Sodium butyrate–induced reactivation of HSV-1 caused significant moderation of specific host epigenetic changes and failed to elicit or suppress immune activity. The relative silencing of immune activity could be valuable in developing novel therapies for HSK and viral gene therapy.
Abstract
Purpose.
To determine host response by gene expression in HSV-1 latent trigeminal ganglia (TG) after sodium butyrate (NaBu) treatment.
Methods.
Corneas of 6-week-old female BALB/c mice were scarified and inoculated with HSV-1 17Syn+ (high phenotypic reactivator) or its mutant 17ΔPst(LAT−) (low phenotypic reactivator) at 104 plaque-forming units/eye. NaBu-induced viral reactivation was by intraperitoneal (IP) administration at postinfection (PI) day 28, followed by euthanasia after 1 hour. NaBu-treated, uninfected mice served as the control. The resultant labeled cRNA from TG isolated total RNA was hybridized to gene microarray chips containing 14,000 mouse genes. Quantitative real-time PCR was performed to confirm gene expression.
Results.
Differential induction of gene expression between 17Syn+ and its mutant 17ΔPst(LAT−) was designated as NaBu-induced gene expression and yielded significant upregulation of 2- to 16-fold of 0.4% (56/14,000) host genes probed, comprising mainly nucleosome assembly and binding, central nervous system structural activity, hormonal activity, and signaling activity. Approximately 0.2% (24/14,000) of the host genes, mainly of the same functional categories were downregulated 3- to 11-fold. Immune activity was minor in comparison to our reports on gene expression during latency and heat stress induction. Euchromatin analysis revealed that the LAT-ICP0 locus is amenable to the effects of NaBu. Histone activity was detected by early transcription of histone cluster 2 H2be (Hist2h2be).
Conclusions.
NaBu-induced reactivation of HSV-1 is twofold: drug action involving significant moderation of specific host epigenetic changes and failure to elicit or suppress immune activity at the early time point of 1 hour.
Sodium butyrate (NaBu), a short-chain fatty acid that functions as an endogenous inhibitor of histone deacetylase (HD) enzymes, is a byproduct of carbohydrate metabolism in the gastrointestinal tract.1,2 HD inhibitors are potent inducers of growth arrest, differentiation, and/or apoptosis of cells in vitro and in vivo.3–5 HSV-1 reactivation in mice with a frequency of 75% to 100% has been induced in vivo using single and multiple doses of NaBu.6 The in vivo changes in the patterns of chromatin structure associated with the HSV-1 genome in mouse trigeminal ganglia (TG) after NaBu treatment have been detected as early as 0.5 hour after NaBu treatment.7 NaBu has been used in vitro to induce HSV-1 reactivation in quiescently infected neuronal PC-12 cells.8
Reactivation from HSV-1 latent sensory ganglionic neurons such as the TG can lead to peripheral shedding of infectious virus that could manifest in the eye as debilitating recurrent, epithelial, ulcerative, and/or stromal keratitis causing corneal scarring, thinning, and neovascularization.9 HSV-1 encephalitis is the most common cause of sporadic fatality and an estimated two thirds of such cases are most likely the result of viral reactivation from latency.10,11 Therapies for HSK are not entirely successful, because many patients either do not respond to intensive combination therapy or have rapid recurrence.12 Therapeutic application of HD inhibitors for central nervous system disorders has also been studied.13 Recently, we have shown that HSV-1 infection of human brain cells induces microRNA, miRNA-146a, and AD-type inflammatory signaling.14 The HSV-1 lifecycle after initial infection alternates between a productive phase of approximately 2 weeks15 and a distinguishable latency phase,11,16–20 in the latter of which the only abundant viral transcript is the non–protein-encoding LAT.21–23 LAT is not crucial for the establishment or maintenance of latency.24–26 It has been reported to enhance latency in rabbits.27 However, it is essential for efficient spontaneous reactivation of HSV-1 from latency.28,29 Reactivation of the latent HSV-1 genome has been linked to a reactivation critical region (RCR) including the LAT core promoter through the LAT 5′ exon/enhancer, because recombinants deficient in this region show greatly reduced reactivation phenotypes.23,30–33 LAT overlaps the infected cell protein 0 (ICP0), an immediate-early (IE) regulatory gene, and is transcribed off the opposite DNA strand. LAT is required for efficient HSV reactivation in animal models. A proposed function of LAT is the suppression of the nearby lytic phase transcripts, ICP0, g34.5 virulence gene, and infected cell protein 4 (ICP4), through unidentified mechanisms, thus promoting the establishment and maintenance of latency.23,34 ICP0 is essential for productive HSV-1 replication35 and is thought to have a role in reactivation from latency.36 LAT could act to suppress IE genes. Apart from putative LAT-mediated suppression of lytic transcripts during latency, host molecular/immune mechanisms could contribute to the establishment of latency.37
HSV infects at least 90% of the world's population,10,11 and infection rates are extremely high; hence, prevention of infection is not at present a viable option. However, blockage or significant reduction of viral reactivation from latency is a logical therapeutic approach. Human hosts can harbor HSV in the latent phase without any herpetic lesions appearing for their entire lives. Recent data have documented a significant frequency of shedding of HSV DNA in tears, saliva, and vaginal secretions in humans without any active herpetic disease.38–42 Our hypothesis is that this random HSV DNA shedding and herpetic disease is caused in part by a dynamic interaction between the genetics and phenotype of the virus coupled with inducible host repressive response mechanisms balancing viral latency and reactivation. However, certain drugs can alter cellular processes, facilitating amenable novel therapeutic approaches.
We recently investigated host gene expression during latency and heat stress–induced reactivation of HSV-1 and noted transient changes to occur at earlier times and to be mainly immune activity driven.43,44 An unanswered question is the nature of the host gene expression during NaBu-induced HSV-1 reactivation from latency. In the present study, we investigated differences of host gene expression in the TG of mice latent with two viruses, LAT-positive 17Syn+, and its mutant, LAT-negative 17ΔPst(LAT−), the latter a recombinant arising from LAT promoter deletion both of which originate from the strain 17 background.45 We deduced a subtractive mechanism in which the panel of gene expression resulting from the mutant 17ΔPst(LAT−) (synonymous with latency) was subtracted from the panel of gene expression resulting from the parent 17Syn+ (synonymous with reactivation) 1 hour after NaBu IP injection into mice. Our analysis revealed that transcript levels of Hist2h2be were elevated at an early time point, paralleling a report in which octamer-containing histone H2B has been shown in vitro to be transactivated by transfection of the HSV-1 virion component, VP16, but not by viral infection.46 We analyzed the activity of the LAT-ICP0 locus of the viral genome to determine whether there are changes in the histone gene expression in vivo.
Materials and Methods
Mice, Viral Inoculation Infection Assay, and NaBu Treatment
All animal procedures followed the Principles of Laboratory Animal Care of the National Institutes of Health in a protocol approved by the LSU Health Sciences Center Institutional Animal Care and Use Committee. All experiments complied with the guidelines in the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research. Identical groups of 6-week-old female BALB/c mice were anesthetized by intramuscular injection with ketamine hydrochloride (1 mg/kg) and xylazine (0.5 mg/kg; Vedco Inc., St Joseph, MO). Corneas were scarified with a three-by-three cross-hatch pattern and inoculated with a suspension of either HSV-1 17Syn+ or its mutant 17ΔPst(LAT−) at 104 plaque–forming units (pfu)/eye in 5 μL. Slit lamp examinations and ocular swab collections were performed at postinfection days (PI) 2 and 3, to verify corneal infection. At 28 days PI, ocular swabs were co-cultured with primary rabbit kidney cells and tested negative for infectious virus. Drug treatment consisted of NaBu/phosphate-buffered saline (1200 mg/kg of body weight in a 100-μL dose) administered IP.6 The mice were euthanatized 1 hour later. TG were aseptically removed and stored (RNA/later; Ambion, Austin, TX) at −80°C until used. We analyzed latently infected mice (six TG pooled together) for each virus: 17Syn+ or its mutant 17ΔPst. Two separate independent experiments for the microarrays (12 latently infected mice, 24 TG) and three separate independent experiments for ChIP (36 latently infected mice, 72 TG, including 0 hour experiments) were performed for each condition. High-quality RNA was required; the extraction protocol yielded sufficient RNA from a minimum of six pooled TG tissues (three mice) per gene array analysis.
Total RNA Extraction
Total RNA was extracted from the TG (RNeasy kit; Qiagen Sciences, Germantown, MD). The total RNA was DNase treated (Ambion), by modifying the manufacturer's protocol, and the pure total RNA was concentrated using a cleanup kit (Qiagen). RNA was spectrophotometrically assayed (ND-8000 spectrophotometer; NanoDrop Technologies, Wilmington, DE), and all the samples had an A260/A280 ≥ 2.0. There were no significant differences in spectral purity, rate of degradation, or yield of RNA from the TG between the groups. The RNA quality was further assayed for integrity by microfiltration on protein chips (2100 Bioanalyzer; Agilent Technologies, Palo Alto, CA), and all the samples agreed with the spectrophotometry and had 18s/28s > 1 and RIN values > 7 (Supplementary Methods Part A, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.09-5019/-/DCSupplemental).
DNA Microarray Experiments
Purified RNA samples (6 μg) were reverse transcribed by T7-(dT)24 oligomer reverse transcriptase (Superscript II; Invitrogen, Carlsbad, CA), and DNA polymerase I (Invitrogen-Gibco, Grand Island, NY) for first- and second-strand cDNA synthesis (Superscript kit; Invitrogen) and purified with a sample cleanup module (GeneChip; Affymetrix Inc., Santa Clara, CA). The cDNA was used for the synthesis of biotin-labeled antisense cRNA (target) in an in vitro transcription (IVT) reaction using an RNA transcript labeling kit (Bioarray HighYield Transcript; Enzo, Farmingdale, NY). The IVT-cRNA was repurified and then fragmented with fragmentation buffer (Affymetrix, Inc.) containing 20 μg in 40 μL. Denatured cRNA was pelleted down, to obtain a condensation fragment and was dissolved in hybridization buffer to a final concentration of 1.1 μg/μL. The cRNA cocktail was incubated at 99°C and 45°C for a total of 10 minutes and hybridized for 16 hours to mouse expression gene chips, representing 14,000 genes (430 2.0 array; Affymetrix). Duplicate chips were used for each of the samples. The gene chips were washed, stabilized, and stained according to the manufacturer's recommendations. Each chip was scanned in the phycoerythrin filter using a confocal laser scanner (Supplementary Methods Part B, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.09-5019/-/DCSupplemental). Data analysis was performed (Microarray suite 5.0; Affymetrix) to generate an absolute analysis for each chip (Supplementary Methods Part C, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.09-5019/-/DCSupplemental).
The relative abundance of individual genes is based on the signal intensities of the corresponding probe sets (analyzed by ArrayAssist 4.0; Stratagene, La Jolla, CA). Data from the experiments were individually normalized using gcRMA.47 The quality of all microarray experiments was assessed by the housekeeping gene probe mouse β-actin to measure the consistency of the hybridization signals from its 3′, middle, and 5′ fragments of the mRNA coding regions.48 A Student's t-test was performed to identify genes with significantly altered expression (P < 0.05). These genes were then filtered to identify genes with a twofold change between the composite data from 17Syn+ latent-infected TG and its mutant 17ΔPst (LAT−) latent TG 1 hour after NaBu IP injection into the mice. The genes with a P ≥ 0.05 and threefold change were also identified. Probe ontology of the genes complemented and annotations on the microarray are accessible through the NetAffx Analysis Center. Genes were clustered according to information available at the National Center for Biotechnology Information (NCBI, National Library of Medicine, National Institutes of Health, Bethesda, MD).
Quantitative Real-Time Polymerase Chain Reactions
HSV-1 DNA Copy Numbers.
DNA from each TG removed from 17Syn+ (or its mutant 17ΔPst(LAT−)) latent mice was extracted with a DNA elute kit according to the manufacturer's instructions (Gentra Puregene; Qiagen). DNA samples in DNA hydration buffer were stored at 4°C and processed for real-time PCR. DNA extracted from TG of naïve, uninfected mice and DNA from naïve uninfected TG spiked with 17Syn+ virus served as negative and positive controls, respectively.
Copy numbers of HSV-1 DNA were determined by calculating the number of HSV-1 polymerase genes per sample. All primer pairs used (Supplementary Table S1, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.09-5019/-/DCSupplemental) were specifically designed and synthesized by Integrated DNA Technologies (IDT, San Diego, CA). The real-time PCR reactions were performed in a 20-μL volume containing a solution of 1× master mix (TaqMan Universal; Applied Biosystems, Inc. [ABI], Foster City, CA), 100 nM of primers and probe, and 5 μL of DNA sample. For each reaction, the 96-well plate (Bio-Rad, Hercules, CA) contained both positive and negative controls, and loaded plates were centrifuged for 30 seconds at room temperature and 1000g in a swing-out rotor (CRU 5000 centrifuge; Damon/IEC, Needham, MA) to eliminate any air bubbles. The reaction protocol used was as follows: denaturation, 10 seconds at 95°C; annealing, 30 seconds at 55°C; and extension, 10 seconds at 72°C (iCycler iQ Multi-Color Real-Time PCR Detection System; Bio-Rad) for 45 cycles. The cosmid containing the HSV-1 DNA polymerase gene used as a standard was obtained from David Bloom (University of Florida, Gainesville, FL). The cosmid contained a copy of 4.8-kb restriction fragment (HindIIIA) encompassing the HSV-1 DNA polymerase gene from 17Syn+. A standard curve was generated from 10- and 2-fold serial dilutions of the pHindIIIA cosmid.
Confirmatory Analysis of Selected Genes.
Reverse-transcribed total RNA of 17 genes was prepared from the TG of the NaBu-treated groups of 17Syn+ and its mutant 17ΔPst (LAT−) latent-infected mice. One-third of these important expressed genes representing all the categorized functional groups were analyzed by real-time PCR to confirm the relative quantitative expression levels (Supplementary Methods Part D, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.09-5019/-/DCSupplemental). Primer pairs (IDT) used included those for the host gene β-actin (Supplementary Table S1, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.09-5019/-/DCSupplemental). Reverse transcription was primed with T7-Oligo (dT) primer (Affymetrix, Inc.). Real-time PCR reactions were performed in a 20 μL volume containing a solution of 1× SYBR green supermix (iQ; Bio-Rad), 0.5 μM of forward primer, 0.5 μM of reverse primer, and 1 μL of cDNA or 20 ng of reverse-transcribed total RNA. A four-step protocol was used: denaturation, 3 minutes at 95°C; amplification and quantification, 40 cycles for 15 seconds at 95°C and for 30 seconds at 60°C; melting curve, 60 to 95°C with a heating rate of 0.5°C per second; followed by cooling using a single-color real-time PCR detection system (MyiQ; Bio-Rad). A single-peak melting curve was observed for each gene product. Relative quantitative expression levels were determined for each gene. All results are displayed as an expression ratio of the 17Syn+ latent TG to its mutant 17ΔPst(LAT−) latent TG 1 hour after mouse NaBu-treatment, normalized against β-actin expression levels using the 2-ΔΔCt method.49
Chromatin Immunoprecipitation
NaBu-treated mice were euthanatized at 0.5 hour, 1 hour, or 2 hours after drug treatment (latently infected non-NaBu-treated were 0 minute). ChIP assays (Upstate Biotechnology, Charlottesville, VA) were performed using anti-acetyl H3 (catalog number 06-5991; Upstate, Lake Placid, NY)50 with minor modifications to accommodate the TG. Validations of the efficiencies of ChIP assays were performed as previously described.50 The primer pairs were specifically designed and synthesized by IDT. A standard curve was generated by using 10-fold serial dilutions of the purified 17Syn+ or its mutant 17ΔPst(LAT−) to control for differences in primer sensitivities. All assays were normalized to β-actin, for comparison between the different treatment groups of mice.
Results
NaBu-Induced Gene Expression in the TG of Latently Infected Mice Is Specific to 17Syn+
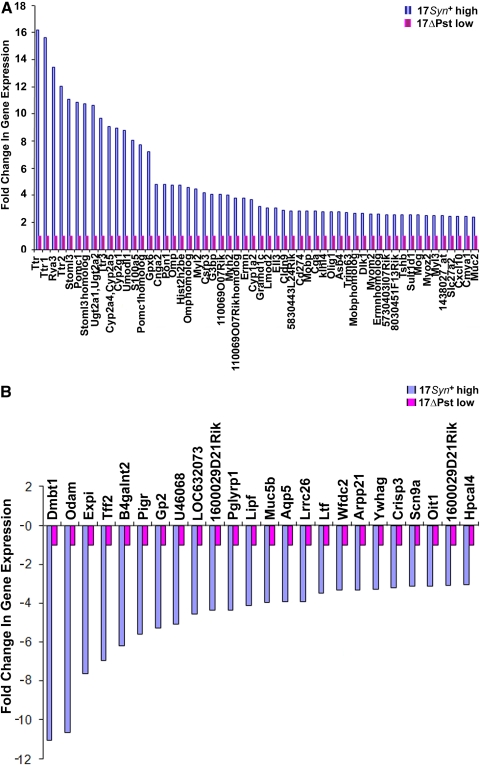
Signal intensity values between 17Syn+ latent TG relative to its mutant 17ΔPst(LAT−) latent TG 1 hour post-NaBu IP injection of mice were normalized (Microarray Suite 5; Affymetrix), resulting in the identification of 56 (0.4%) of 14,000 upregulated genes (twofold expression change; P < 0.05; see Fig. 2A; Supplementary Table S2, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.09-5019/-/DCSupplemental) from the original data of 17Syn+ latent TG and its mutant 17ΔPst(LAT−) (Supplementary Table S3, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.09-5019/-/DCSupplemental). Twenty-four genes were also downregulated (threefold change, P ≥ 0.05; Supplementary Table S4, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.09-5019/-/DCSupplemental). Robust multiarray analysis with correction for GC content (gcRMA) normalization51 was applied, to normalize the data of all experiments.
Figure 2.
(A) Normalized intensities of the 56 genes that were expressed at more than twofold in the 17Syn+ latent TG relative to its mutant 17ΔPst(LAT−) latent TG 1 hour after mouse NaBu treatment. (B) Normalized intensities of the 24 genes (threefold change, P ≥ 0.05) that were downregulated in the 17Syn+ latent TG relative to its mutant 17ΔPst(LAT−) latent TG 1 hour after mouse NaBu treatment. Changes in the expression of genes in the 17Syn+ are in multiples of gene expression of the mutant 17ΔPst(LAT−), with reference set at 1. High: genes that were significantly altered; low: genes that were indistinguishable from the background.
Establishment of Latency as Measured by Mean Viral DNA Copy Numbers Is Equivalent for Pooled TG of Both HSV-1 17Syn+ and Its Mutant 17ΔPst(LAT−)
In two independent experiments, we latently infected identical mice with either 17Syn+ or its mutant 17ΔPst(LAT−) and recovered TG individually from each mouse to quantify HSV-1 DNA copy numbers via the HSV-1 polymerase gene. The purpose was to evaluate establishment of viral loads of 17Syn+ and its mutant 17ΔPst in latently infected TG (Tables 1, 2). In the first experiment, the mean ± SEM copy numbers of six pooled latent TG were 6.183 × 104 ± 1.888 × 104 for 17Syn+ and 6.025 × 104 ± 1.154 × 104 for its mutant 17ΔPst(LAT−). In the second independent experiment, the copy numbers were 1.453 × 105 ± 0.397 × 105 for 17Syn+ and 1.458 × 105 ± 0.314 × 105 for its mutant 17ΔPst(LAT−). Respective similarities in these two separate experiments answer the critical question of established similar viral DNA loads of 17Syn+ and its mutant 17ΔPst, which we used for performing the gene array. A previous mouse study showed that drug (acyclovir), heat stress, and the absence of treatment did not alter viral DNA copy numbers in the TG of latently infected mice52 and that nonalteration of viral load in the establishment of latency has also been shown in rabbits.53,54 Thus, HSV-1 reactivation from latency is not a phenomenon reflecting differences in the number of latently infected neurons or the number of copies of viral genome in individual neurons but the ability of the 17Syn+ higher phenotypic reactivator to trigger specific responses.
Table 1.
Copy Numbers of HSV-1 DNA in Mouse Trigeminal Ganglia in Experiment 1
| Mice (6 TG) (n = 3) | 17Syn+ (High Phenotypic Reactivator) |
Mice (6 TG) (n = 3) | Mutant 17ΔPst(LAT−) (Low Phenotypic Reactivator) |
||
|---|---|---|---|---|---|
| Left TG | Right TG | Left TG | Right TG | ||
| Mouse 1 | 9.0 × 104 | 0.9 × 104 | Mouse 4 | 7.6 × 104 | 7.7 × 104 |
| Mouse 2 | 14.0 × 104 | 3.9 × 104 | Mouse 5 | 8.9 × 104 | 0.95 × 104 |
| Mouse 3 | 4.5 × 104 | 4.8 × 104 | Mouse 6 | 5.7 × 104 | 5.3 × 104 |
| Mean ± SEM | 6.183 × 104 ± 1.888 × 104 | Mean ± SEM | 6.025 × 104 ± 1.154 × 104 | ||
HSV-1 copy numbers of all samples were determined in triplicate analyses. The sum of the copy numbers of the six mutant 17ΔPst(LAT−) latent TG (representative of pooled TG from three mice used per microarray analysis sample) after averaging yielded 97.4% of HSV-1 DNA compared with that of 17Syn+ latent TG in identically infected mice, implying that relatively the same amounts of viral DNA are stably established for 17Syn+ and its mutant during latency.
Table 2.
Copy Numbers of HSV-1 DNA in Mouse Trigeminal Ganglia in Experiment 2
| Mice (6 TG) (n = 3) | 17Syn+ (High Phenotypic Reactivator) |
Mice (6 TG) (n = 3) | Mutant 17ΔPst (LAT−) (Low Phenotypic Reactivator) |
||
|---|---|---|---|---|---|
| Left TG | Right TG | Left TG | Right TG | ||
| Mouse 1′ | 0.90 × 105 | 2.10 × 105 | Mouse 4′ | 0.85 × 105 | 1.30 × 105 |
| Mouse 2′ | 0.75 × 105 | 1.05 × 105 | Mouse 5′ | 0.35 × 105 | 2.40 × 105 |
| Mouse 3′ | 3.15 × 105 | 0.77 × 105 | Mouse 6′ | 1.85 × 105 | 2.00 × 105 |
| Mean ± SEM | 1.453 × 105 ± 0.397 × 105 | Mean ± SEM | 1.458 × 105 ± 0.314 × 105 | ||
Determination of HSV-1 copy numbers was identical to that in Table 1. The sum of the copy numbers of the six 17Syn+ latent TG (representative of pooled TG from three mice used per microarray analysis sample) after averaging yielded 99.7% of HSV-1 DNA compared to that of the mutant 17ΔPst(LAT−) latent TG in identically infected mice, implying that relatively the same amounts of viral DNA are stably established for 17Syn+ and its mutant during latency in the second separate experiment.
Expression of Nucleosome Assembly, Binding, Signaling, Hormone Activity, and Structural Genes in Response to NaBu-Induced Viral Reactivation
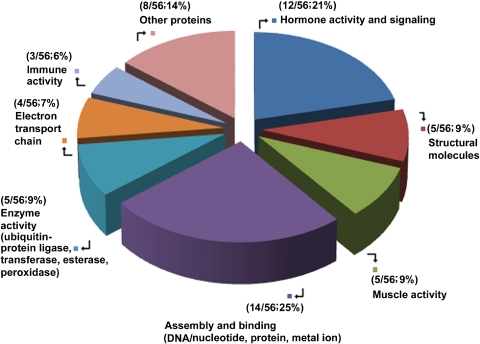
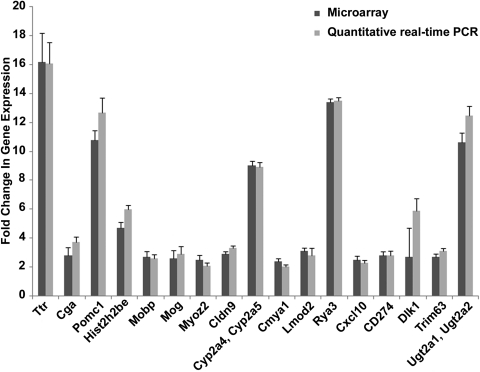
The 56 upregulated genes were broadly categorized into functional activities and comprised nucleosome assembly and binding (14/56; 25%), CNS structural molecules (5/56; 9%), hormone/signaling activity (12/56; 21%), enzyme activity (5/56; 9%), immune activity (3/56; 6%), electron transport (4/56; 7%), heart and skeletal muscle activity (5/56 or 9%), and other proteins (including transport and transcription) (8/56; 14%; Fig. 1). Genes were significantly expressed if their change in expression was twofold or more, yielding a total of 56 genes at 2- to 16-fold (Fig. 2A). NaBu action affected gene expression, mainly in the structural activity of histone, myelin sheath, and blood–brain barrier molecules, as well as hormonal activity and signaling activity (Supplementary Tables S5–S7, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.09-5019/-/DCSupplemental). Twenty-four genes, belonging mainly to the same functional categories as the upregulated ones, were downregulated 3- to 11-fold (Fig. 2B; Supplementary Table S4, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.09-5019/-/DCSupplemental). Transcripts of 17 of the 56 genes that were found to be upregulated in the microarray were analyzed by quantitative real-time PCR and their upregulation confirmed (Fig. 3; Supplementary Table S2, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.09-5019/-/DCSupplemental).
Figure 1.
Broad categories of functional activities of expressed genes in the 17Syn+ high phenotypic reactivator latent TG relative to its mutant 17ΔPst(LAT−) low phenotypic reactivator latent TG, 1 hour after mouse NaBu treatment.
Figure 3.
The ratio of change in gene expression in microarray analysis versus quantitative real-time polymerase chain reaction of the selected 17 genes in the gene expression of the 17Syn+ latent TG relative to its mutant 17ΔPst(LAT−) latent TG 1 hour after mouse NaBu treatment. Error bars, SD of an average of results in two duplicate independent experiments.
Hist2h2be Upregulation in NaBu-Induced Viral Reactivation from Latency
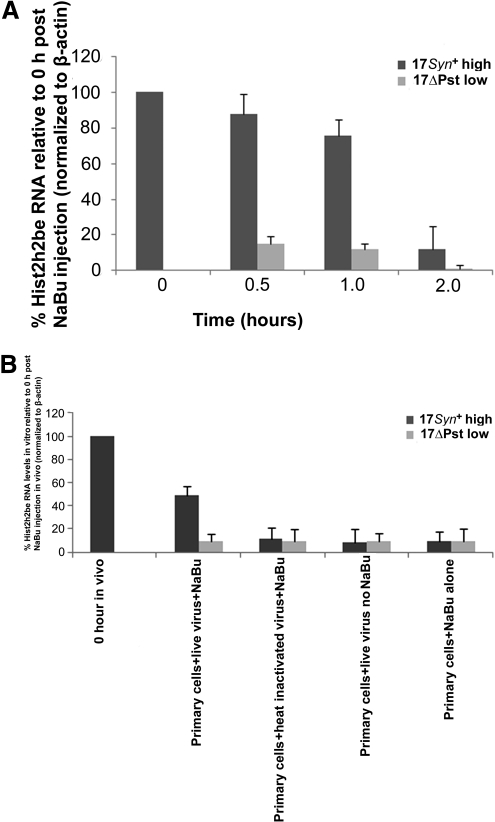
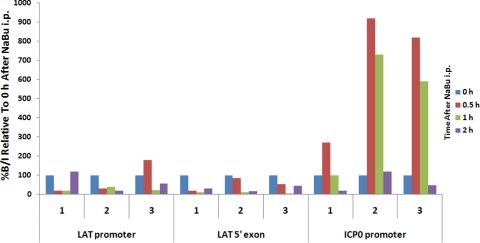
The Hist2h2be gene and its transcript (Fig. 4A) were upregulated by approximately fivefold (Supplementary Table S5, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.09-5019/-/DCSupplemental). To investigate the source of Hist2h2be activity, we prepared primary cultures of TG excised from female BALB/c mice and cells cultured separately with live virus, heat-inactivated virus, or no virus before incubation for 1 hour with NaBu (Supplementary Methods Part E, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.09-5019/-/DCSupplemental). The Hist2h2be gene was analyzed by real-time PCR in each specimen in duplicate, only those infected with live 17Syn+ virus and incubated with NaBu showed a relatively significant increase in histone transcript (Fig. 4B). The data show that changes in chromatin structure occurred in the ICP0 promoter region within 1 to 2 hours of NaBu treatment (Fig. 5).
Figure 4.
(A) The profile of Hist2h2be RNA levels (P < 0.05) at 0.5, 1.0, and 2.0 hours after mouse NaBu treatment. Six TGs in a group per time point were separately pooled before RNA extraction from 17syn+ or its mutant 17ΔPst(LAT−) latent mice. The cDNA was analyzed by real-time PCR in triplicate using Hist2h2be primer pairs. Relative quantities of Hist2h2be RNA were normalized to host β-actin, and the results presented as percent of Hist2h2be expression relative to that at the 0-hour time point. Error bars, the SD of an average of results in three independent experiments. (B) The profile of Hist2h2be RNA levels (P < 0.05) in primary cultures prepared from trigeminal ganglia from six mice. Cells were made into a suspension by trypsinization and plated in six-well plates for 48 hours. Monolayer cell cultures were incubated with virus, and subsequently drug or no drug incubations were performed as stipulated for 1 hour. Treated cells were allowed to recover before RNA extraction. Neuronal cells were identified and estimated (13% ± 1.01% of total cells per well; n = ∼9000 estimated total cell population) by fixing and staining with the neuronal marker β-tubulin-III (with counterstain conjugated to AlexaFluor 594-orange red) and before fixing, viral infection of neuronal cells (∼70%) was monitored by initially incubating with green fluorescent protein–tagged KOS. The cDNA was analyzed in triplicate by real-time PCR with Hist2h2be primer pairs. Relative quantities of Hist2h2be RNA were normalized to host β-actin. Results presented as the percentage of Hist2h2be relative to the 0-hour time point in the in vivo experiment. Error bars, SD of an average of two independent experiments.
Figure 5.
The profile of activity of the LAT promoter and the LAT 5′ exon (enhancer) regions. At 0.5 hour, 1 hour, or 2 hours after NaBu treatment, the mice were killed. All changes were calculated as a percent change relative to that at the 0-hour time point (synonymous to no reactivation or latent), set at 100%. The real-time PCR of deacetylation of the LAT promoter and the LAT 5′ exon (enhancer) regions at an early time point starting at 0.5 hour: Corresponding increase of 2- to 12-fold in acetylation of histones forming the chromatin bound to the ICP0 promoter region. Three independent experiments were performed (1, 2, and 3). In the ChIP assay, three mice or six TG, were used per experiment. Bound/input is abbreviated as B/I.
NaBu Treatment Targets Expression of Genes of Nucleosome Assembly and Binding Molecules of HSV-1 Latent Neurons
Binding molecules for DNA/nucleotide, protein, and metal ions resulted in the largest cumulative upregulation of gene expression (14/56; 25%; Supplementary Table S5, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.09-5019/-/DCSupplemental; Fig 1). This was followed by hormone/signaling activity (12/56; 21%; Supplementary Table S6, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.09-5019/-/DCSupplemental; Fig 1). Twelve (50%) of the 24 genes that were downregulated belonged to the same functional categories as the upregulated genes. The opposing and, at the same time, complementary relationship between binding and signaling provides an important insight into the interdependence of these two disjunct processes in the scheme of viral reactivation from latency and how they program the LAT-ICP0 locus in a major shift to influence the replication processes (Supplementary Fig. S1, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.09-5019/-/DCSupplemental).
Expression of Hormonal Activity Genes in NaBu-Treated HSV-1 Latent Neurons
Ttr originally identified in cells of the choroid plexus had profoundly high expression in the NaBu-treated HSV-1 latent TG (∼16-fold). Ttr, together with its three homologs, also showed substantial gene expression activity (4/56; 7%; Supplementary Table S6, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.09-5019/-/DCSupplemental; Fig. 2). Kent and Fraser55 have shown the Ttr precursor to be downregulated approximately fivefold (∼sevenfold by real-time PCR) and that signaling pathways are also triggered. Therefore, stress-induced or spontaneous viral reactivation probably suppresses or does not directly target Ttr activity. Expressions of Cga (∼3-fold), Pomc1 (∼11-fold), and the Pomc1 homolog were detected in the NaBu-treated HSV-1 latent TG although these genes were originally identified in hypothalamus-pituitary axis tissues (Supplementary Table S6, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.09-5019/-/DCSupplemental; Fig. 2). Pomc1 has been reported to be upregulated ∼6-fold (∼14-fold by real-time PCR).55
Discussion
This study has unraveled changes in host gene expression associated with NaBu induction of HSV-1 LAT positive high phenotypic reactivator 17Syn+ (Fig. 2; Supplementary Table S2, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.09-5019/-/DCSupplemental), in the sensory ganglionic neurons of the TG, known to be an important site for latent viral genomes. The 17Syn+ is a virus that reactivates in rabbits. NaBu action at early times of viral induction suggests no major role of immunity in contrast to our recent gene expression data of pivotal function of mainly innate immunity in HSV-1 latency43,44 and principally interplay of adaptive immunity and LAT-ICP0 locus function during heat stress–induced HSV-1 reactivation from latency, the latter indicating transient upregulation that occurs at earlier times.44
Two genes were expressed by both heat stress–induced viral reactivation44 and NaBu-induced viral reactivation. These two genes were Cga, a hormone metabolism gene and Dlk1, an ion- and protein-binding gene, both of which were upregulated approximately threefold (Supplementary Table S8, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.09-5019/-/DCSupplemental). Cga and Dlk1 expression could be caused by the presence of both molecules in pathways obligatorily triggered during physiological perturbation, irrespective of the types of stimuli involved, or these molecules could be critical to viral reactivation from latency. Both genes belong to assembly, binding, and hormone activity/signaling genes that formed 46% of the genes upregulated and 50% of the genes downregulated by NaBu (Figs. 1, 2; Supplementary Tables S4–S7, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.09-5019/-/DCSupplemental). Assembly, binding, and hormone activity/signaling genes also formed 28% of upregulated genes during heat stress induction.44 The differences in gene expression between heat stress44 and NaBu action could be due to the mode of stimuli endured. Consequentially, the abrupt change to the physiological environment due to the injection of the drug could trigger contrasting responses in comparison to that of heat stress, which entails a noninvasive external warming of the animal. The physical nature of each of these two different treatments could affect the physiological balance differently and produce some characteristic reactions that may offer some plausible explanations for this divergence; however, it does not indicate marked nonspecificity or unidirectional action of NaBu. These two stimuli are relevant to HSV pathogenesis, as butyrate is naturally produced in the gut, and humans are exposed to large differences in external temperature as well as the scorching heat of sunlight.
Both heat stress44 and NaBu-induced gene expression could be relevant to typical stresses or environmental changes capable of influencing the behavior of HSV-1 in neurons and triggering of viral reactivation. Heat stress and NaBu action jolt the virus to counteract the inert phase into an active replicating state. By their nature, viral reactivation and concomitant production of infectious virus are deleterious to the host and must be suppressed. Thus, there is competition between virus reactivation and host suppression that could provide a plausible explanation as to why the virus remains in latency in many individuals. Current data favor host responses that tend to block reactivation of virus and therefore prevent shedding of infectious particles.12,38–47,56–62 Immune profile changes that we observed in our recent gene expression studies43,44 support this role of active host immune mechanisms in suppression of HSV-1 reactivation to enhance neuronal survival and reduce or eliminate shedding of infectious virus in preference to the less deleterious shedding of viral DNA. However, our observation of relatively inactive host immunity at the earlier time point of 1 hour suggests multiple pathways and that during NaBu action HSV-1 reactivation could follow a different pathway not involving immunity (Supplementary Table S8, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.09-5019/-/DCSupplemental). NaBu action indicates its failure to elicit or alternatively the drug's ability to suppress a strong host immune response (Figs. 1, 2; Supplementary Tables S2, S4, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.09-5019/-/DCSupplemental). NaBu action resulted in fewer molecules in the defense and immune functional category to be upregulated (3/56; 6%) and a relatively higher number of molecules to be downregulated (7/24; 29%). Crisp3 which we had previously reported as upregulated during latency,43 was downregulated by NaBu. NaBu has also been shown to have anti-inflammatory effects on human monocytes in vitro.63
A nonimmune pathway of NaBu-induced HSV-1 reactivation could be essential in virus gene therapy using oncolytic (nonpathogenic) HSV against metastatic cancers. NaBu-induced reactivation of oncolytic HSV would increase the efficiency of the virus in the temporary absence of active host immunity, the latter counteractive to virus gene therapy. NaBu could therefore be useful in therapies for ocular and other diseases in which host immunity interferes with the effectiveness of treatment63–66 and some evidence exists66–68 in support of this NaBu role.
We reported upregulation of expression of immunity genes during heat-stress–induced HSV-1 reactivation from latency44 in contrast to NaBu action. There is a likelihood that NaBu action beyond this early time of 1 hour affects immunity differently. However, downregulation of immunity genes (7/24; 29%), coupled with our experience and the report of a previous study,69 suggest no change in the expression change pattern of any groups of genes occurring at later times or in the long term, and that they all return to equilibrium or preinduced levels over longer periods. In support, periods lasting longer have not been shown to significantly influence the molecular process during HSV-1 reactivation from latency, as Sawtell and Thompson70 have evaluated changes in attributes from 2 to 72 hours after heat stress in the HSV-1 latent mouse of selected molecules whose expression reflects a major shift in the physiological state of the neuron. They found no noticeable changes in the levels of these molecules over these longer periods in vivo,70 indicating that molecular processes in vivo occur very early and were more subtle.44
We observed that NaBu action altered expression of genes involved in remodeling of chromatin, which could help in explaining viral reactivation from latency. The chromatin structure of the HSV genome in lytic (productive) and latent states has been reviewed by Knipe and Cliffe.71 They have highlighted the principal events of the lytic-latent cycles. The essential activities are initial nucleocapsid penetration to release the viral genome into the nucleus which is transcribed to sequentially express IE, early (E), and late (L) viral gene products. This expression is followed by fusion with the neuronal membrane at the axonal termini and retrograde transport to the nucleus to be latent as a circular episome associated with nucleosomes. Stress induction or spontaneous reactivation from the latent state involves transportation of viral DNA anterograde from the latent sites to epithelial cells for productive infection. Knipe and Cliffe have proposed that an important factor in the lytic or latent infection decision by HSV is how the virus deals with the host cell response that assembles chromatin on naked viral DNA on entry into cells that were originally devoid of HSV DNA. Host cell mechanisms are implicated in attempting to assemble chromatin on the viral DNA to silence the viral genes. Further, they have suggested how regulation of chromatin could result in lytic infection of epithelial cells or latent infection of sensory neurons.
ChIP assays have revealed that histones are associated with lytic genes,72,73 and we observed expression of Hist2h2be, a member of the histone H2B family. In addition, immune-modulating Cxcl10 and Cd274 were upregulated and were generally coupled with 29% downregulation of defense and immunity genes suggesting potential perturbations in the environment of the neurons. The upregulation of Hist2h2be at the early time point of 0.5 hour (Fig. 4A) parallels the in vivo concurrent stochastic repression and derepression of the LAT and ICP0 promoters, respectively (Fig. 5). First, the suggestion is that ICP0 acts to prevent silencing of the viral genome. Second, it could be that LAT transcription7 and that of ICP0 are tightly modulated.44 Thus, the LAT-ICP0 locus could support congruent host–virus interactions to maintain neuronal survival while permitting virus and/or viral DNA release (Supplementary Fig. S1, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.09-5019/-/DCSupplemental).
The changes in gene expression identified in this study represent variations in many different functional categories including significant contribution by assembly, binding, and hormone activity/signaling molecules. However, it is not certain whether HSV-1 is able to directly manipulate expression of any of these cellular genes. The explanation is that there are two main conditions comprising stress associated with NaBu action and HSV-1 itself which could affect cellular gene transcription. Moreover, these two conditions are complicated by another layer of complexity—that is, injection of the drug— that could profoundly affect outcome. In perspective, the LAT-ICP0 locus background in terms of its activity in the 17Syn+ strain or lack thereof in its mutant 17ΔPst(LAT−) has shed some light on the multifunctional capacity of this locus to interpolate between latency and reactivation in consequence to serious HSV diseases. These changes in gene expression could be useful in studying the component pathways essential to HSV-1 reactivation from latency, by using methods like nonviral RNA interference.74
Acknowledgments
The authors thank Doan Nguyen and Patrick Byrne (Gene Therapy Microarray and Bioinformatics Core-LSUHSC, New Orleans, LA) for invaluable assistance.
Footnotes
Supported, in part, by National Institutes of Health Grants EY006311 (JMH) and EY02377 (LSU Eye Center Core Grant for Vision Research); an unrestricted research grant from LSU Health Sciences Center (PSB, JMH); a Senior Scientific Investigator Award (JMH) and an unrestricted grant to the Louisiana State University Eye Center from Research to Prevent Blindness, New York, NY; unrestricted support from the Louisiana Lions Eye Foundation, New Orleans, Louisiana, and the Vaccine Center and the South Louisiana Institute for Infectious Disease Research sponsored by the Louisiana Board of Regents, and Lions International, USA.
Disclosure: C. Clement, None; P.S. Bhattacharjee, None; M. Kumar, None; T.P. Foster, None; H.W. Thompson, None; J.M. Hill, None
References
- 1. Cummings JH. Short chain fatty acids in the human colon. Gut. 1981;22:763–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang D, Wang Z, Tian B, Li X, Li S, Tian Y. Two hour exposure to sodium butyrate sensitizes bladder cancer to anticancer drugs. Int J Urol. 2008;15:435–441 [DOI] [PubMed] [Google Scholar]
- 3. Wu ZQ, Zhang R, Chao C, Zhang JF, Zhang YQ. Histone deacetylase inhibitor trichostatin A induced caspase-independent apoptosis in human gastric cancer cell. Chin Med J (Engl). 2007;120:2112–2118 [PubMed] [Google Scholar]
- 4. Takai N, Ueda T, Nishida M, Nasu K, Narahara H. Histone deacetylase inhibitors induce growth inhibition, cell cycle arrest and apoptosis in human choriocarcinoma cells. Int J Mol Med. 2008;21:109–115 [PubMed] [Google Scholar]
- 5. Budillon A, Di Gennaro E, Bruzzese F, Rocco M, Manzo G, Caraglia M. Histone deacetylase inhibitors: a new wave of molecular targeted anticancer agents. Recent Pat Anticancer Drug Discov. 2007;2:119–134 [DOI] [PubMed] [Google Scholar]
- 6. Neumann DM, Bhattacharjee PS, Hill JM. Sodium butyrate: a chemical inducer of in vivo reactivation of HSV-1 in the ocular mouse model. J Virol. 2007;81:6106–6110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Neumann DM, Bhattacharjee PS, Giordani NV, Bloom DC, Hill JM. In vivo changes in the patterns of chromatin structure associated with the latent herpes simplex virus type 1 genome in mouse trigeminal ganglia can be detected at early times after butyrate treatment. J Virol. 2007;81:13248–13253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Danaher RJ, Jacob RJ, Steiner MR, Allen WR, Hill JM, Miller CS. Histone deacetylase inhibitors induce reactivation of herpes simplex virus type 1 in a latency-associated transcript-independent manner in neuronal cells. J Neurovirol. 2005;11:306–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kaufman HE, Rayfield MA, Gebhardt BM. Herpes simplex viral infections. In: Kaufman HE, Barron BA, McDonald MB. eds. The Cornea. 2nd ed. Boston: Butterworth-Heinemann; 1998:247–277 [Google Scholar]
- 10. Jones C, Inman M, Peng W, et al. The herpes simplex virus type 1 locus that encodes the latency-associated transcript enhances the frequency of encephalitis in male BALB/c mice. J Virol. 2005;79:14465–14469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sekizawa T, Openshaw H. Encephalitis resulting from reactivation of latent herpes simplex virus in mice. J Virol. 1984;50:263–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hill JM, Clement C. HSV-1 DNA in human corneas: what are the virological and clinical implications? J Infect Dis. 2009;200:1–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kazantsev AG, Thompson LM. Therapeutic application of histone deacetylase inhibitors for central nervous system disorders. Nat Rev Drug Discov. 2008;7:854–868 [DOI] [PubMed] [Google Scholar]
- 14. Toma HS, Murina AT, Areaux RG, Jr., et al. Ocular HSV-1 latency, reactivation and recurrent disease. Semin Ophthalmol. 2008;23:249–273 [DOI] [PubMed] [Google Scholar]
- 15. Hill JM, Zhao Y, Clement C, Neumann DM, Lukiw WJ. HSV-1 infections of human brain cells induce miRNA-146a and Alzheimer-type inflammatory signaling. Neuroreport. 2009;20:1500–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cabrera CV, Wohlenberg C, Openshaw H, Rey-Nendez M, Puga A, Notkins AL. Herpes simplex virus DNA sequences in the CNS of latently infected mice. Nature. 1980;288:288–290 [DOI] [PubMed] [Google Scholar]
- 17. Fraser NW, Lawrence WC, Wroblewska Z, Gilden DH, Koprowski H. Herpes simplex type 1 DNA in human brain tissue. Proc Natl Acad Sci USA. 1981;78:6461–6465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Marsden H. Herpes simplex virus in latent infection. Nature. 1980;288:212–213 [DOI] [PubMed] [Google Scholar]
- 19. Rock DL, Fraser NW. Detection of HSV-1 genome in central nervous system of latently infected mice. Nature. 1983;302:523–525 [DOI] [PubMed] [Google Scholar]
- 20. Sequiera LW, Jennings LC, Carrasco LH, Lord MA, Curry A, Sutton RN. Detection of herpes-simplex viral genome in brain tissue. Lancet. 1979;2(8143):609–612 [DOI] [PubMed] [Google Scholar]
- 21. Roizman B, Knipe DM, Whitley RJ. Herpes simplex viruses. In: Knipe DM, Howley PM. eds. Fields Virology. Baltimore, MD: Lippincott, Williams & Wilkins; 2007:2503–2602 [Google Scholar]
- 22. Bloom DC. HSV LAT and neuronal survival. Int Rev Immunol. 2004;23:187–198 [DOI] [PubMed] [Google Scholar]
- 23. Bloom DC. HSV-1 latency and the roles of the LATs. In: Sandri-Goldin RM. ed. Alpha Herpesviruses: Molecular and Cellular Biology. Norwich, UK: Caister Academic Press; 2006:325–342 [Google Scholar]
- 24. Perng GC, Dunkel EC, Geary PA, et al. The latency-associated transcript gene of herpes simplex virus type 1 (HSV-1) is required for efficient in vivo spontaneous reactivation of HSV-1 from latency. J Virol. 1994;68:8045–8055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ho DY, Mocarski ES. Herpes simplex virus latent RNA (LAT) is not required for latent infection in the mouse. Proc Natl Acad Sci USA. 1989;86:7596–7600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. lzumi KM, McKelvey AM, Devi-Rao G, et al. Molecular and biological characterization of a type 1 herpes simplex virus (HSV-1) specifically deleted for expression of the latency-associated transcript (LAT). Microb Pathogenesis. 1989;7:121–134 [DOI] [PubMed] [Google Scholar]
- 27. Perng GC, Slanina SM, Yukht A, et al. The latency-associated transcript gene enhances establishment of herpes simplex virus type 1 latency in rabbits. J Virol. 2000;73:1885–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Perng GC, Ghiasi H, Slanina SM, et al. The spontaneous reactivation function of the herpes simplex virus type 1 LAT gene resides completely within the first 1.5 kilobases of the 8.3-kilobase primary transcript. J Virol. 1996;70:976–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Perng GC, Slanina SM, Yukht A, et al. A herpes simplex virus type 1 latency-associated transcript mutant with increased virulence and reduced spontaneous reactivation. J Virol. 1999;73:920–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bloom DC, Hill JM, Devi-Rao G, Wagner EK, Feldman LT, Stevens JG. A 348-base-pair region in the latency-associated transcript facilitates herpes simplex virus type 1 reactivation. J Virol. 1996;70:2449–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hill JM, Sedarati F, Javier RT, Wagner EK, Stevens JG. Herpes simplex virus latent phase transcription facilitates in vivo reactivation. Virology. 1990;174:117–125 [DOI] [PubMed] [Google Scholar]
- 32. Ginsberg SD, Hemby SE, Lee VM, Eberwine JH, Trojanowski JQ. Expression profile of transcripts in Alzheimer's disease tangle-bearing CA1 neurons. Ann Neurol. 2000;48:77–87 [PubMed] [Google Scholar]
- 33. Leib DA, Bogard CL, Kosz-Venenchak M, et al. A deletion mutant of the latency-associated transcript of herpes simplex virus type 1 reactivates from the latent state with reduced frequency. J Virol. 1989;63:2893–2900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen SH, Kramer MF, Schaffer PA, Coen DM. A viral function represses accumulation of transcripts from productive-cycle genes in mouse ganglia latently infected with herpes simplex virus. J Virol. 1997;71:5878–5884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Preston CM. Repression of viral transcription during herpes simplex virus latency. J Gen Virol. 2000;81:1–19 [DOI] [PubMed] [Google Scholar]
- 36. Everett RD, Boutel C, Orr A. Phenotype of a herpes simplex virus type 1 mutant that fails to express immediate-early regulatory protein ICP0. J Virol. 2004;78:1763–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Miller CS, Danaher RJ, Jacob RJ. Molecular aspects of herpes simplex virus 1 latency, reactivation, and recurrence. Crit Rev Oral Biol Med. 1998;9:541–562 [DOI] [PubMed] [Google Scholar]
- 38. Miller CS, Danaher RJ. Asymptomatic shedding of herpes simplex virus (HSV) in the oral cavity. Oral Surg Oral Med Oral Pathol Oral Radiol Endodon. 2008;105:43–50 [DOI] [PubMed] [Google Scholar]
- 39. Kaufman HE, Azcuy AM, Varnell ED, Sloop GD, Thompson HW, Hill JM. HSV-1 DNA in tears and saliva of normal adults. Invest Ophthalmol Vis Sci. 2005;46:241–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Aryee EA, Bailey RL, Natividad-Sancho A, Kaye S, Holland MJ. Detection, quantification and genotyping of herpes simplex virus in cervicovaginal secretions by real-time PCR: a cross sectional survey. Virol J. 2005;2:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Legoff J, Bouhlal H, Gresenguet G, et al. Real-time PCR quantification of genital shedding of herpes simplex virus (HSV) and human immunodeficiency virus (HIV) in women coinfected with HSV and HIV. J Clin Microbiol. 2006;44:423–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kumar M, Hill JM, Clement C, Varnell ED, Thompson H, Kaufman HE. A double-blind placebo-controlled study to evaluate valacyclovir alone and with aspirin for asymptomatic HSV-1 DNA shedding in human tears and saliva. Invest Ophthalmol Vis Sci. 2009;50:5601–5608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Clement C, Popp MP, Bloom DC, et al. Microarray analysis of host gene expression for comparison between naïve and HSV-1 latent rabbit trigeminal ganglia. Mol Vis. 2008;14:1209–1221 [PMC free article] [PubMed] [Google Scholar]
- 44. Clement C, Bhattacharjee PS, Kaufman HE, Hill JM. Heat-induced reactivation of HSV-1 in latent mice: upregulation in the TG of CD83 and other immune response genes and their LAT-ICP0 locus. Invest Ophthalmol Vis Sci. 2009;50:2855–2861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bloom DC, Devi-Rao GB, Hill JM, et al. Molecular analysis of herpes simplex virus type 1 during epinephrine-induced reactivation of latently infected rabbits in vivo. J Virol. 1994;68:1283–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Latchman DS, Partidge JF, Estridge JK, Kemp LM. The different competitive abilities of viral TAATGARAT elements and cellular octamer motifs, mediate the induction of viral immediate-early genes and the repression of the histone H2B gene in herpes simplex virus infected cells. Nucleic Acids Res. 1989;17:8533–8542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wu Z, Irizarry RA, Gentleman R, Martinez-Murillo F, Spencer F. A model-based background adjustment for oligonucleotide expression arrays. J Am Stat Assoc. 2004;99:909–917 [Google Scholar]
- 48. Hubbell E, Liu WM, Mei R. Robust estimators for expression analysis. Bioinformatics. 2002;18:1585–1592 [DOI] [PubMed] [Google Scholar]
- 49. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–408 [DOI] [PubMed] [Google Scholar]
- 50. Kubat NJ, Amelio AL, Giordani NV, Bloom DC. The herpes simplex virus type 1 latency-associated transcript (LAT) enhancer/rcr is hyperacetylated during latency independently of LAT transcription. J Virol. 2004;78:12508–12518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Qin LX, Beyer RP, Hudson FN, Linford NJ, Morris DED, Kerr KF. Evaluation of methods for oligonucleotide array data via quantitative real-time PCR. BMC Bioinformatics. 2006;7:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gebhardt BM, Kaufman HE, Hill JM. Effect of acyclovir on thermal stress–induced herpesvirus reactivation. Curr Eye Res. 2004;29:137–144 [DOI] [PubMed] [Google Scholar]
- 53. O'Neil JE, Loutsch JM, Aguilar JS. et al. Wide variations in herpes simplex virus type 1 inoculum dose and latency-associated transcript expression phenotype do not alter the establishment of latency in the rabbit eye model. J Virol. 2004;78:5038–5044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hill JM, Gebhardt BM, Wen R, et al. Quantitation of herpes simplex virus type 1 DNA and latency-associated transcripts in rabbit trigeminal ganglia demonstrates a stable reservoir of viral nucleic acids during latency. J Virol. 1996;70:3137–3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kent JR, Fraser NW. The cellular response to herpes simplex virus type 1 (HSV-1) during latency and reactivation. J Neurovirol. 2005;11:376–383 [DOI] [PubMed] [Google Scholar]
- 56. Steiner I, Splvack JH, Deshmane SL, Ace CI, Preston CM, Fraser NW. A herpes simplex virus type 1 mutant containing a nontransinducing Vmw65 protein establishes latent infection in vivo in the absence of viral replication and reactivates efficiently from explanted trigeminal ganglia. J Virol. 1990;64:1630–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Divito S, Cherpes TL, Hendricks RL. A triple entente: virus, neurons, and CD8+ T cells maintain HSV-1 latency. Immunol Res. 2006;36:119–126 [DOI] [PubMed] [Google Scholar]
- 58. Kummer M, Turza NM, Muhl-Zurbes P, et al. Herpes simplex virus type 1 induces CD83 degradation in mature dendritic cells with immediate-early kinetics via the cellular proteasome. J Virol. 2007;81:6326–6338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Clement C, Tiwari V, Scanlan PM, Valyi-Nagy T, Yue BY, Shukla D. A novel role for phagocytosis-like uptake in herpes simplex virus entry. J Cell Biol. 2006;174:1009–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Openshaw H, McNeill JI, Lin XH, Niland J, Cantin EM. Herpes simplex virus DNA in normal corneas: persistence without viral shedding from ganglia. J Med Virol. 1995;46:75–80 [DOI] [PubMed] [Google Scholar]
- 61. Saemann MD, Bohmig GA, Osterreicher CH, et al. Anti-inflammatory effects of sodium butyrate on human monocytes: potent inhibition of IL-12 and up-regulation of IL-10 production. FASEB J. 2000;14:2380–2382 [DOI] [PubMed] [Google Scholar]
- 62. Mark KE, Wald A, Magaret AS, et al. Rapidly cleared episodes of herpes simplex virus reactivation in immunocompetent adults. J Infect Dis. 2008;198:1141–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lepisto AJ, Frank GM, Hendricks RL. How herpes simplex virus type 1 rescinds corneal privilege. Chem Immunol Allergy. 2007;92:203–212 [DOI] [PubMed] [Google Scholar]
- 64. Beffert U, Bertrand P, Champagne D, Gauthier S, Poirier J. HSV-1 in brain and risk of Alzheimer's disease. Lancet. 1998;351:1330–1331 [DOI] [PubMed] [Google Scholar]
- 65. Bhattacharjee PS, Neumann DM, Foster TP, et al. Effective treatment of ocular HSK with a human apolipoprotein E mimetic peptide in a mouse eye model. Invest Ophthalmol Vis Sci. 2008;49:4263–4268 [DOI] [PubMed] [Google Scholar]
- 66. Russell RG, Nasisse MP, Larsen HS, Rouse BT. Role of T-lymphocytes in the pathogenesis of herpetic stromal keratitis. Invest Ophthalmol Vis Sci. 1984;25:938–944 [PubMed] [Google Scholar]
- 67. Otsuki A, Patel A, Kasai K, et al. Histone deacetylase inhibitors augment antitumor efficacy of herpes-based oncolytic viruses. Mol Ther. 2008;16:1546–1555 [DOI] [PubMed] [Google Scholar]
- 68. Kim SH, Kim KW, Jeong JW. Inhibition of hypoxia-induced angiogenesis by sodium butyrate, a histone deacetylase inhibitor, through hypoxia-inducible factor-1 alpha suppression. Oncol Rep. 2007;17:793–797 [PubMed] [Google Scholar]
- 69. Higaki S, Gebhardt BM, Lukiw WJ, Thompson HW, Hill JM. Gene expression profiling in the HSV-1 latently infected mouse trigeminal ganglia following hyperthermic stress. Curr Eye Res. 2003;26:231–238 [DOI] [PubMed] [Google Scholar]
- 70. Sawtell NM, Thompson RL. Comparison of herpes simplex virus reactivation in ganglia in vivo and in explants demonstrates quantitative and qualitative differences. J Virol. 2004;78:7784–7794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Knipe DM, Cliffe A. Chromatin control of herpes simplex virus lytic and latent infection. Nat Rev Microbiol. 2008;6:211–221 [DOI] [PubMed] [Google Scholar]
- 72. Herrera FJ, Triezenberg SJ. VP16-dependent association of chromatin-modifying coactivators and underrepresentation of histones at immediate-early gene promoters during herpes simplex virus infection. J Virol. 2004;78:9689–9696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kent JR, Zeng PY, Atanasiu D, Gardner J, Fraser NW, Berger SL. During lytic infection herpes simplex virus type 1 is associated with histones bearing modifications that correlate with active transcription. J Virol. 2004;78:10178–10186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Thakker DR, Natt F, Husken D, et al. Neurochemical and behavioral consequences of widespread gene knockdown in the adult mouse brain by using nonviral RNA interference. Proc Natl Acad Sci USA. 2004;101:17270–17275 [DOI] [PMC free article] [PubMed] [Google Scholar]