The authors characterize a new transgenic mouse line, BEST1-cre, that provides RPE-specific ocular cre expression. These mice begin expressing cre at postnatal day 10 and maintain expression into adulthood without causing retinal dysfunction. Cre expression is present in up to 90% of RPE nuclei. Therefore, these mice provide a useful tool for studying the postnatal function of genes within the RPE.
Abstract
Purpose.
To generate and characterize a constitutively active, RPE-specific, cre-expressing transgenic mouse line. This line can be used to create RPE-specific knockouts by crossing with mice harboring loxP-flanked (floxed) genes.
Methods.
A transgene construct was assembled with the BEST1 promoter driving cre expression. Transgenic mice were generated on a C57BL/6 background. Cre expression was assessed by immunofluorescence and Western blot analysis. Cre enzymatic activity was tested by crossing to three lines with floxed DNA regions and detecting deletion of the intervening sequences or through histochemical detection of lacZ activity. Potential cre-mediated toxicity was assessed by retinal histology up to 24 months of age and by electroretinography.
Results.
The BEST1-cre line with expression in the highest percentage of RPE cells displayed a patchy mosaic expression pattern, with 50% to 90% of RPE cells expressing cre. In mice outcrossed to a mixed B6/129 background, expression was consistently found in 90% of RPE cells. Within the eye, only the RPE cells were immunoreactive with an anti-cre antibody. Maximum cre expression quantified by Western blot analysis occurred at P28. Crosses with three lines containing floxed sequences revealed RPE-specific cre activity in the eye and extraocular expression limited to the testes. Histology and electroretinography showed no cre-mediated RPE toxicity.
Conclusions.
This BEST1-cre transgenic line enables generation of RPE-specific knockout mice. The mosaic expression pattern provides an internal control; the non–cre-expressing RPE cells continue to express the floxed genes. These mice should facilitate study of the multifunctional RPE and the generation of mouse models of human retinal disease.
The cre-loxP system has revolutionized mouse genetics by facilitating the creation of cell-type–specific knockout animal models. Cre recombinase is a P1 bacteriophage protein that binds to a 34-bp-long target recognition site known as loxP.1 Through intramolecular recombination, cre has the ability to excise loxP-flanked (floxed) sequences from the genome, leaving a single loxP site in its place. Tissue-specific promoter-driven cre expression makes possible the study of cell-autonomous gene function and permits the examination of genes whose systemic absence is lethal. Such a system is of particular interest in the study of diseases of the eye due to the complex interplay of the various cell types that make up the retina.
The retinal pigment epithelium (RPE) is a monolayer lying between the neural retina and Bruch's membrane. It performs many specialized functions that support and nourish the photoreceptors, such as maintaining the blood–retina barrier by regulating the movement of nutrients, ions, and water between the photoreceptors and the choroid; the phagocytosis of shed photoreceptor outer segments; and the facilitation of retinoid metabolism (visual cycle).2 In addition, human RPE dysfunction has been implicated in the pathogenesis of both wet and dry age-related macular degeneration, the most common cause of irreversible blindness in the elderly populations of developed countries.3–5
Four mouse lines that express cre in the RPE have been reported.6–9 Two of the lines that express it in the RPE—a dopachrome tautomerase (Dct)-cre line7 and a tyrosinase related protein line (TRP)-1-cre6—do so in a noninducible manner and exhibit cre expression and activity during embryonic development. These lines were not assessed for effects of cre expression on retinal morphology or function in adult animals. A third line utilizes an inducible monocarboxylate transporter 3 promoter to drive RPE-specific cre expression.8 When crossed with a Rosa-lacZ line, cre expression occurs in approximately 20% of RPE cells. When crossed with a cre-activated diphtheria toxin line, the number of missing RPE cells suggests a higher percentage of cre-expressing RPE cells, but this has not been directly demonstrated. Retinal structure was shown to be normal 6 months after induction.
Recently, a mouse line that takes advantage of the tetracycline-inducible system to control cre expression has been developed for the knockout of genes from the RPE.9 The reverse Tet-inducible transactivator is under control of a 2.9-kb fragment of the bestrophin 1 (BEST1) promoter, whereas cre is located downstream of the (tetracycline-responsive element [TRE]), which, in theory, should limit cre expression to the RPE and only when the animal has been given doxycycline. Maximal cre activity was achieved after induction at P4, but substantial activity was detected on induction as late as P25. No cre-mediated retinal toxicity was observed up to 10 months of age. This inducible BEST1-cre mouse line is likely to be a useful tool; however, the complication of daily doses of doxycycline, which is done by gavage in animals before weaning, may limit the utility of this line for some applications.
We therefore sought to generate a transgenic mouse line with constitutive RPE-restricted expression of cre beginning after ocular development for use in studying RPE function in the developed eye and age-related retinal disease. We chose to use a fragment of the human BEST1 promoter which has been shown to promote robust ocular expression that is restricted to the RPE in the eye of transgenic mice.10 Herein, we provide analysis of a new BEST1-cre transgenic line in which we study cre expression timing, localization, enzymatic activity, and effect on retinal integrity during the full mouse lifespan.
Materials and Methods
Generation of BEST1-cre Conditional Mouse Lines
The EcoRI/XcmI fragment of the human BEST1 promoter (nucleotides −585 to +38) was isolated and cloned into the SacI/XbaI sites of a vector (pBluescriptKSII; Stratagene, La Jolla, CA).10 Cre recombinase cDNA, SV40 t-antigen intron and HSV-TK polyA were PCR extracted from the pACN vector11 and inserted into the plasmid immediately downstream of the promoter in restriction sites HindIII/XhoI. The BEST1-cre construct was excised from the vector sequence and microinjected into zygotes derived from superovulated C57BL/6 females at the transgenic mouse core facility at The University of Pennsylvania School of Medicine. The mice were screened using PCR analysis of tail tissue DNA with primers LF17 (5′-ATG CCC AAG AAG AAG AGG AAG GTG TCC-3′) and LF21 (5′-TGG CCC AAA TGT TGC TGG ATA GTT TTT A-3′). Founders were crossed to C57BL/6 mice to extend this Tg(BEST1-cre)Jdun line (referred to further as BEST1-cre). Genotype was confirmed by using PCR as described above or by quantitative PCR to distinguish transgene copy number (Embark Scientific, Austin, TX). All procedures were approved by the Institutional Animal Care and Use Committees of the University of Pennsylvania and Stanford University and complied with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research.
PCR Analysis of Cre-Mediated Excision of DNA
To determine in which tissues cre is expressed, the BEST1-cre line was crossed with mice carrying a floxed allele for Tfam12 and, separately, another line carrying a floxed allele for Hephaestin (Heph) C57BL/6-Hephtm1Jdun (generation of which is unpublished). DNA was extracted from RPE, neural retina and other organs (QIAamp DNA Micro Kit; Qiagen, Valencia, CA). Briefly, mice were euthanatized, and the eyes were enucleated and placed in PBS on ice. The anterior segment was removed, and the neural retina was peeled off and placed directly into buffer (Buffer ATL; Qiagen). Then, after neural retina removal, the buffer was pipetted up and down directly in the eye cup to collect RPE cells. PCR for Heph recombination was performed with the following primer sequences: forward primer (5′-GAC AAG AGC TCT AGG AGA GAT GCC A-3′), and reverse primer (5′-CCA AGC ATT CAG TAG ACC TAG GAA GGA-3′). Primers for Tfam genotyping have been previously described.12 DNA was amplified using polymerase and Taq PCR master mix (DreamTaq; Fermentas Life Sciences, Glen Burnie, MD) as recommended by the manufacturer.
Reverse Transcription-PCR and Western Blot Analysis
RNA extraction and reverse transcription-PCR (RT-PCR) were described previously.13 Cell lysates were prepared as described previously.14 Total protein for each sample was quantified with a BSA kit (Roche Applied Science, Indianapolis, IN). Equal amounts of protein from each sample were separated by 12% SDS-PAGE gel. Protein transfer and chemiluminescence detection were performed as described previously.15
Immunofluorescence
Eyes were enucleated immediately after death and fixed for 2 hours in 4% paraformaldehyde. The globes were then rinsed in PBS and prepared as eye cups, cryoprotected in 30% sucrose, and embedded in optimal cutting temperature compound (OCT, Tissue-Tek; Sakura Finetek, Torrance, CA). Immunofluorescence was performed on 10-μm-thick cryosections as described elsewhere.16 The primary antibody was mouse anti-cre recombinase (1:500 dilution; clone 2D8; Millipore, Billerica, MA). The secondary antibody was donkey anti-mouse labeled with Cy3 (Jackson ImmunoResearch, West Grove, PA). FITC-phalloidin (Invitrogen, Carlsbad, CA) labeling was performed according to the manufacturer's instructions.
β-Galactosidase Staining
Albino mice for β-galactosidase staining were generated by mating B6.129S4-Gt(ROSA)26Sortm1Sor/J mice (referred to as Rosa-lacZ; The Jackson Laboratory, Bar Harbor, ME) with B6(Cg)-Tyrc-2J/J (The Jackson Laboratory). The F1 albino Rosa-lacZ offspring were then mated to albino BEST1-cre+/−; Ferroportin flox/+. The offspring were then intercrossed and albino Rosa-lacZ;BEST1-cre+/− mice were used in the β-galactosidase analysis. To perform β-galactosidase staining, eyes from 5-month-old mice were enucleated immediately after death and fixed in 4% paraformaldehyde in PBS on ice for 1 hour. The anterior chambers and lenses were removed, the eye cups were rinsed in PBS and incubated overnight in X-gal (5-bromo-4-chloro-3-idolyl-β-d-galactopyranoside) and mixer solution (10 mM K3Fe(CN)6, 10 mM K4Fe(CN)6 3 H2O, 2 mM MgCl2, 0.01% sodium deoxycholate, and 0.02% NP-40 in PBS). The eyes were then removed from X-gal mixer solution, rinsed in PBS, and processed as just described. Ten-micron cryosections were imaged on an epifluorescence microscope (80i; Nikon, Tokyo, Japan).
Electroretinography and Retinal Morphology
Electroretinography (ERG) recordings followed procedures described previously.17,18 In brief, mice were dark-adapted overnight and then anesthetized with a cocktail containing (in milligrams/kilogram body weight): 25 ketamine, 10 xylazine, and 1000 urethane. In each mouse, the pupils were dilated with 1% tropicamide saline solution (Mydriacyl; Alconox, New York, NY), and the mouse was placed on a stage maintained at 37°C. Two miniature cups with embedded platinum wires made of UV-transparent plastic serving as recording electrodes were placed in electrical contact with the corneas. A platinum wire loop placed in the mouth served as the reference and a ground electrode. ERGs were recorded (Espion Electrophysiology System; Diagnosys LLC, Lowell, MA) on an apparatus modified by the manufacturer for experiments with mice by substituting LEDs with emission maximum at 365 nm for the standard blue ones. A stage with the mouse was positioned such that the mouse's head was located inside the stimulator (ColorDome; Diagnosys), thus ensuring full-field uniform illumination. Methods for light stimulation and calibration of light stimuli were described previously.17 Mice were evaluated at age 4.5 and 12 months.
Retinas from 24-month-old BEST1-cre+/− and wild-type mice were analyzed for outer nuclear layer (ONL) thickness and RPE nuclei number using retinal sections passing through the optic nerve head. The eyes were enucleated immediately after death and fixed overnight in 2% glutaraldehyde/2% paraformaldehyde in PBS. The eye cups were prepared by removal of the anterior segment and embedded in JB-4 according to the manufacturer's protocol (Polysciences, Inc., Warrington, PA). Semithin 3-μm section were cut and stained with toluidine blue O (Sigma-Aldrich, St. Louis, MO). Images of five sections per eye were acquired using a 20× objective on the epifluorescence microscope (80i; Nikon), The 20× images were merged (Photoshop CS 3; Adobe, San Jose, CA) and analyzed (NIS-elements, ver. 3.1; Nikon). Significant differences in photoreceptor nuclei number between wild-type and transgenic mice were analyzed by two-way ANOVA with correction for multiple measures (Prism, ver. 5.0; GraphPad, San Diego, CA).
Results
Generation of RPE-Specific Cre Transgenic Mice
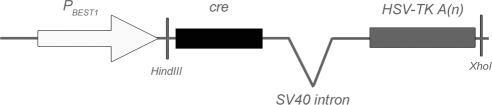
To generate an RPE-specific cre transgene, we used a minimal portion of the human BEST1 promoter that was previously shown to drive RPE-specific expression.10 The −585- to +38-bp fragment of the BEST1 promoter was cloned upstream of an SV-40 intron, cre, and an HSV-TK polyadenylation signal (Fig. 1). The purified BEST1-cre transgene DNA fragment was used to generate transgenic mice on a pure C57BL/6 background. Retinal sections from F2 mice were examined for cre immunoreactivity in the RPE (data not shown). One line of six germline transmitters containing the highest percentage of cre immunoreactivity was selected for further analysis.
Figure 1.
Schematic representation of the transgenic construct. PBEST1, promoter of human BEST1 gene; SV40, simian virus 40; HSV-TK A(n), herpes simplex virus thymidine kinase polyadenylation signal.
Timing and Localization of Cre Expression
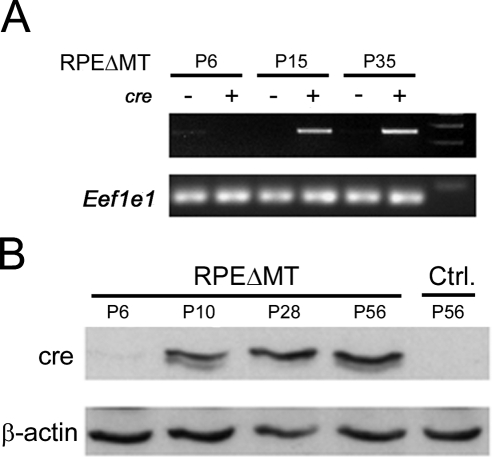
To determine the onset of cre expression within the RPE, RT-PCR, and Western blot analysis for cre were performed on eye cups from mice with genotype of BEST1-cre/+; Tfamloxp/loxp (termed RPEΔMT mice hereafter). RT-PCR detected cre mRNA at P15 but not at P6 (Fig. 2A). Western blot analysis revealed cre protein beginning at P10, increasing at P28, and remaining relatively constant at least until P56 (Fig. 2B).
Figure 2.
Timing of cre expression. RT-PCR (A) and Western blot analysis (B) detecting cre in RPEΔMT mouse eye cups beginning at P10. +, the mice carrying the BEST1-cre transgene; −, the absence of the transgene. Eef1e1 and β-actin serve as housekeeping controls.
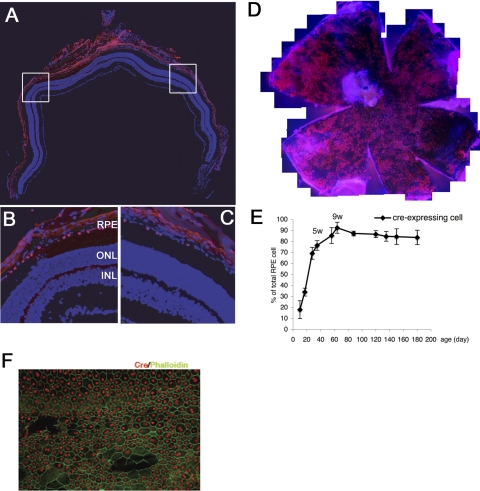
We analyzed localization of cre in the mouse retina by using immunofluorescence with a mouse monoclonal antibody. In the retinas of adult (8-week-old) mice, cre was detected only in RPE nuclei (Figs. 3A–C). There was no cre immunoreactivity in any other ocular cell type. The cre expression pattern was mosaic, with 50% to 90% of RPE cells immunolabeling with anti-cre (Fig. 3D). Depending on the genetic background, the degree of mosaicism was altered; in RPEΔMT mice that have a mixed B6/129 background, cre immunoreactivity was consistently observed in 90% of the RPE cells at 9 weeks (Figs. 3E, 3F).
Figure 3.
Immunofluorescence localization of cre to the RPE. (A–C). Fluorescence photomicrographs from 8 weeks BEST1-cre (C57BL/6) transgenic mouse retinas labeled with mouse anti-cre (red) and DAPI (blue). Images in (B) and (C) are enlargements of the boxed areas in (A) and show mosaic expression of cre. (D) An RPE/choroid/sclera flat mount from a 3-month-old BEST1-cre (C57BL/6) transgenic mouse labeled with mouse anti-cre (red) and DAPI (blue). The flat mount shows areas where cre was not detectable by immunofluorescence, demonstrating the patchy, mosaic expression of the transgene. (E) Quantification of cre-expressing RPE cells in RPEΔMT mice (B6/129) at various ages showing an increase in postnatal cre expression peaking at ∼9 weeks. Error bars, SD from three eyes at each time point. (F) Staining for cre (red) and phalloidin (green) in RPEΔMT retina flat mount at 6 weeks of age.
Cre Recombinase Activity is Limited to the RPE
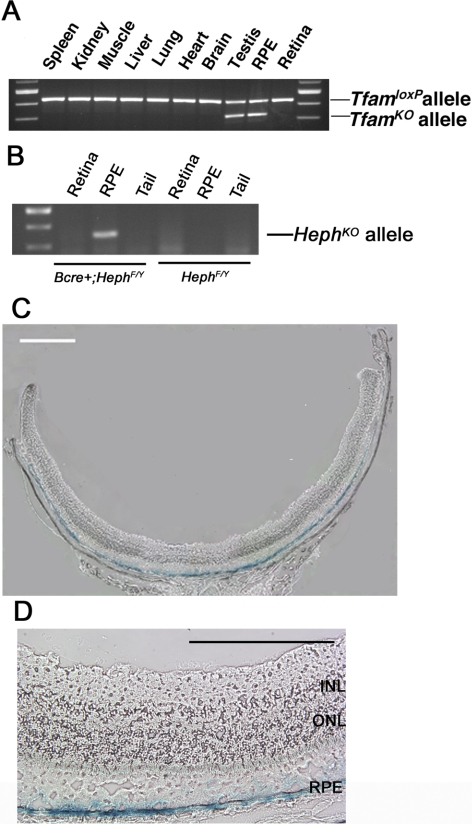
Cre recombinase activity was analyzed using PCR analysis of DNA from different tissues. In 12-week-old RPEΔMT mice, the TfamKO allele, an indicator of cre activity, was detected in DNA derived from the RPE and testis (Fig. 4A), but not in spleen, kidney, liver, brain, heart, muscle, lung, or neural retina. The recombination observed in the testis is likely due to BEST1 promoter activity within the Sertoli cells.19 Similar to the RPEΔMT mice, 12-week-old BEST1-cre; HephF/Y mice, contain the deleted allele in DNA derived from RPE but not from retina or tail (Fig. 4B). We confirmed the localization of cre activity by analyzing β-galactosidase activity in retinal cryosections from albino BEST1-cre; Rosa-lacZ mice. Within the eye, X-gal staining of these sections produced a blue reaction product only in the RPE (Figs. 4C, 4D).
Figure 4.
Cre recombinase activity in the RPE. (A) PCR genotyping of Tfam in various tissues from a RPEΔMT mouse (3 months). The TfamKO allele was detected only in DNA derived from RPE and testis. (B) PCR genotyping of Heph in RPE, retina, and tail from a 3-month-old BEST1-cre;Hephflox/flox mouse. The HephKO allele was detected in DNA derived from RPE only. (C, D) Localization of cre-activated β-galactosidase activity in the RPE of a 5-month-old albino BEST1-cre;Rosa-lacZ mouse. Cre activity was limited to the RPE. Scale bar: (C) 500 μm; (D) 200 μm. RPE, retinal pigment epithelium; INL, inner nuclear layer.
Expression of Cre in the RPE Is Nontoxic to the Retina
Cre is toxic to photoreceptors when expressed at high levels,20 but not at lower levels.21 However, inducible BEST1-cre within RPE has been shown to be nontoxic when expressed for 10 months.9 Since cre is expressed in a noninducible manner in the BEST1-cre mice reported herein, it can be expressed from P10 until the end of the mouse's life and could eventually cause retinal toxicity. Therefore, we assessed retinal function by ERG and retinal structure using morphometry.
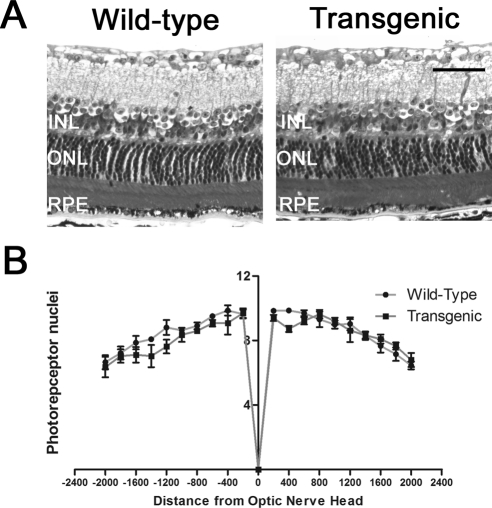
We used ERG to test retinal function in BEST1-cre transgenic mice at 4.5 months and at 12 months. At 4.5 months, there were no statistically significant differences in saturating scotopic (rod a-wave and rod b-wave) or photopic (cone b-wave) responses between wild-type mice and BEST1-cre transgenic mice (Table 1). Furthermore, at 12 months, no differences in responses to saturating scotopic or photopic ERGs were found (Table 1). Consistent with this result, at 2 years of age, the number of photoreceptor nuclei per row in optic nerve–containing sections was not significantly different at any of the points measured at 400-μm intervals in wild-type and BEST1-cre transgenic mice (Fig. 5). Taken together, the ERG and morphometry data demonstrate that retinal function and structure are not adversely affected by the BEST1-cre transgene.
Table 1.
Scotopic and Photopic Maximum ERG Amplitudes in 4.5- and 12-Month Wild-Type and BEST1-Cre Mice
| Scotopic ERG |
Photopic ERG b-Wave | ||
|---|---|---|---|
| a-Wave | b-Wave | ||
| 4.5 Months (n = 6 eyes) | |||
| Wild-type | −332.7 ± 11.0 | 354.4 ± 20.0 | 164.7 ± 9.0 |
| Transgenic | −333.9 ± 12.5 | 352.7 ± 14.0 | 177.1 ± 5.8 |
| P | 0.956 | 0.945 | 0.274 |
| 12 Months (n = 6 eyes) | |||
| Wild-type | −196.3 ± 15.4 | 209.1 ± 14.8 | 148.8 ± 5.0 |
| Transgenic | −198.9 ± 10.4 | 210.7 ± 47.3 | 162.1 ± 11.9 |
| P | 0.905 | 0.975 | 0.327 |
Data are expressed as mean microvolts ± SD.
Figure 5.
Retinal morphology in 2-year-old wild-type and transgenic mice. (A) Semithin plastic sections of retinas, 400 μm from the optic nerve. Scale bar, 50 μm. RPE, retinal pigment epithelium; INL, inner nuclear layer. (B) Quantification of photoreceptor nuclei number from 200 to 2000 μm from the optic nerve head (n = 3 eyes for each group). Negative values are inferior. The graph shows no difference in photoreceptor nuclei number between wild-type and transgenic mice.
Discussion
RPE-specific expression of cre is useful for generating gene knockouts limited to the RPE. This facilitates study of the RPE-cell autonomous function of genes, some of which cause embryonic lethality when knocked out ubiquitously. It also facilitates generation of mouse models of human retinopathies in which the RPE harbors the primary defect. Herein, we describe generation of an RPE-specific transgenic mouse line in which maximum cre expression begins after eye development. The expressed cre is enzymatically active and nontoxic to the RPE up to at least 2 years of age.
Previous RPE cre-expressing lines (Table 2) using Dct or Trp-1 promoters demonstrate cre activity in the embryo.6,7 While this is advantageous for developmental studies, this embryonic deletion may cause developmental abnormalities, and therefore analysis of phenotypes in adulthood is more complicated. The BEST1-cre transgenic mice presented herein have low-level cre expression beginning at P10 (Fig. 2). At this time all retinal layers are developed. At P10, no new photoreceptors are born, although photoreceptor outer segments continue to elongate until P17.22,23 The RPE cells are proliferating at P10, as 25% more are found at P17.24 Therefore, gene knockouts within the RPE at this stage could only lead to developmental abnormalities if the effect of gene deletion is very rapid. Further, since the Trp-1 lines express cre in several different cell types within the eye, our BEST1-cre transgenic mice are better suited for studies involving the cell autonomous functions of the RPE.
Table 2.
Comparison of Transgenic Mice Expressing Cre in the RPE
| Strain |
|||||
|---|---|---|---|---|---|
| Dct7 | Trp-16 | Tet-on VMD2-cre9 | Mct3-creER8 | BEST1-cre | |
| Onset of expression | E12.5 | E10.5 | 5 days after induction | 1 week after induction | P10 |
| Restricted to RPE in ocular tissue | Yes | No | Yes | Yes | Yes |
| Requires induction | No | No | Yes | Yes | No |
| Age to which function/structure is normal | Not reported | Not reported | ERG and structure, 10 mo after induction | Structure, 6 mo after induction | ERG, 12 mo; structure, 24 mo |
| % RPE Cre positive | Not reported | Not reported | Not reported; greatest activity occurs with induction at P4 | 20% when induced at P0; 5% when induced at 6 wk | 50%–90% depending on strain |
Immunofluorescence of cre within the RPE of adult BEST1-cre mice displays a patchy expression pattern. The percentage of cre-positive cells is influenced by genetic background. On the pure C57BL/6 background in which these mice were generated, the percentage of cre-positive cells varies from 50% to 90%. On a mixed B6/129 background, expression is more consistently 90% of RPE cells. This mosaic expression pattern provides an internal control for studies of RPE morphology; immunolabeling with the anti-cre antibody identifies cells that will delete the gene of interest, with cre-negative neighbors serving as controls. This mosaicism may represent a slight impediment for biochemical studies, but in genetic backgrounds with 90% cre-positive RPE cells, most RPE cells will have deleted the floxed gene. In contrast, the monocarboxylate transporter 3-cre line, cre is expressed in as few as 5% to 20% of RPE cells, depending on genetic background and timing of induction.
Silencing of transgenes has been observed in some transgenic mouse lines, especially when the transgene copy number is over 100.25 Our BEST1-cre line has a transgene copy number of 20, based on qPCR analysis (data not shown). Occasional BEST1-cre mice on the pure C57BL/6 background have complete silencing of cre expression in the RPE. Progeny of silenced mice have exhibited cre expression in 90% of RPE cells when crossed to 129 background mice. Among 23 BEST1-cre mice on a C57BL/6 background, q-RT-PCR for cre expression in RNA isolated from RPE/choroid revealed strong correlation in the cre expression level between the eyes of each mouse. The three silenced mice had silencing in both eyes. Thus, it is imperative in studies with our BEST1-cre line to assess the extent of cre expression in at least one eye from each mouse. This assessment is easily accomplished by immunofluorescence or qPCR.
A previous line using the BEST1 promoter has been generated that expresses cre in response to doxycycline induction. In addition to the complication of using oral gavage before weaning, this line may be limited by inability to detect cre-positive RPE nuclei by immunofluorescence. Lack of immunofluorescence makes it difficult to use a technique such as flat mounting to identify RPE cells that may have changes associated with gene deletion.
Functional studies described herein show that the cre in our line is not only immunoreactive but also enzymatically active, as it caused recombination between loxP sites in three floxed lines: Rosa26-lacZ, Tfam, and Heph. The Tfam26 and Heph lines have interesting retinal degeneration phenotypes.
This transgenic mouse with RPE-limited expression of cre beginning in the late stages of postnatal eye development and a mosaic cre expression pattern should prove useful for future studies on the multifaceted functions of the RPE.
Acknowledgments
The authors thank Jean Richa and the University of Pennsylvania transgenic core for microinjection of zygotes and Maithili Navaratnarajah for technical assistance.
Note Added in Proof
It is not recommended that floxed mice be maintained for multiple generations with the Best1-cre allele, as occasional mice bred in this manner have recently had germline deletion of the floxed allele.
Footnotes
Supported by Grants EY015240, EY014650, EY12910, and T-32 EY007035-30 (JI) from the National Institutes of Health Grants; the International Retina Research Foundation; the American Health Assistance Foundation; the Foundation Fighting Blindness; the Thome Memorial Foundation; the Rosanne Silbermann Foundation; and a Stanford Medical School Dean's Postdoctoral Fellowship.
Disclosure: J. Iacovelli, None; C. Zhao, None; N. Wolkow, None; P. Veldman, None; K. Gollomp, None; P. Ojha, None; N. Lukinova, None; A. King, None; L. Feiner, None; N. Esumi, None; D.J. Zack, None; E. Pierce, None; D. Vollrath, None; J.L. Dunaief, None
References
- 1. Davey RA, MacLean HE. Current and future approaches using genetically modified mice in endocrine research. Am J Physiol Endocrinol Metab. 2006;291:E429–E438 [DOI] [PubMed] [Google Scholar]
- 2. Saari JC. Biochemistry of visual pigment regeneration: the Friedenwald lecture. Invest Ophthalmol Vis Sci. 2000;41:337–348 [PubMed] [Google Scholar]
- 3. Sunness JS. The natural history of geographic atrophy, the advanced atrophic form of age-related macular degeneration. Mol Vis. 1999;5:25. [PubMed] [Google Scholar]
- 4. Zarbin MA. Current concepts in the pathogenesis of age-related macular degeneration. Arch Ophthalmol. 2004;122:598–614 [DOI] [PubMed] [Google Scholar]
- 5. Anderson DH, Radeke MJ, Gallo NB, et al. The pivotal role of the complement system in aging and age-related macular degeneration: hypothesis re-visited. Prog Retin Eye Res. 2010;29:95–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mori M, Metzger D, Garnier JM, Chambon P, Mark M. Site-specific somatic mutagenesis in the retinal pigment epithelium. Invest Ophthalmol Vis Sci. 2002;43:1384–1388 [PubMed] [Google Scholar]
- 7. Guyonneau L, Rossier A, Richard C, Hummler E, Beermann F. Expression of Cre recombinase in pigment cells. Pigment Cell Res. 2002;15:305–309 [DOI] [PubMed] [Google Scholar]
- 8. Longbottom R, Fruttiger M, Douglas R, Martinez-Barbera J, Greenwood J, Moss S. Genetic ablation of retinal pigment epithelial cells reveals the adaptive response of the epithelium and impact on photoreceptors. Proc Natl Acad Sci U S A. 2009;106:18728–18733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Le YZ, Zheng W, Rao PC, et al. Inducible expression of cre recombinase in the retinal pigmented epithelium. Invest Ophthalmol Vis Sci. 2008;49:1248–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Esumi N, Oshima Y, Li Y, Campochiaro PA, Zack DJ. Analysis of the VMD2 promoter and implication of E-box binding factors in its regulation. J Biol Chem. 2004;279:19064–19073 [DOI] [PubMed] [Google Scholar]
- 11. Bunting M, Bernstein KE, Greer JM, Capecchi MR, Thomas KR. Targeting genes for self-excision in the germ line. Genes Dev. 1999;13:1524–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Larsson NG, Wang J, Wilhelmsson H, et al. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat Genet. 1998;18:231–236 [DOI] [PubMed] [Google Scholar]
- 13. Zhao C, Bellur DL, Lu S, et al. Autosomal-dominant retinitis pigmentosa caused by a mutation in SNRNP200, a gene required for unwinding of U4/U6 snRNAs. Am J Hum Genet. 2009;85:617–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Strick DJ, Feng W, Vollrath D. Mertk drives myosin II redistribution during retinal pigment epithelial phagocytosis. Invest Ophthalmol Vis Sci. 2009;50:2427–2435 [DOI] [PubMed] [Google Scholar]
- 15. Liu Y, Vollrath D. Reversal of mutant myocilin non-secretion and cell killing: implications for glaucoma. Hum Mol Genet. 2004;13:1193–1204 [DOI] [PubMed] [Google Scholar]
- 16. Dunaief JL, Dentchev T, Ying GS, Milam AH. The role of apoptosis in age-related macular degeneration. Arch Ophthalmol. 2002;120:1435–1442 [DOI] [PubMed] [Google Scholar]
- 17. Lyubarsky AL, Falsini B, Pennesi ME, Valentini P, Pugh EN., Jr UV- and midwave-sensitive cone-driven retinal responses of the mouse: a possible phenotype for coexpression of cone photopigments. J Neurosci. 1999;19:442–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lyubarsky AL, Lem J, Chen J, Falsini B, Iannaccone A, Pugh EN., Jr Functionally rodless mice: transgenic models for the investigation of cone function in retinal disease and therapy. Vision Res. 2002;42:401–415 [DOI] [PubMed] [Google Scholar]
- 19. Masuda T, Esumi N. SOX9, through interaction with MITF and OTX2, regulates BEST1 expression in the retinal pigment epithelium. J Biol Chem. 2010;285:26933–26944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jimeno D, Feiner L, Lillo C, et al. Analysis of kinesin-2 function in photoreceptor cells using synchronous Cre-loxP knockout of Kif3a with RHO-Cre. Invest Ophthalmol Vis Sci. 2006;47:5039–5046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Le YZ, Zheng L, Zheng W, et al. Mouse opsin promoter-directed Cre recombinase expression in transgenic mice. Mol Vis. 2006;12:389–398 [PubMed] [Google Scholar]
- 22. Cepko CL, Austin CP, Yang X, Alexiades M, Ezzeddine D. Cell fate determination in the vertebrate retina. Proc Natl Acad Sci U S A. 1996;93:589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. LaVail MM. Kinetics of rod outer segment renewal in the developing mouse retina. J Cell Biol. 1973;58:650–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Strauss O. The retinal pigment epithelium in visual function. Physiol Rev. 2005;85:845–881 [DOI] [PubMed] [Google Scholar]
- 25. Garrick D, Fiering S, Martin DI, Whitelaw E. Repeat-induced gene silencing in mammals. Nat Genet. 1998;18:1:56–59 [DOI] [PubMed] [Google Scholar]
- 26. Zhao C, Yasumura D, Li X, et al. mTOR-mediated dedifferentiation of the retinal pigment epithelium initiates photoreceptor degeneration in mice. J Clin Invest. 2011;121:369–383 [DOI] [PMC free article] [PubMed] [Google Scholar]