Abstract
Background
There is increasing interest in the role of immune activation and inflammation in HIV disease, but data on direct effects of HIV replication on immune cell activation are limited.
Methods
High sensitivity multiplex bead array assays (MBAAs) were used to measure changes in plasma cytokines and chemokines [interleukin (IL)-1β, IL-2, IL-6, IL-7, IL-8, IL-12p70, IL-17, tumor necrosis factor-α (TNFα), interferon-γ, granulocyte macrophage colony-stimulating factor, IL-4, IL-5, IL-10, IL-13, CXCL10] from randomization (month 0) to month 2 in a random sample of 200 patients from both the drug conservation (DC) and viral suppression (VS) arms of the Strategies for Management of Antiretroviral Therapy (SMART) trial. IL-6 was also measured by ELISA. Data were evaluated using nonparametric correlation and censored parametric analysis of covariance and associations were declared as statistically significant when the Bonferroni-adjusted P-value was less than 0.003.
Results
Compared with the VS arm, significant increases were seen in the DC arm for TNFα (+0.34 loge pg/ml, P = 0.0001), IL-10 (+0.33 loge pg/ml, P = 0.00001) and CXCL10 (+0.66 loge pg/ml, P = 0.00001). IL-6 ELISA poorly correlated with IL-6 MBAA (Spearman's rho = 0.29, P = 0.0001).
Conclusion
Resumption of HIV replication after ceasing antiretroviral therapy is associated predominantly with an increase of monocyte/macrophage-derived cytokines. Measurement of IL-6 levels may be affected by assay method and this should be considered in future studies of biomarkers.
Keywords: antiretroviral therapy, chemokines, clinical trial, cytokines, interruption
Introduction
The role of immune activation and inflammation in HIV disease has become increasingly of interest to the field of HIV pathogenesis and clinical care. A number of studies have shown that cellular markers of immune activation, including CD38 expression on CD8+ T cells, are stronger predictors of HIV disease progression than plasma viral load [1–8]. Soluble markers of immune activation, including neopterin and β2-microglobulin have been shown to predict HIV infection progression to AIDS or death [9–12], although these studies were carried out for the most part in the HAART era. More recent studies focused on individuals starting HAART have shown that baseline immune activation/inflammation markers are predictive of subsequent CD4+ T-cell responses [13–15]. In addition, for some who experience viral suppression while on HAART, there is a persistence of immune activation/inflammation that may correlate with the development of non-AIDS morbidity and mortality, including cardiovascular disease (CVD), metabolic disease, neurocognitive decline, bone disease and cancer [16]. Recently, there has been speculation that measurement of biomarkers can even improve risk prediction of AIDS over and above what can be done using CD4+ T-cell counts and plasma viral load levels and, if so, whether they should be used in routine HIV monitoring [17].
The Strategies for Management of Antiretroviral Therapy (SMART) trial randomized patients to a drug conservation (DC) arm, in which they stopped therapy, or to a viral suppression (VS) arm, in which they maintained therapy [18]. The findings of this trial have focused the field's attention on patients who developed non-AIDS-defining comorbidities that were significantly associated with inflammatory markers including interleukin (IL)-6, high sensitivity C-reactive protein and D-dimer [19]. A number of additional analyses have evaluated the association between microbial translocation markers and clinical outcomes [20,21], cystatin C and its role in renal disease [22] and hyaluronic acid and the risk of non-AIDS mortality in HIV/hepatitis co-infected patients [23].
Activation of various immune cells can be examined by assaying cytokines and chemokines produced by these cells in plasma. Examples of such cytokines include interferon-γ (IFNγ) and IL-2 produced by type 1 CD4+ helper T (Th1) cells, IL-4 and IL-5 produced by Th2 cells and IL-17 produced Th17 cells [24]. In addition, monocytes/macrophages are a source of cytokines and chemokines when activated including pro-inflammatory mediators such as IL-1, IL-6, tumor necrosis factor-α (TNFα) and CXCL10 [25]. In contrast, the regulatory cytokine IL-10 is produced by multiple cell types including monocytes/macrophages and regulatory T cells.
The current analysis was undertaken to evaluate the effects of HIV replication after ceasing antiretroviral therapy (ART) on changes in plasma cytokines and chemokines produced by peripheral blood lymphocytes or monocytes/macrophages. The specific cytokines and chemokines were selected in order to determine whether we could identify pathways that may be involved in driving immune activation/inflammation and CD4+ T cell decline in the DC vs. VS arms of the SMART Study.
Patients and methods
Patient characteristics
We evaluated stored plasma samples obtained from a subset of patients enrolled in the SMART trial. The methods and results of the SMART trial have been shown in detail elsewhere [18]. Specimens collected at randomization and 2 months after randomization were identified for a random sample of 200 participants from each treatment group – the DC and VS arms – who at time of randomization consented to storing blood for future research and reported no history of CVD at randomization. In addition, all patients had to be receiving ART at randomization with a viral load suppressed to 400 copies/ml or less. More in detail, of the 5472 patients enrolled in SMART, 1330 (24%, 666 in the DC and 664 in VS arm) satisfied the following criteria: consented to have a sample stored both at randomization and at month 2, did not have a history of CVD at randomization and had a suppressed viral load of 400 copies/ml or less at randomization. For 95 in the DC (14%) and 109 in the VS arm (16%) of these 1330 patients, samples had been already been analyzed using ELISA [19] and are included in this investigation so that results could be compared. The remaining 105 patients in the DC arm and 91 in the VS arm were randomly chosen from the remaining 571 and 555 patients in the two arms, respectively. Patients with a known history of CVD were excluded because they had already developed a morbidity that was shown to be associated with increased values of soluble factors. In contrast, at the time of developing the analysis plan, there was no knowledge of what the distribution of these factors would be in CVD-free patients with one or more risk factors for CVD. However, previous analyses indicated that, in order to increase the power to detect differences, the study population should be restricted to patients with suppressed viral load at baseline and the analysis focused on changes at month 2. The institutional review board at the University of Minnesota approved plans for analysis of stored specimens for consenting participants.
Biomarkers analysis
Plasma specimens were collected using EDTA (lavender top) blood collection tubes and were shipped frozen to the INSIGHT specimen repository multiplex bead array assays (MBAAs) were performed strictly according to the manufacturer's protocol for plasma samples. Prior to adding samples to an assay plate, aliquots were gently vortexed and then centrifuged at 13 200 revolutions/min for 10 min at 48°C. Dilutions of plasma samples and standards were prepared according to the manufacturers’ instructions and assayed in duplicate wells on each plate. Each sample was tested in a plate of the same lot number of each manufacturer's kit. Two separate multiplex kits were used: a High Sensitivity Human Cytokine LINCOplex Immunoassay Kit (13-plex, Linco/Millipore, Billerica, Massachusetts, USA) and a Human Cytokine Milliplex Map Immunoassay Kit (2-plex, Linco/Millipore). The biomarkers that were measured included IL-1β, IL-2, IL-6, IL-7, IL-8, IL-12p70, IL-17, TNFα, IFNγ, granulocyte macrophage colony-stimulating factor (GM-CSF), IL-4, IL-5, IL-10, IL-13 and CXCL10. A fluorescent bead-based instrument (Luminex-100; Luminex Corporation, Austin, Texas, USA) was used to read each multiplex plate. Luminex bead-based data were analyzed using Milliplex Analyst software v3.4 (Millipore; VigeneTech Inc., Boston, Massachusetts, USA) and a five parameter logistic curve fit. For a subset of patients, IL-6 was also measured by the Laboratory for Clinical Biochemistry Research at the University of Vermont using a standard ELISA assay both at baseline and 1 month after randomization: these values were used in a previous analysis of the SMART trial data [19]. To avoid confusion, IL-6 measured using MBAA is indicated with ‘IL6-M’, whereas the ELISA measurement with ‘IL6-E’. Classical ELISA-based cytokine assays are robust, easy to use and very well suited for measurement of single cytokines. The rationale for using MBAA was that multiple cytokines and chemokines could be assayed on a single serum sample. This made the approach much more cost-effective and also allowed for a much more comprehensive interrogation of cytokine and chemokine changes [26]. A potential drawback is that there may not be concordance of results for the two methods, particularly if there is no uniformity of assay reagents [27,28].
Statistical analysis
Some patients had biomarker values that were below the level of quantification. We adopted two separate approaches to deal with censored values: setting levels below the limit of quantification equal to the limit of quantification or, alternatively, a censored analysis – using PROC LIFETEST and LIFEREG in SAS (SAS Institute, Cary, North Carolina, USA), the latter with the normality assumption in the natural logarithmic scale (loge) [29].
The correlation between IL6-E and IL6-M (in the natural logarithmic scale, in the subset of patients for whom they were available) was analyzed using linear correlation analysis and Spearman's rank-correlation coefficient (rho statistic). The median (range) levels of the biomarkers at randomization (raw scale) and at month 2 were described in both the DC and VS arm, and we also calculated the mean change in biomarkers (natural logarithmic scale) by month 2 after randomization within each arm. In contrast, the Kaplan–Meier method and log-rank test were employed to compare medians when using the alternative approach. The difference in biomarker changes after 2 months between the DC and VS arm were estimated using analysis of covariance (ANCOVA) with a separate model for each biomarker and after controlling for the value at randomization of the biomarker under analysis. We performed a sensitivity ANCOVA after excluding patients with a viral load of 51–400 copies/ml at randomization. For IL6-E, month 2 values were unavailable, so the change after 1 month was analyzed instead. A parametric censored ANCOVA was used as an alternative analysis (results available as supplemental online materials http://links.lww.com/QAD/A137). For biomarkers showing more than 10% censored values (Table 1), the proportion of values below the lower limit of quantification were also compared using a χ2-test.
Table 1.
Median (interquartile range) cytokine and chemokine values and percentages of patients with a value below the limit of detection at randomization and the second time point.
| Randomization |
Month 2 (Month 1 for IL6-E) |
|||||
|---|---|---|---|---|---|---|
| Cytokines, pg/ml | DC arm | VS arm | P-valuea | DC arm | VS arm | P-valuea |
| IL-1β | ||||||
| Median (range) | 0.64 (0.02, 39.22) | 0.77 (0.02, 19.30) | 0.60 (0.02, 13.32) | 0.67 (0.02, 10.98) | ||
| n (%) with value below limit | 22 (11.0%) | 24 (12.0%) | 0.50 | 18 (9.0%) | 21 (10.6%) | 0.65 |
| IL-2 | ||||||
| Median (range) | 3.19 (0.02, 219.9) | 4.27 (0.01, 145.1) | 2.47 (0.01, 65.32) | 3.56 (0.08, 81.04) | ||
| n (%) with value below limit | 9 (4.5%) | 9 (4.5%) | 1.00 | 9 (4.5%) | 11 (5.5%) | 0.64 |
| IL6-M | ||||||
| Median (range) | 4.72 (0.02, 136.1) | 5.54 (0.06, 100.2) | 5.43 (0.05, 139.1) | 5.74 (0.09, 147.1) | ||
| n (%) with value below limit | 2 (1.0%) | 1 (0.5%) | 0.56 | 1 (0.5%) | 2 (1.0%) | 0.56 |
| IL6-E | ||||||
| Median (range) | 2.37 (0.16, 14.79) | 2.54 (0.27, 88.63) | 3.52 (0.45, 15.50) | 2.61 (0.59, 163.4) | ||
| IL-7 | ||||||
| Median (range) | 10.47 (0.02, 77.63) | 11.71 (0.90, 111.9) | 9.05 (0.17, 33.53) | 11.37 (0.82, 51.89) | ||
| n (%) with value below limit | 3 (1.5%) | 1 (0.5%) | 0.04 | 0 (0.0%) | 0 (0.0%) | 0.24 |
| IL-8 | ||||||
| Median (range) | 5.40 (0.02, 53.55) | 5.63 (0.13, 71.61) | 5.85 (1.03, 54.14) | 5.50 (0.06, 47.08) | ||
| n (%) with value below limit | 1 (0.5%) | 0 (0.0%) | 1 (0.5%) | 1 (0.5%) | ||
| IL-12 p70 | ||||||
| Median (range) | 2.15 (0.03, 798.4) | 3.46 (0.04, 476.6) | 1.50 (0.03, 99.70) | 2.30 (0.03, 302.7) | ||
| n (%) with value below limit | 32 (16.0%) | 32 (16.0%) | 0.19 | 23 (11.5%) | 21 (10.6%) | 0.11 |
| IL-17 | ||||||
| Median (range) | 3.00 (0.07, 302.2) | 3.04 (0.16, 631.6) | 2.98 (0.07, 162.1) | 3.04 (0.17, 762.0) | ||
| n (%) with value below limit | 108 (54.0%) | 137 (68.5%) | 0.92 | 107 (53.5%) | 101 (50.8%) | 0.001 |
| TNFα | ||||||
| Median (range) | 10.92 (0.10, 101.2) | 11.48 (1.37, 120.9) | 15.61 (2.28, 94.94) | 11.08 (1.12, 77.28) | ||
| n (%) with value below limit | 1 (0.5%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | ||
| IFNγ | ||||||
| Median (range) | 5.47 (0.09, 696.7) | 7.17 (0.04, 293.0) | 4.30 (0.08, 142.2) | 5.43 (0.03, 301.2) | ||
| n (%) with value below limit | 18 (9.0%) | 16 (8.0%) | 0.86 | 17 (8.5%) | 19 (9.5%) | 0.58 |
| GM-CSF | ||||||
| Median (range) | 1.23 (0.05, 367.3) | 1.42 (0.05, 169.1) | 0.86 (0.01, 71.86) | 1.46 (0.01, 110.7) | ||
| n (%) with value below limit | 34 (17.0%) | 52 (26.0%) | 1.00 | 34 (17.0%) | 31 (15.6%) | 0.01 |
| IL-4 | ||||||
| Median (range) | 22.99 (0.01, 1567) | 30.12 (0.01, 1743) | 19.13 (0.01, 1361) | 25.73 (0.01, 1541) | ||
| n (%) with value below limit | 15 (7.5%) | 21 (10.5%) | 0.85 | 14 (7.0%) | 14 (7.0%) | 0.22 |
| IL-5 | ||||||
| Median (range) | 0.49 (0.01, 38.41) | 0.67 (0.02, 30.62) | 0.42 (0.01, 65.74) | 0.59 (0.01, 16.40) | ||
| n (%) with value below limit | 28 (14.0%) | 29 (14.5%) | 0.22 | 20 (10.0%) | 19 (9.5%) | 0.13 |
| IL-10 | ||||||
| Median (range) | 23.63 (0.08, 478.2) | 25.85 (1.54, 651.1) | 28.16 (4.47, 438.9) | 24.69 (0.18, 402.8) | ||
| n (%) with value below limit | 3 (1.5%) | 0 (0.0%) | 0.04 | 0 (0.0%) | 1 (0.5%) | 0.24 |
| IL-13 | ||||||
| Median (range) | 4.60 (0.01, 676.3) | 5.67 (0.01, 601.9) | 2.69 (0.01, 685.5) | 5.89 (0.01, 611.0) | ||
| n (%) with value below limit | 62 (31.0%) | 77 (38.5%) | 0.38 | 54 (27.0%) | 58 (29.1%) | 0.05 |
| CXCL1 | ||||||
| Median (range) | 269.4 (50.48, 2251) | 271.8 (31.09, 3193) | 522.3 (95.70, 8728) | 273.7 (51.75, 2903) | ||
| n (%) with value below limit | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | ||
For the calculation of the medians, values below detectable levels were set equal to the lower limit. DC, drug conservation; GM-CSF, granulocyte macrophage colony-stimulating factor; IFN, interferon; IL, interleukin; TNF, tumor necrosis factor; VS, viral suppression.
χ2-test.
A number of general patients’ characteristics were collected (sex, age, race, mode of HIV infection, hepatitis virus co-infection, prior AIDS, current and prior smoking status, diabetes and treatment with blood pressure or lipid lowering therapy) and we checked that patients in the two arms were balanced for these factors. Co-infection with hepatitis B virus (HBV) and hepatitis C virus (HCV) were defined on the basis of serology tests alone. In addition, for a selected subset of biomarkers, we evaluated whether the differences in change between the DC and VS arm varied according to some of these demographic and treatment factors by stratification and formally performing an interaction test.
The association between biomarker changes (natural logarithmic scale), HIV-RNA (log10 scale) at month 2 and CD4+ T-cell count change at 2 months from randomization was also evaluated by linear regression analysis using the data of the DC arm alone. The correlation between biomarkers was studied using linear regression and Spearman's rho statistic separately in the DC arm. This was done only for the subset of biomarkers showing an association with randomization arm CD4+ T-cell count and viral load.
As 15 biomarkers were concomitantly evaluated, associations were declared to be statistically significant when showing a P-value below 0.05/15 = 0.003 (i.e. using a conservative Bonferroni-type adjustment for the inflation of type I error). Statistical analyses were performed using SAS 9.1.3 (SAS Institute, 2002–2003). All reported P-values are two-sided.
Results
Baseline cytokine/chemokine levels
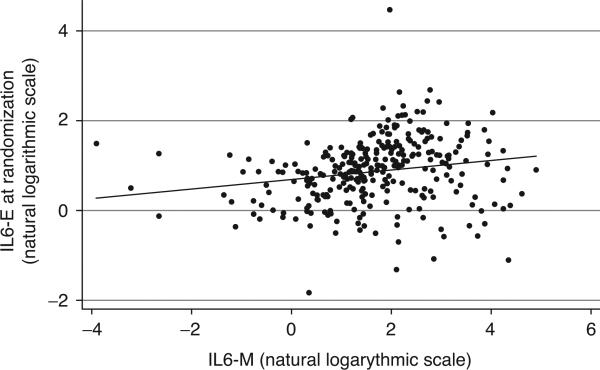
Table 1 shows the comparison between the median plasma levels of cytokines and chemokines both at randomization and at the second time point. At randomization the two groups had similar levels for all biomarkers except IL-2 (3.19 vs. 4.27 pg/ml, P = 0.02) and IL-12p70 (2.15 vs. 3.46 pg/ml, P = 0.01), which were nominally significantly higher in the VS compared with the DC arm. The median and range of values of IL6-M (4.94 pg/ml, range 0.02–136.11) and IL6-E (2.47 pg/ml, range 0.16–88.63), both measured at randomization, differed substantially and were weakly correlated with each other (Fig. 1).
Fig. 1.
Loge IL6-E (by ELISA) vs. loge IL6-M (by multiplex bead array assay) at randomization in a subset of patients with both baseline measurement available (n = 296 patients). Adjusted R2 = 0.03 (P = 0.001). Spearman's rank statistic rho = 0.29 (P = 0.0001).
Patient's characteristics at randomization
Characteristics at randomization of participants assigned to the DC and VS arm in this substudy of the SMART trial were well balanced (Table 2). By inclusion criteria, at randomization, all patients had a HIV-RNA plasma viral load suppressed to a value of 400 copies/ml or less on ART and none of the patients had previously experienced CVD events. In four out of 10 (40%) HBV surface antigen-positive patients, HBV-DNA was detectable at more than 357 copies/ml, whereas HCV-RNA was available only for 62 HCV antibody-positive patients and 46 of these (74%) had a value of more than 615 copies/ml.
Table 2.
Main characteristics of random sample of discontinuation and viral suppression participants at randomization.
| DC (n = 200) | VS (n = 200) | P-valuea | |
|---|---|---|---|
| Characteristics age (years) | |||
| Median (IQR) | 45 (39, 51) | 46 (40, 50) | 0.408 |
| ≥45 years, n (%) | 96 (48.0) | 110 (55.0) | 0.161 |
| Sex, n (%) | 0.820 | ||
| Female | 51 (25.5) | 53 (26.5) | |
| Race, n (%) | 0.766 | ||
| Black | 93 (46.5) | 86 (43.0) | |
| White | 81 (40.5) | 85 (42.5) | |
| Other | 26 (13.0) | 29 (14.5) | |
| Co-infected, n (%) | 0.114 | ||
| Yes | 41 (20.5) | 29 (14.5) | |
| HCV alone | 35 (17.5) | 25 (12.5) | |
| HBV alone | 5 (2.5) | 3 (1.5) | |
| HCV/HBV | 1 (0.5) | 1 (0.5) | |
| Smoker, n (%) | 0.604 | ||
| Yes | 71 (35.5) | 76 (38.0) | |
| Diabetes, n (%) | 0.749 | ||
| Yes | 21 (10.5) | 23 (11.5) | |
| Lipid-lowering drugs, n (%) | 0.908 | ||
| Yes | 51 (25.5) | 50 (25.0) | |
| Treatment received | 0.890 | ||
| PI without NNRTI | 86 (43.0) | 82 (41.0) | |
| NNRTI without PI | 78 (39.0) | 88 (44.0) | |
| Neither PI or NNRTI | 36 (18.0) | 30 (15.0) |
DC, drug conservation; HBV, hepatitis B virus; HCV, hepatitis C virus; IQR, interquartile range; NNRTI, nonnucleoside reverse transcriptase inhibitor; PI, protease inhibitor; VS, viral suppression.
Likelihood ratio χ2-test or Wilcoxon test whichever was appropriate.
Change in biomarkers between randomization and month 2 within each arm and between arms
Within the VS arm, biomarkers levels remained fairly stable between randomization and month 2 with a tendency to slight decrease, with the exception of perhaps GM-CSF that instead showed a slight increase (Table 3a). In contrast in the DC arm, large increases were observed for IL6-E (+0.43 loge pg/ml) and IL6-M (+0.21 loge pg/ml), TNFα (+0.35 loge pg/ml), IL-10 (+0.34 loge pg/ml) and CXCL10 (+0.62 loge pg/ml). ANCOVA demonstrated that by 2 months after randomization, there was a significantly greater increase in the DC vs. the VS arm in the following biomarkers: TNFa [mean difference +0.34 loge pg/ml, 95% confidence interval (CI) 0.24–0.45, P = 0.0001], IL-10 (mean difference +0.33 loge pg/ml, 95% CI 0.18–0.49, P = 0.0001) and CXCL10 (mean difference +0.66 loge pg/ml, 95% CI 0.53–0.79, P = 0.00001). Consistent with previous observation in the SMART study, IL6-E levels showed a positive difference (+0.26 loge pg/ml, 95% CI 0.06–0.46, P = 0.01), although the association did not reach significance at the Bonferroni-adjusted level of 0.003 for IL6-M (mean difference +0.20 loge mg/dl, 95% CI 0.00–0.40, P = 0.05). Results were similar after restricting to patients who had a viral load of 50 copies/ml or less at randomization (+0.40 loge mg/dl, 95% CI 0.24–0.56 for TNFα; +0.32 loge mg/dl, 95% CI 0.09–0.55 for IL-10; +0.71 loge mg/dl, 95% CI 0.52–0.90 for CXCL10; and +0.28 loge mg/dl 95% CI 0.00–0.56 for IL6-E). In the alternative analysis replacing the censored values with the value of the limit of quantification, these results were also confirmed.
Table 3.
Summary results for drug conservation vs. viral suppression at month 2 compared with baseline from fitting an analysis of covariance.
| DC arm (N = 200) Mean loge change (SD) |
VS arm (N = 200) Mean loge change (SD) |
Difference in mean loge change DC/VSa (95% CI) | P-value | |
|---|---|---|---|---|
| (a) Overall results | ||||
| Cytokines | ||||
| IL-1β | –0.02 (1.25) | –0.16 (1.16) | 0.04 (–0.17, 0.25) | 0.708 |
| IL-2 | –0.16 (1.59) | –0.16 (1.34) | –0.16 (–0.41, 0.09) | 0.206 |
| IL6-M | 0.21 (1.26) | –0.05 (0.97) | 0.20 (0.00, 0.41) | 0.049 |
| IL6-E (month 1) | 0.43 (0.76) | 0.06 (0.78) | 0.26 (0.06, 0.46) | 0.010 |
| IL-7 | –0.05 (1.02) | –0.10 (0.90) | –0.07 (–0.22, 0.09) | 0.400 |
| IL-8 | 0.08 (0.77) | –0.03 (0.69) | 0.09 (–0.04, 0.21) | 0.168 |
| IL-12 p70 | –0.29 (1.82) | –0.16 (1.49) | –0.34 (–0.63, –0.05) | 0.022 |
| IL-17 | –0.41 (1.43) | –0.02 (1.03) | –0.44 (–0.65, –0.23) | <0.001 |
| TNFα | 0.35 (0.70) | –0.08 (0.62) | 0.34 (0.24, 0.45) | <0.001 |
| IFNγ | –0.17 (1.63) | –0.25 (1.46) | –0.03 (–0.29, 0.24) | 0.854 |
| GM-CSF | –0.30 (1.76) | 0.05 (1.43) | –0.40 (–0.68, –0.12) | 0.006 |
| IL-4 | –0.20 (2.51) | –0.28 (1.89) | –0.06 (–0.45, 0.34) | 0.785 |
| IL-5 | –0.13 (1.45) | –0.11 (1.16) | –0.13 (–0.36, 0.10) | 0.270 |
| IL-10 | 0.34 (1.08) | –0.13 (0.88) | 0.33 (0.18, 0.49) | <0.001 |
| IL-13 | –0.26 (2.48) | –0.19 (2.08) | –0.11 (–0.54, 0.31) | 0.599 |
| CXCL10 | 0.62 (0.87) | –0.04 (0.65) | 0.66 (0.53, 0.79) | <0.001 |
| DC vs. VS; adjusted differencesb (95% CI) |
||||||
|---|---|---|---|---|---|---|
| Biomarker |
||||||
| IL6-M | IL-12 | IL-17 | TNFα | IL-10 | CXCL10 | |
| (b) Biomarkers levels according to baseline subgroups | ||||||
| Characteristics | ||||||
| Sex | ||||||
| Male | 0.18 (–0.06, 0.42) | –0.35 (–0.68, –0.01) | –0.52 (–0.76, –0.27) | 0.36 (0.24, 0.49) | 0.31 (0.13, 0.49) | 0.61 (0.46, 0.76) |
| Female | 0.27 (–0.13, 0.67) | –0.31 (–0.88, 0.25) | –0.24 (–0.66, 0.17) | 0.29 (0.08, 0.50) | 0.40 (0.09, 0.70) | 0.80 (0.54, 1.06) |
| Interaction test P-value | 0.71 | 0.92 | 0.26 | 0.54 | 0.63 | 0.21 |
| Age, years | ||||||
| <45 | 0.31 (0.01, 0.60) | –0.37 (–0.79, 0.05) | –0.43 (–0.73, –0.12) | 0.40 (0.25, 0.55) | 0.43 (0.20, 0.65) | 0.71 (0.52, 0.90) |
| >45 | 0.12 (–0.17, 0.40) | –0.29 (–0.70, 0.11) | –0.47 (–0.77, –0.18) | 0.32 (0.17, 0.46) | 0.27 (0.05, 0.48) | 0.63 (0.44, 0.81) |
| Interaction test P-value | 0.36 | 0.79 | 0.83 | 0.43 | 0.31 | 0.56 |
| Ethnicity | ||||||
| Black | 0.11 (–0.21, 0.42) | –0.33 (–0.78, 0.12) | –0.64 (–0.97, –0.31) | 0.37 (0.21, 0.54) | 0.31 (0.07, 0.55) | 0.63 (0.42, 0.83) |
| White/other | 0.27 (0.01, 0.54) | –0.35 (–0.73, 0.03) | –0.30 (–0.58, –0.02) | 0.32 (0.19, 0.46) | 0.35 (0.15, 0.55) | 0.68 (0.51, 0.86) |
| Interaction test P-value | 0.43 | 0.95 | 0.12 | 0.66 | 0.81 | 0.69 |
| Class of third drugc | ||||||
| PI based | 0.12 (–0.18, 0.42) | –0.58 (–1.00, –0.17) | –0.38 (–0.69, –0.07) | 0.29 (0.13, 0.45) | 0.42 (0.18, 0.65) | 0.69 (0.48, 0.89) |
| NNRTI based | 0.32 (0.02, 0.62) | 0.10 (–0.32, 0.52) | –0.49 (–0.80, –0.18) | 0.38 (0.22, 0.54) | 0.31 (0.07, 0.54) | 0.67 (0.46, 0.88) |
| Interaction test P-value | 0.35 | 0.02 | 0.64 | 0.43 | 0.52 | 0.90 |
| Co-infected | ||||||
| No | 0.21 (–0.02, 0.43) | –0.27 (–0.59, 0.04) | –0.44 (–0.68, –0.21) | 0.36 (0.24, 0.48) | 0.38 (0.20, 0.55) | 0.75 (0.60, 0.89) |
| Yes | 0.14 (–0.35, 0.63) | –0.73 (–1.43, –0.03) | –0.39 (–0.90, 0.13) | 0.24 (–0.02, 0.50) | 0.08 (–0.29, 0.46) | 0.20 (–0.12, 0.52) |
| Interaction test P-value | 0.81 | 0.25 | 0.84 | 0.40 | 0.17 | 0.002 |
CI, confidence interval; DC, drug conservation; GM-CSF, granulocyte macrophage colony-stimulating factor; IFN, interferon; IL, interleukin; NNRTI, nonnucleoside reverse transcriptase inhibitor; PI, protease inhibitor; TNF, tumor necrosis factor; VS, viral suppression.
Model includes baseline level of marker (natural logarithmic scale).
Adjusted for baseline using analysis of covariance.
Thirty-three patients receiving both/neither NNRTI and PI were excluded.
Of interest, serum levels of IL-12p70, IL-17 and GM-CSF were found to be significantly decreased in the DC vs. the VS arm in the main analysis (Table 3a), but not in the censored analysis (Table S2, http://links.lww.com/QAD/A137). However, for these biomarkers, the percentage of censored values was high (13, 53 and 17%, respectively) and, therefore, likely to have affected the results of the censored analysis. Indeed, for, IL-17 and GM-CSF, the proportion of patients with a value below the lower limit of detection at month 2 was also significantly higher in the DC arm compared with the VS arm (Table 1).
After stratifying for demographics and treatment factors, we found that the difference between the DC and VS arm in CXCL10 could be seen only in HIV mono-infected patients, but not in the hepatitis virus (either HCV or HBV) co-infected patients and this was confirmed by a significant interaction test (P = 0.002). In the HIV mono-infected, the mean (95% CI) loge change in CXCL10 within the DC arm was 0.75 (0.88) and -0.07 (0.64) within the VS arm, whereas the corresponding estimates in the hepatitis co-infected patients were 0.29 (0.72) in the DC arm vs. 0.11 (0.70) in the VS arm, again suggesting that the difference in the increase between arms was driven by a large increase in HIV mono-infected patients in the DC arm. Moreover, mean (SD) CXCL10 levels at randomization were significantly higher in co-infected patients 6.12 loge pg/ml (0.74) compared with mono-infected 5.55 loge pg/ml (0.74) (t-test P = 0.0001).
Association between change in biomarkers and change in CD4 cell count and viral load
Median viral load at month 2 in the DC arm was 4.35 log10 copies/ml (interquartile range 3.52–4.86), and 172 patients (86%) of patients in the VS arm remained with viral load suppression of 400 copies/ml or less at month 2. The estimates for the change in CD4+ T-cell count were -186 cells/μl (186) and -10 cells/μl in the DC and VS arms, respectively. Table 4a/b shows the association between change in biomarkers over 0–2 months from randomization and their association with the concomitant change in CD4+ T-cell count and month 2 viral load from fitting a linear regression analysis using only patients allocated to the DC arm. In univariable analyses, each loge picogram per milliliter (pg/ml) increase in IL-17 was associated with a 0.11 log10 copies/ml lower month 2 viral load (95% CI -0.21 to -0.01, P = 0.03). In contrast, increases in loge pg/ml of TNFα, IL-10 and CXCL10 showed a large and positive correlation with month 2 viral load (+0.26 log10 copies/ml, P = 0.02; +0.17 log10 copies/ml, P = 0.01; and +0.35 log10 copies/, P = 0.001, respectively, Table 4a) and CD4+ T-cell count decreases (-40 cells/μl, P = 0.04; -20 cells/μl, P = 0.10; and -36 cells/μl, P = 0.02, respectively, Table 4b). However, although the association between IL-10 and CXCL10 change and CD4+ T-cell count decrease appeared to be entirely mediated by month 2 viral load, this was less so for TNFα (adjusted mean change -23 cells/μl per 1 loge increase in TNFα, P = 0.20, Table 4b).
Table 4.
Estimated mean month 2 viral load [log10 copies/ml – part (a)] and CD4 cell count change by month 2 [cells/μl – part (b)] associated with 1 loge higher values in biomarkers from fitting a linear regression model (drug conservation army only).
| Per loge higher | Unadjusted analysis Mean (95% CI) | P-value | Adjusted analysisa Mean (95% CI) | P-value |
|---|---|---|---|---|
| (a) Viral load at month 2 (log10 copies/ml) | ||||
| IL-1β | 0.02 (–0.09, 0.14) | 0.713 | 0.03 (–0.08, 0.13) | 0.632 |
| IL-2 | 0.05 (–0.04, 0.14) | 0.301 | 0.05 (–0.03, 0.14) | 0.190 |
| IL6-M | –0.01 (–0.12, 0.11) | 0.930 | –0.01 (–0.12, 0.09) | 0.812 |
| IL-7 | 0.02 (–0.12, 0.16) | 0.786 | 0.05 (–0.08, 0.17) | 0.483 |
| IL-8 | 0.02 (–0.16, 0.21) | 0.796 | 0.02 (–0.15, 0.19) | 0.800 |
| IL-12 | 0.02 (–0.07, 0.10) | 0.709 | 0.01 (–0.06, 0.08) | 0.767 |
| IL-17 | –0.11 (–0.21, –0.01) | 0.033 | –0.10 (–0.19, –0.01) | 0.038 |
| TNFα | 0.26 (0.05, 0.47) | 0.015 | 0.16 (–0.03, 0.35) | 0.104 |
| IFNγ | 0.03 (–0.06, 0.12) | 0.455 | 0.03 (–0.05, 0.11) | 0.466 |
| GM-CSF | –0.01 (–0.10, 0.07) | 0.739 | –0.02 (–0.09, 0.06) | 0.621 |
| IL-4 | 0.01 (–0.04, 0.07) | 0.612 | 0.02 (–0.04, 0.07) | 0.560 |
| IL-5 | –0.02 (–0.12, 0.08) | 0.696 | –0.03 (–0.12, 0.06) | 0.530 |
| IL-10 | 0.17 (0.04, 0.30) | 0.012 | 0.12 (0.00, 0.24) | 0.047 |
| IL-13 | –0.00 (–0.06, 0.06) | 0.935 | –0.02 (–0.07, 0.04) | 0.540 |
| CXCL10 | 0.35 (0.18, 0.51) | <0.001 | 0.26 (0.11, 0.41) | <0.001 |
| (b) Change in CD4+ T-cell count (cells/μl) | ||||
| IL-1β | 1.85 (–19.0, 22.68) | 0.861 | 6.29 (–12.7, 25.26) | 0.514 |
| IL-2 | 2.74 (–13.7, 19.18) | 0.743 | 7.21 (–7.77, 22.20) | 0.344 |
| IL6-M | –3.01 (–23.7, 17.72) | 0.775 | –2.04 (–20.9, 16.81) | 0.831 |
| IL-7 | 10.46 (–15.0, 35.89) | 0.418 | 14.21 (–8.90, 37.31) | 0.227 |
| IL-8 | –1.78 (–35.7, 32.09) | 0.917 | 2.68 (–28.2, 33.51) | 0.864 |
| IL-12 | –0.69 (–15.2, 13.84) | 0.925 | 2.40 (–10.9, 15.65) | 0.722 |
| IL-17 | 7.15 (–11.1, 25.36) | 0.439 | –2.78 (–19.8, 14.19) | 0.747 |
| TNFα | –39.5 (–76.6, –2.41) | 0.037 | –22.5 (–57.2, 12.17) | 0.202 |
| IFNγ | –1.02 (–17.0, 14.99) | 0.900 | 3.00 (–11.6, 17.64) | 0.686 |
| GM-CSF | –1.70 (–16.5, 13.14) | 0.821 | 0.61 (–12.9, 14.14) | 0.929 |
| IL-4 | 0.54 (–9.85, 10.94) | 0.918 | 1.99 (–7.48, 11.46) | 0.679 |
| IL-5 | –3.04 (–21.0, 14.92) | 0.739 | –2.37 (–18.7, 14.00) | 0.775 |
| IL-10 | –19.9 (–43.8, 4.01) | 0.102 | –5.39 (–27.7, 16.94) | 0.634 |
| IL-13 | –5.49 (–16.0, 5.05) | 0.306 | –5.61 (–15.2, 3.97) | 0.249 |
| CXCL10 | –35.8 (–65.7, –5.97) | 0.019 | –11.2 (–39.8, 17.49) | 0.443 |
CI, confidence interval; GM-CSF, granulocyte macrophage colony-stimulating factor; IFN, interferon; IL, interleukin; TNF, tumor necrosis factor.
Adjusted for change in CD4+ T-cell count.
bAdjusted for month 2 log10 viral load.
Correlation between biomarker changes in the DC arm
The Spearman's correlation analysis identified a single strong correlation between changes in TNFα and IL-10 (rho = 0.57, P = 0.00001, Fig. S1, http://links.lww.com/QAD/A137). The only other correlation showing a significant P-value was the association between changes in IL-10 and IL-17, although the degree of correlation was weak (rho = -0.35, P = 0.018).
Discussion
This analysis, designed to identify changes in plasma levels of cytokines and chemokines in patients randomly assigned to the DC and VS arms of the SMART study, might provide information about activation of immune cells associated with resumption of HIV replication in the DC arm. We found that significant increases occurred in the DC arm in IL6-E (by ELISA), TNFa, IL-10 and CXCL10 levels. These changes were significantly larger than the changes observed in the VS arm. In addition, there was correlation between the changes in TNFα, IL-10 and CXCL10 (strong in the case of TNFα and IL-10), suggesting that there was a common driver for the increased production. We hypothesize that this common cause is monocyte/macrophage activation, which typically determines increased production of these particular cytokines and chemokines.
We also examined the association of increased cytokine and chemokine production with changes in CD4+ T-cell numbers in patients allocated to the DC arm only in whom any differences are likely to be due to the major stimulus induced by stopping ART. After adjustment for the effect of month 2 viral load level on CD4+ T-cell count, increased plasma levels of IL-10 and CXCL10 were no longer associated with decreased CD4+ T-cell count. TNFα also was no longer associated, although it retained the smaller P-value (P = 0.20), suggesting that TNFα production might be associated with depletion of CD4+ T cells by mechanisms other than viral load increase (e.g. inducing apoptosis). Indeed, TNFα is known to be an important mediator of apoptosis through engagement of TNFα receptors I and II [30–33]. Furthermore, it has been previously shown that genetic polymorphism in TNFα can impact CD4+ T-cell recovery on HAART and may also be an important mediator of CD4+ T-cell loss as seen in the current analysis [34,35]. In addition, serum TNFα has been associated with HIV disease progression in a number of studies [36–40]. A recent analysis has demonstrated a significant association of soluble TNF receptor 1 and HIV disease progression, including development of AIDS associated malignancies [41]. Another potential role for TNF in HIV pathogenesis is its ability to drive CD8+ T cells to a senescent phenotype [42]. This suggests that high levels of TNF may be linked to the non-AIDS comorbidities seen in the SMART study, as cells with a senescent phenotype have been linked to CVD, neurocognitive decline and osteoporosis [43].
The planned treatment interruption in the DC arm also provided an opportunity to determine changes in cytokine/chemokine levels during a period of a significant increase in HIV replication. This period of intense viral replication is similar to what is seen during acute HIV infection. A study by Norris et al. [44] demonstrated significant increases in TNFα, IL-10 and IFNγ during acute HIV. Similar results were seen in a more recent study by Stacey et al. [45] who compared cytokine changes in acute HIV, HCV and HBV infection. These investigators found early changes in IL-15 and IFNα with a greater proportion of individuals showing an increase in CXCL10. They also noted that over 80% of individuals had significant increases in IL-10 and TNFα. These same levels of cytokine changes were not seen in samples from either HCV or HBV acutely infected individuals. Rychert et al. [46] have recently evaluated the correlation of plasma gp120 with cytokine changes in early HIV infection and found that gp120 levels correlated with an increase in TNFα, IL-10 and IL-6. These changes are consistent with our data. We did see a significant increase in CXCL10, which is an IFN-inducible protein in which production is known to be predominantly stimulated by IFN [47]. All of these data on changes in cytokines and chemokines early in HIV seroconversion point to a critical role for modulation of the immune response against HIV and impact on immune function.
Our analyses also showed a weak but negative correlation between the change in IL-10 levels and change in IL-17 levels over a period of 2 months after randomization when restricting the analysis to the DC arm alone. IL-10 has been shown to suppress Th17 cells in several, though not all, studies [48–51]. The role of Th17 cells in HIV infection has not been fully established, but Th17 cell depletion may contribute to the defect of gut mucosal immune responses induced by HIV infection [52]. Furthermore, IL-10 reduces Th17-mediated immune responses in an animal model of influenza virus infection [53] and, therefore, could affect other antiviral responses.
Of note, we observed that CXCL10 levels at 2 months only increased in patients who did not have HCV co-infection, possibly because serum CXCL10 levels are increased by HCV co-infection itself [54]. Indeed, our data show that CXCL10 levels at randomization were elevated in co-infected patients compared with mono-infected patients and that in the co-infected, there was a similar increase by month 2 in CXCL10 levels regardless of whether they interrupted therapy or not. We cannot, however, rule out that this is a chance finding because of the large number of subgroup analysis performed. In addition, hepatitis co-infection was defined on the basis of serology tests alone and 26% of our HCV-positive population at serology had cleared the infection.
There are other limitations to our analysis. We utilized a MBAA to evaluate plasma levels of cytokines and chemokines at baseline and following 2 months of the intervention because this assay method has the advantage of being able to measure a large number of analytes on the same plasma sample with reduction in time of execution and costs. However, it is recognized that there can be poor concordance between MBAA and ELISA in the assay of some cytokines, which to a large degree reflects differences in the activity of capture and reporter antibodies [27]. This is the probable explanation for our finding of a weak correlation between IL-6 levels assayed by MBAA or ELISA (Fig. 1). For IL-6, differences in the ability these assays to detect the effects of ‘regulatory’ molecules (e.g. sIL-6R or gp130 or both) might also be a factor [55]. It is, therefore, important to take these factors into consideration when comparing the results of studies that have used different assay methods. Despite these caveats, our data have clearly shown changes in plasma cytokine and chemokine levels within and between patient groups using the same assay method (MBAA).
Despite the high sensitivity of the MBAAs employed in this analysis, IL-12p70, IL-13, IL-17 and GM-CSF were not detectable in a large percentage of samples (Table 1). This produced a discrepancy in results between the main analysis and the alternative analysis. It is known that when the censoring is heavy, the censored ANCOVA can produce unreliable results [56]. Nevertheless, only cytokine/chemokine changes that were significantly different by randomization arm in both the main and the alternative analysis were considered to be unlikely to have occurred by chance alone. Finally, because monocyte/macrophage activation was not assessed directly, we cannot rule out the possibility that the observed changes in serum cytokine and chemokine levels were an effect of HIV replication on another source of these molecules.
In conclusion, the results of our analysis demonstrate a highly significant correlation of viral load increase following ART interruption with both CD4+ T-cell decline and increased production of IL6-E (by ELISA), TNFα, CXCL10 and IL-10, which are all known to be monocyte/macrophage derived. These results suggest that the effect of HIV replication on monocyte/ macrophages may constitute a critical pathway in the induction of HIV-mediated immune activation/inflammation, as well as immune deficiency. Similar studies should be performed to confirm the hypothesis generated by these data.
Supplementary Material
Acknowledgements
This work was supported by the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH) (U01-AI068641, U01-AI042170 and U01-AI046362). The funding source had no role in data collection, data analysis or the decision to publish the results.
None of the authors has any financial or personal relationships with people or organizations that could inappropriately influence this work, although most members of the group have, at some stage in the past, received funding from a variety of pharmaceutical companies for research, travel grants, speaking engagements or consultancy fees.
Specific contribution of single authors:
Conception and design (A.L., J.N.).
Acquisition of data (M.A.F., J.B., P.O., M.P.).
Analysis and interpretation of the data (A.C.-L., A.L., M.A.F.).
Drafting of the manuscript (A.C.-L., A.L., M.A.F.).
Critical revision of the manuscript (A.C.-L., M.A.F., J.B., P.O., M.P., J.N., A.L., INSIGHT SSC).
Statistical analysis (A.C.-L.).
The INSIGHT Scientific Steering Committee (SSC) participated in the conception and design of the study. Full access to all data and responsibility for integrity and accuracy of data/analysis (A.L., J.N.).
References
- 1.Giorgi JV, Lyles RH, Matud JL, Yamashita TE, Mellors JW, Hultin LE, et al. Multicenter AIDS Cohort Study Predictive value of immunologic and virologic markers after long or short duration of HIV-1 infection. J Acquir Immune Defic Syndr. 2002;29:346–355. doi: 10.1097/00126334-200204010-00004. [DOI] [PubMed] [Google Scholar]
- 2.Liu Z, Cumberland WG, Hultin LE, Prince HE, Detels R, Giorgi JV. Elevated CD38 antigen expression on CD8+ T cells is a stronger marker for the risk of chronic HIV disease progression to AIDS and death in the Multicenter AIDS Cohort Study than CD4+ cell count, soluble immune activation markers, or combinations of HLA-DR and CD38 expression. J Acquir Immune Defic Syndr Hum Retrovirol. 1997;16:83–92. doi: 10.1097/00042560-199710010-00003. [DOI] [PubMed] [Google Scholar]
- 3.Bürgisser P, Hammann C, Kaufmann D, Battegay M, Rutschmann OT. Expression of CD28 and CD38 by CD8+ T lymphocytes in HIV-1 infection correlates with markers of disease severity and changes towards normalization under treatment. The Swiss HIV Cohort Study. Clin Exp Immunol. 1999;115:458–463. doi: 10.1046/j.1365-2249.1999.00818.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Froebel KS, Raab GM, D'Alessandro C. A single measurement of CD38CD8 cells in HIV+, long-term surviving injecting drug users distinguishes those who will progress to AIDS from those who will remain stable. Clin Exp Immunol. 2000;122:72–78. doi: 10.1046/j.1365-2249.2000.01348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giorgi JV, Liu Z, Hultin LE, Cumberland WG, Hennessey K, Detels R. Elevated levels of CD38+ CD8+ T cells in HIV infection add to the prognostic value of low CD4+ T cell levels: results of 6 years of follow-up. The Los Angeles Center, Multicenter AIDS Cohort Study. J Acquir Immune Defic Syndr. 1993;6:904–912. [PubMed] [Google Scholar]
- 6.Bofill M, Mocroft A, Lipman M, Medina E, Borthwick NJ, Sabin CA, et al. Increased numbers of primed activated CD8+CD38+ CD45RO+ T cells predict the decline of CD4+ T cells in HIV-1-infected patients. AIDS. 1996;10:827–834. doi: 10.1097/00002030-199607000-00005. [DOI] [PubMed] [Google Scholar]
- 7.Wilson CM, Ellenberg JH, Douglas SD, Moscicki AB, Holland CA, Reach Project Of The Adolescent Medicine HIV/AIDS Research Network CD8+CD38+ T cells but not HIV type 1 RNA viral load predict CD4+ T cell loss in a predominantly minority female HIV+ adolescent population. AIDS Res Hum Retroviruses. 2004;20:263–269. doi: 10.1089/088922204322996482. [DOI] [PubMed] [Google Scholar]
- 8.Hazenberg MD, Otto SA, van Benthem BH, Roos MT, Coutinho RA, Lange JM, et al. Persistent immune activation in HIV-1 infection is associated with progression to AIDS. AIDS. 2003;17:1881–1888. doi: 10.1097/00002030-200309050-00006. [DOI] [PubMed] [Google Scholar]
- 9.Fahey JL, Taylor JM, Detels R, Hofmann B, Melmed R, Nishanian P, Giorgi JV. The prognostic value of cellular and serologic markers in infection with human immunodeficiency virus type 1. N Engl J Med. 1990;322:166–172. doi: 10.1056/NEJM199001183220305. [DOI] [PubMed] [Google Scholar]
- 10.Mildvan D, Spritzler J, Grossberg SE, Fahey JL, Johnston DM, Schock BR, Kagan J. Serum neopterin, an immune activation marker, independently predicts disease progression in advanced HIV-1 infection. Clin Infect Dis. 2005;40:853–858. doi: 10.1086/427877. [DOI] [PubMed] [Google Scholar]
- 11.Bedini JL, García F, Miró JM, Aznar E, Serrano J, Lozano L, et al. Serum levels of beta2-microglobulin, neopterin, TNF-alpha and soluble receptors of TNF-alpha and interleukin-2 in intravenous drug abusers according to HIV-1 status and stage of the HIV-1 infection. Clin Microbiol Infect. 1998;4:4–10. doi: 10.1111/j.1469-0691.1998.tb00327.x. [DOI] [PubMed] [Google Scholar]
- 12.Martínez-Brú C, Cortés M, Planella T, Barrio J, Cadafalch J, Domingo P, et al. Beta 2-microglobulin and immunoglobulins are more useful markers of disease progression in HIV than neopterin and adenosine deaminase. Ann Clin Biochem. 1999;36(Pt 5):601–608. doi: 10.1177/000456329903600506. [DOI] [PubMed] [Google Scholar]
- 13.Hunt PW, Martin JN, Sinclair E, Bredt B, Hagos E, Lampiris H, Deeks SG. T cell activation is associated with lower CD4+ T cell gains in human immunodeficiency virus-infected patients with sustained viral suppression during antiretroviral therapy. J Infect Dis. 2003;187:1534–1543. doi: 10.1086/374786. [DOI] [PubMed] [Google Scholar]
- 14.French MA, King MS, Tschampa JM, da Silva BA, Landay AL. Serum immune activation markers are persistently increased in patients with HIV infection after 6 years of antiretroviral therapy despite suppression of viral replication and reconstitution of CD4+ T cells. 1. J Infect Dis. 2009;200:1212–1215. doi: 10.1086/605890. [DOI] [PubMed] [Google Scholar]
- 15.Valdez H, Connick E, Smith KY, Lederman MM, Bosch RJ, Kim RS. Limited immune restoration after 3 years’ suppression of HIV-1 replication in patients with moderately advanced disease. AIDS. 2002;16:1859–1866. doi: 10.1097/00002030-200209270-00002. [DOI] [PubMed] [Google Scholar]
- 16.Philips AN, Carr A, Neuhaus J, Visnegarwala F, Prineas R, Burman WJ, et al. Interruption of antiretroviral therapy and risk of cardiovascular disease in persons with HIV-1 infection: exploratory analyses from the SMART trial. Antivir Ther. 2008;13:177–187. doi: 10.1177/135965350801300215. [DOI] [PubMed] [Google Scholar]
- 17.Rodger AJ, Fox Z, Lundgren JD, Kuller LH, Boesecke C, Gey D, et al. INSIGHT Strategies for Management of Antiretroviral Therapy (SMART) Study Group Activation and coagulation biomarkers are independent predictors of the development of opportunistic disease in patients with HIV infection. J Infect Dis. 2009;200:973–983. doi: 10.1086/605447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.El-Sadr WM, Lundgren JD, Neaton JD, Gordin F, Abrams D, Arduino RC, et al. Strategies for Management of Antiretroviral Therapy (SMART) Study Group CD4+ count-guided interruption of antiretroviral treatment. N Engl J Med. 2006;355:2283–2296. doi: 10.1056/NEJMoa062360. [DOI] [PubMed] [Google Scholar]
- 19.Kuller LH, Tracy R, Belloso W, De Wit S, Drummond F, Lane HC, et al. Inflammatory and coagulation biomarkers and mortality in patients with HIV infection. PLoS Med. 2008;5:1496–1498. doi: 10.1371/journal.pmed.0050203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sandler NG, Wand H, Roque A, Law M, Nason MC, Nixon DE, et al. for the INSIGHT SMART Study Group Plasma levels of soluble CD14 independently predict mortality in HIV infection. J Infect Dis. 2011;203:780–790. doi: 10.1093/infdis/jiq118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marchetti G, Cozzi-Lepri A, Bellistrì GM, Merlini E, Chiodera A, Soscia F, et al. for the Icona Foundation Study Group Role of microbial translocation and immune hyperactivation in disease progression of HIV+ patients with preserved CD4 count in the absence of ART [abstract 333].. 17th Conference on Retroviruses and Opportunistic Infections; San Francisco, California. 16–19 February 2010. [Google Scholar]
- 22.Mocroft A, Wyatt C, Szczech L, Neuhaus J, El-Sadr W, Tracy R, et al. Interruption of antiretroviral therapy is associated with increased plasma cystatin C. AIDS. 2009;23:71–82. doi: 10.1097/QAD.0b013e32831cc129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lars P, Jacquie N, Amanda M, Vincent S, Juergen R, Greg D, et al. for the INSIGHT SMART Study Group Interruption of ART and changes in hyaluronic acid as marker of liver fibrosis progression in strategies for management of ART viral hepatitis-co-infected participants and matched controls. Antivir Ther. in press. [Google Scholar]
- 24.Zhu J, Paul WE. Heterogeneity and plasticity of T helper cells. Cell Res. 2010;20:4–12. doi: 10.1038/cr.2009.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fantuzzi L, Belardelli F, Gessani S. Monocyte/macrophage-derived CC chemokines and their modulation by HIV-1 and cytokines: a complex network of interactions influencing viral replicagtion and AIDS pathogenesis. J Leukoc Biol. 2003;74:719–725. doi: 10.1189/jlb.0403175. [DOI] [PubMed] [Google Scholar]
- 26.de Jager W, Rijkers GT. Solid-phase and bead-based cytokine immunoassay: a comparison. Methods. 2006;38:294–303. doi: 10.1016/j.ymeth.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 27.Elshal MF, McCoy JP. Multiplex bead array assays: performance evaluation and comparison of sensitivity to ELISA. Methods. 2006;38:317–323. doi: 10.1016/j.ymeth.2005.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jenmalm MC, Malit M, Torrence J, Zhang A. Bio-plex cytokine immunoassays and ELISA: comparison of two methodologies in testing samples from asthmatic and healthy children. Bio-Rad Inc.; Hercules, California: [March 2011]. Cytokine assays, techinal note 305. http://www.biocompare.com/Articles/ApplicationNote/1156/Bio-Plex-Cytokine-Immunoassays-And-ELISA-Comparison-Of-Two-Methodologies-In-Testing-Samples-From-Asthmatic-And-Healthy-Children-Rev-A.html. [Google Scholar]
- 29.Marschner IC, Betensky RA, DeGruttola V, Hammer SM, Kuritzkes DR. Clinical trials using HIV-1 RNA-based primary endpoints: statistical analysis and potential biases. J Acquir Immune Defic Syndr Hum Retrovirol. 1999;20:220–227. doi: 10.1097/00042560-199903010-00002. [DOI] [PubMed] [Google Scholar]
- 30.Hsu H, Xiong J, Goeddel DV. The TNF receptor 1-associated protein TRADD signals cell death and NF-kappa B activation. Cell. 1995;81:495–504. doi: 10.1016/0092-8674(95)90070-5. [DOI] [PubMed] [Google Scholar]
- 31.Carpentier I, Coornaert B, Beyaert R. Function and regulation of tumor necrosis factor type 2. Curr Med Chem. 2004;11:2205–2212. doi: 10.2174/0929867043364694. [DOI] [PubMed] [Google Scholar]
- 32.Aggarwal S, Gollapudi S, Gupta S. Increased TNF-alpha-induced apoptosis in lymphocytes from aged humans: changes in TNF-alpha receptor expression and activation of caspases. J Immunol. 1999;162:2154–2161. [PubMed] [Google Scholar]
- 33.Taylor JA, Cummins NW, Bren GD, Rizza SA, Kolbert CP, Behrens MD, et al. Casp8p41 expression in primary T cells induces a proinflammatory response. AIDS. 2010;24:1251–1258. doi: 10.1097/QAD.0b013e3283389e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haas DW, Geraghty DE, Andersen J, Mar J, Motsinger AA, D'Aquila RT, et al. Immunogenetics of CD4 lymphocyte count recovery during antiretroviral therapy: an AIDS Clinical Trials Group study. J Infect Dis. 2006;194:1098–1107. doi: 10.1086/507313. [DOI] [PubMed] [Google Scholar]
- 35.Fernandez S, Rosenow AA, James IR, Roberts SG, Nolan RC, French MA, Price P. Recovery of CD4+ T Cells in HIV patients with a stable virologic response to antiretroviral therapy is associated with polymorphisms of interleukin-6 and central major histocompatibility complex genes. J Acquir Immune Defic Syndr. 2006;41:1–5. doi: 10.1097/01.qai.0000188990.57760.e3. [DOI] [PubMed] [Google Scholar]
- 36.Aukrust P, Müller F, Lien E, Nordoy I, Liabakk NB, Kvale D, et al. Tumor necrosis factor (TNF) system levels in human immunodeficiency virus-infected patients during highly active antiretroviral therapy: persistent TNF activation is associated with virologic and immunologic treatment failure. J Infect Dis. 1999;179:74–82. doi: 10.1086/314572. [DOI] [PubMed] [Google Scholar]
- 37.Reddy MM, Sorrell SJ, Lange M, Grieco MH. Tumor necrosis factor and HIV P24 antigen levels in serum of HIV-infected populations. J Acquir Immune Defic Syndr. 1988;1:436–440. [PubMed] [Google Scholar]
- 38.Godfried MH, van der Poll T, Jansen J, Romijin JA, Schattenkerk JK, Endert E, et al. Soluble receptors for tumor necrosis factor are markers for clinical course but not for major metabolic changes in human immunodeficiency virus infection. Metabolism. 1995;44:1564–1569. doi: 10.1016/0026-0495(95)90076-4. [DOI] [PubMed] [Google Scholar]
- 39.Stein DS, Lyles RH, Graham NM, Tassoni CJ, Margolick JB, Phair JP, et al. Predicting clinical progression or death in subjects with early-stage human immunodeficiency virus (HIV) infection: a comparative analysis of quantification of HIV RNA, soluble tumor necrosis factor type II receptors, neopterin, and beta2-microglobulin. Multicenter AIDS Cohort Study. J Infect Dis. 1997;176:1161–1167. doi: 10.1086/514108. [DOI] [PubMed] [Google Scholar]
- 40.Fahey JL, Taylor JM, Manna B, Nishanian P, Aziz N, Giorgi JV, Detels R. Prognostic significance of plasma markers of immune activation, HIV viral load and CD4 T-cell measurements. AIDS. 1998;12:1581–1590. doi: 10.1097/00002030-199813000-00004. [DOI] [PubMed] [Google Scholar]
- 41.Kalayjian RC, Machekano RN, Rizk N, Robbins GK, Gandhi RT, Rodriguez BA, et al. Pretreatment levels of soluble cellular receptors and interleukin-6 are associated with HIV disease progression in subjects treated with highly active antiretroviral therapy. J Infect Dis. 2010;201:1796–1805. doi: 10.1086/652750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parish ST, Wu JE, Effros RB. Modulation of T lymphocyte replicative senescence via TNF-{alpha} inhibition: role of caspase-3. J Immunol. 2009;182:4237–4243. doi: 10.4049/jimmunol.0803449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Burton DG, Matsubara H, Ikeda K. Pathophysiology of vascular calcification: pivotal role of cellular senescence in vascular smooth muscle cells. Exp Gerontol. 2010;45:819–824. doi: 10.1016/j.exger.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 44.Norris PJ, Pappalardo BL, Custer B, Spotts G, Hecht FM, Busch MP. Elevations in IL-10, TNF-alpha, and IFN-gamma from the earliest point of HIV Type 1 infection. AIDS Res Hum Retroviruses. 2006;22:757–762. doi: 10.1089/aid.2006.22.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stacey AR, Norris PJ, Qin L, Haygreen EA, Taylor E, Heitman J, et al. Induction of a striking systemic cytokine cascade prior to peak viremia in acute human immunodeficiency virus type 1 infection, in contrast to more modest and delayed responses in acute hepatitis B and C virus infections. J Virol. 2009;83:3719–3733. doi: 10.1128/JVI.01844-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rychert J, Strick D, Bazner S, Robinson J, Rosenberg E. Detection of HIV gp120 in plasma during early HIV infection is associated with increased proinflammatory and immunoregulatory cytokines. AIDS Res Hum Retroviruses. 2010;26:1139–1145. doi: 10.1089/aid.2009.0290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stylianou E, Aukrust P, Bendtzen K, Müller F, Frøland SS. Interferons and interferon (IFN)-inducible protein 10 during highly active antiretroviral therapy (HAART)-possible immunosuppressive role of IFN-a in HIV infection. Clin Exp Immunol. 2000;119:479–485. doi: 10.1046/j.1365-2249.2000.01144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gu Y, Yang J, Ouyang X, Liu W, Li H, Yang J, et al. Interleukin 10 suppresses Th17 cytokines secreted by macrophages and T cells. Eur J Immunol. 2008;38:1807–1813. doi: 10.1002/eji.200838331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mills KH. Induction, function and regulation of IL-17-producing T cells. Eur J Immunol. 2008;38:2636–2649. doi: 10.1002/eji.200838535. [DOI] [PubMed] [Google Scholar]
- 50.Heo YJ, Joo YB, Oh HJ, Park MK, Heo YM, Cho ML, et al. IL-10 suppresses Th17 cells and promotes regulatory T cells in the CD4+ T cell population of rheumatoid arthritis patients. Immunol Lett. 2010;127:150–156. doi: 10.1016/j.imlet.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 51.Naundorf S, Schröder M, Höflich C, Suman N, Volk HD, Grütz G. IL-10 interferes directly with TCR-induced IFN-gamma but not IL-17 production in memory T cells. Eur J Immunol. 2009;39:1066–1077. doi: 10.1002/eji.200838773. [DOI] [PubMed] [Google Scholar]
- 52.Klatt NR, Brenchley JM. Th17 cell dynamics in HIV infection. Curr Opin HIV AIDS. 2010;5:135–140. doi: 10.1097/COH.0b013e3283364846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McKinstry KK, Strutt TM, Buck A, Curtis JD, Dibble JP, Huston G, et al. IL-10 deficiency unleashes an influenza-specific Th17 response and enhances survival against high-dose challenge. J Immunol. 2009;182:7353–7363. doi: 10.4049/jimmunol.0900657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roe B, Coughlan S, Hassan J, Grogan A, Farrell G, Norris S, et al. Elevated serum levels of interferon-gamma-inducible protein-10 in patients coinfected with hepatitis C virus and HIV. J Infect Dis. 2007;196:1053–1057. doi: 10.1086/520935. [DOI] [PubMed] [Google Scholar]
- 55.Stone SF, Price P, Brochier J, French MA. Plasma bioavailable interleukin-6 is elevated in human immunodeficiency virus-infected patients who experience herpesvirus-associated immune restoration disease after start of highly active anti-retroviral therapy. J Infect Dis. 2001;184:1073–1077. doi: 10.1086/323599. [DOI] [PubMed] [Google Scholar]
- 56.Flandre P, Durier C, Descamps D, Launay O, Joly V. On the use of magnitude of reduction in HIV-1 RNA in clinical trials: statistical analysis and potential biases. J Acquir Immune Defic Syndr. 2002;30:59–64. doi: 10.1097/00042560-200205010-00007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.