A decline in the quality-control machinery causes damaged proteins to accumulate as cells age. Metabolic signals such as insulin/IGF-1 may counteract this and help protect against age-associated diseases such as Alzheimer's.
Abstract
Aging cells accumulate damaged and misfolded proteins through a functional decline in their protein homeostasis (proteostasis) machinery, leading to reduced cellular viability and the development of protein misfolding diseases such as Alzheimer’s and Huntington’s. Metabolic signaling pathways that regulate the aging process, mediated by insulin/IGF-1 signaling, dietary restriction, and reduced mitochondrial function, can modulate the proteostasis machinery in many ways to maintain a youthful proteome for longer and prevent the onset of age-associated diseases. These mechanisms therefore represent potential therapeutic targets in the prevention and treatment of such pathologies.
PATHWAYS THAT INFLUENCE THE AGING PROCESS
Aging was once regarded as a stochastic, progressive decline. However, the discovery of metabolic pathways able to modulate the aging process has challenged this view. Three main signaling pathways have been identified that can influence the rate of aging (Fig. 1) (Wolff and Dillin 2006). The first, dietary restriction (DR) has been shown to extend lifespan in multiple species (Mair and Dillin 2008). The reduction of dietary intake below unlimited or “ad libitum” levels causes an increase in lifespan, to an optimum point of consumption, typically around 60% of ad libitum food intake. However, although this extension of lifespan by DR has been observed for several decades, its mechanistic basis remains unclear. Evidence suggests that the amino-acid-sensing serine/threonine kinase mTOR, and the energy status-dependent kinase AMPK, are involved in DR signaling (Kapahi and Zid 2004; Kahn et al. 2005). In addition, recent studies in Caenorhabditis elegans have suggested that two transcription factors, PHA-4 and SKN-1, are essential for this process, and act specifically in lifespan extension by DR (Panowski et al. 2007; Bishop and Guarente 2007).
Figure 1.
Pathways that regulate aging. The insulin-signaling pathway and dietary restriction pathway induce longevity through mechanisms that are at least partially understood. Reduced mitochondrial function also increases lifespan, but the signaling components are not clear.
Another pathway mediating lifespan extension, the insulin/IGF-1-like signaling pathway (IIS), has been remarkably well characterized in recent years. Reduced IIS activity extends lifespan in both invertebrate and vertebrate species (Bartke 2008). The role of the IIS system in aging was first identified in C. elegans, and it has been best characterized in that species (Panowski and Dillin, 2008). Following ligand binding, the C. elegans IIS receptor DAF-2 recruits the insulin receptor substrate homolog IST-1, and the PI3K AGE-1 (Wolkow and Ruvkun 2002; Morris et al. 1996). Production of PI3 through AGE-1 activity, opposed by the phosphatase DAF-18, activates AKT kinases, which phosphorylate the transcription factor DAF-16 (Hertweck et al. 2004; Paradis and Ruvkun 1998; Ogg and Ruvkun 1998; Henderson and Johnson 2001; Lee et al. 2001; Lin et al. 2001). Phosphorylation anchors DAF-16 in the cytosol through interaction with 14-3-3 proteins, whereas reduced IIS activity allows it to enter the nucleus, activating a diverse transcriptional profile that promotes extended lifespan (Lin et al. 1997; Ogg et al. 1997; Cahill et al. 2001). DAF-16 is also regulated by mechanisms other than localization, and requires cofactors such as SMK-1, HCF-1, and SIR-2 for extension of longevity (Wolff et al. 2006; Tissenbaum and Guarente 2001). At least one other transcription factor, the heat shock factor HSF-1, is also required for the IIS pathway to extend lifespan, although how the pathway regulates this transcription factor is unclear (Hsu et al. 2003). In addition, in turn, the IIS pathway also regulates localization of the SKN-1 transcription factor that is essential for the long lifespan of DR animals (Tullet et al. 2008).
The third pathway enabling extension of lifespan acts through reduction in the activity of the mitochondrial electron transport chain (ETC). This was first shown, again, in C. elegans, in which reduced expression of several mitochondrial genes including components of ETC complexes I, III, IV, and V by RNAi is sufficient to extend lifespan (Feng et al. 2001; Dillin et al. 2002b; Lee et al. 2003). The role of mitochondria in lifespan has since been shown in Drosophila and rodents (Liu et al. 2005; Copeland et al. 2009). However, the mechanisms and signaling pathways involved remain unknown.
Several lines of evidence suggest that these lifespan extension pathways are separate. For one, genetic epistasis experiments show that combining different lifespan extension pathways leads to additive increases in lifespan (Dillin et al. 2002b; Lakowski and Hekimi 1998). Additionally, temporal requirements for the pathways are different, reduction of IIS or DR has important lifespan extension effects in adulthood, whereas the mitochondrial pathway acts during development (Dillin et al. 2002a; Dillin et al. 2002b; Mair et al. 2003), and the core components act separately. DAF-16 is dispensable for DR-induced lifespan extension, whereas PHA-4 plays a role specifically in DR and not the IIS and ETC pathways (Lakowski and Hekimi 1998; Panowski et al. 2007).
THE IMPORTANCE OF PROTEOSTASIS IN AGING
Although these lifespan extension pathways act independently, it is logical to hypothesize that their underlying downstream mechanisms might be the same. The phenotypes of long-lived animals overlap substantially, including increased stress resistance, altered metabolism, and delayed reproduction and development (Hekimi and Guarente 2003). One possibility is that a major downstream function of all these aging pathways is to change the way that the cellular proteome is maintained. Cellular proteins are challenged throughout life by a multitude of factors that cause protein misfolding and aggregation, including translational error, the presence of polymorphisms, and stresses that lead to covalent modifications such as oxidation. Misfolded proteins can have a range of negative consequences for the cell. Mutated and destabilized proteins with hydrophobic regions inappropriately exposed tend to aggregate with the hydrophobic regions of other proteins, and these protein aggregates can be directly cytotoxic, through disruption of membranes and interaction with cellular components (Chiti et al. 2003; Stefani and Dobson 2003). They may also create increased demands on the cell’s protein homeostasis (proteostasis) machinery, titrating away components of this network and consequently leading to further misfolding of other proteins. The consequences of even a low level of misfolded protein in the cell can be catastrophic—the expression of a metastable or misfolded protein in organismal models of proteotoxic stress has been shown to destabilize other, unrelated proteins in the proteome (Gidalevitz et al. 2006; Gidalevitz et al. 2009).
The importance of preventing protein misfolding has been shown in evolutionary terms in a model of molecular evolution, studying synonymous codon substitutions (Drummond and Wilke 2008). Translational inaccuracy is a major source of mutation, missense errors in translation are believed to occur once every 103–104 codons, suggesting that around 18% of translated proteins will contain an error, and some codons are translated more accurately than others. Mutation in turn leads to higher levels of protein misfolding and aggregation. The model found that the avoidance of translational error through the selection of the most accurately translated codons, driven by the negative consequences of misfolded mutant proteins, was sufficient to account for observed rates of codon evolution. Consistent with this hypothesis, the most conserved proteins are not those most essential for viability, but the ones expressed at the highest rate, and therefore the most damaging if mistranslated. Proteins expressed in long-lived postmitotic cells, in which the formation of misfolded proteins has the most potential to do harm, were also extremely slow evolving. The fact that the avoidance of protein misfolding seems to have been such a driving force in evolution indicates in turn the danger that protein misfolding presents to the cell.
Cells possess a complex network of mechanisms designed to prevent and eliminate protein misfolding, termed the proteostasis network, encompassing regulators of translation, protein folding, trafficking and secretion, and protein degradation (Fig. 2) (Balch et al. 2008). However, despite the presence of these mechanisms, unwanted protein products accumulate with age. Older cells contain more proteins bearing oxidative modifications such as carbonylation, oxidized methionine, and glycation, as well as accumulating cross-linked and aggregated proteins, and less catalytically active enzyme populations (Berlett and Stadtman 1997; Soskic et al. 2008). This accumulation of misfolded proteins seen in elderly organisms seems highly likely to contribute to tissue decline and loss of viability.
Figure 2.
The life cycle of a protein. After synthesis, proteins require correct folding and trafficking to the correct cellular location. Components of the cell’s proteostasis machinery mediate these functions, as well as detoxifying damaged and misfolded proteins. Pathways that regulate aging can modulate many aspects of proteostasis, through transcription factors such as DAF-16 and HSF-1, and effector molecules such as the kinase mTOR.
The implication is that a loss of function of the proteostasis machinery occurs with age, and recent studies have supported this hypothesis. The causes of age-associated decline in proteostasis capacity are still largely unclear, but there is mounting evidence that aging pathways can directly modulate elements of the proteostasis machinery to extend healthy lifespan (Fig. 2). Given this connection between proteome maintenance and the aging process, understanding how pathways that can regulate aging affect the proteostasis machinery is necessary in gaining an understanding of how to increase healthy lifespan. It is this intersection between pathways that modulate aging, and protein homeostasis, that will be discussed in the rest of this review.
RATES OF TRANSLATION
Modulating rates of protein synthesis is one major way in which aging pathways can alter proteostasis. Control of translation rate is generally achieved at two stages of translation initiation—the recruitment of the 40S ribosomal subunit, and the loading of this subunit with the initiator tRNA (Gebauer and Hentze 2004). Recruitment of the 40S subunit is controlled by the eIF4E translation initiation factor, which is inhibited by the 4EBP binding proteins (Richter and Sonenberg 2005). tRNA loading is regulated by the eIF2a subunit, which controls the recycling of eIF2 (Gebauer and Hentze 2004).
The IIS and DR pathways both affect translation rates. Protein synthesis is reduced under DR, possibly through reduced activation of the mTOR kinase (Zid et al. 2009; Wullschleger et al. 2006). mTOR regulates translation by phosphorylation of its targets, 4EBP-1, and the ribosomal kinase S6K. Phosphorylation of 4EBP-1 dissociates it from the translation initiation complex, whereas phosphorylation of S6K activates this kinase, allowing it to phosphorylate translational targets that include ribosomal protein S6 and the elongation factor 2 kinase. Both events promote translation, and conversely, reduction of mTOR activity by DR decreases translation initiation. In Drosophila, d4EBP-1 is required for lifespan extension under DR, and is under the transcriptional control of the FOXO transcription factor (Zid et al. 2009; Puig et al. 2003). Insulin can regulate TOR activity through modifying the activity of the upstream TSC1/TSC2 regulatory complex and phosphorylation of S6K, and reduced Insulin/IGF-1 signaling in long-lived Snell and Ames dwarf mice leads to reduced translation (Manning 2004; Hsieh and Papaconstantinou 2004; Sharp and Bartke 2005).
Indeed, reducing translation rates alone is sufficient to extend lifespan. Reducing TOR function directly increases longevity in multiple species, as does reduced S6K activity (Vellai et al. 2003; Kapahi et al. 2004; Kaeberlein et al. 2005; Jia et al. 2004; Hansen et al. 2007; Selman et al. 2009). In yeast, deletion or inhibition of ribosomal protein levels increases replicative lifespan (Chiocchetti et al. 2007). In addition, in C. elegans, an RNAi screen for longevity-enhancing genes identified many translation regulators (Curran and Ruvkun, 2007). Additionally, reduction of the worm homologs of translation initiation factors eIF4E (ife-2), eIF-4G (ifg-1), eIF-2B (iftb-1), and a range of ribosomal proteins, increases lifespan, whereas ifg-1 is reduced in long-lived dauers (Hansen et al. 2007; Pan et al. 2007; Syntichaki et al. 2007). Initiation factor knockdown may be dependent on DAF-16 to increase lifespan, whereas ribosomal proteins and S6K are independent of DAF-16, suggesting a complicated relationship between IIS and the control of translation. In addition, lifespan extension by translational inhibition and DR or mTOR reduction are additive, suggesting that these signaling pathways also invoke other mechanisms to extend lifespan (Hansen et al. 2007).
It seems that reduction in translation is an important mechanism in the extension of lifespan. What therefore underlies this mechanistic importance? One explanation might be that reduced protein synthesis reduces the load on the remainder of the proteostasis machinery, allowing more efficient protein folding and degradation, for example, and consequently reducing the load of misfolded and damaged proteins. This is substantiated by the observation that reducing translation rates renders cells tolerant to elevated temperature, a stress that causes protein misfolding (Hansen et al. 2007).
Reducing translation also causes preferential translation of certain mRNAs. DR in flies allows preferential translation of mitochondrial genes, including components of complexes I and IV, dependent on 4EBP1 and therefore reduced translation. This altered translational profile enhances mitochondrial activity and facilitates lifespan extension (Zid et al. 2009). In yeast, deletion of 60S ribosomal subunits increases lifespan through the preferential translation of the GCN4 transcription factor, inducing a transcriptional profile necessary for full lifespan extension by DR (Steffen et al. 2008). And finally, recent work suggests that reduced IIS may increase intrinsic thermotolerance and lifespan through preferential translation of specific transcripts (McColl et al. 2010).
PROTEIN FOLDING
Once proteins have been synthesized, the majority do not fold spontaneously into the appropriate conformation. They require assistance, provided in the form of molecular chaperones, and this assisted protein folding is crucial to maintaining protein homeostasis (Morimoto 2008). The binding of chaperones to nascent polypeptides both prevents folding into inappropriate forms, and facilitates the assembly of the correct structure. This assembly involves the stabilization of folding-competent intermediate states, and can also involve active ATP-driven folding into the native form, accomplished by the HSP70 family of chaperones (Buckau et al. 2006). In the case of proteins that have formed unwanted and potentially toxic aggregates, some chaperones, such as HSP104, are able to disassemble these aggregates into intermediate forms that can then be assisted in refolding to the native state. On the other hand, at high concentrations of aggregative proteins, HSP104, TRiC, and potentially other chaperones, can actively aggregate these toxic species into less toxic types of aggregates (Behrends et al. 2006; Shorter and Lindquist 2004).
Chaperones represent many of the transcriptional targets of the C. elegans DAF-16 IIS transcription factor. Heat shock proteins, particularly of the HSP-16 and HSP-12 families, are prominently up-regulated by reduction of daf-2 signaling as determined by microarray, SAGE, CHIP, and mass spectrometry analysis (Murphy et al. 2003; McElwhee et al. 2003; Halaschek-Wiener et al. 2005; Oh et al. 2006; Dong et al. 2007). Heat shock proteins are in fact required for full lifespan extension in daf-2 mutants, and over-expression of HSP-16 is sufficient to extend lifespan in both wild type and long-lived mutant worms (Murphy et al. 2003; Morley and Morimoto 2004; Walker and Lithgow 2003). Expression of small heat shock proteins is also sufficient to extend lifespan in Drosophila (Morrow et al. 2004). This suggests that the ability to correctly fold proteins is important in maintaining youthful cells, and is a target of lifespan extension pathways.
STRESS RESPONSES
The level of chaperones in a cell is believed to be finely tuned to that cells individual protein folding requirements. Introduction of a new, folding-sensitive protein to cells can result in a subsequent destabilization and decreased folding of the rest of the proteome, suggesting that normal chaperone capacity can be easily exceeded by increases in demand (Gidalevitz et al. 2006; Gidalevitz et al. 2009). Many stresses perturb protein folding, necessitating a dramatic up-regulation of chaperones and other proteostasis components if cells are to survive this challenge to their proteome. The responses to different cellular stresses are coordinated by upstream transcription factors that respond to distinct stimuli, and several of these stress responses have been implicated in aging and the extension of longevity (Morimoto 2008).
THE HEAT SHOCK RESPONSE
The heat shock response coordinates the rapid induction of HSPs in response to thermal stress, and is controlled by the HSF transcription factor family. Mammals possess HSFs 1, 2, and 4, with HSF1 acting in a ubiquitous stress-inducible manner, whereas Drosophila, C. elegans, and yeast possess only HSF1 orthologs (Pirkkala et al. 2001; Anckar and Sistonen 2007; McMillan et al. 1998).
In C. elegans the HSF-1 transcription factor is required for lifespan extension by both the IIS and DR pathways, whereas its reduction by RNAi in wild type worms causes a premature aging phenotype (Hsu et al. 2003; Morley and Morimoto 2004; Steinkraus et al. 2008). DAF-16 and HSF-1 function in a very connected way—as well as a requirement for HSF-1 in mediating DAF-16-dependent lifespan extension, DAF-16 activation confers heat shock resistance, and both appear to function in concert with the sirtuin SIRT1 (Hsu et al. 2003; Morley and Morimoto 2004; Lithgow et al. 1995; Tissenbaum et al. 2001; Westerheide et al. 2009). In turn, over-expression of HSF-1 enhances the folding of aggregative proteins, and can extend lifespan, as can administration of a sublethal dose of heat shock in both C. elegans and Drosophila (Hsu et al. 2003; Morley and Morimoto 2004; Lithgow et al. 1995; Hercus et al. 2003). The importance of the heat shock response in lifespan is reflected in a screen for thermal stress resistant worms, a high percentage of which also showed a lifespan extension of more than 15% (Munoz and Riddle 2003).
The heat shock response therefore seems to form an important component of lifespan extension pathways, and without the HSR, animals age at an accelerated rate. The ability to induce the HSR declines with age and this appears to be a relatively early event in the aging process, occurring shortly after the reproductive period in C. elegans (Ben-Zvi et al. 2009). The mechanisms underlying the loss of the response are unclear, but it seems likely that losing the ability to induce the HSR and therefore cope with increases in protein misfolding might underlie deterioration associated with aging.
THE HYPOXIA RESPONSE
The stress response to low oxygen conditions, termed hypoxia, is regulated by the HIF1 transcription factor (Lendahl et al. 2009). Under conditions of normoxia, prolyl hydroxylases hydroxylate HIF1 at specific proline residues. This hydroxylation targets HIF1 for proteosomal degradation through recognition by the VHL E3 ubiquitin ligase. In hypoxia HIF1 is not targeted for degradation and is able to activate a transcriptional program required for survival in low oxygen conditions. This program includes the up-regulation of HSPs, which is in turn partly dependent on up-regulation of HSF1 transcription by HIF1 (Baird et al. 2006).
Recent data suggests that HIF-1 plays a role in regulating longevity in C. elegans. The picture, however, is confusing. Although one report has suggested that increasing active HIF-1 by knockdown of its negative regulator VHL-1 increases lifespan, independent of other lifespan extension pathways, another report suggests the opposite—that null mutations in HIF-1 increase lifespan, in a manner not additive to the lifespan extension observed in S6K mutants, suggesting that it lies downstream of the TOR pathway (Mehta et al. 2009; Chen et al. 2009). In this study, knockdown of the HIF-1-specific PH family member EGL-9, increasing levels of active HIF-1, has no effect on wild type lifespan, but decreases lifespan extension by DR. The discrepancies might be explained by technical differences between the studies, including the different temperatures at which the lifespan assays were performed. In addition, a third study found that both HIF-1 over-expression, and hif-1 loss of function mutations, extend lifespan, through different pathways (Zhang et al. 2009). Therefore, although the HIF-1 hypoxia pathway seems to influence lifespan, the nature of this influence and it is interaction with aging pathways is not understood, and may lie in a complex interaction between hypoxia, nutrient availability, and temperature.
THE UPRER
Although many proteins require chaperones for folding, this is particularly true of proteins trafficking through the secretory pathway. The endoplasmic reticulum (ER) maintains a set of chaperones to guide the folding of secreted and membrane proteins. In addition, the ER has its own stress response, enabling the organelle to cope with increased flux, for example during development or in cell types with heavy secretory loads, or during stresses such as heat shock that increase protein misfolding (Schroder and Kaufman 2005). This ER stress response (the unfolded protein response, or UPR) is regulated by three upstream sensors, IRE1, PERK, and ATF-6, and has several outcomes, including reduced translation rates; mRNA degradation; transcriptional up-regulation of a range of genes, including many chaperones; and after excessive or prolonged ER stress, apoptosis.
It was recently shown that in C. elegans, the ability to mount a response to ER stress declines with age, at a similar rate to the loss of activity in the HSR (Ben-Zvi et al. 2009). It seems likely, therefore, that the UPR might also play an important role in aging and lifespan extension. Indeed, recent findings suggest that in C. elegans loss of IRE-1 in the worm abolishes lifespan extension by DR and by loss of hif-1 at elevated temperatures, and that IRE-1 and XBP-1 are both required for full lifespan extension by reduced IIS (Chen et al. 2009; Henis-Korenblit et al. 2010). It will be interesting to explore further the roles that this stress response pathway plays in longevity.
PROTEIN TRAFFICKING
Another important aspect of protein homeostasis is the delivery of proteins to their correct compartments. Although it is currently unclear whether the protein trafficking machinery changes with age, there is evidence that components of this machinery are crucial to the lifespan extension induced by reductions in IIS activity. In a study of genes required for daf-2 mediated lifespan extension, a large number of the genes identified were involved in endolysosomal trafficking and vesicle membrane fusion, including subunits of the HOPS and ESCRT complexes (Samuelson et al. 2007). Knockdown of most of these genes alter vesicular trafficking. Genes acting in the fusion of vesicles with lysosomes appeared to function within the IIS pathway, whereas other genes functioned in parallel, but converging with the IIS system. Interestingly, inactivation of some of the genes affecting vesicle-lysosome fusion also prevented the up-regulation of a known DAF-16 target, SOD-3, suggesting that these genes might influence IIS signaling itself. Trafficking to the lysosome therefore seems important in maintaining a youthful proteome. This may reflect the importance of being able to degrade misfolded secreted or membrane proteins through endosomal uptake, maturation and fusion with the lysosome. These results are also interesting in light of the importance of the autophagy pathway in lifespan extension, discussed below.
PROTEIN DEGRADATION
Despite the mechanisms in place to prevent protein misfolding, some proteins slip through this safety net, or become irrevocably damaged over time. To remove and recycle these terminally altered proteins, the cell has pathways that mediate protein degradation. The best characterized are the ubiquitin-proteasome system, in which proteins are specifically tagged with chains of ubiquitin and directed to the proteasome for degradation, and autophagy, a process of cellular “eating” in which cellular constituents are engulfed in membrane-bound autophagosomes that are then targeted to lysosomes for proteolytic breakdown of their constituents (Ciechanover 2005). Both pathways have been implicated in the aging process.
AUTOPHAGY
The role of autophagy in aging has been closely examined in recent years (Cuervo 2008). There have been three main types of autophagy characterized thus far. In chaperone-mediated autophagy, specific proteins are recognized by chaperones and delivered to the lysosome, where they bind to the lysosomal LAMP2A receptor and are translocated into the lysosome for degradation. Microautophagy is a poorly characterized process, involving the engulfment of regions of the cytosol by the lysosomal membrane itself. Macroautophagy, the most studied form of autophagy, involves the formation of a novel membrane that encloses cytosol, damaged proteins, and sometimes organelles, to form an autophagic vesicle that then fuses with the lysosome. Formation of the autophagosome is regulated by the beclin-VPS34 and the mTOR kinase complexes (Yorimitsu and Klionsky 2005).
The best characterized of these processes in the context of aging is macroautophagy. It has been known for some time that in older cells, lysosome function and autophagy decrease (Vittorini et al. 1999; Terman 1995). It is not entirely clear what underlies this decrease. Studies in fly neurons have shown a decline in levels of several autophagy proteins with age (Simonsen et al. 2008). In addition, mammalian studies have indicated a decreased ability to clear autophagosomes, and to respond to hormonal autophagy cues, in older cells (Terman 1995; Brunk and Terman 2002; Donati et al. 2001; Donati et al. 2008).
The interactions between macroautophagy and aging pathways have been largely elucidated in C. elegans. Macroautophagy is necessary for both lifespan extension and formation of the long-lived dauer life cycle stage in daf-2 IIS mutants, and large numbers of autophagosomes are observed in the cells of these animals (Melendez et al. 2003). This autophagy is independent of the DAF-16 transcription factor, however, and given that DAF-16 is essential for lifespan extension in this model, this suggests that increased autophagy is not sufficient for lifespan extension (Hansen et al. 2008).
Macroautophagy is also required for DR-induced longevity (Jia and Levine 2007; Hansen et al. 2008). Worms subjected to DR have higher rates of protein turnover, reduced levels of oxidatively damaged proteins, and larger numbers of autophagosomes. This elevated autophagy is dependent on the RAB-10 small GTPase and the PHA-4 transcription factor, but the specific downstream targets of PHA-4 involved in the regulation of autophagy are not clear (Hansen et al. 2008). The inhibition of mTOR under low nutrient conditions also results in enhanced autophagy, and this autophagy is required for the longevity of animals with reduced mTOR activity (Hansen et al. 2008).
The role of macroautophagy in lifespan regulation is not specific to C. elegans promoting autophagy in neurons of Drosophila through enhanced expression of autophagy proteins that decline with age, leads to increased longevity, and reduction of autophagy in this species may cause premature aging (Simonsen et al. 2008). Perhaps the most persuasive indication of the importance of macroautophagy in aging has been the increased longevity seen folowing application of autophagy-inducing drugs. The application of rapamycin, a natural inhibitor of mTOR, to mice at a relatively late age (600 days) significantly increased longevity (Harrison et al. 2009). In a study of many different potentially lifespan-extending agents, using large cohorts of mice, this was the only one with such a profound effect on lifespan. In addition, a separate study has shown that treatment with spermidine, a polyamine the cellular levels of which decrease during aging, increases longevity of yeast, worms, flies, and human immune cells, and this enhanced longevity is dependent on the induction of autophagy (Eisenberg et al. 2009).
Finally, chaperone-mediated autophagy (CMA) has also proven to be important in the aging process. Rates of CMA decrease with age, because of decreased levels of the lysosomal LAMP-2A receptor through impaired transit to the lysosomal membrane, and reduced receptor activation (Cuervo and Dice 2000; Kiffin et al. 2007). Rescue of LAMP-2A levels in mouse livers at 10 months old, the age at which LAMP-2A levels begin to decline, maintains youthful liver function in these animals, with an increased ability to respond to stress-induced damage (Zhang and Cuervo 2008). An interesting aspect of this study was that the function of other aspects of the quality control machinery, including macroautophagy and the ubiquitin-proteasome system, were also improved, suggesting that the repair of individual elements of the proteostasis machinery could have indirect effects in preserving other components.
PROTEASOMAL DEGRADATION
The ubiquitin-proteasosome system (UPS) targets proteins for degradation by the proteasome, a large proteolytic complex, and the importance of this degradation pathway in the removal of misfolded proteins and in cellular signaling have been well characterized. The effects of aging on proteasome activity, however, are only just becoming clear. Conflicting reports have previously suggested that proteasome activity is increased, decreased, or constant with increasing age (Carrard et al. 2002; Ferrington et al. 2005; Keller et al. 2004). These studies have been complicated by tissue-specific differences, and a comprehensive look at proteasome activity with age in different tissues has been lacking. A new tool in C. elegans, however, holds promise in redressing this, and suggests that there are indeed significant tissue-specific differences in the effect of age on UPS activity. Using a photoconvertible fluorescent moiety, Dendra 2, conjugated to a mutant form of ubiquitin, Hamer et al. have shown that neurons of aging worms show a decreased level of UPS-mediated proteolysis, whereas muscle cells remain unaffected in this regard (Hamer et al. 2010). This line of research seems likely to contribute substantially to understanding the effects of age on UPS activity.
The ubiquitin-proteasome degradation pathway also interacts directly with aging signaling pathways through the targeting of their components for degradation by specific E3 ligases. As discussed already, the VHL-1 E3 ligase affects longevity through its effects on HIF-1 (Mehta et al. 2009). Also, the DAF-16 transcription factor is ubiquitinated and targeted for degradation by an E3 ligase, RLE-1 (Li et al. 2007). Mutations in rle-1 result in lifespan extension, dependent on an increase in DAF-16 protein levels. In addition, another C. elegans E3 ligase, WWP-1, along with an E2 ubiquitin-conjugating enzyme, UBC-18, have been identified as factors essential for the extension of lifespan through DR (Carrano et al. 2009). Over-expression of WWP-1 is able to increase lifespan by about 20%, and this lifespan extension is dependent on PHA-4 and the catalytic E3 ligase activity of WWP-1. The effect of age-dependent changes in UPS activity on signaling within these pathways might therefore represent an interesting area for future research.
DISEASES OF PROTEIN MISFOLDING
An important consequence of declining proteostasis capacity is the emergence of a variety of protein misfolding maladies at advanced ages. These diseases are characterized by the aggregation of a particular protein, or proteins, in specific cell types. Despite the involvement of a multitude of disparate toxic proteins, the late age of onset is remarkably similar in many of these conditions, including Alzheimer’s, Parkinson’s, and Huntington’s diseases. It is also becoming increasingly clear that signaling pathways that regulate aging can alter susceptibility to these diseases, and that this may be through their regulation of the proteostasis machinery.
NEURODEGENERATIVE DISEASE
Neurons seem to be particularly vulnerable to diseases of protein misfolding (Drummond and Wilke 2008; Lee et al. 2006). Major breakthroughs in understanding the influence of aging on neurodegenerative protein misfolding diseases have come about through the study of invertebrate models of these pathologies. Using organisms such as C. elegans or Drosophila allows the study of basic cellular disease processes in the context of postmitotic cells within a multicellular organism. Many studies have shown that the expression of human disease proteins in C. elegans or Drosophila can be toxic, and results in scoreable physiological and behavioral readouts (Morley et al. 2002; Cohen et al. 2006; Kerr et al. 2009). Aging and proteostasis pathways can be easily manipulated in these systems, to study their effects on proteotoxicity phenotypes.
The first such study used C. elegans expressing polyQ repeats, seen in CAG expansion disorders such as Huntington’s disease, fused to YFP in body wall muscle cells (Morley et al. 2002). This shows that polyQ becomes aggregated in a length-dependent manner, with aggregation accompanied by loss of the animal’s motility. Aggregation was also age-dependent. At a threshold length of 35-40 polyQ, aggregates accumulate progressively throughout life. In this study, reducing IIS pathway activity was able to delay aggregation and toxicity, suggesting that this aging pathway can modify the proteostasis machinery sufficiently to enable cells to deal with highly toxic proteins to a much later age. Subsequent studies have shown that HSF-1 over-expression is also highly protective against polyQ aggregation, and that many other components of the proteostasis network are modulators of polyQ toxicity, including genes involved in RNA metabolism and protein synthesis, folding, trafficking, and degradation (Hsu et al. 2003; Nollen et al. 2004; Jia et al. 2007). Interestingly, similar gene categories have also been identified as regulators of α-synuclein toxicity in C. elegans models of Parkinson’s disease (Hamamichi et al. 2008; van Ham et al. 2008).
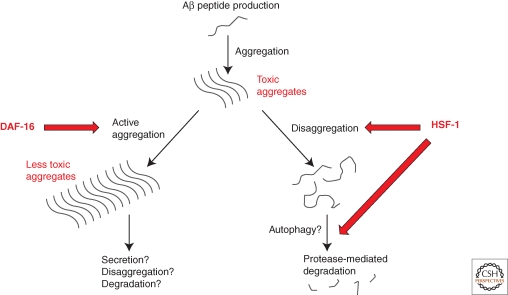
Further mechanistic insights have come from studies of a model of Alzheimer’s disease in C. elegans. In this system, the toxic disease-associated Aβ1-42 peptide is expressed in the body wall muscle cells, producing a progressive paralysis phenotype (Link 1995; Cohen et al. 2006). Again, reduced function of the IIS pathway can protect against toxicity, dependent on the DAF-16 and HSF-1 transcription factors (Cohen et al. 2006). Interestingly, although daf-2 knockdown decreased toxicity, it increased the amount of high molecular weight Aβ aggregates, whereas daf-16 knockdown increased toxicity and decreased the presence of these high molecular weight species. And hsf-1 knockdown both dramatically increased toxicity, and greatly increased the amount of both large and small aggregates. This has suggested a model in which HSF-1 and DAF-16 both transcriptionally regulate detoxification activities, with HSF-1 mediating a disaggregation and degradation activity, and DAF-16 inducing a secondary active aggregation activity that assembles small oligomers into larger, less toxic structures (Fig. 3). Studies showing that HSP104 and TRiC can actively aggregate proteins suggest that similar chaperones might be employed under the transcriptional control of DAF-16 (Behrends et al. 2006; Shorter and Lindquist 2004). Recent work has also separated the HSF-1-controlled activity into separate Aβ disaggregation and proteolytic degradation activities, moving closer to an identification of the molecular components involved (Bieschke et al. 2009).
Figure 3.
Detoxification of Aβ aggregates by DAF-16 and HSF-1. The Aβ is produced throughout life by proteolysis of the amyloid precursor protein. In youthful animals, Aβ aggregates are believed to be detoxified by an active aggregation activity, regulated by the transcription factor DAF-16, and a disaggregation/proteolytic degradation pathway downstream of the transcription factor HSF-1.
Other studies on the Aβ worm have shown that DR is also protective against Aβ toxicity, and that HSF-1 is again required for this protection (Steinkraus et al. 2008). However, it is interesting to note that DR was not protective against Aβ neurotoxicity in a Drosophila model (Kerr et al. 2009). It has also been suggested that autophagy is required for the protective effect of daf-2 knockdown in Aβ-expressing worms (Florez-McClure et al. 2007). Autophagic vesicles are seen in Aβ-expressing cells, and knockdown of autophagy regulators enhances toxicity. Curiously, in this study, reduced daf-2 expression actually reduced the number of autophagic vesicles in Aβ-expressing cells, suggesting a model in which IIS reduction increases the efficiency of autophagosome-vacuole fusion, which is consistent with a proposed role for increased vesicle-lysosome fusion efficiency in daf-2 lifespan extension (Samuelson et al. 2007).
These results have been substantiated in rodents, in which a heterozygous mutation in the IGF-1 receptor is substantially protective in a mouse model of AD (Cohen et al. 2009). Although behavioral phenotypes of AD are substantially delayed, there appears to be no decrease in Aβ production. Instead, Aβ fibrils are packed into dense areas, leaving surrounding brain tissue intact. This is consistent with the existence of a conserved Aβ-aggregative activity downstream of reduced IIS. These results are, however, inconsistent with other data suggesting that increased IGF signaling can increase Aβ and polyQ clearance (Carro et al. 2002; Yamamoto et al. 2006). The answer to this anomaly may lie in the different effects of different levels of IIS in different organs, as discussed elsewhere (Cohen and Dillon 2008). Regardless, the observation that interference with the aging process through reduction in IIS can modify Aβ toxicity in mammals suggests that this may represent a promising avenue for future therapeutics.
OTHER DISEASES
Although the links between neurodegenerative diseases and age-associated proteostasis decline are the best characterized, it is possible that other late onset diseases are also diseases of proteostasis failure. Type II diabetes is one example. A component of the group of late-onset diseases known as “metabolic syndrome,” diabetes and insulin resistance have been associated with a decline in UPR and autophagy capacity (Ozcan et al. 2004; Jung and Lee 2010). The decline of these proteostasis functions with age could therefore predispose toward diabetes. In turn, this suggests that enhancement of proteostasis in older individals could protect against a variety of different but associated protein misfolding diseases.
The temptation to see increased proteostasis capacity as a panacea for all diseases of old age, however, should be avoided. The restricted expression of proteostasis regulators may actually help to limit the development of cancers. Cancer cells have been shown to depend on the activity of the HSF1 and UPR pathways for viability, and susceptibility to Alzheimer’s disease and cancer show an inverse correlation (Bi et al. 2005; Dai et al. 2007; Roe et al. 2010). This suggests that increasing the capacity of these pathways, while protecting against some diseases, could also potentiate tumorigenesis.
EVOLUTIONARY TRADEOFFS?
It is interesting to ask why, given the important role of proteostasis in longevity and protection against disease, components of the proteostasis machinery are not constitutively expressed at high levels. The potentiation of cancerous growth in mammals might be one explanation; however, restricted capacity of the proteostasis machinery has been shown throughout the evolutionary tree, for example in C. elegans (Gidalevitz et al. 2006; Gidalevitz et al. 2009). The answer might lie in the concept of a trade-off between somatic maintenance and reproduction. On an evolutionary level, faced with finite resources, organisms must balance the costly demands of reproduction and somatic maintenance, including maintenance of appropriate protein homeostasis (Kirkwood 2005). This balance takes into account extrinsic mortality, the chance that an organism will die because of external factors such as predation, to create the best evolutionary strategy for maximizing offspring. Evidence suggests that altering the proteostasis machinery to mimic conditions that extend lifespan, for example, overexpression of chaperones, activation of stress responses, or reduction of translation rates, reduces fertility and increases development time. Indeed, reduced IIS, DR, or reduced mitochondrial function, have all been shown to reduce fecundity as well as extending lifespan, although it is important to note that increased lifespan and reduced fecundity are not inseparable (Dillin et al. 2002a; Partridge et al. 2005; Grandison et al. 2009). This concept may explain the restrained expression of many components of the proteostasis machinery.
CONCLUSIONS
The many and intimate connections between pathways that regulate aging, and the cellular protein homeostasis machinery, suggest that maintenance of a healthy proteostasis state is essential for longevity. It also seems that these pathways may play an important role in the prevention of age-associated protein misfolding diseases. This makes pathways that regulate aging, as well as the proteostasis machinery itself, promising targets for future therapeutics.
In fact, several drugs targeting various components of the proteostasis machinery are currently in clinical trials (Balch et al. 2008). However, it is clear is that the agents and pathways that influence proteostasis act as an integrated network. Affecting one component has effects throughout the entire system. This presents both opportunities, and problems. As the expression of one misfolded protein has a destabilizing effect on proteostasis throughout the proteome, it could be that by aiding the folding of just one protein, a range of related pathologies, and symptoms could be alleviated. In addition, an understanding of upstream proteostasis regulators can enable intervention in the whole network simultaneously. In these contexts, invertebrate models should prove useful tools in screening drugs and interventions that can protect against proteotoxic diseases.
However, given the potential negative effects that manipulation of aging and proteostasis pathways can have on, as mentioned, growth and fertility, caution is required in interfering with these programs. In addition, although many age-associated diseases seem likely to benefit from an increase in proteostasis capacity, this may also play into the hands of other maladies, including cancer. With these caveats in mind, however, modulation of the protein homeostasis network is a promising avenue for the treatment of some of the biggest medical problems facing our aging society.
REFERENCES
- Anckar J, Sistonen L 2007. Heat shock factor 1 as a coordinator of stress and developmental pathways. Adv Exp Med Biol 594: 78–88 [DOI] [PubMed] [Google Scholar]
- Baird NA, Turnbull DW, Johnson EA 2006. Induction of the heat shock pathway during hypoxia requires regulation of heat shock factor by hypoxia-inducible factor-1. J Biol Chem 281: 38675–38681 [DOI] [PubMed] [Google Scholar]
- Balch WE, Morimoto RI, Dillin A, Kelly JW 2008. Adapting proteostasis for disease intervention. Science 319: 916–919 [DOI] [PubMed] [Google Scholar]
- Bartke A 2008. Insulin and aging. Cell Cycle 7: 3338–3343 [DOI] [PubMed] [Google Scholar]
- Behrends C, Langer CA, Boteva R, Bottcher UM, Stemp MJ, Schaffar G, Rao BV, Giese A, Kretzschmar H, Siegers K, et al. 2006. Chaperonin TRiC promotes the assembly of polyQ expansion proteins into nontoxic oligomers. Mol Cell 23: 887–897 [DOI] [PubMed] [Google Scholar]
- Ben-Zvi A, Miller EA, Morimoto RI 2009. Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. Proc Natl Acad Sci 106: 14914–14919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlett BS, Stadtman ER 1997. Protein oxidation in aging, disease, and oxidative stress. J Biol Chem 272: 20313–20316 [DOI] [PubMed] [Google Scholar]
- Bi M, Naczki C, Koritzinsky M, Fels D, Blais J, Hu N, Harding H, Novoa I, Varia M, Raleigh J, et al. 2005. ER stress-regulated translation increases tolerance to extreme hypoxia and promotes tumor growth. EMBO J 24: 3470–3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieschke J, Cohen E, Murray A, Dillin A, Kelly JW 2009. A kinetic assessment of the C. elegans amyloid disaggregation activity enables uncoupling of disassembly and proteolysis. Protein Sci 18: 2231–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop NA, Guarente L 2007. Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature 447: 545–549 [DOI] [PubMed] [Google Scholar]
- Brunk UT, Terman A 2002. Lipofuscin: mechanisms of age-related accumulation and influence on cell function. Free Radic Biol Med 33: 611–619 [DOI] [PubMed] [Google Scholar]
- Bukau B, Weissman J, Horwich A 2006. Molecular chaperones and protein quality control. Cell 125: 443–451 [DOI] [PubMed] [Google Scholar]
- Cahill CM, Tzivion G, Nasrin N, Ogg S, Dore J, Ruvkun G, Alexander-Bridges M 2001. Phosphatidylinositol 3-kinase signaling inhibits DAF-16 DNA binding and function via 14-3-3-dependent and 14-3-3-independent pathways. J Biol Chem 276: 13402–13410 [DOI] [PubMed] [Google Scholar]
- Carrano AC, Liu Z, Dillin A, Hunter T 2009. A conserved ubiquitination pathway determines longevity in response to diet restriction. Nature 460: 396–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrard G, Bulteau AL, Petropoulos I, Friguet B 2002. Impairment of proteasome structure and function in aging. Int J Biochem Cell Biol 34: 1461–1474 [DOI] [PubMed] [Google Scholar]
- Carro E, Trejo JL, Gomez-Isla T, LeRoith D, Torres-Aleman I 2002. Serum insulin-like growth factor I regulates brain amyloid-β levels. Nat Med 8: 1390–1397 [DOI] [PubMed] [Google Scholar]
- Chen D, Thomas EL, Kapahi P 2009. HIF-1 modulates dietary restriction-mediated lifespan extension via IRE-1 in Caenorhabditis elegans. PLoS Genet 5: e1000486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiocchetti A, Zhou J, Zhu H, Karl T, Haubenreisser O, Rinnerthaler M, Heeren G, Oender K, Bauer J, Hintner H, et al. 2007. Ribosomal proteins Rpl10 and Rps6 are potent regulators of yeast replicative life span. Exp Gerontol 42: 275–286 [DOI] [PubMed] [Google Scholar]
- Chiti F, Stefani M, Taddei N, Ramponi G, Dobson CM 2003. Rationalization of the effects of mutations on peptide and protein aggregation rates. Nature 424: 805–808 [DOI] [PubMed] [Google Scholar]
- Ciechanover A 2005. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat Rev Mol Cell Biol 6: 79–87 [DOI] [PubMed] [Google Scholar]
- Cohen E, Dillin A 2008. The insulin paradox: aging, proteotoxicity and neurodegeneration. Nat Rev Neurosci 9: 759–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen E, Bieschke J, Perciavalle RM, Kelly JW, Dillin A 2006. Opposing activities protect against age-onset proteotoxicity. Science 313: 1604–1610 [DOI] [PubMed] [Google Scholar]
- Cohen E, Paulsson JF, Blinder P, Burstyn-Cohen T, Du D, Estepa G, Adame A, Pham HM, Holzenberger M, Kelly JW, et al. 2009. Reduced IGF-1 signaling delays age-associated proteotoxicity in mice. Cell 139: 1157–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland JM, Cho J, Lo T Jr, Hur JH, Bahadorani S, Arabyan T, Rabie J, Soh J, Walker DW 2009. Extension of Drosophila life span by RNAi of the mitochondrial respiratory chain. Curr Biol 19: 1591–1598 [DOI] [PubMed] [Google Scholar]
- Cuervo AM 2008. Autophagy and aging: keeping that old broom working. Trends Genet 24: 604–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo AM, Dice JF 2000. Age-related decline in chaperone-mediated autophagy. J Biol Chem 275: 31505–31513 [DOI] [PubMed] [Google Scholar]
- Dai C, Whitesell L, Rogers AB, Lindquist S 2007. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell 130: 1005–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillin A, Crawford DK, Kenyon C 2002a. Timing requirements for insulin/IGF-1 signaling in C. elegans. Science 298: 830–834 [DOI] [PubMed] [Google Scholar]
- Dillin A, Hsu AL, Arantes-Oliveira N, Lehrer-Graiwer J, Hsin H, Fraser AG, Kamath RS, Ahringer J, Kenyon C 2002b. Rates of behavior and aging specified by mitochondrial function during development. Science 298: 2398–2401 [DOI] [PubMed] [Google Scholar]
- Ding Q, Dimayuga E, Markesbery WR, Keller JN 2004. Proteasome inhibition increases DNA and RNA oxidation in astrocyte and neuron cultures. J Neurochem 91: 1211–1218 [DOI] [PubMed] [Google Scholar]
- Donati A, Cavallini G, Paradiso C, Vittorini S, Pollera M, Gori Z, Bergamini E 2001. Age-related changes in the regulation of autophagic proteolysis in rat isolated hepatocytes. J Gerontol A Biol Sci Med Sci 56: B288–B293 [DOI] [PubMed] [Google Scholar]
- Donati A, Recchia G, Cavallini G, Bergamini E 2008. Effect of aging and anti-aging caloric restriction on the endocrine regulation of rat liver autophagy. J Gerontol A Biol Sci Med Sci 63: 550–555 [DOI] [PubMed] [Google Scholar]
- Dong MQ, Venable JD, Au N, Xu T, Park SK, Cociorva D, Johnson JR, Dillin A, Yates JR 3rd 2007. Quantitative mass spectrometry identifies insulin signaling targets in C. elegans. Science 317: 660–663 [DOI] [PubMed] [Google Scholar]
- Drummond DA, Wilke CO 2008. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell 134: 341–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg T, Knauer H, Schauer A, Buttner S, Ruckenstuhl C, Carmona-Gutierrez D, Ring J, Schroeder S, Magnes C, Antonacci L, et al. 2009. Induction of autophagy by spermidine promotes longevity. Nat Cell Biol 11: 1305–1314 [DOI] [PubMed] [Google Scholar]
- Feng J, Bussiere F, Hekimi S 2001. Mitochondrial electron transport is a key determinant of life span in Caenorhabditis elegans. Dev Cell 1: 633–644 [DOI] [PubMed] [Google Scholar]
- Ferrington DA, Husom AD, Thompson LV 2005. Altered proteasome structure, function, and oxidation in aged muscle. FASEB J 19: 644–646 [DOI] [PubMed] [Google Scholar]
- Florez-McClure ML, Hohsfield LA, Fonte G, Bealor MT, Link CD 2007. Decreased insulin-receptor signaling promotes the autophagic degradation of β-amyloid peptide in C. elegans. Autophagy 3: 569–580 [DOI] [PubMed] [Google Scholar]
- Gebauer F, Hentze MW 2004. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol 5: 827–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidalevitz T, Ben-Zvi A, Ho KH, Brignull HR, Morimoto RI 2006. Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science 311: 1471–1474 [DOI] [PubMed] [Google Scholar]
- Gidalevitz T, Krupinski T, Garcia S, Morimoto RI 2009. Destabilizing protein polymorphisms in the genetic background direct phenotypic expression of mutant SOD1 toxicity. PLoS Genet 5: e1000399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandison RC, Piper MD, Partridge L 2009. Amino-acid imbalance explains extension of lifespan by dietary restriction in Drosophila. Nature 462: 1061–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halaschek-Wiener J, Khattra JS, McKay S, Pouzyrev A, Stott JM, Yang GS, Holt RA, Jones SJ, Marra MA, Brooks-Wilson AR, et al. 2005. Analysis of long-lived C. elegans daf-2 mutants using serial analysis of gene expression. Genome Res 15: 603–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ham TJ, Thijssen KL, Breitling R, Hofstra RM, Plasterk RH, Nollen EA 2008. C elegans model identifies genetic modifiers of α-synuclein inclusion formation during aging. PLoS Genet 4: e1000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamamichi S, Rivas RN, Knight AL, Cao S, Caldwell KA, Caldwell GA 2008. Hypothesis-based RNAi screening identifies neuroprotective genes in a Parkinson’s disease model. Proc Natl Acad Sci 105: 728–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamer G, Matilainen O, Holmberg CI 2010. A photoconvertible reporter of the ubiquitin-proteasome system in vivo. Nat Methods 7: 473–478 [DOI] [PubMed] [Google Scholar]
- Hansen M, Chandra A, Mitic LL, Onken B, Driscoll M, Kenyon C 2008. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet 4: e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Taubert S, Crawford D, Libina N, Lee SJ, Kenyon C 2007. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell 6: 95–110 [DOI] [PubMed] [Google Scholar]
- Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, et al. 2009. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature 460: 392–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hekimi S, Guarente L 2003. Genetics and the specificity of the aging process. Science 299: 1351–1354 [DOI] [PubMed] [Google Scholar]
- Henderson ST, Johnson TE 2001. daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Curr Biol 11: 1975–1980 [DOI] [PubMed] [Google Scholar]
- Henis-Korenblit S, Zhang P, Hansen M, McCormick M, See SJ, Cary M, Kenyon C 2010. Insulin/IGF-1 signaling mutants reprogram ER stress response regulators to promote longevity. Proc Natl Acad Sci 107: 9730–9735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hercus MJ, Loeschcke V, Rattan SI 2003. Lifespan extension of Drosophila melanogaster through hormesis by repeated mild heat stress. Biogerontology 4: 149–156 [DOI] [PubMed] [Google Scholar]
- Hertweck M, Gobel C, Baumeister R 2004. C elegans SGK-1 is the critical component in the Akt/PKB kinase complex to control stress response and life span. Dev Cell 6: 577–588 [DOI] [PubMed] [Google Scholar]
- Hsieh CC, Papaconstantinou J 2004. Akt/PKB and pp38 MAPK signaling, translational initiation and longevity in Snell dwarf mouse livers. Mech Ageing Dev 125: 785–798 [DOI] [PubMed] [Google Scholar]
- Hsu AL, Murphy CT, Kenyon C 2003. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science 300: 1142–1145 [DOI] [PubMed] [Google Scholar]
- Jia K, Levine B 2007. Autophagy is required for dietary restriction-mediated life span extension in C. elegans. Autophagy 3: 597–599 [DOI] [PubMed] [Google Scholar]
- Jia K, Chen D, Riddle DL 2004. The TOR pathway interacts with the insulin signaling pathway to regulate C. elegans larval development, metabolism and life span. Development 131: 3897–3906 [DOI] [PubMed] [Google Scholar]
- Jia K, Hart AC, Levine B 2007. Autophagy genes protect against disease caused by polyglutamine expansion proteins in Caenorhabditis elegans. Autophagy 3: 21–25 [DOI] [PubMed] [Google Scholar]
- Jung HS, Lee MS 2010. Role of autophagy in diabetes and mitochondria. Ann N Y Acad Sci 1201: 79–83 [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, Powers RW 3rd, Steffen KK, Westman EA, Hu D, Dang N, Kerr EO, Kirkland KT, Fields S, Kennedy BK 2005. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science 310: 1193–1196 [DOI] [PubMed] [Google Scholar]
- Kahn BB, Alquier T, Carling D, Hardie DG 2005. AMP-activated protein kinase: ancient energy gauge provides clues to modern understanding of metabolism. Cell Metab 1: 15–25 [DOI] [PubMed] [Google Scholar]
- Kapahi P, Zid B 2004. TOR pathway: linking nutrient sensing to life span. Sci Aging Knowledge Environ 2004: PE34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapahi P, Zid BM, Harper T, Koslover D, Sapin V, Benzer S 2004. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr Biol 14: 885–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller JN, Dimayuga E, Chen Q, Thorpe J, Gee J, Ding Q 2004. Autophagy, proteasomes, lipofuscin, and oxidative stress in the aging brain. Int J Biochem Cell Biol 36: 2376–2391 [DOI] [PubMed] [Google Scholar]
- Kerr F, Augustin H, Piper MD, Gandy C, Allen MJ, Lovestone S, Partridge L 2009. Dietary restriction delays aging, but not neuronal dysfunction, in Drosophila models of Alzheimer's disease. Neurobiol Aging 4: T225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiffin R, Kaushik S, Zeng M, Bandyopadhyay U, Zhang C, Massey AC, Martinez-Vicente M, Cuervo AM 2007. Altered dynamics of the lysosomal receptor for chaperone-mediated autophagy with age. J Cell Sci 120: 782–791 [DOI] [PubMed] [Google Scholar]
- Kirkwood TB 2005. Understanding the odd science of aging. Cell 120: 437–447 [DOI] [PubMed] [Google Scholar]
- Lakowski B, Hekimi S 1998. The genetics of caloric restriction in Caenorhabditis elegans. Proc Natl Acad Sci 95: 13091–13096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RY, Hench J, Ruvkun G 2001. Regulation of C. elegans DAF-16 and its human ortholog FKHRL1 by the daf-2 insulin-like signaling pathway. Curr Biol 11: 1950–1957 [DOI] [PubMed] [Google Scholar]
- Lee JW, Beebe K, Nangle LA, Jang J, Longo-Guess CM, Cook SA, Davisson MT, Sundberg JP, Schimmel P, Ackerman SL 2006. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature 443: 50–55 [DOI] [PubMed] [Google Scholar]
- Lee SS, Lee RY, Fraser AG, Kamath RS, Ahringer J, Ruvkun G 2003. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat Genet 33: 40–48 [DOI] [PubMed] [Google Scholar]
- Li W, Gao B, Lee SM, Bennett K, Fang D 2007. RLE-1, an E3 ubiquitin ligase, regulates C. elegans aging by catalyzing DAF-16 polyubiquitination. Dev Cell 12: 235–246 [DOI] [PubMed] [Google Scholar]
- Lin K, Dorman JB, Rodan A, Kenyon C 1997. daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science 278: 1319–1322 [DOI] [PubMed] [Google Scholar]
- Lin K, Hsin H, Libina N, Kenyon C 2001. Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nat Genet 28: 139–145 [DOI] [PubMed] [Google Scholar]
- Link CD 1995. Expression of human β-amyloid peptide in transgenic Caenorhabditis elegans. Proc Natl Acad Sci 92: 9368–9372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lithgow GJ, White TM, Melov S, Johnson TE 1995. Thermotolerance and extended life-span conferred by single-gene mutations and induced by thermal stress. Proc Natl Acad Sci 92: 7540–7544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Jiang N, Hughes B, Bigras E, Shoubridge E, Hekimi S 2005. Evolutionary conservation of the clk-1-dependent mechanism of longevity: loss of mclk1 increases cellular fitness and lifespan in mice. Genes Dev 19: 2424–2434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair W, Dillin A 2008. Aging and survival: the genetics of life span extension by dietary restriction. Annu Rev Biochem 77: 727–754 [DOI] [PubMed] [Google Scholar]
- Mair W, Goymer P, Pletcher SD, Partridge L 2003. Demography of dietary restriction and death in Drosophila. Science 301: 1731–1733 [DOI] [PubMed] [Google Scholar]
- Manning BD 2004. Balancing Akt with S6K: implications for both metabolic diseases and tumorigenesis. J Cell Biol 167: 399–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McColl G, Rogers AN, Alavez S, Hubbard AE, Melov S, Link CD, Bush AI, Kapahi P, Lithgow GJ 2010. Insulin-like signaling determines survival during stress via posttranscriptional mechanisms in C. elegans. Cell Metab 12: 260–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElwee J, Bubb K, Thomas JH 2003. Transcriptional outputs of the Caenorhabditis elegans forkhead protein DAF-16. Aging Cell 2: 111–121 [DOI] [PubMed] [Google Scholar]
- McMillan DR, Xiao X, Shao L, Graves K, Benjamin IJ 1998. Targeted disruption of heat shock transcription factor 1 abolishes thermotolerance and protection against heat-inducible apoptosis. J Biol Chem 273: 7523–7528 [DOI] [PubMed] [Google Scholar]
- Mehta R, Steinkraus KA, Sutphin GL, Ramos FJ, Shamieh LS, Huh A, Davis C, Chandler-Brown D, Kaeberlein M 2009. Proteasomal regulation of the hypoxic response modulates aging in C. elegans. Science 324: 1196–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melendez A, Talloczy Z, Seaman M, Eskelinen EL, Hall DH, Levine B 2003. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science 301: 1387–1391 [DOI] [PubMed] [Google Scholar]
- Morimoto RI 2008. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev 22: 1427–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley JF, Morimoto RI 2004. Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Mol Biol Cell 15: 657–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley JF, Brignull HR, Weyers JJ, Morimoto RI 2002. The threshold for polyglutamine-expansion protein aggregation and cellular toxicity is dynamic and influenced by aging in Caenorhabditis elegans. Proc Natl Acad Sci 99: 10417–10422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JZ, Tissenbaum HA, Ruvkun G 1996. A phosphatidylinositol-3-OH kinase family member regulating longevity and diapause in Caenorhabditis elegans. Nature 382: 536–539 [DOI] [PubMed] [Google Scholar]
- Morrow G, Samson M, Michaud S, Tanguay RM 2004. Overexpression of the small mitochondrial Hsp22 extends Drosophila life span and increases resistance to oxidative stress. FASEB J 18: 598–599 [DOI] [PubMed] [Google Scholar]
- Muñoz MJ, Riddle DL 2003. Positive selection of Caenorhabditis elegans mutants with increased stress resistance and longevity. Genetics 163: 171–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon C 2003. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424: 277–283 [DOI] [PubMed] [Google Scholar]
- Nollen EA, Garcia SM, van Haaften G, Kim S, Chavez A, Morimoto RI, Plasterk RH 2004. Genome-wide RNA interference screen identifies previously undescribed regulators of polyglutamine aggregation. Proc Natl Acad Sci 101: 6403–6408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogg S, Ruvkun G 1998. The C. elegans PTEN homolog, DAF-18, acts in the insulin receptor-like metabolic signaling pathway. Mol Cell 2: 887–893 [DOI] [PubMed] [Google Scholar]
- Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, Ruvkun G 1997. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature 389: 994–999 [DOI] [PubMed] [Google Scholar]
- Oh SW, Mukhopadhyay A, Dixit BL, Raha T, Green MR, Tissenbaum HA 2006. Identification of direct DAF-16 targets controlling longevity, metabolism and diapause by chromatin immunoprecipitation. Nat Genet 38: 251–257 [DOI] [PubMed] [Google Scholar]
- Ozcan U, Cao Q, Yilmaz E, Lee AH, Iwakoshi NN, Ozdelen E, Tuncman G, Görgün C, Glimcher LH, Hotamisligil GS 2004. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science 306: 457–461 [DOI] [PubMed] [Google Scholar]
- Pan KZ, Palter JE, Rogers AN, Olsen A, Chen D, Lithgow GJ, Kapahi P 2007. Inhibition of mRNA translation extends lifespan in Caenorhabditis elegans. Aging Cell 6: 111–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panowski SH, Dillin A 2009. Signals of youth: Endocrine regulation of aging in Caenorhabditis elegans. Trends Endocrinol Metab 20: 259–264 [DOI] [PubMed] [Google Scholar]
- Panowski SH, Wolff S, Aguilaniu H, Durieux J, Dillin A 2007. PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature 447: 550–555 [DOI] [PubMed] [Google Scholar]
- Paradis S, Ruvkun G 1998. Caenorhabditis elegans Akt/PKB transduces insulin receptor-like signals from AGE-1 PI3 kinase to the DAF-16 transcription factor. Genes Dev 12: 2488–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge L, Gems D, Withers DJ 2005. Sex and death: What is the connection? Cell 120: 461–472 [DOI] [PubMed] [Google Scholar]
- Pirkkala L, Nykanen P, Sistonen L 2001. Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB J 15: 1118–1131 [DOI] [PubMed] [Google Scholar]
- Puig O, Marr MT, Ruhf ML, Tjian R 2003. Control of cell number by Drosophila FOXO: Downstream and feedback regulation of the insulin receptor pathway. Genes Dev 17: 2006–2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter JD, Sonenberg N 2005. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature 433: 477–480 [DOI] [PubMed] [Google Scholar]
- Roe CM, Fitzpatrick AL, Xiong C, Sieh W, Kuller L, Miller JP, Williams MM, Kopan R, Behrens MI, Morris JC 2010. Cancer linked to Alzheimer disease but not vascular dementia. Neurology 74: 106–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelson AV, Carr CE, Ruvkun G 2007. Gene activities that mediate increased life span of C. elegans insulin-like signaling mutants. Genes Dev 21: 2976–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder M, Kaufman RJ 2005. The mammalian unfolded protein response. Annu Rev Biochem 74: 739–789 [DOI] [PubMed] [Google Scholar]
- Selman C, Tullet JM, Wieser D, Irvine E, Lingard SJ, Choudhury AI, Claret M, Al-Qassab H, Carmignac D, Ramadani F, et al. 2009. Ribosomal protein S6 kinase 1 signaling regulates mammalian life span. Science 326: 140–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp ZD, Bartke A 2005. Evidence for down-regulation of phosphoinositide 3-kinase/Akt/mammalian target of rapamycin (PI3K/Akt/mTOR)-dependent translation regulatory signaling pathways in Ames dwarf mice. J Gerontol A Biol Sci Med Sci 60: 293–300 [DOI] [PubMed] [Google Scholar]
- Shorter J, Lindquist S 2004. Hsp104 catalyzes formation and elimination of self-replicating Sup35 prion conformers. Science 304: 1793–1797 [DOI] [PubMed] [Google Scholar]
- Simonsen A, Cumming RC, Brech A, Isakson P, Schubert DR, Finley KD 2008. Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy 4: 176–184 [DOI] [PubMed] [Google Scholar]
- Soskic V, Groebe K, Schrattenholz A 2008. Nonenzymatic posttranslational protein modifications in ageing. Exp Gerontol 43: 247–257 [DOI] [PubMed] [Google Scholar]
- Stefani M, Dobson CM 2003. Protein aggregation and aggregate toxicity: New insights into protein folding, misfolding diseases and biological evolution. J Mol Med 81: 678–699 [DOI] [PubMed] [Google Scholar]
- Steffen KK, MacKay VL, Kerr EO, Tsuchiya M, Hu D, Fox LA, Dang N, Johnston ED, Oakes JA, Tchao BN, et al. 2008. Yeast life span extension by depletion of 60s ribosomal subunits is mediated by Gcn4. Cell 133: 292–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinkraus KA, Smith ED, Davis C, Carr D, Pendergrass WR, Sutphin GL, Kennedy BK, Kaeberlein M 2008. Dietary restriction suppresses proteotoxicity and enhances longevity by an hsf-1-dependent mechanism in Caenorhabditis elegans. Aging Cell 7: 394–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syntichaki P, Troulinaki K, Tavernarakis N 2007. eIF4E function in somatic cells modulates ageing in Caenorhabditis elegans. Nature 445: 922–926 [DOI] [PubMed] [Google Scholar]
- Terman A 1995. The effect of age on formation and elimination of autophagic vacuoles in mouse hepatocytes. Gerontology 41 Suppl 2: 319–326 [DOI] [PubMed] [Google Scholar]
- Tissenbaum HA, Guarente L 2001. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature 410: 227–230 [DOI] [PubMed] [Google Scholar]
- Tullet JM, Hertweck M, An JH, Baker J, Hwang JY, Liu S, Oliveira RP, Baumeister R, Blackwell TK 2008. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell 132: 1025–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellai T, Takacs-Vellai K, Zhang Y, Kovacs AL, Orosz L, Muller F 2003. Genetics: influence of TOR kinase on lifespan in C. elegans. Nature 426: 620. [DOI] [PubMed] [Google Scholar]
- Vittorini S, Paradiso C, Donati A, Cavallini G, Masini M, Gori Z, Pollera M, Bergamini E 1999. The age-related accumulation of protein carbonyl in rat liver correlates with the age-related decline in liver proteolytic activities. J Gerontol A Biol Sci Med Sci 54: B318–B323 [DOI] [PubMed] [Google Scholar]
- Walker GA, Lithgow GJ 2003. Lifespan extension in C. elegans by a molecular chaperone dependent upon insulin-like signals. Aging Cell 2: 131–139 [DOI] [PubMed] [Google Scholar]
- Wang X, Li W, Williams M, Terada N, Alessi DR, Proud CG 2001. Regulation of elongation factor 2 kinase by p90(RSK1) and p70 S6 kinase. EMBO J 20: 4370–4379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerheide SD, Anckar J, Stevens SM Jr, Sistonen L, Morimoto RI 2009. Stress-inducible regulation of heat shock factor 1 by the deacetylase SIRT1. Science 323: 1063–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff S, Dillin A 2006. The trifecta of aging in Caenorhabditis elegans. Exp Gerontol 41: 894–903 [DOI] [PubMed] [Google Scholar]
- Wolff S, Ma H, Burch D, Maciel GA, Hunter T, Dillin A 2006. SMK-1, an essential regulator of DAF-16-mediated longevity. Cell 124: 1039–1053 [DOI] [PubMed] [Google Scholar]
- Wolkow CA, Munoz MJ, Riddle DL, Ruvkun G 2002. Insulin receptor substrate and pp55 orthologous adaptor proteins function in the Caenorhabditis elegans daf-2/insulin-like signaling pathway. J Biol Chem 277: 49591–49597 [DOI] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN 2006. TOR signaling in growth and metabolism. Cell 124: 471–484 [DOI] [PubMed] [Google Scholar]
- Yamamoto A, Cremona ML, Rothman JE 2006. Autophagy-mediated clearance of huntingtin aggregates triggered by the insulin-signaling pathway. J Cell Biol 172: 719–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T, Klionsky DJ 2005. Autophagy: Molecular machinery for self-eating. Cell Death Differ 12 Suppl 2: 1542–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Cuervo AM 2008. Restoration of chaperone-mediated autophagy in aging liver improves cellular maintenance and hepatic function. Nat Med 14: 959–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Shao Z, Zhai Z, Shen C, Powell-Coffman JA 2009. The HIF-1 Hypoxia-Inducible Factor Modulates Lifespan in C. elegans. PLoS ONE 4: e6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zid BM, Rogers AN, Katewa SD, Vargas MA, Kolipinski MC, Lu TA, Benzer S, Kapahi P 2009. 4E-BP extends lifespan upon dietary restriction by enhancing mitochondrial activity in Drosophila. Cell 139: 149–160 [DOI] [PMC free article] [PubMed] [Google Scholar]