The age-related decrease in relative proliferative capacity observed in human corneal endothelial cells (HCECs) results, at least in part, from nuclear oxidative DNA damage. The authors explored the ability of HCECs to protect themselves against oxidative stress and DNA damage by comparing the expression of oxidative stress and DNA damage-signaling genes and by immunostaining for nuclear DNA damage foci in cells from young and older donors.
Abstract
Purpose.
Nuclear oxidative DNA damage increases with age in human corneal endothelial cells (HCECs) and contributes to their decreased proliferative capacity. These studies investigated whether HCECs respond to this damage by upregulating their expression of oxidative stress and DNA damage-signaling genes in an age-dependent manner.
Methods.
HCECs were dissected from the corneas of young (30 years and younger) and older (50 years and older) donors. Total RNA was isolated and reverse-transcribed. Oxidative stress and DNA damage-signaling gene expression were analyzed using commercial PCR-based microarrays. Western blot analyses were conducted on selected proteins to verify the microarray results. Nuclear DNA damage foci were detected in the endothelium of ex vivo corneas by immunostaining for H2AX-Ser139.
Results.
Four of 84 genes showed a statistically significant age-related difference in the expression of oxidative stress-related genes; however, Western blot analysis demonstrated an age-related increase in only 2 (cytoglobin and GPX-1) of 11 proteins tested. No age-related differences were detected in the expression of DNA damage-signaling genes. Western blot analysis of seven DNA damage-related proteins verified this finding. Intense nuclear staining of DNA damage foci was observed in nuclei within the central endothelium of older donors. Central endothelium from young donors consistently showed a low level of positive staining.
Conclusions.
HCECs respond to age-related increases in oxidative nuclear DNA damage by forming DNA damage repair foci; however, they do not vigorously defend against or repair this damage by upregulating the expression of multiple oxidative stress or DNA damage-signaling genes.
Human corneal endothelial cells (HCECs) exhibit both age-dependent and topographically dependent reduction in proliferative capacity.1–3 This reduction is mediated, at least in part, by an age-related increase in the expression of the G1-phase inhibitors, p21Cip1, and p16INK4a.4–6 Of importance to an understanding of the molecular basis for these differences in growth capacity is the fact that oxidative DNA damage can induce a p53-dependent increase in the expression of p21Cip1.7 Recent studies from this laboratory8 explored the relationship between oxidative DNA damage and reduced proliferation in HCECs. ELISA for 8-hydroxy-2′-deoxyguanosine (8-OHdG), a marker of oxidative DNA damage,9 found that 8-OHdG levels were significantly higher (P = 0.0031) in the endothelia of older donors than in young donors. Immunolocalization of 8-OHdG in ex vivo corneas showed intense staining in the nuclei of many, but not all, cells located in the central endothelia of older donors (50 years and older), and this staining was more intense than in central endothelia of young donors (30 years and younger). Nuclear 8-OHdG staining was not observed in peripheral cells, regardless of donor age; however, 8-OHdG was present in a punctate cytoplasmic pattern suggesting that, in peripheral HCECs, oxidative DNA damage is located primarily in mitochondria. Results of 8-OHdG immunolocalization studies in cultured HCECs paralleled those obtained in ex vivo corneas. Importantly, HCECs cultured from young donors and treated with increasing concentrations of hydrogen peroxide, an inducer of oxidative stress, exhibited a dose-dependent decrease in proliferative capacity very similar to that observed in untreated cells of older donors. Together, these data provide evidence that oxidative nuclear DNA damage plays a role in the age-dependent and topographically related decrease in proliferative capacity observed in HCECs.
The current studies investigated whether HCECs respond to oxidative nuclear DNA damage by upregulating their expression of oxidative stress and DNA damage-signaling genes in an age-dependent manner. For these studies, commercially available real-time PCR microarrays were used to compare the relative expression of oxidative stress and antioxidant genes and genes involved in the DNA damage-signaling pathway in HCECs directly isolated from the corneas of young and older donors. Western blot analysis analyzed the relative expression of a subset of proteins to validate the microarray results. Immunostaining for the phosphorylated histone, H2AX-Ser139, was used to visualize nuclear DNA damage foci in the endothelia of corneas of young and older donors.
Materials and Methods
Human Donor Corneas
Corneas were obtained from National Disease Research Interchange (NDRI; Philadelphia, PA) according to the exclusion criteria reported previously10 and were maintained in corneal storage medium (Optisol; Chiron Ophthalmics, Inc., Irvine, CA) at 4°C until immediately before the experiment. Corneas handled by the eye bank, NDRI, and this laboratory strictly observed the tenets of the Declaration of Helsinki for the protection of donor confidentiality. Donor information for all studies is presented in Table 1. Donors were divided into two age groups: young (30 years and younger) and older (50 years and older).
Table 1.
Donor Information
| Age (y) | Hours* | Days† | Cause of Death | Experiment |
|---|---|---|---|---|
| 3 | 4:30 | 3 | Undetermined | Superarrays |
| 14 | 9:30 | 3 | Subdural hematoma | Superarrays |
| 15 | 8:30 | 2 | Gastroenteritis | Superarrays |
| 57 | 2:30 | 3 | Breast cancer | Superarrays |
| 73 | 10:45 | 4 | Myocardial infarction | Superarrays |
| 74 | 14:00 | 3 | Myocardial infarction | Superarrays |
| 11 | 9:20 | 5 | Respiratory deficiency | Western blot analysis |
| 16 | 11:00 | 2 | Motor vehicle accident | Western blot analysis |
| 19 | 10:30 | 2 | Cardiomyopathy | Western blot analysis |
| 25 | 4:30 | 7 | Motor vehicle accident | Western blot analysis |
| 28 | 15:30 | 3 | Cardiac arrest | Western blot analysis |
| 56 | 2:00 | 5 | Cardiac arrest | Western blot analysis |
| 63 | 17:00 | 2 | Myocardial infarction | Western blot analysis |
| 69 | 8:00 | 2 | Myocardial infarction | Western blot analysis |
| 70 | 13:30 | 3 | Myocardial infarction | Western blot analysis |
| 74 | 8:00 | 2 | Breast cancer | Western blot analysis |
| 5 | 4:55 | 9 | Motor vehicle accident | H2AX-Ser139 ICC |
| 23 | 1:30 | 2 | Cardiac arrest | H2AX-Ser139 ICC |
| 30 | 2:00 | 7 | Traumatic injury | H2AX-Ser139 ICC |
| 63 | 18:23 | 14 | Myocardial infarction | H2AX-Ser139 ICC |
| 64 | 10:05 | 7 | Myocardial infarction | H2AX-Ser139 ICC |
| 68 | 14:20 | 9 | Myocardial infarction | H2AX-Ser139 ICC |
Number of hours between death and corneal preservation.
Number of days of corneal preservation in corneal storage medium at 4°C.
Total RNA Extraction and Reverse Transcription
Corneas from three donors per age group were quickly washed three times in ice-cold phosphate-buffered saline (PBS; Invitrogen/Life Technologies, Carlsbad, CA). Descemet's membrane with associated endothelium was stripped from the corneas. The tissue strips were quickly placed in 1.0 mL reagent (TRIzol; Invitrogen) and were homogenized for 1 minute at room temperature. Chloroform was added, and the sample was mixed for 15 seconds, followed by incubation at room temperature for 3 minutes and centrifugation at 14,000g for 30 minutes at 4°C. The upper aqueous phase was collected, and a kit was used for the purification of total RNA from animal and human tissue (RNeasy Micro Kit; Qiagen, Valencia, CA) according to the manufacturer's instructions. All samples were stored at −20°C before use.
Real-Time PCR-Based Array Analysis
The relative expression of genes involved in oxidative stress was determined in each of the six RNA samples (RT2 Profiler Human Oxidative Stress and Antioxidant Defense PCR Array; SABiosciences Corp., Frederick, MD). The same six RNA samples were tested for the relative expression of genes involved in DNA damage signaling (RT2 Profiler Human DNA Damage Signaling PCR Array; SABiosciences Corp.). Total RNA concentration for each sample was first determined by 260:280 nm absorbance ratio, and then equal amounts of RNA were reverse transcribed to form cDNA. Samples were diluted in qPCR master mix (RT2 SYBR Green; SABiosciences Corp.) according to the supplier's directions and pipetted into 96-well PCR array plates to evaluate the expression of 84 oxidative stress or DNA damage-signaling genes. Real-time PCR was performed in technical duplicates (ABI Prism 7900HT Sequence Detection System; Applied Biosystems, Foster City, CA). Raw data from the real-time PCR was uploaded using a PCR array data analysis template available at http://www.sabiosciences.com/pcr/arrayanalysis.php. (A large number of gene expression analyses have been performed and the results published using the arrays and Web-based automated analysis method [RT2 Profiler] available through this supplier at http://www.sabiosciences.com/support_publication.php). Quality controls included within the array plates confirmed the lack of DNA contamination and successfully tested for RNA quality and PCR performance. The integrated Web-based software package for the PCR array system automatically performed all comparative threshold cycle (ΔΔCt)–based fold-change calculations from the uploaded data. For these calculations, the average expression of three housekeeping genes (β2-microglobulin, β-actin, and glyceraldehyde-3-phosphate dehydrogenase) was used for normalization of the data. After normalization, the relative expression of each gene was averaged for the three samples in each age group. Fold changes in average gene expression were expressed as the difference in expression of HCECs from older donors compared with those of young (control) donors. A twofold or greater change in expression with P ≤ 0.05 was considered significant. These parameters have been used previously11 to analyze results of microarrays from the same supplier.
Western Blot Analysis
Descemet's membrane, together with the intact endothelium, was carefully isolated under a dissecting microscope to avoid cell loss and stromal tissue contamination and then washed in 10 mM HEPES buffer (Invitrogen). Total protein was solubilized (Sequential Extraction Reagent 3 [ER3]; Bio-Rad, Hercules, CA) with 1% TBP (tributylphosphine) reducing agent (Bio-Rad). Equal amounts of protein from five young and five older donors were pooled to form one sample per age group. A total of 20 μg protein was loaded per well for all blots, except for detection of apoptosis-inducing factor (AIF). In that case, 5 μg total protein was loaded to obtain readable bands for image analysis, and 10% Bis-Tris gels (NuPAGE; Invitrogen) were chosen for the detection of GADD45Aα and all oxidative stress-related proteins because of their expected low relative molecular weights. Most protein samples related to the DNA damage-signaling pathway were run on 3% to 8% Tris-Acetate gels (NuPAGE; Invitrogen). All gels were run at constant 200 V at room temperature until the bottom line of the prestained molecular weight markers approached the bottom of the gel. Prestained high molecular weight protein standard (HiMark; Invitrogen) was used for high molecular weight (>100 kDa) proteins. Another protein standard (Novex Sharp; Invitrogen) was used for proteins lower than 100 kDa. Proteins were then transferred to polyvinylidine difluoride membranes (Millipore, Bedford, MA). For high molecular weight proteins, gel transfers were performed at constant 70 V at 4°C for 2 hours to ensure complete transfer. The remaining protein samples were transferred at constant 25 V at room temperature for 2 hours. Nonspecific binding was blocked by incubation of the membranes for 1 hour at room temperature in 5% milk diluted in 0.1% Tween diluted with Tris-buffered saline (TTBS; Sigma-Aldrich, St. Louis, MO). Table 2 provides information regarding the primary antibodies used for all Western blot analyses. Both primary and secondary antibodies were diluted in TTBS. Blots were incubated with primary antibody at 4°C overnight on a shaker. After the membranes were washed in TTBS, they were incubated in secondary antibody diluted 1/1000 and incubated at room temperature for 1 hour. Peroxidase-conjugated secondary antibodies, including donkey anti–rabbit IgG, donkey anti–mouse IgG, and donkey anti–goat IgG, were purchased from Jackson ImmunoResearch (West Grove, PA). After washing, blots were incubated in chemiluminescent substrate (SuperSignal West Pico or Femto Chemiluminescent Substrate; Pierce, Rockford, IL). Protein bands were identified based on the expected relative molecular weight and by comparing band position using positive controls recommended by primary antibody suppliers (data not shown). Blots were run at least twice for each protein analyzed. Images were digitally scanned and analyzed using ImageJ software (developed by Wayne Rasband, National Institutes of Health, Bethesda, MD; available at http://rsb.info.nih.gov/ij/index.html). β-Actin (rabbit anti–β-actin: Sigma-Aldrich) was used for normalization of all protein bands. The normalized relative intensity for each band was averaged between the duplicate samples. Data are presented as the relative fold difference in protein expression in HCECs from older donors compared with those of young donors.
Table 2.
Primary Antibodies Used for Western Blot Analyses
| Antibody | Source | Catalog No. | Isotype | Dilution |
|---|---|---|---|---|
| Oxidative stress-related proteins | ||||
| Catalase | Abcam* | ab1877 | Rabbit | 1/1000 |
| Cytoglobin | Abcam | ab57713 | Mouse | 1/1000 |
| Glutathione peroxidase-1 | Abcam | ab50427 | Goat | 1/1000 |
| Glutathione peroxidase-3 | R&D Systems† | AF4199 | Goat | 1/1000 |
| Glutathione peroxidase-7 | Abcam | ab51948 | Goat | 1/1000 |
| Glutathione reductase | Abcam | ab84963 | Rabbit | 1/1000 |
| IPCEF-1 (PIP3E) | Abcam | ab80817 | Rabbit | 1/1000 |
| Peroxiredoxin-2 | Millipore‡ | 07–610 | Rabbit | 1/1000 |
| Peroxiredoxin-3 | Abcam | ab16753 | Mouse | 1/1000 |
| Peroxiredoxin-5 | Abcam | ab16944 | Mouse | 1/1000 |
| Superoxide dismutase-1 | Abcam | ab52950 | Rabbit | 1/500 |
| DNA damage signaling pathway proteins | ||||
| AIF | Santa Cruz§ | sc-5586 | Rabbit | 1/200 |
| ATM | Cell Signaling‖ | 2873 | Rabbit | 1/1000 |
| DNA-PKcs | Santa Cruz | sc-56090 | Mouse | 1/200 |
| GADD45a | Santa Cruz | sc-796 | Mouse | 1/1000 |
| OGG1/2 | Santa Cruz | sc-12074 | Goat | 1/200 |
| PCNA | Santa Cruz | sc-7907 | Rabbit | 1/200 |
| p53 | Cell Signaling | 9282 | Rabbit | 1/1000 |
Abcam, Inc., Cambridge, MA.
R&D Systems, Inc., Minneapolis, MN.
Millipore Corp., Billerica, MA.
Santa Cruz Biotechnology, Inc., Santa Cruz, CA.
Cell Signaling Technology, Inc., Danvers, MA.
Immunocytochemistry
Whole corneas from three young and three older donors were washed in PBS, fixed for 10 minutes in 100% methanol at −20°C, washed in PBS, and cut into central and peripheral areas using a 6.0-mm corneal vacuum punch (Barron; Katena Products, Denville, NJ). The resultant corneal tissues were immunostained according to established protocols.1 Briefly, tissue was incubated for 10 minutes in blocking buffer containing 4% bovine serum albumin (BSA; Thermo Fisher Scientific, Waltham, MA) diluted in PBS, and incubated overnight in rabbit anti–phospho-histone H2A.X-Ser139 (Cell Signaling Technology, Inc., Danvers, MA), diluted 1/100. Tissue samples were then washed in PBS and incubated for 1 hour with a 1/100 dilution of FITC donkey anti–rabbit IgG (Jackson ImmunoResearch, West Grove, PA). Both antibodies were diluted in blocking buffer. Corneal tissues incubated in secondary antibody alone acted as negative controls. To visualize all nuclei, tissue was incubated for 15 minutes at room temperature in iodide (TO-PRO-3; Invitrogen) diluted 1/1000 in PBS. Corneal pieces were then washed and placed endothelial-side up in tissue mounting medium (Vectashield; Vector Laboratories, Burlingame, CA). Fluorescence confocal microscopy was used to visualize positive staining. Laser images (0.5 μm) were digitally recorded.
Results
Age-Related Expression of Oxidative Stress and Antioxidant Genes and Proteins
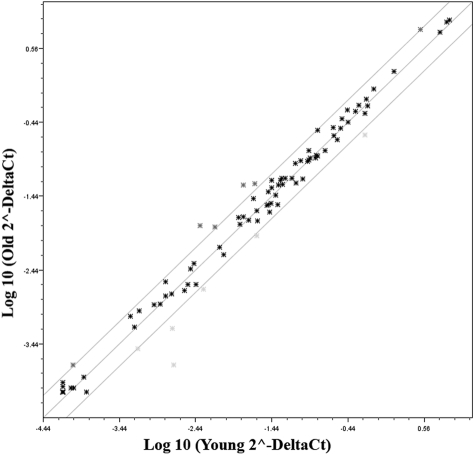
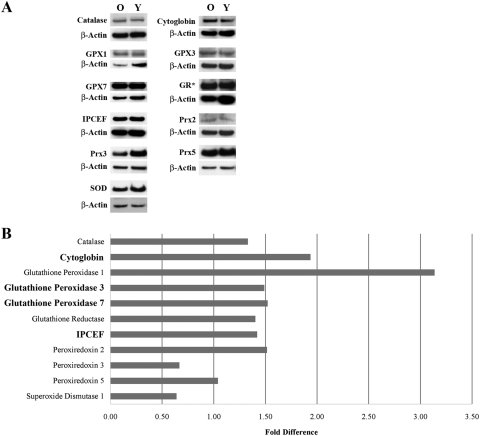
A real-time PCR-based microarray was used to compare the relative expression of 84 oxidative stress and antioxidant-related genes in HCECs isolated from the corneas of three young and three older donors. Data from the older donors were then compared with those from the young (control) donors. As indicated, a twofold or greater change in expression with P ≤ 0.05 was considered statistically significant. A scatter plot of the data is presented in Figure 1. Supplementary Table S1, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.10-6492/-/DCSupplemental, presents the results for each gene, with genes arranged according to function (as defined by SABiosciences). Only 4 of the 84 genes in the array were differentially expressed at statistically significant levels in HCECs from older donors. Those genes are highlighted in Supplementary Table S1, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.10-6492/-/DCSupplemental, and include GPX3 (glutathione peroxidase-3 [plasma]), GPX7 (glutathione peroxidase-7), CYGB (cytoglobin), and IPCEF-1 (interaction protein for cytohesin exchange factors-1, also known as phosphoinositide-binding protein PIP3E). Genes coding for CYGB and GPX3 were expressed at 3.4-fold and 2.5-fold higher levels, respectively. IPCEF-1 and GPX7 were expressed at 10-fold and 2.3-fold lower levels, respectively, compared with young donors. Six genes (GPX2, PXDN, MT3, NOS2, NOX5, AOX1) appeared to be differentially expressed by at least twofold higher or lower levels in HCECs from older donors; however, P values for those genes did not indicate statistical significance. Semiquantitative Western blot analyses were then conducted to determine the relative protein level of the four genes found by microarray analysis to be expressed at significantly different levels in HCECs of young and older donors. Seven additional oxidative stress-related proteins were chosen for analysis because of their relative importance in neutralizing the effects of hydrogen peroxide and other reactive oxygen species. These proteins included catalase, glutathione peroxidase-1, glutathione reductase, peroxiredoxin-2, peroxiredoxin-3, peroxiredoxin-5, and superoxide dismutase-1. As indicated, equal amounts of protein from five young and five older donors were pooled to form one sample per age group (see Table 1), and these samples were used for all blots. Figure 2A shows representative blots for each oxidative stress-related protein and the corresponding β-actin bands. Figure 2B presents results of the analysis of the expression of each protein in HCECs of older donors compared with those of young (control) donors. In HCECs of older donors, cytoglobin was expressed at a 1.94-fold higher level, glutathione peroxidase-3 at a 1.49-fold higher level, IPCEF-1 (PIP3E) at a 1.42-fold higher level, and glutathione peroxidase-7 at a 1.52-fold higher level than in cells of young donors. These results differed somewhat from those of the microarray analysis in that protein levels of the four genes whose expression was found to differ significantly in HCECs of older donors differed only in the two age groups by 1.4- to 1.94-fold. In addition, all four proteins showed an increase in expression level, whereas the microarray data indicated that GPX7 and IPCEF-1 expression was lower in HCECs of older donors. Analysis of the seven additional oxidative stress proteins showed that the protein level of glutathione peroxidase-1 was increased in HCECs of older donors by 3.14-fold over those of young donors. This result appeared to differ from the microarray analysis in that GPX1 gene expression was reduced by 1.38-fold in HCECs of older donors; however, the P value did not indicate statistical significance. Most of the remaining proteins were expressed at or only slightly above levels observed in young donors. Interestingly, superoxide dismutase-1 and peroxiredoxin-3 protein levels were expressed at somewhat lower levels (0.640- and 0.668-fold, respectively) in cells of older donors.
Figure 1.
Scatter plot showing the average relative expression (ΔΔCt) of oxidative stress and antioxidant defense genes in HCECs of older donors compared with those from young (control) donors. Three genes—β2-microglobulin, β-actin, and glyceraldehyde-3-phosphate dehydrogenase—were used for normalization of the data. The two outer lines on the plot indicate a twofold change in gene expression from that of the control, indicated by the middle line in the plot.
Figure 2.
Western blot results of 11 proteins related to oxidative stress and antioxidant defense. Equal amounts of protein, pooled from five young (Y) and five older (O) donors, were used for each blot. Results were normalized to β-actin. (A) Representative images of the blots for each protein plus the corresponding β-actin band. (B) Graph showing the fold difference in expression for each of the 11 oxidative stress-related proteins. Results are highlighted for the four proteins whose genes showed a statistically significant difference in relative gene expression. Results were calculated from the densitometric analysis of the Western blot analysis and are expressed as the fold difference in HCECs from older donors compared with young (control) donors. GR*, glutathione reductase.
Age-Related Expression of DNA Damage-Signaling Pathway Genes and Proteins
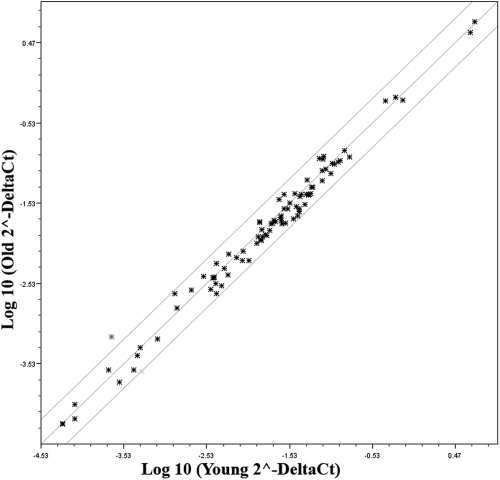
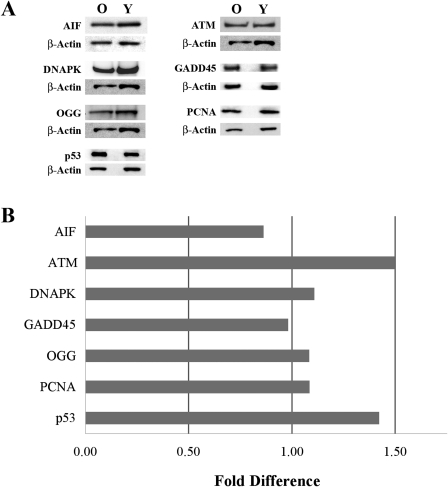
A real-time PCR-based microarray was also used to compare age-related changes in the expression of 84 genes involved in the DNA damage-signaling pathway. The same RNA samples from three young and three older donors used for the oxidative stress array were used for these studies (see Table 1). A scatter plot of the array data are presented in Figure 3. Results are presented in Supplementary Table S2, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.10-6492/-/DCSupplemental, with genes arranged according to function (as indicated by SABiosciences). One gene, GML (glycosylphosphatidylinositol anchored molecule-like protein), was not consistently detected. Expression of the remaining 83 genes was detectable in the HCEC samples. As can be seen from the data, 80 of the total 83 detectable genes showed less than a twofold change in expression. SEMA4A (sema domain, immunoglobulin domain [Ig], transmembrane domain [TM], and short cytoplasmic domain [semaphorin] 4A), XRCC3 (x-ray repair complementing defective repair in Chinese hamster cells 3), and BTG2 (BTG family, member 2) appeared to be differentially expressed by at least twofold higher or lower levels in HCECs of older donors; however, P values for these genes were greater than 0.05. Overall, results indicated that there was no significant age-related change in the relative expression of any gene included in the DNA damage-signaling pathway array. To verify the PCR array results, a limited number of proteins was chosen for semiquantitative Western blot analysis, based on their having different relative functions in the DNA damage-signaling pathway. The same two pooled protein samples representing the young and older donor age groups used for analysis of the oxidative stress proteins were used for these blots (see Table 1). Proteins studied included AIF, ATM (ataxia telangiectasia mutated kinase), DNAPK (DNA protein kinase catalytic subunit), GADD45α (growth arrest and DNA-damage-inducible-α), OGG1/2 (8-oxoguanine DNA glycosylase), PCNA (proliferating cell nuclear antigen), and p53 (p53 tumor suppressor protein). Representative blots for each protein are shown in Figure 4A. Results of the densitometric analyses are in Figure 4B. As can be seen, there was little evidence of an age-related difference in the expression of any of the proteins examined, thereby verifying the overall results of the microarray.
Figure 3.
Scatter plot showing the average relative expression (ΔΔCt) of DNA damage-signaling pathway genes in HCECs of older donors compared with those of young (control) donors. Three genes—β2-microglobulin, β-actin, and glyceraldehyde-3-phosphate dehydrogenase—were used for normalization of the data. The two outer lines on the plot indicate a twofold change in gene expression from that of the control, indicated by the middle line in the plot.
Figure 4.
Western blot results of seven proteins related to the DNA damage-signaling pathway. Equal amounts of protein, pooled from five young (Y) and five older (O) donors, were used for each blot. Results were normalized to β-actin. (A) Representative images of the blots for each protein plus the corresponding β-actin band. (B) Graph showing the fold difference in expression for each of the seven DNA damage-signaling pathway proteins. Results were calculated from the densitometric analysis of the Western blot analysis and are expressed as the fold difference in HCECs of older donors compared with young (control) donors.
Detection of DNA Damage Foci
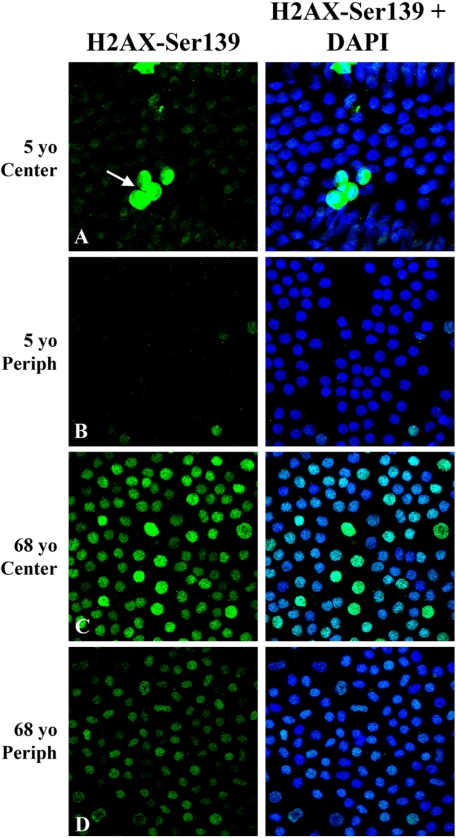
Because results of the two microarray and Western blot studies strongly suggest that HCECs of older donors do not vigorously upregulate the expression of oxidative stress proteins or proteins involved in the DNA damage-signaling pathway, studies were conducted to determine whether HCECs are capable of detecting nuclear DNA damage. To answer this question, ex vivo corneas from three young and three older donors were immunostained for phosphorylated histone H2AX. H2AX is a histone variant found in the nucleosome. Within 1 to 3 minutes after DNA double-strand breakage, H2AX becomes rapidly phosphorylated on Ser139, and the relative number of phosphorylated H2AX molecules increases linearly with the severity of the DNA damage.12,13 This specific phosphorylation results in the focal recruitment of repair factors to the site of DNA damage and may result in the induction of cell cycle checkpoint regulatory factors.12–14 Figure 5 presents representative results of the immunolocalization studies. Regardless of donor age or location within the endothelium, it was possible to observe occasional nuclei with very intense stain in a plane just above that of the endothelial monolayer (Fig. 5A, arrow), indicating the presence of apoptotic cells. Central (6.0-mm diameter) endothelia of young donors consistently showed a very low level of positive nuclear staining. In contrast, individual nuclei in the central endothelia of older donors consistently exhibited more intense punctate staining, with the relative intensity varying from nucleus to nucleus. In the peripheral (6.0- to 9.5-mm rim) endothelia of young donors, little to no positive stain was observed, whereas light nuclear staining was observed in many peripheral cells of older donors. No staining was observed in negative controls (data not shown). Positive nuclear staining for H2AX phosphorylated on Ser139 provided evidence that HCECs are capable of detecting nuclear DNA damage, and the punctate staining pattern strongly suggested that HCECs responded to this damage by forming DNA damage foci.
Figure 5.
Identification of DNA damage foci in ex vivo corneal endothelia of young and older donors. Representative images show immunostaining for phosphorylated histone, H2AX-Ser139, a recognized marker for DNA damage foci. Left: H2AX-Ser139 staining alone; right: H2AX-Ser139 staining plus iodide staining to indicate all nuclei. Final magnification, 40×; zoom, 2.
Discussion
Previous studies from this laboratory8 have provided evidence that oxidative nuclear DNA damage increases in HCECs in an age-dependent and topographically related manner and that increased oxidative stress may be responsible for the age-related decrease in proliferative capacity observed in HCECs. The present studies explored the ability of HCEC to upregulate gene and protein expression as a protective or reparative mechanism in response to age-dependent increases in oxidative stress and DNA damage. In these studies, real-time PCR-based microarrays compared gene expression in HCECs isolated from three young and three older donors. Western blot studies were conducted using protein extracted from five young and five older donors. Equal amounts of protein were then pooled to form two samples representing each of the two age groups. This type of analysis has been used previously for Western blot studies from this laboratory15–17 and other laboratories.18–20 The benefit to using pooled samples was to eliminate donor-to-donor variation and to obtain data that reflected consistent age-related changes in protein expression.
Expression of Genes and Proteins Related to Oxidative Stress and Antioxidant Defense
Results of the real-time PCR microarray provide evidence that HCECs express multiple genes involved in the response to oxidative stress. Western blot analysis provided a semiquantitative analysis of the protein expression of a limited number of these genes. When results of the microarray and Western blot analysis were compared for certain proteins, some differences were observed. These differences may be attributed to the fact that the analyses were conducted using RNA or protein samples isolated from different donors. In addition, there could have been relative differences in protein turnover compared with overall gene expression. Both analysis methods indicated an age-related upregulation in the expression of cytoglobin, which facilitates the diffusion of oxygen through tissues, scavenges nitric oxide and other reactive oxygen species, and serves a protective function during oxidative stress.21,22 Results of the microarray and Western blot analyses of glutathione peroxidase-1 differed in that no significant age-related difference in gene expression was indicated; however, protein expression was found to be increased threefold in older donors. Both glutathione peroxidase-3 and -7 proteins appeared to be expressed in HCECs of older donors at, or slightly above, levels found in young donors. Glutathione peroxidases are an important family of enzymes whose main function is to catalyze the reduction of endogenously produced hydrogen peroxide and lipid hydroperoxides in the presence of glutathione.23 Gene expression of IPCEF-1 was found by microarray analysis to be at a 10-fold lower level in HCECs of older donors; however, Western blot analysis indicated that IPCEF-1 protein levels were 1.42-fold higher in HCECs of older donors, indicating that this protein is expressed at similar levels regardless of donor age. IPCEF-1 is involved in translocating cytohesins to the plasma membrane and in membrane receptor signaling.24 Glutathione reductase, catalase, peroxiredoxin-2, and peroxiredoxin-5 proteins were all expressed in HCECs of older donors at or slightly above the levels observed in HCECs of young donors. Interestingly, superoxide dismutase-1 and peroxiredoxin-3 were expressed at somewhat lower levels in HCECs of older donors. The expression of one or more of these oxidative stress-related proteins has previously been reported in corneal endothelia of rabbits,25–27 rats,25,28–30 dogs,25,31 and humans.16,17,25,31 However, to our knowledge, this is the first time relative expression levels of these proteins have been compared in an age-dependent fashion in human corneal endothelial cells.
Together, these results indicate that there is only a limited response of HCECs to age-related increases in oxidative stress. This suggests that, with donor age, there is no significant increase in protection of these cells from accumulating oxidative damage. Previous studies8 indicate that cells in the central 6.0-mm diameter of the endothelium exhibit a significantly higher level of oxidative nuclear DNA damage than cells in the surrounding 6.0- to 9.5-mm peripheral rim. It is possible that significant increases in the levels of oxidative stress-related proteins in central endothelium could have been masked because both the RNA and the protein samples analyzed in these studies were prepared from the entire endothelial monolayer, therefore reflecting an average of the overall expression levels. It should be noted that these studies have investigated potential changes in the relative expression of oxidative stress-related proteins; however, they did not explore the relative activity of these proteins. Previous studies of corneal endothelial cells from rabbits26 and rats28 indicate that the relative activity of several antioxidant enzymes, including superoxide dismutase, glutathione peroxidase, and catalase, do not increase with increasing age or as the result of acute oxidative stress, suggesting that there may be a similar lack of increased activity in HCECs with donor age.
Expression of Genes and Proteins Related to the DNA Damage-Signaling Pathway
Results of the real-time PCR microarray provide evidence that multiple genes involved in the DNA damage-signaling pathway are expressed in HCECs; however, analysis of the data revealed no statistically significant difference in the expression of any gene represented in the microarray as a function of donor age. Results of the Western blot analysis verified those of the microarray. The seven proteins analyzed were chosen because they are known to have different and important functions in the response of cells to DNA damage. AIF is released from mitochondria during the apoptotic process. Among its functions is the induction of DNA fragmentation and chromatin condensation.32,33 Of interest is the fact that, when gels were being prepared for Western blot analysis of AIF, the protein load had to be reduced from the usual 20 μg to 5 μg because the relative expression of this protein was high in both samples. ATM is a serine/threonine kinase that becomes autophosphorylated in response to DNA double-strand breaks34,35 and helps regulate cell cycle checkpoints and DNA repair.36 Among its downstream targets are Chk2 and p53.37 DNA-PK is a phosphatidylinositol kinase composed of two DNA-binding subunits, collectively known as Ku, and a single 450-kDa catalytic subunit (DNA-PKcs). DNA-PK appears to selectively regulate the p53-dependent apoptosis pathway.38,39 GADD45α is a p53 tumor suppressor target gene and is also inducible in a p53-independent manner.40–42 GADD45 proteins have been implicated in regulating the G2/M-phase cell cycle checkpoint, DNA repair, and apoptosis. OGG1 is a DNA glycosylase that initiates base excision repair of oxidized purine bases.43,44 PCNA functions in cell cycle progression, DNA replication, and DNA repair.45,46 The PCNA gene is induced by p53, and PCNA protein interacts with p53-controlled proteins, such as GADD45 and p21, in the process of deciding cell fate. The tumor suppressor, p53, plays a major role in the cellular response to DNA damage and other stresses.47,48 DNA damage leads to the phosphorylation of p53 through the activity of several transducing proteins, including ATM and DNA-PK. This phosphorylation promotes the retention of p53 within the cell and promotes its downstream induction of several cell cycle regulatory proteins, including p21Cip1 and GADD45, as well as pro-apoptotic proteins, such as Bax; p53 also plays a role in DNA repair, including nucleotide excision repair after UV-induced damage.41,48 Of these seven proteins, only two (PCNA and p53) were previously identified in corneal endothelium.2,49–52 As has been noted, the activity of some proteins in the DNA damage-signaling pathway is dependent on posttranslational modification, such as phosphorylation; however, this type of modification would not have been detected with the specific antibodies used in this study. Other studies have identified genes expressed in HCECs that are related to DNA damage. Sakai et al.53 constructed a human corneal endothelial cDNA library and identified genes expressed in HCECs from donors averaging 59.9 ± 5.9 years old. Damage-specific DNA-binding protein 2 (DDB2), which helps protect cells against DNA damage after ultraviolet radiation, was one of the genes detected. Inoki et al.54 found that DDB2 is produced efficiently in cultured HCECs on exposure to UV irradiation. Neither of these studies compared age-related expression levels of this protein, and the DDB2 gene was not included in the microarray used for the present studies.
Detection of DNA Damage Foci
The immunodetection of histone H2AX phosphorylated on Ser139 provides strong evidence that HCECs are capable of detecting and responding to DNA damage by forming nuclear DNA damage foci. The relative number and staining intensity of H2AX-Ser139–associated DNA damage foci increased with donor age and was greatest in the central endothelia of older donors. Similar results were obtained in studies of H2AX-Ser139 immunostaining in senescent mouse embryo fibroblasts and in cells of the lung, spleen, dermis, liver, and gut of aging mice.55
Relevance of the Data
The lack of a significant age-related upregulation in the expression of either oxidative stress-related or DNA damage pathway proteins in HCECs is consistent with results found in other differentiated cell types.56–58 This lack of upregulation appears to correlate with the fact that terminally differentiated cells do not normally divide. Thus, surveillance and repair of the entire genome, as is required for dividing cells, may not be needed, and DNA repair is limited to the transcribed genome to preserve the function and specificity of long-lived tissues. Previous studies from this laboratory5,6,8 have provided evidence for an important connection between oxidative nuclear DNA damage, increased expression of p21Cip1, and decreased proliferative capacity in HCECs. Taken together, results of these studies suggest that HCECs protect themselves against oxidative stress and respond to DNA damage not by upregulating the expression of multiple oxidative stress-related or DNA damage pathway genes but by increasing the expression of the G1-phase inhibitor, p21Cip1, thereby restricting their ability to divide. At the same time, the more limited repair of transcriptionally active DNA domains would ensure that the cells remain functionally viable for a long period. It should be kept in mind that the current studies focused on the response of HCECs to oxidative stress and DNA repair as a means of further understanding the basis for the age-related decrease in proliferative capacity. Further study is needed to determine the age-dependent effect of oxidative stress on mitochondrial DNA and overall mitochondrial function in these very metabolically active and physiologically important cells.
Footnotes
Supported by National Eye Institute Grant R01 EY12700 (NCJ).
Disclosure: N.C. Joyce, None; D.L. Harris, None; C.C. Zhu, None
References
- 1. Senoo T, Joyce NC. Cell cycle kinetics in corneal endothelium from old and young donors. Invest Ophthalmol Vis Sci. 2000;41:660–666 [PubMed] [Google Scholar]
- 2. Zhu CC, Joyce NC. Proliferative response of corneal endothelial cells from young and older donors. Invest Ophthalmol Vis Sci. 2004;45:1743–1751 [DOI] [PubMed] [Google Scholar]
- 3. Mimura T, Joyce NC. Replication competence and senescence in central and peripheral human corneal endothelium. Invest Ophthalmol Vis Sci. 2006;47:1387–1396 [DOI] [PubMed] [Google Scholar]
- 4. Kikuchi M, Zhu C, Senoo T, Obara Y, Joyce NC. p27kip1 siRNA induces age-dependent proliferation of human corneal endothelial cells. Invest Ophthalmol Vis Sci. 2006;47:4803–4809 [DOI] [PubMed] [Google Scholar]
- 5. Enomoto K, Mimura T, Harris DL, Joyce NC. Age-related differences in cyclin-dependent kinase inhibitor expression and retinoblastoma hyperphosphorylation in human corneal endothelial cells. Invest Ophthalmol Vis Sci. 2006;47:4330–4340 [DOI] [PubMed] [Google Scholar]
- 6. Joyce NC, Harris DL. Decreasing expression of the G1-phase inhibitors, p21Cip1 and p16INK4a, promotes division of corneal endothelial cells from older donors. Mol Vis. 2010;16:897–906 [PMC free article] [PubMed] [Google Scholar]
- 7. Cox LS, Lane DP. Tumour suppressors, kinases and clamps: how p53 regulates the cell cycle in response to DNA damage. Bioessays. 1995;17:501–508 [DOI] [PubMed] [Google Scholar]
- 8. Joyce NC, Zhu CC, Harris DL. Relationship between oxidative stress, DNA damage, and proliferative capacity in human corneal endothelium. Invest Ophthalmol Vis Sci. 2009;50:2116–2122 [DOI] [PubMed] [Google Scholar]
- 9. Melov S. Mitochondrial oxidative stress: physiologic consequences and potential for a role in aging. Ann N Y Acad Sci. 2000;908:219–225 [DOI] [PubMed] [Google Scholar]
- 10. Joyce NC, Zhu CC. Human corneal endothelial cell proliferation: potential for use in regenerative medicine. Cornea. 2004;23(suppl. 1):S8–S19 [DOI] [PubMed] [Google Scholar]
- 11. Hoffman AE, Zheng T, Ba Y, Zhu Y. The circadian gene NPAS2, a putative tumor suppressor, is involved in DNA damage response. Mol Cancer Res. 2008;6:1461–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rogakou EP, Pilch DR, Orr H, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868 [DOI] [PubMed] [Google Scholar]
- 13. Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol. 2000;10:886–895 [DOI] [PubMed] [Google Scholar]
- 14. Pilch DR, Sedelnikova OA, Redon C, Celeste A, Nussenzweig A, Bonner WM. Characteristics of γ-H2AX foci at DNA double-strand break sites. Biochem Cell Bio. 2003;81:123–129 [DOI] [PubMed] [Google Scholar]
- 15. Zhu C, Rawe I, Joyce NC. Differential protein expression in human corneal endothelial cells cultured from young and older donors. Mol Vis. 2008;14:1805–1814 [PMC free article] [PubMed] [Google Scholar]
- 16. Jurkunas UV, Bitar MS, Rawe I, Harris DL, Colby K, Joyce NC. Increased clusterin expression in Fuchs' endothelial dystrophy. Invest Ophthalmol Vis Sci. 2008;49:2946–2955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jurkunas UV, Rawe I, Zhu C, et al. Decreased expression of peroxiredoxins in Fuchs' endothelial dystrophy. Invest Ophthalmol Vis Sci. 2008;49:2956–2963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Donahue MP, Rose K, Hochstrasser D, et al. Discovery of proteins related to coronary artery disease using industrial-scale proteomics analysis of pooled plasma. Am Heart J. 2006;152:478–485 [DOI] [PubMed] [Google Scholar]
- 19. Weinkauf M, Hiddemann W, Dreyling M. Sample pooling in 2-D gel electrophoresis: a new approach to reduce nonspecific expression background. Electrophoresis. 2006;27:4555–4558 [DOI] [PubMed] [Google Scholar]
- 20. Laimer M, Kocher T, Chiocchetti A, et al. Proteomic profiling reveals a catalogue of new candidate proteins for human skin aging. Exp Dermatol. 2010;19:912–918 [DOI] [PubMed] [Google Scholar]
- 21. Hodges NJ, Innocent N, Dhanda S, Graham M. Cellular protection from oxidative DNA damage by over-expression of the novel globin cytoglobin in vitro. Mutagenesis. 2008;23:293–298 [DOI] [PubMed] [Google Scholar]
- 22. Li D, Chen XQ, Li WJ, Yang YH, Wang JZ, Yu AC. Cytoglobin up-regulated by hydrogen peroxide plays a protective role in oxidative stress. Neurochem Res. 2007;32:1375–1380 [DOI] [PubMed] [Google Scholar]
- 23. Brigelius-Flohe R. Glutathione peroxidases and redox-regulated transcription factors. Biol Chem. 2006;387:1329–1335 [DOI] [PubMed] [Google Scholar]
- 24. Venkateswarlu K. Interaction protein for cytohesin exchange factors 1 (IPCEF1) binds cytohesin 2 and modifies its activity. J Biol Chem. 2003;278:43460–43469 [DOI] [PubMed] [Google Scholar]
- 25. Redmond TM, Duke EJ, Coles WH, Simson JA, Crouch RK. Localization of corneal superoxide dismutase by biochemical and histocytochemical techniques. Exp Eye Res. 1984;38:369–378 [DOI] [PubMed] [Google Scholar]
- 26. Cejkova J, Vejrazka M, Platenik J, Stipek S. Age-related changes in superoxide dismutase, glutathione peroxidase, catalase and xanthine oxidoreductase/xanthine oxidase activities in the rabbit cornea. Exp Gerontol. 2004;39:1537–1543 [DOI] [PubMed] [Google Scholar]
- 27. Atalla LR, Sevanian A, Rao NA. Immunohistochemical localization of peroxidative enzymes in ocular tissue. CLAO J. 1990;16:S30–S33 [PubMed] [Google Scholar]
- 28. Crouch RK, Patrick J, Goosey J, Coles WH. The effect of age on corneal and lens superoxide dismutase. Curr Eye Res. 1984;3:1119–1123 [DOI] [PubMed] [Google Scholar]
- 29. Rao NA, Thaete LG, Delmage JM, Sevanian A. Superoxide dismutase in ocular structures. Invest Ophthalmol Vis Sci. 1985;26:1778–1781 [PubMed] [Google Scholar]
- 30. Fujii T, Mori K, Takahashi Y, et al. Immunohistochemical study of glutathione reductase in rat ocular tissues at different developmental stages. Histochem J. 2001;33:267–272 [DOI] [PubMed] [Google Scholar]
- 31. Ostojic J, Grozdanic S, Syed NA, et al. Neuroglobin and cytoglobin distribution in the anterior eye segment: comparative immunohistochemical study. J Histochem Cytochem. 2008;56:863–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Susin SA, Lorenzo HK, Zamzami N, et al. Molecular characterization of mitochondrial apoptosis-inducing factor. Nature. 1999;397:441–446 [DOI] [PubMed] [Google Scholar]
- 33. Joza N, Pospisilik JA, Hangen E, et al. AIF: not just an apoptosis-inducing factor. Ann N Y Acad Sci. 2009;1171:2–11 [DOI] [PubMed] [Google Scholar]
- 34. Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer association. Nature. 2003;421:499–506 [DOI] [PubMed] [Google Scholar]
- 35. Yajima H, Lee KJ, Zhang S, Kobayashi J, Chen BP. DNA double-strand break formation upon UV-induced replication stress activates ATM and DNA-PKcs kinases. J Mol Biol. 2009;385:800–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–611 [DOI] [PubMed] [Google Scholar]
- 37. Reinhardt HC, Yaffe MB. Kinases that control the cell cycle in response to DNA damage: Chk1, Chk2, and MK2. Curr Opin Cell Biol. 2009;21:245–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yang J, Yu Y, Hamrick HE, Duerksen-Hughes PJ. ATM, ATR and DNA-PK: initiators of the cellular genotoxic stress response. Carcinogenesis. 2003;24:1571–1580 [DOI] [PubMed] [Google Scholar]
- 39. Wang S, Guo M, Ouyang H, et al. The catalytic subunit of DNA-dependent protein kinase selectively regulates p53-dependent apoptosis but not cell-cycle arrest. Proc Natl Acad Sci U S A. 2000;97:1584–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang XW, Zhan Q, Coursen JD, et al. GADD45 induction of a G2/M cell cycle checkpoint. Proc Natl Acad Sci U S A. 1999;96:3706–3711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Smith ML, Ford JM, Hollander MC, et al. p53-mediated DNA repair responses to UV radiation: studies of mouse cells lacking p53, p21, and/or gadd45 genes. Mol Cell Biol. 2000;20:3705–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gao M, Guo N, Huang C, Song L. Diverse roles of GADD45alpha in stress signaling. Curr Protein Pept Sci. 2009;10:388–394 [DOI] [PubMed] [Google Scholar]
- 43. de Souza-Pinto NC, Maynard S, Hashiguchi K, Hu J, Muftuoglu M, Bohr VA. The recombination protein RAD52 cooperates with the excision repair protein OGG1 for the repair of oxidative lesions in mammalian cells. Mol Cell Biol. 2009;29:4441–4454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hegde ML, Hazra TK, Mitra S. Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell Res. 2008;18:27–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Prosperi E. Multiple roles of the proliferating cell nuclear antigen: DNA replication, repair and cell cycle control. Prog Cell Cycle Res. 1997;3:193–210 [DOI] [PubMed] [Google Scholar]
- 46. Paunesku T, Mittal S, Protic M, et al. Proliferating cell nuclear antigen (PCNA): ringmaster of the genome. Int J Radiat Biol. 2001;77:1007–1021 [DOI] [PubMed] [Google Scholar]
- 47. El-Deiry WS. Regulation of p53 downstream genes. Cancer Biol. 1998;8:345–357 [DOI] [PubMed] [Google Scholar]
- 48. Smith ML, Seo YR. p53 regulation of DNA excision repair pathways. Mutagenesis. 2002;17:149–156 [DOI] [PubMed] [Google Scholar]
- 49. Gan L, Fagerholm P, Ekenbark S. Expression of proliferating cell nuclear antigen in corneas kept in long term culture. Acta Ophthalmol Scand. 1998;76:308–313 [DOI] [PubMed] [Google Scholar]
- 50. Slettedal JK, Lyberg T, Roger M, Beraki K, Ramstad H, Nicolaissen B. Regeneration with proliferation of the endothelium of cultured human donor corneas with extended postmortem time. Cornea. 2008;27:212–219 [DOI] [PubMed] [Google Scholar]
- 51. Paull AC, Whikehart DR. Expression of the p53 family of proteins in central and peripheral human corneal endothelial cells. Mol Vis. 2005;11:328–334 [PubMed] [Google Scholar]
- 52. Song Z, Wang Y, Xie L, Zang X, Yin H. Expression of senescence-related genes in human corneal endothelial cells. Mol Vis. 2008;14:161–170 [PMC free article] [PubMed] [Google Scholar]
- 53. Sakai R, Kinouchi T, Kawamoti S, et al. Construction of human corneal endothelial cDNA library and identification of novel active genes. Invest Ophthalmol Vis Sci. 2002;43:1749–1756 [PubMed] [Google Scholar]
- 54. Inoki T, Endo H, Inoki Y, et al. Damaged DNA-binding protein 2 accelerates UV-damaged DNA repair in human corneal endothelium. Exp Eye Res. 2004;79:367–376 [DOI] [PubMed] [Google Scholar]
- 55. Wang C, Jurk D, Maddick M, Nelson G, Martin-Ruiz C, von Zglinicki T. DNA damage response and cellular senescence in tissues of aging mice. Aging Cell. 2009;8:311–323 [DOI] [PubMed] [Google Scholar]
- 56. Gaubatz JW, Tan BH. Aging affects the levels of DNA damage in postmitotic cells. Ann N Y Acad Sci. 1994;719:97–107 [DOI] [PubMed] [Google Scholar]
- 57. Nouspikel T. DNA repair in differentiated cells: some new answers to old questions. Neuroscience. 2007;145:1213–1221 [DOI] [PubMed] [Google Scholar]
- 58. Fortini P, Dogliotti E. Mechanisms of dealing with DNA damage in terminally differentiated cells. Mutat Res. 2010;685:38–44 [DOI] [PubMed] [Google Scholar]