ABSTRACT
Bacterial biofilms often form multispecies communities in which complex but ill-understood competition and cooperation interactions occur. In light of the profound physiological modifications associated with this lifestyle, we hypothesized that the biofilm environment might represent an untapped source of natural bioactive molecules interfering with bacterial adhesion or biofilm formation. We produced cell-free solutions extracted from in vitro mature biofilms formed by 122 natural Escherichia coli isolates, and we screened these biofilm extracts for antiadhesion molecules active on a panel of Gram-positive and Gram-negative bacteria. Using this approach, we showed that 20% of the tested biofilm extracts contained molecules that antagonize bacterial growth or adhesion. We characterized a compound, produced by a commensal animal E. coli strain, for which activity is detected only in biofilm extract. Biochemical and genetic analyses showed that this compound corresponds to a new type of released high-molecular-weight polysaccharide whose biofilm-associated production is regulated by the RfaH protein. We demonstrated that the antiadhesion activity of this polysaccharide was restricted to Gram-positive bacteria and that its production reduced susceptibility to invasion and provided rapid exclusion of Staphylococcus aureus from mixed E. coli and S. aureus biofilms. Our results therefore demonstrate that biofilms contain molecules that contribute to the dynamics of mixed bacterial communities and that are not or only poorly detected in unconcentrated planktonic supernatants. Systematic identification of these compounds could lead to strategies that limit pathogen surface colonization and reduce the burden associated with the development of bacterial biofilms on medical devices.
IMPORTANCE
We sought to demonstrate that bacterial biofilms are reservoirs for unknown molecules that antagonize bacterial adhesion. The use of natural strains representative of Escherichia coli species biodiversity showed that nonbiocidal antiadhesion polysaccharides are frequently found in mature biofilm extracts (bacterium-free suspensions which contain soluble molecules produced within the biofilm). Release of an antiadhesion polysaccharide confers a competitive advantage upon the producing strain against clinically relevant pathogens such as Staphylococcus aureus. Hence, exploring the biofilm environment provides a better understanding of bacterial interactions within complex communities and could lead to improved control of pathogen colonization.
Introduction
The development of matrix-encased biofilm communities on medical implants challenges most treatments of bacterial infections due to high biofilm tolerance to antibiotics (1). To reduce trauma and cost associated with physical removal of recalcitrant biofilms, several approaches have been proposed to eliminate already formed biofilms, including the use of dispersion molecules (2, 3), antimicrobial compounds active against biofilm bacteria (4), bacterial communication inhibitors (5), and even biological entities such as bacteriophages (6) or grazing organisms that feed on biofilm bacteria (7).
Many of these strategies are impractical in medical settings, and early prevention of the initial adhesion stages of biofilm formation by coating patient care equipment with antibiotics or other biocides is thus considered a promising, straightforward approach (8, 9). However, toxicity issues and the onset of resistance associated with biocidal coatings have fostered interest in nonbiocidal solutions using tensioactive molecules or materials having antiadhesion properties (8). These approaches could represent a safe and effective means of controlling infection, since a decrease in bacterial pathogen colonization on critical surfaces would also impede a crucial step in bacterial pathogenesis (10).
Several nonbiocidal surface-active agents produced by microorganisms were shown to modify microbial interactions with the environment (11). For instance, rhamnolipids produced by the opportunistic pathogen Pseudomonas aeruginosa impair bacterial biofilms (12), several biosurfactants or bioemulsifiers produced by marine bacteria display antibiofilm activity against pathogenic bacteria (13), and we have previously shown that soluble polysaccharides released by extraintestinal Escherichia coli induce physicochemical surface modifications, preventing biofilm formation by a wide range of Gram-positive and Gram-negative bacteria (14). Hence, microbe-derived compounds that affect wettability, detergency, and other amphipath-related properties could potentially be used to regulate the attachment or detachment of microorganisms to or from surfaces (15).
Although biofilms represent an ideal environment for antagonistic and synergistic interactions, production and release of such molecules have been studied mainly under liquid planktonic conditions. However, the biofilm lifestyle is known to trigger physiological and metabolic adjustments, potentially leading to production of biofilm-associated molecules (16, 17). Therefore, biofilms could represent untapped sources of natural bioactive molecules that antagonize adhesion or biofilm formation, potentially influencing the dynamics of bacterial populations in mixed-species contexts (18).
Here we hypothesized that soluble molecules could accumulate within mature biofilms formed by natural commensal and pathogenic isolates representative of the biodiversity of the E. coli species. We identified several such biofilm-associated molecules displaying antiadhesion properties, including a high-molecular-weight polysaccharide active only against Gram-positive bacteria, the activity of which is otherwise poorly detected in unconcentrated planktonic supernatants. We provide genetic and biochemical evidence that this biofilm-associated molecule corresponds to a newly released polysaccharide regulated by the RfaH protein. Our results therefore experimentally demonstrate that high-cell-density biofilms constitute unexplored reservoirs of bacterial interference molecules that could limit biofilm formation and control of pathogen biofilm colonization.
RESULTS
Biofilms formed by natural E. coli isolates contain soluble antiadhesion molecules.
To identify E. coli biofilm-associated molecules that potentially prevent bacterial adhesion, we produced mature biofilms from 122 commensal and pathogenic E. coli strains of human and animal origin representative of the ecological and phylogenetic biodiversity of the E. coli species (see Table S1 in the supplemental material). We recovered the biofilm biomass formed after 72 h of growth and filtered the resuspension solution. This bacterium-free suspension, which contains soluble molecules produced within the biofilm, will here be referred to as biofilm extract (Fig. S1A). We then tested each biofilm extract for potential antibiofilm activity using microtiter plate wells, containing a 1:1 ratio of biofilm extract to growth medium, and inoculated it with a panel of biofilm-forming Gram-negative and Gram-positive bacteria, including E. coli K-12 MG1655 carrying an F conjugative plasmid and strain O42, Enterobacter cloacae 1092, Pseudomonas aeruginosa PAK ∆retS, Klebsiella pneumoniae 21, Staphylococcus aureus 15981, Staphylococcus epidermidis O-47, and Enterococcus faecalis 54 (Fig. S1B).
This screen led to the identification of 24 natural E. coli isolates with biofilm extracts active against at least one of the biofilm-forming bacteria included in our test panel (Table 1; see also Fig. S1B in the supplemental material). Among these 24 active strains, we discarded 8 strains whose genomes contained genes responsible for production of the previously demonstrated antiadhesive group 2 capsule (14), and most of them lost antibiofilm activity upon deletion of kpsD, a gene essential for group 2 capsule secretion (Table 1).
TABLE 1 .
E. coli strains displaying antibiofilm activity in biofilm extracts
| Straina | Pathovarf | Phylogenetic group |
Group II capsuleb |
Activity spectrum |
|
|---|---|---|---|---|---|
| Biofilm extract |
Planktonic supernatant |
||||
| K-12 MG1655 | NDc | A | ND | ND | ND |
| DAEC18 | DAEC | B2 | Dc |
S. aureus, E. faecalis, E. coli K-12, E. cloacae, K. pneumoniae |
S. aureus, E. coli K-12, E. cloacae, K. pneumoniae |
| IAI37 | UPEC | A1 | D |
E. faecalis, E. coli K-12, E. cloacae, K. pneumoniae |
E. coli K-12, E. cloacae, K. pneumoniae |
| IAI44 | UPEC | A1 | ND | E. coli K-12 | E. coli K-12 |
| IAI71 | UPEC | B22 | D | E. coli K-12 | E. coli K-12 |
| IAI73 | UPEC | B23 | ND |
S. aureus, E. coli K-12, K. pneumoniae |
S. aureus, E. coli K-12, K. pneumoniae |
|
Commensal | B1 | ND | S. aureus | ND |
| ROAR212 | Commensal | A | ND | S. aureus | S. aureus |
|
Commensal | A | ND |
S. aureus, S. epidermidis, E. faecalis |
ND |
| ROAR319 | Commensal | B1 | ND | S. aureus, E. cloacae | S. aureus |
| ROAR381 | Commensal | A | D | E. coli K-12, K. pneumoniae | E. coli K-12, K. pneumoniae |
| DAEC9 | DAEC | D | D | E. coli K-12, K. pneumoniae | E. coli K-12, K. pneumoniae |
| ROAR002 | Commensal | A | ND | E. coli K-12, K. pneumoniae | E. coli K-12, K. pneumoniae |
| ROAR029 | Commensal | D | ND | E. coli K-12 | ND |
| ROAR194 | Commensal | A1 | ND | S. aureus, E. cloacae | S. aureus, E. cloacae |
| ROAR057 | Commensal | B1 | D | E. coli K-12 | E. coli K-12 |
| ROAR248 | Commensal | D | ND | S. aureus |
S. aureus, E. faecalis, E. coli K-12 |
| H-19 | EHEC | B1 | ND | E. coli K-12, S. epidermidis | E. coli K-12, S. epidermidis |
| ROAR094 | Commensal | D | ND | E. coli K-12 | E. coli K-12 |
| ROAR206 | Commensal | D | ND | E. coli K-12, K. pneumoniae | K. pneumoniae |
| C1845 | DAEC | B2 | D | S. aureus, E. coli K-12 | S. aureus, E. coli K-12 |
| 289KH89 | NPEC | A | ND | E. coli K-12 | E. coli K-12 |
| ROAR066 | Commensal | B1 | ND | S. aureus | S. aureus |
| ROAR208 | Commensal | C | ND | E. coli K-12 | E. coli K-12 |
| RS218 | ExPEC | B2 | D | S. aureus, E. coli K-12 | S. aureus, E. coli K-12 |
See Table S1 in the supplemental material.
Presence of genes involved in biosynthesis of group 2 capsule.
ND, not detected; D, detected.
Commensal strain from the B1 phylogenetic group isolated from a roe deer (Capreolus capreolus) in the Fontainebleau forest (France) (Table S1).
Commensal strain of the A phylogenetic group collected from a dog (Canis familiaris) in the Pyrenees mountains (France) (Table S1).
DAEC, diffusely adhering E. coli; UPEC, uropathogenic E. coli; EHEC, enterohemorrhagic E. coli; NPEC, nonpathogenic E. coli; ExPEC, extraintestinal pathogenic E. coli.
Inhibition of biofilm formation by the remaining 16 selected active strains could result from the limitation of bacterial adhesion, bacteriostatic or bacteriocidal activity, or biofilm dispersion. We first tested potential bacteriostatic or bacteriocidal activity by spotting aliquots of active biofilm extracts on bacterial lawns formed by the Gram-negative and Gram-positive strains included in our test panel. With the exception of E. coli ROAR029, which displayed growth-inhibiting activity against E. coli K-12, no other growth inhibition zone could be detected (data not shown). This suggested that biofilm inhibition induced by the 15 remaining biofilm extracts was not the consequence of a growth inhibition (Table 1 and data not shown). Finally, addition of prepared biofilm extracts did not lead to significant biofilm dispersion when added to 24-h biofilms formed in microtiter plates by all tested bacteria (data not shown). These results therefore indicated that 15 out of 122 (12%) of the tested biofilm extracts exhibited antiadhesion activities that could not be attributed to the soluble molecules previously described in E. coli.
Identification of two new biofilm-associated antiadhesion polysaccharides.
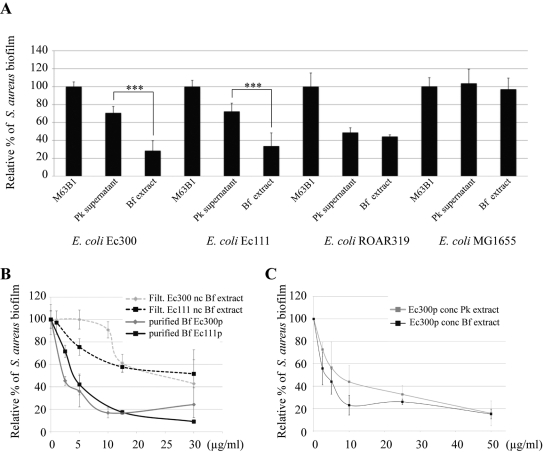
Among the 15 active extracts identified by our screen, biofilm extracts from E. coli Ec300 and Ec111, two commensal animal E. coli strains belonging to the A and B1 phylogenetic groups, respectively (Table 1; see also Table S1 in the supplemental material), displayed significantly higher antiadhesion activities than their respective planktonic supernatants (Fig. 1A). This observation contrasted both with strains that displayed antiadhesion activity in biofilm and planktonic extracts (Fig. 1A, strain ROAR319) and with inactive E. coli strains (Fig. 1A, strain MG1655). This indicated that the E. coli Ec300 and Ec111 antiadhesion molecules were specifically associated with mature biofilm conditions (Fig. 1A). Both Ec300 and Ec111 biofilm extract antiadhesion activities were retained in fractions corresponding to a molecular mass cutoff above 100 kDa, and both were highly resistant to proteinase K and heat treatment, three properties indicative of high-molecular-weight polysaccharide polymers (data not shown). Indeed, as shown in Fig. 1B, concentrated and purified (see Materials and Methods) Ec300 and Ec111 polysaccharides (termed Ec300p and Ec111p, respectively) exhibited comparable antibiofilm activities. Both polysaccharides displayed molecular masses above 500, as indicated by gel filtration analysis (data not shown), and although purified Ec111p contains more glucosamine and galactosamine than Ec300p, both molecules were composed of mannose, glucose, galactose, and glucuronic acid, indicating that they were distinct from the previously described group 2 capsule, which contained galactose, glycerol, phosphate, and acetate at a molar ratio of 1:2:1:1, respectively (Table 2) (14). The differences between Ec300p and Ec111p composition may account for the observed difference of activity, and here, we focused the rest of the study on the E. coli Ec300 antiadhesion polysaccharide, which displayed a wider activity spectrum than that of the Ec111 antiadhesion polysaccharide (Table 1).
FIG 1 .
Effect of planktonic and biofilm extracts upon S. aureus biofilm formation. (A) S. aureus biofilm inhibition upon addition of planktonic (Pk) or biofilm (Bf) extracts from indicated E. coli strains. M63B1, control in which only M63B1 minimal medium was added. Statistical t tests were used to evaluate the significance of planktonic (Pk) and biofilm (Bf) extract inhibition. ***, P < 0.001. (B) S. aureus biofilm inhibition with nonconcentrated E. coli Ec300 and Ec111 biofilm extracts (nc Bf extract) or with purified polysaccharides present in E. coli Ec300 and Ec111 biofilm extracts (purified Bf Ec300p and Ec111p, respectively). (C) Comparison of specific antibiofilm activities over S. aureus of Ec300 polysaccharide concentrated from the planktonic supernatant (conc Ec300p Pk) or biofilm extract (conc Ec300p Bf). The x axis units correspond to the polysaccharidic concentration (compared to a glucose standard) in µg/ml. Experiments were performed at least in triplicate; error bars represent standard deviations of the means.
TABLE 2 .
Composition of purified antibiofilm polysaccharides by gas-liquid chromatography and high-pressure liquid chromatography
| Polysaccharide | Compositiona |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mannose | Glucose | Galactose | Glucosamine | Phosphate | Glycerol | Galactosamine | Glucuronic acid |
Acetate | Pyruvate | Fucose | |
| Group 2 capsule (14) |
— | — | 1 | — | 1 | 2 | — | — | 1 | — | — |
| Colanic acid (33) |
— | 1 | 1.8 | — | — | — | — | 1 | 1 | 1 | 1.9 |
| Retained fraction Ec111 Bf |
1 | 0.67 | 0.52 | 0.35 | 0.07 | — | 0.45 | 0.18 | ND | — | — |
| Retained fraction Ec300 Bf |
1 | 0.26 | 0.24 | 0.06 | 0.06 | — | 0.02 | 0.19 | ND | — | — |
| Ec300 planktonic |
1 | 0.33 | 0.28 | 0.03 | 0.02 | — | — | 0.12 | ND | — | — |
| Ec300 ∆galF-his | 1 | 1.92 | 1.05 | 0.82 | 0.18 | — | — | — | ND | ND | — |
| Ec300∆12208 | 1 | 0.19 | 0.08 | — | 0.02 | — | — | — | ND | ND | — |
| K. pneumoniae 342 | 1 | 0.27 | 0.52 | 0.03 | 0.04 | — | — | 0.23 | ND | ND | — |
—, not detected; ND, not determined. The composition of purified retained biofilm fractions from strains Ec111 and Ec300 was tested twice, with no significant differences in the results. The composition of Ec300 mutants and K. pneumoniae 342 was done once. The results are presented in the form of ratios, using mannose, the most abundant monosaccharide in Ec300p and Ec111p, as the standard.
E. coli Ec300 biofilm produces more antiadhesion polysaccharides than comparable planktonic cultures.
To investigate the origin of reduced antiadhesion activity in the E. coli Ec300 planktonic extract, we precipitated, purified, and quantified total soluble polysaccharides extracted either from 24-h stationary-phase planktonic cultures or from corresponding biofilm extracts. We then compared the specific activities of these extracts against that of biofilm formed by S. aureus in microtiter plates and observed that similar quantities of purified planktonic or biofilm polysaccharide led to a comparable reduction in S. aureus biofilm formation (Fig. 1C). This therefore demonstrated that E. coli Ec300 released small amounts of active Ec300p in planktonic supernatant. Moreover, we determined that the amount of antiadhesion polysaccharide produced per an equivalent number of bacteria was 2-fold higher in E. coli Ec300 biofilm extracts than in the corresponding planktonic supernatants (data not shown). Hence, our results indicated that production of Ec300p is not stricto sensu biofilm specific, but its activity is poorly detectable in unconcentrated planktonic extracts.
Antiadhesion activity of Ec300p on representative Gram-positive bacteria is independent of cell wall composition.
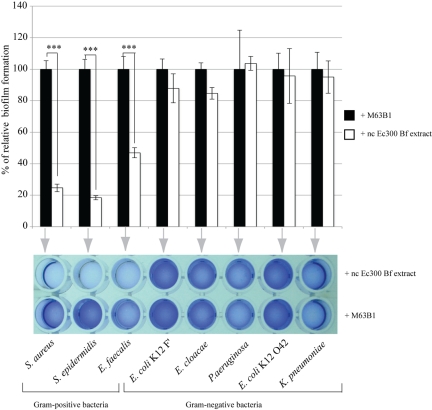
Whereas Ec300p is not active against tested Gram-negative bacteria, it displays significant antiadhesion activity against representative Gram-positive bacteria, including S. aureus, S. epidermidis, and E. faecalis, without a detectable bactericidal effect (Fig. 2 and Table 1; see also Fig. S2 in the supplemental material). In order to determine whether this restricted range of Ec300p antiadhesion activity was due to Gram-positive bacterial cell wall characteristics, we determined Ec300p activity on biofilm formed by two S. aureus mutants with altered cell wall properties. We used HG001 ∆tagO, which lacks teichoic acids, and HG001 ∆dltA, in which teichoic acids have no d-alanine esters, leading to a net negative charge increase (19). Although these two mutants displayed reduced biofilm formation capacity compared to that of the wild-type HG001 strain, addition of E. coli Ec300 biofilm extract nonetheless further reduced the corresponding biofilm formation (data not shown). We then tested whether the activity of Ec300p depended on the nature of adhesion factors expressed by different S. aureus strains. We took advantage of the fact that S. aureus 15981 biofilm formation is based on production of the poly-N-acetyl glucosamine (PNAG) polymer in the presence of glucose or is protein based in the presence of sodium chloride (20). However, under both conditions, addition of Ec300p (50 µg/ml final) still led to a similar reduction in S. aureus biofilm formation (Fig. S3). These results therefore demonstrated that E. coli Ec300 biofilm extract inhibits representative Gram-positive bacterial biofilms independently of tested adhesion factors or cell wall net charge alterations.
FIG 2 .
E. coli Ec300 polysaccharide is only active on Gram-positive bacteria. Biofilm inhibition was determined in the presence of nonconcentrated E. coli Ec300 biofilm extract (nc Ec300 Bf extract). Tested representative Gram-positive bacteria include Staphylococcus aureus 15981, Staphylococcus epidermidis 047, and Enterococcus faecalis 54. Gram-negative bacteria tested include Escherichia coli K-12 MG1655 F′ tet ΔtraD, Enterobacter cloacae 1092, Pseudomonas aeruginosa PAK ΔretS, E. coli 042, and Klebsiella pneumoniae 21. Statistical t tests were used to evaluate the significance of biofilm inhibition compared to that of fresh media. ***, P < 0.001.
Identification of genetic determinants of the E. coli Ec300 antiadhesion molecule.
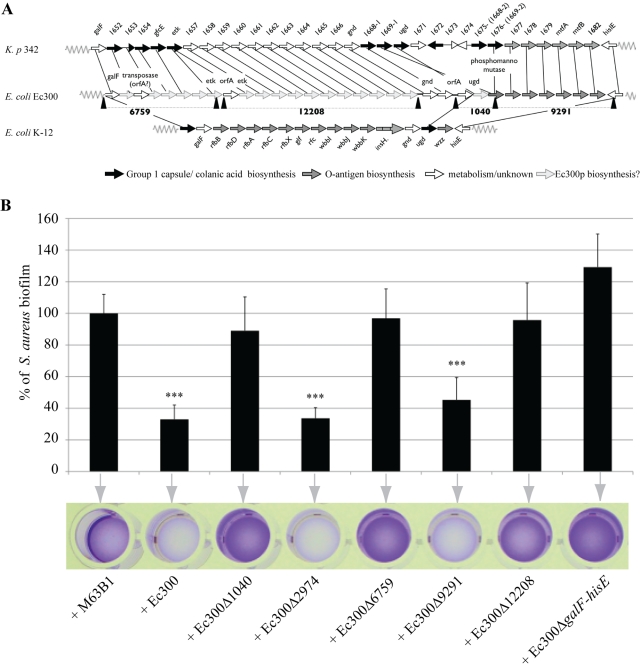
To investigate the nature of Ec300p and genetic regulation behind increased production in the E. coli Ec300 biofilm, we attempted to identify genes involved in the production of Ec300p. We hypothesized that Ec300p genetic determinants could be localized on genomic regions absent in the inactive E. coli K-12 MG1655 strain (Table 1). Sequencing of the E. coli Ec300 genome using Solexa (Illumina) technology revealed 145 contigs containing E. coli Ec300 genes absent from the MG1655 genome. We subjected these 145 contigs to a BLAST search performed on a sliding search window of 1 kb, and we identified 5 candidate contigs carrying genes potentially involved in the biosynthesis, metabolism, and export of Ec300-specific polysaccharides (see Table S2 in the supplemental material). The homology between 4 of these 5 candidate contigs (6759, 12208, 1040, and 9291) and the galF-his region, often encoding colanic acid, the group 1 capsule, and lipopolysaccharide O antigen (LPS O antigen) (21), suggested that all four contigs were located within the E. coli Ec300 galF-his region (Fig. 3A). Consistently, deletion of the E. coli Ec300 galF-hisE region abolished biofilm formation and prevented us from testing the antiadhesion activities of the E. coli Ec300 galF-hisE biofilm extracts (Fig. S4). However, polysaccharides extracted from the planktonic supernatant of Ec300 ∆galF-hisE displayed no antiadhesive activity, therefore demonstrating that this region is involved in E. coli Ec300 antiadhesive activity. Moreover, deletion of contig 2974, which is not included in the E. coli Ec300 galF-hisE region, did not impact the activity of the Ec300 supernatant (Fig. 3; see also Table S2 in the supplemental material).
FIG 3 .
The E. coli Ec300 galF-his region is involved in Ec300 polysaccharide production. (A) Schematic alignment of the galF-his region of E. coli Ec300, K. pneumoniae 342, and E. coli K-12, representing predicted roles of genes in biosynthesis of different polysaccharides. (B) S. aureus biofilm inhibition upon addition of equivalent nonconcentrated biofilm extracts from indicated E. coli Ec300 derivative strains corresponding to deletions of the contigs potentially involved in Ec300 polysaccharide biosynthesis or upon addition of concentrated planktonic extracts from the E. coli Ec300 galF-hisE region (Table 2; Fig. 3A). Of note, one of the identified contigs, contig 2974, does not belong to the galF-his locus. M63B1, control in which only M63B1 minimal medium was added. Experiments were performed at least in triplicate; error bars represent standard deviations of the means. Statistical t tests were used to evaluate the significance of biofilm inhibition from various extracts. ***, P < 0.001.
The 40-kb galF-his region of K. pneumoniae strain 342 shared homology and synteny with identified E. coli Ec300 contigs and was the only bacterial strain displaying homology with contig 12208 (Fig. 3A; see also Table S2 in the supplemental material). Consistently, polysaccharide from K. pneumoniae 342 biofilm extract inhibited S. aureus biofilm formation and had a composition similar to that of Ec300p (Table 2 and data not shown).
To confirm the implication of the identified contigs in the synthesis of Ec300p, we produced biofilm extracts from E. coli Ec300 mutants carrying deletions corresponding to all 4 remaining identified contigs and tested their ability to inhibit S. aureus biofilm formation. By homology with K. pneumoniae 342, contig 9291 is believed to encode LPS O antigen (Fig. 3A; see also Table S2 in the supplemental material), and its deletion did not affect the the antiadhesive activity of the E. coli Ec300 biofilm extract (Fig. 3B). In contrast, biofilm extracts from E. coli Ec300 mutants corresponding to deletion of contigs 6759, 12208, and 1040 lost their activity, therefore associating these three regions with the antiadhesive activity of Ec300 supernatant (Fig. 3B). The K. pneumoniae 342 genomic regions corresponding to contigs 6759, 12208, and 1040 contained genes implicated in colanic acid and group 1 capsule secretion (Fig. 3A). However, neither analysis of E. coli Ec300 unassembled genomic contigs nor use of specific PCR primers (see Table S4) enabled us to detect the presence of genes typically associated with group 1 capsule or colanic acid biosynthesis, such as wzz and wzy (21, 22). Furthermore, the osidic composition ratio of Ec300p did not match with published compositions of colanic acid and the group 1 capsule, particularly in glucuronic acid, galactose, and glucose content (Table 2). Concentrated total polysaccharides, purified from biofilm extracts or the planktonic supernatant of E. coli Ec300 deleted from contig 12208, consistently displayed altered osidic composition and no antiadhesion activity. In addition, contig 12208 does not contain genes previously described as being involved in acid colanic or group 1 capsule production (except for etk, interrupted by a transposase in Ec300) (Table 2 and data not shown). Taken together, these results indicated that the antiadhesive polysaccharide produced by E. coli Ec300 did not correspond to colanic acid or to the group 1 capsule; this therefore suggested that Ec300p corresponded to a new antiadhesive polysaccharide.
Regulation of genes involved in Ec300p synthesis.
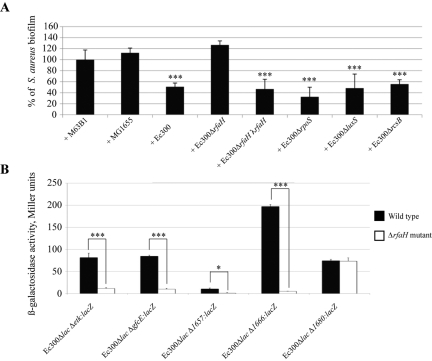
In order to study the regulation of Ec300p production, we analyzed the antiadhesion activities of biofilm extracts obtained from strains carrying mutations corresponding to regulators previously associated with biofilm and polysaccharide regulation, including rscB, luxS, rpoS, oxyR, crr, crp, rpoN, phoP, hns, and rfaH (23). E. coli Ec300 rfaH was the only mutant to impair Ec300 antiadhesion activity against S. aureus biofilm (Fig. 4A). Consistently, introduction of a wild-type rfaH allele in trans at the lambda attachment site restored biofilm-associated antiadhesion activity in Ec300∆rfaH λrfaH (Fig. 4A). RfaH is an antiterminator that positively controls expression of long operons coding for extracellular components such as secreted virulence factors, lipopolysaccharide, and conjugation proteins. Although RfaH binds 12-bp JUMPstart motifs (5′-RGGCGGTAGYNT-3′) often located in the promoter-proximal 5′ region, analysis of the DNA region located in the E. coli Ec300 galF-hisE region corresponding to contigs 6759, 12208, and 1040 did not reveal any convincing JUMPstart motifs.
FIG 4 .
Production of antiadhesion E. coli Ec300 polysaccharide is regulated by rfaH. (A) Effect of nonconcentrated biofilm extracts from indicated strains on S. aureus biofilm formation. M63B1, control in which only M63B1 minimal medium was added. (B) Beta-galactosidase activity measurements of lacZ transcriptional fusions in genes located in contigs essential for production of E. coli Ec300 antiadhesion polysaccharide in a wild-type and ∆rfaH background. ∆1680, control; region not involved in Ec300p synthesis. Experiments were performed in triplicate; error bars represent standard deviations of the means. Statistical t tests were used to evaluate the significance of biofilm inhibition from various extracts compared to that of addition of M63B1. *, P < 0.05; ***, P < 0.001.
To evaluate the potential regulatory role of RfaH, we introduced lacZ transcriptional reporter fusions in front of promoter regions corresponding to genes located in contigs involved in the production of Ec300p, including gfcE, etk (contig 6759), genes homologous to kp1657, and kp1666 in contig 12208 and kp1680 in uninvolved contig 9291 (Fig. 4B; see also Table S2 in the supplemental material). All fusions were introduced into a wild-type and E. coli Ec300ΔrfaH ∆lacIZ background; comparison of beta-galactosidase activities in 24-h planktonic cultures showed that genes located in contigs involved in the synthesis of E. coli Ec300p were regulated by rfaH. In contrast, expression of the kp1680-lacZ fusion in nonessential contig 9291 was unaffected by the rfaH mutation (Fig. 4B).
In order to elucidate increased production of Ec300p in E. coli Ec300 biofilms, we compared the expression levels of genes involved in its synthesis in bacteria harvested during the exponential and late stationary phases as well as under biofilm conditions, using lacZ transcriptional fusions. We first observed that expression of etk, 1666, and rfaH increased between exponential phase and stationary growth phase but did not further increase in mature biofilm (see Fig. S5 in the supplemental material). Moreover, although this increased expression was also observed in the nonessential and rfaH-independent gene kp1680, genes involved in Ec300p synthesis, such as gfcE and 1657, were not regulated upon entry into biofilm phase (Table S2 and Fig. S5).
Taken together, these results demonstrated that, while all tested genes involved in Ec300p synthesis are regulated by RfaH, some but not all of them are upregulated upon entry into stationary and biofilm phases.
E. coli Ec300 antiadhesion polysaccharide induces increased surface hydrophilicity and impairs S. aureus initial adhesion.
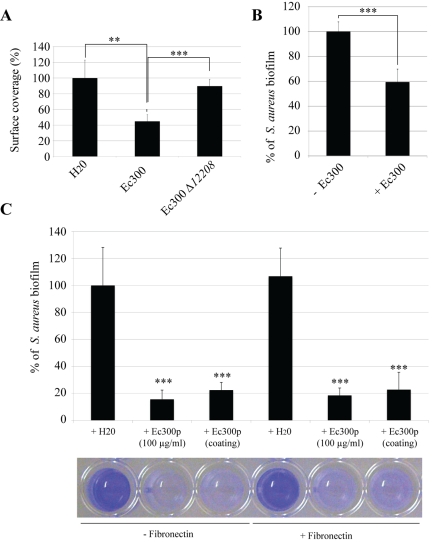
The antiadhesion properties of Ec300p towards representative Gram-positive bacteria could result from a limitation in initial adhesion, prevention of bacterial/bacterial interactions, or both. To investigate this, we first coated glass slides with different concentrations of Ec300p, and we observed that this coating resulted in a strong increase in surface hydrophilicity, even compared to that resulting from coating with the group 2 capsule (see Fig. S6 in the supplemental material). This increased hydrophilicity was not observed with polysaccharides extracted from the E. coli Ec300∆12208 strain (Fig. S6). We then tested S. aureus 15981 adhesion on an Ec300p-coated glass slide and showed that bacterial surface coverage was severely reduced after 5 min of incubation in static inoculation suspension, whereas a similar treatment performed with inactive polysaccharide extracted from an E. coli Ec300∆12208 biofilm had no effect (Fig. 5A). We then tested the impact of E. coli Ec300 polysaccharide pretreatment on S. aureus 15981 biofilm formation under dynamic continuous-flow culture conditions and showed that the coating of an internal glass slide with Ec300p also reduced by 40% the amount of biofilm biomass formed in a microfermentor after 24 h of incubation (Fig. 5B). To determine whether Ec300p also interfered with S. aureus adhesion to components of the eukaryotic cell extracellular matrix, such as fibronectin, we used polystyrene microtiter plate wells coated or not with fibronectin. Addition of Ec300p at a 1:1 ratio (50 µg/ml final concentration) right before bacterial inoculation drastically reduced S. aureus adhesion; an overnight coating of wells with Ec300p prior to bacterial inoculation had the same effect (Fig. 5C). Precoating of wells with fibronectin did not interfere with the antiadhesion activity of Ec300p (Fig. 5C).
FIG 5 .
Effect of Ec300 antiadhesion polysaccharide on S. aureus intitial adhesion. (A) S. aureus surface coverage of a glass slide when the surface was pretreated with either polysaccharides extracted from E. coli Ec300 or E. coli Ec300∆12208 at a final concentration of 250 µg/ml. (B) Reduction in S. aureus biofilm formation in a continuous-flow microfermentor on a glass surface pretreated in a solution of 500 µg/ml of concentrated Ec300 polysaccharide. (C, top) Interference effect of addition of Ec300p over S. aureus biofilm formation in microtiter plates coated or not with fibronectin. (Bottom) Corresponding microtiter well biofilm stained with crystal violet. The figure of a representative experiment is shown. Experiments were performed in triplicate; error bars represent standard deviations. Statistical t tests were used to evaluate the significance of biofilm inhibition. **, P < 0.01; ***, P < 0.001.
The ability of purified Ec300p to reduce S. aureus bacterial/bacterial interactions was evaluated by monitoring its capacity to modulate S. aureus multicellular aggregate formation. S. aureus aggregation was determined by measuring its sedimentation rate in static liquid solutions. In contrast to its effect on S. aureus initial adhesion, addition of Ec300p did not reduce formation of bacterial aggregates formed by either the mildly or strongly aggregating strains S. aureus 15981 or S. aureus HG001∆tagO, respectively (data not shown). Taken together, these results strongly suggested that the Ec300p antibiofilm effect relies on its capacity to impair initial adhesion of Gram-positive bacteria, possibly due to its ability to increase surface hydrophilicity.
Production of the E. coli Ec300 antiadhesion polysaccharide impairs initial S. aureus/E. coli interactions in mixed biofilms.
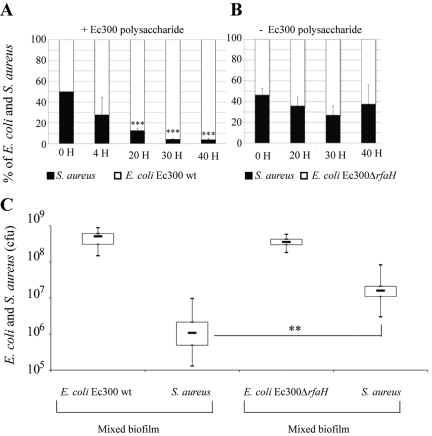
To assess whether production of Ec300p also modulates E. coli/S. aureus interactions, we evaluated the potential consequences of production of the E. coli Ec300 antiadhesion molecule in mixed S. aureus/E. coli Ec300 biofilms. Mixed cultures adjusted to a 1:1 ratio of E. coli Ec300 or Ec300∆rfaH to S. aureus were inoculated into several continuous-flow biofilm microfermentors. We then determined the proportions of the 2 strains in the biofilm biomass formed in individual microfermentors and arrested at different time points. Although S. aureus formed considerably more biofilm than E. coli Ec300 when inoculated individually (data not shown), we observed that the proportion of S. aureus in the resulting E. coli Ec300/S. aureus mixed biofilm was rapidly reduced, attaining only 5% after 40 h of biofilm culture (Fig. 6A). In contrast, when S. aureus biofilm was mixed with the inactive E. coli Ec300∆rfaH mutant at a 1:1 ratio, S. aureus still represented half of the biofilm biomass after 50 h of biofilm growth (Fig. 6B). This capacity of E. coli Ec300 to exclude S. aureus from mixed biofilms was due neither to a growth competitive advantage, as demonstrated by competition experiments performed under planktonic conditions, nor to the presence of residual amounts of antiadhesion polysaccharide present in the E. coli Ec300 inoculum, as revealed by experiments performed with washed and unwashed E. coli Ec300 bacterial inoculum (data not shown). Considering Ec300p’s inability to impair S. aureus bacterial/bacterial aggregation under liquid conditions (see above), these results suggested that the reduction in S. aureus proportions in mixed S. aureus/E. coli Ec300 biofilm was due to the production of Ec300p during E. coli Ec300 biofilm formation. We then studied the impact of production of Ec300p on S. aureus ability to adhere to preformed E. coli biofilm bacteria by inoculating S. aureus onto E. coli Ec300 or E. coli Ec300∆rfaH biofilms grown for 8 h in microfermentors. After 24 h, we observed a 15-fold reduction in the ability of S. aureus to colonize and settle into E. coli Ec300 biofilm compared to its ability to colonize an E. coli Ec300∆rfaH biofilm (Fig. 6C). These results therefore indicated that the competitive advantage exhibited by E. coli Ec300 compared to S. aureus in mixed biofilms correlates with production of Ec300p, which inhibits the initial S. aureus surface adhesion and alters S. aureus/E. coli interactions in mixed biofilms.
FIG 6 .
Effect of Ec300 antiadhesion polysaccharide production in mixed S. aureus/E. coli Ec300 biofilms. (A) Evolution of a mixed Ec300 and S. aureus biofilm under continuous-flow microfermentor conditions. Percentage of Ec300 and S. aureus in mixed microfermentors, as revealed by CFU plating in differential media at different time points. wt, wild type. (B) Evolution of mixed E. coli Ec300∆rfaH and S. aureus biofilms under continuous-flow microfermentor conditions. Percentages were obtained as described above. (C) Ability of S. aureus to colonize preformed E. coli Ec300 or E. coli Ec300∆rfaH biofilms formed under continuous-flow microfermentor conditions, as determined by CFU counting of the different strains. Experiments were performed in triplicate; error bars represent standard deviations of the means. Statistical t tests were used to evaluate the effect of biofilm exclusion upon Ec300p production. **, P < 0.01; ***, P < 0.001.
DISCUSSION
Bacterial biofilms undergo significant gene expression and metabolic modifications, potentially leading to production of biofilm-specific metabolites or polymers (17, 24, 25). Some of these molecules could display antagonist activities against other microorganisms in mixed-species contexts (14, 18). In this study, we screened mature biofilms formed by commensal and pathogenic natural E. coli isolates so as to identify such antiadhesion molecules associated with biofilm environments.
Using continuous-flow biofilm microfermentors, our analysis showed that 20% of tested strains representative of the biodiversity of the E. coli species produce antiadhesion molecules, and at least 10% of which correspond to polysaccharides. Hence, beyond previously described group 2 capsule antiadhesion molecules, our results demonstrate the widespread occurrence of polysaccharides potentially interfering with the bacterial capacity to interact with surfaces or other bacteria (14). We focused our study on the E. coli Ec300 polysaccharide (Ec300p) produced by the commensal isolate E. coli Ec300, since Ec300p displays original properties in terms of its spectrum of activity, composition, and genetic determinants.
A key aspect of our screen resided in the identification of antiadhesion molecules specific to the biofilm lifestyle. While many identified antiadhesion molecules were seemingly also produced under both planktonic and biofilm conditions, in the case of E. coli Ec300, a commensal isolate of the A phylogenetic group, we detected Ec300p antibiofilm activity within thick mature biofilm extracts. Hence, our results support the hypothesis that biofilms could constitute a molecular sink in which polymers and biologically active molecules are trapped, and indeed sequestered, compared to planktonic cultures (26, 27). We showed that the activity of the concentrated purified planktonic E. coli Ec300 polysaccharide was similar to that of Ec300p from biofilm extracts, therefore demonstrating that E. coli Ec300p production is not strictly biofilm specific. However, resuspension of mature biofilms used to produce biofilm extracts consistently contained more bacteria than the equivalent volume of planktonic culture, and the high cell density achieved within thick mature biofilms could explain the detection of Ec300p activity in biofilm extract compared to nondetection of activity in unconcentrated planktonic supernatants, as previously shown in the case of amino acid valine accumulation within biofilms (16).
Nevertheless, we also determined that the amount of antiadhesion polysaccharide produced per bacterium was 2-fold higher in E. coli Ec300 biofilm extracts than in corresponding planktonic stationary-phase supernatants. This indicated that biofilm-specific metabolism or regulation could also contribute to the observed biofilm-associated production of the E. coli Ec300 polysaccharide. We show that Ec300p synthesis is positively regulated by RfaH, which is a transcriptional antiterminator involved in the regulation of various surface structures (22, 23). However, since there is no significant difference in the rfaH expression level between planktonic stationary and biofilm growth phases, other regulatory mechanisms could be involved in biofilm-associated production of Ec300p, including, for instance, growth-phase-dependent regulation. Interestingly, a previous study showed that genes involved in colanic acid production, an RfaH-regulated process, are also induced upon E. coli biofilm formation as part of a stress response, suggesting that Ec300p production could result from increased stress in late stationary phase and biofilms (24).
We determined that genes located in the galF-hisE region encode Ec300p. This locus is highly variable and complex and is involved in the synthesis of surface polysaccharides in Enterobacteriaceae, including the group 1 capsule, colanic acid, and lipopolysaccharide O antigen (21, 28). Although neither Ec300p composition nor its genetic determinants match with the O antigen, group 1 capsule, or colanic acid, we nonetheless identified genes involved in group 1 capsule translocation (wzb-wzc) in a region essential to Ec300p production corresponding to contig 6759 (Fig. 3A). This suggests that E. coli Ec300 produces a new type of released polysaccharide, potentially exported though heterologous export machinery.
The galF-his region is known to be a stronghold of E. coli chromosome polymorphism (29), with hot spots of both recombination and gene acquisition and loss (30). Interestingly, whereas most galF-his regions identified in E. coli strains extend over 16 kb only, the E. coli Ec300 region is 40 kb long and displays a high level of synteny and homology with the galF-his region of K. pneumoniae 342, which also exhibited significant antiadhesion activity. The presence of several transposases in E. coli Ec300 suggests that genes coding for Ec300p were acquired by horizontal gene transfer into the galF-his locus, potentially from K. pneumoniae 342. In support of this hypothesis, genes flanking contigs involved in synthesis of antiadhesion Ec300p revealed the presence of transposition-associated genes, and recombination between the O9-antigen strain and the Klebsiella strain has been previously reported (31).
Our analysis indicated that the E. coli Ec300 galF-his region contains genes not involved in the synthesis of Ec300p. Furthermore, the Ec300ΔgalF-his mutant lost its ability to produce E. coli Ec300p and was also unable to form biofilms. This suggested that the Ec300 galF-hisE region could also encode other nonidentified polysaccharides, potentially including O antigen.
The E. coli species releases several high-molecular-weight extracellular polysaccharides, contributing to biofilm structure as a component of the extracellular matrix (14, 28, 32, 33). Our results further document that, in addition to functions usually assigned to bacterial capsules, including promotion of adhesion, masking of surface adhesins, and resistance to host and environmental stresses, the reduction in bacterial adhesion by surface-active bacterial polysaccharides is more widespread than previously considered.
Biofilms represent an ideal environment in which the study of antagonistic and synergistic interactions could be particularly relevant due to biofilm prevalence in nature. Although much interest has been focused on communication behavior in multispecies biofilms, the investigation of competitive interference affecting population dynamics in this context represents a relatively novel area of interest (18).
In addition to well-known extracellular bacterial antagonist molecules such as colicins, toxins, and phages (6, 34), surface-active agents are also likely to affect the dynamics of population growth on surfaces by regulating the attachment or detachment of microorganisms to or from surfaces (15). However, the nature and roles of surfactants potentially produced in mixed or complex bacterial community contexts are still poorly understood. Recent reports have shown that high-molecular-weight bacterial polysaccharidic surfactants could modify population dynamics in a mixed biofilm context (14). Here we show that, besides Ec300p antiadhesion activity in vitro, production of E. coli Ec300p results in very rapid exclusion of S. aureus from mixed E. coli and S. aureus biofilms.
Reduction in susceptibility to invasion, or colonization resistance, is a very well-known but undefined community property shown to strongly depend on species composition (35). Here we also showed that E. coli Ec300 biofilms producing Ec300p are significantly protected from further colonization by incoming S. aureus. Hence, the increased release of antiadhesion extracellular polysaccharides such as Ec300p in a biofilm community could constitute a molecular basis for colonization resistance and could provide a direct advantage at the initial colonization stage compared to that of strong colonizers, such as biofilm-forming S. aureus.
Although the role of such antiadhesion molecules, which interfere with the settling of colonizing bacteria, has been poorly investigated, their widespread occurrence could play a significant role in bacterial competition and niche exclusion in multispecies communities. For instance, Ec300p activity could provide a direct advantage at the initial colonization stage compared to that of strong colonizers, such as biofilm-forming S. aureus.
A particularly intriguing aspect of our study is the restricted activity spectrum of Ec300p toward Gram-positive bacteria. As previously considered for the broad-host-range antiadhesion activity of the group 2 capsule, interactions with negatively charged Ec300p macromolecules could result in repulsive electrostatic forces on negatively charged bacterial surfaces. However, S. aureus mutants with an altered cell wall (tagO and dltA), leading to different global net surface charges, are still unable to form a biofilm in the presence of Ec300p. Other effects may therefore be involved, including surface hydration, steric repulsion, hydrophobicity, or electron donor-electron acceptor (acid-base) properties, reported to be 10 to 100 times more important for bacterial adhesion than other interactions (36). Alternatively, Gram-positive bacteria generally display more pronounced hydrophobic properties than Gram-negative bacteria and could be more sensitive to the rise in surface hydrophilicity induced by Ec300p coating.
Surface-active molecules were proposed to be a promising means of preventing infection by interfering with pathogen adhesion (8, 14, 15). However, use of broad-host-range antiadhesion molecules could have undesirable effects. In this respect, the narrow host range of Ec300p could provide an interesting alternative for preventing colonization and biofilm infection by S. aureus and, potentially, other Gram-positive bacteria. Moreover, as opposed to antibiotics and other toxins targeting vital cellular functions, use of antiadhesion molecules could lower the selective pressure upon bacterial fitness, therefore limiting the arrival of resistant mutants as selective pressure is significantly lowered.
In conclusion, our results demonstrate that biofilms constitute a reservoir of molecules that could play significant roles in the dynamic of commensal/pathogen interactions within mixed bacterial communities. Screening biofilm extracts could be extended to other biofilm-forming organisms and reveal activities that would otherwise go undetected. This not only would lead to a better understanding of biofilm biology but also could uncover new interference molecules of potential biomedical interest.
MATERIALS AND METHODS
Bacterial strains, plasmids, and liquid growth conditions.
The E. coli natural isolates used in this study are listed in Table S1 in the supplemental material and originate from previously published collections (37–40). The other bacterial strains and plasmids used are listed in Table S3 in the supplemental material. Gram-positive strains were cultured in tryptic soy broth (TSB) supplemented with 0.25% or 0.5% glucose (E. faecalis). All other experiments were performed in 0.4% glucose-M63B1 minimal medium (M63B1) or in lysogenic broth (LB) medium at 37°C, with appropriate antibiotics when required.
Purification of planktonic and biofilm extracts. (i) Planktonic supernatants.
Bacteria were cultured for 24 to 48 h in M63B1 and then adjusted to an optical density (OD) of 3 (OD at 600 nm [OD600], 3). Cultures were centrifuged for 10 min at 7500 rpm at 4°C, and the supernatants were filtered through a 0.2-µm filter.
(ii) Biofilm extracts.
Continuous-flow microfermentors containing a removable glass spatula were used as described (http://www.pasteur.fr/recherche/unites/Ggb/matmet.html) to maximize biofilm development and minimize planktonic growth. Inoculation was performed by dipping the glass spatula for 5 min in a culture adjusted to an OD600 of 1 from overnight bacterial cultures grown in M63B1 supplemented with the appropriate antibiotics. The spatula was then reintroduced into the microfermentor. After 72 h at 37°C, microfermentor medium (OD600 < 0.1) was carefully discarded, and biofilm biomass was rapidly resuspended in the remaining 15 ml medium for 30 s. Under these particular conditions, most tested E. coli strains developed thick mature biofilms (1- to 4-fold biomass variance), and occasional poor-biofilm-forming strains were excluded from the analysis.
(iii) Nonconcentrated extract.
The final OD600 value was measured and adjusted to an OD600 of 3, and the extract was centrifuged and filtered through a 0.2-µm filter (see Fig. S1 in the supplemental material).
(iv) Concentrated extract.
Nonconcentrated extract was ethanol precipitated and concentrated.
(v) Purified extract.
Concentrated extract was purified on a DEAE anion-exchange column, as described in reference 14.
Biofilm-related assays. (i) Biofilm inhibition assays.
Overnight cultures were adjusted to an OD600 of 0.1 before inoculating 50 µl into 96-well polyvinyl chloride (PVC) plates (Falcon; Becton Dickinson Labware, Oxnard, CA) and added at a 1:1 ratio to 50 µl of filter-sterilized biofilm or planktonic supernatant equilibrated to an OD600 of 3. Biofilms were left to grow for either 8 or 16 h at 37°C before quantification as described in reference 14.
(ii) Microtiter plate precoating assays.
One hundred microliters of biofilm extract were inoculated into a 96-well PVC microtiter plate and left to incubate for 24 h at 4°C. Biofilm extract was removed, and microtiter plates were washed 3 times with sterile deionized water. Exponential-phase cultures were then adjusted to an OD600 of 0.06, inoculated into precoated microtiter plates, and left to grow for 16 and 24 h at 37°C before quantification as described in reference 14.
(iii) Growth inhibition test.
The inhibitory effect of filter-sterilized supernatants and extracts was evaluated as decribed in reference 16.
Molecular characterization of the inhibitory compound.
Molecular cutoff analyses and proteinase K treatments were conducted as described in reference 14. To determine the polysaccharide composition, biofilm extracts were filtered through a 0.2-µm filter, precipitated with 3 volumes of ethanol, and dialyzed against deionized water (10-kDa cassettes; Pierce, Rockford, IL). Total amounts of neutral sugars were quantified by phenol-sulfuric acid methods, using glucose as a standard. Extract active fractions were purified and submitted to either sizing chromatography or composition analysis, as described in reference 14.
Generation of deletion and transcriptional fusions.
Gene deletions and replacement with an antibiotic resistance cassette in the different strains of E. coli and lacZ-zeo fusion mutants in E. coli Ec300 were generated by the λ red linear DNA gene replacement system and the 3-step PCR procedure described previously (41, 42).
Beta-galactosidase assays.
Beta-galactosidase activity on exponential- and stationary-phase cells and 72-h biofilms were performed as described in reference 43.
Mixed biofilm assays. (i) Competition.
Overnight LB cultures of S. aureus, E. coli Ec300, and E. coli Ec300ΔrfaH strains were mixed in a 1:1 ratio (OD600 = 1) before inoculation in a microfermentor (LB, 60 ml/h flow rate). Dilutions of the biomass collected at different time points were plated in LB with and without suitable antibiotics.
(ii) Biofilm colonization.
E. coli Ec300 biofilms were pregrown for 8 h. The spatula was then carefully removed and dipped in an S. aureus culture adjusted to an OD600 of 1 for 5 min. The mixed biofilm was allowed to grow 24 h more at 37°C in LB before recovery, and dilutions of the biomass were plated in LB and in plates supplemented with suitable antibiotics.
Functional analysis of polysaccharide activity. (i) Initial adhesion on glass slides.
Overnight LB cultures of fluorescent S. aureus were diluted to an OD600 of 1 before being mixed in a 1:1 ratio with either different E. coli Ec300 strains or 100 µg/ml of E. coli Ec300 filtered sterilized biofilm extracts. The glass slides were incubated for 5 min and rinsed once in deionized water before microscopic observation. At least 10 pictures were taken of each slide. Experiments were performed in triplicate. Bacteria adhering to the glass surface were counted, and the total surface area covered was estimated with ImageJ64 software developed by the NIH (http://imagej64.mac.informer.com/).
(ii) Aggregation assay.
S. aureus cultures were adjusted to an OD600 of 3 by dilution with spent medium, and the aggregation assay was conducted as described in reference 14.
SUPPLEMENTAL MATERIAL
Natural E. coli isolates used in this study.
Description of genomic contigs potentially involved in the biosynthesis of the E. coli Ec300 antiadhesion polysaccharide.
Bacterial strains used in this study.
Primers used in this study.
(A) Schematic representation of the procedure used to produce biofilm extract from biofilms grown under continuous-flow conditions (see also http://www.pasteur.fr/recherche/unites/Ggb/matmet.html). (B) Panel of Gram-negative and Gram-positive bacteria used to test the activity of biofilm extracts. (a) Representative example of biofilm formation in microtiter plate wells in the absence of biofilm extract. (b) Representative example of broad-range antibiofilm activity. (c) Representative example of narrow-range antibiofilm activity. Download Figure S1, PDF file, 1.000 MB.
Nonbiocidal effect of nonconcentrated Ec300 and Ec111 biofilm extracts. On an S. aureus bacterial lawn grown on LB agar plates (A) or in liquid LB medium in microtiter plates (B). Download Figure S2, PDF file, 0.994 MB.
Activity of E. coli Ec300 polysaccharide compared to that of PNAG-dependent and PNAG-independent biofilm formation. Biofilm inhibition was determined in the presence of E. coli Ec300 polysaccharide added at 50 g/ml (final concentration in each well). Experiments were performed at least in triplicate; error bars represent standard deviations of the means. Statistical t tests were used to evaluate the significance of biofilm inhibition compared to that of H2O. ***, P < 0.001. PNAG, poly-N-acetyl glucosamine. Download Figure S3, PDF file, 0.156 MB.
Biofilm formation of E. coli Ec300 and the ΔgalF-hisE mutant. (A) Image of a typical microfermentor after 72 h at 37°. (B) Relative biofilm formation of each strain. Download Figure S4, PDF file, 1.444 MB.
Comparision of beta-galactosidase activity measurements of lacZ transcriptional fusions at different growth stages (exponential phase, late stationary phase, and biofilm conditions) of the indicated genes. Experiments were performed in triplicate; error bars represent standard deviations of the means. Statistical t tests were used to evaluate the significance of biofilm inhibition from various extracts compared to that of addition of M63B1. *, P < 0.05; ***, P < 0.001. Download Figure S5, PDF file, 0.233 MB.
Determination of the surface contact angle of a drop of water in untreated (double-distilled water [dH2O]), group 2 capsule (G2CPS), and Ec300p-treated microscopy slides. Contact angle measurements. Microscope glass slides were washed with detergent and extensively rinsed with distilled water. Washed slides were dried under a laminar flow hood, coated with 300 μl of 100 or 500 μg/ml of ethanol-precipitated cell-free extracts for 5 min, and rinsed with distilled water. Four-microliter drops of water were deposited using a DigiDrop machine on the surfaces of coated and uncoated slides, and contact angles were then measured. For each slide, 5 drops were measured, and each drop was measured 3 times. Download Figure S6, PDF file, 0.418 MB.
ACKNOWLEDGMENTS
We thank C. Bouchier, P. Glaser, and L. Frangeul (Genopole, Institut Pasteur, Paris, France), Zoe Rouy (Genoscope, Évry, France), and C. Buchrieser (Intracellular Bacteria Biology Unit) for their help during genome sequencing and analyses. We are grateful to N. Henry for her help in the determination of surface contact angles. We thank C. Beloin, S. Létoffé, J. Valle, S. Chalabaev, and C. Forestier for helpful discussions and critical reading of the manuscript.
O.R. was supported by a fellowship from the Network of Excellence EuroPathogenomics, European Community grant LSHB-CT-2005-512061. L.T. was an ANR fellow (ERA-NET EuroPathogenomics). This work was supported by a grant from the Région Île-de-France (DIM Malinf).
Footnotes
Citation Rendueles O, et al. 2011. Screening of Escherichia coli species biodiversity reveals new biofilm-associated antiadhesion polysaccharides. mBio 2(3):e00043-11. doi:10.1128/mBio.00043-11.
REFERENCES
- 1. Lynch AS, Robertson GT. 2008. Bacterial and fungal biofilm infections. Annu. Rev. Med. 59:415–428 [DOI] [PubMed] [Google Scholar]
- 2. Kaplan JB. 2010. Biofilm dispersal: mechanisms, clinical implications, and potential therapeutic uses. J. Dent. Res. 89:205–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kolodkin-Gal I, et al. 2010. d-Amino acids trigger biofilm disassembly. Science 328:627–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fey PD. 2010. Modality of bacterial growth presents unique targets: how do we treat biofilm-mediated infections? Curr. Opin. Microbiol. 13:610–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Njoroge J, Sperandio V. 2009. Jamming bacterial communication: new approaches for the treatment of infectious diseases. EMBO Mol. Med. 1:201–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Donlan RM. 2009. Preventing biofilms of clinically relevant organisms using bacteriophage. Trends Microbiol. 17:66–72 [DOI] [PubMed] [Google Scholar]
- 7. Kadouri D, O’Toole GA. 2005. Susceptibility of biofilms to Bdellovibrio bacteriovorus attack. Appl. Environ. Microbiol. 71:4044–4051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Danese PN. 2002. Antibiofilm approaches: prevention of catheter colonization. Chem. Biol. 9:873–880 [DOI] [PubMed] [Google Scholar]
- 9. Francolini I, Donelli G. 2010. Prevention and control of biofilm-based medical-device-related infections. FEMS Immunol. Med. Microbiol. 59:227–238 [DOI] [PubMed] [Google Scholar]
- 10. Vasilev K, Cook J, Griesser HJ. 2009. Antibacterial surfaces for biomedical devices. Expert Rev. Med. Devices 6:553–567 [DOI] [PubMed] [Google Scholar]
- 11. Mukherjee AK, Das K. 2010. Microbial surfactants and their potential applications: an overview. Adv. Exp. Med. Biol. 672:54–64 [DOI] [PubMed] [Google Scholar]
- 12. Davey ME, Caiazza NC, O’Toole GA. 2003. Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas aeruginosa PAO1. J. Bacteriol. 185:1027–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kiran GS, Sabarathnam B, Selvin J. 2010. Biofilm disruption potential of a glycolipid biosurfactant from marine Brevibacterium casei. FEMS Immunol. Med. Microbiol. 59:432–438 [DOI] [PubMed] [Google Scholar]
- 14. Valle J, et al. 2006. Broad-spectrum biofilm inhibition by a secreted bacterial polysaccharide. Proc. Natl. Acad. Sci. U. S. A. 103:12558–12563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Banat IM, et al. 2010. Microbial biosurfactants production, applications and future potential. Appl. Microbiol. Biotechnol. 87:427–444 [DOI] [PubMed] [Google Scholar]
- 16. Valle J, et al. 2008. The amino acid valine is secreted in continuous-flow bacterial biofilms. J. Bacteriol. 190:264–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Whiteley M, et al. 2001. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413:860–864 [DOI] [PubMed] [Google Scholar]
- 18. Hibbing ME, Fuqua C, Parsek MR, Peterson SB. 2010. Bacterial competition: surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 8:15–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gross M, Cramton SE, Götz F, Peschel A. 2001. Key role of teichoic acid net charge in Staphylococcus aureus colonization of artificial surfaces. Infect. Immun. 69:3423–3426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mack D, Siemssen N, Laufs R. 1992. Parallel induction by glucose of adherence and a polysaccharide antigen specific for plastic-adherent Staphylococcus epidermidis: evidence for functional relation to intercellular adhesion. Infect. Immun. 60:2048–2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Whitfield C. 2006. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu. Rev. Biochem. 75:39–68 [DOI] [PubMed] [Google Scholar]
- 22. Rahn A, Whitfield C. 2003. Transcriptional organization and regulation of the Escherichia coli K30 group 1 capsule biosynthesis (cps) gene cluster. Mol. Microbiol. 47:1045–1060 [DOI] [PubMed] [Google Scholar]
- 23. Beloin C, et al. 2006. The transcriptional antiterminator RfaH represses biofilm formation in Escherichia coli. J. Bacteriol. 188:1316–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Beloin C, et al. 2004. Global impact of mature biofilm lifestyle on Escherichia coli K-12 gene expression. Mol. Microbiol. 51:659–674 [DOI] [PubMed] [Google Scholar]
- 25. Ciornei CD, et al. 2010. Biofilm-forming Pseudomonas aeruginosa bacteria undergo lipopolysaccharide structural modifications and induce enhanced inflammatory cytokine response in human monocytes. Innate Immun. 16:288–301 [DOI] [PubMed] [Google Scholar]
- 26. Flemming HC, Wingender J. 2010. The biofilm matrix. Nat. Rev. Microbiol. 8:623–633 [DOI] [PubMed] [Google Scholar]
- 27. Sutherland I. 2001. Biofilm exopolysaccharides: a strong and sticky framework. Microbiology 147:3–9 [DOI] [PubMed] [Google Scholar]
- 28. Corbett D, Roberts I. 2008. Capsular polysaccharides in Escherichia coli. Adv. Appl. Microbiol. 65:1–26 [DOI] [PubMed] [Google Scholar]
- 29. Milkman R, Jaeger E, McBride RD. 2003. Molecular evolution of the Escherichia coli chromosome. VI. Two regions of high effective recombination. Genetics 163:475–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Touchon M, et al. 2009. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 5:e1000344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sugiyama T, et al. 1998. Generation of Escherichia coli O9a serotype, a subtype of E. coli O9, by transfer of the wb* gene cluster of Klebsiella O3 into E. coli via recombination. J. Bacteriol. 180:2775–2778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Da Re S, Ghigo JM. 2006. A CsgD-independent pathway for cellulose production and biofilm formation in Escherichia coli. J. Bacteriol. 188:3073–3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Grant WD, Sutherland IW, Wilkinson JF. 1969. Exopolysaccharide colanic acid and its occurrence in the Enterobacteriaceae. J. Bacteriol. 100:1187–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tait K, Sutherland IW. 2002. Antagonistic interactions amongst bacteriocin-producing enteric bacteria in dual species biofilms. J. Appl. Microbiol. 93:345–352 [DOI] [PubMed] [Google Scholar]
- 35. Robinson C, Bohannan B, Young V. 2010. From structure to function: the ecology of host-associated microbial communities. Microbiol. Mol. Biol. Rev. 74:453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. van Oss CJ. 1989. Energetics of cell-cell and cell-biopolymer interactions. Cell Biophys. 14:1–16 [DOI] [PubMed] [Google Scholar]
- 37. Escobar-Paramo P, et al. 2004. A specific genetic background is required for acquisition and expression of virulence factors in Escherichia coli. Mol. Biol. Evol. 21:1085–1094 [DOI] [PubMed] [Google Scholar]
- 38. Escobar-Páramo P, et al. 2006. Identification of forces shaping the commensal Escherichia coli genetic structure by comparing animal and human isolates. Environ. Microbiol. 8:1975–1984 [DOI] [PubMed] [Google Scholar]
- 39. Picard B, et al. 1999. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect. Immun. 67:546–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Skurnik D, et al. 2006. Effect of human vicinity on antimicrobial resistance and integrons in animal faecal Escherichia coli. J. Antimicrob. Chemother. 57:1215–1219 [DOI] [PubMed] [Google Scholar]
- 41. Chaveroche MK, Ghigo JM, d’Enfert C. 2000. A rapid method for efficient gene replacement in the filamentous fungus Aspergillus nidulans. Nucleic Acids Res. 28:E97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Derbise A, Lesic B, Dacheux D, Ghigo JM, Carniel E. 2003. A rapid and simple method for inactivating chromosomal genes in Yersinia. FEMS Immunol. Med. Microbiol. 38:113–116 [DOI] [PubMed] [Google Scholar]
- 43. Miller JH. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Natural E. coli isolates used in this study.
Description of genomic contigs potentially involved in the biosynthesis of the E. coli Ec300 antiadhesion polysaccharide.
Bacterial strains used in this study.
Primers used in this study.
(A) Schematic representation of the procedure used to produce biofilm extract from biofilms grown under continuous-flow conditions (see also http://www.pasteur.fr/recherche/unites/Ggb/matmet.html). (B) Panel of Gram-negative and Gram-positive bacteria used to test the activity of biofilm extracts. (a) Representative example of biofilm formation in microtiter plate wells in the absence of biofilm extract. (b) Representative example of broad-range antibiofilm activity. (c) Representative example of narrow-range antibiofilm activity. Download Figure S1, PDF file, 1.000 MB.
Nonbiocidal effect of nonconcentrated Ec300 and Ec111 biofilm extracts. On an S. aureus bacterial lawn grown on LB agar plates (A) or in liquid LB medium in microtiter plates (B). Download Figure S2, PDF file, 0.994 MB.
Activity of E. coli Ec300 polysaccharide compared to that of PNAG-dependent and PNAG-independent biofilm formation. Biofilm inhibition was determined in the presence of E. coli Ec300 polysaccharide added at 50 g/ml (final concentration in each well). Experiments were performed at least in triplicate; error bars represent standard deviations of the means. Statistical t tests were used to evaluate the significance of biofilm inhibition compared to that of H2O. ***, P < 0.001. PNAG, poly-N-acetyl glucosamine. Download Figure S3, PDF file, 0.156 MB.
Biofilm formation of E. coli Ec300 and the ΔgalF-hisE mutant. (A) Image of a typical microfermentor after 72 h at 37°. (B) Relative biofilm formation of each strain. Download Figure S4, PDF file, 1.444 MB.
Comparision of beta-galactosidase activity measurements of lacZ transcriptional fusions at different growth stages (exponential phase, late stationary phase, and biofilm conditions) of the indicated genes. Experiments were performed in triplicate; error bars represent standard deviations of the means. Statistical t tests were used to evaluate the significance of biofilm inhibition from various extracts compared to that of addition of M63B1. *, P < 0.05; ***, P < 0.001. Download Figure S5, PDF file, 0.233 MB.
Determination of the surface contact angle of a drop of water in untreated (double-distilled water [dH2O]), group 2 capsule (G2CPS), and Ec300p-treated microscopy slides. Contact angle measurements. Microscope glass slides were washed with detergent and extensively rinsed with distilled water. Washed slides were dried under a laminar flow hood, coated with 300 μl of 100 or 500 μg/ml of ethanol-precipitated cell-free extracts for 5 min, and rinsed with distilled water. Four-microliter drops of water were deposited using a DigiDrop machine on the surfaces of coated and uncoated slides, and contact angles were then measured. For each slide, 5 drops were measured, and each drop was measured 3 times. Download Figure S6, PDF file, 0.418 MB.