between disease severity and chromosome 9p21. Third is a study by Muehlschlegel and colleagues reporting that the major allele of 9p21 SNP rs10116277 (in LD with rs1333049, r2 of 0.9 based on Hapmap Caucasians) was associated with lower risk for all-cause mortality within 5 years after coronary artery bypass graft surgery.25 Lastly, a recent study by Reilly and colleagues demonstrated that the SNPs predisposing to MI in patients with coronary atherosclerosis are distinct from those that associated with the development of coronary atherosclerosis, with a 9p21 SNP being an example.26
We found a haplotype in a less commonly reported LD block (block 1) to be associated with better prognosis (lower risk for cardiovascular outcome) in INVEST among Caucasians (OR=0.60, 0.47–0.77, p=4.0*10−5), but not in African Americans (OR=0.81, 0.46–1.43, p=0.475) or Hispanics (0.80, 0.55–1.17, p=0.25). We also found the complementary haplotype in this block that was associated with worse prognosis (higher risk for cardiovascular outcome) among both Caucasians (OR=1.52, 1.20–1.93, p =0.0006) and African Americans (OR=4.11, 1.55–10.88, p=0.004), but not Hispanics (OR=0.98, 0.59–1.62, p =0.93). Unfortunately, none of the previously mentioned studies evaluated any SNPs in LD block 1, which would be of interest to see if the associations we observed hold true.15, 16, 25
Given that our associations with prognosis were in the opposite direction of the association with disease risk and that many of the alleles in this region have frequencies very close to 50% in Caucasians, it is obviously important to ensure proper coding of the alleles. This fact becomes especially critical given that the actual causal allele has not yet been agreed upon and a given allele has even been reported to both increase and decrease regulatory activity in the region.27–30 Taken together, we are left with a dangerous scenario whereby “risk” alleles may be mis-reported because of slight differences in frequency and LD patterns. To further complicate the matter, many of the SNPs involve complementary alleles (e.g. G/C and A/T SNPs such as rs1333049 and rs7044859) making allele calling even more problematic. Given that INVEST-GENES is ethnically diverse and that the minor allele frequencies in African Americans and Hispanics do not approach 50% as they do in Caucasians (e.g. 13% and 32% in blacks and Hispanics, respectively, versus 45% in whites for rs2393207, supplemental Table S3), it was far easier for us to identify errors in genotype calling generated from “strand issues” than it would be for groups with large European populations. Difficulties in precisely and reproducibly identifying genotypes, particularly when there is a similar prevalence of high- and low-risk alleles in a population, is an important issue that needs to be brought to the attention of investigators and clinicians, particularly because direct-to-consumer genotyping companies are already giving patients an estimate of their heart disease risk based on the chromosome 9p21 SNP genotyping results.
Our study is the first to report the analysis of the 9p21 locus with adverse outcomes in African Americans and Hispanics patients with hypertension and CAD, although underpowered and hypothesis generating in nature in these groups. Given that the LD is much lower in African Americans than in Caucasians, evaluating the same SNP associations across different race/ethnicity groups may allow us to identify functionally important SNPs. Indeed, we found a haplotype that was associated with risk for adverse outcome in both Caucasians and African Americans. Interestingly, despite having intermediate LD between Caucasians and African Americans, we observed no SNPs associated with adverse outcomes in Hispanic individuals (mostly from Puerto Rico). It is possible that environmental factors may play a larger role in CVD outcomes in these Hispanic individuals or that there are relative differences in the penetrance of genetic factors.
LIMITATIONS
Our study has some limitations that worth noting: 1). The sample size of the replication cohort INFORM patients was relatively small, compared to INVEST-GENEs Caucasians, therefore we cannot conclude whether lack of replication of block 1 SNPs in INFORM Caucasians was due to suboptimal power or true negative associations. The trends toward significance and consistency of point estimates suggest power may have been an issue. 2) Additionally, the INVEST and INFORM populations had some important phenotypic differences between them (e.g. stable CAD versus ACS patients) that may have contributed to the lack of replication of the block 1 associations. 3). Our analyses in African Americans in both INVEST and INFORM are underpowered and are therefore hypothesis-generating in nature.
CONCLUSION
The well documented chromosome 9p21 SNPs associated with higher risk for incident CAD/MI did not appear to be associated with increased risk for adverse outcomes among patients with established CAD. In fact, our findings suggest trends toward protection from those SNPs widely published for association with incident CAD/MI risk. Several other studies have also noted no association between 9p21 and prognosis14, 15 or association in the opposite direction.16, 25 Whether the 9p21 haplotype in a different LD block (LD block 1) than the most often reported block is associated with prognosis should be further investigated in populations with established CAD. Caution is warranted when interpreting direct-to-consumer genetics tests of this locus as the risk allele may differ depending on the patient population. As more becomes understood regarding the mechanisms by which these SNPs modulate CAD pathogenesis, we will be better able to assess how their effects may differ in disease initiation versus progression.
To date, the chromosome 9p21region is the best replicated independent genetic risk factor for development of coronary artery disease (CAD), usually as incident myocardial infarction (MI), among Caucasians. However, whether this genetic marker is a predictor of adverse outcomes among patients with established CAD is unclear. Given that our ability to predict which individuals will develop adverse outcomes is limited and direct-to-consumer genetics companies are offering genotyping of this locus to patients, an understanding of how well previously noted associations apply to different patient populations is important. Accordingly, we investigated whether alleles of 9p21 SNPs, previously associated with increased risk for incident CAD, were also associated with increased risk for adverse cardiovascular outcomes among patients with previously documented CAD. We utilized data from 2 cohorts: one enrolled into the study with chronic stable CAD (INternational VErapamil SR Trandolapril STudy, INVEST) and the other enrolled at the time of an acute coronary syndrome (INvestigation oF Outcomes from acute coronary syndRoMes study, INFORM). Both cohorts were followed for at least 2 years for cardiovascular outcomes. We found the 9p21 SNPs previously associated with higher risk for incident CAD/MI were not associated with long-term outcomes among patients with established CAD. Several other studies have also noted no association between 9p21 and prognosis and some suggest a protective effect associated with these SNPs among those with established CAD. Caution is therefore warranted when interpreting genetic tests of 9p21 locus as the risk associated with an allele may differ depending on the patient population.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Robert Roberts for helpful early communications related to the analyses performed here, Lynda Stauffer for processing and genotyping samples, Robert Kolb for coordinating INVEST sites for INVEST-GENES participation, and INVEST patients and investigators who agreed to participate in the genetic substudy.
Funding Sources: This project was funded by NIH grants HL074730, HL69758, HL077113, GM074492 and RR017568, a grant from Abbott Pharmaceuticals and Florida Opportunity Fund (JAJ). ALB is supported by K23HL091120; RCD is supported by K23 HL086558.
Footnotes
DISCLOSURES Drs. Johnson, Pepine, Cooper-DeHoff, and Langaee received grant funding from Abbott Laboratories. Drs. Cooper-DeHoff and Pepine along with the University of Florida hold U.S. Patent No. 5,991,731 related to INVEST. Dr. Pepine has been a consultant for Abbott Laboratories.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Helgadottir A, Thorleifsson G, Manolescu A, Gretarsdottir S, Blondal T, Jonasdottir A, Jonasdottir A, Sigurdsson A, Baker A, Palsson A, Masson G, Gudbjartsson DF, Magnusson KP, Andersen K, Levey AI, Backman VM, Matthiasdottir S, Jonsdottir T, Palsson S, Einarsdottir H, Gunnarsdottir S, Gylfason A, Vaccarino V, Hooper WC, Reilly MP, Granger CB, Austin H, Rader DJ, Shah SH, Quyyumi AA, Gulcher JR, Thorgeirsson G, Thorsteinsdottir U, Kong A, Stefansson K. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 2.McPherson R, Pertsemlidis A, Kavaslar N, Stewart A, Roberts R, Cox DR, Hinds DA, Pennacchio LA, Tybjaerg-Hansen A, Folsom AR, Boerwinkle E, Hobbs HH, Cohen JC. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Consortium WTCC. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, Dixon RJ, Meitinger T, Braund P, Wichmann HE, Barrett JH, Konig IR, Stevens SE, Szymczak S, Tregouet DA, Iles MM, Pahlke F, Pollard H, Lieb W, Cambien F, Fischer M, Ouwehand W, Blankenberg S, Balmforth AJ, Baessler A, Ball SG, Strom TM, Braenne I, Gieger C, Deloukas P, Tobin MD, Ziegler A, Thompson JR, Schunkert H. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Samani NJ, Raitakari OT, Sipila K, Tobin MD, Schunkert H, Juonala M, Braund PS, Erdmann J, Viikari J, Moilanen L, Taittonen L, Jula A, Jokinen E, Laitinen T, Hutri-Kahonen N, Nieminen MS, Kesaniemi YA, Hall AS, Hulkkonen J, Kahonen M, Lehtimaki T. Coronary artery disease-associated locus on chromosome 9p21 and early markers of atherosclerosis. Arterioscler Thromb Vasc Biol. 2008;28:1679–1683. doi: 10.1161/ATVBAHA.108.170332. [DOI] [PubMed] [Google Scholar]
- 6.Schunkert H, Gotz A, Braund P, McGinnis R, Tregouet DA, Mangino M, Linsel-Nitschke P, Cambien F, Hengstenberg C, Stark K, Blankenberg S, Tiret L, Ducimetiere P, Keniry A, Ghori MJ, Schreiber S, El Mokhtari NE, Hall AS, Dixon RJ, Goodall AH, Liptau H, Pollard H, Schwarz DF, Hothorn LA, Wichmann HE, Konig IR, Fischer M, Meisinger C, Ouwehand W, Deloukas P, Thompson JR, Erdmann J, Ziegler A, Samani NJ. Repeated replication and a prospective meta-analysis of the association between chromosome 9p21.3 and coronary artery disease. Circulation. 2008;117:1675–1684. doi: 10.1161/CIRCULATIONAHA.107.730614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shen GQ, Rao S, Martinelli N, Li L, Olivieri O, Corrocher R, Abdullah KG, Hazen SL, Smith J, Barnard J, Plow EF, Girelli D, Wang QK. Association between four snps on chromosome 9p21 and myocardial infarction is replicated in an italian population. J Hum Genet. 2008;53:144–150. doi: 10.1007/s10038-007-0230-6. [DOI] [PubMed] [Google Scholar]
- 8.Kathiresan S, Voight BF, Purcell S, Musunuru K, Ardissino D, Mannucci PM, Anand S, Engert JC, Samani NJ, Schunkert H, Erdmann J, Reilly MP, Rader DJ, Morgan T, Spertus JA, Stoll M, Girelli D, McKeown PP, Patterson CC, Siscovick DS, O'Donnell CJ, Elosua R, Peltonen L, Salomaa V, Schwartz SM, Melander O, Altshuler D, Ardissino D, Merlini PA, Berzuini C, Bernardinelli L, Peyvandi F, Tubaro M, Celli P, Ferrario M, Fetiveau R, Marziliano N, Casari G, Galli M, Ribichini F, Rossi M, Bernardi F, Zonzin P, Piazza A, Mannucci PM, Schwartz SM, Siscovick DS, Yee J, Friedlander Y, Elosua R, Marrugat J, Lucas G, Subirana I, Sala J, Ramos R, Kathiresan S, Meigs JB, Williams G, Nathan DM, MacRae CA, O'Donnell CJ, Salomaa V, Havulinna AS, Peltonen L, Melander O, Berglund G, Voight BF, Kathiresan S, Hirschhorn JN, Asselta R, Duga S, Spreafico M, Musunuru K, Daly MJ, Purcell S, Voight BF, Purcell S, Nemesh J, Korn JM, McCarroll SA, Schwartz SM, Yee J, Kathiresan S, Lucas G, Subirana I, Elosua R, Surti A, Guiducci C, Gianniny L, Mirel D, Parkin M, Burtt N, Gabriel SB, Samani NJ, Thompson JR, Braund PS, Wright BJ, Balmforth AJ, Ball SG, Hall AS, Schunkert H, Erdmann J, Linsel-Nitschke P, Lieb W, Ziegler A, Konig I, Hengstenberg C, Fischer M, Stark K, Grosshennig A, Preuss M, Wichmann HE, Schreiber S, Schunkert H, Samani NJ, Erdmann J, Ouwehand W, Hengstenberg C, Deloukas P, Scholz M, Cambien F, Reilly MP, Li M, Chen Z, Wilensky R, Matthai W, Qasim A, Hakonarson HH, Devaney J, Burnett MS, Pichard AD, Kent KM, Satler L, Lindsay JM, Waksman R, Epstein SE, Rader DJ, Scheffold T, Berger K, Stoll M, Huge A, Girelli D, Martinelli N, Olivieri O, Corrocher R, Morgan T, Spertus JA, McKeown P, Patterson CC, Schunkert H, Erdmann E, Linsel-Nitschke P, Lieb W, Ziegler A, Konig IR, Hengstenberg C, Fischer M, Stark K, Grosshennig A, Preuss M, Wichmann HE, Schreiber S, Holm H, Thorleifsson G, Thorsteinsdottir U, Stefansson K, Engert JC, Do R, Xie C, Anand S, Kathiresan S, Ardissino D, Mannucci PM, Siscovick D, O'Donnell CJ, Samani NJ, Melander O, Elosua R, Peltonen L, Salomaa V, Schwartz SM, Altshuler D. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat Genet. 2009;41:334–341. doi: 10.1038/ng.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wahlstrand B, Orho-Melander M, Delling L, Kjeldsen S, Narkiewicz K, Almgren P, Hedner T, Melander O. The myocardial infarction associated cdkn2a/cdkn2b locus on chromosome 9p21 is associated with stroke independently of coronary events in patients with hypertension. J Hypertens. 2009;27:769–773. doi: 10.1097/HJH.0b013e328326f7eb. [DOI] [PubMed] [Google Scholar]
- 10.Hinohara K, Nakajima T, Takahashi M, Hohda S, Sasaoka T, Nakahara K, Chida K, Sawabe M, Arimura T, Sato A, Lee BS, Ban JM, Yasunami M, Park JE, Izumi T, Kimura A. Replication of the association between a chromosome 9p21 polymorphism and coronary artery disease in japanese and korean populations. J Hum Genet. 2008;53:357–359. doi: 10.1007/s10038-008-0248-4. [DOI] [PubMed] [Google Scholar]
- 11.Zhou L, Zhang X, He M, Cheng L, Chen Y, Hu FB, Wu T. Associations between single nucleotide polymorphisms on chromosome 9p21 and risk of coronary heart disease in chinese han population. Arterioscler Thromb Vasc Biol. 2008;28:2085–2089. doi: 10.1161/ATVBAHA.108.176065. [DOI] [PubMed] [Google Scholar]
- 12.Palomaki GE, Melillo S, Bradley LA. Association between 9p21 genomic markers and heart disease: A meta-analysis. Jama. 2010;303:648–656. doi: 10.1001/jama.2010.118. [DOI] [PubMed] [Google Scholar]
- 13.Buysschaert I, Carruthers KF, Dunbar DR, Peuteman G, Rietzschel E, Belmans A, Hedley A, De Meyer T, Budaj A, Van de Werf F, Lambrechts D, Fox KA. A variant at chromosome 9p21 is associated with recurrent myocardial infarction and cardiac death after acute coronary syndrome: The grace genetics study. Eur Heart J. 2010;31:1132–1141. doi: 10.1093/eurheartj/ehq053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ellis KL, Pilbrow AP, Frampton CM, Doughty RN, Whalley GA, Ellis CJ, Palmer BR, Skelton L, Yandle TG, Palmer SC, Troughton RW, Richards AM, Cameron VA. A common variant at chromosome 9p21.3 is associated with age of onset of coronary disease but not subsequent mortality. Circ Cardiovasc Genet. 2010;3:286–293. doi: 10.1161/CIRCGENETICS.109.917443. [DOI] [PubMed] [Google Scholar]
- 15.Dandona S, Stewart AF, Chen L, Williams K, So D, O'Brien E, Glover C, Lemay M, Assogba O, Vo L, Wang YQ, Labinaz M, Wells GA, McPherson R, Roberts R. Gene dosage of the common variant 9p21 predicts severity of coronary artery disease. J Am Coll Cardiol. 2010;56:479–486. doi: 10.1016/j.jacc.2009.10.092. [DOI] [PubMed] [Google Scholar]
- 16.Horne BD, Carlquist JF, Muhlestein JB, Bair TL, Anderson JL. Association of variation in the chromosome 9p21 locus with myocardial infarction versus chronic coronary artery disease. Circ Cardiovasc Genet. 2008;1:85–92. doi: 10.1161/CIRCGENETICS.108.793158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pepine CJ, Handberg EM, Cooper-DeHoff RM, Marks RG, Kowey P, Messerli FH, Mancia G, Cangiano JL, Garcia-Barreto D, Keltai M, Erdine S, Bristol HA, Kolb HR, Bakris GL, Cohen JD, Parmley WW. A calcium antagonist vs a non-calcium antagonist hypertension treatment strategy for patients with coronary artery disease. The international verapamil-trandolapril study (invest): A randomized controlled trial. JAMA. 2003;290:2805–2816. doi: 10.1001/jama.290.21.2805. [DOI] [PubMed] [Google Scholar]
- 18.Pepine CJ, Handberg-Thurmond E, Marks RG, Conlon M, Cooper-DeHoff R, Volkers P, Zellig P. Rationale and design of the international verapamil sr/trandolapril study (invest): An internet-based randomized trial in coronary artery disease patients with hypertension. J Am Coll Cardiol. 1998;32:1228–1237. doi: 10.1016/s0735-1097(98)00423-9. [DOI] [PubMed] [Google Scholar]
- 19.Lanfear DE, Jones PG, Marsh S, Cresci S, McLeod HL, Spertus JA. Beta2-adrenergic receptor genotype and survival among patients receiving beta-blocker therapy after an acute coronary syndrome. Jama. 2005;294:1526–1533. doi: 10.1001/jama.294.12.1526. [DOI] [PubMed] [Google Scholar]
- 20.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 21.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: Analysis and visualization of ld and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 22.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. Plink: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.SCHOENFELD D. Partial residuals for the proportional hazards regression model. Biometrika. 1982;69:239–241. [Google Scholar]
- 25.Muehlschlegel JD, Liu KY, Perry TE, Fox AA, Collard CD, Shernan SK, Body SC. Chromosome 9p21 variant predicts mortality after coronary artery bypass graft surgery. Circulation. 2010;122:S60–65. doi: 10.1161/CIRCULATIONAHA.109.924233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reilly MP, Li M, He J, Ferguson JF, Stylianou IM, Mehta NN, Burnett MS, Devaney JM, Knouff CW, Thompson JR, Horne BD, Stewart AF, Assimes TL, Wild PS, Allayee H, Nitschke PL, Patel RS, Martinelli N, Girelli D, Quyyumi AA, Anderson JL, Erdmann J, Hall AS, Schunkert H, Quertermous T, Blankenberg S, Hazen SL, Roberts R, Kathiresan S, Samani NJ, Epstein SE, Rader DJ. Identification of adamts7 as a novel locus for coronary atherosclerosis and association of abo with myocardial infarction in the presence of coronary atherosclerosis: Two genome-wide association studies. Lancet. 2011;377:383–392. doi: 10.1016/S0140-6736(10)61996-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Visel A, Zhu Y, May D, Afzal V, Gong E, Attanasio C, Blow MJ, Cohen JC, Rubin EM, Pennacchio LA. Targeted deletion of the 9p21 non-coding coronary artery disease risk interval in mice. Nature. 2010;464:409–412. doi: 10.1038/nature08801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Y, Sanoff HK, Cho H, Burd CE, Torrice C, Mohlke KL, Ibrahim JG, Thomas NE, Sharpless NE. Ink4/arf transcript expression is associated with chromosome 9p21 variants linked to atherosclerosis. PLoS One. 2009;4:e5027. doi: 10.1371/journal.pone.0005027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jarinova O, Stewart AF, Roberts R, Wells G, Lau P, Naing T, Buerki C, McLean BW, Cook RC, Parker JS, McPherson R. Functional analysis of the chromosome 9p21.3 coronary artery disease risk locus. Arterioscler Thromb Vasc Biol. 2009;29:1671–1677. doi: 10.1161/ATVBAHA.109.189522. [DOI] [PubMed] [Google Scholar]
- 30.Cunnington MS, Santibanez Koref M, Mayosi BM, Burn J, Keavney B. Chromosome 9p21 snps associated with multiple disease phenotypes correlate with anril expression. PLoS Genet. 2010;6:e1000899. doi: 10.1371/journal.pgen.1000899. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.