Abstract
The years 2000 through mid-2010 marked a transformational period in understanding of the biosynthesis of marine natural products. During this decade the field emerged from one largely dominated by chemical approaches to understanding biosynthetic pathways to one incorporating the full force of modern molecular biology and bioinformatics. Fusion of chemical and biological approaches yielded great advances in understanding the genetic and enzymatic basis for marine natural product biosynthesis. Progress was particularly pronounced for marine microbes, especially actinomycetes and cyanobacteria. During this single decade, both the first complete marine microbial natural product biosynthetic gene cluster sequence was released as well as the first entire genome sequence for a secondary metabolite-rich marine microbe. The decade also saw tremendous progress in recognizing the key role of marine microbial symbionts of invertebrates in natural product biosynthesis. Application of genetic and enzymatic knowledge led to genetic engineering of novel “unnatural” natural products during this time, as well as opportunities for discovery of novel natural products through genome mining. The current review highlights selected seminal studies from 2000 through to June 2010 that illustrate breakthroughs in understanding of marine natural product biosynthesis at the genetic, enzymatic, and small-molecule natural product levels. A total of 154 references are cited.
1 Introduction
Beginning with identification of the first marine natural products, chemists have been fascinated by the immense structural diversity and complexity of metabolites isolated from organisms ranging from marine plants and invertebrates to microbes. The wide spectrum of metabolites isolated from these organisms,1,2 as well as the potent biomedical activities observed for many of these marine natural products,3-5 led to interest in understanding the biochemical pathways by which these molecules are assembled in nature.
Initial studies of natural product biosynthesis for both terrestrial and marine organisms relied primarily upon isotopic labeling studies, tracking the incorporation of specific small-molecule building blocks into natural products.6 These investigations were critical in establishing chemical building block paradigms for common natural product classes including polyketides, nonribosomal peptides, terpenes, alkaloids, and others. These labeling-focused studies were followed by early studies of the enzymatic basis for biosynthesis as well as identification of the first genes encoding these biosynthetic enzymes.7-11 Such initial genetic and enzymatic studies of natural product biosynthesis focused primarily upon molecules from terrestrial microbes and plants. The chemical uniqueness and potent biological activities observed for many molecules from the sea then sparked interest in exploring the genetic and enzymatic basis for biosynthesis of marine natural products.
The last decade was a revolutionary period for the field of marine natural product biosynthesis. During this time, the field emerged from one technologically dominated by chemical labeling methods to one capitalizing on advancements in molecular biology, genetics, bioinformatics, and other biological disciplines. Increases in speed and decreases in cost of DNA sequencing,12 population of public databases with vast amounts of gene and protein information,13-15 and other advances in the biological sciences made possible great strides in understanding the biosynthesis of marine molecules during 2000 through mid-2010. As a result of such biological advances, knowledge of marine natural product biosynthesis metamorphosized during the last decade from a predominantly chemically-based perspective to now encompass the entirety of the Central Dogma, from transcription of DNA into RNA and translation into enzymes catalyzing chemical reactions essential in formation of marine natural products.
Explorations at all levels of the marine natural product “Dogma”—from genes, to enzymes, to natural products—have revealed insights into the common threads and differentiating features of biosynthetic pathways that lead to the immense structural diversity of marine natural products. This multifaceted approach has offered insights into the genetic capabilities of marine organisms to produce natural products,16-18 provided evidence for a microbial origin of several natural products originally ascribed to macroorganisms,19 afforded understanding of the role of individual enzymes in biosynthesis of natural products,20 and provided the ability to engineer genes and corresponding enzymes for production of so-called “unnatural” natural products.20 As the field of marine natural product biosynthesis further evolves in the molecular age, knowledge gained from biosynthetic studies will play increasingly prominent roles in medicine, chemical ecology, green organic synthesis, and numerous other applications.
Several reviews during the past decade offered broad coverage of advancements in understanding the biosynthesis of marine and/or terrestrial natural products for selected groups of organisms;21-23 other reviews focused on biosynthetic developments for selected classes of metabolites including polyketides,24-33 nonribosomal peptides,31,32,34-36 halogenated natural products,37 terpenoids,38,39 and others.40-42 In light of these outstanding reviews, herein we do not attempt to offer a comprehensive review of progress in the field of marine natural product biosynthesis during the past ten years. Instead, we highlight selected “firsts” during the past decade, focusing on studies of marine biosynthesis at the genetic, enzymatic, and small-molecule levels. In so doing, we aim to illustrate how each of these levels of the natural product “Dogma” are inherently intertwined and synergistic, with progress at one level stimulating understanding at other levels. Highlighted studies aim to illuminate exciting transformations in this field during the past decade and predict what lies on the horizon in this evolving area.
2 2000–2010: From the first marine natural product biosynthetic gene cluster to entire genome sequences
At the genetic level, marine natural product biosynthesis has been best explored for prokaryotic microorganisms including actinomycetes and cyanobacteria, while understanding of eukaryotic systems largely lags behind. Genetic-level understanding of marine prokaryote biosynthesis advanced rapidly relative to understanding of eukaryotic biosynthesis in part because prokaryotic genomes are often smaller in size than eukaryotic genomes, thus speeding identification and sequencing of targeted DNA. Advancement in the comprehension of prokaryotic biosynthesis was also facilitated tremendously by biosynthetic gene organization. In prokaryotes, genes encoding the biosynthesis of a natural product are typically clustered into a single region of DNA ranging from a few kilobases to 100 or more kilobases in size. This compartmentalization of biosynthetic genes not only aids identification of a complete set of genes for prokaryotic natural product biosynthesis, but also greatly facilitates heterologous expression and other molecular experiments. Among eukaryotes, genes for natural product biosynthesis are instead typically scattered across relatively large regions of the genome, making identification of a complete set of genes for biosynthesis of a targeted natural product much more difficult.
The last decade was particularly transformative for understanding the genetic basis of marine prokaryotic natural product biosynthesis. It began with emergence of complete gene sequences for the biosynthesis of several marine microbial natural products (Table 1). By the conclusion of the decade, the first multi-million base pair entire genome sequences of marine microbes were released (Section 2.3) and identification of specific biosynthetic genes from large microbial assemblages was achieved (Section 2.4). This explosion of genetic information afforded valuable insights into the metabolic capabilities of marine microbes, made possible both in vivo and in vitro explorations of natural product biosynthesis, facilitated discovery of novel natural products, and allowed engineering and improved understanding of individual genes involved in natural product biosynthesis.
Table 1.
Marine natural product gene clusters identified and characterized during the previous decade
| Year published | Molecule | Approximate gene cluster size (kb) |
Organism | Molecule class |
|---|---|---|---|---|
| 2010 | ML-449142 | 83 | Streptomyces sp. | Polyketide |
| 2010 | Rifamycin/saliniketal143 | 92 | Salinispora arenicola | Polyketide |
| 2010 | Tirandamycin144 | 56 | Streptomyces sp. | Polyketide/nonribosomal peptide |
| 2010 | TP-1161145 | 16 | Streptomyces sp. | Ribosomal peptide |
| 2009 | BE-14106146 | 85 | Streptomyces sp. | Polyketide |
| 2009 | Psymberin93 | 62 | Uncultivated prokaryotic symbiont of Psammocinia aff. bulbosa (sponge) |
Polyketide |
| 2008 | Cyclomarin/cyclomarazine147 | 47 | Salinispora arenicola | Nonribosomal peptide |
| 2008 | Napyradiomycin112 | 43 |
Streptomyces aculeolatus NRRL 18422 and CNQ525 |
Polyketide/terpenoid |
| 2007 | Bryostatin148 | 65 | Uncultivated prokaryotic symbiont of Bugula neritina (bryozoan) |
Polyketide |
| 2007 | Hectochlorin149 | 38 | Lyngbya majuscula | Polyketide/nonribosomal peptide |
| 2007 | Salinosporamide70 | 41 | Salinispora tropica | Polyketide/nonribosomal peptide |
| 2007 | Sporolide70 | 50 | Salinispora tropica | Polyketide (enediyne) |
| 2005 | Patellamide99 | 11 |
Prochloron didemni, uncultivated cyanobacterial symbiont of Lissoclinum patella (ascidian) |
Ribosomal peptide |
| 2004 | Curacin57 | 64 | Lyngbya majuscula | Polyketide/nonribosomal peptide |
| 2004 | Jamaicamide126 | 58 | Lyngbya majuscula | Polyketide/nonribosomal peptide |
| 2004 | Lyngbyatoxin150 | 11 | Lyngbya majuscula | Nonribosomal peptide/terpenoid |
| 2004 | Nodularin151 | 48 | Nodularia spumigena | Polyketide/nonribosomal peptide |
| 2004 | Onnamide/theopedrin85 | >36 | Uncultivated prokaryotic symbiont of Theonella swinhoei (sponge) |
Polyketide/nonribosomal peptide |
| 2003 | Barbamide59 | 26 | Lyngbya majuscula | Polyketide/nonribosomal peptide |
| 2002 | Eicosapentaenoic acid152 | 17 | Photobacterium profundum | Polyketide |
| 2002 | Griseorhodin153 | 34 | Streptomyces sp. | Polyketide |
| 2000 | Docosahexaenoic acid154 | >20 | Moritella marina | Polyketide |
| 2000 | Enterocin/wailupemycin | 21 | Streptomyces maritimus | Polyketide |
2.1 The enterocin and wailupemycin gene cluster marks the first completed DNA sequence for biosynthesis of a marine actinomycete natural product
The enterocins (1–3) and wailupemycins (4–10) were the first marine actinomycete natural products for which a complete gene cluster was identified, sequenced, and verified (Fig. 1).43 These molecules comprise more than half a dozen structurally-related aromatic polyketides isolated from a sediment-derived marine actinomycete identified as “Streptomyces maritimus”.44 The enterocins and wailupemycins drew the interest of researchers in part because of their unique polyketide carbon skeletons, which labeling studies during the mid-1970s revealed were derived from an uncommon benzoate starter unit along with seven malonate extender units that underwent a Favorskii-like rearrangement to yield the observed range of carbon skeletons.45 More than twenty years after labeling-based insights were obtained, the genetic basis for biosynthesis of these natural products was finally unveiled and revealed information unattainable through strictly chemical-based studies.
Fig. 1.
Enterocin and wailupemycin biosynthesis. (A) Organization of the enterocin and wailupemycin gene cluster, the first completed marine natural product biosynthetic gene cluster.43 (B) Elucidation of this gene cluster allowed in vitro and in vivo studies of the function of cluster genes and the encoded enzymes, supporting the current pathway for enterocin and wailupemycin biosynthesis.43,47,51-53
Piel et al. hypothesized that marine natural product biosynthesis followed genetic paradigms analogous to terrestrial actinomycetes and that the enterocins were derived from a type II polyketide synthase (PKS). Type II polyketide synthases are multi-enzyme complexes minimally consisting of two discreet ketosynthase units (KSα and KSβ) and an acyl carrier protein (ACP), which are employed iteratively in assembly of aromatic polyketides, as reviewed by Hertweck et al.27 Screening of a S. maritimus genomic DNA clone library for type II PKS-encoding gene sequences led to the identification and complete sequencing of the biosynthetic gene cluster for enterocins and wailupemycins (1–10, Fig. 1). The function of this gene cluster was confirmed by heterologous expression of the pathway and further explored by a wide range of gene knockouts and enzymatic studies made possible by this complete gene sequence information.46-53
Bioinformatic analysis of the ~21 kb enterocin and wailupemycin cluster (Fig. 1) revealed gene sequences encoding several proteins homologous to those with roles in terrestrial biosynthesis of aromatic polyketides via type II PKSs. Specifically, the enterocin and wailupemycin biosynthetic machinery was proposed to include proteins for polyketide core assembly, including the KSα subunit EncA, the KSβ subunit EncB, the ACP EncC, and a ketoreductase (KR), EncD.43 The proposed function of these enzymes was later confirmed by heterologous expression and mutagenesis studies, which interestingly suggested the KR reduces the enterocin polyketide chain during chain elongation in contrast to most KRs from type II PKS systems that act during the post-PKS stage of the biosynthesis.47
The gene cluster for enterocins and wailupemycins (Fig. 1) also afforded clues as to the molecular basis for the unusual Favorskii-like carbon skeleton rearrangement,43 which labeling studies had suggested was key in yielding the diverse cyclization modes observed for these compounds.45 From the gene cluster, a flavin-dependent oxygenase protein product, EncM, was identified as the prime candidate for catalyzing oxidative carbon-carbon bond rearrangements.43 Later biochemical characterization of EncM confirmed its role in the observed Favorskii-type rearrangement, providing evidence that this remarkable enzyme was responsible for not only carbon bond rearrangement, but also played key roles in two aldol condensations and reactions forming two heterocycles within this group of marine metabolites (Fig. 1).51 In total, the remarkable flavoprotein EncM was implicated in formation of at least four rings and five chiral centers within these molecules.51
Sequencing of the enterocin and wailupemycin gene cluster also revealed plausible pathways for biosynthesis of the highly unusual benzoate starter unit observed for these molecules.43 Analysis of gene sequences within the cluster most strongly suggested a plant-like benzoate biosynthetic pathway, in which benzoyl-CoA was formed from l-phenylalanine through a β-oxidative process (Fig. 1). Experimental evidence for this unprecedented bacterial pathway was then provided by a series of site-directed mutagenesis experiments.53-55
Overall, elucidation of the gene cluster for enterocin and wailupemycin biosynthesis opened the biochemical door to a treasure trove of marine natural product biosynthetic discoveries ranging from insights into oxidative carbon skeleton rearrangements to a novel pathway for bacterial biosynthesis of benzoyl-CoA that mirrors that of plants.46-53 Furthermore, as the first sequenced marine actinomycete biosynthetic gene cluster, this cluster set the stage for elucidation and evaluation of numerous other polyketide biosynthetic clusters, both terrestrial and marine.
2.2 Elucidation of the barbamide gene cluster marks the first complete marine cyanobacterial biosynthetic gene cluster
The early 2000s saw not only identification and sequencing of the first biosynthetic gene clusters from marine actinomycetes, but also publication of the first complete biosynthetic gene clusters for complex cyanobacterial natural products. The first completed freshwater and brackish cyanobacterial natural product gene cluster coded for the microcystins,56 while the first gene cluster identified from a truly marine cyanobacterium was that of barbamide (11) from Lyngbya majuscula strain 19L.57
Barbamide (11) drew the attention of biosynthetic researchers due to unique structural features including an N-methyl-dolaphenine group, a thiazole ring, both O- and N-methylation, as well as a highly unusual trichloroisovaleric acid unit. Labeling studies showing incorporation of acetate, S-adenosylmethionine, l-phenylalanine, l-leucine, and l-cysteine, as well as the overall structure of barbamide, were strongly suggestive of a mixed type I PKS and nonribosomal peptide synthase (NRPS) biosynthetic origin.58 Type I PKSs and NRPSs are responsible for assembling specific carboxylate and amino acid-based building blocks into final polyketide and nonribosomal peptides products via a pathway resembling an enzyme-driven chemical assembly line, as reviewed by Walsh.32
Type I PKS and NRPS megasynthases include multiple modules of catalytic and carrier protein domains, with each module responsible for adding and modifying a single specific chemical building block on growing natural product chains.32 The order of these modules is generally co-linear with the sequence of individual building blocks appearing in the final polyketide or polypeptide.32 For NRPSs, an amino acid starter unit is activated by an adenylation (A) domain within the first module, and subsequently loaded onto a phosphopantetheinyl prosthetic group of the peptidyl carrier protein (PCP) domain within the module. Additional amino acid residues to be added to the polypeptide chain are then loaded onto distinct PCP domains of different modules within the assembly line, and condensation (C) domains within these modules catalyze formation of an amide bond between the thioester group of a peptide chain from the previous module with the amino group of the current module, leaving the extended peptide attached to the PCP-domain of the current module.34,35 Some modules include combined condensation-cyclization (Cy) domains, which catalyze both amide bond formation and cyclization of serine, threonine, or cysteine side chains with the nitrogen atom of the amide. Modules may also include optional domains for modification of individual amino acid residues, such as methyl transferase (MT) and epimerization (E) domains. A nonribosomal peptide chain is terminated by a final thioesterase (TE) domain, which hydrolyzes the polypeptide chain from the ACP-domain and may also cyclize the final molecule.34,35
Type I PKSs act as molecular assembly lines analogous to NRPSs. The first module of a type I PKS minimally consists of an acyltransferase (AT) and acyl carrier protein (ACP), with the acyltransferase activating and transferring an acyl-CoA precursor (e.g. acetyl-CoA, malonyl-CoA) to the phosphopantetheinyl prosthetic group on the ACP.11,25 This unit is then transferred to the ketosynthase (KS) of the next module, where it reacts in a Claisen condensation with another monomeric unit bound to an ACP within this module. As a result, the polyketide chain is extended by one unit and the elongated chain is left bound to this ACP. This chain can then be passed to the KS of the next module within the assembly line, where it undergoes another round of Claisen condensation with an additional monomer.11,25 At the end of the assembly line, termination and release of the polyketide is catalyzed by a TE domain. Within individual modules, domains for modification of individual units may also be found, including ketoreductase (KR), dehydratase (DH), and enoyl reductase (ER) domains.11,25
Cloning and sequencing of the ~26 kb barbamide (11) gene cluster confirmed the mixed PKS-NRPS biosynthetic origin predicted by chemical labeling studies and resulted in a proposed route to barbamide biosynthesis, which includes both a distinct peptidyl carrier protein (PCP) encoded by barA as well as a thioesterase (TE) encoded by barC (Fig. 2).59 This separation of PCP and TE-encoding genes from the remaining PKS-NRPS encoding genes (barE, barF, and barG) deviates from the typical co-linearity of PKS-NRPS gene sequences and corresponding natural products. Another relatively uncommon feature of the barbamide gene cluster is that a PKS biosynthetic module is split across two ORFs, barE and barF (Fig. 2),59 suggesting the complexity of protein–protein interactions required for 11 biosynthesis.
Fig. 2.
Barbamide biosynthesis. (A) Organization of the L. majuscula barbamide gene cluster, the first completed natural product biosynthetic gene cluster from a marine cyanobacterium.59 (B) Proposed scheme for biosynthesis of barbamide, including domain organization of PKS/NRPS enzymes encoded by genes from (A).59,61,141 A, adenylation domain; ACP, acyl carrier protein; AT, acyltransferase domain; C, condensation domain; Cy, condensation/cyclization domain; KS, ketosynthase domain; MT, methyltransferase domain; PCP, peptidyl carrier protein; TE, thioesterase domain.
The identity of amino acid-derived substrates processed by adenylation domains within BarD, BarE, and BarG was explored through overexpression of these domains followed by application of an ATP-pyrophosphate (PPi) exchange assay,59 measuring ATP-PPi exchange activity and indicating processing of specific amino acids by adenylation domains. The predicted substrates of these domains, as illustrated within Fig. 2, also served as further confirmation of the role of the bar cluster in barbamide biosynthesis. One of the most intriguing structural features of 11 is the trichloroisovaline residue, which both the ATP-pyrophosphate exchange assay and labeling studies suggested is derived from trichloroleucine.58,59 Later enzyme-focused studies further illuminated the intriguing mechanism of 11 chlorination (Fig. 2),60,61 discussed in Section 3.2.
2.3 Whole-genome sequencing advances understanding of the metabolic potential of marine microorganisms
At the start of the decade, studies of the genetic basis for marine natural product biosynthesis focused on identification and sequencing of individual gene clusters, as highlighted for the enterocins/wailupemycins (1–10) and barbamide (11) in Sections 2.1 and 2.2, respectively. While these and analogous studies provided valuable insights into the biosynthesis of selected natural products, they offered little indication of the total quantity and diversity of biosynthetic pathways from marine microbes. Thus, the true genetic capabilities of marine microbes remained largely a mystery.
As the decade progressed, improvements in DNA sequencing technologies resulted in the emergence of genome sequencing as a viable means to explore the biosynthetic potential of marine microbes. Relative to sequencing of individual biosynthetic pathways, genome sequencing carries a variety of advantages. By sequencing entire genomes, the entirety of an organism’s natural product potential comes to light. Not only does this expedite identification of gene clusters for all known molecules from an organism, it also illuminates biosynthetic gene clusters for natural products not yet identified from the organism.62 From this genetic level information, scientists may then attempt to tap into the total potential of an organism to produce novel chemistry via genome mining,63,64 highlighted in Section 4.2.
2.3.1 The first marine actinomycete genome sequence reveals unprecedented genetic devotion to natural product biosynthesis
Terrestrial actinomycetes are unmatched in their ability to produce clinically useful antibiotics, the majority of which come from members of the genus Streptomyces.65 Among members of this genus, entire genome sequences were first released for S. coelicolor strain A3(2) and S. avermitilis, and revealed that these soil-dwelling microbes harbored far more biosynthetic gene clusters than recognized from their laboratory-based chemical profiles.66,67 This raised the intriguing possibility that marine actinomycete genera may also harbor a rich variety of unrealized biosynthetic capabilities. A recent review summarizes progress on whole-genome sequencing of both terrestrial and marine actinomycetes.62
Among marine actinomycetes, members of the chemically-rich, obligate marine genus Salinispora provided intriguing model organisms for whole-genome sequencing. Three distinct Salinispora species have been proposed: S. tropica, S. aerinicola, and “S. pacifica”. 68 S. tropica, producer of intriguing natural products that include the clinically promising salinosporamide A (12),69 was the first target for genome sequencing. Of the 5,183,331 bp circular genome of S. tropica, nearly 10% of genetic material was predicted through bioinformatics to encode natural product biosynthesis, an even greater proportion than for sequenced terrestrial actinomycetes.70 This included a total of at least 17 putative natural product biosynthetic gene clusters. These clusters were strikingly diverse, with predicted involvement in biosynthesis of polyketides, nonribosomal peptides, melanins, terpenoids, and aminocylitols.70 The majority of these gene clusters were predicted to encode metabolites not previously reported from S. tropica, suggesting the immense untapped metabolic potential of this marine actinomycete. Near the end of the 2000s, these cryptic natural products began to be searched for through genome mining,63 a strategy for identifying compounds encoded by novel gene clusters and highlighted in Section 4.2.
In addition to shedding light on the unrealized metabolic potential of S. tropica, genome sequencing also facilitated identification of gene clusters for biosynthesis of molecules known from S. tropica, including the salinosporamides (12), sporolides (13,14), and lymphostin (15).70 Subsequent genetic and enzymatic studies of the salinosporamide pathway provided further insight into salinosporamide assembly, including elucidation of a new route to chlorination of this γ-lactam-β-lactone proteasome inhibitor, highlighted in Section 3.1, as well as the ability to access novel salinosporamides through genetic engineering.71-75 Likewise, exploration of the sporolide pathway revealed insights into the enediyne origin of this metabolite as well as information on the biosynthesis of the cyclohexenone building block of this molecule.76

The 5,786,361 bp circular genome of S. arenicola was also recently sequenced and found to harbor an impressive range of biosynthetic capabilities.77 Comparison of genomes between S. tropica and S. arenicola suggested that Salinispora species are distinguishable via genomic islands containing species-specific natural product pathways as well as the bulk of other species-specific gene sequences.77 Association of natural product biosynthetic genes with regions unique to individual species suggests that acquisition of natural product pathways through horizontal gene transfer may represent an important, yet not widely recognized force in bacterial diversification.77 As sequencing technologies continue to improve and evolve, comparative genomics will surely continue to grow as an invaluable tool for understanding bacterial evolution at the molecular level.
2.3.2 Genome sequencing among marine cyanobacteria: current status and future possibilities
In addition to marine actinomycetes, marine cyanobacteria have also drawn recent interest in the genome sequencing arena. Draft or finished genome sequences have been released for more than 35 strains of six different species of marine cyanobacteria thus far.21 However, these studies have revealed only a handful of gene clusters for polyketide and nonribosomal peptide biosynthesis, most of which come from Crocosphaera watsonii WH8501, although to date no natural products have been reported from this cyanobacterium.21 Future targeting of whole-genome sequencing toward cyanobacterial genera already known as rich sources of natural products (e.g. Lyngbya spp.) may help further understanding of the true genetic capabilities of chemically talented cyanobacteria.
2.4 Genetic approaches advance understanding of the role of microbial symbionts in marine natural product biosynthesis
Each of the studies discussed thus far illustrated advances in understanding the biosynthetic capabilities of microbes cultivable under standard laboratory conditions. However, the vast majority of microbes are not cultivable under typical laboratory conditions. Thus, there is a potential abundance of microbial biosynthetic capabilities inaccessible through cultivation-dependent natural product isolation approaches. Many structurally unique marine natural products have been isolated from invertebrates, including sponges, bryozoans, and tunicates.2,78 These invertebrate-derived compounds include numerous polyketides, nonribosomal peptides, and hybrid molecules bearing striking structural similarities to bacterial metabolites.2,78 Such similarities provided circumstantial evidence that these natural products were derived not from invertebrates themselves but instead from microbial symbionts. Using classical methods including cultivation, immunolocalization, in situ hybridization, and cell separation, researchers attempted for decades to establish the true origin of many marine invertebrate-associated natural products.78 Unfortunately, these attempts often failed to provide conclusive evidence for the source of these metabolites. During the last decade, however, metagenomic-based approaches provided convincing evidence of a microbial symbiont origin for several metabolites originally ascribed to invertebrate hosts.19,79 To illustrate the power of genetic tools in identifying the critical role of uncultured microbes in marine natural product biosynthesis, we focus on the first biosynthetic gene clusters identified from sponge and tunicate symbionts. Multiple recent reviews also summarize selected areas of progress in this field.19,80,81
2.4.1 Elucidation of the first biosynthetic gene cluster from an uncultured sponge symbiont
More natural products with therapeutic potential have been isolated from marine sponges than from any other invertebrate source,82,83 making them a particularly intriguing system for exploration of symbiont biosynthetic hypotheses. However, sponges also represent an incredibly complex system for study, as they are notorious for the large amount of microbial biodiversity harbored within their tissues.82,84 Hence, a single sponge sample contains an enormous diversity of genetic material, making localization of specific biosynthetic gene sequences analogous to searching for a needle in a haystack.
Piel et al. provided some of the first genetic evidence supporting a microbial origin of structurally intriguing natural products from uncultured sponge-associated microbes.85 The onnamides (16) and theopederins (17) are molecules of mixed polyketide and nonribosomal peptide biosynthetic origin originally isolated from the marine sponge Theonella swinhoei,86,87 and bear strong similarity to pederin (18) isolated from the beetle Paederus fuscipes.88 Structural features and chemical labeling evidence suggested that all of these molecules were assembled in part by a type I polyketide synthase.89
Hypothesizing a type I PKS origin for the beetle-associated pederin and sponge-associated onnamides (16) and theopederins (17), Piel et al. explored metagenomic DNA from these complex biotic mixtures for genes with predicted involvement in polyketide biosynthesis.85,90 Large clone libraries of 60,000+ members were constructed with fragments of metagenomic DNA and systematically screened for ketosynthase (KS) sequences of interest.85,90 Sequencing of clones positive for targeted KS sequences revealed the onnamide/theopederin biosynthetic gene cluster (Fig. 3), which showed substantial homology to the cluster proposed for pederin biosynthesis.85,90 For the majority of both the pederin (18) and onnamide/theopederin (16, 17) clusters, bioinformatic analysis of the encoded type I PKS proteins strongly predicted observed polyketide structures.85,90 Both of these gene clusters were unique from previously elucidated clusters in that the encoded type I PKS megasynthases lacked sequences for acyltransferase (AT) domains. Instead, AT-encoding genes were found separately within these gene clusters.85,90 This paradigm has since been reported for several other type I PKS systems.29,91
Fig. 3.
Onnamide and theopederin gene cluster, revealed through metagenomic analysis of the sponge T. swinhoei.85 This gene cluster exhibits significant homology to the pederin and psymberin gene clusters, also identified through metagenomic approaches.90,93
Unfortunately, without the ability to culture proposed microbial sources of these natural products, it remains impossible to directly confirm a microbial origin and conclusively link these metabolites to their candidate gene clusters. However, all pieces of available molecular evidence point to microbial biosynthesis. First, stark similarities observed for the pederin (18) and onnamide/theopederin (16, 17) gene clusters, despite distinct terrestrial and marine origins of these natural products, are strongly suggestive of a microbial origin.85,90 Also, a complete pathway for psymberin (19), a metabolite with structural analogies to the theopederins, onnamides, and pederins,92 was recently identified from an uncultured microbial symbiont of the sponge Psammocinia aff. bulbosa and found to include genes with significant homology to theopederin, onnamide, and pederin genes,93 further suggesting the importance of prokaryotic symbionts in biosynthesis of these polyketides. The distinct, largely clustered organization of the biosynthetic genes from all of these clusters also implicates a prokaryotic origin,85,90 as this type of genetic organization is not typical of eukaryotic systems. For beetle-associated pederin (18), 16S rRNA analyses also correlated the presence of an abundant Pseudomonas aeruginosa strain with production of pederins.90 A Pseudomonas sp. may also be responsible for production of the onnamides (16) and theopederins (17) in the sponge T. swinhoei; however, the presence of any bacterial species and metabolite production have not yet been correlated, perhaps owing to the considerable complexity of the microbial community associated with these sponges. Finally, recent heterologous expression studies demonstrated that individual proteins encoded by these gene clusters were capable of acting upon pederin, onnamide, and theopederin derivatives;94 these reconstitution experiments provided biochemical evidence suggesting the involvement of these proteins in biosynthesis of these molecules. These experiments also yielded successful diversification of polyketides originating from uncultivated marine microbes, an accomplishment with implications for development of complex symbiotic marine metabolites as drugs.
2.4.2 Molecular evidence supports the role of symbiotic cyanobacteria in marine tunicate natural product chemistry
Like sponges, tunicates are a rich source of marine natural products for which the true metabolic origin has been questioned.95 The patellamides (20, 21) represent one such group of tunicate natural products whose origin has been particularly widely contested since the discovery of the first examples of this group of metabolites from the Didemnidae family ascidian Lissoclinum patella in the early 1980s.96 The patellamides, pseudosymmetricial cyclic peptide dimers, are comprised of modified d-amino acids and typified by an unusual fused oxazoline-thiazole unit.96
Patellamides (20, 21) and related metabolites have been isolated from multiple species of Didemnidae family ascidians.95-98 Interestingly, these ascidians were observed to harbor Prochloron spp. cyanobacterial symbionts, often in quantities large enough to render them visible to the naked eye and comprising the bulk of microbial biomass of these ascidians.99 This circumstantial evidence led investigators to long suspect Prochloron spp. as the true source of patellamides. Despite more than thirty years of efforts, however, Prochloron spp. remained completely resistant to laboratory cultivation.99 Thus, using traditional microbiological approaches, it was not possible to directly explore the hypothesis that Prochloron spp. were the true source of patellamides. In the pre-molecular age, researchers thus turned to cell separation and dissection methods to assess the metabolic origin of these molecules. However, results from these studies were inconsistent, with some studies localizing patellamides to the cyanobacteria,95 and others finding these metabolites throughout symbiont and host.100 With molecular age tools in hand, Schmidt et al. tackled the patellamide origin challenge by employing genetic sequencing and heterologous expression, providing experimental evidence for the true producer of these metabolites.99
Schmidt et al. hypothesized that if the patellamides (20, 21) were cyanobacterial natural products, then biosynthesis would likely proceed through ribosomal or nonribosomal peptide pathways analogous to those previously reported for bacterial systems. The nonribosomal pathway hypothesis was explored first, and screening of a library of Prochloron spp. DNA for sequences associated with nonribosomal peptide biosynthesis revealed a nonribosomal peptide gene cluster within some Prochloron spp.101 However, the presence of this gene cluster did not correlate with patellamide production, suggesting these genes were not responsible for patellamide production. Furthermore, bioinformatic analyses of this nonribosomal peptide cluster predicted molecular features inconsistent with those observed for the patellamides.101
Thus, the possibility of a ribosomal biosynthetic origin for the patellamides (20, 21) was next investigated in Prochloron spp. by employing the emerging tool of genome sequencing.99 The cyanobacterium P. didemni, harvested from samples of the ascidian L. patella, was genome-sequenced and the draft sequence scanned for peptide-encoding sequences predicted to yield the patellamide core. A single coding sequence was identified within the draft genome of P. didemni that contained the sequence required for assembly of the amino acids found in 20 and 21. This putative coding sequence for the patellamide core was flanked by other genes predicted to play roles in processing the linear amino acid chain into a mature patellamide molecule (Fig. 4).99 The organization of these patellamide genes was similar to that observed for ribosomally encoded natural products from other bacterial species, further suggesting a role of the P. didemni sequence in patellamide biosynthesis.
Fig. 4.

The patellamide gene cluster from P. didemni.99 The substantial structural diversity observed in the patellamide family of natural products originates primarily from mutation within the patE gene.104
To confirm this biosynthetic origin of the patellamides (20, 21), the putative ribosomal pathway was expressed in an Escherichia coli host and heterologous patellamide biosynthesis confirmed by mass spectrometry.99 Prochloron spp. were independently demonstrated as the true producers of patellamides by Long et al., who achieved production of patellamides by expressing randomly cloned DNA from Prochloron sp. in E. coli bacterial artificial chromosomes.102 With Prochloron spp. shown by multiple studies as the true source of patellamides, these cyanobacterial metabolites marked the first class of marine natural products whose biosynthetic origin was conclusively linked to a microbial symbiont without traditional cultivation-based approaches. This seminal discovery also brought over twenty years of debate over the true biosynthetic origin of these fascinating molecules to an end and brought to light the importance of ribosomal pathways for natural product biosynthesis in cyanobacteria. Ribosomally-encoded natural product biosynthesis in relation to nonribosomal peptide biosynthesis was recently reviewed by McIntosh et al.41
With the importance of microbial symbionts in marine natural product biosynthesis firmly established, essential groundwork was laid for understanding the evolution and ecological function of symbiont biosynthetic pathways.103 Donia et al. embarked on such an exploration, studying how the patellamide pathway evolved to yield >50 known structural analogs.104 Patellamide-encoding gene sequences were evaluated for Prochloron spp. from >40 different ascidian samples, with patellamide structural diversity found to result exclusively from modifications to short regions of DNA within the patE gene (Fig. 4).104 These highly mutated sequences were those responsible for encoding the peptide precursor to mature patellamide, with point mutations within this sequence responsible for changes to the final patellamide structure. Outside of peptide-encoding region, patE genes were nearly identical across Prochloron spp. samples bio-synthesizing different patellamide analogs. Additionally, the remainder of this gene cluster was highly conserved between these Prochloron spp. samples.104 Thus, the diverse range of observed patellamide structures resulted exclusively from mutation of short, targeted DNA sequences within an otherwise highly conserved gene cluster. The diverse range of patellamide structures resulting from modification to a very targeted region of this pathway suggested that patellamide diversification carries a significant, but as yet unknown ecological advantage.104 With a complete set of chemical and genetic tools now in hand, the patellamide system represents a particularly promising target for further exploration of the ecological roles of symbiont natural products.
3 Biosynthetic breakthroughs at the enzymatic level
As the above seminal works illustrate, genetic level understanding of marine natural product biosynthesis improved tremendously during the first decade of this century. These advancements illuminated the molecular basis for marine natural product biosynthesis and identified novel enzymatic reactions involved in their construction and diversification. In continuing to follow the “Dogma” of natural product biosynthesis, we next explore in more detail some biosynthetic “firsts” at the enzymatic level. While there are many more examples that could be highlighted, we focus on two enzymatic processes, halogenation and polyketide assembly, in which marine systems greatly contributed to our mechanistic understanding of natural product biosynthesis.
3.1 Enzymatic biohalogenation strategies first deduced in marine organisms
Over 4500 organohalogens have been naturally described with the majority isolated from marine organisms.105 Nature has evolved several enzymatic strategies for the incorporation of halogen atoms into organic substrates, and the past decade has witnessed significant advances in our mechanistic understanding of how these biochemical reactions unfold.106-108 Marine systems aided this collective understanding of biohalogenation, and seminal contributions from the marine field are highlighted in this section.
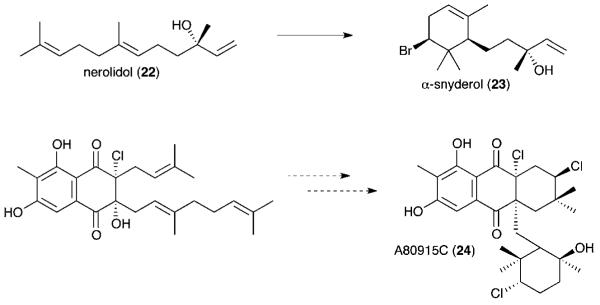
Halogenation is common amongst marine algal natural products and is largely thought to derive primarily from vanadium-dependent haloperoxidases (V-HPO).109 Structural studies of marine algal V-dependent bromoperoxidase (V-BPO) and terrestrial fungal V-dependent chloroperoxidase (V-CPO) in the late 1990s and early 2000s supported the mechanism of hydrogen peroxide activation of the conserved vanadate metal center followed by halide binding to generate a halonium intermediate whose structure still remains unknown.110 Simple to complex organohalogen structures are consequently formed by the interception of the hypohalite species with electron-rich organic substrates such as phenols or olefins. Seminal work by Butler and coworkers with Corallina officinalis V-BPO on the in vitro reconstitution of snyderol (23) biosynthesis from the sesquiterpene nerolidol (22) revealed the regio and stereoselectivity of this class of red algal halogenating enzymes that was previously thought to lack such specificity traits (Fig. 5).111 Recently, three marine streptomycete genes with homology to V-CPOs were identified in the napyradiomycin biosynthetic gene cluster whose products are proposed to facilitate the oxidative cyclization of two terpenoid side chains attached to the naphthoquinone core of A80915C (24).112 Not only is this the first such example of bacterial V-HPOs, but also the first in which V-HPOs were characterized in a dedicated natural product biosynthesis gene cluster.
Fig. 5.
Proposed V-dependent haloperoxidase-mediated biosynthesis of the brominated α-snyderol (23) and the chlorinated A80915C (24).
Examination of the marine actinomycete S. tropica recently uncovered a novel route to biochlorination orthogonal to all known chlorinating enzymes. In the context of salinosporamide A (12) biosynthesis, the SAM-dependent chlorinase SalL utilizes nucleophilic chloride to displace methionine to generate 5′-chloro-5′-deoxyadenosine (Fig. 6).113 SalL is related to a fluorinase from Streptomyces cattleya,114 which instead utilizes fluoride, and their high-resolution crystal structures revealed the molecular mechanisms behind halide desolvation and nucleophilic displacement.115 The SalL enzymatic product 5′-chloro-5′-deoxyadenosine is then converted in a seven-step biosynthetic pathway to chloroethylmalonyl-CoA,71,116 a novel halogenated PKS extender unit unique to salinosporamide A (12). This discovery allowed for the engineered biosynthesis of a focused library of salinosporamide derivatives, as discussed in Section 4.1.
Fig. 6.
Biosynthesis of the chlorinated proteasome inhibitor salinosporamide A (12) involves the SAM-dependent chlorinase SalL.
Halogenation of unactivated carbon centers requires a distinct enzymatic strategy involving a radical-type mechanism employing α-ketoglutarate dependent non-heme Fe(ii) halogenases.21 Sequence analysis of numerous L. majuscula gene clusters associated with the biosynthesis of halogenated natural products suggested that marine cyanobacteria routinely employ this halogenation strategy. The mechanistic and structural basis of non-heme iron halogenases was first elucidated in Pseudomonas spp. associated with the biosynthesis of the chlorothreonine residue of syringomycin E117,118 and the cyclopropane ring of coronamic acid.119 The distinct trichloromethyl group of barbamide (11) was similarly shown to be constructed by two non-heme iron halogenases, BarB1 and BarB2, that function sequentially on a PCP-bound leucine substrate prior to its incorporation into the natural product.60,61 A novel departure was characterized in the mixed PKS/NRPS-catalyzed pathways to curacin A (25) and jamaicamide (26) in which the associated non-heme iron halogenases were not discrete enzymes, as previously observed, but rather imbedded domains (Hal) in multi-functional synthases that give rise to the cyclopropane ring in curacin A via a chlorinated intermediate and the vinyl chloride residue in the jamaicamides (Fig. 7).120 Recent crystal structure analysis of the excised curacin A halogenase revealed a conformational switch triggered by the binding of α-ketoglutarate and chloride and their coordination to iron to support substrate recognition of the (closed) holoenzyme.121
Fig. 7.
Proposed biosynthesis of the L. majuscula metabolites curacin A (25) and jamaicamide (26) proceeds via a parallel β-branch pathway that deviates with the decarboxylase ECH2.
3.2 Fascinating assembly line biosynthetic transformations catalyzed by marine cyanobacteria
In addition to rare halogenase domains associated with L. majuscula modular PKS/NRPS assembly line systems, two additional trans operating biosynthetic processes are noteworthy for their mechanistic details first elucidated for marine cyanobacterial enzymes. These include the newly-described chain initiation process catalyzed by the bifunctional GCN5-related N-acetyltransferase (GNAT) domain of CurA122 and elongating processes involving polyketide chain β-branching to diverse structural variants as catalyzed by HMG-CoA synthase (HCS)-like gene cassettes.120
The loading module of the curacin A (25) PKS contains an atypical arrangement of three domains consisting of an adapter domain (AD), GNAT and ACP122 that are also found in other PKSs such as the sponge symbiont theopederin synthase. Biochemical analysis of the recombinant CurA AD-GNAT-ACP tridomain along with analysis of crystal structures of the excised GNAT domain in ligand-free and malonyl-CoA-bound forms revealed an unprecedented mechanism for the CurA GNAT scaffold.122 The bifunctional GNAT first catalyzes the decarboxylation of malonyl-CoA to acetyl-CoA and then directs, with the assistance of the N-terminal AD, the S-acetyl transfer of acetyl-CoA to the adjacent ACP domain (Fig. 8). Distinct substrate binding tunnels for malonyl-CoA and holo-ACP were clearly viewed in the high-resolution crystal structures and helped establish a new function for the GNAT protein superfamily.
Fig. 8.

Proposed chain initiation of curacin A (25) PKS catalyzed by the AD-GNAT-ACP tridomain.
Another common feature of marine polyketides, particularly those produced by marine cyanobacterial cis-AT PKSs and marine invertebrate symbiont trans-AT PKSs, involves branching at the β position relative to the respective β-ketoacylthioester PKS intermediates.123 The first β-branch to be mechanistically elucidated at the biochemical level involved the pksX product bacillaene from Bacillus subtilus,124,125 revealing five core proteins for β-branch incorporation, namely an ACP, a KS homolog, a HMG-CoA synthase (HCS) homolog, and two enoyl-CoA hydratase (ECH) homologs that function as a dehydratase and a decarboxylase. Numerous natural deviations are common that account for the wide range of observed chemistry in marine metabolites,123 including the cyclopropane ring of curacin A (25),57 the vinyl chloride residue of the jamaicamides (26),126 the exomethylene of the pederin family,85 and the β-methoxylacylidene groups of the bryostatins127 (Fig. 7). In the case of 25 and 26 biosynthesis, the non-heme iron halogenases (Hal) discussed in Section 3.1 are novel components encoded within the HCS cassettes that similarly function via parallel paths to give rise to the distinctive β-branching of these cyanobacterial products. Biochemical analysis of the Cur and Jam ECH2 decarboxylases provided divergent products, an α,β-enoyl thioester intermediate in the Cur pathway and a β,γ-enoyl thioester product in the Jam pathway.120,128 Only the Cur α,β-enoyl thioester was a substrate of the Cur enoyl reductase (ER) domain that resulted in the unprecedented cyclopropanation reaction of the chlorinated substrate to give the distinctive cyclopropyl group of curacin A. This subtle evolutionary diversification in the Cur and Jam pathways captured in this study at the biochemical level is an elegant example of functional evolution in secondary metabolism.
4 From genes and enzymes to small molecules: employing molecular age knowledge toward natural product manipulation and discovery
Within the natural product “Dogma”, the past decade yielded a wealth of information at both the genetic and enzymatic levels. These genetic and enzymatic breakthroughs translated into application of this molecular knowledge toward discovery of new marine natural products. With identification of gene clusters for several natural products (Sections 2.1–2.3) came the opportunity to manipulate individual biosynthetic genes for purposes of better understanding the function of these genes and for producing modified “unnatural” natural products. During the period 2000–2010, this approach emerged as a method of promise for accessing libraries of structurally related marine natural products, offering an alternative to development of sometimes complex and/or costly organic synthesis schemes for generation of natural product analogs. In addition to genetic engineering-based approaches providing access to new derivatives of known compounds, genome sequencing and the resulting emergence of novel biosynthetic pathways encoding unknown natural products (Section 2.3) opened an avenue to access novel natural products through genome mining. Together, genetic engineering and genome mining represent two prominent ways in which molecular age knowledge was translated into novel marine natural products over the last ten years.
4.1 Genetic engineering affords new molecules from old pathways
Just as the enterocin and wailupemycin gene cluster was the first fully sequenced marine biosynthetic gene cluster (Section 2.1, Fig. 1),43 the first examples of genetically engineered marine products resulted from engineering of this gene cluster in the marine actinomycete “S. maritimus”.49 The enterocins (1–3) and wailupemycins (4–10), biosynthesized by a type II iterative PKS, drew interest in part because of the incorporation of an unprecedented benzoyl CoA-derived starter unit.53,54,129 Xiang and Moore first observed that inactivation of encP, a phenylalanine ammonia-lyase encoding gene implicated in starter unit biosynthesis, resulted in abolition of 1–10 biosynthesis.54 However, biosynthesis could be restored by supplementing the ΔencP gene deletion mutant with cinnamic or benzoic acid starter units,54 suggesting this knockout mutant might provide the opportunity for mutasynthesis of enterocin and wailupemycin analogs with altered starter units. Thus, a series of aryl acids and cinnamates analogous to benzoic acid were administered to ΔencP mutant S. maritimus cultures. While the majority of evaluated substrate mimics did not result in formation of enterocin and wailupemycin analogs, p-fluorobenzoic acid, 2- and 3-thiophenecarboxylic acids, and cyclohex-1-enecarboxylic acid were successfully incorporated into mutasynthetic products.49
Transitioning from an in vivo approach to an in vitro approach, the complete enterocin pathway was reconstituted in vitro with purified recombinant enzymes that facilitated the successful chemoenzymatic of eighteen additional novel wailupemycin and enterocin analogs differing in the starter unit.46,48 The in vitro system afforded much broader accommodation of benzoic acid analogs as starter units than the in vivo system, suggesting the presence of additional mechanisms for substrate “gate-keeping” in vivo.48 Successful mutasynthesis of novel analogs within the enterocin and wailupemycin family of natural products provided the first example of both in vivo and in vitro mutasynthesis of a marine actinomycete metabolite.48,49 Mutasynthesis of enterocins and wailupemycins also represented the first in vivo example with a type II PKS from any terrestrial or marine microbe.49 Following successful mutasynthesis of enterocin and wailupemycin natural products, in vivo mutasynthetic approaches were successfully applied with other marine actinomycete natural products including the salinosporamides (12) from S. tropica and the cyclomarins from S. arenicola.72,73,75,130,131
Together, these studies illustrate the potential of both in vivo and in vitro metabolic engineering in the generation of novel structural analogs with utility both in understanding the function of individual components of biosynthetic pathways as well as providing derivatives for better understanding of the relationship between natural product structure and biological activity. Continued efforts toward engineering of novel natural products will further develop capabilities for production of targeted “unnatural” natural products during the next decade.
4.2 Genome mining: applying information from genome sequencing in marine natural product discovery
The first released genome sequences for marine microbes (Section 2.3) revealed that some of these organisms harbor genes for biosynthesis of far more natural products than previously recognized through cultivation-based studies. These orphan pathways, biosynthetic pathways for which corresponding natural products are unknown, drew interest of researchers for many reasons. From ecological and evolutionary perspectives, researchers were intrigued by selective pressures favoring evolution of large numbers of biosynthetic pathways, many of which appeared either inactive or expressed at low levels under laboratory conditions. From an applied perspective, discovery of orphan pathways among sequenced microbes brought forth the intriguing possibility that these pathways might be accessed for discovery of structurally unique, biomedically useful compounds.18 Considering the growing need for novel classes of antibiotics and other drugs, the potential of orphan pathways as an avenue for drug discovery and improved basic biosynthetic understanding drew considerable interest during the later portion of the last decade.
Genome mining, exploiting information from biosynthetic gene clusters toward discovery of novel natural products, was first accomplished with terrestrial microbes.18,63,64 The terrestrial actinomycete S. coelicolor A3(2) became one of the first targets of genome mining, drawing attention when genome sequencing suggested at least 20 biosynthetic gene clusters, the majority of which could not be linked to known compounds.66 Applying a full arsenal of genetic, enzymatic, bioinformatic, and chemical approaches, researchers were able to successfully “mine” for a variety of novel compounds from this and other sequenced terrestrial actinomycetes, as detailed in several excellent reviews.18,63,64 Near the middle of the last decade, scientists began to explore the metabolic potential of genome-sequenced marine microbes through genome mining.
4.2.1 Unlocking new chemistry from marine actinomycetes through genome mining
The first completed marine actinomycete genome sequence was for S. tropica (Section 2.2.1),70 and sequencing of this organism opened the door for the first success in marine actinomycete genome mining. While sequencing of the S. tropica genome revealed at least 17 biosynthetic gene clusters, corresponding natural products were known for only three of these gene clusters,70 suggesting the substantial untapped metabolic potential of this marine actinomycete. Initial genome mining efforts in S. tropica centered on the 80 kb slm secondary metabolic gene cluster, encoding a 10-module type I PKS as well as other accessory enzymes. Bioinformatic analyses of this highly repetitive PKS suggested the slm cluster was responsible for production of a polyene macrolactam, and characteristic polyene UV chromophores were recognized among chemical extracts from S. tropica cultures. Isolation and identification of molecules with this unique UV profile revealed a series of novel polyene macrolactam natural products, named salinilactams.70 Complete structure elucidation of the first representative of this class, salinilactam A (27), proved challenging by traditional analytical methods (i.e. NMR spectroscopy, MS) due to significant overlap of NMR signals from the many conjugated olefinic protons. In the end, final structure assembly relied strongly on bioinformatic analyses of type I PKS architecture, which predicted specifics of the repetitive polyene skeletal structure. Further, just as gene cluster information was critical in assigning the complete structure of salinilactam A, the structure of 27 was also important in completion of the S. tropica genome sequence. The slm gene cluster included highly repetitive DNA sequences within the type I PKS, making it difficult to complete final assembly of the genome in this region of the circular chromosome. However, the molecular formula of 27 suggested the number of type I PKS modules required for 27 assembly and thereby facilitated assembly of this repetitive region of the S. tropica genome sequence. In this way, genomic information, chemical methods, and bioinformatics worked synergistically toward both elucidation of the salinilactam structure and finishing of the S. tropica genome. Further genome mining with the completed genome sequence of S. tropica as well as genome sequences of several S. aerinicola and “S. pacifica” strains is underway, and will likely yield additional novel chemistry from this biosynthetically rich genus of marine actinomycetes.

4.2.2 Genome mining for novel natural products from marine cyanobacteria
Like marine actinomycete genome mining, cyanobacterial genome mining also represents an emerging strategy for discovery of novel natural products and understanding the genetic basis for biosynthesis. Kalaitzis et al. recently reviewed progress in cyanobacterial genome mining, so we will highlight only one seminal study.17
The first example of successful genome mining among marine cyanobacteria yielded trichamide (28), a cyclic peptide from Trichodesmium erythraeum.132 Following discovery of the ribosomally-encoded pathway for patellamide (20, 21) production from uncultivated Prochloron didemni (Section 2.4.2),99 Sudek et al. searched the GenBank database for genes homologous to those of the patellamide pathway.132 Within the draft genome of T. erythraeum, they located genes with striking homology to the patellamide genes, including a precursor peptide gene, a heterocylization gene, and protease genes. From the precursor peptide gene sequence, the linear sequence of the mature peptide was predicted and cultures of T. erythraeum explored for the presence of such a metabolite. Employing Fourier transform mass spectrometry, the bioinformatics-predicted structure of 28 was identified. Given the striking similarities between the trichamide and patellamide gene clusters,132 trichamide biosynthesis was proposed to closely parallel the patellamide pathway discussed in Section 2.4.2.99

Genome mining represents a relatively recent approach to discovery of marine natural products, and remains largely in its infancy. However, the abundance of orphan pathways observed so far from both draft and complete marine microbial genome sequences suggests that genome mining may hold great potential for unlocking novel chemistry from the sea. One current stumbling block for genome mining appears to be low yields of natural products from many orphan pathways. Thus, for the biosynthetic potential of these organisms to be realized, strategies for improving titers of metabolites produced by these pathways must be developed or analytical methods with lower limits of metabolite detection applied.
5 The next decade: diving deeper in a sea of biosynthesis
As illustrated by the highlighted seminal studies, the years 2000 through mid-2010 were an extremely fruitful period for understanding biosynthesis of marine natural products. The fusion of chemical and biological approaches in addressing marine biosynthetic questions during the last decade was critical toward this immense progress. Within a single decade, the field evolved from sequencing and analysis of individual biosynthetic gene clusters to the sequencing of entire genomes and recognition of the impressive number of biosynthetic pathways harbored by some marine prokaryotes. During the decade, molecular era advances also finally allowed uncultivated marine microbial symbionts to receive recognition as important natural products chemists. Biosynthetic insights at the genetic and enzymatic levels also paved the way for genetic engineering of novel “unnatural” natural products as well as the discovery of novel natural products predicted by genome sequences. Despite remarkable progress in the field during the past decade, there remains tremendous room for advancement in coming years. The tools, techniques, and discoveries of the past decade have set the stage for breakthroughs that will influence far-reaching fields including medicine, enzymology, synthetic organic chemistry, ecology, and others.
One area of potential advancement is in better understanding mechanisms regulating marine natural product biosynthesis. The expression of genes involved in natural product biosynthesis is controlled by complex mechanisms that are not yet well understood,133 although regulatory genes are a common element amongst the majority of characterized marine natural product biosynthetic gene clusters. These regulatory genes are thought to be largely pathway specific, governing expression of genes and production of natural products within a single pathway, and studies among terrestrial actinomycetes have demonstrated roles for conserved families of regulatory proteins in regulation of individual biosynthetic pathways.134 In contrast to local regulatory genes found within and acting primarily upon single gene clusters, other regulatory genes have been proposed as global regulators acting upon many biosynthetic pathways.134 At present, however, much remains to be known about both local and global regulation of natural product biosynthesis, especially among marine microorganisms. Improved understanding of the function of individual marine biosynthetic regulatory elements, as well as systematic ways to manipulate these elements for control of natural product production, hold immense potential for discovery and practical application of marine natural products. It is thought one major reason that not all natural products predicted by genome sequencing have been observed is because some pathways are not adequately expressed under laboratory conditions. Through improved understanding of biosynthetic regulatory mechanisms, it may someday be possible to systematically turn on or upregulate expression of biosynthetic genes, a success that would certainly facilitate efforts for discovery of novel natural products predicted by genome sequencing, as well as solve supply limitations for biomedically-promising, low-yielding marine microbial natural products. In terrestrial actinomycetes, dramatic enhancements in natural product titers have been observed through manipulation of local regulatory pathways,135,136 and such approaches may be plausible for marine actinomycetes as well. Realizing the full biosynthetic potential of marine microbes during this decade will necessitate efforts to improve understanding of mechanisms regulating expression of biosynthetic genes as well as the development of reliable strategies for upregulating expression to stimulate biosynthesis.
Improved methods for effective heterologous expression of large biosynthetic pathways represents another area of promise for the field during the next decade. With the important role of uncultivated marine microbial symbionts in production of biomedically interesting, structurally unprecedented natural products now established (Section 2.4),19 effective expression of these and other entire pathways in heterologous hosts holds much potential for solving issues of natural product supply limitations that have historically posed a major roadblock to development of such metabolites as human therapeutics. Several studies have already shown promise for production of actinomycete natural products via heterologous expression, with one recent example being successful expression of actinomycete natural product gene clusters of up to ~75 kb in size in a genetically engineered Streptomyces avermitilis strain whose genome was systematically minimized by ~1.5 Mbp.137 Future studies will certainly build on past successes that are summarized in existing reviews.138,139
If DNA-sequencing technologies continue to improve in speed and decrease in cost, it is quite plausible that sequencing of individual gene clusters may become obsolete during next decade, being replaced with entire genome sequencing as a routine operation for evaluation of biosynthetic gene clusters within marine microbes. Already, an enormous quantity of data from whole-genome sequencing of both terrestrial and marine organisms has populated public databases. With this increasing amount of genome sequence information arises the need for improved technologies for compiling and interpreting this abundance of information.140 While databases may become populated with enormous quantities of sequences, the practical value of these sequences will ultimately be determined by the quality of analysis and annotation of these DNA sequences.
Overall, the horizon of marine natural product biosynthesis appears very promising. Building on success of the first decade of the 21st century, the next decade should see researchers diving even deeper into the sea of biosynthesis.
6 Acknowledgements
A.L.L. acknowledges support from an NIH IRACDA postdoctoral fellowship (GM068524). Work in the B.S.M. lab is supported by grants from the NIH (CA127622, GM085770, and AI47818).
Biography

Amy L. Lane Amy L. Lane graduated summa cum laude with a B.S. in chemistry from Indiana State University in 2003. She then completed graduate research in the laboratory of Prof. Julia Kubanek at Georgia Institute of Technology, exploring the ecological functions and drug discovery potential of marine macroalgal natural products and earning a Ph.D. in chemistry in 2008. From 2008–2010, she pursued a National Institutes of Health (NIH) postdoctoral fellowship at Scripps Institution of Oceanography in the laboratory of Prof. Bradley S. Moore, where she completed studies on the biosynthesis of marine actinomycete natural products. She is presently an assistant professor of chemistry at the University of North Florida. Her current research interests involve application of genetic and chemical tools toward discovery of marine natural products and understanding their ecological functions.

Bradley S. Moore Bradley S. Moore is currently Professor of Oceanography and Pharmaceutical Sciences at the Scripps Institution of Oceanography and the Skaggs School of Pharmacy and Pharmaceutical Sciences at University of California, San Diego. He holds degrees in chemistry from the Universities of Hawaii (B.S. 1988) and Washington (Ph.D. 1994), was a postdoc at the University of Zurich (1994–1995), and held prior faculty appointments at the Universities of Washington (1996–1999) and Arizona (1999–2005). His research interests involve exploring and exploiting marine microbial genomes to discover new biosynthetic enzymes, secondary metabolic pathways, and natural products for drug discovery and development. Dr Moore is a member of the NPR Editorial Board (since 2005) and in 2011 became the NPR Editorial Board Chair.
Footnotes
This paper is part of an NPR themed issue on Marine Natural Products.
7 References
- 1.Blunt JW, Copp BR, Munro MHG, Northcote PT, Prinsep MR. Nat. Prod. Rep. 2010;27:165–237. doi: 10.1039/b906091j. [DOI] [PubMed] [Google Scholar]
- 2.Faulkner DJ. Nat. Prod. Rep. 2000;17:1–6. doi: 10.1039/a909113k. [DOI] [PubMed] [Google Scholar]
- 3.Molinski TF, Dalisay DS, Lievens SL, Saludes JP. Nat. Rev. Drug Discovery. 2009;8:69–85. doi: 10.1038/nrd2487. [DOI] [PubMed] [Google Scholar]
- 4.Newman DJ, Cragg GM. J. Nat. Prod. 2004;67:1216–1238. doi: 10.1021/np040031y. [DOI] [PubMed] [Google Scholar]
- 5.Mayer AMS, Glaser KB, Cuevas C, Jacobs RS, Kem W, Little RD, McIntosh JM, Newman DJ, Potts BC, Shuster DE. Trends Pharmacol. Sci. 2010;31:255–265. doi: 10.1016/j.tips.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 6.Herbert RB, editor. The Biosynthesis of Secondary Metabolites. 2nd edn Chapman & Hall; London: 1989. [Google Scholar]
- 7.Harrison DM. Nat. Prod. Rep. 1990;7:459–484. doi: 10.1039/np9900700459. [DOI] [PubMed] [Google Scholar]
- 8.Beale MH. Nat. Prod. Rep. 1991;8:441–454. [Google Scholar]
- 9.O’Hagan D. Nat. Prod. Rep. 1992;9:447–479. doi: 10.1039/np9920900447. [DOI] [PubMed] [Google Scholar]
- 10.Dewick PM. Nat. Prod. Rep. 1995;12:101–133. [Google Scholar]
- 11.Staunton J, Weissman KJ. Nat. Prod. Rep. 2001;18:380–416. doi: 10.1039/a909079g. [DOI] [PubMed] [Google Scholar]
- 12.Kircher M, Kelso J. BioEssays. 2010;32:524–536. doi: 10.1002/bies.200900181. [DOI] [PubMed] [Google Scholar]
- 13.Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Eddy SR, Griffiths-Jones S, Howe KL, Marshall M, Sonnhammer ELL. Nucleic Acids Res. 2002;30:276–280. doi: 10.1093/nar/30.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haft DH, Loftus BJ, Richardson DL, Yang F, Eisen JA, Paulsen IT, White O. Nucleic Acids Res. 2001;29:41–43. doi: 10.1093/nar/29.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Besemer J, Lomsadze A, Borodovsky M. Nucleic Acids Res. 2001;29:2607–2618. doi: 10.1093/nar/29.12.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmidt EW, Donia MS. Complex Enzymes in Microbial Natural Product Biosynthesis, Part A: Overview Articles and Peptides. Methods Enzymol. 2009;458:575–595. [Google Scholar]
- 17.Kalaitzis JA, Lauro FM, Neilan BA. Nat. Prod. Rep. 2009;26:1447–1465. doi: 10.1039/b817074f. [DOI] [PubMed] [Google Scholar]
- 18.Gross H. Curr. Opin. Drug Discovery Dev. 2009;12:207–219. [PubMed] [Google Scholar]
- 19.Piel J. Nat. Prod. Rep. 2009;26:338–362. doi: 10.1039/b703499g. [DOI] [PubMed] [Google Scholar]
- 20.Gulder TAM, Moore BS. Curr. Opin. Microbiol. 2009;12:252–260. doi: 10.1016/j.mib.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones AC, Monroe EA, Eisman EB, Gerwick L, Sherman DH, Gerwick WH. Nat. Prod. Rep. 2010;27:1048–1065. doi: 10.1039/c000535e. [DOI] [PubMed] [Google Scholar]
- 22.Moore BS. Nat. Prod. Rep. 2005;22:580–593. doi: 10.1039/b404737k. [DOI] [PubMed] [Google Scholar]
- 23.Moore BS. Nat. Prod. Rep. 2006;23:615–629. doi: 10.1039/b508781n. [DOI] [PubMed] [Google Scholar]
- 24.Moore BS, Hertweck C. Nat. Prod. Rep. 2002;19:70–99. doi: 10.1039/b003939j. [DOI] [PubMed] [Google Scholar]
- 25.Hertweck C. Angew. Chem., Int. Ed. 2009;48:4688–4716. doi: 10.1002/anie.200806121. [DOI] [PubMed] [Google Scholar]
- 26.Chan YA, Podevels AM, Kevany BM, Thomas MG. Nat. Prod. Rep. 2009;26:90–114. doi: 10.1039/b801658p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hertweck C, Luzhetskyy A, Rebets Y, Bechthold A. Nat. Prod. Rep. 2007;24:162–190. doi: 10.1039/b507395m. [DOI] [PubMed] [Google Scholar]
- 28.Hill AM. Nat. Prod. Rep. 2006;23:256–320. doi: 10.1039/b301028g. [DOI] [PubMed] [Google Scholar]
- 29.Piel J. Nat. Prod. Rep. 2010;27:996–1047. doi: 10.1039/b816430b. [DOI] [PubMed] [Google Scholar]
- 30.Zhou H, Li YR, Tang Y. Nat. Prod. Rep. 2010;27:839–868. doi: 10.1039/b911518h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Du L, Lou L. Nat. Prod. Rep. 2010;27:255–278. doi: 10.1039/b912037h. [DOI] [PubMed] [Google Scholar]
- 32.Walsh CT. Science. 2004;303:1805–1810. doi: 10.1126/science.1094318. [DOI] [PubMed] [Google Scholar]
- 33.Olano C, Mendez C, Salas JA. Nat. Prod. Rep. 2010;27:571–616. doi: 10.1039/b911956f. [DOI] [PubMed] [Google Scholar]
- 34.Koglin A, Walsh CT. Nat. Prod. Rep. 2009;26:987–1000. doi: 10.1039/b904543k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Finking R, Marahiel MA. Annu. Rev. Microbiol. 2004;58:453–488. doi: 10.1146/annurev.micro.58.030603.123615. [DOI] [PubMed] [Google Scholar]
- 36.Sieber SA, Marahiel MA. Chem. Rev. 2005;105:715–738. doi: 10.1021/cr0301191. [DOI] [PubMed] [Google Scholar]
- 37.Butler A, Carter-Franklin JN. Nat. Prod. Rep. 2004;21:180–188. doi: 10.1039/b302337k. [DOI] [PubMed] [Google Scholar]
- 38.Kuzuyama T, Seto H. Nat. Prod. Rep. 2003;20:171–183. doi: 10.1039/b109860h. [DOI] [PubMed] [Google Scholar]
- 39.Christianson DW. Chem. Rev. 2006;106:3412–3442. doi: 10.1021/cr050286w. [DOI] [PubMed] [Google Scholar]
- 40.Barry SM, Challis GL. Curr. Opin. Chem. Biol. 2009;13:205–215. doi: 10.1016/j.cbpa.2009.03.008. [DOI] [PubMed] [Google Scholar]
- 41.McIntosh JA, Donia MS, Schmidt EW. Nat. Prod. Rep. 2009;26:537–559. doi: 10.1039/b714132g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liang ZX. Nat. Prod. Rep. 2010;27:499–528. doi: 10.1039/b908165h. [DOI] [PubMed] [Google Scholar]
- 43.Piel J, Hertweck C, Shipley PR, Hunt DM, Newman MS, Moore BS. Chem. Biol. 2000;7:943–955. doi: 10.1016/s1074-5521(00)00044-2. [DOI] [PubMed] [Google Scholar]
- 44.Sitachitta N, Gadepalli M, Davidson BS. Tetrahedron. 1996;52:8073–8080. [Google Scholar]
- 45.Seto H, Sato T, Urano S, Uzawa J, Yonehara H. Tetrahedron Lett. 1976;17:4367–4370. [Google Scholar]
- 46.Cheng Q, Xiang L, Izumikawa M, Meluzzi D, Moore BS. Nat. Chem. Biol. 2007;3:557–558. doi: 10.1038/nchembio.2007.22. [DOI] [PubMed] [Google Scholar]
- 47.Hertweck C, Xiang L, Kalaitzis JA, Cheng Q, Palzer M, Moore BS. Chem. Biol. 2004;11:461–468. doi: 10.1016/j.chembiol.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 48.Kalaitzis JA, Cheng Q, Thomas PM, Kelleher NL, Moore BS. J. Nat. Prod. 2009;72:469–472. doi: 10.1021/np800598t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kalaitzis JA, Izumikawa M, Xiang L, Hertweck C, Moore BS. J. Am. Chem. Soc. 2003;125:9290–9291. doi: 10.1021/ja035973o. [DOI] [PubMed] [Google Scholar]
- 50.Piel J, Hoang K, Moore BS. J. Am. Chem. Soc. 2000;122:5415–5416. [Google Scholar]
- 51.Xiang L, Kalaitzis JA, Moore BS. Proc. Natl. Acad. Sci. U. S. A. 2004;101:15609–15614. doi: 10.1073/pnas.0405508101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xiang L, Kalaitzis JA, Nilsen G, Chen L, Moore BS. Org. Lett. 2002;4:957–960. doi: 10.1021/ol0255155. [DOI] [PubMed] [Google Scholar]
- 53.Xiang L, Moore BS. J. Bacteriol. 2003;185:399–404. doi: 10.1128/JB.185.2.399-404.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xiang L, Moore BS. J. Biol. Chem. 2002;277:32505–32509. doi: 10.1074/jbc.M204171200. [DOI] [PubMed] [Google Scholar]
- 55.Xiang L, Moore BS. J. Bacteriol. 2005;187:4286–4289. doi: 10.1128/JB.187.12.4286-4289.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tillett D, Dittmann E, Erhard M, von Dohren H, Borner T, Neilan BA. Chem. Biol. 2000;7:753–764. doi: 10.1016/s1074-5521(00)00021-1. [DOI] [PubMed] [Google Scholar]
- 57.Chang ZX, Sitachitta N, Rossi JV, Roberts MA, Flatt PM, Jia JY, Sherman DH, Gerwick WH. J. Nat. Prod. 2004;67:1356–1367. doi: 10.1021/np0499261. [DOI] [PubMed] [Google Scholar]
- 58.Sitachitta N, Marquez BL, Williamson RT, Rossi J, Robert MA, Gerwick WH, Nugyen V-A, Willis CL. Tetrahedron. 2000;56:9103–9133. [Google Scholar]
- 59.Chang ZX, Flatt P, Gerwick WH, Nguyen VA, Willis CL, Sherman DH. Gene. 2002;296:235–247. doi: 10.1016/s0378-1119(02)00860-0. [DOI] [PubMed] [Google Scholar]
- 60.Galonic DP, Vaillancourt FH, Walsh CT. J. Am. Chem. Soc. 2006;128:3900–3901. doi: 10.1021/ja060151n. [DOI] [PubMed] [Google Scholar]
- 61.Flatt PM, O’Connell SJ, McPhail KL, Zeller G, Willis CL, Sherman DH, Gerwick WH. J. Nat. Prod. 2006;69:938–944. doi: 10.1021/np050523q. [DOI] [PubMed] [Google Scholar]
- 62.Nett M, Ikeda H, Moore BS. Nat. Prod. Rep. 2009;26:1362–1384. doi: 10.1039/b817069j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Challis GL. J. Med. Chem. 2008;51:2618–2628. doi: 10.1021/jm700948z. [DOI] [PubMed] [Google Scholar]
- 64.Corre C, Challis GL. Nat. Prod. Rep. 2009;26:977–986. doi: 10.1039/b713024b. [DOI] [PubMed] [Google Scholar]
- 65.Watve MG, Tickoo R, Jog MM, Bhole BD. Arch. Microbiol. 2001;176:386–390. doi: 10.1007/s002030100345. [DOI] [PubMed] [Google Scholar]
- 66.Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, Bateman A, Brown S, Chandra G, Chen CW, Collins M, Cronin A, Fraser A, Goble A, Hidalgo J, Hornsby T, Howarth S, Huang CH, Kieser T, Larke L, Murphy L, Oliver K, O’Neil S, Rabbinowitsch E, Rajandream MA, Rutherford K, Rutter S, Seeger K, Saunders D, Sharp S, Squares R, Squares S, Taylor K, Warren T, Wietzorrek A, Woodward J, Barrell BG, Parkhill J, Hopwood DA. Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 67.Ikeda H, Ishikawa J, Hanamoto A, Shinose M, Kikuchi H, Shiba T, Sakaki Y, Hattori M, Omura S. Nat. Biotechnol. 2003;21:526–531. doi: 10.1038/nbt820. [DOI] [PubMed] [Google Scholar]
- 68.Jensen PR, Williams PG, Oh DC, Zeigler L, Fenical W. Appl. Environ. Microbiol. 2007;73:1146–1152. doi: 10.1128/AEM.01891-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Feling RH, Buchanan GO, Mincer TJ, Kauffman CA, Jensen PR, Fenical W. Angew. Chem., Int. Ed. 2003;42:355–357. doi: 10.1002/anie.200390115. [DOI] [PubMed] [Google Scholar]
- 70.Udwary DW, Zeigler L, Asolkar RN, Singan V, Lapidus A, Fenical W, Jensen PR, Moore BS. Proc. Natl. Acad. Sci. U. S. A. 2007;104:10376–10381. doi: 10.1073/pnas.0700962104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Eustáquio AS, McGlinchey RP, Liu Y, Hazzard C, Beer LL, Florova G, Alhamadsheh MM, Lechner A, Kale AJ, Kobayashi Y, Reynolds KA, Moore BS. Proc. Natl. Acad. Sci. U. S. A. 2009;106:12295–12300. doi: 10.1073/pnas.0901237106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Eustaquio AS, Moore BS. Angew. Chem., Int. Ed. 2008;47:3936–3938. doi: 10.1002/anie.200800177. [DOI] [PubMed] [Google Scholar]
- 73.Eustaquio AS, O’Hagan D, Moore BS. J. Nat. Prod. 2010;73:378–382. doi: 10.1021/np900719u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu Y, Hazzard C, Eustaquio AS, Reynolds KA, Moore BS. J. Am. Chem. Soc. 2009;131:10376–10377. doi: 10.1021/ja9042824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McGlinchey RP, Nett M, Eustaquio AS, Asolkar RN, Fenical W, Moore BS. J. Am. Chem. Soc. 2008;130:7822–7823. doi: 10.1021/ja8029398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McGlinchey RP, Nett M, Moore BS. J. Am. Chem. Soc. 2008;130:2406–2407. doi: 10.1021/ja710488m. [DOI] [PubMed] [Google Scholar]
- 77.Penn K, Jenkins C, Nett M, Udwary DW, Gontang EA, McGlinchey RP, Foster B, Lapidus A, Podell S, Allen EE, Moore BS, Jensen PR. ISME J. 2009;3:1193–1203. doi: 10.1038/ismej.2009.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Piel J. Nat. Prod. Rep. 2004;21:519–538. doi: 10.1039/b310175b. [DOI] [PubMed] [Google Scholar]
- 79.Gurgui C, Piel J. Methods Mol. Biol. 2010;668:247–264. doi: 10.1007/978-1-60761-823-2_17. [DOI] [PubMed] [Google Scholar]
- 80.Piel J. Curr. Med. Chem. 2006;13:39–50. [PubMed] [Google Scholar]
- 81.Schmidt EW. Trends Biotechnol. 2005;23:437–440. doi: 10.1016/j.tibtech.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 82.Faulkner J, Unson MD, Bewley CA. Pure Appl. Chem. 1994;66:1983–1990. [Google Scholar]
- 83.Matsunaga S, Fusetani N. Curr. Org. Chem. 2003;7:945–966. [Google Scholar]
- 84.Hentschel U, Hopke J, Horn M, Friedrich AB, Wagner M, Hacker J, Moore BS. Appl. Environ. Microbiol. 2002;68:4431–4440. doi: 10.1128/AEM.68.9.4431-4440.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Piel J, Hui DQ, Wen GP, Butzke D, Platzer M, Fusetani N, Matsunaga S. Proc. Natl. Acad. Sci. U. S. A. 2004;101:16222–16227. doi: 10.1073/pnas.0405976101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fusetani N, Sugawara T, Matsunaga S. J. Org. Chem. 1992;57:3828–3832. [Google Scholar]
- 87.Sakemi S, Ichiba T, Kohmoto S, Saucy G, Higa T. J. Am. Chem. Soc. 1988;110:4851–4853. [Google Scholar]
- 88.Matsumoto T, Yanagiya M, Maeno S, Yasuda S. Tetrahedron Lett. 1968;9:6297–6300. doi: 10.1016/s0040-4039(01)98905-1. [DOI] [PubMed] [Google Scholar]
- 89.Cardani C, Fuganti C, Ghiringh D, Grassell P, Pavan M, Valcuron Md. Tetrahedron Lett. 1973;14:2815–2818. [Google Scholar]
- 90.Piel J. Proc. Natl. Acad. Sci. U. S. A. 2002;99:14002–14007. doi: 10.1073/pnas.222481399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Irschik H, Kopp M, Weissman KJ, Buntin K, Piel J, Muller R. ChemBioChem. 2010;11:1840–1849. doi: 10.1002/cbic.201000313. [DOI] [PubMed] [Google Scholar]
- 92.Cichewicz RH, Valeriote FA, Crews P. Org. Lett. 2004;6:1951–1954. doi: 10.1021/ol049503q. [DOI] [PubMed] [Google Scholar]
- 93.Fisch KM, Gurgui C, Heycke N, van der Sar SA, Anderson SA, Webb VL, Taudien S, Platzer M, Rubio BK, Robinson SJ, Crews P, Piel J. Nat. Chem. Biol. 2009;5:494–501. doi: 10.1038/nchembio.176. [DOI] [PubMed] [Google Scholar]
- 94.Zimmermann K, Engeser M, Blunt JW, Munro MHG, Piel J. J. Am. Chem. Soc. 2009;131:2780–2781. doi: 10.1021/ja808889k. [DOI] [PubMed] [Google Scholar]
- 95.Degnan BM, Hawkins CJ, Lavin MF, McCaffrey EJ, Parry DL, Vandenbrenk AL, Watters DJ. J. Med. Chem. 1989;32:1349–1354. doi: 10.1021/jm00126a034. [DOI] [PubMed] [Google Scholar]
- 96.Ireland CM, Durso AR, Newman RA, Hacker MP. J. Org. Chem. 1982;47:1807–1811. [Google Scholar]
- 97.Fu X, Do T, Schmitz FJ, Andrusevich V, Engel MH. J. Nat. Prod. 1998;61:1547–1551. doi: 10.1021/np9802872. [DOI] [PubMed] [Google Scholar]
- 98.Carroll AR, Coll JC, Bourne DJ, MacLeod JK, Zabriskie TM, Ireland CM, Bowden BF. Aust. J. Chem. 1996;49:659–667. [Google Scholar]
- 99.Schmidt EW, Nelson JT, Rasko DA, Sudek S, Eisen JA, Haygood MG, Ravel J. Proc. Natl. Acad. Sci. U. S. A. 2005;102:7315–7320. doi: 10.1073/pnas.0501424102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Salomon CE, Faulkner DJ. J. Nat. Prod. 2002;65:689–692. doi: 10.1021/np010556f. [DOI] [PubMed] [Google Scholar]
- 101.Schmidt EW, Sudek S, Haygood MG. J. Nat. Prod. 2004;67:1341–1345. doi: 10.1021/np049948n. [DOI] [PubMed] [Google Scholar]
- 102.Long PF, Dunlap WC, Battershill CN, Jaspars M. ChemBioChem. 2005;6:1760–1765. doi: 10.1002/cbic.200500210. [DOI] [PubMed] [Google Scholar]
- 103.Schmidt EW. Nat. Chem. Biol. 2008;4:466–473. doi: 10.1038/nchembio.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Donia MS, Hathaway BJ, Sudek S, Haygood MG, Rosovitz MJ, Ravel J, Schmidt EW. Nat. Chem. Biol. 2006;2:729–735. doi: 10.1038/nchembio829. [DOI] [PubMed] [Google Scholar]
- 105.Gribble GW. J. Chem. Educ. 2004;81:1441–1449. [Google Scholar]
- 106.Vaillancourt FH, Yeh E, Vosburg DA, Garneau-Tsodikova S, Walsh CT. Chem. Rev. 2006;106:3364–3378. doi: 10.1021/cr050313i. [DOI] [PubMed] [Google Scholar]
- 107.Blasiak LC, Drennan CL. Acc. Chem. Res. 2009;42:147–155. doi: 10.1021/ar800088r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Butler A, Sandy M. Nature. 2009;460:848–854. doi: 10.1038/nature08303. [DOI] [PubMed] [Google Scholar]
- 109.Butler A, Carter-Franklin JN. Nat. Prod. Rep. 2004;21:180–188. doi: 10.1039/b302337k. [DOI] [PubMed] [Google Scholar]
- 110.Winter JM, Moore BS. J. Biol. Chem. 2009;284:18577–18581. doi: 10.1074/jbc.R109.001602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Carter-Franklin JN, Butler A. J. Am. Chem. Soc. 2004;126:15060–15066. doi: 10.1021/ja047925p. [DOI] [PubMed] [Google Scholar]
- 112.Winter JM, Moffitt MC, Zazopoulos E, McAlpine JB, Dorrestein PC, Moore BS. J. Biol. Chem. 2007;282:16362–16368. doi: 10.1074/jbc.M611046200. [DOI] [PubMed] [Google Scholar]
- 113.Eustáquio AS, Pojer F, Noel JP, Moore BS. Nat. Chem. Biol. 2008;4:69–74. doi: 10.1038/nchembio.2007.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dong CJ, Huang FL, Deng H, Schaffrath C, Spencer JB, O’Hagan D, Naismith JH. Nature. 2004;427:561–565. doi: 10.1038/nature02280. [DOI] [PubMed] [Google Scholar]
- 115.Deng H, O’Hagan D. Curr. Opin. Chem. Biol. 2008;12:582–592. doi: 10.1016/j.cbpa.2008.06.036. [DOI] [PubMed] [Google Scholar]
- 116.Kale AJ, McGlinchey RP, Moore BS. J. Biol. Chem. 2010;285:33710–33717. doi: 10.1074/jbc.M110.153833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Vaillancourt FH, Yin J, Walsh CT. Proc. Natl. Acad. Sci. U. S. A. 2005;102:10111–10116. doi: 10.1073/pnas.0504412102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Blasiak LC, Vaillancourt FH, Walsh CT, Drennan CL. Nature. 2006;440:368–371. doi: 10.1038/nature04544. [DOI] [PubMed] [Google Scholar]
- 119.Vaillancourt FH, Yeh E, Vosburg DA, O’Connor SE, Walsh CT. Nature. 2005;436:1191–1194. doi: 10.1038/nature03797. [DOI] [PubMed] [Google Scholar]
- 120.Gu L, Wang B, Kulkarni A, Geders TW, Grindberg RV, Gerwick L, Hakansson K, Wipf P, Smith JL, Gerwick WH, Sherman DH. Nature. 2009;459:731–735. doi: 10.1038/nature07870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Khare D, Wang B, Gu L, Razelun J, Sherman DH, Gerwick WH, Hakansson K, Smith JL. Proc. Natl. Acad. Sci. U. S. A. 2010;107:14099–14104. doi: 10.1073/pnas.1006738107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gu L, Geders TW, Wang B, Gerwick WH, Hakansson K, Smith JL, Sherman DH. Science. 2007;318:970–974. doi: 10.1126/science.1148790. [DOI] [PubMed] [Google Scholar]
- 123.Calderone CT. Nat. Prod. Rep. 2008;25:845–853. doi: 10.1039/b807243d. [DOI] [PubMed] [Google Scholar]
- 124.Calderone CT, Kowtoniuk WE, Kelleher NL, Walsh CT, Dorrestein PC. Proc. Natl. Acad. Sci. U. S. A. 2006;103:8977–8982. doi: 10.1073/pnas.0603148103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Butcher RA, Schroeder FC, Fischbach MA, Straight PD, Kolter R, Walsh CT, Clardy J. Proc. Natl. Acad. Sci. U. S. A. 2007;104:1506–1509. doi: 10.1073/pnas.0610503104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Edwards DJ, Marquez BL, Nogle LM, McPhail K, Goeger DE, Roberts MA, Gerwick WH. Chem. Biol. 2004;11:817–833. doi: 10.1016/j.chembiol.2004.03.030. [DOI] [PubMed] [Google Scholar]
- 127.Buchholz TJ, Rath CM, Lopanik NB, Gardner NP, Hakansson K, Sherman DH. Chem. Biol. 2010;17:1092–1100. doi: 10.1016/j.chembiol.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Geders TW, Gu L, Mowers JC, Liu H, Gerwick WH, Hakansson K, Sherman DH, Smith JL. J. Biol. Chem. 2007;282:35954–35963. doi: 10.1074/jbc.M703921200. [DOI] [PubMed] [Google Scholar]
- 129.Hertweck C, Jarvis AP, Xiang LK, Moore BS, Oldham NJ. ChemBioChem. 2001;2:784–786. doi: 10.1002/1439-7633(20011001)2:10<784::AID-CBIC784>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 130.Nett M, Guider TAM, Kale AJ, Hughes CC, Moore BS. J. Med. Chem. 2009;52:6163–6167. doi: 10.1021/jm901098m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Schultz AW, Lewis CA, Luzung MR, Baran PS, Moore BS. J. Nat. Prod. 2010;73:373–377. doi: 10.1021/np9006876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sudek S, Haygood MG, Youssef DTA, Schmidt EW. Appl. Environ. Microbiol. 2006;72:4382–4387. doi: 10.1128/AEM.00380-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Rokem JS, Lantz AE, Nielsen J. Nat. Prod. Rep. 2007;24:1262–1287. doi: 10.1039/b617765b. [DOI] [PubMed] [Google Scholar]
- 134.Martin JF, Liras P. Curr. Opin. Microbiol. 2010;13:263–273. doi: 10.1016/j.mib.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 135.Niraula NP, Kim SH, Sohng JK, Kim ES. Appl. Microbiol. Biotechnol. 2010;87:1187–1194. doi: 10.1007/s00253-010-2675-3. [DOI] [PubMed] [Google Scholar]
- 136.Chen YH, Smanski MJ, Shen B. Appl. Microbiol. Biotechnol. 2010;86:19–25. doi: 10.1007/s00253-009-2428-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Komatsua M, Uchiyama T, Omura S, Cane DE, Ikeda H. Proc. Natl. Acad. Sci. U. S. A. 2010;107:2646–2651. doi: 10.1073/pnas.0914833107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Rodriguez E, Menzella HG, Gramajo H. Complex Enzymes in Microbial Natural Product Biosynthesis, Part B: Polyketides, Aminocoumarins and Carbohydrates. Methods Enzymol. 2009;459:339–365. doi: 10.1016/S0076-6879(09)04625-4. [DOI] [PubMed] [Google Scholar]
- 139.Fujii I. Nat. Prod. Rep. 2009;26:155–169. doi: 10.1039/b817092b. [DOI] [PubMed] [Google Scholar]
- 140.Schadt EE, Linderman MD, Sorenson J, Lee L, Nolan GP. Nat. Rev. Genet. 2010;11:647–657. doi: 10.1038/nrg2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Galonic DP, Vaillancourt FH, Walsh CT. J. Am. Chem. Soc. 2006;128:3900–3901. doi: 10.1021/ja060151n. [DOI] [PubMed] [Google Scholar]
- 142.Jorgensen H, Degnes KF, Dikiy A, Fjaervik E, Klinkenberg G, Zotchev SB. Appl. Environ. Microbiol. 2010;76:283–293. doi: 10.1128/AEM.00744-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wilson MC, Gulder TAM, Mahmud T, Moore BS. J. Am. Chem. Soc. 2010;132:12757–12765. doi: 10.1021/ja105891a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Carlson JC, Fortman JL, Anzai Y, Li SY, Burr DA, Sherman DH. ChemBioChem. 2010;11:564–572. doi: 10.1002/cbic.200900658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Engelhardt K, Degnes KF, Zotchev SB. Appl. Environ. Microbiol. 2010;76:7093–7101. doi: 10.1128/AEM.01442-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Jorgensen H, Degnes KF, Sletta H, Fjaervik E, Dikiy A, Herfindal L, Bruheim P, Klinkenberg G, Bredholt H, Nygard G, Doskeland SO, Ellingsen TE, Zotchev SB. Chem. Biol. 2009;16:1109–1121. doi: 10.1016/j.chembiol.2009.09.014. [DOI] [PubMed] [Google Scholar]
- 147.Schultz AW, Oh DC, Carney JR, Williamson RT, Udwary DW, Jensen PR, Gould SJ, Fenical W, Moore BS. J. Am. Chem. Soc. 2008;130:4507–4516. doi: 10.1021/ja711188x. [DOI] [PubMed] [Google Scholar]
- 148.Sudek S, Lopanik NB, Waggoner LE, Hildebrand M, Anderson C, Liu HB, Patel A, Sherman DH, Haygood MG. J. Nat. Prod. 2007;70:67–74. doi: 10.1021/np060361d. [DOI] [PubMed] [Google Scholar]
- 149.Ramaswamy AV, Sorrels CM, Gerwick WH. J. Nat. Prod. 2007;70:1977–1986. doi: 10.1021/np0704250. [DOI] [PubMed] [Google Scholar]
- 150.Edwards DJ, Gerwick WH. J. Am. Chem. Soc. 2004;126:11432–11433. doi: 10.1021/ja047876g. [DOI] [PubMed] [Google Scholar]
- 151.Moffitt MC, Neilan BA. Appl. Environ. Microbiol. 2004;70:6353–6362. doi: 10.1128/AEM.70.11.6353-6362.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Allen EE, Bartlett DH. Microbiology-SGM. 2002;148:1903–1913. doi: 10.1099/00221287-148-6-1903. [DOI] [PubMed] [Google Scholar]
- 153.Li AY, Piel J. Chem. Biol. 2002;9:1017–1026. doi: 10.1016/s1074-5521(02)00223-5. [DOI] [PubMed] [Google Scholar]
- 154.Morita N, Tanaka M, Okuyama H. Biochem. Soc. Trans. 2000;28:943–945. [PubMed] [Google Scholar]