Abstract
An approximately 12.5-kbp region of DNA sequence from beyond the end of the previously described clavulanic acid gene cluster was analyzed and found to encode nine possible open reading frames (ORFs). Involvement of these ORFs in clavulanic acid biosynthesis was assessed by creating mutants with defects in each of the ORFs. orf12 and orf14 had been previously reported to be involved in clavulanic acid biosynthesis. Now five additional ORFs are shown to play a role, since their mutation results in a significant decrease or total absence of clavulanic acid production. Most of these newly described ORFs encode proteins with little similarity to others in the databases, and so their roles in clavulanic acid biosynthesis are unclear. Mutation of two of the ORFs, orf15 and orf16, results in the accumulation of a new metabolite, N-acetylglycylclavaminic acid, in place of clavulanic acid. orf18 and orf19 encode apparent penicillin binding proteins, and while mutations in these genes have minimal effects on clavulanic acid production, their normal roles as cell wall biosynthetic enzymes and as targets for β-lactam antibiotics, together with their clustered location, suggest that they are part of the clavulanic acid gene cluster.
Among prokaryotes, the Streptomyces spp. are unusual for both their morphological and biochemical versatility. When these filamentous soil bacteria grow on solid substrates, they exhibit a complex life cycle which progresses from spores through vegetative and aerial mycelia and back to spores. Often, coincident with these morphological changes, the organisms produce a diverse array of exocellular enzymes and secondary metabolites.
Streptomyces clavuligerus is notable in this regard for its ability to produce β-lactam-type compounds. In particular, it is the species used industrially for the production of clavulanic acid, a multi-billion-dollar/annum product useful for its β-lactamase inhibitory activity. While the biosynthesis of clavulanic acid has been the subject of intense investigation in recent years, the details of its production are still not completely worked out.
Previously Jensen et al. characterized an approximately 15-kb stretch of chromosomal DNA located adjacent to the cephamycin gene cluster in S. clavuligerus and demonstrated that the open reading frames (ORFs) were involved in clavulanic acid production (12). Genes located in this region include ceaS (encoding carboxyethylarginine synthase) (14), bls (β-lactam synthetase) (3, 18), pah (proclavaminate amidinohydrolase) (2, 11, 34), cas2 (clavaminate synthase) (4, 17, 26, 27), and cad (clavulanic acid dehydrogenase) (21), all enzymes for recognized steps in the biosynthetic pathway. In addition, oat, a gene encoding a protein with ornithine acetyltransferase activity, has been identified (13), but its role in clavulanic acid is not yet understood, and claR, a gene encoding a transcriptional regulator that controls the late steps in clavulanic acid biosynthesis, is also located in this region (22, 24). Two additional genes, orf7 and orf10, encode an apparent peptide transport protein and a cytochrome P450 type protein, respectively (11, 12), as judged by BLAST analysis. While both have been shown to be essential for clavulanic acid biosynthesis, their precise roles are unclear.
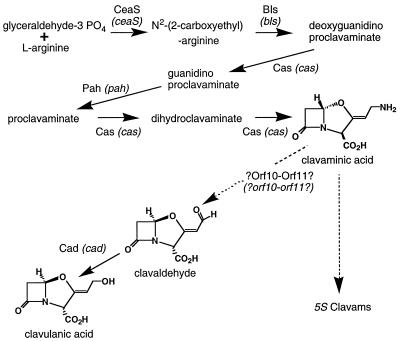
Genes within this region account for all of the early steps and for the final step in clavulanic acid biosynthesis (Fig. 1). However, the late step(s) of the pathway from clavaminic acid to clavaldehyde remains unaccounted for. While it has been suggested that the cytochrome P450 protein encoded by orf10 might carry out this conversion (16), no demonstration of activity in this regard has followed. Furthermore, it is not clear that a single protein could accomplish both the inversion of stereochemistry of the clavam nucleus and the side chain modifications at C2 that this step involves. Since other candidate genes encoding enzymes capable of bringing about these changes were not obvious within this 15-kb region, several groups, including our own, looked beyond the end of the known clavulanic acid biosynthetic gene cluster for additional genes involved in biosynthesis of this important metabolite.
FIG. 1.
The biosynthetic pathway leading to clavulanic acid and the 5S clavams.
Recently, Li et al. (16) provided evidence for two additional genes, orf11 and orf12, extending the clavulanic acid gene cluster; they encode an apparent ferredoxin and a protein showing some similarity to β-lactamases, respectively. While the ferredoxin is presumed to work in concert with the cytochrome P450 encoded by orf10, the role of the β-lactamase-like protein is again unclear, although gene disruption showed it to be essential for clavulanic acid biosynthesis. Similarly, Mellado et al. (19) provided additional sequence information giving evidence for the presence of orf13 to orf19 in this region. Through gene disruption they demonstrated an involvement of orf14 in clavulanic acid biosynthesis, but the rest of the ORFs remain uncharacterized. While it is not immediately apparent from BLAST similarity searches how any of the ORFs would be involved in the clavaminate to clavaldehyde conversion, it is also not clear what type of ORFs would be expected for this conversion. Furthermore, the close spacing of the ORFs does suggest operon arrangements indicating that the genes might be involved in related processes. Here we describe a mutational analysis of orf11 to orf19 and provide evidence to suggest that all of these ORFs, with the possible exception of orf18 and orf19, are part of the clavulanic acid biosynthetic gene cluster.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Bacterial strains and plasmids used in this study are described in Table 1. Escherichia coli strains were maintained on Luria broth (28) agar plates and grown in Luria broth liquid medium at 37°C. Plasmid-bearing cultures were supplemented with ampicillin (100 μg/ml) or apramycin (25 μg/ml) as appropriate.
TABLE 1.
Bacterial strains and plasmids used in this study
| Bacterial strains and plasmids | Relevant featuresa | Source or reference |
|---|---|---|
| Strains | ||
| S. clavuligerus | ||
| NRRL 3585 | Wild type; cephamycin and clavulanic acid producer | NRRL |
| E. coli | ||
| XLI-blue | General cloning host | Stratagene |
| ET12567(pUZ8002) | Methylation deficient; transfer functions from pUZ8002 | 15 |
| Plasmids | ||
| Cloning vectors | ||
| pBluescript II KS(+) | E. coli general cloning vector, Ampr | Stratagene |
| pSL1180 | E. coli general cloning vector, Ampr | Pharmacia |
| pTZ18R | E. coli general cloning vector, Ampr | US Biochemicals |
| pUC119 | E. coli general cloning vector, Ampr | 32 |
| pSET152 | E. coli replicon, Streptomyces φC31 attachment site, aptr | NRRL |
| pCR 2.1-TOPO | E. coli vector for cloning PCR products, Ampr | Invitrogen |
| pJV326 | par-defective, oriT-containing, Streptomyces-E. coli shuttle | 9 |
| pIJ486 | Streptomyces pIJ101 replicon, Tsrr | 33 |
| pMT3226 | pSET152 derivative, xylE controlled by gylP1P2 promoters | 10, 31 |
| Antibiotic resistance cassettes | ||
| pUC120apr | Aprr cassette in pUC120 | 23 |
| pSKNeo | Neor cassette in pBluescript II KS(+) | 20 |
| Intermediate constructs | ||
| K6L2 | E. coli cosmid carrying S. clavuligerus genomic DNA | 2 |
| p667-3PB | middle of orf18 to end of orf19 in pUC119 | This study |
| pCEC001 | Start of orf10 to middle of orf18 in pTZ18R | This study |
| pCEC004 | Middle of orf13 to middle of orf16 in pBluescript II SK(+) | This study |
| pCEC028 | Middle of orf12 to end of orf15 in pBluescript II SK(+) | This study |
| pCEC062 | End of orf17 to middle of orf18 in pBluescript II KS(+) | This study |
| Gene disruption constructs | ||
| pLOG221 | orf11::apr disruption construct | This study |
| pLOG 240 | orf12::apr disruption construct | This study |
| pCEC046 | orf14::apr disruption construct | This study |
| pCEC047 | orf13::apr disruption construct | This study |
| pCEC063 | orf15::apr disruption construct | This study |
| pCEC068 | orf16::apr disruption construct | This study |
| pCEC076 | orf17::apr disruption construct | This study |
| pCEC085 | orf18::neo disruption construct | This study |
| pCEC086 | orf15-Δfs replacement construct | This study |
| pCEC089 | orf19::apr disruption construct | This study |
| pCEC179 | Δorf16 replacement construct | This study |
| Gene insertion construct | ||
| pMT8.34 | pgyl::orf18 construct in pSET152 | This study |
Abbreviations: Amp, ampicillin; Apr, apramycin; Neo, neomycin; Tsr, thiostrepton.
S. clavuligerus wild-type and mutant strains were maintained on either MYM agar (29) or ISP #4 medium (Difco) at 28°C, except for orf18::apr mutants of the S. clavuligerus pgyl-orf18 strain, which were maintained on ISP #4 supplemented with 1% glycerol. For assessment of metabolite production, cultures were grown as previously described (20). Briefly, glycerol spore stocks of S. clavuligerus were used to inoculate Trypticase soy broth-1% soluble starch seed medium. Seed cultures (40 to 48 h) were used to inoculate both starch asparagine (SA) and soy flour (Soy) media to 2% (vol/vol), and samples were removed at 72 and 96 h for analysis. All cultures were incubated at 28°C and 250 rpm. Representative mutants were grown in duplicate cultures, and all experiments were repeated at least twice in two separate laboratories. Plasmid-carrying cultures were supplemented with apramycin (25 μg/ml), thiostrepton (5 μg/ml), or neomycin (50 μg/ml) as appropriate.
DNA manipulations.
Plasmid and genomic DNA preparations from S. clavuligerus were isolated using standard techniques (15). Plasmid DNA was typically introduced into Streptomyces spp. by transformation of protoplasts. Protoplasts were prepared and used for transformation procedures as described previously (23).
Where indicated, plasmids were also introduced into S. clavuligerus spores through the use of the conjugation procedure described by Kieser et al. (15), with E. coli ET12567 (pUZ8002) as the donor strain. Exconjugants were isolated on AS-1 (5) (with appropriate antibiotic selection) rather than on soy-mannitol medium.
Plasmid DNA isolation from E. coli cultures, restriction endonuclease digestions, ligations, PCRs, Southern analyses, and transformations of E. coli were all performed using standard techniques (28).
DNA sequence analysis.
Cosmid clone K6L2, which carries an approximately 30-kb fragment of S. clavuligerus chromosomal DNA in the vector pLAFR3 (2), was digested with PstI and EcoRI to generate a unique DNA fragment of 11.2 kb. This fragment, now known to extend from the beginning of orf10 to near the 5′ end of orf18 in the clavulanic acid gene cluster, was subcloned into plasmid pTZ18R to yield plasmid pCEC001. Restriction analysis of K6L2 indicated that a 2.8-kb PstI-BamHI fragment is located adjacent to the 11.2-kb PstI-EcoRI fragment. This 2.8-kb PstI-BamHI fragment, now known to contain the 5′ end of orf18 and all of orf19, was subcloned from cosmid K6L2 into pUC119 to yield plasmid p667-3PB. The DNA sequence for part of the pCEC001 insert was already known from previous studies (12). The DNA sequence for the remaining part, and for the entire insert of p667-3PB, was determined essentially as described previously (12). Subclones generated by restriction endonuclease digestion as well as ordered sets of deletion subclones were sequenced from universal primers. Sequence-specific oligonucleotide primers were used for regions which were not obtained from analyses using universal primers. All sequence information was verified on both strands, and the arrangement of restriction fragments was confirmed by obtaining sequence information for cross-fragment junctions. Sequence analyses were performed using a DYEnamic ET Terminator cycle sequencing kit (Amersham Pharmacia, Baie d'Urfe, Quebec, Canada) by the Molecular Biology Service Unit, University of Alberta.
Gene disruption of orf11 to orf19.
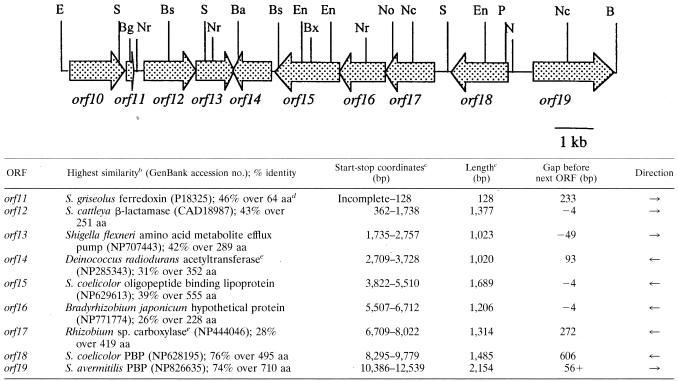
The region of the S. clavuligerus chromosome encompassing orf10 to orf19 is represented diagrammatically in Table 2, and restriction sites of relevance to the gene disruption studies are indicated. In preparing the disruption constructs described below, whenever DNA fragments with incompatible ends were ligated they were first made blunt by digestion with either the Klenow fragment of DNA polymerase I or with T4 DNA polymerase.
TABLE 2.
Characteristics and arrangement of the newly described ORFs from the clavulanic acid gene clustera
The top panel represents the ORFs found in the inserts from pCEC001 and p667-3PB. B, BamHI; Ba, BalI; Bg, BglII; Bs, BstEII; Bx, BstXI; E, EcoRI; En, EcoNI; N, NaeI; Nc, NcoI; No, NotI; Nr, NruI; P, PstI; S, SphI. Only sites mentioned in the text are shown.
Protein with highest level of similarity to ORF-encoded protein.
Includes stop codon.
aa, amino acids.
Protein was annotated as a hypothetical protein, but conserved domains were detected, suggesting that the protein was an acetyltransferase (orf14) or carboxylase (orf17).
Disruption of orf11.
A 2.2-kb SphI fragment of S. clavuligerus DNA which extends from near the 3′ end of orf10 to within orf13 was subcloned into pBluescript II SK(+).
The resulting plasmid was linearized by digestion with BglII at the unique site centrally located in orf11 and ligated to a 1.45-kb NcoI fragment carrying the apramycin resistance gene (aprr) from pUC120apr (23). The disrupted orf11 gene (aprr in the same orientation as orf11) was then excised as a BamHI-HindIII fragment and inserted into the Streptomyces vector pIJ486 to give pLOG221. pLOG221 was transformed through S. lividans and into wild-type S. clavuligerus. Apramycin- and thiostrepton-resistant transformants were subjected to two rounds of sporulation on solid ISP #4 medium with no antibiotic supplementation and then replica plated onto antibiotic-supplemented medium. The presence of putative gene replacement mutants (apramycin resistant and thiostrepton sensitive) was confirmed by Southern analysis.
Disruption of orf12.
A 1.8-kb NruI-SphI fragment of S. clavuligerus DNA which extends from just downstream of orf11 to within orf13 was subcloned into pBluescript II SK(+). The resulting plasmid was linearized at the BstEII site located within orf12 and ligated to the aprr cassette. The disrupted orf12 gene (aprr in the orientation opposite to orf12) was transferred to pIJ486 as a BamHI-HindIII fragment to yield pLOG240. pLOG240 was transformed through S. lividans and into wild-type S. clavuligerus, after which putative gene replacement mutants were selected as described above and then confirmed by Southern analysis.
Disruption of orf13.
A 2.9-kb BstEII fragment of S. clavuligerus DNA carrying part of orf12, all of orf13 and orf14, and the 3′ end of orf15 was subcloned into pBluescript II SK(+). The resulting plasmid, pCEC028, was linearized at the unique NruI site within orf13 and ligated to the aprr cassette. pCEC028 was linearized with HindIII and ligated to HindIII-digested pIJ486 to give a shuttle plasmid, pCEC047 (aprr in the orientation opposite to orf13). pCEC047 and the transformation protocol described for orf11 were used to generate mutants of S. clavuligerus.
Disruption of orf14.
pCEC028 was linearized by digestion at the unique BalI site located within orf14 and ligated to the aprr cassette. The resulting plasmid was fused to pIJ486 at the HindIII site to give a shuttle plasmid, pCEC046 (an aprr fragment in the same orientation as orf14), and pCEC046 and the transformation protocol described for orf11 were used to generate mutants of S. clavuligerus.
Disruption of orf15.
A 4-kb NruI fragment of S. clavuligerus DNA carrying part of orf13, all of orf14 and orf15, and part of orf16 was subcloned into pBluescript II SK(+) to generate pCEC004. pCEC004 was digested with SacI and XbaI and religated to delete the BstXI site from the multiple cloning site. The resulting plasmid was linearized at the remaining BstXI site within orf15, ligated to the aprr cassette, and then fused to pIJ486 at the HindIII site. The resulting shuttle plasmid, pCEC063 (aprr in the orientation opposite to orf15), and the transformation protocol described for orf11 were used to generate mutants of S. clavuligerus.
A second version of the orf15 mutant was prepared in which the aprr cassette was removed and replaced by a deletion frameshift mutation. To prepare these orf15-Δfs mutants, pCEC004 was linearized by digestion with EcoNI, which removes a 0.75-kb fragment from within orf15. The 0.75-kb EcoNI fragment encompasses the BstXI site that is the site of the disruption in the original orf15::apr mutant. The linearized plasmid was made blunt and then religated, generating a deletion frameshift mutation in orf15. The resulting plasmid was fused to pIJ486 at the SacI site to give the shuttle plasmid, pCEC086. pCEC086 was transformed through S. lividans and into the orf15::apr mutant. Since the desired orf15-Δfs mutants lack a selectable marker, presumptive mutants were chosen on the basis of their loss of apramycin resistance after sporulation on nonselective medium; the mutations were confirmed by Southern analysis.
Disruption of orf16.
A 2.7-kb BstXI-NcoI fragment of S. clavuligerus DNA extending from the middle of orf15 to the middle of orf17 was subcloned into pSL1180. The resulting plasmid was linearized by digestion at the unique NruI site within orf16, ligated to the aprr cassette, and then fused to pIJ486 at the HindIII site. The resulting shuttle plasmid, pCEC068 (aprr in the same orientation as orf16), and the transformation protocol described for orf11 were used to generate mutants of S. clavuligerus.
A second version of the orf16 mutant was prepared in which the aprr cassette was removed and replaced by a deletion spanning most of orf16. To prepare these Δorf16 mutants, two 1-kb fragments from the upstream and downstream flanks of orf16 were amplified by the PCR. The upstream fragment, PCR I, included DNA sequence encoding the first 14 amino acid residues of orf16 and was generated using primers 5′-CCCCGACCCCGAGGCCGTGG and 5′-ATCGATCGGCAGCGCGTCCGTCAC (italicized residues represent a ClaI digestion site introduced for cloning). The downstream fragment, PCR II, included a DNA sequence encoding the last 13 amino acid residues and the stop codon of orf16 and was generated using primers 5′-GCGCGGCGTACGTGCACATTG and 5′-ATCGATGGCACCACCGACACCGATCAGA (italicized residues represent a ClaI digestion site introduced for cloning). Both PCR I and PCR II fragments were cloned into pCR2.1-TOPO through the use of a TOPO TA cloning kit (Invitrogen) and sequenced to confirm fidelity. The two fragments were then assembled, and the resulting 2-kb PCR I-PCR II fragment was transferred into the partition-defective E. coli-Streptomyces shuttle vector, pJV326, to yield pCEC179. pCEC179 was then transformed into E. coli ET12567 (pUZ8002) and from there introduced into the S. clavuligerus orf16::apr mutant by conjugation. Thiostrepton-resistant exconjugants were subjected to two rounds of sporulation on ISP #4 medium with no antibiotic supplementation, and presumptive mutants were chosen on the basis of their loss of both apramycin and thiostrepton resistance after sporulation on nonselective medium. The identity of the mutants was confirmed by Southern analysis.
Disruption of orf17.
An approximately 3-kb NotI-PstI fragment of S. clavuligerus DNA including most of orf17 and orf18 was subcloned into pBluescript II KS(+) to give pCEC062. pCEC062 was subjected to partial digestion with NcoI to obtain a singly cut plasmid as the major digestion product. DNA fragments obtained upon partial NcoI digestion were then ligated with the aprr cassette to give plasmids in which aprr had inserted into one of the three NcoI sites present on the plasmid. Screening by restriction analysis identified one plasmid that contained aprr inserted at the NcoI site within orf17 (in the orientation opposite to orf17), and that plasmid was fused to pIJ486 at the HindIII site. The resulting shuttle plasmid, pCEC076, and the transformation protocol described for orf11 were used to generate mutants of S. clavuligerus.
Disruption of orf18.
pCEC062 was linearized by digestion at the unique EcoNI site within orf18 and ligated to the neomycin resistance gene cassette (neor) isolated from pSKNeo (20) as a 1-kb AccI fragment. The resulting plasmid was fused to pIJ486 at the SstI site to give the shuttle plasmid, pCEC085 (neor in the orientation opposite to orf18). pCEC085 was transformed through S. lividans and into S. clavuligerus as described for orf11. Despite repeated attempts, no orf18 mutants were isolated.
For an alternative host strain in which to attempt the disruption of orf18, an additional copy of orf18 under the transcriptional control of the glycerol-inducible promoter was introduced into the S. clavuligerus chromosome to create a strain diploid with respect to orf18. To accomplish this, the pSET152-based vector pMT3226 (10, 31) was digested with BamHI and XbaI to release the xylE gene and a 1.6-kb SphI-NaeI fragment of S. clavuligerus DNA extending from 39 bp upstream to 109 bp downstream of orf18 was inserted in its place. The resulting plasmid, pMT8.34, carries the orf18 gene positioned in the correct orientation just downstream from the gylP1P2 promoters. pMT8.34 was then introduced into wild-type S. clavuligerus by transformation, and apramycin-resistant transformants were analyzed by PCR to confirm that the plasmid had integrated at the ΦC31 attachment site rather than by recombination with the native orf18 gene. The resulting diploid strain was designated the pgyl::orf18 strain.
pCEC085 was then transformed into the pgyl::orf18 strain with selection for neomycin resistance, and putative mutants were isolated from transformants after two rounds of sporulation on nonselective ISP #4 medium supplemented with 1% glycerol. Southern analysis was carried out on these putative disruptants to determine whether the neor cassette had disrupted the native copy of orf18 or the ectopic pgyl::orf18.
Disruption of orf19.
A 1.6-kb DNA fragment internal to orf19 was amplified by PCR using oligonucleotides 5′-GCATAGGATCCGGATACGGCTCGTGGTCGTCCAGATACTGG (italicized residues represent a BamHI digestion site introduced for cloning) and 5′-GCATCGAATTCGAGGGCCGCGTAGATCGTCGCCATCTGGATG (italicized residues represent a EcoRI digestion site introduced for cloning) as primers. The PCR product was cloned into pSL1180 and then linearized by digestion at the unique NcoI site located midway through the orf19 fragment and ligated to the aprr cassette. The resulting plasmid was fused to pIJ486 at the EcoRI site, and the resulting shuttle plasmid, pCEC089 (aprr in the same orientation as orf19), and the transformation protocol described for orf11 were used to generate mutants of S. clavuligerus.
HPLC and mass spectrometric analyses.
Culture filtrates were routinely analyzed for the presence of clavulanic acid and the other clavam metabolites by high-pressure liquid chromatography (HPLC) after imidazole derivatization, as described previously (20). In those cases in which new clavam metabolites were detected, the imidazole-derivatized culture supernatants were reanalyzed by an alternative HPLC procedure employing a volatile buffer system; effluents were then analyzed by electrospray ionization mass spectrometry. Derivatized samples (5 μl) were analyzed on an XTerra column (Waters Scientific, Milford, Mass.) (0.21 by 10 cm) at a flow rate of 0.25 ml/min. The mobile phase consisted of solvent A (10 mM ammonium bicarbonate, pH 10) and solvent B (acetonitrile) used in a binary gradient system as follows: 100% solvent A for 5 min, linear gradient to 85% solvent A over 10 min, 85% solvent A for 5 min, linear gradient to 100% solvent A over 1 min, and 100% solvent A for 9 min. Eluant was monitored at 311 nm to detect the imidazole derivatives, and mass spectra were acquired using a ZMD-2 single-quadrupole instrument (Waters Scientific).
Bioassays.
In an indirect bioassay for β-lactamase inhibitors, clavulanic acid was detected using Klebsiella pneumoniae ATCC 29665 as the indicator organism growing on Trypticase soy agar (Becton Dickinson) containing penicillin G at 6 μg/ml. Alanylclavam was detected by a bioassay using Bacillus sp. ATCC 27860 as the indicator organism growing on a defined agar medium (25). Cephamycin C was detected in culture filtrates by a bioassay using the indicator organism E. coli ESS (1) growing on Trypticase soy agar.
Nucleotide sequence accession number.
The nucleotide sequence of this region of the S. clavuligerus chromosome has been deposited in GenBank (accession number AY258009).
RESULTS
DNA sequence information from the clavulanic acid gene cluster.
Previously, we reported the DNA sequence for an approximately 15-kb region of the S. clavuligerus chromosome (GenBank accession number AF205427) encoding a series of structural genes involved in clavulanic acid production. While these genes accounted for many of the expected activities, it was not clear that all of the genetic information needed to form clavulanic acid was present in this region. In view of the possibility that additional genes might be required for the production of clavulanic acid, we investigated the DNA sequence of the region beyond this already reported area. Beginning immediately adjacent to the sequence information reported previously (12), we have determined an additional 12,595 bp of DNA sequence ending at a BamHI site. The previously deposited sequence information terminated partway through what is now recognized as orf11. Examination of the new DNA sequence information indicates the presence of the rest of orf11 as well as of eight additional complete open reading frames, orf12 to orf19 (carrying on with the numbering established for the previous sequence information) (Table 2). Two of these ORFs (orf11 and orf12) have been reported previously by Li et al. (16). Recently, Mellado et al. (19) also reported additional DNA sequence information extending from orf10 through orf18 and into orf19.
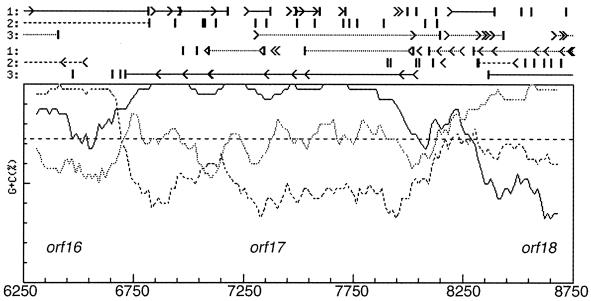
The sequence information presented in this study covers largely the same region as that reported by Mellado et al. but extends further to give the complete sequence for orf19. With the benefit of their published sequence information for comparison, we were able to identify several areas where our sequence data differed. These differences are noted in Table 3. Careful reexamination of our data in these regions did not show any ambiguous or unclear bases, and so these may represent differences between isolates or they may represent sequencing errors. None of these sequence discrepancies affected the reading frame of the putatively encoded proteins, but they did result in a number of amino acid changes. Analysis of the sequenced region by database searching allowed us to recognize the same similarities between the proteins encoded by orf11 to orf19 and other sequenced proteins reported by Mellado et al. (Table 2) (19). However, we did choose different start codon possibilities for several of the ORFs in the region, resulting in different sizes for the encoded proteins (Table 2). Because of the presence of a more convincing ribosome binding site, we designated the ATG codon at nucleotide (nt) 362 the start of orf12, making the ORF 1,377 bp in length (including the stop codon), 24 bp shorter than that suggested by Mellado et al. The predicted ORF12 protein shows only limited similarity to other proteins in the databases, and no useful similarities which could help to clarify the start codon were noted in the extreme 5′ end of the ORF. Similarly, we chose a different start site for orf16 (nt 6712), making the gene 1,206 bp in length, 24 bp longer than that suggested by Mellado et al., and providing a 4-bp overlap and presumed translational coupling with orf17. This is consistent with the Frame analysis of the region (http://www.nih.go.jp/∼jun/cgi-bin/frameplot.pl) (Fig. 2), which shows an abrupt transition from orf16 to orf17 indicative of translational coupling with no intergenic region. We also chose a different start site for orf17 (nt 8022), making the gene 1,314 bp in length, 276 bp shorter than that suggested by Mellado et al., and introducing an intergenic region in place of the proposed 4-bp overlap. Once again this change is consistent with the Frame analysis of the region, which shows clear evidence of an intergenic region between the two genes (Fig. 2). In addition, examination of the amino acids that would be encoded by this otherwise intergenic region shows a predominance of arginine and alanine residues quite unlike that seen with the rest of the ORF17 protein (data not shown). While the ultimate determination of start codons for the various ORFs will require biochemical analyses, the existence of these various possible alternatives should be considered if expression studies are planned in the meantime.
TABLE 3.
Differences between newly determined sequence and published sequence
| orf | Location in new sequencea (nt) | Comparison of sequences from indicated sources
|
|
|---|---|---|---|
| Publishedb | This workb | ||
| 16 | 6374-6382 | GGC CCC GGG | GGC CTC GGG |
| A G P | A E P | ||
| 16 | 6590-6598 | GAG AGC CGG | GAG ATC CGG |
| L A P | L D P | ||
| 17 | 6853-6861 | GCG CTT AAG | GCG CTG GAG |
| R K L | R O L | ||
| 17 | 6865-6873 | CTT GAC GCC | CTT GTC GCC |
| D V G | K D G | ||
| 19 | 10521-10538 | TAC CGC AGA AGG CCG TCG | TAC GCG GAG AAG GCG TCG |
| V RRRP S | V AEKA S | ||
FIG. 2.
Frame analysis of the orf16-orf18 region. nt 6250 to 8750 of the deposited sequence were subjected to Frame analysis. Frame analysis examines percentages of G+C content of DNA as a function of codon position and is useful for the prediction of protein coding regions in organisms with genomic DNA with high levels of G+C content (see http://www.nih.go.jp/∼jun/cgi-bin/frameplot.pl). Genes within organisms with genomic DNA with high levels of G+C content exhibit biased codon usage, resulting in extremely high levels of G+C distribution at the third-letter position of each codon. Lines marked with arrowheads and bars above the main graph indicate the presence of potential start and stop codons in each of the six possible reading frames.
In addition to the sequence information, Li et al. also presented evidence based on gene disruption mutation to indicate that orf12 is essential for clavulanic acid production (16). Similarly, Mellado et al. presented evidence based on mutant studies suggesting that orf14 is important but not essential for clavulanic acid production (19). Furthermore, they reported an unpublished result from Lorenzana and Liras indicating that orf15 was essential for clavulanic acid production. No information was presented regarding the involvement of the remaining ORFs in clavulanic acid production except to note that the close spacing of the ORFs suggested an operon arrangement consistent with their involvement in clavulanic acid biosynthesis.
Targeted disruption of orf11 to orf19.
To assess the involvement of these newly described ORFs in clavulanic acid production, each was mutated using established procedures. In each case, the isolated gene was disrupted by insertion of an antibiotic resistance gene into the open reading frame; the disrupted gene was then introduced into the wild-type strain and allowed to replace the resident wild-type copy of the gene by homologous recombination. Presumptive mutants were initially isolated on the basis of their antibiotic resistance phenotypes and then confirmed by Southern analysis. Disruption mutants were prepared successfully for all of the ORFs except orf18. In each case, the size of the fragment(s) observed upon Southern analysis of the disruption mutant was consistent with the insertion of the antibiotic resistance marker at the intended site.
Clavulanic acid production by disruption mutants.
Each disruption mutant was then characterized to determine the effect of the mutation on clavulanic acid production. Mutants were cultivated on both Soy and SA media, and samples were withdrawn after 72 and 96 h of incubation and assessed for metabolite production by bioassay and HPLC analysis. All mutant cultures were compared to similarly grown wild-type S. clavuligerus reference cultures. Effects of the mutations on clavulanic acid production are shown in Table 4.
TABLE 4.
Effect of gene disruption mutations on clavulanic acid production
| Mutant gene (similaritya) | Reduction in clavulanic acid productionb (% relative to wild type) in indicated medium
|
|
|---|---|---|
| Soy | SA | |
| orf11 (ferredoxin) | 70-80 | 25-65 |
| orf12 (β-lactamase) | 100 | 100 |
| orf13 (efflux pump) | 75-95 | 75-95 |
| orf14 (acetyltransferase) | 99.5 | 100 |
| orf15 (peptide binding protein) | 100 | 100 |
| orf16 (hypothetical protein) | 100 | 100 |
| orf17 (carboxylase) | 100 | 100 |
| orf18 (PBP) | Not done | Not done |
| orf19 (PBP) | 0 | 0 |
Protein with highest level of similarity to that encoded by indicated ORF.
Representative mutants were cultured in duplicate and growth experiments were repeated on two or more occasions in two different locations. Typical clavulanic acid production levels for the wild type were 225 μg/ml in Soy medium and 20 μg/ml in SA medium. The limit of detection for clavulanic acid was about 1 μg/ml in Soy medium grown cultures.
Disruption of orf11 resulted in a decreased ability of the mutant strains to produce clavulanic acid. When grown on SA medium, clavulanic acid levels were reduced by 25 to 65%; when grown on Soy medium, mutant strains showed a 70 to 80% reduction in clavulanic acid production. Mutants disrupted in orf13 were severely compromised but not completely defective in their ability to produce clavulanic acid on both Soy and SA media. Levels of clavulanic acid were reduced by 75 to 95% relative to those of the wild-type control strain. Mutants disrupted in orf14 were even more severely affected such that clavulanic acid production was at the limits of detection. However, both bioassays and HPLC analyses indicated the presence of trace amounts of clavulanic acid. We estimate that production was at about 0.5% of wild-type levels (about 0.8 to 1.0 μg/ml). In contrast, mutants with a defect in orf19 showed no demonstrable effects on either growth or clavulanic acid production. Finally, mutants with defects in orf12, orf15, orf16, or orf17 were unable to produce detectable clavulanic acid (detection limit, about 1 μg/ml) when grown on either growth medium at either of the time points tested.
Production of a new clavam metabolite by orf15 and orf16 mutants.
S. clavuligerus has been shown to produce at least four other clavam metabolites, clavam-2-carboxylate (C2C), 2-hydroxymethylclavam (2HMC), 2-formyloxymethylclavam, and alanylclavam, in addition to clavulanic acid when grown on Soy medium (20). These other clavam metabolites have no β-lactamase inhibitory activity and differ from clavulanic acid in their stereochemistry and side chain modifications at C-2 and C-3. Clavulanic acid displays 3R,5R stereochemistry in contrast to the 5S stereochemistry of the other clavam metabolites. These other clavam metabolites will hereinafter be referred to as 5S clavams. When imidazole-derivatized samples of Soy grown culture supernatants were examined by HPLC, the various disruption mutants were all found to produce at least some levels of the 5S clavam metabolites. Production levels of the 5S clavam metabolites are typically more variable than those of clavulanic acid even in the wild-type strain, making small changes in production difficult to follow (30). However, obvious changes in the levels of production of the 5S clavam metabolites were apparent for orf11::apr mutants, which showed a 50 to 90% decrease in the production of C-2-C and 2-HMC (as detected by HPLC) and only traces of alanylclavam (as detected by bioassays). Similarly, orf13::apr mutants showed a 90 to 100% decrease in production of these 5S clavams. The remaining mutants showed variable amounts of the 5S clavams, but for all of the mutant types, at least some of the 5S clavams were present even in those strains in which no clavulanic acid production could be detected.
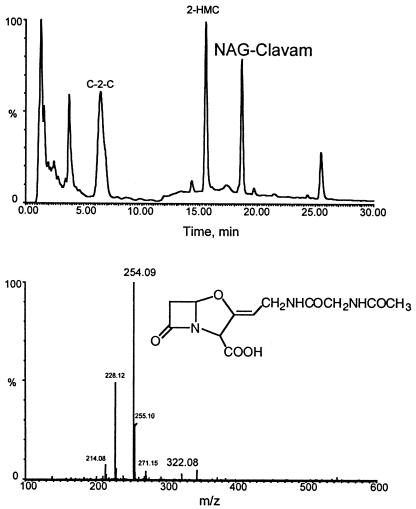
Although neither orf15::apr nor orf16::apr mutant cultures produced any clavulanic acid, both of them showed evidence of the presence of a new metabolite when examined by HPLC. This peak was only seen in culture supernatants derivatized with imidazole, suggesting that the compound had a clavam structure. Furthermore, the UV spectrum of the new peak showed an absorption maximum at 312 nm consistent with the presence of a clavam structure (data not shown). Previously, Elson et al. (8) had investigated an uncharacterized non-clavulanic-acid-producing mutant of S. clavuligerus (designated dcl8) and found that it accumulated a new clavam compound, N-acetylglycylclavaminic acid (NAG-clavam). To gain additional information about the new metabolite accumulated by the orf15::apr and orf16::apr mutant cultures, culture filtrates were reanalyzed by HPLC using a volatile solvent system so that effluents could be subjected to electrospray ionization mass spectrometric analyses. Under these conditions, the new imidazole-derivatized metabolite had a retention time of 18.6 min and gave a spectrum with a parent ion of mass 322.08 and a major fragment of mass 254.09 (Fig. 3). The total mass, together with the mass of the major fragment which resulted from removal of the imidazole group, was consistent with the new peak representing NAG-clavam. When authentic NAG-clavam was subjected to HPLC and mass spectrometric analysis, it showed results with respect to retention time, UV absorption spectrum, mass, and fragmentation pattern identical to those of the unknown peak in the orf15::apr and 16::apr mutants.
FIG. 3.
HPLC and mass spectrometric analyses of culture filtrates from an orf15::apr mutant. (A) HPLC elution profile with detection at 311 nm. 2-Hydroxymethyl clavam (2-HMC) and clavam 2-carboxylate (C-2-C) are two of the 5S clavams produced by S. clavuligerus; NAG-clavam is the new 5S clavam found in orf15 and orf16 mutants. (B) Mass spectrum of the NAG-clavam peak at 18.6 min.
Although NAG-clavam was observed in the orf15::apr mutant, it is the orf14 gene located just downstream which encodes a protein showing similarity to acetyltransferases. In view of the orientation and close spacing of the ORFs (Table 2), it seemed possible that the production of NAG-clavam might also be due to a polar effect of the orf15 disruption on orf14. To explore this further, we constructed a new orf15 mutant in which the apramycin disruption cassette was removed and replaced with a simple frameshift deletion mutation less likely to affect the expression of a downstream gene. When this new orf15-Δfs mutant was examined it showed the same non-clavulanic-acid-producer phenotype and accumulation of NAG-clavam as the original orf15::apr mutant, suggesting that the accumulation of this new metabolite is truly a result of mutation of orf15. By the same argument, the ability of the orf16 mutation to cause accumulation of NAG-clavam might also be a reflection of a polar effect of the orf16 mutation on the downstream orf15 gene. To eliminate this possibility, a second version of the orf16 mutant was also constructed in which the aprr cassette was removed and replaced by an in-frame deletion of the entire orf16 gene. Once again, the phenotype of the new Δorf16 mutant was unchanged compared with that of the original mutant, suggesting that mutations in both orf15 and orf16 have the effect of causing the mutant strains to accumulate NAG-clavam.
Mutation of the orf18 gene.
The deduced amino acid sequences encoded by orf18 and orf19 show high similarity to those of penicillin binding proteins (PBPs) from S. coelicolor and other species. While the disruption of orf19 gave rise to mutants on the first attempt, however, we were unable to disrupt orf18 in either the wild-type background or in any of a variety of non-antibiotic-producer mutants. On the assumption that orf18 might be an essential gene, the integrating plasmid, pSET152, was used to introduce a second copy of orf18 into the chromosome. This second copy of orf18 was first engineered to replace most of the native upstream sequences with a glycerol-regulated promoter from S. coelicolor (10, 31). Following insertion of the glycerol-regulated copy of orf18, the resulting pgyl-orf18 strain was subjected to the same gene replacement strategy that was unsuccessful in the wild-type strain. Presumptive gene replacement mutants were selected on the basis of their antibiotic resistance phenotypes, and Southern analysis confirmed that disruption of orf18 had taken place in each case. Of the five mutants analyzed, four had the disruption in the extra copy of orf18 inserted ectopically at the att site via pSET152 (pgyl-orf18::neo) and one was mutant in the native chromosomal copy of orf18 (nat-orf18::neo).
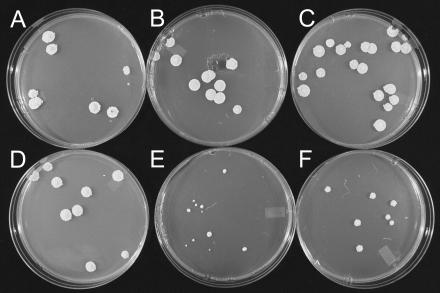
To test whether orf18 was essential in S. clavuligerus, the nat-orf18::neo mutant was grown on medium in the absence of added glycerol to suppress expression of the glycerol-regulated orf18 gene, which still remained intact. When cultivated on either Soy or SA liquid medium with no added glycerol, no difference was seen between the extent of growth of the nat-orf18::neo mutant and that of either the wild type or a control strain in which pSET152 with no insert was integrated at the att site. While these results suggested that orf18 is not essential, a second control strain transformed with pSET152 carrying an xylE reporter gene under the control of the same glycerol-regulated promoter (pgyl-xylE) showed low levels of catechol 2,3-dioxygenase (C23O) activity when grown on both Soy and SA liquid media even without added glycerol. Similarly, cultures of the pgyl-xylE strain grown on Soy medium agar plates without added glycerol still showed low levels of C23O activity. However, when the pgyl-xylE strain was grown on SA agar plates, C23O activity was absent. Under these same conditions, growth of the nat-orf18::neo mutant was noticeably inhibited and the resulting colonies were of various sizes (Fig. 4E). Addition of glycerol to the SA solid medium stimulated growth of the nat-orf18::neo mutant but not back to the level of that of wild type (Fig. 4F).
FIG. 4.
Response of orf18 mutants and control strains to growth on media with and without added glycerol. (A) S. clavuligerus wild type growing on SA-1% glycerol. (B) S. clavuligerus pgyl-orf18::neo mutant growing on SA-1% glycerol. (C) S. clavuligerus pgyl-orf18 strain growing on SA-1% glycerol. (D) S. clavuligerus pgyl-xylE strain growing on SA-1% glycerol. (E) S. clavuligerus nat-orf18::neo strain growing on SA with no added glycerol. (F) S. clavuligerus nat-orf18::neo strain growing on SA-1% glycerol.
All of these strains produced clavulanic acid; any changes seen in the levels of metabolite production were difficult to attribute to the orf18 mutation, because glycerol affects clavulanic acid production even in the wild type.
DISCUSSION
The clavulanic acid gene cluster has been extended by the addition of new DNA sequence information to give a total size of 25,344 bp (from the start codon of ceaS to the BamHI site down stream from orf19) for the cluster. While many of the ORFs added as a result of this study do not appear to encode enzymes directly involved in the biosynthesis of clavulanic acid, i.e., transport proteins, β-lactamase-type proteins, and PBPs, evidence is presented from gene disruption studies to indicate that all (with the possible exception of orf18 and orf19) are involved in some aspect of clavulanic acid biosynthesis. From previous studies, orf10, encoding a putative cytochrome P450, was known to be essential for clavulanic acid biosynthesis. orf11 lies immediately downstream and in the same orientation as orf10, and its high degree of similarity to ferredoxins has led to the suggestion that it works in concert with ORF10 as a electron transport partner. In this regard, our observation that disruption of orf11 causes a decrease but not a loss of clavulanic acid production and that disruption of orf10 causes a complete loss of clavulanic acid production suggests that ORF11 cannot be the only possible electron transport partner for ORF10.
The ORF10-ORF11 pair of proteins has been suggested to be a likely candidate for the enzymes needed to carry out the oxidative deamination and enantiomerization involved in the conversion of clavaminic into clavulanic acid. However, it is of interest that disruption of genes encoding proteins (such as Cad and ClaR) that function in the late stages of the clavulanic acid biosynthetic pathway (after the branching point from clavaminate to clavulanic acid) do not have major effects on the amount of the 5S clavam metabolites formed (12). In contrast, disruption of both orf10 and orf11 does cause marked decreases in the ability of the mutant strains to form the 5S clavam metabolites as well as similar or even more dramatic effects on production of clavulanic acid. If the sole function of the ORF10-ORF11 pair was to catalyze the conversion of clavaminic acid to clavaldehyde, then it is not clear why mutation of these proteins should decrease production of the 5S clavam metabolites. If anything, mutation of ORF10-ORF11 might be expected to channel more of the clavaminic acid in the direction of the 5S clavam metabolites, resulting in increased production levels. Alternatively, de la Fuente et al. (7) recently observed that mutants disrupted in orf10 showed elevated levels of production of the unrelated antibiotic holomycin, suggesting a complex interregulation between these two antibiotic gene clusters and making interpretation of mutant phenotypes more complex.
orf12 apparently encodes a protein with similarities to β-lactamases, although only some of the conserved motifs which characterize these proteins are present. Our studies agree with those of Li et al. (16) showing that disruption of orf12 causes a complete loss of clavulanic acid production ability. Similarly, orf13, which encodes an export pump-type protein, also appears to be important, although not completely essential, for production of both clavulanic acid and the 5S clavams. Mutants with disruptions in orf13 produce only about 5% of the wild-type levels of clavulanic acid and the 5S clavams. Since the ORF13 protein resembles other transport-type proteins, it may represent an export system involved in the excretion of all clavam metabolites.
In our hands, disruption of orf14 produced mutants with a phenotype that differs from those described by Mellado et al. (19). In that study, disruption of orf14, which encodes an apparent acetyltransferase activity, caused only a partial loss of production (about 33% of wild-type levels), whereas our mutants were almost completely (>99%) blocked in the production of clavulanic acid. This discrepancy cannot easily be explained by differences in disruption methodology, since Mellado et al. replaced an internal SalI fragment of the orf14 gene with the resistance gene, resulting in a disruption starting about 250 bp into the ORF, whereas our disruption was closer to the 3′ end of the ORF. If anything, our mutant should have been less severely affected.
Disruption of both orf15 and orf16 caused the complete loss of clavulanic acid production but the appearance of a new metabolite, NAG-clavam. The putative ORF15 protein is highly similar (about 50% identity) to the ORF7 protein of the clavulanic acid cluster. There is a growing body of evidence to suggest that genes encoding enzymes from early in the biosynthetic pathway to clavulanic acid are duplicated in S. clavuligerus (12). However, orf7 and orf15 cannot be considered paralogues in the same way that other genes such as cas1 and cas2 are because the putative proteins that they encode are not functionally equivalent. Mutants with defects in either orf7 or orf15 are unable to produce clavulanic acid; therefore, neither gene product can compensate for a mutation in the other. Furthermore, the orf7 mutant does not make NAG-clavam whereas the orf15 mutant does.
The orf16 mutant shows limited similarity to other proteins in the databases and then only to hypothetical proteins. Nonetheless, in similarity to the results seen with the orf15 mutant, clavulanic acid production was lost and NAG-clavam was produced in orf16 mutants. Because of the close spacing of the orf14, orf15, and orf16 genes, additional care was taken to prepare gene disruption mutants, with simple deletion or frame shift mutations replacing the antibiotic resistance gene insertions to minimize the possibility of polar effects. No evidence of polarity was seen, and so we conclude that accumulation of NAG-clavam is a genuine feature of mutation of either orf15 or orf16. Accumulation of this metabolite has been seen previously in non-clavulanic-acid-producer mutants of S. clavuligerus. Elson et al. (8) reported that a non-clavulanic-acid-producer mutant, dcl8, accumulated novel acylated derivatives of clavaminic acid (including NAG-clavam) in amounts equivalent to that seen with the clavulanic acid produced by the wild-type parent as well as minor amounts of N-acetylclavaminic acid and trace amounts of N-glycylclavaminic acid. Our orf15 mutant produced NAG-clavam but in amounts equivalent to only about 10% of the clavulanic acid amount produced by the wild type. On initial analysis, we did not see evidence of the other two acylated metabolites, but on closer scrutiny, we noted a small peak at retention time 17.5 min in orf15::apr (Fig. 3) and orf16:apr culture filtrates which eluted at about the same position as the clavaminic acid peak in the wild type. Upon mass spectrometric analysis, this peak was concluded to consist of a mixture of two compounds, clavaminic acid (fragmented mass, 155) and N-glycylclavaminic acid (fragmented mass, 212). In orf16::apr mutants, the N-glycylclavaminic peak was even more prominent (data not shown). No evidence of N-acetylclavaminic acid production was noted in the orf15::apr and orf16:apr culture filtrates, although the chromatographic conditions may not have been suitable for its detection.
Elson et al. suggested that the dcl8 mutant was likely blocked in the conversion of clavaminic acid to clavulanic acid, and the resultant accumulation of clavaminic acid in the mutants led to the formation of various acylated derivatives of clavaminic acid. This would imply that NAG-clavam is a shunt metabolite rather than a biosynthetic intermediate. However, in view of similarity that the product of orf14 shows to acetyltransferases, if ORF14 functions immediately before ORF15 and ORF16 in the pathway to form NAG-clavam as a product then mutation of orf15 and orf16 could result in accumulation of NAG-clavam.
ORF18 and ORF19 show strong similarities to PBPs. While orf19 was disrupted with no apparent effect on viability or metabolite production, mutation of orf18 was more complex. Attempts to disrupt the gene in the wild type or in any of a variety of non-antibiotic-producing mutant backgrounds were unsuccessful. When a second copy of the orf18 gene was introduced on an integrating plasmid, however, disruptions could be obtained in either copy, but not in both copies, of the orf18 gene. The introduced second copy of orf18 was designed to be regulatable (under the control of a glycerol-regulated promoter) to demonstrate unequivocally that the gene was essential. However, the promoter was found to be leaky in S. clavuligerus such that promoter activity could be detected in most growth media even in the absence of added glycerol. The tightest regulation by the glycerol-regulated promoter was observed on solid SA medium, on which growth of the nat-orf18::neo mutant was severely inhibited but not totally abolished. These results suggest that orf18 is essential but that even low levels of expression are sufficient to support growth and that the glycerol-regulated promoter does not give tight enough control to see an absolute response.
Since orf18 and orf19 encode PBPs, proteins normally associated with primary metabolism in other species, and since disruption of these genes had no marked effects on metabolite production, this could be taken as evidence that orf18 and orf19 are not part of the cephamycin C-clavulanic acid gene supercluster. However, since β-lactam metabolites such as clavulanic acid and cephamycin C specifically target PBPs and related enzymes, it seems unlikely that genes encoding two such enzymes should be located at the boundary of the cephamycin C-clavulanic acid gene supercluster just by chance. If orf18 and orf19 truly encoded primary metabolic enzymes, then they might be expected to be distributed on the S. clavuligerus chromosome in a way similar to that seen with the corresponding genes from S. coelicolor, since a considerable degree of synteny has been noted in actinomycete genomes that have been analyzed to date (6). The S. coelicolor genome contains 13 genes which are annotated as potential PBPs, and these are quite widely distributed throughout the chromosome (http://www.sanger.ac.uk/Projects/S_coelicolor/), extending from 0.879 Mb to 5.773 Mb. In particular, the two PBPs from S. coelicolor which show the greatest similarity (72 to 76% identity at the amino acid level) to orf18 and orf19 are located at 2.831 Mb (similar to orf18) and 4.409 Mb (similar to orf19) on the S. coelicolor chromosome. Therefore, we conclude that orf18 and orf19 are likely to be part of the cephamycin-clavulanic acid supercluster and that additional genes will have to be analyzed before it can be concluded that the end of the cluster has been reached.
Acknowledgments
This study was supported by Glaxo SmithKline and by grants from the Natural Sciences and Engineering Research Council of Canada and the Canadian Institutes of Health Research.
A.S. Paradkar was supported by a postdoctoral fellowship from the Alberta Heritage Foundation for Medical Research.
REFERENCES
- 1.Aharonowitz, Y., and A. L. Demain. 1978. Carbon catabolite regulation of cephamycin production in Streptomyces clavuligerus. Antimicrob. Agents Chemother. 14:159-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aidoo, K. A., A. Wong, D. C. Alexander, R. A. Rittammer, and S. E. Jensen. 1994. Cloning, sequencing and disruption of a gene from Streptomyces clavuligerus involved in clavulanic acid biosynthesis. Gene 147:41-46. [DOI] [PubMed] [Google Scholar]
- 3.Bachmann, B. O., R. Li, and C. A. Townsend. 1998. β-Lactam synthetase: a new biosynthetic enzyme. Proc. Natl. Acad. Sci. USA 95:9082-9086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baldwin, J. E., R. M. Adlington, J. S. Bryans, A. O. Bringhen, J. B. Coates, N. P. Crouch, M. D. Lloyd, C. J. Schofield, S. W. Elson, K. H. Baggaley, R. Cassels, and N. H. Nicholson. 1991. Isolation of dihydroclavaminic acid, an intermediate in the biosynthesis of clavulanic acid. Tetrahedron 47:4089-4100. [Google Scholar]
- 5.Baltz, R. H. 1980. Genetic recombination by protoplast fusion in Streptomyces. Dev. Industrial Microbiol. 21:43-54. [Google Scholar]
- 6.Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, H. J., T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. L., L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 7.de la Fuente, J. L., L. M. Lorenzana, J. F. Martin, and P. Liras. 2002. Mutants of Streptomyces clavuligerus with disruptions in different genes for clavulanic acid biosynthesis produce large amounts of holomycin: possible cross-regulation of two unrelated secondary metabolic pathways. J. Bacteriol. 184:6559-6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elson, S. W., J. Gillett, N. H. Nicholson, and J. W. Tyler. 1988. N-acyl derivatives of clavaminic acid produced by a mutant of Streptomyces clavuligerus. J. Chem. Soc. Chem. Commun. 1988:979-980. [Google Scholar]
- 9.He, J., N. Magarvey, M. Piraee, and L. C. Vining. 2001. The gene cluster for chloramphenicol biosynthesis in Streptomyces venezuelae ISP5230 includes novel shikimate pathway homologues and a monomodular non-ribosomal peptide synthetase gene. Microbiology 147:2817-2829. [DOI] [PubMed] [Google Scholar]
- 10.Hindle, Z., and C. P. Smith. 1984. Substrate induction and catabolite repression of the Streptomyces coelicolor glycerol operon are mediated through the GylR protein. Mol. Microbiol. 12:737-745. [DOI] [PubMed] [Google Scholar]
- 11.Hodgson, J. E., A. P. Fosberry, N. S. Rawlinson, H. N. M. Ross, R. J. Neal, J. C. Arnell, A. J. Earl, and E. J. Lawlor. 1995. Clavulanic acid biosynthesis in Streptomyces clavuligerus: gene cloning and characterization. Gene 166:49-55. [DOI] [PubMed] [Google Scholar]
- 12.Jensen, S. E., K. J. Elder, K. A. Aidoo, and A. S. Paradkar. 2000. Enzymes catalyzing the early steps of clavulanic acid biosynthesis are encoded by two sets of paralogous genes in Streptomyces clavuligerus. Antimicrob. Agents Chemother. 44:720-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kershaw, N. J., H. J. McNaughton, K. S. Hewitson, H. Hernandez, J. Griffen, C. Hughes, P. Greaves, B. Barton, C. V. Robinson, and C. J. Schofield. 2002. ORF6 from the clavulanic acid gene cluster of Streptomyces clavuligerus has ornithine acetyltransferase activity. Eur. J. Biochem. 269:2052-2059. [DOI] [PubMed] [Google Scholar]
- 14.Khaleeli, N., R. F. Li, and C. A. Townsend. 1999. Origin of the β-lactam carbons in clavulanic acid from an unusual thiamine pyrophosphate-mediated reaction. J. Am. Chem. Soc. 121:9223-9224. [Google Scholar]
- 15.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. John Innes Foundation, Norwich, England.
- 16.Li, R., N. Khaleeli, and C. A. Townsend. 2000. Expansion of the clavulanic acid gene cluster: identification and in vivo functional analysis of three new genes required for biosynthesis of clavulanic acid by Streptomyces clavuligerus. J. Bacteriol. 182:4087-4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marsh, E. N., M. D. Chang, and C. A. Townsend. 1992. Two isozymes of clavaminate synthase central to clavulanic acid formation: cloning and sequencing of both genes from Streptomyces clavuligerus. Biochemistry 31:12648-12657. [DOI] [PubMed] [Google Scholar]
- 18.McNaughton, H. J., J. E. Thirkettle, Z. H. Zhang, C. J. Schofield, S. E. Jensen, B. Barton, and P. Greaves. 1998. β-lactam synthetase: implications for β-lactamase evolution. Chem. Commun. 1998:2325-2326. [Google Scholar]
- 19.Mellado, E., L. M. Lorenzana, M. Rodriguez-Saiz, B. Diez, P. Liras, and J. L. Barredo. 2002. The clavulanic acid biosynthetic cluster of Streptomyces clavuligerus: genetic organization of the region upstream of the car gene. Microbiology 149:1427-1438. [DOI] [PubMed] [Google Scholar]
- 20.Mosher, R. H., A. S. Paradkar, C. Anders, B. Barton, and S. E. Jensen. 1999. Genes specific for the biosynthesis of clavam metabolites antipodal to clavulanic acid are clustered with the gene for clavaminate synthase 1 in Streptomyces clavuligerus. Antimicrob. Agents Chemother. 43:1215-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicholson, N. H., K. H. Baggaley, R. Cassels, M. Davison, S. W. Elson, M. Fulston, J. W. Tyler, and S. R. Woroniecki. 1994. Evidence that the immediate biosynthetic precursor of clavulanic acid is its N-aldehyde analog. J. Chem. Soc. Chem. Commun. 1281-1282.
- 22.Paradkar, A. S., K. A. Aidoo, and S. E. Jensen. 1998. A pathway-specific transcriptional activator regulates late steps of clavulanic acid biosynthesis in Streptomyces clavuligerus. Mol. Microbiol. 27:831-843. [DOI] [PubMed] [Google Scholar]
- 23.Paradkar, A. S., and S. E. Jensen. 1995. Functional analysis of the gene encoding the clavaminate synthase 2 isoenzyme involved in clavulanic acid biosynthesis in Streptomyces clavuligerus. J. Bacteriol. 177:1307-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perez-Redondo, R., A. Rodriguez-Garcia, J. F. Martin, and P. Liras. 1998. The claR gene of Streptomyces clavuligerus, encoding a LysR-type regulatory protein controlling clavulanic acid biosynthesis, is linked to the clavulanate-9-aldehyde reductase (car) gene. Gene 211:311-321. [DOI] [PubMed] [Google Scholar]
- 25.Pruess, D. L., and M. Kellett. 1983. Ro-22-5417, a new clavam antibiotic from Streptomyces clavuligerus I. Discovery and biological activity. J. Antibiot. (Tokyo) 36:208-212. [DOI] [PubMed] [Google Scholar]
- 26.Salowe, S. P., W. J. Krol, D. Iwata-Reuyl, and C. A. Townsend. 1991. Elucidation of the order of oxidations and identification of an intermediate in the multistep clavaminate synthase reaction. Biochemistry 30:2281-2292. [DOI] [PubMed] [Google Scholar]
- 27.Salowe, S. P., E. N. Marsh, and C. A. Townsend. 1990. Purification and characterization of clavaminate synthase from Streptomyces clavuligerus: an unusual oxidative enzyme in natural product biosynthesis. Biochemistry 29:6499-6508. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Stuttard, C. 1982. Temperate phages of Streptomyces venezuelae: lysogeny and host specificity shown by phages SV1 and SV2. J. Gen. Microbiol. 128:115-121. [Google Scholar]
- 30.Thai, W., A. S. Paradkar, and S. E. Jensen. 2001. Construction and analysis of β-lactamase-inhibitory protein (BLIP) non-producer mutants of Streptomyces clavuligerus. Microbiology 147:325-335. [DOI] [PubMed] [Google Scholar]
- 31.Trepanier, N. K., S. E. Jensen, D. C. Alexander, and B. K. Leskiw. 2002. The positive activator of cephamycin C and clavulanic acid production in Streptomyces clavuligerus is mistranslated in a bldA mutant. Microbiology 148:643-656. [DOI] [PubMed] [Google Scholar]
- 32.Vieira, J., and J. Messing. 1987. Production of single-stranded plasmid DNA. Methods Enzymol. 153:3-11. [DOI] [PubMed] [Google Scholar]
- 33.Ward, J. M., G. R. Janssen, T. Kieser, and M. J. Bibb. 1986. Construction and characterisation of a series of multi-copy promoter-probe plasmid vectors for Streptomyces using the aminoglycoside phosphotransferase gene from Tn5 as indicator. Mol. Gen. Genet. 203:468-478. [DOI] [PubMed] [Google Scholar]
- 34.Wu, T. K., R. W. Busby, T. A. Houston, D. B. McIlwaine, L. A. Egan, and C. A. Townsend. 1995. Identification, cloning, sequencing, and overexpression of the gene encoding proclavaminate amidino hydrolase and characterization of protein function in clavulanic acid biosynthesis. J. Bacteriol. 177:3714-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]