Abstract
A chromosome-encoded β-lactamase gene from a Shewanella oneidensis reference strain was cloned and expressed in Escherichia coli. It encoded a carbapenem-hydrolyzing Ambler class D β-lactamase, OXA-54, that shared 92% amino acid identity with the plasmid-encoded carbapenem-hydrolyzing oxacillinase OXA-48 from Klebsiella pneumoniae. This work suggests that Shewanella spp. may produce the progenitor of oxacillinases compromising the efficacy of imipenem in clinically relevant gram-negative pathogens.
Several carbapenem-hydrolyzing oxacillinases have been characterized recently, mostly from Acinetobacter baumannii isolates (14, 19). These Ambler class D β-lactamases that possess a weak activity against imipenem are divided into a first group, made of OXA-24, OXA-25, OXA-26, and OXA-40, which differ by a few amino acid substitutions (1, 4, 10), and a second group of enzymes, consisting of OXA-23 and OXA-27, that share 99% amino acid identity (3, 6, 15), with the two groups of oxacillinases sharing only 60% identity. Recently we have characterized β-lactamase OXA-48 from a Klebsiella pneumoniae isolate that was resistant to all β-lactams including carbapenems (16). This oxacillinase had a strong activity against imipenem, and its gene was not part of a class 1 integron as most oxacillinase genes are but was plasmid located downstream of the insertion sequence element, IS1999 (16). The β-lactamase OXA-48 was weakly related to class D β-lactamases, with less than 46% amino acid identity with any other oxacillinases.
The entire genome sequence analysis of Shewanella oneidensis strain MR-1 (a ca. 5-Mb chromosome and a ca. 160-kb plasmid) was published recently (9) (GenBank accession no. AE014299). In silico analysis of the chromosome sequence of this bacterial strain identified an open reading frame that shared 84% nucleotide identity with that of blaOXA-48. Thus, we have cloned the corresponding gene and characterized biochemically this protein in order to look for its ability to hydrolyze β-lactams, including carbapenems.
S. oneidensis MR-1 had been isolated from a lake sediment and was previously named Shewanella putrefaciens (21). Escherichia coli reference strain DH10B and plasmid pCR-Blunt II-TOPO (Invitrogen, Life Technologies, Cergy-Pontoise, France) were used for cloning and expression experiments. Whole-cell DNA of S. oneidensis MR-1 was extracted as described previously (18). The PCR experiment was performed using primers PREOXA-54A (5′-GGCGATGGCAAACGAATGGC-3′) and PREOXA-54B (5′-GTGATGGCTTGACGTAGCCC-3′), designed from the DNA sequence of S. oneidensis MR-1 in the databases to amplify by PCR the entire β-lactamase gene. The corresponding 820-bp PCR product was subsequently cloned into the kanamycin-resistant plasmid pCR-Blunt II-TOPO (Invitrogen, Life Technologies), giving rise to the recombinant plasmid pSON-1, and transformed in E. coli DH10B, as recommended by the manufacturer. Selection was performed on Mueller-Hinton plates containing 50 μg of amoxicillin and 30 μg of kanamycin per ml. Once cloned, the PCR product was sequenced on both strands as described previously (17), and the oxacillinase gene sequence revealed total identity with that found in the databases corresponding to S. oneidensis MR-1. Antibiogram obtained by disk diffusion was performed with E. coli DH10B harboring plasmid pSON-1. Results suggested that the cloned gene encoded a functional β-lactamase named OXA-54. Susceptibility testing of S. oneidensis MR-1 was performed at 30°C on blood-containing trypticase soy agar plates, as recommended (8), with the E-test technique (inoculum MacFarland 1, incubation for 24 h at 20°C; AB Biodisk, Solna, Sweden), since this species is unable to grow on Mueller-Hinton-containing plates. S. oneidensis MR-1 was susceptible to all β-lactams (Table 1). Specific activity measured with a culture extract of S. oneidensis MR-1 obtained as described previously (2) and a 100 μM concentration of imipenem as the substrate revealed that this strain hydrolyzed imipenem significantly (2.1 mU · mg of protein−1). Using subinhibitory concentrations of imipenem and cefoxitin as inducers as previously described (20), we were unable to demonstrate any induction of OXA-54 production from S. oneidensis MR-1 (data not shown).
TABLE 1.
MICs (μg/ml) of β-lactams for S. oneidensis MR-1, E. coli DH10B harboring recombinant plasmid pSON-1 expressing OXA-54 from S. oneidensis MR-1, E. coli DH10B harboring recombinant plasmid pVT-1 expressing OXA-48 from K. pneumoniae 11978, and the E. coli DH10B reference strain
| β-Lactam(s)a | MIC (μg/ml) of drug for strain
|
|||
|---|---|---|---|---|
| S. oneidensis MR-1 | E. coli DH10B (pSON-1) (OXA-54) | E. coli DH10B (pVT-1) (OXA-48)b | E. coli DH10B | |
| Amoxicillin | 0.25 | >512 | >512 | 4 |
| Amoxicillin + CLA | 0.25 | >512 | >512 | 4 |
| Ticarcillin | 0.5 | >512 | >512 | 4 |
| Ticarcillin + CLA | <0.02 | >512 | >512 | 4 |
| Piperacillin | 0.12 | 32 | 128 | 2 |
| Piperacillin + TZB | <0.02 | 32 | 128 | 2 |
| Cephalothin | 4 | 4 | 64 | 4 |
| Cefuroxime | 0.5 | 4 | 8 | 4 |
| Cefoxitin | 0.5 | 4 | 8 | 4 |
| Ceftazidime | 0.03 | 0.12 | 0.12 | 0.06 |
| Cefotaxime | 0.01 | 0.12 | 0.25 | 0.06 |
| Cefepime | 0.01 | 0.06 | 0.06 | 0.06 |
| Cefpirome | 0.01 | 0.25 | 0.25 | 0.06 |
| Moxalactam | 0.12 | 2 | 2 | 0.06 |
| Aztreonam | 0.03 | 0.06 | 0.06 | 0.12 |
| Imipenem | 0.12 | 1 | 2 | 0.06 |
| Meropenem | 0.05 | 0.12 | 0.25 | 0.06 |
CLA, clavulanic acid at a fixed concentration of 2 μg/ml; TZB, tazobactam at a fixed concentration of 4 μg/ml.
Data are from reference 16.
E. coli DH10B (pSON-1) gave an oxacillinase phenotype, conferring resistance to penicillins that was unchanged after clavulanic acid addition and a reduced susceptibility to cephalothin and sparing expanded-spectrum cephalosporins. MICs were determined by an agar dilution technique as previously reported (18), and results were interpreted according to the guidelines of the National Committee for Clinical Laboratory Standards (13). The MIC of imipenem was 1 μg/ml for E. coli DH10B (pSON-1), indicating that the β-lactamase OXA-54 was able to hydrolyze imipenem (Table 1).
Isoelectric focusing analysis revealed that E. coli DH10B (pSON-1) produced a β-lactamase with a pI of 6.8. Purification of the β-lactamase OXA-54 was performed with a culture of E. coli DH10B harboring recombinant plasmid pSON-1, as described previously (10). Briefly, the culture extracts were subjected to two purification steps, including use of ion-exchange chromatography. First, we used a Q-Sepharose column and a 20 mM Tris-HCl buffer (pH 9.0), and the enzyme was recovered in the flow-through. Then, this extract was loaded on an S-Sepharose column preequilibrated with 100 mM sodium phosphate buffer (pH 5.8) and β-lactamase was eluted with a linear K2SO4 gradient in order to prevent potential inhibitory activity of NaCl.
After purification from culture extracts of E. coli DH10B (pSON-1), the specific activity of the β-lactamase OXA-54 against cephalothin was determined to be 1 U/mg of protein, and its purification factor was 75-fold (with 1 U of enzyme activity being defined as the activity that hydrolyzed 1 μmol of cephalothin per min). Protein purity was estimated to be >95% by sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis (data not shown). The β-lactamase OXA-54 had a broad-spectrum hydrolysis profile, including penicillins, cephalothin, cefpirome, and imipenem and to a lesser extend cefuroxime, cefotaxime, and cefepime (Table 2). Catalytic activity (kcat/Km) of OXA-54 for imipenem was similar to that reported for OXA-48 (0.250 versus and 0.145 mM−1 · s−1, respectively) (Table 2), and 20-fold higher than that reported for the carbapenem-hydrolyzing β-lactamase OXA-40 (10). Hydrolysis of meropenem was at a low level, as observed for the other carbapenem-hydrolyzing oxacillinases (1, 10). OXA-54 did not hydrolyze ceftazidime and aztreonam, whereas hydrolysis of cefotaxime and cefepime was detected at low levels (Table 2). Oxacillin was well hydrolyzed by OXA-54, as reported for OXA-48, whereas this substrate is weakly hydrolyzed by the other carbapenem-hydrolyzing oxacillinases (Table 2). Activity inhibition measured by determination of 50% inhibitory concentrations after 3 min of preincubation with the inhibitor showed that OXA-54 was weakly inhibited by clavulanic acid (50 μM), tazobactam (2 μM), and sulbactam (150 μM), as reported for most of the oxacillinases (12). OXA-54 activity was inhibited by NaCl (50% inhibitory concentration, 11 mM), as observed for other oxacillinases except for those that have an FGN motif at class D β-lactamase positions 144 to 146 (DBL 144 to 146), i.e., most of the carbapenem-hydrolyzing oxacillinases (1, 3, 10).
TABLE 2.
Kinetic parameters of purified β-lactamase OXA-54 from S. oneidensis compared to those for OXA-48 from K. pneumoniae 11978a
| Substrate | OXA-54
|
OXA-48
|
||||
|---|---|---|---|---|---|---|
| kcat (S−1) | Km (μM) | kcat/Km (M−1 · s−1) | kcat (s−1) | Km (μM) | kcat/Km (M−1 · s−1) | |
| Benzylpenicillin | 120 | 60 | 2 | 245 | 40 | 6 |
| Ampicillin | 540 | 4,300 | 0.125 | 340 | 5,200 | 0.065 |
| Ticarcillin | 30 | 240 | 0.125 | 45 | 55 | 0.8 |
| Piperacillin | 20 | 240 | 0.085 | 75 | 410 | 0.2 |
| Cephalothin | 3 | 230 | 0.015 | 3 | 20 | 0.15 |
| Cephaloridin | 1 | 45 | 0.02 | 2 | 27 | 0.075 |
| Cefotaxime | 15 | 1,600 | 0.01 | 11 | 190 | 0.06 |
| Ceftazidime | NDb | —c | — | 4 | 5,100 | 0.001 |
| Cefepime | 1 | 180 | 0.005 | 1 | 160 | 0.005 |
| Cefpirome | 4 | 110 | 0.035 | 8 | 390 | 0.02 |
| Oxacillin | 35 | 75 | 0.5 | 25 | 30 | 0.85 |
| Aztreonam | ND | — | — | ND | — | — |
| Imipenem | 1 | 4 | 0.25 | 2 | 14 | 0.15 |
| Meropenem | 0.1 | 125 | 0.001 | 0.1 | 200 | 0.0005 |
Data are the means from three independent experiments. Standard deviations were within 10% of the means. Kinetic data for OXA-48 are from reference 16.
ND, no detectable hydrolysis (<0.01 s−1).
—, not determinable.
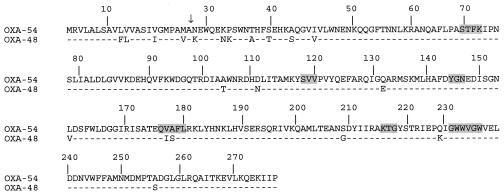
A detailed analysis of the amino acid sequence of β-lactamase OXA-54 did not identify specific residues that may be shared only by carbapenem-hydrolyzing oxacillinases (Fig. 1). The S-T-F-K tetrad was found at positions 70 to 73 (DBL numbering [5]), and the other structural elements characteristic of Ambler class D β-lactamases were found also (Fig. 1) (10, 11). The typical motif YGN of oxacillinases was found at positions DBL 144 to 146, and the SVV motif was found at positions DBL 118 to 120. OXA-54 shared 92% amino acid identity with OXA-48 (Fig. 1) but is weakly related to other oxacillinases, sharing 47, 36, 33, and 21% amino acid identity with OXA-10, OXA-23 and OXA-27, OXA-24, OXA-25, OXA-26, and OXA-40 group, and OXA-1, respectively (data not shown).
FIG. 1.
Comparison of the amino acid sequence of the carbapenem-hydrolyzing oxacillinase β-lactamase OXA-54 of reference strain S. oneidensis MR-1 to that of OXA-48 of the K. pneumoniae clinical isolate 11978. Shaded amino acid residues are conserved regions in the oxacillinase family. Numbering of β-lactamases is according to DBL (5). The vertical arrow indicates the putative cleavage site of the leader peptide of the mature β-lactamases.
Since the insertion sequence IS1999 was found upstream of the plasmid-located blaOXA-48 gene in K. pneumoniae 11978 (16), PCR experiments using whole-cell DNA of S. oneidensis MR-1 as the template and primers located inside the IS1999 transposase gene were performed. A negative result indicated that IS1999 was, at least, not always present in S. oneidensis.
Comparison of the surrounding sequences of blaOXA-48 to those of blaOXA-54 suggested strongly the origin of blaOXA-48 as being a Shewanella spp. Indeed, 21 out of 26 bp that separated IS1999 from the start codon of blaOXA-48 were identical to the DNA sequence located upstream of the blaOXA-54 gene. Noncoding sequences of 236 and 303 bp found downstream of the blaOXA-54 and blaOXA-48 genes, respectively, did not share nucleotide identity. However further downstream, the 252 bp of the 3′ end of a gene encoding a putative LysR-type transcriptional regulator (9) was found in S. oneidensis. This 252-bp sequence shared 88% nucleotide identity with the sequence located further downstream of blaOXA-48.
Conclusion.
This study demonstrates that a gram-negative facultative aerobe, S. oneidensis, which is an environmental species, may be a source of carbapenem-hydrolyzing oxacillinases. Interestingly, the finding of this β-lactamase in this species was unexpected according to phenotype analysis of its antibiotic susceptibility pattern.
This is the first characterization of a potential reservoir of carbapenem-hydrolyzing oxacillinases, whereas acquired carbapenem-hydrolyzing oxacillinases are reported increasingly in clinically relevant gram-negative species such as A. baumannii (14). The blaOXA-54 gene was not part of an integron in S. oneidensis, whereas a gene encoding an IntI-like tyrosine recombinase acting as an integron integrase has been detected in the genome of S. oneidensis MR-1 (7).
Acknowledgments
This work was funded by a grant from the Ministère de l'Education Nationale et de la Recherche (UPRES-EA3539), Université Paris XI, Paris, France. L.P. is a researcher from the INSERM, Paris, France.
We thank Vincent Méjean for the gift of the S. oneidensis MR-1 reference strain.
REFERENCES
- 1.Afzal-Shah, M., N. Woodford, and D. M. Livermore. 2001. Characterization of OXA-25, OXA-26, and OXA-27, molecular class D β-lactamases associated with carbapenem resistance in clinical isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 45:583-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aubert, D., L. Poirel, J. Chevalier, S. Léotard, J.-M. Pagès, and P. Nordmann. 2001. Oxacillinase-mediated resistance to cefepime and susceptibility to ceftazidime in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 45:1615-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonnet, R., H. Marchandin, C. Chanal, D. Sirot, R. Labia, C. De Champs, E. Jumas-Bilak, and J. Sirot. 2002. Chromosome-encoded class D β-lactamase OXA-23 in Proteus mirabilis. Antimicrob. Agents Chemother. 46:2004-2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bou, G., A. Oliver, and J. Martinez-Beltran. 2000. OXA-24, a novel class D β-lactamase with carbapenemase activity in an Acinetobacter baumannii clinical strain. Antimicrob. Agents Chemother. 44:1556-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Couture, F., J. Lachapelle, and R. C. Lévesque. 1992. Phylogeny of LCR-1 and OXA-5 with class A and class D β-lactamases. Mol. Microbiol. 6:1693-1705. [DOI] [PubMed] [Google Scholar]
- 6.Donald, H. M., W. Scaife, S. G. Amyes, and H. K. Young. 2000. Sequence analysis of ARI-1, a novel OXA β-lactamase responsible for imipenem resistance in Acinetobacter baumannii 6B92. Antimicrob. Agents Chemother. 44:196-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drouin, F., J. Melancon, and P. H. Roy. 2002. The IntI-like tyrosine recombinase of Shewanella oneidensis is active as an integron integrase. J. Bacteriol. 184:1811-1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gon, S., J. C. Patte, J.-P. Dos Santos, and V. Méjean. 2002. Reconstitution of the trimethylamine oxide reductase regulatory elements of Shewanella oneidensis in Escherichia coli. J. Bacteriol. 184:1262-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heidelberg, J. F., I. T. Paulsen, K. E. Nelson, E. J. Gaidos, W. C. Nelson, T. D. Read, J. A. Eisen, R. Seshadri, N. Ward, B. Methe, R. A. Clayton, T. Meyer, A. Tsapin, J. Scott, M. Beanan, L. Brinkac, S. Daugherty, R. T. DeBoy, R. J. Dodson, A. S. Durkin, D. H. Haft, J. F. Kolonay, R. Madupu, J. D. Peterson, L. A. Umayam, O. White, A. M. Wolf, J. Vamathevan, J. Weidman, M. Impraim, K. Lee, K. Berry, C. Lee, J. Mueller, H. Khouri, J. Gill, T. R. Utterback, L. A. McDonald, T. V. Feldblyum, H. O. Smith, J. C. Venter, K. H. Nealson, and C. M. Fraser. 2002. Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nat. Biotechnol. 20:1118-1123. [DOI] [PubMed] [Google Scholar]
- 10.Héritier, C., L. Poirel, D. Aubert, and P. Nordmann. 2003. Genetic and functional analysis of the chromosome-encoded carbapenem-hydrolyzing oxacillinase OXA-40 of Acinetobacter baumannii. Antimicrob. Agents Chemother. 47:268-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ledent, P., X. Raquet, B. Joris, J. Van Beeumen, and J.-M. Frère. 1993. A comparative study of class-D β-lactamases. Biochem. J. 292:555-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naas, T., and P. Nordmann. 1999. OXA-type β-lactamases. Curr. Pharm. Des. 5:865-879. [PubMed] [Google Scholar]
- 13.National Committee for Clinical Laboratory Standards. 2001. Performance standards for antimicrobial susceptibility testing. Eleventh informational supplement. M100-S11. NCCLS, Wayne, Pa.
- 14.Nordmann, P., and L. Poirel. 2002. Emerging carbapenemases in Gram-negative aerobes. Clin. Microbiol. Infect. 8:321-331. [DOI] [PubMed] [Google Scholar]
- 15.Paton, R., R. S. Miles, J. Hood, and S. G. B. Amyes. 1993. ARI-1: β-lactamase mediated imipenem resistance in Acinetobacter baumannii. Int. J. Antimicrob. Agents. 2:81-88. [DOI] [PubMed] [Google Scholar]
- 16.Poirel, L., C. Héritier, V. Tolün, and P. Nordmann. 2004. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 48:15-22. [DOI] [PMC free article] [PubMed]
- 17.Poirel, L., D. Girlich, T. Naas, and P. Nordmann. 2001. OXA-28, an extended-spectrum variant of OXA-10 β-lactamase from Pseudomonas aeruginosa and its plasmid- and integron-located gene. Antimicrob. Agents Chemother. 45:447-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poirel, L., T. Naas, M. Guibert, E. B. Chaibi, R. Labia, and P. Nordmann. 1999. Molecular and biochemical characterization of VEB-1, a novel class A extended-spectrum beta-lactamase encoded by an Escherichia coli integron gene. Antimicrob. Agents Chemother. 43:573-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poirel, L., and P. Nordmann. 2002. Acquired carbapenem-hydrolyzing β-lactamases and their genetic support. Curr. Pharm. Biotechnol. 3:117-127. [DOI] [PubMed] [Google Scholar]
- 20.Poirel, L., M. Guibert, D. Girlich, T. Naas, and P. Nordmann. 1999. Cloning, sequence analyses, expression, and distribution of ampC-ampR from Morganella morganii clinical isolates. Antimicrob. Agents Chemother. 43:769-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Venkateswaran, K., D. P. Moser, M. E. Dollhopf, D. P. Lies, D. A. Saffarini, B. J. MacGregor, D. B. Ringelberg, D. C. White, M. Nishijima, H. Sano, J. Burghardt, E. Stackebrandt, and K. H. Nealson. 1999. Polyphasic taxonomy of the genus Shewanella and description of Shewanella oneidensis sp. nov. Int. J. Syst. Bacteriol. 49:705-724. [DOI] [PubMed] [Google Scholar]