The authors show that genetic variants associated with an optic nerve quantitative trait, vertical cup-to-disc ratio, are associated with primary open angle glaucoma and that this effect can be modified by factors controlling optic nerve area.
Abstract
Purpose.
Genetically complex disorders, such as primary open angle glaucoma (POAG), may include highly heritable quantitative traits as part of the overall phenotype, and mapping genes influencing the related quantitative traits may effectively identify genetic risk factors predisposing to the complex disease. Recent studies have identified SNPs associated with optic nerve area and vertical cup-to-disc ratio (VCDR). The purpose of this study was to evaluate the association between these SNPs and POAG in a US Caucasian case-control sample.
Methods.
Five SNPs previously associated with optic disc area, or VCDR, were genotyped in 539 POAG cases and 336 controls. Genotype data were analyzed for single SNP associations and SNP interactions with VCDR and POAG.
Results.
SNPs associated with VCDR rs1063192 (CDKN2B) and rs10483727 (SIX1/SIX6) were also associated with POAG (P = 0.0006 and P = 0.0043 for rs1063192 and rs10483727, respectively). rs1063192, associated with smaller VCDR, had a protective effect (odds ratio [OR] = 0.73; 95% confidence interval [CI], 0.58–0.90), whereas rs10483727, associated with larger VCDR, increased POAG risk (OR = 1.33; 95% CI, 1.08–1.65). POAG risk associated with increased VCDR was significantly influenced by the C allele of rs1900004 (ATOH7), associated with increased optic nerve area (P-interaction = 0.025; OR = 1.89; 95% CI, 1.22–2.94).
Conclusions.
Genetic variants influencing VCDR are associated with POAG in a US Caucasian population. Variants associated with optic nerve area are not independently associated with disease but can influence the effects of VCDR variants suggesting that increased optic disc area can significantly contribute to POAG risk when coupled with risk factors controlling VCDR.
Primary open angle glaucoma (POAG), which affects 2% of Caucasians and up to 12% of persons of African race older than 60, is a leading cause of blindness worldwide.1,2 Patients affected by POAG have a range of phenotypes, from ocular hypertension and minimal optic nerve disease to significant optic nerve deterioration in the setting of normal IOP. A family history of glaucoma is a major risk factor for POAG, and genetic risk factors are likely to contribute to both elevated IOP and optic nerve/retinal ganglion cell degeneration.3
Although POAG has a significant overall heritability,4 the genetic contributions to the disorder are complex and involve multiple genetic factors susceptible to the influence of environmental exposures.5,6 Complex disorders, such as POAG, may include highly heritable quantitative traits as part of the overall phenotype. Mapping genes that influence the related quantitative trait (often also called endophenotypes) rather than the complete complex phenotype has several important advantages, including objective definitions of the phenotype, identification of genes that are important risk factors for the disease, and possible reduction in the underlying molecular heterogeneity.
Several ocular quantitative traits with high heritability including IOP, central corneal thickness (CCT), and size and configuration of the optic nerve, are attributes that are structural features related to POAG. Given the complexity of the disease process, the genes associated with these features are reasonable candidates for POAG. Recently there has been considerable interest in genetic factors influencing optic nerve area and vertical cup-to-disc ratio (VCDR). Larger optic nerves may be more susceptible to damage related to IOP,7,8 and persons with larger VCDR may be more likely to have progressive enlargement of the cup.9,10 Heritability of the optic disc area and of the VCDR is estimated to be approximately 52% to 59% and 48% to 80%, respectively.11–14
Using quantitative analytical approaches, two recent genomewide studies have identified an association between a single-nucleotide polymorphism (SNP) near the ATOH7 gene (rs1900004) and the optic disc area (Rotterdam study,15 Australia study16). The Rotterdam study also identified several genomic regions showing association with VCDR.15 SNPs located in the two regions with the best evidence for association with VCDR (rs10483727 near the SIX1/SIX6 gene complex on 14q22–23; rs1063192 within the CDKN2B gene on 9p21) were also marginally associated with POAG in a sample of 188 cases (P = 0.017 for rs1063192; P = 0.021 for rs10483727). The purpose of this study was to evaluate the association between POAG and these SNPs influencing the optic nerve parameters of optic nerve size and VCDR in a US Caucasian case-control sample.
Patients and Methods
Patients and Control Subjects
This study was approved by the institutional review boards of the Massachusetts Eye and Ear Infirmary. The study adhered to the tenets of the Declaration of Helsinki. Informed consent was obtained from all patients and family members after explanation of the nature and possible consequences of the study.
Patients with POAG were recruited from the Glaucoma Consultation Service at the Massachusetts Eye and Ear Infirmary. POAG was defined as glaucomatous optic nerve damage in both eyes and visual field loss in at least one eye. Intraocular pressure was not included in the criteria for diagnosis of POAG. Glaucomatous optic nerve damage was defined as VCDR higher than 0.7 or focal loss of the nerve fiber layer (notch) associated with a specific visual field defect, or both. Visual fields were performed using automated perimetry. Control subjects had no evidence of glaucoma (IOP <22 mm Hg OU, VCDR ≤0.7, cup asymmetry <0.2) and no family history of glaucoma. VCDR was determined by visual examination of the optic nerve using either a 90-diopter lens or a direct ophthalmoscope.
A cohort of 539 unrelated patients with POAG and 336 unrelated control subjects without glaucoma were included in the present study. Demographic and clinical features of the study subjects are summarized in Table 1. In the POAG group, age at diagnosis ranged from 30 to 87 years (61.4 ± 11.2 years [mean ± SD]), and VCDR ranged from 0.20 to 0.99 (0.81 ± 0.13). Age at enrollment in the control group ranged from 40 to 89 years (65.2 ± 10.7 years), and VCDR ranged from 0.10 to 0.70 (0.30 ± 0.11). Optic disc area was not measured in this patient group.
Table 1.
Demographic and Clinical Features of Study Subjects
| Group | n | Female (%) | Age at Diagnosis (y)* |
Vertical Cup-to-Disc Ratio |
||
|---|---|---|---|---|---|---|
| Range | Mean ± SD | Range | Mean ± SD | |||
| POAG | 539 | 51.2 | 30–87 | 61.4 ± 11.2† | 0.20–0.99 | 0.81 ± 0.13† |
| Control | 336 | 54.5 | 40–89 | 65.2 ± 10.7 | 0.10–0.70 | 0.30 ± 0.11 |
For the control subjects, age at diagnosis referred to age at enrollment.
P < 0.0001 compared with the control group.
All study subjects were Caucasian and were from the United States. Control subjects were from the same geographic regions and had similar ethnic backgrounds as did patients with POAG. Cases and control subjects were balanced for sex, with 51.2% and 54.5% female in POAG patients and control subjects, respectively (P > 0.05; Table 1).
Polymorphisms and Genotyping
Three SNPs (rs7916697, rs1900004, rs3858145) in the 5′ region of the ATOH7 gene that have been associated with optic disc size15,16 and two SNPs (rs1063192 and rs10483727) on chromosomes 9 and 14 that have been associated with vertical cup-to-disc ratio15 were genotyped by SNP genotyping assays (TaqMan; Applied Biosystems [ABI], Foster City, CA). Genomic DNA from peripheral blood was prepared from all persons by using standard techniques (Gentra, Minneapolis, MN). Oligonucleotide primers and probes were ordered from ABI (assay by demand). The assays were performed on a PCR system (7500 Real-Time PCR; ABI) according to the manufacturer's instructions.
Statistical Analysis
Statistical analyses were performed using PLINK (version 1.07).17 Hardy-Weinberg equilibrium was assessed by using the χ2 test. The minor allele frequencies of each SNP between patients with POAG and control subjects were compared by using Fisher's exact test. Odds ratios (ORs) and 95% confidence intervals (CIs) were calculated using logistic regression. The association of the SNPs with POAG was further evaluated by using logistic regression after adjusting for age and sex. An additive effects model was applied to analysis of allele dosage in which the genotypes AA, AB, and BB were coded as 0, 1, and 2, respectively, where A represented the minor allele and B represented the common allele. Interaction effects between the SNPs were analyzed by including an interaction term in the logistic regression model. Haplotype frequencies for the ATOH7 gene were estimated using the standard E-M algorithm and were tested using the χ2 test. Association of the SNPs with VCDR was evaluated by using linear regression. Multiple comparisons were corrected using the Bonferroni method.
Results
VCDR Association Analyses
Characteristics and genotype counts for the five SNPs evaluated in this study are summarized in Table 2. All SNPs followed Hardy-Weinberg equilibrium in both the POAG cases and the control subjects (P > 0.05).
Table 2.
Characteristics and Genotype Counts for SNPs Investigated in This Study
| SNP | Chromosome | Position (bp)* | Nearest Gene | Location | Minor Allele | Genotype Count (AA/AB/BB)† |
|
|---|---|---|---|---|---|---|---|
| POAG | Control Subjects | ||||||
| rs7916697 | 10 | 69661859 | ATOH7 | 5′UTR | A | 41/201/285 | 28/122/174 |
| rs1900004 | 10 | 69670887 | ATOH7 | Intergenic | T | 43/188/296 | 20/125/176 |
| rs3858145 | 10 | 69681844 | ATOH7 | Intergenic | A | 47/205/258 | 29/136/148 |
| rs1063192 | 9 | 21993367 | CDKN2B | 3′UTR | G | 47/172/170 | 51/155/102 |
| rs10483727 | 14 | 60142628 | SIX1/SIX6 | Intergenic | T | 93/174/120 | 54/132/118 |
Chromosome position was based on NCBI Build 36.3 (National Center for Biotechnology, Bethesda, MD).
A, minor allele; B, common allele.
To investigate the association between these SNPs and VCDR in this population, we stratified VCDR measurements for the entire study sample (cases and controls) by genotype for each SNP. The G allele of SNP rs1063192 was significantly associated with a smaller VCDR (P-trend = 0.0013; Table 3), with a mean VCDR of 0.48, 0.51, and 0.58 in those carrying genotypes GG, GA, and AA, respectively. The T allele of SNP rs10483727 was significantly associated with a larger VCDR (P-trend = 0.0013; Table 3), with a mean VCDR of 0.60, 0.53, and 0.49 in those carrying genotypes TT, TC, and CC, respectively. These associations survived the Bonferroni correction for multiple testing (corrected P < 0.01) and are similar to the results obtained by the Rotterdam group.15 All three SNPs in the 5′region of the ATOH7 gene (rs7916697, rs1900004, rs3858145), previously associated with optic nerve area in both the Rotterdam and Australian studies, were not significantly associated with VCDR (P-trend > 0.45; Table 3) in this US Caucasian population.
Table 3.
Association with Vertical Cup-to-Disc Ratio
| SNP | Genotype | n (%) | Vertical Cup-to-Disc Ratio (mean ± SD) |
|---|---|---|---|
| rs7916697 | AA | 59 (8.7) | 0.57 ± 0.29 |
| AG | 261 (38.4) | 0.58 ± 0.28 | |
| GG | 360 (52.9) | 0.58 ± 0.28 | |
| Total | 680 (100.0) | P-trend = 0.74 | |
| rs1900004 | TT | 52 (7.6) | 0.62 ± 0.28 |
| TC | 256 (37.5) | 0.57 ± 0.28 | |
| CC | 375 (54.9) | 0.59 ± 0.28 | |
| Total | 683 (100.0) | P-trend = 0.87 | |
| rs3858145 | AA | 65 (9.8) | 0.59 ± 0.28 |
| AG | 273 (41.4) | 0.57 ± 0.28 | |
| GG | 322 (48.8) | 0.59 ± 0.27 | |
| Total | 660 (100.0) | P-trend = 0.45 | |
| rs1063192 | GG | 72 (13.5) | 0.48 ± 0.26 |
| GA | 251 (47.0) | 0.51 ± 0.27 | |
| AA | 211 (39.5) | 0.58 ± 0.28 | |
| Total | 534 (100.0) | P-trend = 0.0013 | |
| rs10483727 | TT | 118 (22.3) | 0.60 ± 0.27 |
| TC | 229 (43.3) | 0.53 ± 0.28 | |
| CC | 182 (34.4) | 0.49 ± 0.27 | |
| Total | 529 (100.0) | P-trend = 0.0013 |
P-trend was obtained from linear regression. Bonferroni corrected significance level was 0.01 (0.05/5).
POAG Association Analyses
To investigate the association between these SNPs and POAG, we compared the genotype and allelic distribution for each SNP between POAG cases and controls. Both SNPs associated with VCDR (rs1063192 and rs10483727) were also associated with POAG (P = 0.0045 and P = 0.010, respectively; Table 4). SNP rs1063192, associated with a smaller VCDR, had a protective effect, with a lower frequency for the risk allele G in POAG patients than in the control subjects (34.2% vs. 41.7%; OR = 0.73; 95% CI, 0.58–0.90). The T allele of SNP rs10483727, associated with a larger VCDR, was more frequent in patients with POAG than in controls (46.5% vs. 39.5%; OR = 1.33; 95% CI, 1.08–1.65). After adjusting for age and sex, these associations became even stronger (P = 0.0006 and P = 0.0043 for rs1063192 and rs10483727, respectively; Table 4) and survived Bonferroni correction for multiple testing (corrected P < 0.01).
Table 4.
Association with POAG
| SNP | Minor Allele | Minor Allele Frequencies |
Unadjusted |
Adjusted for Age and Sex |
|||
|---|---|---|---|---|---|---|---|
| POAG | Control Subjects | P* | OR (95% CI)† | P‡ | OR (95% CI)‡ | ||
| rs7916697 | A | 0.269 | 0.275 | 0.78 | 0.97 (0.78–1.21) | 0.79 | 0.97 (0.78–1.21) |
| rs1900004 | T | 0.260 | 0.257 | 0.91 | 1.02 (0.81–1.27) | 0.96 | 1.01 (0.80–1.26) |
| rs3858145 | A | 0.293 | 0.310 | 0.47 | 0.92 (0.74–1.15) | 0.41 | 0.91 (0.73–1.14) |
| rs1063192 | G | 0.342 | 0.417 | 0.0045 | 0.73 (0.58–0.90) | 0.0006 | 0.67 (0.53–0.84) |
| rs10483727 | T | 0.465 | 0.395 | 0.010 | 1.33 (1.08–1.65) | 0.0043 | 1.37 (1.10–1.69) |
Obtained from Fisher's exact test. Bonferroni corrected significance level was 0.01 (0.05/5).
Obtained from logistic regression.
Obtained from logistic regression after adjustment for age and sex.
All three SNPs in the 5′ region of the ATOH7 gene (rs7916697, rs1900004, rs3858145) previously associated with optic nerve area were not significantly associated with POAG (P > 0.47; Table 4). Haplotype analysis also did not find a significant association between these three SNPs and POAG (P > 0.42; data not shown).
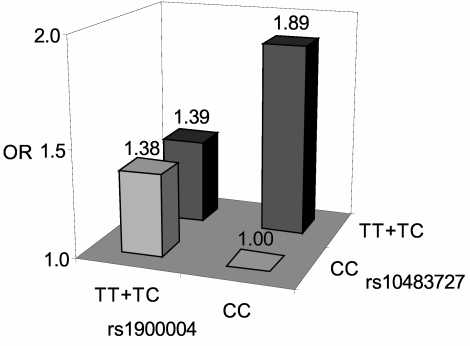
A measure of optic nerve area was not available for our study population; hence, we were unable to assess the relationship between rs1900004 and optic nerve area in this study. However, we were able to identify a significant interaction effect on POAG risk between SNP rs1900004 and rs10483727 (P-interaction = 0.025; Fig. 1), where SNP rs1900004 can modify the disease risk associated with rs10483727. Persons with rs1900004 genotype CC (associated with larger optic nerves) who also carried rs10483727 risk genotypes TT or TC had a significantly increased risk for POAG (OR = 1.89; 95% CI, 1.22–2.94) compared with those who had the rs10483727 risk genotypes and rs1900004 genotypes TT and TC (OR = 1.39; 95% CI, 0.88–2.19). A similar analysis performed with SNP rs1063192 did not show an interactive effect (P = 0.49). These results suggest that persons with larger optic nerves and larger VCDR are at increased risk for POAG than persons with increased VCDR without increased optic nerves.
Figure 1.
Interaction effects between rs10483727 and rs1900004. Logistic regression modeling showed that the joint effects between rs10483727 and rs1900004 were interactive (P-interaction = 0.025). Persons with rs1900004 genotype CC and those with rs10483727 risk genotype TT or TC are at significantly increased risk for POAG (OR = 1.89; 95% CI, 1.22–2.94) compared with persons with rs10483727 risk genotypes and rs1900004 genotype TT or TC, which are associated with smaller optic nerve area (OR = 1.39; 95% CI, 0.82–2.33).
Discussion
POAG is a genetically and phenotypically complex disease. Genetic factors—through interactions with other genes, environmental factors, or both—may influence both IOP and optic nerve susceptibility. One approach to identify genes that predispose to complex traits such as POAG is to define phenotypic subtypes or endophenotypes that are phenotypically homogeneous and may be caused by a smaller set of susceptibility genes. Ocular quantitative traits are a type of endophenotype that have the added advantages of precise measurement and continuous trait analysis, features that enhance gene discovery by minimizing misclassification and increasing study power.
In this study, we have shown that SNPs located in two VCDR quantitative trait loci on 9p22 and 14q22–23 (rs1063192 and rs10483727, respectively) are also associated with POAG in a US Caucasian population. The T allele of rs10483727, located near the SIX1/SIX6 gene complex on chromosome 14q22–23, is associated with a larger VCDR and an increased risk for POAG, whereas the G allele of rs1063192, located within the CDNKN2B gene on chromosome 9p22, is associated with a smaller VCDR and a decreased risk for POAG. Collectively, these results suggest that genetic variants influencing normal variation in VCDR are risk factors for the progressive increase in VCDR found in persons with POAG.
SNP rs1900004, located near the ATOH7 gene, is a major factor controlling optic nerve area.15,16 Although this SNP was not independently associated with POAG, we identified a significant interaction with SNP rs10483727 such that persons who carry the rs10483727 risk genotype TT or TC were more likely to develop POAG if they also carried the rs1900004 genotype associated with larger optic nerve area (CC). Previous studies have suggested that persons with larger disc areas are at increased risk for POAG and other forms of glaucoma.18 Our results suggest that optic disc area alone is not necessarily a risk factor for glaucoma but that it can significantly increase disease risk when coupled with risk factors controlling vertical cup-to-disc ratio.
The ATOH7 gene codes for Math5, a protein known to regulate retinal ganglion cell histogenesis in vertebrate model organisms,19 and recent analyses from both the Rotterdam study15 and the Australian group16 indicate that this gene is a major factor controlling optic disc area. The gene products of both the SIX1 and SIX6 genes are also developmentally regulated transcription factors.20–22 Mutations in SIX1 are a cause of the brachio-oto-renal syndrome,22 whereas mutations in SIX6 have been shown to cause anophthalmia.21 Our results showing that rs10483727 is associated with POAG as well as VCDR suggest that one or both of the SIX1/SIX6 genes may also contribute to optic nerve or retinal ganglion cell development. In addition, the interactive effect with rs1900004 could suggest that the SIX1/SIX6 gene complex may participate in developmental pathways that include ATOH7.
SNP rs1063192 is located on chromosome 9p21 within the CDKN2B gene and adjacent to CDKN2A, a tumor suppressor gene. The CDKN2B protein is a cyclin-dependent kinase that may regulate cell growth and is induced by transforming growth factor beta (TGF-β). TGF-β is a multifunctional cytokine that modulates developmental and repair processes in several tissues, including the retina and trabecular meshwork.23,24 The CDKN2B gene has also been associated with type 2 diabetes mellitus (T2DM) in several genomewide association studies.25–27 T2DM has been implicated as a risk factor for POAG,28 suggesting that further investigation among CDKN2B, POAG, T2DM, and glycemic load could be of interest. In this study, we confirm that the G allele is associated with a smaller VCDR and also show that carriers of the G allele are less likely to develop POAG. Collectively, these results add further support for a role of TGF-β in glaucoma and suggest that other proteins in the TGF-β signaling pathways may also contribute to glaucoma pathogenesis.
The SNPs associated with POAG and optic nerve parameters are all connected with genes that can influence optic nerve quantitative parameters through developmental processes. Although genes regulating ocular development may not be obvious candidates as risk modulators for a progressive late-onset disease such as POAG, our results suggest that such genes may, in fact, be one set of risk factors predisposing to POAG. Further study will be needed to determine whether these genetic risk factors participate in optic nerve and ganglion cell repair or have other metabolic roles in addition to regulating optic nerve size and structure.
Our study provides confirmation that a quantitative trait-mapping approach can be used to identify genetic risk factors for a genetically and phenotypically complex disease such as POAG. The identification of additional genetic factors influencing other ocular quantitative traits that could contribute to POAG will help further define the genetic architecture of this important blinding disease.
Footnotes
Supported in part by National Institutes of Health Grants EY015872 and P30EY014104, the Massachusetts Lions Eye Research Fund, and Research to Prevent Blindness.
Disclosure: B.J. Fan, None; D.Y. Wang, None; L.R. Pasquale, None; J.L. Haines, None; J.L. Wiggs, None
References
- 1. Friedman DS, Jampel HD, Muñoz B, West SK. The prevalence of open-angle glaucoma among blacks and whites 73 years and older: the Salisbury Eye Evaluation Glaucoma Study. Arch Ophthalmol. 2006;124:1625–1630 [DOI] [PubMed] [Google Scholar]
- 2. Leske MC, Connell AM, Schachat AP, Hyman L. The Barbados Eye Study: prevalence of open angle glaucoma. Arch Ophthalmol. 1994;112:821–829 [DOI] [PubMed] [Google Scholar]
- 3. Wiggs JL. Genetic etiologies of glaucoma. Arch Ophthalmol. 2007;125:30–37 [DOI] [PubMed] [Google Scholar]
- 4. Charlesworth J, Kramer PL, Dyer T, et al. The path to open-angle glaucoma gene discovery: endophenotypic status of intraocular pressure, cup-to-disc ratio, and central corneal thickness. Invest Ophthalmol Vis Sci. 2010;51:3509–3514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kang JH, Willett WC, Rosner BA, Hankinson SE, Pasquale LR. Caffeine consumption and the risk of primary open-angle glaucoma: a prospective cohort study. Invest Ophthalmol Vis Sci. 2008;49:1924–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kang JH, Wiggs JL, Rosner BA, et al. Endothelial nitric oxide synthase gene variants and primary open-angle glaucoma: interactions with sex and postmenopausal hormone use. Invest Ophthalmol Vis Sci. 2010;51:971–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bellezza AJ, Hart RT, Burgoyne CF. The optic nerve head as a biomechanical structure: initial finite element modeling. Invest Ophthalmol Vis Sci. 2000;41:2991–3000 [PubMed] [Google Scholar]
- 8. Hoffmann EM, Zangwill LM, Crowston JG, Weinreb RN. Optic disk size and glaucoma. Surv Ophthalmol. 2007;52:32–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Keltner JL, Johnson CA, Anderson DR, et al. The association between glaucomatous visual fields and optic nerve head features in the Ocular Hypertension Treatment Study. Ophthalmology. 2006;113:1603–1612 [DOI] [PubMed] [Google Scholar]
- 10. Miglior S, Pfeiffer N, Torri V, et al. Predictive factors for open-angle glaucoma among patients with ocular hypertension in the European Glaucoma Prevention Study. Ophthalmology. 2007;114:3–9 [DOI] [PubMed] [Google Scholar]
- 11. Klein BE, Klein R, Lee KE. Heritability of risk factors for primary open angle glaucoma: the Beaver Dam Eye Study. Invest Ophthalmol Vis Sci. 2004;45:59–62 [DOI] [PubMed] [Google Scholar]
- 12. van Koolwijk LM, Despriet DD, van Duijn CM, et al. Genetic contributions to glaucoma: heritability of intraocular pressure, retinal nerve fiber layer thickness, and optic disc morphology. Invest Ophthalmol Vis Sci. 2007;48:3669–3676 [DOI] [PubMed] [Google Scholar]
- 13. Chang TC, Congdon NG, Wojciechowski R, et al. Determinants and heritability of intraocular pressure and cup-to-disc ratio in a defined older population. Ophthalmology. 2005;112:1186–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schwartz JT, Reuling FH, Feinleib M. Size of the physiologic cup of the optic nerve head: hereditary and environmental factors. Arch Ophthalmol. 1975;93:776–778 [DOI] [PubMed] [Google Scholar]
- 15. Ramdas WD, van Koolwijk LM, Ikram MK, et al. A genome-wide association study of optic disc parameters. PLoS Genet. 2010;6:e1000978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Macgregor S, Hewitt AW, Hysi PG, et al. Genome-wide association identifies ATOH7 as a major gene determining human optic disc size. Hum Mol Genet. 2010;19:2716–2724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zangwill LM, Weinreb RN, Beiser JA, et al. Baseline topographic optic disc measurements are associated with the development of primary open-angle glaucoma: the Confocal Scanning Laser Ophthalmoscopy Ancillary Study to the Ocular Hypertension Treatment Study. Arch Ophthalmol. 2005;123:1188–1197 [DOI] [PubMed] [Google Scholar]
- 19. Brown NL, Dagenais SL, Chen CM, Glaser T. Molecular characterization and mapping of ATOH7, a human atonal homolog with a predicted role in retinal ganglion cell development. Mamm Genome. 2002;13:95–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ghanbari H, Seo HC, Fjose A, Brändli AW. Molecular cloning and embryonic expression of Xenopus Six homeobox genes. Mech Dev. 2001;101:271–277 [DOI] [PubMed] [Google Scholar]
- 21. Gallardo ME, Lopez-Rios J, Fernaud-Espinosa I, et al. Genomic cloning and characterization of the human homeobox gene SIX6 reveals a cluster of SIX genes in chromosome 14 and associates SIX6 hemizygosity with bilateral anophthalmia and pituitary anomalies. Genomics. 1999;61:82–91 [DOI] [PubMed] [Google Scholar]
- 22. Nolen LD, Amor D, Haywood A, et al. Deletion at 14q22–23 indicates a contiguous gene syndrome comprising anophthalmia, pituitary hypoplasia, and ear anomalies. Am J Med Genet A. 2006;140:1711–1718 [DOI] [PubMed] [Google Scholar]
- 23. Kirwan RP, Leonard MO, Murphy M, Clark AF, O'Brien CJ. Transforming growth factor-beta-regulated gene transcription and protein expression in human GFAP-negative lamina cribrosa cells. Glia. 2005;52:309–324 [DOI] [PubMed] [Google Scholar]
- 24. Robertson JV, Golesic E, Gauldie J, West-Mays JA. Ocular gene transfer of active TGF-beta induces changes in anterior segment morphology and elevated IOP in rats. Invest Ophthalmol Vis Sci. 2010;51:308–318 [DOI] [PubMed] [Google Scholar]
- 25. Duesing K, Fatemifar G, Charpentier G, et al. Strong association of common variants in the CDKN2A/CDKN2B region with type 2 diabetes in French Europids. Diabetologia. 2008;51:821–826 [DOI] [PubMed] [Google Scholar]
- 26. Moore AF, Jablonski KA, McAteer JB, et al. Diabetes Prevention Program Research Group: extension of type 2 diabetes genome-wide association scan results in the diabetes prevention program. Diabetes. 2008;57:2503–2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ruchat SM, Elks CE, Loos RJ, et al. Association between insulin secretion, insulin sensitivity and type 2 diabetes susceptibility variants identified in genome-wide association studies. Acta Diabetol. 2009;46:217–226 [DOI] [PubMed] [Google Scholar]
- 28. Pasquale LR, Kang JH, Manson JE, Willett WC, Rosner BA, Hankinson SE. Prospective study of type 2 diabetes mellitus and risk of primary open-angle glaucoma in women. Ophthalmology. 2006;113:1081–1086 [DOI] [PubMed] [Google Scholar]