Abstract
The activation and regulation of target genes by the tumour-suppressor p53 dictates the fate of a cell, with cell cycle arrest or apoptosis being two distinct outcomes. PERP (p53 apoptosis effector related to PMP-22), a p53 transcriptional target, is induced specifically during apoptosis but not cell cycle arrest. Downregulation of PERP is associated with the aggressive, monosomy 3-type of uveal melanoma (UM), the most common primary intraocular tumour in adults, and increased PERP expression has a pro-apoptotic effect in UM cells. Here, we identify a novel effect of PERP expression, as elevated PERP protein positively influences active levels of its own transcriptional regulator, p53. Using fluorescent fusion proteins of PERP, p53 and MDM2, we demonstrate in single living UM cells that PERP expression significantly enhances p53 activity and its nuclear localization, increases p53-dependent transcription (including that of MDM2) while allowing oscillatory nucleo-cytoplasmic shuttling of p53/MDM2 complexes. Phosphorylation of p53 serine residues that interfere with the interaction between p53 and its negative regulator MDM2 and enhance pro-apoptotic gene transcription also occurs subsequent to PERP expression. These results implicate a role for PERP in amplifying functional p53 levels that promote p53-dependent apoptosis, and reveal a potential target for exploitation in enhancing p53 activity.
Keywords: PERP, apoptosis, uveal melanoma, p53, fluorescent protein
The regulation of p53 and of its impressive array of interacting and target genes holds a central place among the molecular mechanism(s) influencing the choice between cell cycle arrest and apoptosis.1 The ability to engage in apoptosis is critical to the tumour-suppressor role of p53 that is strongly supported by the presence of p53 mutations in over half of human cancers2, 3 and the compromised p53 activity by other mechanisms in the majority of other cancers.3 The latter scenario points towards the deregulation of downstream p53 targets as a mechanism employed by tumour cells to evade apoptosis. PERP (p53 apoptosis effector related to PMP-22) was identified as a p53 transcriptional target that is distinctively induced during apoptosis, and not cell cycle arrest,4 and consequently emerged as a prime candidate effector in the p53-dependent apoptotic pathway. Subsequent studies confirmed the pro-apoptotic role for PERP in a variety of cell types and tissues,5, 6, 7, 8 with reported engagement of caspase-dependent pathways.5, 8 However, the precise function of PERP – a tetraspan protein primarily localized at the plasma membrane – in eliciting an apoptotic response remains unknown.
Cellular p53 levels are principally determined by the rate of degradation – mostly but not entirely linked to the E3 ligase activity of MDM2 – rather than synthesis.3, 9 Increased p53 stability in response to stress is achieved primarily through decreased MDM2 protein levels and reduced p53-MDM2 interaction, while the ensuing elevated levels of transcriptionally active p53 result in increased MDM2 transcription.10, 11 This p53-MDM2 regulatory feedback loop is essential in maintaining tight regulation of p53 levels both in unstressed and stressed cells, with additional control through posttranslational modifications, most notably phosphorylation of key p53 serine (Ser) residues,12 some of which influence the p53-MDM2 interaction13 and the nature of p53-target genes transcribed.14 (Co)localization and nuclear-cytoplasmic shuttling of p53 and MDM2 also contribute to regulation of p53 activity.15, 16
We have shown that PERP is an important molecular determinant of apoptosis in primary uveal melanoma (UM) tumours that is significantly downregulated in the aggressive monosomy-3 type, compared with less aggressive disomy-3 type of UM.5, 17 Downregulation of PERP (THW18) was also reported in tumours of the ovary, uterus and breast, and in cutaneous melanoma, pancreas and mammary carcinoma cell lines, compared with the respective normal tissues and non-metastasizing cell lines.18
The aim of this study was to investigate the effect of increased PERP expression on its own upstream transcriptional regulator, p53. For this purpose, we used the UM cell line MEL20219 that provides a biological background of low endogenous PERP protein with intact cell death machinery downstream of PERP.5 Our results show that PERP expression causes nuclear localization of p53 and increases the level of transcriptionally active p53 protein, which also presents posttranslational modifications known to influence the p53–MDM2 interaction and to enhance the pro-apoptotic gene transcription. Together, these results propose a novel role for PERP in enhancing functional p53 levels and reveal a potential new target for exploitation in the development of new therapeutic agents aimed at increasing the endogenous p53 protein pool in neoplastic cells.
Results
PERP expression augments p53 protein level
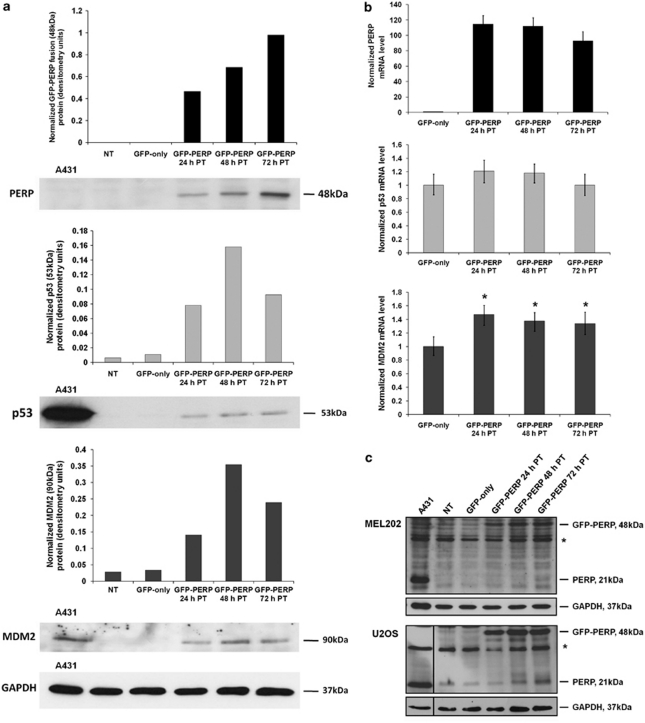
To investigate the effect of increased levels of PERP protein on its transcriptional regulator p53, lysates of MEL202 cells expressing green fluorescent protein (GFP)–PERP were analyzed by western blotting alongside lysates from control non-transfected (NT) and GFP-only-transfected cells. Significantly increased levels of p53 protein were observed in MEL202 cells expressing GFP–PERP compared with control cells, in which p53 was barely detectable (Figure 1a). Furthermore, the increased p53 protein levels in response to PERP expression were accompanied by an increase in the p53-negative regulator MDM2. As PERP is a transcriptional target of p53, the increase in p53 protein in response to PERP expression suggested a role for PERP in the positive feedback regulation of p53. To determine if the observed increase in p53 and MDM2 proteins was transcription-driven, p53 and MDM2 transcriptional levels were determined by real-time quantitative PCR (Q-PCR) in control and GFP–PERP-expressing cells. There was no significant increase in p53 mRNA in cells expressing PERP compared with GFP-only-expressing cells (T-test, P≥0.19). However, MDM2 mRNA was significantly higher in cells expressing PERP at the three time points analyzed (T-test, P≤0.004; Figure 1b). Taken together, the results suggested that the increased p53 protein levels detected in response to PERP expression may be a consequence of increased p53 protein stability rather than transcription, and that the ensuing p53 pool is likely to be transcriptionally active resulting in increased MDM2 at both the transcriptional and protein level.
Figure 1.
PERP expression augments p53 and MDM2 protein level in MEL202 cells. (a) p53 and MDM2 proteins are increased in cells expressing GFP–PERP. MEL202 cells were transfected with GFP–PERP and lysates prepared at 24-, 48- and 72-h PT and analyzed by western blotting alongside NT and GFP-only-transfected cells that served as controls. A431 cell lysate served as a positive control for the respective antibodies. PERP, p53 and MDM2 proteins were detected with appropriate antibodies and their relative levels were quantified by densitometry and normalized to GAPDH. (b) Elevated PERP expression leads to increased MDM2 transcription. RNA was extracted from MEL202 cells transfected with GFP-only or GFP–PERP expression plasmids at the indicated times PT. The levels of PERP, p53 and MDM2 mRNA were determined by Q-PCR and normalized to the respective endogenous level of GAPDH. Normalized mRNA levels are expressed relative to the respective levels in GFP-only-transfected cells that were given an arbitrary value of 1. The mean of three independent experiments along with S.D. is presented. (*T-test, P≤0.004 compared with GFP-only-transfected cells). (c) Expression of exogenous PERP increases endogenous PERP protein. Representative western blots showing endogenous PERP protein level in control cells and cells expressing GFP–PERP in MEL202 and U2OS cell lines; increased levels of endogenous PERP are detectable at 72-h PT in MEL202 cells and 48–72 h in U2OS cells. *a nonspecific protein that is consistently detected with the anti-PERP antibody
There was also evidence that endogenous PERP protein (21 kDa) was slightly elevated in MEL202 cells at 72 h following transfection with GFP–PERP (Figure 1c). Owing to the low basal level of endogenous PERP in MEL202 cells and similar intensities of nonspecific proteins, the level of endogenous PERP in response to GFP–PERP expression was further investigated in the wild-type p53-expressing cell line U2OS. An increase in endogenous PERP was apparent in U2OS cells at 48 and 72 h following transfection with GFP–PERP (Figure 1c). It was conceivable that the observed increase in endogenous PERP was due to the GFP–PERP-induced increased activity of its transcriptional regulator, p53. Consequently, it was important to characterize the functional status of the raised p53 protein following GFP–PERP expression.
PERP expression influences the nuclear translocation and the p53-driven expression of MDM2
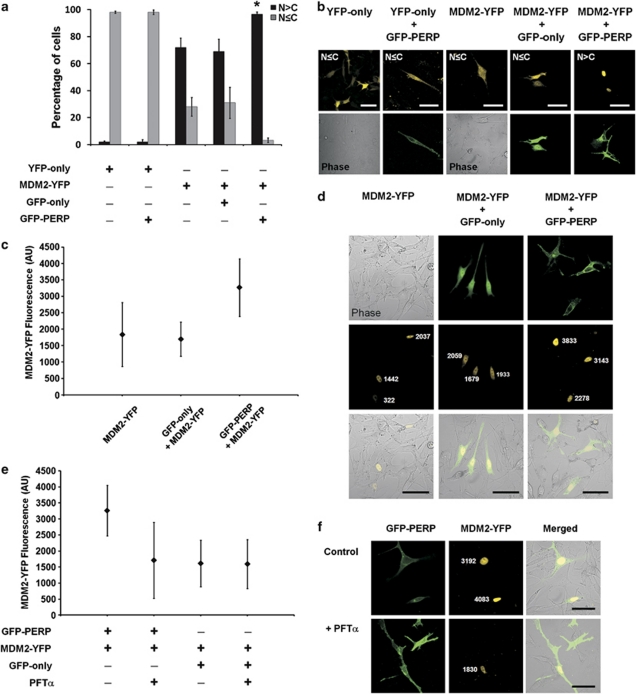
The effect of increased PERP expression described above was found in transiently transfected cell populations where, on average, the transfection efficiency was 13%. To refine our findings, we assessed the effect of PERP expression on the subcellular localization and expression of the p53-transcriptional target MDM2 in individual cells by co-transfecting GFP–PERP with an expression plasmid in which MDM2 was fused to yellow fluorescent protein (YFP) under the control of the human MDM2 native promoter that was previously shown to mirror the kinetic behaviour of endogenous MDM2.20 The first striking observation was the almost exclusive nuclear localization of MDM2–YFP when co-expressed with GFP–PERP (in 96.8% of co-transfected cells; T-test, P=0.02 and 0.04 versus MDM2–YFP or MDM2–YFP and GFP-only; Figures 2a and b). In contrast, MDM2–YFP expression alone or in combination with GFP-only expression showed an additional diffuse cytoplasmic localization of MDM2 in many cells (28 and 31%, respectively; Figures 2a and b). Control cells transfected with YFP-only presented a diffuse YFP expression throughout the cytoplasm and nucleus, which was maintained following co-expression of GFP–PERP (98% cells; Figures 2a and b).
Figure 2.
PERP expression influences the nuclear translocation and the p53-driven expression of MDM2. (a) PERP expression leads to predominantly nuclear localization of MDM2. MEL202 cells transfected with YFP-only, YFP-only and GFP–PERP, MDM2–YFP, MDM2–YFP and GFP-only, or MDM2–YFP and GFP–PERP were monitored by confocal fluorescence microscopy and the analysis of the intracellular distribution of proteins of interest was undertaken at 20-h PT. The number of cells exhibiting predominantly nuclear MDM2 localization (N>C) or more even distribution in the nucleus and cytoplasm (N≤C) were counted. Results are presented as the mean percentage of transfected cells from three independent transfections in each scenario with S.D., with 200 cells counted for each transfection. MDM2 was predominantly nuclear in a significantly higher proportion of cells when co-expressed with GFP–PERP (*T-test, P=0.02 and P=0.04 versus MDM2–YFP or MDM2–YFP and GFP-only-transfected cells, respectively). (b) Differential MDM2 subcellular distribution in the presence and absence of GFP-PERP. A predominantly nuclear MDM2–YFP localization (yellow) is evident in cells co-expressing GFP-PERP, in contrast to the diffuse cytoplasmic/nuclear localization of MDM2–YFP in the absence of GFP–PERP and of YFP alone in control cells. Green fluorescence shows the characteristic distribution of GFP or GFP–PERP proteins. The phase image of cells lacking GFP fluorescence is shown. Scale bar=50 μm. (c) Single cell analysis verified that PERP expression leads to increased MDM2 expression. MEL202 cells were transfected and monitored as described in (a). Images were taken from three independent transfection experiments and YFP fluorescence was measured in cells in arbitrary units (AU). Mean YFP fluorescence is indicated (♦) with S.D. MDM2–YFP fluorescence was significantly higher in cells co-expressing GFP–PERP (T-test, P<0.0001), compared with cells expressing GFP-only and MDM2–YFP, or MDM2–YFP-only. Expression of GFP alone did not affect the levels of MDM2 (T-test, P=0.55). (d) Representative images showing increased MDM2 protein in cells expressing PERP. MDM2–YFP fluorescence (yellow) is indicated in AU by the corresponding nucleus. GFP-only and, respectively, GFP–PERP expression are shown in green. The corresponding phase image of cells expressing MDM2–YFP-only is also presented. Scale bar=50 μm. (e) PERP-induced MDM2 expression is p53 dependent. MEL202 cells were transfected with GFP–PERP, MDM2–YFP and GFP-only as indicated, in the presence (+) for at least 14 h, or absence (−) of 30 μM PFTα. YFP fluorescence was measured in cells from three independent transfections. Mean YFP fluorescence is indicated (♦) with S.D. PFTα significantly reduced the level of MDM2–YFP expression in cells co-expressing GFP–PERP (T-test, P<0.0001). (f) Representative images of MEL202 cells co-expressing GFP–PERP (green) and MDM2-YFP (yellow) in the presence and absence of PFTα. YFP fluorescence (AU) measured in the nuclei of each cell is indicated. Scale bar=50 μm
To determine the effect of PERP on the expression of MDM2, YFP fluorescence was measured in cells co-expressing GFP–PERP and MDM2–YFP and compared with that in control cells (MDM2–YFP-transfected and GFP-only plus MDM2–YFP co-transfected cells). The level of YFP fluorescence – and therefore MDM2 expression – was significantly higher in cells co-expressing GFP–PERP compared with cells co-expressing GFP-only and MDM2–YFP (T-test, P<0.0001), or expressing MDM2–YFP alone (T-test, P<0.0001; Figures 2c and d).
To directly assess the role of p53-driven transcription of MDM2 in response to PERP expression, cells co-transfected with GFP–PERP and MDM2–YFP were treated with pifithrin-α (PFTα), a reversible inhibitor of p53-dependent gene transcription.21 The level of MDM2–YFP fluorescence in cells co-expressing GFP–PERP was significantly reduced in cells treated with PFTα (T-test, P<0.0001; Figures 2e and f), indicating that the induction of MDM2 expression in cells expressing GFP–PERP was likely through p53-dependent transcriptional activation. No significant difference was seen in control cells co-expressing GFP-only and MDM2–YFP with or without PFTα treatment (T-test, P=0.95).
PERP expression localizes p53 in the nucleus with evidence of oscillatory activity
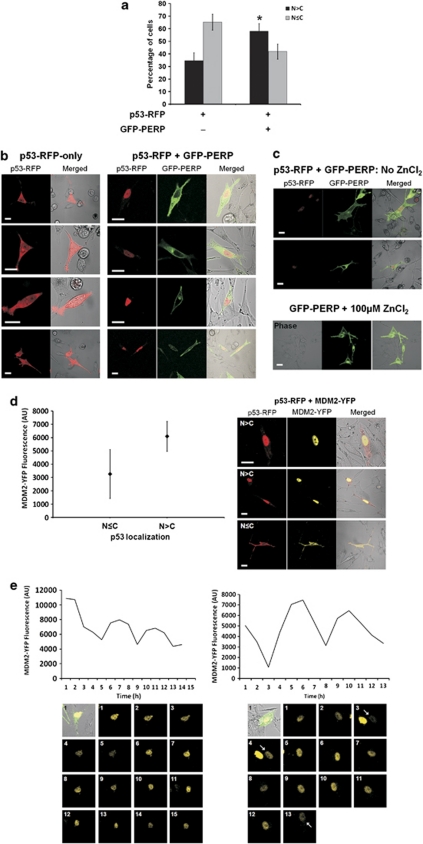
To monitor the effect of PERP expression on the cellular localization of p53, MEL202 cells were co-transfected with GFP–PERP and an expression plasmid in which p53 was fused to red fluorescent protein (RFP) placed under the control of a zinc-inducible promoter. Cells transfected with p53–RFP-only served as controls. In the absence of GFP–PERP expression, p53–RFP was localized primarily in the cytoplasm with low expression in the nucleus (65% of transfected cells; Figures 3a and b), compared with 35% of cells showing nuclear-only p53–RFP expression. However, following GFP–PERP expression, p53–RFP was localized predominantly in the nucleus in a significantly higher proportion of co-transfected cells (58% T-test, P=0.04; Figures 3a and b). Nuclear expression of p53–RFP when co-expressed with GFP–PERP was also detected in the absence of ZnCl2 induction, albeit at much lower expression levels, and ZnCl2 had no effect on the expression or localization of GFP–PERP (Figure 3c). The PERP-induced nuclear localization of p53 suggests that p53 is likely to wield its transcriptional activity – a scenario that is also supported by the p53-dependent upregulation of MDM2 transcription in the presence of elevated PERP (Figure 2). Furthermore, the highest nuclear MDM2–YFP fluorescence in cells co-transfected with MDM2–YFP and p53–RFP occurred when p53–RFP was also localized primarily in the nucleus, while cytoplasmic p53 was associated (fluorescence colocalized) with cytoplasmic MDM2–YFP (Figure 3d), illustrating the nuclear-cytoplasmic shuttling of both p53 and MDM2 that is central to their activities.15
Figure 3.
p53 cellular localization is influenced by PERP expression and is subject to oscillatory regulation. (a) p53 localizes primarily in the nucleus following PERP expression. MEL202 cells transfected with p53–RFP or p53-RFP and GFP–PERP were monitored by confocal fluorescence microscopy and the analysis of the intracellular distribution of proteins of interest was undertaken at 20-h PT. The number of cells exhibiting a predominant nuclear localization (N>C) or a more even p53 distribution in the nucleus and cytoplasm (N≤C) were counted and are presented as the mean percentage of transfected cells from three independent transfections with S.D. p53–RFP was predominantly nuclear in a significantly higher proportion of cells when co-expressed with GFP–PERP compared with cells expressing p53-RFP only (*T-test, P=0.04). (b) Representative images showing the predominant cytoplasmic localization of p53 (red) in the absence of GFP–PERP expression (left panel), and the prevalent nuclear p53 localization (red) in the presence of GFP–PERP (green; right panel), following induction of the pMT promoter of p53–RFP with 100 μM ZnCl2. Scale bar=20 μm. (c) ZnCl2 does not influence PERP expression or p53 localization. Low basal expression of p53–RFP observed in the absence of ZnCl2 induction had a predominant nuclear localization in the presence of GFP–PERP expression, whilst 100 μM ZnCl2 had no effect on GFP–PERP expression or localization. Scale bar=20 μm. (d) Increased MDM2 expression occurs when p53 is primarily localized in nucleus. MEL202 cells were co-transfected with p53–RFP and MDM2-YFP and images were taken from three independent transfections at 20-h PT. YFP fluorescence was measured in arbitrary units (AU) in cells expressing p53 predominantly in the nucleus (N>C) or more evenly in the nucleus and cytoplasm (N≤C). Mean fluorescence is indicated (♦) with S.D. MDM2-YFP fluorescence was significantly higher in cells expressing p53 primarily in the nucleus (T-test, P<0.0005). Representative images of the differential localization of p53 (red) and MDM2 (yellow) are shown. Scale bar=20 μm. (e) PERP-induced elevated MDM2 expression exhibits oscillations. MEL202 cells co-transfected with GFP–PERP (green in bright field) and MDM2-YFP (yellow) were monitored by time-lapse fluorescence microscopy. Nuclear MDM2–YFP fluorescence was measured (AU) in single cells over time (hours) and data are presented graphically alongside corresponding time point images. An arrow indicates the relevant cell where necessary
As we had shown that the increase in MDM2 expression in the presence of elevated PERP was p53-dependent (Figure 2e), we next addressed the behaviour of p53 in the presence of elevated PERP over time in cells co-expressing GFP–PERP and MDM2–YFP. These cells were monitored by time-lapse fluorescence confocal microscopy and the MDM2–YFP fluorescence was assessed as a measure of the kinetic behaviour of p53.20 Full acquisition of YFP fluorescence measurements in a single cell over the entire time-lapse period (usually at least 18 h) proved difficult because of the high motility of MEL202 cells, which resulted in many cells leaving the field of view. However, we were able to detect dampened oscillations in MDM2–YFP fluorescence at regular 5–6 h intervals (Figure 3e), which corresponded to previous findings albeit in response to DNA damage.20 We also observed cell death in approximately 31% of cells expressing GFP–PERP and MDM2–YFP over the time frame of imaging, which corresponded to approximately 30–48-h post-transfection (PT). These levels of cell death were comparable with our previous findings5 and confirmed that the observations of MDM2–YFP expression in this study reflect the endogenous response to GFP–PERP expression.
Endogenous MDM2 and p53 are predominantly nuclear in the presence of elevated PERP expression
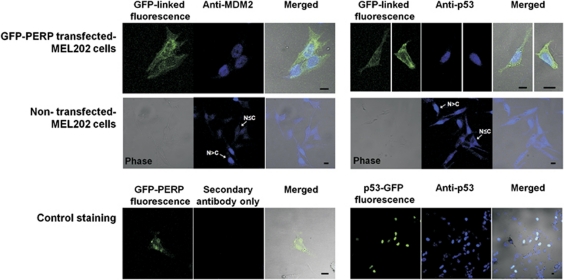
To verify that the prevalent nuclear localization of MDM2–YFP and p53–RFP seen in MEL202 cells following GFP–PERP expression reflected the subcellular localization of endogenous MDM2 and p53 in the presence of elevated PERP, MEL202 cells were transfected with GFP–PERP and fixed 24-h PT. Endogenous MDM2 and p53 were detected by immunofluorescence. Every cell-expressing GFP–PERP that was detected (52 cells) exhibited nuclear localization of MDM2 and p53, whereas a mixture of nuclear, nuclear/cytoplasmic or cytoplasmic expression of MDM2 and p53 was seen in NT cells (Figure 4). The fixation and immunofluorescence procedure caused a significant detachment of GFP–PERP-transfected cells. This is likely due to the pro-apoptotic effect of PERP on MEL202 cells5 that causes more readily detachment during fixation and subsequent immunodetection.
Figure 4.
Endogenous MDM2 and p53 are primarily nuclear in the presence of GFP–PERP expression. Endogenous MDM2 (blue; left panel) and p53 (blue; right panel) proteins were detected in fixed GFP–PERP-transfected MEL202 cells and NT cells within the same cell population by immunofluorescence. NT cells displayed a mixture of cytoplasmic (N≤C) and nuclear (N>C) endogenous MDM2 and p53, while every detected cell expressing GFP–PERP (green) presented a nuclear MDM2 or p53 localization. Cells incubated with secondary antibody only verified anti-MDM2 antibody specificity and MEL202 cells transfected with p53–GFP (green) and subjected to UV treatment (20 mJ/cm2) served as a positive control for anti-p53 antibody. Scale bar=200 μm
P53 elevated by PERP expression is modified on key phosphorylation sites
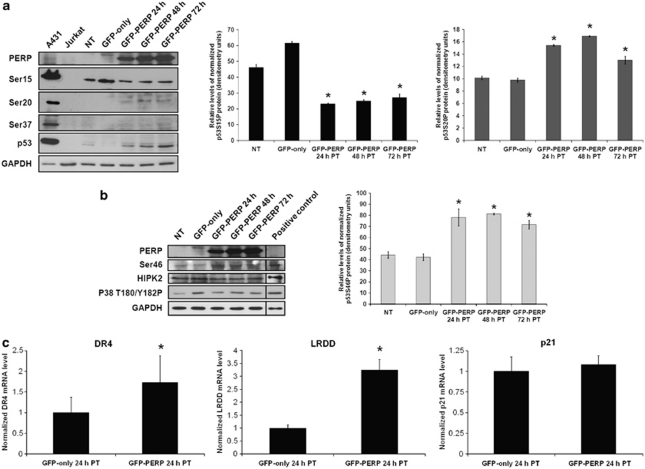
To determine if expression of PERP results in specific p53 posttranslational modification, western blotting was used to assess the phosphorylation status of key p53 Ser residues in the presence of enhanced PERP expression. Phosphorylation at Ser15 was detected in control cells (NT and GFP-only-transfected), but was significantly reduced in GFP–PERP-expressing cells (T-test, P≤0.003 versus GFP-only-transfected cells; Figure 5a). No significant change in phosphorylation at Ser37 was detected. In response to DNA damage, phosphorylation by ataxia telangiectasia mutated and ataxia telangiectasia and Rad3 related at Ser15 and Ser37 can impair the interaction between p53 and MDM2, promoting the accumulation and activation of p53.12, 22 Consequently, reduction of phosphorylation at Ser15, and insignificant detection of Ser37P suggest that impairment of the p53–MDM2 interaction by phosphorylation at these two Ser residues does not contribute to the increased p53 protein seen in response to PERP expression. However, a significant increase in the level of Ser20 phosphorylation was observed in cells expressing GFP–PERP (T-test, P≤0.004 versus GFP-only-transfected cells; Figure 5a). As p53Ser20P is known to interfere with p53 binding to MDM2,23, 24 it is possible that this modification may contribute to the PERP-related increased p53 accumulation.
Figure 5.
p53 elevated by PERP expression is modified on key phosphorylation sites. (a) Differential phosphorylation of p53 residues involved in MDM2 interaction in cells expressing PERP. MEL202 cells were transfected with GFP–PERP and lysates prepared at 24-, 48- and 72-h PT and analyzed by western blotting alongside NT and GFP-only-transfected cells that served as controls. A431 and Jurkat cell lysates served as positive controls for the respective antibodies. Phospho-p53 proteins, phosphorylated on specific serine (Ser) residues as indicated, were detected with appropriate antibodies (see Materials and methods section) and their relative levels were quantified by densitometry and normalized to GAPDH. In cells expressing GFP–PERP, p53Ser15P was significantly reduced (*T-test, P≤0.003) whereas p53Ser20P was significantly increased (*T-test, P≤0.004), compared with cells transfected with GFP–only. Total p53 protein detected with anti-p53 antibody (clone 7F5) is also presented. (b) Phosphorylation at p53Ser46 is increased in cells expressing PERP. Control and transfected MEL202 cells were analyzed as in (a) for phosphorylation of p53 at Ser46. UV-treated HeLa cell lysate served as a positive control for the anti-p53Ser46 antibody. P53Ser46P was significantly higher in cells transfected with GFP–PERP compared with GFP-only-transfected cells (*T-test, P≤0.03). No significant changes in HIPK2 or phospho-p38 (Thr180/Tyr182) protein levels were detected. PC-12 and UV-treated COS cell lysates served as controls for anti-HIPK2 and anti-phospho-p38/anti-GAPDH, respectively. (c) Elevated p53 induced by PERP expression may enhance activation of specific pro-apoptotic p53 target genes. The levels of p53 target genes DR4 and LRDD (pro-apoptotic genes) and p21 (involved in cell cycle arrest) in MEL202 cells expressing GFP-only (control) and GFP–PERP were determined by Q-PCR at 24-h PT. mRNA levels normalized to the endogenous level of GAPDH are expressed relative to the respective levels of mRNA in GFP-only-expressing cells that were given an arbitrary value of 1. The mean of three independent experiments along with S.D. is presented. The level of DR4 and LRDD mRNA was significantly higher in GFP-PERP-transfected cells compared with control cells (*T-test, P=0.04 and P=0.005, respectively). There was no significant difference in the level of p21 (T-test, P=0.50)
Total p53 protein level, detected using anti-p53 antibody (clone 7F5) confirmed the upregulation of p53 protein in cells transfected with GFP–PERP detected previously with a different anti-p53 antibody (clone DO-1; Figure 1a), albeit with a slightly higher p53 level in NT cells (Figure 5a).The detection of p53Ser15P in cells in which total p53 was low/undetectable (NT and GFP-only-transfected) is likely due to differences in antibody specificity.
Phosphorylation of p53 at Ser46 was characterized as a specific phosphorylation event that irreversibly commits cells to apoptosis.14, 25 We detected the presence of p53Ser46P in control MEL202 cells with significantly higher levels in GFP–PERP-expressing cells (T-test, P≤0.03 versus GFP-only-transfected cells; Figure 5b), indicating that the p53 protein elevated in response to PERP expression is likely to be active in apoptosis regulation. The effect of PERP expression on homeodomain-interacting protein kinase 2 (HIPK2) and p38 mitogen-activated protein kinase (MAPK) (p38), both previously implicated in the induction of p53 Ser46 phosphorylation,26, 27, 28, 29 was also assessed by western blotting. No significant changes were detected in HIPK2 expression or in the level of phospho-p38 (Thr180/Tyr182), suggesting the possibility that different pathway(s) may be involved.
Q-PCR analysis of p53 target genes revealed a statistically significant upregulation of death receptor 4 (DR4) and leucine-rich repeats and death domain containing (LRDD), both pro-apoptotic genes30 in MEL202 cells expressing GFP–PERP compared with GFP-only-expressing cells (Figure 5c). No significant changes occurred in the level of cyclin-dependent kinase inhibitor 1A (p21) gene, involved in cell cycle arrest.30
Discussion
The initial findings that elevated PERP expression resulted in increased p53 protein levels in MEL202 cells suggested a role for PERP in the positive feedback regulation of its own transcriptional regulator, with potential implications for the regulation of p53 and the vast array of p53-targets involved in the tight control of cell cycle arrest and apoptosis. Consequently, the study aimed to investigate, in real-time and live cells, the nature of the observed p53 upregulation and the functionality of the elevated p53.
The active status of the p53 elevated in response to PERP expression was initially indicated by the simultaneous increase in the p53-negative regulator protein, MDM2. Q-PCR analysis confirmed increased MDM2 transcription, which most likely occurs through the well-characterized autoregulatory p53–MDM2 feedback loop.10, 11 Single cell analysis corroborated that elevated levels of PERP protein positively influenced the transcription level of MDM2, which was p53 dependent. Thus, the p53 that is augmented in response to PERP expression is transcriptionally active and able to function in its transcriptional regulatory role.
In the absence of exogenous PERP, p53 was localized primarily in the cytoplasm of MEL202 cells. P53 localization outside the nucleus occurs in many tumour cells3 and was proposed as a mechanism employed by neoplastic cells to inactivate p53 transactivation function.2 Cytoplasmic retention of p53 renders tumour cells less responsive to genotoxic stress15 and correlates with resistance to chemo- and radiotherapy in many tumour types,15, 31, 32 although this does not preclude the transcriptional-independent, mitochondrial-related p53 function leading to apoptosis.33 Notably, GFP–PERP expression resulted in predominantly nuclear localization of p53 and MDM2, as well as higher MDM2 transcription. Colocalization of p53 and MDM2, in the cytoplasm or the nucleus, can promote the MDM2-mediated proteasomal degradation of p53 without nuclear to cytoplasmic transport.34 However, cytoplasmic localization of MDM2 in control cells lacking GFP–PERP expression suggested that nuclear-cytoplasmic shuttling of MDM2 is also occurring.16 Furthermore, time-lapse analysis of individual cells co-expressing GFP–PERP and MDM2–YFP provided evidence of the oscillatory nature of p53 regulation, manifested in regular pulses in MDM2 at 5–6-h intervals, analogous with previously reported oscillatory behaviour of p53 in DNA-damaged cells.20
Cell death following co-expression of GFP–PERP with MDM2–YFP was consistent with the previously observed level of apoptosis in GFP–PERP-expressing MEL202 cells,5 substantiating the finding that elevated PERP is sufficient to induce apoptosis in MEL202 cells via the caspase-dependent pathway,5 downstream of the p53/MDM2 feedback loop junction. Consistent apoptotic levels also confirm that the transcriptional changes in MDM2–YFP following GFP–PERP expression are representative of endogenous processes in MEL202 cells. Taken together, current findings indicate that the pro-apoptotic effect of PERP involves and may be amplified by the effect PERP expression has on its own transcriptional regulator p53. In line with our previous observations, PERP was targeted to the plasma membrane of MEL202 cells.5 A cytoplasmic and nuclear distribution of PERP has been recently described in renal tubular epithelial cells8 and the potential for interaction of PERP with p53 and/or MDM2 in these subcellular compartments cannot be ruled out at present.
Evidence that differential phosphorylation of key Ser residues of p53 occurs following PERP expression indicates that the elevated p53 is subject to posttranslational modifications that associate with p53 activation. Phosphorylation at p53Ser15, Ser20 and Ser37 can lower the affinity between p53 and MDM2, resulting in their reduced interaction and the subsequent accumulation of p53.13, 22, 23 Our results showed that p53Ser15 phosphorylation is reduced in cells expressing PERP, however, a slight but significant increase in the level of p53Ser20 phosphorylation was detected in the presence of elevated PERP. Phosphorylation at Ser20 enhances p53 tetramerization, nuclear accumulation, stability and transcriptional activity,23, 24 and therefore it is likely that this modification contributes to the stability and increased levels of nuclear p53 seen in response to PERP expression.
Of particular interest was the increased level of p53Ser46 phosphorylation; this modification is specifically involved in regulating the ability of p53 to induce apoptosis25 by preferentially activating the transcription of pro-apoptotic p53-target genes.14, 33 The p53 affinity switch from promoters of cell cycle arrest-related genes to apoptotic-related ones usually requires severe, non-repairable DNA damage.14, 25, 35 The increase in p53Ser46P induced by PERP expression is likely insufficient to drive the switch of gene–promoter targets to the level seen in response to genotoxic stress. However, we observed an upregulation of DR4 and LRDD, both p53-induced pro-apoptotic genes,30 with no change in the cell cycle arrest-related gene, p21. Detection of p53Ser46P in control cells (NT and GFP-only-transfected) suggests that other cell signals are contributing to p53 phosphorylation induction. PERP expression may enhance these signals or may render p53 more susceptible to Ser46 phosphorylation. Initial experiments suggest that p53Ser46 phosphorylation in response to PERP may occur by processes other than HIPK2 or p38 activation. Further studies will be required to elucidate the precise mechanism of p53 phosphorylation in response to PERP expression.
Our findings that PERP can act by increasing p53 protein, which is both transcriptionally active and subject to apoptosis-linked phosphorylation events, suggest the existence of an amplifying positive feedback loop at the functional level of p53 that may tip the balance in favour of p53-dependent apoptosis rather than cell cycle arrest and/or lead to increased PERP levels that in turn promote apoptosis. The finding that PERP can only partially reconstitute the level of apoptosis in p53-null cells4 also supports the existence of a PERP–p53 positive regulatory loop. Consequently, cells lacking functional PERP may also be impaired in their ability to regulate levels of p53.
Downregulation of PERP correlates with the aggressive monosomy-3 type of UM,17 a disease in which the upstream signalling to p53 is reportedly intact, but functional defects downstream of p53 were inferred.36 Many tumours express p53 mutants with a reduced ability to activate p53-target gene expression that normally respond to wild-type p53 activation.3, 37 This study demonstrates the transcriptional activity of p53 in MEL202 cells and reflects the reported low frequency of p53 mutations in primary UMs.36, 38, 39 Our previous findings that UM cells, including MEL202 cells, readily commit to apoptosis in response to upregulation of PERP expression5 imply that the downstream cell death pathway required for PERP-induced apoptosis is intact and functional in UM cells. The scenario of a downstream target of p53, such as PERP, influencing levels of its own transcriptional regulator is conceivable because loss of PERP function in these cells would not only have a detrimental effect on downstream signalling involving caspase-mediated apoptosis,5 but may also adjust p53 levels that influence the fine balance between cell death and survival. Deregulation of PERP appears as a mechanism engaged by UM cells to escape death by apoptosis.5, 17
In summary, this study identifies a novel role for PERP in stabilizing active p53 levels and places PERP at a signalling junction that can influence both upstream and downstream pathways of p53. Although interpretation of experiments involving manipulation of expression levels of p53-linked proteins should always be made with caution because of potentially abnormal or unregulated interactions with other proteins,3 our data show that increased PERP expression affects several aspects of p53 regulation, including increased protein stability, apoptosis-promoting posttranslational modifications, enhanced nuclear accumulation, and oscillatory regulation. Our findings highlight a potentially important target for development of cancer therapies aimed at enhancing p53 apoptotic activity and reconstituting the p53-dependent apoptotic pathway.
Materials and Methods
Cell culture
Tissue culture media and supplements were from Invitrogen (Paisley, UK), unless otherwise stated. The UM cell line MEL20219 was grown in RPMI-1640 medium with 2 mM -glutamine and 25 mM HEPES supplemented with 10% v/v heat-inactivated fetal calf serum (FCS; Biosera, East Sussex, UK), 1 mM sodium pyruvate and 1% v/v non-essential amino acids. U2OS cells (ATCC No: HTB-96; LGC Standards, Teddington, Middlesex, UK) were grown in Dulbecco's modified Eagle's medium with -glutamine and 10% FCS. Cells were grown at 37 °C in controlled humidity and 5% CO2 atmosphere.
Plasmid constructs and transient transfection of cell lines
The pcDNA-DEST53 plasmid containing GFP fused to the N-terminus of the full-length open-reading frame (ORF) of human PERP (GFP–PERP) under the CMV promoter was constructed as described previously.5 The pEGFP-N1 vector (Clontech, Palo Alto, CA, USA) encoding GFP-only and pG-A-EYFP vector encoding YFP-only (kindly supplied by Louise Ashall, University of Liverpool, UK) served as controls. The pU293 plasmid encoding human MDM2 fused to enhanced YFP under the control of the human MDM2 native promoter (MDM2–YFP20) and pU265 containing p53 ORF fused to DsRed-Express RFP under a zinc-inducible pMT promoter (p53-RFP) were kind gifts from Galit Lahav (Weizmann Institute of Science, Israel) and Rachel Nelson (University of Liverpool, UK), respectively. To induce p53–RFP expression, zinc chloride (Sigma-Aldrich, Poole, UK) was added to cell media in a final concentration of 100 μM, 6-h PT. The pp53-EGFP Vector (Clontech) encoded p53 fused to enhance GFP (p53–GFP) under the CMV promoter. Transient transfections were performed using GeneJuice Transfection Reagent (Novagen, Merck Biosciences Ltd., Darmstadt, Germany). Cells were transfected with 1 μg plasmid DNA per 35 mm dish. For co-transfections, the total amount of plasmid DNA used was always 1 μg.
Laser scanning confocal fluorescence microscopy of live transfected cells
Live cell imaging of transfected cells was performed in an incubator XL (PeCon GnbH) on the microscope stage that was humidity controlled at 37 °C with 5% CO2. Cells were imaged using a LSM 5 EXCITER Laser Scanning Microscope (Carl Zeiss, Welwyn Garden City, Hertfordshire, UK) using a Fluar 40X/1.3 NA oil immersion objective. Images were taken from 20-h PT. Excitation of EGFP was achieved with a 488 nm laser, and the emitted light was collected through a 505–530 nm bandpass filter from a 545 nm dichroic mirror (515 nm dichroic mirror when in combination with YFP); YFP was excited at 514 nm (or at 488 nm when in combination with RFP) and its emission detected through a 515 nm dichroic mirror and a 505–550 nm BP filter; RFP was excited at 543 nm and its emission detected through a 545 nm dichroic mirror and a 560 nm longpass filter. Data acquisition and analysis were carried out with LSM510 AIM software, version 4.2 (Carl Zeiss) and ImageJ 1.43h.40 Cell death was assessed as described previously.5
PFTα treatment
Cells were treated with 30 μM PFTα (Sigma-Aldrich) at 6-h PT and images were taken from 20-h PT.
Western blot analysis and antibodies
Lysates were prepared from cells at the indicated times as described previously5 and resolved by SDS-PAGE before transfer to nitrocellulose membrane. The membranes were incubated overnight at 4 °C with the appropriate primary antibody diluted with 5% w/v non-fat dry milk or 5% bovine serum albumin (BSA) in tris-buffered saline-Tween-20 buffer (TBS-T) buffer (20 mM Tris-HCl, 150 mM NaCl, pH 7.6 with 0.1% v/v Tween-20) as recommended by the manufacturer. The primary antibody dilutions used were as follows: anti-PERP polyclonal (1 : 1000 dilution) and anti-GAPDH monoclonal (clone 6C5, 1 : 10 000) (Abcam, Cambridge, UK); anti-p53 monoclonal (clone DO-1, 1 : 1000) and anti-MDM2 monoclonal (clone 2A10, 1 : 1000) (Calbiochem, Merck Biosciences Ltd., Darmstadt, Germany); anti-HIPK2 monoclonal (clone F-189, 1 : 200) (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA); anti-phospho-p38 MAPK monoclonal (Thr180/Tyr182) (clone 3D7, 1 : 1000) and Phospho-p53 Antibody Sampler Kit including anti-phospho-p53 (Ser15 monoclonal; Ser20, Ser37, Ser46, polyclonal; 1 : 1000) and anti-p53 monoclonal (clone 7F5, 1 : 1000) (Cell Signaling Technology, Beverly, MA, USA). Immunocomplexes were detected with horseradish peroxidase-conjugated secondary antibody (Sigma-Aldrich) by a chemiluminescent method (Amersham ECL Western Blotting Detection Reagents, GE Healthcare Life Sciences, Little Chalfont, Buckinghamshire, UK), with 2-min exposure to autoradiography films unless otherwise stated. Sequential probing of blots with different antibodies was achieved by incubation in stripping solution (62.5 mM Tris-HCl, pH 6.8, 100 mM β-mercaptoethanol, 2% w/v SDS) at 65oC for 30 min, followed by 2 × 10 min washes in TBS-T at room temperature, before blocking in 5% milk/TBS-T and reprobing with antibody. Detection of phospho-p53 products was carried out using separate blots of identical samples rather than sequential probing of the same blot to prevent carry-over of previously detected p53 products. Blots were probed with anti-GAPDH to confirm equal loading of samples. Relative intensities of the protein bands were obtained by densitometry using ImageJ (Rasband, WS, ImageJ, US National Institutes of Health, Bethesda, MD, USA, http://rsb.info.nih.gov/ij/, 1997–2007) and normalized to endogenous GAPDH. Western blot images presented are representative of three independent experiments.
Immunofluorescence
MEL202 cells were seeded in eight-well Lab-Tek Chamber Slide (Nunc, Rochester, NY, USA) 24 h before transfection. Media was removed 24 h following transfection and cells were washed in phosphate-buffered saline (PBS). Cells were fixed with freshly prepared 4% paraformaldehyde in PBS for 10 min followed by 3 × 5 min washes. Following permeabilisation with 0.3% Triton X-100 in PBS for 5 min and a 5 min PBS wash, cells were incubated in blocking buffer (5% normal goat serum in PBS) for 60 min before incubating with primary antibody diluted in antibody dilution buffer (1% BSA in PBS) for 60 min. Cells were washed for 3 × 5 min in PBS before addition, where necessary, of diluted fluorochrome-conjugated secondary antibody for 45 min, followed by 3 × 5 min washes in PBS. All incubations were carried out at room temperature. Cells were coverslipped with Fluorescence Mounting Medium (Dako, Cambridgeshire, UK) and examined using a LSM 5 EXCITER Laser Scanning Microscope (Carl Zeiss) with excitation at 633 nm. The primary antibody dilutions used were: anti-MDM2 monoclonal (clone 2A10, 1 : 100) (Calbiochem, Merck Biosciences); anti-p53 Alexa Fluor 647 Conjugate monoclonal (clone 1C12, 1 : 50) (Cell Signaling Technology). The secondary antibody used for detection of MDM2 was Alexa Fluor 633 goat anti-mouse IgG (H+L; 1 : 500) (Invitrogen).
Real-time quantitative PCR
RNA was isolated from cells using RNeasy Kit (Qiagen Ltd. West Sussex, UK) with on-column DNase I treatment for removal of genomic DNA. First-strand cDNA was synthesized from 1 μg RNA using Superscript II Reverse Transcriptase (Invitrogen). Real-time Q-PCR was performed using qPCR Core Kit for SYBR Green I (Eurogentec, Southampton, UK) as described previously.5 The gene-specific primer set used for PERP was 5′-GGCTTCATCATCCTGGTGAT-3′ and 5′-ACAGCAGCCAAGGCAAGGAG-3′ for p53, 5′-ATCTACAAGCAGTCACAGCAC-3′ and 5′-TTCCTTCCACTCGGATAAGATGC-3′ and for MDM2, 5′-CCTGGCTCTGTGTGTAATAAG-3′ and 5′- ATCCAACCAATCACCTGAATG-3′. Primer sets for the p53-target genes30 PUMA, DR4, DR5, LRDD and p21 were purchased from SABiosciences (Qiagen). Q-PCR data were normalized to GAPDH (primer set: 5′-AACAGCCTCAAGATCATCAG-3′ and 5′-TGAGTCCTTCCACGATACC-3′). Amplifications were performed in triplicate using a Stratagene MX3000P QPCR System (Stratagene, La Jolla, CA, USA). Q-PCR data are expressed relative to a calibrator sample assigned an arbitrary quantity of 1.
Acknowledgments
We are grateful to Bruce Ksander and Martine Jager for the gift of the MEL202 cell line, Galit Lahav and Rachel Nelson for the generous gifts of MDM2-YFP and p53-RFP plasmid constructs, respectively, and Louise Ashall for the YFP-only plasmid. The technical support given by Daniel Brotchie is acknowledged. This work was generously supported by The Humane Research Trust, UK.
Glossary
- PERP
p53 apoptosis effector related to PMP-22
- Ser
serine
- NT
non-transfected
- GFP
green fluorescent protein
- Q-PCR
real-time quantitative PCR
- YFP
yellow fluorescent protein
- PFTα
pifithrin-α
- RFP
red fluorescent protein
- HIPK2
homeodomain-interacting protein kinase 2
- p38
p38 mitogen-activated protein kinase (MAPK)
- DR4
death receptor 4
- LRDD
leucine-rich repeats and death domain containing
- p21
cyclin-dependent kinase inhibitor 1A
- UM
uveal melanoma
- ORF
open-reading frame
- PT
post-transfection
- BSA
bovine serum albumin
- TBS-T buffer
tris-buffered saline-Tween-20 buffer
- PBS
phosphate-buffered saline
The authors declare no conflict of interest.
Footnotes
Edited by A Stephanou
References
- Levine AJ, Hu W, Feng Z. The P53 pathway: what questions remain to be explored. Cell Death Differ. 2006;13:1027–1036. doi: 10.1038/sj.cdd.4401910. [DOI] [PubMed] [Google Scholar]
- Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- Attardi LD, Reczek EE, Cosmas C, Demicco EG, McCurrach ME, Lowe SW, et al. PERP, an apoptosis-associated target of p53, is a novel member of the PMP-22/gas3 family. Genes Dev. 2000;14:704–718. [PMC free article] [PubMed] [Google Scholar]
- Davies L, Gray D, Spiller D, White MR, Damato B, Grierson I, et al. P53 apoptosis mediator PERP: localization, function and caspase activation in uveal melanoma. J Cell Mol Med. 2009;13:1995–2007. doi: 10.1111/j.1582-4934.2008.00590.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihrie RA, Reczek E, Horner JS, Khachatrian L, Sage J, Jacks T, et al. Perp is a mediator of p53-dependent apoptosis in diverse cell types. Curr Biol. 2003;13:1985–1990. doi: 10.1016/j.cub.2003.10.055. [DOI] [PubMed] [Google Scholar]
- Nowak M, Koster C, Hammerschmidt M. Perp is required for tissue-specific cell survival during zebrafish development. Cell Death Differ. 2005;12:52–64. doi: 10.1038/sj.cdd.4401519. [DOI] [PubMed] [Google Scholar]
- Singaravelu K, Devalaraja-Narashimha K, Lastovica B, Padanilam BJ. PERP, a p53 proapoptotic target, mediates apoptotic cell death in renal ischemia. Am J Physiol Renal Physiol. 2009;296:F847–F858. doi: 10.1152/ajprenal.90438.2008. [DOI] [PubMed] [Google Scholar]
- Harris SL, Levine AJ. The p53 pathway: positive and negative feedback loops. Oncogene. 2005;24:2899–2908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]
- Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- Appella E, Anderson CW. Post-translational modifications and activation of p53 by genotoxic stresses. Eur J Biochem. 2001;268:2764–2772. doi: 10.1046/j.1432-1327.2001.02225.x. [DOI] [PubMed] [Google Scholar]
- Shieh SY, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- Mayo LD, Seo YR, Jackson MW, Smith ML, Rivera Guzman J, Korgaonkar CK, et al. Phosphorylation of human p53 at serine 46 determines promoter selection and whether apoptosis is attenuated or amplified. J Biol Chem. 2005;280:25953–25959. doi: 10.1074/jbc.M503026200. [DOI] [PubMed] [Google Scholar]
- O'Brate A, Giannakakou P. The importance of p53 location: nuclear or cytoplasmic zip code. Drug Resist Updat. 2003;6:313–322. doi: 10.1016/j.drup.2003.10.004. [DOI] [PubMed] [Google Scholar]
- Tao W, Levine AJ. Nucleocytoplasmic shuttling of oncoprotein Hdm2 is required for Hdm2-mediated degradation of p53. Proc Natl Acad Sci USA. 1999;96:3077–3080. doi: 10.1073/pnas.96.6.3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraoan L, Gray D, Hiscott P, Ebrahimi B, Damato B, Grierson I. Expression of p53-induced apoptosis effector PERP in primary uveal melanomas: downregulation is associated with aggressive type. Exp Eye Res. 2006;83:911–919. doi: 10.1016/j.exer.2006.04.016. [DOI] [PubMed] [Google Scholar]
- Hildebrandt T, Preiherr J, Tarbe N, Klostermann S, Van Muijen GN, Weidle UH. Identification of THW, a putative new tumor suppressor gene. Anticancer Res. 2000;20:2801–2809. [PubMed] [Google Scholar]
- Verbik DJ, Murray TG, Tran JM, Ksander BR. Melanomas that develop within the eye inhibit lymphocyte proliferation. Int J Cancer. 1997;73:470–478. doi: 10.1002/(sici)1097-0215(19971114)73:4<470::aid-ijc3>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Lahav G, Rosenfeld N, Sigal A, Geva-Zatorsky N, Levine AJ, Elowitz MB, et al. Dynamics of the p53-Mdm2 feedback loop in individual cells. Nat Genet. 2004;36:147–150. doi: 10.1038/ng1293. [DOI] [PubMed] [Google Scholar]
- Komarov PG, Komarova EA, Kondratov RV, Christov-Tselkov K, Coon JS, Chernov MV, et al. A chemical inhibitor of p53 that protects mice from the side effects of cancer therapy. Science. 1999;285:1733–1737. doi: 10.1126/science.285.5434.1733. [DOI] [PubMed] [Google Scholar]
- Tibbetts RS, Brumbaugh KM, Williams JM, Sarkaria JN, Cliby WA, Shieh SY, et al. A role for ATR in the DNA damage-induced phosphorylation of p53. Genes Dev. 1999;13:152–157. doi: 10.1101/gad.13.2.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chehab NH, Malikzay A, Stavridi ES, Halazonetis TD. Phosphorylation of Ser-20 mediates stabilization of human p53 in response to DNA damage. Proc Natl Acad Sci USA. 1999;96:13777–13782. doi: 10.1073/pnas.96.24.13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh SY, Taya Y, Prives C. DNA damage-inducible phosphorylation of p53 at N-terminal sites including a novel site, Ser20, requires tetramerization. EMBO J. 1999;18:1815–1823. doi: 10.1093/emboj/18.7.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda K, Arakawa H, Tanaka T, Matsuda K, Tanikawa C, Mori T, et al. p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell. 2000;102:849–862. doi: 10.1016/s0092-8674(00)00073-8. [DOI] [PubMed] [Google Scholar]
- Perfettini JL, Castedo M, Nardacci R, Ciccosanti F, Boya P, Roumier T, et al. Essential role of p53 phosphorylation by p38 MAPK in apoptosis induction by the HIV-1 envelope. J Exp Med. 2005;201:279–289. doi: 10.1084/jem.20041502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulavin DV, Saito S, Hollander MC, Sakaguchi K, Anderson CW, Appella E, et al. Phosphorylation of human p53 by p38 kinase coordinates N-terminal phosphorylation and apoptosis in response to UV radiation. EMBO J. 1999;18:6845–6854. doi: 10.1093/emboj/18.23.6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Orazi G, Cecchinelli B, Bruno T, Manni I, Higashimoto Y, Saito S, et al. Homeodomain-interacting protein kinase-2 phosphorylates p53 at Ser 46 and mediates apoptosis. Nat Cell Biol. 2002;4:11–19. doi: 10.1038/ncb714. [DOI] [PubMed] [Google Scholar]
- Hofmann TG, Moller A, Sirma H, Zentgraf H, Taya Y, Droge W, et al. Regulation of p53 activity by its interaction with homeodomain-interacting protein kinase-2. Nat Cell Biol. 2002;4:1–10. doi: 10.1038/ncb715. [DOI] [PubMed] [Google Scholar]
- Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- Bosari S, Viale G, Roncalli M, Graziani D, Borsani G, Lee AK, et al. p53 gene mutations, p53 protein accumulation and compartmentalization in colorectal adenocarcinoma. Am J Pathol. 1995;147:790–798. [PMC free article] [PubMed] [Google Scholar]
- Moll UM, LaQuaglia M, Benard J, Riou G. Wild-type p53 protein undergoes cytoplasmic sequestration in undifferentiated neuroblastomas but not in differentiated tumors. Proc Natl Acad Sci USA. 1995;92:4407–4411. doi: 10.1073/pnas.92.10.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousden KH. Outcomes of p53 activation – spoilt for choice. J Cell Sci. 2006;119:5015–5020. doi: 10.1242/jcs.03293. [DOI] [PubMed] [Google Scholar]
- Xirodimas DP, Stephen CW, Lane DP. Cocompartmentalization of p53 and Mdm2 is a major determinant for Mdm2-mediated degradation of p53. Exp Cell Res. 2001;270:66–77. doi: 10.1006/excr.2001.5314. [DOI] [PubMed] [Google Scholar]
- Rinaldo C, Prodosmo A, Mancini F, Iacovelli S, Sacchi A, Moretti F, et al. MDM2-regulated degradation of HIPK2 prevents p53Ser46 phosphorylation and DNA damage-induced apoptosis. Mol Cell. 2007;25:739–750. doi: 10.1016/j.molcel.2007.02.008. [DOI] [PubMed] [Google Scholar]
- Sun Y, Tran BN, Worley LA, Delston RB, Harbour JW. Functional analysis of the p53 pathway in response to ionizing radiation in uveal melanoma. Invest Ophthalmol Vis Sci. 2005;46:1561–1564. doi: 10.1167/iovs.04-1362. [DOI] [PubMed] [Google Scholar]
- Yee KS, Vousden KH. Complicating the complexity of p53. Carcinogenesis. 2005;26:1317–1322. doi: 10.1093/carcin/bgi122. [DOI] [PubMed] [Google Scholar]
- Brantley MA, Jr, Harbour JW. Deregulation of the Rb and p53 pathways in uveal melanoma. Am J Pathol. 2000;157:1795–1801. doi: 10.1016/s0002-9440(10)64817-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishore K, Ghazvini S, Char DH, Kroll S, Selle J. p53 gene and cell cycling in uveal melanoma. Am J Ophthalmol. 1996;121:561–567. doi: 10.1016/s0002-9394(14)75431-5. [DOI] [PubMed] [Google Scholar]
- Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics Int. 2004;11:36–42. [Google Scholar]