Conditionally immortalized mouse Müller glia respond to growth factor stimulation by forming nonadherent spheres that can be induced to upregulate genes characteristic of differentiated retinal neurons. Culturing in three-dimensional peptide hydrogels alters patterns of gene expression and offers an approach that can be exploited to promote differentiation in vitro.
Abstract
Purpose.
Müller glia in the mammalian retina have some stem cell-like characteristics, although their capacity for neurogenesis remains limited both in vivo and in vitro. In vitro studies to date have used traditional two-dimensional (2D) cell culture to assess neuronal differentiation of Müller glia. The purpose of this study was to compare the effects of 2D and three-dimensional (3D) environments on Müller glial gene expression after growth factor stimulation.
Methods.
Conditionally immortalized mouse Müller glia cells (ImM10) were cultured under nonimmortalizing conditions with EGF/FGF2 to generate spheres that were differentiated in vitro on uncoated culture dishes (2D) or encapsulated in self-assembling, RADA-16 peptide hydrogels (3D) under identical media and growth factor supplementation conditions. Gene expression was analyzed using quantitative RT-PCR and immunocytochemistry. Cellular morphology was analyzed with light and confocal microscopy; sphere ultrastructure was analyzed with transmission electron microscopy.
Results.
ImM10 Müller cells express numerous genes associated with neural stem cells and retinal progenitors in both normal growth conditions and sphere-forming conditions. When encapsulated in the 3D hydrogel, cells can migrate and send processes into the hydrogel. Many genes associated with neurogenesis, as well as retinal neuron–specific genes, are differentially expressed in 2D and 3D differentiation conditions.
Conclusions.
ImM10 Müller glia upregulate genes characteristic of retinal neurons after growth factor stimulation in vitro, and gene expression patterns are altered in 3D hydrogel cultures.
There is increasing interest in developing stem cell–based therapies for retinal disease. Sources for stem-like cells under investigation include embryonic stem cells, induced pluripotent stem cells, and a variety of tissue-specific stem cells including those from bone marrow, embryonic and fetal retina, ciliary body and iris, and Müller glia.1–5 The potential advantages of Müller-derived retinal stem cells include that they arise from the same progenitor pool as retinal neurons, which should bias them toward generation of retinal cell types. Because Müller glia are resident within the retina, they may provide an endogenous stem cell source. Alternatively, Müller glia from adult retinas can be expanded in vitro for transplantation, thereby avoiding ethical concerns associated with the use of embryonic and fetal cells.
In teleost fish, Müller glia are the source of retinal stem cells capable of regenerating all types of retinal neurons.6 In birds7 and, to a lesser extent, in rats8 and mice,9 Müller glia proliferate after retinal injury, and a subset of Müller cells upregulates genes characteristic of retinal neurons. Müller glia in the periphery of the human retina also show some retinal progenitor cell characteristics, including expression of the genes Sox2, Notch1, Chx10, Shh, and Nestin.10 Müller glial dedifferentiation and subsequent neurogenesis can be enhanced in vitro using specific media conditions, supplements, growth factors, and pharmacologic agents.11–15 However, only a fraction of Müller glia in the mammalian retina appear capable of generating cells with neuronal characteristics either in vivo or in vitro.
The mechanisms restricting the neurogenic potential of mammalian Müller glia are not well understood. In the adult retina, an inhibitory environment potentially maintains Müller cells in a differentiated state and represses their stem cell characteristics. Most in vitro studies of retinal stem cells have focused on identification of growth factors and signaling molecules to promote neurosphere formation and differentiation. Less is known about the role of the physical environment on retinal stem cell growth and differentiation. During normal development, proliferation, cell migration, and differentiation, all occur in three dimensions (3D) with cells interacting with neighboring cells and the extracellular matrix. In vitro, cells are typically maintained on flat, two-dimensional (2D) surfaces that lack the 3D architecture of the normal retinal environment. Biocompatible hydrogel polymers can provide a 3D environment for cultured cells and have been shown to promote the proliferation and differentiation of embryonic16 and neural stem cells17 and to enhance neurite outgrowth and synapse formation.18 Polymer scaffolds can also enhance survival, migration, and differentiation of postnatal day (P) 0 to P3 mouse retinal progenitors after transplantation compared with the injection of dissociated cells.19–22
This study assesses the effects of RADA-16 hydrogel scaffolds on the neurogenic response of conditionally immortalized mouse Müller cells (ImM10 cell line23). RADA-16 hydrogels consist of 10-nm diameter peptides of repeating amino acids (arginine [R]-alanine [A]-aspartate [D]-alanine [A]) that self-assemble into nanoscale scaffolds with pores between 5 and 200 nm, similar to the extracellular matrix (ECM) in vivo.17,24,25 RADA-16 hydrogels provide a chemically defined 3D environment that avoids the influence of undefined and potentially variable components including growth factors and structural and adhesive molecules and that eliminates the risk of animal-borne contaminants or disease associated with native ECM preparations.26,27 Using quantitative RT-PCR and immunocytochemistry, we show that genes associated with neurogenesis and differentiation of multiple classes of retinal neurons are differentially expressed by ImM10 Müller glia cultured in 2D vs. 3D conditions after identical growth factor stimulation.
Materials and Methods
ImM10 Cell Line
The isolation and characterization of the conditionally immortalized mouse Müller glial cell line, ImM10, has been previously described.23 ImM10 Müller cells carry a transgene for a temperature-sensitive, interferon-gamma (IFNγ)–inducible SV40 large T antigen (TAg) that immortalizes the cells when maintained at 33°C in the presence of IFNγ.28 Removal of IFNγ from the culture medium stops TAg production, and any residual TAg is inactivated by culturing the cells at or above normal body temperatures, enabling the cells to differentiate. For maintenance, ImM10 cells were cultured in immortalizing conditions in growth media (NeuroBasal medium, 2% fetal bovine serum, B27 supplement, 20 mM l-glutamine, 50U/mL IFNγ [PeproTech; Rocky Hill, NJ], penicillin/streptomycin) at 33°C. Cells used in this study ranged from passage 5 to 16. Cell culture media and supplements were obtained from Gibco/Invitrogen (Carlsbad, CA) or Sigma/Aldrich (St. Louis, MO) unless otherwise noted.
Sphere and Differentiation Cultures
All experiments were performed in nonimmortalizing conditions (39°C, without IFNγ) in defined, serum-free media. To generate spheres, ImM10 cells were harvested by trypsinization and cultured for 7 days in sphere-forming media (NeuroBasal, supplemented with fibroblast growth factor-2 [FGF-2], 20 ng/mL [PeproTech]; epidermal growth factor [EGF], 20 ng/mL [PeproTech]; B27 supplement; l-glutamine, 20 mM; biotin, 100 mg/L; human transferrin, 5000 mg/L; insulin, 500 mg/L; hydrocortisone, 0.36 mg/L; selenite, 0.52 mg/L; and penicillin/streptomycin).
For 2D cultures, spheres were transferred to uncoated 100-mm dishes/plates and grown for 5 days in priming medium (Neurobasal, FGF-2, 20 ng/mL; l-glutamine, 20 mM; biotin, 100 mg/L; transferrin, 5000 mg/L; insulin, 500 mg/L; hydrocortisone, 0.36 mg/L; selenite, 0.52 mg/L; and penicillin/streptomycin). To promote differentiation, priming media were replaced with differentiation media (Neurobasal, B27 supplement, l-glutamine and penicillin/streptomycin) for 5 days.
For sphere formation in 3D cultures, ImM10 cells were harvested, embedded in 0.15% RADA-16 hydrogels (Puramatrix; Becton Dickinson, Franklin Lakes, NJ) at 1 × 105 cells/mL, and cultured in sphere-forming medium for 7 days. For differentiation in 3D cultures, spheres grown in 100-mm dishes for 7 days were embedded in 0.15% RADA-16 hydrogel, cast in hanging transwell inserts with 8-μm pores (Millipore, Billerica, MA) in 24-well plates. RADA-16 hydrogel cures and becomes firm in the presence of physiological salt concentrations (i.e., culture media). Cells encapsulated as spheres were cultured in priming medium for 5 days before transfer to differentiation medium for an additional 5 days.
Sphere Measurements
To assess changes in sphere size, ImM10 cells were plated ∼1 × 104/mL in sphere-forming media in 100-mm plates and incubated for 9 days at 39°C. Wells were imaged in 5 to 10 random fields per plate using a 10× objective on an inverted microscope (Olympus, Center Valley, PA) equipped with a cooled CCD digital camera (Rolera; QImaging, Surrey, BC, Canada). Using image-processing software (Image-Pro; Media Cybernetics, Bethesda, MD), the diameters of nonadherent spheres (≥4 cells) was measured across the longest axis; if two distinct spheres were touching, each sphere was measured separately. Images from four independent experiments were analyzed, with each time point represented by ≥20 spheres from at least one experiment. For statistical analysis, sphere diameters were log converted to achieve a normal distribution before analysis by Pearson correlation (SPSS for Windows 9.0; SPSS, Inc., Chicago, IL), with P ≤ 0.05 considered statistically significant.
RNA Isolation
RNA was isolated from ImM10 cells in 2D cultures using affinity spin columns according to manufacturer's protocol (RNeasy Mini; Qiagen, Valencia, CA). Cells in 3D cultures were removed from the matrix by gentle trituration followed by centrifugation and trypsinization. The cell/matrix mixture was washed and triturated again, cells were pelleted by centrifugation, and RNA was isolated using affinity columns (RNeasyPlus Micro; Qiagen). RNA yields were quantified by spectrophotometry (ND-1000; NanoDrop Products, Wilmington, DE) and RNA quality was assessed by microfluidics gel electrophoresis (RNA Nano LabChip; 2100 Bioanalyzer; Agilent Technologies, Santa Clara, CA). Although the overall RNA yield was lower for cells from 3D cultures, all samples had an RNA integrity number of 9.7 to 10.
Quantitative Reverse Transcriptase PCR
First-strand cDNA was synthesized from total RNA using the oligo dT primers (Stratascript; Agilent, Santa Clara, CA) and was amplified using optimized primers (Supplementary Table S1, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.10-6400/-/DCSupplemental) with cyanine dye chemistry (Brilliant SYBR Green QPCR Master Mix; Stratagene). PCR reactions were performed in triplicate on a thermocycler (MX3005p; Stratagene) using the following cycling conditions: denature at 95°C for 10 minutes; 40 cycles of 95°C for 30 seconds, 60°C for 30 seconds, 72°C for 30 seconds; and a dissociation curve. All qRT-PCR results were normalized to acidic ribosomal phosphoprotein P0 (Rplp0), a reference gene that is stably expressed in the retina.29 Fold-differences in gene expression were calculated relative to control samples (ImM10 cells in growth conditions) using the ΔΔCT method, corrected for amplification efficiency. Statistical analysis was performed using the pairwise fixed reallocation randomization test (REST [relative expression software tool]; http://www.gene-quantification.de/rest.html).30 For each treatment condition, three biological replicates were analyzed.
Neurogenesis and Neural Stem Cell–Targeted PCR Arrays
Targeted neurogenesis and neural stem cell PCR arrays (SABiosciences; Qiagen, Frederick, MD) were used to analyze the expression of 84 genes involved in neurogenesis (see www.sabiosciences.com for array gene list). First-strand cDNA was synthesized, and qRT PCR reactions were performed according to the manufacturer's instructions. Array data were analyzed (RT2 Profiler PCR Array Data Analysis software; SABiosciences) and normalized to three reference genes: glucuronidase β (Gusb), hypoxanthine guanine phosphoribosyl transferase 1 (Hprt1), and heat shock protein 1 β (Hspcb). For each treatment condition, three biological replicates were analyzed.
Immunocytochemistry
2D Cultured Cells.
Immunocytochemistry was performed using previously established protocols.23 Supplementary Table S2, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.10-6400/-/DCSupplemental, lists all primary antibodies and dilutions. Cells were fixed in 4% paraformaldehyde (0.1 M cacodylate buffer, pH 7.4) for 30 minutes. Fixed spheres were embedded in optimal cutting temperature medium (OCT; TissueTek), frozen, and sectioned on a cryostat before immunostaining; differentiated cells were fixed and immunostained in their culture dishes. Fixed cells were incubated in blocking buffer (10% normal goat serum, 0.5% Triton-X, 1% fish gelatin, 5% bovine serum albumin) for 2 hours at room temperature (RT) before application of primary antibodies and overnight incubation at 4°C. Secondary antibodies conjugated to AlexaFluor 488 or AlexaFluor 543 (Molecular Probes, Eugene, OR) were diluted in blocking buffer (1:200) and applied for 1 hour RT. Specificity of labeling was confirmed by omitting primary antibody or substituting normal serum for the species used to generate the primary antibody. Nuclei were counterstained with bis-benzimide (Hoechst 33342; Invitrogen) and imaged by epifluorescence microscopy.
3D Cultures.
Cells were fixed with 4% paraformaldehyde in 0.1 M cacodylate buffer for 1 hour in the hydrogel matrix on transwell inserts, washed in PBS, and incubated in blocking buffer for 16 hours with multiple changes. Blocker was removed, and primary antibodies were applied for 3 days at 4°C to allow full penetration of the antibodies into the hydrogel. Cells were incubated in secondary antibodies for 4 hours at RT, and nuclei were counterstained with 1,5-bis{[(2-di-methylamino) ethyl]amino}-4, 8-dihydroxyanthracene-9,10-dione (Draq-5; Biostatus Limited, Shepshed, Leicestershire, UK). For imaging, the transwell insert containing the matrix-encapsulated cells was placed directly onto a coverslip, with mounting medium (Prolong Gold; Molecular Probes, Eugene, OR) applied to both sides of the matrix to retard bleaching of the fluorescent labels and were imaged using confocal microscopy.
Phalloidin Labeling
RADA-16 encapsulated cells were fixed in 4% paraformaldehyde in 0.1 M cacodylate buffer for 1 hour and incubated overnight with rhodamine-conjugated phalloidin (1:300) at room temperature (Invitrogen). Cells were washed, nuclei were counterstained (Draq-5; Biostatus Limited), and encapsulated cells within the matrix were imaged using confocal microscopy.
Confocal Microscopy
Cells in 3D cultures were imaged by confocal microscopy (TCS-SP2; Leica Microsystems, Exton, PA). Bleed-through between fluorescence channels was eliminated by scanning channels sequentially. Images were captured with a 63× oil immersion objective and frame averaged to reduce noise. In all cases, image scale was calibrated, and brightness and contrast were adjusted as necessary to highlight specific labeling.
Electron Microscopy
Nonadherent spheres were pelleted and fixed in 0.1 M sodium cacodylate buffer, pH 7.2, containing 2.5% glutaraldehyde for 2 hours and postfixed in 5% tannic acid for 5 minutes, followed by 1% osmium tetroxide. Spheres were dehydrated in a graded alcohol series and embedded in Araldite resin. Thin sections (80–100 nm) were cut on an ultramicrotome and imaged by transmission electron microscopy (Tecnai Spirit; FEI, Hillsboro, OR).
Live/Dead Stain
Cell viability was determined by staining with calcein AM (10 μM), ethidium bromide homodimer 1 (10 μM; Live/Dead; Invitrogen) and bis-benzimide (Hoechst 33342; 1:2000; Invitrogen) for 30 minutes, followed by PBS washes before imaging using epifluorescence illumination.
Results
Sphere Formation and Gene Expression by ImM10 Müller Cells
We have previously characterized the conditionally immortalized ImM10 Müller glial cell line.23 When cultured under immortalizing, growth media conditions, ImM10 Müller cells adhered to the dish, extended processes and proliferated (Figs. 1A–D). When cultured under nonimmortalizing conditions in sphere-forming medium containing EGF and FGF-2, ImM10 cells detached from the dish and formed nonadherent spheres that increased in size over time (Figs. 1E–K, 1Q). Although the sphere size varied at all time points analyzed, there was a statistically significant correlation between sphere diameter and days in culture (Pearson correlation = 0.425; P < 0.001). Although other groups have demonstrated the formation of spheres after exogenous growth factor stimulation of cultured Müller cells,12–15 the ultrastructure of Müller glial-derived spheres has not been previously examined. Electron microscopy of spheres revealed the presence of junctional complexes, including numerous desmosomes (Figs. 1M, 1N, arrowheads) and gap junctions (Fig. 1O, bracket, enlarged in 1P). The desmosomes (arrowheads, Figs. 1M, 1N) showed the typical gap between the plasma membranes of the juxtaposed cells with underlying electron-dense plaques in the cytoplasm. In contrast, the gap junctions were characterized by fused membranes with a pentilaminar structure that resulted from fusion of the two phospholipid bilayers (Fig. 1P).
Figure 1.
ImM10 Müller cell growth and morphology in growth media and sphere-forming cultures. (A–D) Growth of ImM10 cells in immortalizing growth media conditions with IFNγ at 33°C over 6 days in culture. (E–K) Parallel cultures of ImM10 cells plated at the same density under nonimmortalizing conditions (39°C, without IFNγ) in serum-free sphere-forming media containing EGF and FGF2. (A–K) Number in lower right corner indicates the days after plating. Scale bars, 100 μm (A–K). (L) Low-magnification electron micrograph of a neurospheres at 7 days (N, nucleus). (M, N) High-magnification electron micrograph showing the presence of numerous desmosomes within the neurospheres (arrowheads). (O) Gap junctions are also present within the neurospheres (arrow/bracket). Arrowheads: condensed chromatin adjacent to the nuclear envelope. (P) High-magnification micrograph of the gap junction bracketed in (O), showing the pentilaminar structure typical of gap junctions. Arrow: central electron-dense, central line that consists of the juxtaposed head groups of the phospholipids within the outer leaflets of the plasma membranes of the adjacent cells. (Q) Box plots: changes in sphere diameter over time in culture. Gray boxes: interquartile range, with the median indicated by the horizontal bar. Whiskers: range of the data with outliers indicated by open circles and extreme values by asterisk. n, number of spheres measured at each time point. y-axis, sphere diameter, plotted on a log scale.
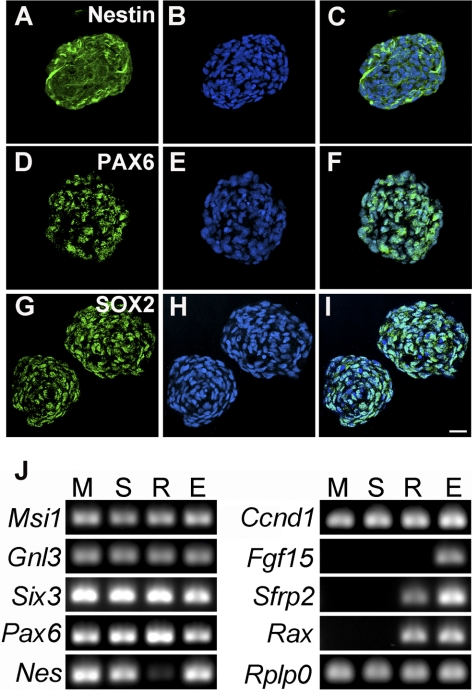
Consistent with previous studies,14,15 most cells in spheres generated from EGF/FGF2-treated ImM10 cells expressed nestin, SOX2, and PAX6 (Figs. 2A–I). By RT-PCR, sphere cells also expressed many of the genes characteristically coexpressed in retinal progenitor cells, including Msi1, Gnl3, Pax6, Nestin, Ccnd1, and Six3 (Fig. 2J). In contrast, Fgf15, Sfrp2, and Rax were not expressed. qPCR showed that Nestin was significantly downregulated in spheres (fold change = −8.04; P = 0.025 vs. growth conditions). Compared with ImM10 cells in immortalizing growth conditions, there were no statistically significant changes in mRNA expression in sphere cultures for Pax6 (fold change = −1.64; P = 0.726), Notch1 (fold change = 1.75; P = 0.759), Msi1 (fold change = 1.51; P = 0.967), Hes1 (fold change = 1.25; P = 0.991), or Gnl3 (fold change = −1.27; P = 0.962).
Figure 2.
Expression of stem cell genes in sphere cultures of ImM10 Müller cells. (A–I) Immunolabeled Müller glia–derived neurospheres. (A) There is broad immunoreactivity for Nestin in cells within the neurospheres. (B) Hoechst labeling of all nuclei. (C) Merged image of (A) and (B). (D) PAX6 immunoreactivity in the nuclei of sphere cells. (E) Hoechst labeling. (F) Merge of (D) and (E) showing expression of PAX6 in most cells. (G) SOX2 immunoreactivity in the nuclei of sphere cells. (H) Hoechst labeling. (I) Merge of (G) and (H) showing expression of SOX2 in most cells. Scale bar, 50 μm (A–I). (J) RT-PCR reveals expression of many, but not all, genes associated with neural stem cells and retinal progenitors in both neurospheres (S) and untreated Müller cells (M). Adult C57BL/6 retina (R) and E12.5 whole embryo (E) served as controls. The following genes were analyzed: musashi 1 (Msi1), nucleostemin (guanine nucleotide binding protein-like 3 [Gnl3]), sine oculis-related homeobox homolog 3 (Six3), paired box gene 6 (Pax6), nestin (Nes), cyclin D1 (Ccnd1), fibroblast growth factor 15 (Fgf15), secreted frizzled related protein 2 (Sfrp2), retinal and anterior neural fold homeobox (Rax). Acidic ribosomal phosphoprotein P0 (Rplp0) was used as normalizing gene.
Morphology and Gene Expression in 2D Differentiation Conditions
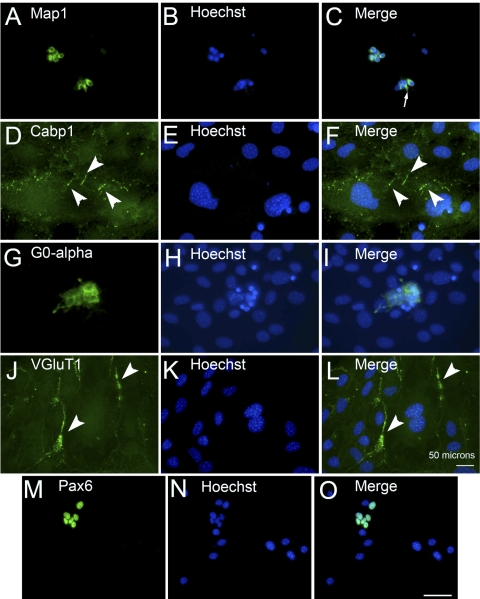
After sphere formation, cells were transferred to FGF-2 containing priming medium for 5 days and then to differentiation medium for 5 days (Fig. 3A) based on the culture conditions described by Merhi-Soussi and colleagues.31 Spheres in priming medium readhered to the dish, and individual cells migrated out (Fig. 3C, arrowhead). After subsequent incubation in differentiation medium for 5 days (2D conditions), a subset of cells acquired a nonglial morphology, characterized by small cell bodies, compact nuclei with dense chromatin, and thin, branching neurite-like processes (Fig. 3D, arrowheads). Under differentiation conditions, some cells upregulated the expression of general neuronal and retinal cell-specific markers. MAP1, a marker of differentiated neurons that is expressed by retinal ganglion cells, was present in a small number of differentiated ImM10 cells that often appeared in clusters (Figs. 4A–C). CaBP1, expressed in OFF cone bipolars and subsets of amacrine cells,32 showed diffuse cytoplasmic immunostaining in cells that retained a glial morphology (Fig. 4D). In addition, robust CaBP1 immunoreactivity was visible in distinctly linear structures within the cytoplasm, potentially associated with the cytoskeleton (Figs. 4D–F, arrowheads). In differentiation conditions, small clusters of cells with small nuclei were robustly stained by antibodies against Goα (Figs. 4G–I), a gene expressed in rod bipolar cells and ON cone bipolar cells.33 Some cells with diffusely stained nuclei cells expressed vesicular glutamate transporter 1 (VGluT1), which often appeared to be localized to the membranes between juxtaposed cells (Figs. 4J–L, arrowheads). Under both immortalizing growth conditions23 and nonimmortalizing, sphere-forming conditions (Fig. 3), all ImM10 cells expressed PAX6. In contrast, under differentiation conditions, only a subset of cells continued to express PAX6 (Figs. 4M–O).
Figure 3.
Morphology of ImM10 cells in 2D and 3D Differentiation cultures. (A) Experimental design for differentiation cultures in 2D and 3D cultures. (B–D) Morphology of cells in 2D differentiation cultures. (E–G) Morphology of cells in 3D differentiation cultures. (B) DIC image of cells in sphere-forming media for 7 days. (C) After transfer to priming media, spheres adhere to the dish and cells begin to migrate out (arrowhead). (D) After 5 days in differentiation conditions, some cells acquire a more neuronal morphology and extend thin, neurite-like processes (arrowheads). (E) One day after encapsulation in the 3D matrix and culturing in priming media conditions, cells at the margins of the spheres extend processes (arrowhead). (F) After 5 days in priming media, many cells have migrated from the neurospheres (arrowheads). (G) After 5 days in differentiation conditions, most, but not all, cells have migrated from the neurospheres (arrowheads). Scale bars, 100 μm (B–G).
Figure 4.
Immunostaining of ImM10 cells in 2D differentiation cultures. (A) Immunolabeling reveals small subsets of cells that express the neuronal gene MAP1. (B) Hoechst labeling. (C) Merge of (A) and (B). (D) Subsets of cells immunopositive for CaBP1 (arrowheads), a gene expressed in OFF cone bipolar cells and subsets of amacrine cells. (E) Hoechst labeling. (F) Merge of (D) and (E). (G) Subsets of cells immunopositive for Goα, a gene expressed in rod bipolar cells and ON cone bipolars. (H) Hoechst labeling. (I) Merge of (G) and (H). (J) Subsets of cells immunopositive for VGLUT1. Intense VGluT1 immunoreactivity is localized at the plasma membranes of juxtaposed cells (arrowheads). (K) Hoechst labeling. (L) Merge of (J) and (K). (A–L) Scale bar (L), 50 μm. (M) Subsets of cells express PAX6. (N) Hoechst labeling. (O) Merge of (M) and (N). (M–O) Scale bar (O), 50 μm.
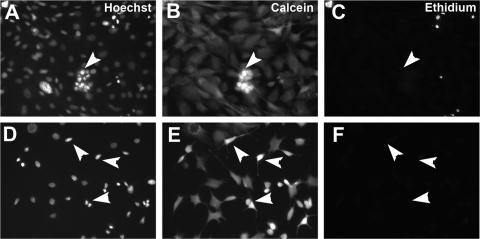
Many of the cells expressing differentiated neuronal markers had small nuclei with intensely stained chromatin (Figs. 4B, 4G). To determine whether these were pyknotic cells, differentiation cultures were stained using calcein AM to label live cells, ethidium bromide homodimer to label dead cells, and bis-benzimide (Hoechst 33342) to label all nuclei, both live and dead. Whereas scattered ethidium-positive nuclei were detected under these conditions, the majority of cells were calcein AM positive and ethidium negative live cells (Fig. 5). Cells with typical glial morphologies (large, flat cells with large nuclei containing diffusely stained chromatin; Figs. 5A–D, arrowheads) and those with nonglial morphologies (small cell bodies with thin processes and small nuclei containing densely stained chromatin [Figs. 5E–H, arrowheads]), were calcein AM positive and ethidium bromide negative.
Figure 5.
Live/Dead assays of ImM10 cells after 2D differentiation conditions. (A) Hoechst staining labels all nuclei present, both live and dead. In 2D differentiation conditions, cells expressing neuronal genes often appear in clusters of cells with small, brightly staining nuclei (arrowhead). (B) Calcein AM labels all live cells in the dish, including the cluster of cells identified in (A) (arrowhead). (C) Ethidium bromide homodimer 1 labels all dead cells. Although some dead cells are present, the cluster of cells identified in (A) is not labeled with ethidium (arrowhead). (D) Hoechst staining showing that smaller, brightly staining nuclei do not always appear in clusters (arrowheads). (E) Cells identified in (D) are calcein AM–positive live cells (arrowheads). (F) Cells identified in (D) are not labeled with ethidium (arrowheads). Scale bar, 100 μm.
Morphology in 3D Cultures
DIC imaging of live cells at the light microscope level allowed gross examination of cell morphology and migration in 3D culture during differentiation conditions (Figs. 3E–G). To examine the morphology, process extension, and cytoskeletal arrangement of cells within the hydrogel, rhodamine-conjugated phalloidin was used to stain F-actin. ImM10 cells encapsulated within the RADA-16 hydrogels and grown for 2 days with growth media (immortalizing conditions) extended processes into the matrix (Figs. 6A, 6B). When ImM10 cells were cast as single cells in RADA-16 hydrogels and then cultured in sphere-forming media containing EGF and FGF2 under nonimmortalizing conditions, a large number of cells formed spheres (Figs. 6C, 6D). For differentiation experiments, ImM10 spheres were encapsulated within the RADA-16 hydrogel and cultured in FGF-2 containing priming media for 5 days, followed by differentiation media for 5 days. At 1 day after encapsulation, cells at the margins of the spheres began to extend processes (Fig. 3E, arrowhead); after 5 days, many cells had migrated from the sphere (Fig. 3F, arrowheads). By day 10, most, but not all, cells had migrated from the neurospheres (Fig. 3G, arrowheads).
Figure 6.
Morphology and gene expression in 3D differentiation cultures. (A–D) Rhodamine-conjugated phalloidin labels cytoskeletal F-actin. (A) ImM10 cells encapsulated in a 3D hydrogel and grown in growth media for 5 days send processes into the matrix. (B) High-magnification image of a process extending into the 3D matrix. (C) Many, but not all, ImM10 cells encapsulated within the 3D matrix as dissociated cells form spheres in EGF/FGF2 containing sphere-forming media for 7 days. (D) Same image as (C), merged with Draq5 nuclear staining. Scale bars: 100 μm (A, C, D); 20 μm (B). (E) RT-PCR analysis of gene expression in ImM10 cells in different culture conditions. U, untreated Müller cells in growth media conditions; S, spheres; 2D, cells that were primed and differentiated in 2D conditions; 3D, cells that were primed and differentiated in 3D conditions. Recoverin (Rcvrn), melanopsin (Opn4), and calcium-binding protein 5 (Cabp5) are expressed only in 2D differentiation conditions. M-cone opsin (Opn1mw) is expressed only in differentiation conditions and is upregulated in 3D conditions. Rhodopsin kinase (Rhok), cone-rod homeobox (Crx), and metabotropic glutamate receptor 6 (Grm6, commonly known as mGluR6) are expressed only in 3D differentiation conditions. (F–K) Immunostaining of cells in 3D differentiation cultures. (F) Many cells in 3D differentiation conditions express the neuronal marker MAP1. (G) Draq5 labels all nuclei. (H) Merge of (F) and (G). (I) Cells in 3D differentiation conditions express the photoreceptor gene CRX. (J) Draq5 labeling. (K) Merge of (H) and (I) shows some nuclear localization of CRX immunoreactivity (arrows), though significant staining is cytoplasmic. Scale bar, 100 μm (F–K).
Gene Expression in 2D and 3D Differentiation Cultures
ImM10 Müller cells grown under nonimmortalizing, differentiation conditions in 2D and 3D cultures showed significant changes in expression of multiple genes characteristically expressed in neural stem cells or during neurogenesis compared with ImM10 Müller cells cultured in growth media under immortalizing conditions (Table 1). Many of these genes were differentially expressed between 2D and 3D differentiation conditions. The most highly upregulated genes in 2D cultures were apolipoprotein E (Apoe; 14.27-fold), neuronal pentraxin 1 (Nptx1; 13.2-fold), fibroblast growth factor 2 (Fgf2; 6.64-fold), and brain-specific angiogenesis inhibitor 1 (Bai1; 5.47-fold). Some genes were expressed only in 2D differentiation conditions, but not in any other treatment conditions or growth media conditions; therefore, a fold-change could not be calculated. These included odd Oz/tenascin-m homolog 1 (Odz1), hairy/enhancer-of-split related with YRPW motif-like protein (Heyl), bone morphogenetic protein 15 (Bmp15), and SRY (sex-determining region Y) box 3 (Sox3). Genes that were expressed in all conditions but were significantly upregulated only in 2D differentiation conditions included Apoe, Bai1, ionotropic glutamate (NMDA) receptor 1 (Grin1), amyloid beta A4 precursor protein-binding family B member 2 (Apbb2), paired box gene 6 (Pax6), midkine (Mdk), and Egf.
Table 1.
Genes with Statistically Significant Changes in Gene Expression in 2D or 3D Differentiation Cultures Compared with ImM10 Cells in Growth Conditions Using Neurogenesis qRT-PCR Arrays
| Gene | Accession No. | 2D Fold Change | P | 3D Fold Change | P |
|---|---|---|---|---|---|
| Apbb1 | NM_009685 | 3.3 | 0.002 | + | NS |
| Apoe | NM_009696 | 14.27 | 0.015 | + | NS |
| Arnt2 | NM_007488 | + | NS | −2.25 | 0.05 |
| Bai1 | NM_174991 | 5.47 | 0.03 | + | NS |
| Bdnf | NM_007540 | + | NS | −2.32 | 0.049 |
| Bmp15 | NM_009757 | ++ | — | ND | — |
| Bmp2 | NM_007553 | ND | — | ++ | — |
| Cdk5rap2 | NM_145990 | −1.66 | 0.003 | + | NS |
| Cdk5rap3 | NM_030248 | 1.56 | 0.002 | + | NS |
| Cxcl1 | NM_008176 | −1.63 | 0.029 | 6.9 | 0.006 |
| Dlg4 | NM_007864 | 2.76 | 0.003 | 1.49 | 0.01 |
| Dll1 | NM_007865 | + | NS | 5.87 | 0.032 |
| Egf | NM_010113 | 2.01 | 0.041 | −3.63 | 0.006 |
| Erbb2 | NM_001003817 | 1.35 | 0.041 | + | NS |
| Fez1 | NM_183171 | 2.69 | 0.011 | 3.27 | 0.001 |
| Fgf2 | NM_008006 | 6.64 | 0.006 | 6.03 | 0.012 |
| Flna | NM_010227 | + | NS | −2.96 | 0.029 |
| Gdnf | NM_010275 | −8.84 | 0.009 | + | NS |
| Gnao1 | NM_010308 | 2.82 | 0.002 | 2.48 | 0.003 |
| Grin1 | NM_008169 | 3.58 | 0.035 | + | NS |
| Hey1 | NM_010423 | + | NS | −8 | 0.015 |
| Heyl | NM_013905 | ++ | — | ND | — |
| Mdk | NM_010784 | 2.1 | 0.025 | + | NS |
| Mef2c | NM_025282 | ND | — | ++ | — |
| Notch2 | NM_010928 | 2.41 | 0.023 | 3.55 | 0.022 |
| Nptx1 | NM_008730 | 13.2 | 0.019 | 165.4 | 0.028 |
| Nrcam | NM_176930 | ++ | — | ++ | — |
| Nrp1 | NM_008737 | 2.39 | 0.004 | 2.14 | 0.004 |
| Ntn1 | NM_008744 | + | NS | −7.36 | 0.03 |
| Odz1 | NM_011855 | ++ | — | ND | — |
| Pard6b | NM_021409 | 1.49 | 0.024 | + | NS |
| Pax6 | NM_013627 | 2.88 | 0.009 | + | NS |
| Ptn | NM_008973 | + | NS | 3.93 | 0.008 |
| Sox3 | NM_009237 | ++ | — | ND | — |
+, expressed; NS, no significant change vs. growth conditions; ++, expressed in differentiation conditions only, not growth, thus no fold change calculated; —, no P can be calculated for genes not expressed in ImM10 growth cultures; ND, not detected.
In 3D conditions, the most highly upregulated genes were Nptx1 (165.4-fold), chemokine (C-X-C motif) ligand 1 (Cxcl1; 6.9-fold), Fgf2 (6.03-fold), and delta-like 1 (Dll1; 5.87-fold). Myocyte enhancer factor 2C (Mef2C) and bone morphogenic protein 2 (Bmp2) were expressed only in 3D conditions, whereas delta-like 1 (Dll1) and pleiotrophin (Ptn) were expressed in all cells but upregulated only in 3D differentiation conditions.
Although many genes were differentially expressed in 2D and 3D differentiation conditions, several genes were upregulated in both conditions. These genes included Nrcam, the neuronal cellular adhesion molecule involved in axonal pathfinding,34 which was detected only in differentiation conditions. Nptx1, a gene involved in synaptic refinement,35 was upregulated in both but was more robustly increased in 3D conditions. Other genes were upregulated to similar levels in both 2D and 3D cultures, including Fgf2, guanine nucleotide-binding protein G(o) subunit alpha (Gnao1), disks large homolog 4 (Dlg4, a postsynaptic scaffolding gene that is expressed in neuronal processes of the retina36), fasciculation and elongation protein ζ-1 (Fez1, a gene involved in axon outgrowth and fasciculation37), Notch2, and neuropilin-1 (Nrp1, a coreceptor for class 3 semaphorins that is essential for neural development and axon guidance38,39).
Only a few genes were downregulated, and there was no overlap in the genes that were significantly downregulated in 2D and 3D differentiation conditions. Genes downregulated in 2D conditions included glial-derived neurotrophic factor (Gdnf), cyclin-dependent kinase 5 regulatory subunit associated protein 2 (Cdk5rap2), and chemokine (C-X-C motif) ligand 1 (Cxcl1). Genes downregulated in 3D conditions were hairy/enhancer-of-split related with YRPW motif protein 1 (Hey1), netrin-1 (Ntn1), epidermal growth factor (Egf), filamin-A (Flna), brain derived neurotrophic factor (Bdnf), and aryl hydrocarbon receptor nuclear translocator 2 (Arnt2).
We also compared changes in mRNA expression of a panel of retinal neuron-specific genes in ImM10 cells in growth, sphere, and differentiation cultures (2D and 3D) using RT-PCR (Fig. 6E). With the exception of recoverin (Rcvrn), which was also expressed at low levels in growth and sphere cultures, genes characteristic of differentiated retinal neurons were detected only in differentiation cultures and some were differentially expressed in 2D and 3D cultures. By qRT-PCR, melanopsin (Opn4), and calcium binding protein 5 (Cabp5), rhodopsin kinase (Rhok), cone-rod homeobox (Crx), and mGluR6 (Grm6) were expressed only in 3D differentiation conditions (Fig. 6E). By immunostaining, Goα, VGluT1, and CaBP1 were expressed in 2D differentiation conditions (Fig. 4) but were not detected in 3D conditions (data not shown). M-cone opsin (Opn1mw) was expressed at low levels in 2D differentiation conditions but was highly upregulated in 3D conditions (Fig. 6E). Using immunocytochemistry, MAP1 was expressed by a large number of cells in 3D conditions (Figs. 6F–H). CRX was expressed only in 3D conditions (Figs. 6I–K), although only a fraction of the immunoreactivity in each cell appeared to be localized to the nuclei.
Discussion
Our results demonstrate that ImM10 conditionally immortalized Müller cells have some characteristics of retinal stem cells, in that they express many genes associated with neuronal stem cells and retinal progenitors and, after FGF2 and EGF growth factor stimulation, generate spheres that are morphologically similar to the neurospheres generated in culture by neural stem cells,40,41 retinal progenitors, and Müller glia.13,14,42 Ultrastructural analysis of ImM10-derived spheres revealed the presence of numerous macula adherens and gap junctions. Gap junctions are critical in neurosphere formation and in the dedifferentiation of embryonic stem cells43 and cortical neural progenitors44 and could play a role in the generation of Müller glia-derived spheres. When cultured in sphere-forming conditions, ImM10 cells expressed many genes typically associated with neural stem cells and retinal progenitors, including Nestin, Sox2, Six3, Msi1, Pax6, Gnl3, and Ccnd1.45–49 Interestingly, these genes are also expressed by ImM10 cells cultured in immortalizing growth conditions (Fig. 2J). Further, with the exception of nestin, we found no statistically significantly differences in mRNA expression of many genes typically coexpressed in retinal progenitors between growth and sphere cultures using quantitative RT-PCR. Gene expression in the ImM10 cell line is consistent with expression patterns in primary mouse Müller glia determined using serial analysis of gene expression,46 single-cell microarray analysis,50 and immunocytochemistry.10,51,52 Our results differ somewhat from reports of gene expression in other Müller glial cell lines. Spontaneously immortalized human Müller glia express Pax6, Sox2, Chx10, and Notch1 mRNA under growth conditions (DMEM + 10% fetal calf serum), although only a fraction of cells are PAX6 immunopositive.14 Rat Müller cells cultured in DMEM/F12 with N2 supplement, EGF, and FGF2 generate spheres that robustly express SOX2, Nestin, and Musashi-1; however, expression was not reported for the same cells cultured in adherent growth conditions.15 Therefore, it is unclear whether sphere conditions specifically upregulate retinal stem cell genes in rat Müller glia or simply maintain expression of genes already transcribed in these cells.
Using identical media and growth factor stimulation on parallel cultures, we found that induction of neuronal gene expression is differentially regulated by growth on 2D versus 3D substrates. Mdk, a heparin-binding growth factor that is important in neurogenesis, was expressed in all conditions but was upregulated only in 2D cultures. In zebrafish, mdk-a and mdk-b are expressed in retinal progenitors and upregulated in proliferating Müller glia during the early stages of retinal regeneration.53 Sox3 also regulates neurogenesis and neuronal differentiation in zebrafish54 and was expressed only in 2D conditions. In contrast, Mef2c was robustly expressed only in 3D conditions. Mef2c is involved in early neuronal differentiation, and mouse embryonic stem cells that express a constitutively active form of Mef2c differentiate into a nearly pure population of neurons.55
Consistent with the morphologic changes we observed in differentiation cultures, some genes associated with axon outgrowth, guidance, and synapse formation were also significantly upregulated in both 2D and 3D cultures (compared with growth conditions) consistent with a progression from neurogenesis toward differentiation. Among these, Bmp2 and Ptn are involved in dendrite formation and neurite outgrowth56–58 and were upregulated only in 3D conditions. Nptx1, a gene involved in synaptic refinement,35 was upregulated in 2D conditions, but was more robustly upregulated in 3D conditions. APOE is synthesized by cultured Müller cells and is secreted into the vitreous by Müller cells in vivo59 and was expressed in all conditions, though it was upregulated only in 2D differentiation cultures. Apart from its well-described role in cholesterol transport, APOE modulates long-term potentiation in the hippocampus through its involvement in NMDA and AMPA receptor regulation.60 Grin1, the ionotropic glutamate (NMDA) receptor 1, was also upregulated in 2D conditions. In the rodent retina, NMDA receptor subunits are expressed primarily on inner retinal amacrine and ganglion cells.61
In addition to changes in gene expression associated with neural stem cells and neurogenesis genes in general, several genes specific to retinal neurons were expressed in differentiation conditions. The cone-rod homeobox transcription factor (Crx) is critical to the development of photoreceptors62 and was detected in only cells in 3D cultures using both RT-PCR and immunohistochemistry. Unexpectedly, CRX immunoreactivity in 3D cultures was both nuclear and cytoplasmic. Because antibody penetration and subsequent removal of unbound antibodies from the 3D matrix was more difficult than in 2D cultures, one possible explanation for the cytoplasmic staining is increased nonspecific staining. We cannot exclude the possibility that at least some of the immunoreactivity reflects mislocalization of the CRX protein. Other studies of in vitro differentiation of retinal stem cells and Müller glia have shown aberrant cytoplasmic localization of proteins including Chx1014 and Hes1.12
Some Crx regulatory targets were expressed in 2D cultures in the absence of detectible Crx expression. Recoverin is a calcium binding protein expressed in photoreceptors that is involved in the inhibition of rhodopsin kinase and the adaptation to light.63 Recoverin was expressed at low levels by ImM10 in all conditions but was robustly upregulated in 2D differentiation conditions. Cells in both 2D and 3D conditions expressed the transcripts for m-cone opsin, but it was more strongly upregulated in 3D conditions. Upregulation of recoverin in 2D cultures in the absence of Crx expression most likely reflects positive regulation by other transcriptional regulatory genes because, in the retinas of Crx null mice, recoverin expression is downregulated but still detectable.64 Although no m-opsin expression is detected in Crx null retinas by serial analysis of gene expression,65 the nuclear receptor TRbeta2 is a regulator of M-opsin.66 Therefore, the expression of TRbeta2 or other positive regulators of m-opsin could contribute to low levels of transcription.
Our culture conditions were modeled on a previously published study in which FGF2 priming followed by B27 differentiation resulted in the upregulation of photoreceptor markers in approximately 30% of retinal progenitors cultured from the neonatal (P0-P2) mouse retina.31 We observed the upregulation of some genes characteristic of rod photoreceptors in both 2D and 3D cultures but did not observe cells with the distinct morphology of photoreceptors. Further, although markers for multiple classes of retinal neurons were expressed, there was no obvious bias toward gene expression patterns associated with any single retinal cell type in either 2D or 3D conditions. Variability in the neuronal genes that are upregulated in Müller glia in vivo after retinal injury has also been reported. Injection of neurotoxic agents or the gliotoxic α-aminoadipate results in the upregulation of genes characteristic of late-born cell types (photoreceptor and bipolar cells) in a subset of Müller glia.8,11,67 In contrast, after intraocular NMDA injection, some Müller cells acquired amacrine cell phenotypes suggesting that Müller cells can also generate early-born retinal cell types.9 Consistent with this, ImM10 cells in 2D cultures upregulated melanopsin, which is expressed in a subset of retinal ganglion cells. The more robust generation of rod photoreceptors in vitro from neonatal retinal progenitors may reflect an intrinsic bias in progenitors isolated from a developmental stage when they are actively generating rod photoreceptors in vivo.
Mammalian Müller glia can upregulate genes characteristic of retinal neurons after injury in vivo or growth factor stimulation in vitro. However, in all studies published to date, only a subset of Müller glia adopt a neuronal-like morphology. ImM10 cells also showed a persistence of glial morphologies and a relatively low frequency of differentiation. We also found that some of the cells that upregulated neuronal genes, such as CaBP1 and VGluT1, retained a distinctly glial morphology. This is consistent with reports showing that after in vitro differentiation of human Müller glia, protein kinase C immunoreactivity was present in cells with a distinctly glial morphology.14 It remains to be determined whether the changes in gene expression observed in Müller glia in vitro represent aberrant expression of neuronal genes in glial cells or incomplete differentiation into neurons.
To our knowledge, there are no previous studies examining the effect of 3D substrates on Müller glia. We found that the general behavior of ImM10 Müller glia in RADA-16 hydrogels was similar to those cultured on tissue culture plastic. When embedded as single cells and treated with EGF/FGF2 sphere-forming medium, ImM10 cells generated spheres within the gels. When embedded as spheres and cultured in priming and differentiation conditions, cells would often completely migrate from the spheres and extend processes, similar to their behavior in 2D. Other researchers have shown that retinal progenitor cells isolated from the perinatal mouse retina can migrate into the network of pores within biodegradable scaffolds made of poly lactic-co-glycolic acid (PLGA) or poly-L-lactic acid (PLLA).19,20 In contrast, in preliminary studies using a thiol-modified sodium hyaluronate and gelatin-based hydrogel (Extracel; Glycosan Biosystems, Salt Lake City, UT), we observed minimal proliferation, migration, or process extension (data not shown). Similarly, when seeded onto glycerol sebacate polymers with precast 50-μm pores, most perinatal retinal progenitors remained within the pores and did not migrate into the dense surrounding polymer.22 Retinal progenitors cultured on micropatterned polycaprolactone films failed to form clusters and retained a circular or glial morphology, despite increased expression of recoverin and rhodopsin.68 Perinatal retinal progenitors cultured on PLGA/PLLA gels and then subsequently transplanted subretinally into rhodopsin null mice showed increased survival and expression of recoverin compared with cells transplanted without the gels. Differential migration likely reflects the physical and chemical structures of each type of polymer as well as differences in culture conditions and the types of cells.
It is notable that the RADA-16 hydrogels used in our studies were not modified to add specific adhesion molecules, attachment motifs, or growth factors, and both 2D and 3D cultures used identical culture conditions. Thus, the changes in gene expression can be attributed to the presence of the 3D matrix. Because largely unique sets of neuronal genes were upregulated in 2D and 3D cultures, we cannot conclude that one method is superior to the other for promoting neuronal differentiation of Müller cells. Future studies comparing the effects of different matrices in combination with specific modifications will be necessary to identify the optimal substrate to enhance the differentiation and promote the generation of specific retinal cell types. The ability to cast cells into the RADA-16 hydrogel could offer advantages for generating sheets of cells for transplantation, and our finding that cells can readily migrate within the gels would permit transplanted cells to integrate within the host tissue. For tissue engineering purposes, it may ultimately prove useful to develop combination scaffolds that incorporate patterned structures and chemical modifications to direct cell growth and provide specific growth factors or cell adhesion molecules.
Our results add to the mounting evidence that Müller glia express many genes characteristic of retinal progenitors and support the proposal that adult Müller glia could function as retinal progenitors or stem cells. However, the low numbers of differentiated cells that have thus far been generated from mammalian Müller glia, either in vitro or in vivo, remain insufficient for therapeutic applications. Development of cell replacement therapies will require additional research to identify and overcome the restrictions that prevent mammalian Müller glia from achieving the robust regenerative ability they have in lower vertebrates.
Acknowledgments
The authors thank Francoise Haeseleer (University of Washington, Seattle, WA) for the generous gift of CaBP1 antibody, Alan Burns (College of Optometry, University of Houston, Houston, TX) for guidance on the electron microscopy, and Evelyn Brown, Mary Guirguis, and Laila Pillai for expert technical assistance.
Footnotes
Presented in part at the annual meeting of the Association for Research in Vision and Ophthalmology, Fort Lauderdale, Florida, April 2008 and May 2009.
Supported in part by National Institutes of Health/National Eye Institute Grant T32 EY07024 and Ezell Fellowship; the American Optometric Foundation (MJP); a New Faculty Grant from University of Houston; and unrestricted funds from the University of Houston, College of Optometry (DCO) and EY07751-CORE.
Disclosure: M.J. Phillips, None; D.C. Otteson, None
References
- 1. Baker PS, Brown GC. Stem-cell therapy in retinal disease. Curr Opin Ophthalmol. 2009;20:175–181 [DOI] [PubMed] [Google Scholar]
- 2. Limb GA, Daniels JT. Ocular regeneration by stem cells: present status and future prospects. Br Med Bull. 2008;85:47–61 [DOI] [PubMed] [Google Scholar]
- 3. Enzmann V, Yolcu E, Kaplan HJ, Ildstad ST. Stem cells as tools in regenerative therapy for retinal degeneration. Arch Ophthalmol. 2009;127:563–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ohta K, Ito A, Tanaka H. Neuronal stem/progenitor cells in the vertebrate eye. Dev Growth Differ. 2008;50:253–259 [DOI] [PubMed] [Google Scholar]
- 5. Amabile G, Meissner A. Induced pluripotent stem cells: current progress and potential for regenerative medicine. Trends Mol Med. 2009;15:59–68 [DOI] [PubMed] [Google Scholar]
- 6. Bernardos RL, Barthel LK, Meyers JR, Raymond PA. Late-stage neuronal progenitors in the retina are radial Müller glia that function as retinal stem cells. J Neurosci. 2007;27:7028–7040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Reh TA, Fischer AJ. Stem cells in the vertebrate retina. Brain Behav Evol. 2001;58:296–305 [DOI] [PubMed] [Google Scholar]
- 8. Ooto S, Akagi T, Kageyama R, et al. Potential for neural regeneration after neurotoxic injury in the adult mammalian retina. Proc Natl Acad Sci U S A. 2004;101:13654–13659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Karl MO, Hayes S, Nelson BR, Tan K, Buckingham B, Reh TA. Stimulation of neural regeneration in the mouse retina. Proc Natl Acad Sci U S A. 2008;105:19508–19513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bhatia B, Singhal S, Lawrence JM, Khaw PT, Limb GA. Distribution of Müller stem cells within the neural retina: evidence for the existence of a ciliary margin-like zone in the adult human eye. Exp Eye Res. 2009;89:373–382 [DOI] [PubMed] [Google Scholar]
- 11. Takeda M, Takamiya A, Jiao JW, et al. alpha-Aminoadipate induces progenitor cell properties of Müller glia in adult mice. Invest Ophthalmol Vis Sci. 2008;49:1142–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nickerson PE, Da Silva N, Myers T, Stevens K, Clarke DB. Neural progenitor potential in cultured Müller glia: effects of passaging and exogenous growth factor exposure. Brain Res. 2008;1230:1–12 [DOI] [PubMed] [Google Scholar]
- 13. Florian C, Langmann T, Weber BH, Morsczeck C. Murine Müller cells are progenitor cells for neuronal cells and fibrous tissue cells. Biochem Biophys Res Commun. 2008;374:187–191 [DOI] [PubMed] [Google Scholar]
- 14. Lawrence JM, Singhal S, Bhatia B, et al. MIO-M1 cells and similar Müller glial cell lines derived from adult human retina exhibit neural stem cell characteristics. Stem Cells. 2007;25:2033–2043 [DOI] [PubMed] [Google Scholar]
- 15. Das AV, Mallya KB, Zhao X, et al. Neural stem cell properties of Müller glia in the mammalian retina: regulation by Notch and Wnt signaling. Dev Biol. 2006;299:283–302 [DOI] [PubMed] [Google Scholar]
- 16. Levenberg S, Huang NF, Lavik E, Rogers AB, Itskovitz-Eldor J, Langer R. Differentiation of human embryonic stem cells on three-dimensional polymer scaffolds. Proc Natl Acad Sci U S A. 2003;100:12741–12746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gelain F, Bottai D, Vescovi A, Zhang S. Designer self-assembling peptide nanofiber scaffolds for adult mouse neural stem cell 3-dimensional cultures. PLoS ONE. 2006;1:e119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Holmes TC, de Lacalle S, Su X, Liu G, Rich A, Zhang S. Extensive neurite outgrowth and active synapse formation on self-assembling peptide scaffolds. Proc Natl Acad Sci U S A. 2000;97:6728–6733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lavik EB, Klassen H, Warfvinge K, Langer R, Young MJ. Fabrication of degradable polymer scaffolds to direct the integration and differentiation of retinal progenitors. Biomaterials. 2005;26:3187–3196 [DOI] [PubMed] [Google Scholar]
- 20. Tomita M, Lavik E, Klassen H, Zahir T, Langer R, Young MJ. Biodegradable polymer composite grafts promote the survival and differentiation of retinal progenitor cells. Stem Cells. 2005;23:1579–1588 [DOI] [PubMed] [Google Scholar]
- 21. Redenti S, Neeley WL, Rompani S, et al. Engineering retinal progenitor cell and scrollable poly(glycerol-sebacate) composites for expansion and subretinal transplantation. Biomaterials. 2009;30:3405–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Neeley WL, Redenti S, Klassen H, et al. A microfabricated scaffold for retinal progenitor cell grafting. Biomaterials. 2008;29:418–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Otteson DC, Phillips MJ. A conditional immortalized mouse Müller glial cell line expressing glial and retinal stem-cell genes. Invest Ophthalmol Vis Sci. 2010;51:5991–6000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yokoi H, Kinoshita T, Zhang S. Dynamic reassembly of peptide RADA16 nanofiber scaffold. Proc Natl Acad Sci U S A. 2005;102:8414–8419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gelain F, Horii A, Zhang S. Designer self-assembling peptide scaffolds for 3-d tissue cell cultures and regenerative medicine. Macromolecular Biosci. 2007;7:544–551 [DOI] [PubMed] [Google Scholar]
- 26. Kleinman HK, Martin GR. Matrigel: basement membrane matrix with biological activity. Semin Cancer Biol. 2005;15:378–386 [DOI] [PubMed] [Google Scholar]
- 27. Vukicevic S, Kleinman HK, Luyten FP, Roberts AB, Roche NS, Reddi AH. Identification of multiple active growth factors in basement membrane Matrigel suggests caution in interpretation of cellular activity related to extracellular matrix components. Exp Cell Res. 1992;202:1–8 [DOI] [PubMed] [Google Scholar]
- 28. Jat PS, Noble MD, Ataliotis P, et al. Direct derivation of conditionally immortal cell lines from an H-2Kb-tsA58 transgenic mouse. Proc Natl Acad Sci U S A. 1991;88:5096–5100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Simpson DA, Feeney S, Boyle C, Stitt AW. Retinal VEGF mRNA measured by SYBR green I fluorescence: a versatile approach to quantitative PCR. Mol Vis. 2000;6:178–183 [PubMed] [Google Scholar]
- 30. Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Merhi-Soussi F, Angenieux B, Canola K, et al. High yield of cells committed to the photoreceptor fate from expanded mouse retinal stem cells. Stem Cells. 2006;24:2060–2070 [DOI] [PubMed] [Google Scholar]
- 32. Haeseleer F, Sokal I, Verlinde CL, et al. Five members of a novel Ca(2+)-binding protein (CABP) subfamily with similarity to calmodulin. J Biol Chem. 2000;275:1247–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dhingra A, Lyubarsky A, Jiang M, et al. The light response of ON bipolar neurons requires G[alpha]o. J Neurosci. 2000;20:9053–9058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stoeckli ET, Landmesser LT. Axon guidance at choice points. Curr Opin Neurobiol. 1998;8:73–79 [DOI] [PubMed] [Google Scholar]
- 35. Bjartmar L, Huberman AD, Ullian EM, et al. Neuronal pentraxins mediate synaptic refinement in the developing visual system. J Neurosci. 2006;26:6269–6281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Koulen P, Fletcher EL, Craven SE, Bredt DS, Wassle H. Immunocytochemical localization of the postsynaptic density protein PSD-95 in the mammalian retina. J Neurosci. 1998;18:10136–10149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fujita T, Maturana AD, Ikuta J, et al. Axonal guidance protein FEZ1 associates with tubulin and kinesin motor protein to transport mitochondria in neurites of NGF-stimulated PC12 cells. Biochem Biophys Res Commun. 2007;361:605–610 [DOI] [PubMed] [Google Scholar]
- 38. Chen C, Li M, Chai H, Yang H, Fisher WE, Yao Q. Roles of neuropilins in neuronal development, angiogenesis, and cancers. World J Surg. 2005;29:271–275 [DOI] [PubMed] [Google Scholar]
- 39. Pellet-Many C, Frankel P, Jia H, Zachary I. Neuropilins: structure, function and role in disease. Biochem J. 2008;411:211–226 [DOI] [PubMed] [Google Scholar]
- 40. Deleyrolle LP, Reynolds BA. Isolation, expansion, and differentiation of adult mammalian neural stem and progenitor cells using the neurosphere assay. Methods Mol Biol. 2009;549:91–101 [DOI] [PubMed] [Google Scholar]
- 41. Reynolds BA, Tetzlaff W, Weiss S. A multipotent EGF-responsive striatal embryonic progenitor cell produces neurons and astrocytes. J Neurosci. 1992;12:4565–4574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Monnin J, Morand-Villeneuve N, Michel G, Hicks D, Versaux-Botteri C. Production of neurospheres from mammalian Müller cells in culture. Neurosci Lett. 2007;421:22–26 [DOI] [PubMed] [Google Scholar]
- 43. Todorova MG, Soria B, Quesada I. Gap junctional intercellular communication is required to maintain embryonic stem cells in a non-differentiated and proliferative state. J Cell Physiol. 2008;214:354–362 [DOI] [PubMed] [Google Scholar]
- 44. Duval N, Gomes D, Calaora V, Calabrese A, Meda P, Bruzzone R. Cell coupling and Cx43 expression in embryonic mouse neural progenitor cells. J Cell Sci. 2002;115:3241–3251 [DOI] [PubMed] [Google Scholar]
- 45. Trimarchi JM, Stadler MB, Cepko CL. Individual retinal progenitor cells display extensive heterogeneity of gene expression. PLoS ONE. 2008;3:e1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Blackshaw S, Harpavat S, Trimarchi J, et al. Genomic analysis of mouse retinal development. PLoS Biol. 2004;2:E247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Livesey FJ, Young TL, Cepko CL. An analysis of the gene expression program of mammalian neural progenitor cells. Proc Natl Acad Sci U S A. 2004;101:1374–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bazan E, Alonso FJ, Redondo C, et al. In vitro and in vivo characterization of neural stem cells. Histol Histopathol. 2004;19:1261–1275 [DOI] [PubMed] [Google Scholar]
- 49. Zhang L, Mathers PH, Jamrich M. Function of rx, but not pax6, is essential for the formation of retinal progenitor cells in mice. Genesis. 2000;28:135–142 [PubMed] [Google Scholar]
- 50. Roesch K, Jadhav AP, Trimarchi JM, et al. The transcriptome of retinal Müller glial cells. J Comp Neurol. 2008;509:225–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fischer AJ, Reh TA. Potential of Müller glia to become neurogenic retinal progenitor cells. Glia. 2003;43:70–76 [DOI] [PubMed] [Google Scholar]
- 52. Lin YP, Ouchi Y, Satoh S, Watanabe S. Sox2 plays a role in the induction of amacrine and Müller glial cells in mouse retinal progenitor cells. Invest Ophthalmol Vis Sci. 2009;50:68–74 [DOI] [PubMed] [Google Scholar]
- 53. Calinescu AA, Vihtelic TS, Hyde DR, Hitchcock PF. Cellular expression of midkine-a and midkine-b during retinal development and photoreceptor regeneration in zebrafish. J Comp Neurol. 2009;514:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dee CT, Hirst CS, Shih YH, Tripathi VB, Patient RK, Scotting PJ. Sox3 regulates both neural fate and differentiation in the zebrafish ectoderm. Dev Biol. 2008;320:289–301 [DOI] [PubMed] [Google Scholar]
- 55. Li H, Radford JC, Ragusa MJ, et al. Transcription factor MEF2C influences neural stem/progenitor cell differentiation and maturation in vivo. Proc Natl Acad Sci U S A. 2008;105:9397–9402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hocking JC, Hehr CL, Chang RY, Johnston J, McFarlane S. TGFbeta ligands promote the initiation of retinal ganglion cell dendrites in vitro and in vivo. Mol Cell Neurosci. 2008;37:247–260 [DOI] [PubMed] [Google Scholar]
- 57. Kerrison JB, Lewis RN, Otteson DC, Zack DJ. Bone morphogenetic proteins promote neurite outgrowth in retinal ganglion cells. Mol Vis. 2005;11:208–215 [PubMed] [Google Scholar]
- 58. Yanagisawa H, Komuta Y, Kawano H, Toyoda M, Sango K. Pleiotrophin induces neurite outgrowth and upregulates growth-associated protein (GAP)-43 mRNA through the ALK/GSK3beta/beta-catenin signaling in developing mouse neurons. Neurosci Res. 2009;66:111–116 [DOI] [PubMed] [Google Scholar]
- 59. Amaratunga A, Abraham CR, Edwards RB, Sandell JH, Schreiber BM, Fine RE. Apolipoprotein E is synthesized in the retina by Müller glial cells, secreted into the vitreous, and rapidly transported into the optic nerve by retinal ganglion cells. J Biol Chem. 1996;271:5628–5632 [DOI] [PubMed] [Google Scholar]
- 60. Rogers JT, Weeber EJ. Reelin and apoE actions on signal transduction, synaptic function and memory formation. Neuron Glia Biol. 2008;4:259–270 [DOI] [PubMed] [Google Scholar]
- 61. Fletcher EL, Hack I, Brandstatter JH, Wassle H. Synaptic localization of NMDA receptor subunits in the rat retina. J Comp Neurol. 2000;420:98–112 [PubMed] [Google Scholar]
- 62. Hennig AK, Peng GH, Chen S. Regulation of photoreceptor gene expression by Crx-associated transcription factor network. Brain Res. 2008;1192:114–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Chen CK. Recoverin and rhodopsin kinase. Adv Exp Med Biol. 2002;514:101–107 [DOI] [PubMed] [Google Scholar]
- 64. Furukawa T, Morrow EM, Li T, Davis FC, Cepko CL. Retinopathy and attenuated circadian entrainment in Crx-deficient mice. Nat Genet. 1999;23:466–470 [DOI] [PubMed] [Google Scholar]
- 65. Blackshaw S, Fraioli RE, Furukawa T, Cepko CL. Comprehensive analysis of photoreceptor gene expression and the identification of candidate retinal disease genes. Cell. 2001;107:579–589 [DOI] [PubMed] [Google Scholar]
- 66. Ng L, Hurley JB, Dierks B, et al. A thyroid hormone receptor that is required for the development of green cone photoreceptors. Nat Genet. 2001;27:94–98 [DOI] [PubMed] [Google Scholar]
- 67. Wan J, Zheng H, Chen ZL, Xiao HL, Shen ZJ, Zhou GM. Preferential regeneration of photoreceptor from Müller glia after retinal degeneration in adult rat. Vision Res. 2008;48:223–234 [DOI] [PubMed] [Google Scholar]
- 68. Steedman M, Tao S, Klassen H, Desai T. Enhanced differentiation of retinal progenitor cells using microfabricated topographical cues. Biomed Microdevices. 12:363–369 [DOI] [PMC free article] [PubMed] [Google Scholar]